Abstract
Allostery in proteins plays an important role in regulating protein activities and influencing many biological processes such as gene expression, enzyme catalysis, and cell signaling. The process of allostery takes place when a signal detected at a site on a protein is transmitted via a mechanical pathway to a functional site and, thus, influences its activity. The pathway of allosteric communication consists of amino acids that form a network with covalent and non-covalent bonds. By mutating residues in this allosteric network, protein engineers have successfully established novel allosteric pathways to achieve desired properties in the target protein. In this review, we highlight the most recent and state-of-the-art techniques for allosteric communication engineering. We also discuss the challenges that need to be overcome and future directions for engineering protein allostery.
Introduction
Allostery is a common phenomenon in biological macromolecules, where distant parts of a molecule are energetically coupled to produce a functional response [1–3]. The original view of allostery is based on conformational change of the functional site of a protein when perturbation is introduced at a distal site (allosteric site) [4–6]. Cooper and Dryden [7] further proposed a dynamics-based model of allostery, in which the active and inactive states are structurally similar but the dynamics of the active site is changed. Their model extends the original theory of allostery as it shifts the model of two distinguishable states to probabilistic description of ensembles that represent these states. Using the ensemble-based theory, allostery can be formulated regarding the conformational free energies of each domain in a protein and the negative or positive coupling interactions between them [8,9]. In addition to the above-mentioned mechanisms of allostery, Banerjee-Ghosh et al. [10] reported a novel allosteric mechanism based on charge. They demonstrated that charge injection at the distal site redistributed the charge within the antibody, which affected its interaction with the antigen [10].
Allosteric communication plays a pivotal role in various essential biological processes including gene regulation, cell signaling, and metabolism [11]. Genetic mutations that disrupt allosteric communications within a protein may lead to a loss of function and, in some cases, to disease. For instance, mutations in the S-loop of human glutathione synthetase that affect allosteric communication disrupt normal biological processes, resulting in hemolytic anemia, metabolic acidosis, premature death, and neurological disorders [12]. Presenilin 1 catalyzes the cleavage of transmembrane proteins including the amyloid precursor protein [13]. Genetic mutations in presenilin 1 that alter allosteric communication can alter the cleavage pattern and lead to aberrant β-amyloid peptides that have been linked to Alzheimer’s disease [13]. Therefore, understanding allosteric communications and the change in protein function caused by perturbations is critical for clarifying disease etiology and developing effective therapeutics.
Understanding of allosteric communication makes it feasible to design and engineer novel allosteric networks to serve desired purposes. In the past two years, scientists have developed innovative methods to engineer allosteric networks to improve enzyme activity, confer biosensors with novel ligand binding, and construct complex regulatory systems for synthetic biology [14]. In this review, we summarize recent advances in the design and engineering of allosteric communications by addressing the following three topics: (i) the experimental and computational approaches that we can use to elucidate allosteric communication networks, (ii) how do we use this information to engineer hybrid long-range allosteric routes, (iii) the general methods developed to design alternative allosteric communications.
The state-of-the-art methods developed to map allosteric communication pathways
To rationally design and engineer allosteric communications, a prerequisite is to decode the signal transduction pathways and critical residues within the pathway for individual proteins. In the past two years, we have seen great advances in experimental, computational, and integrative methods to meet this challenge.
Site-directed mutagenesis is a commonly used experimental approach to identify essential residues for allosteric communication [15]. Mutations that disrupt allosteric signaling pathways can affect protein structure and function, which can be distinguished from mutated residues that are not important for allosteric signaling. Based on this, Leander et al. [15] used deep mutational scanning, a systematic mutagenesis method, to study the functional landscape of allostery. Solution NMR is another popular experimental method applied to identify residues on the allosteric communication pathway. Residues with changes in chemical shift when the signaling pathway is perturbed are potential targets. Rennella et al. [16] applied methyl-transverse relaxation optimized spectroscopy (TROSY)-based NMR to investigate an extensive allosteric communication pathway initiating from the end of the barrel-like structure to the catalytic center of the proteasome. This methodology can be used to dissect allosteric communication pathways in homo-oligomeric complexes with high molecular weight [16]. Solution NMR has been combined with other experimental methods to enhance the discovery of allosteric communications. Through the combination of NMR and surface plasmon resonance, Kohler et al. [17] demonstrated that ligands with subtle changes in structure share the same allosteric pathways, while ligands with distinct pharmacophores rewire the allosteric communication network.
Compared to experimental methods, computational approaches used to investigate allosteric communications can be more time-efficient and cost-effective. Molecular dynamics (MD) simulations are powerful tools for studying allosteric communications in biological systems [18], such as CRISPR-Cas9 [19], tyrosine phosphatase [20], HBV Capsid [21], β-lactamases [22], and G protein-coupled receptor (GPCR) [23]. Besides MD simulations, another group of computational methods based on spectral graph or information theory has been developed [24]. In this method, the three-dimensional structure of a protein is converted to a network whereby each node represents an amino acid and the edge indicates a covalent or non-covalent bond between residues [25]. Combining this network modeling approach with physics and perturbation propagation algorithm, Wang et al. developed Ohm [26], a comprehensive and user-friendly platform for allosteric communication analysis. This innovative approach relies solely on protein structure and minimizes the calculation time, which is more rapid and cost-effective than MD simulations. Westerlund et al. [27] extended the framework of residue interacting network to include lipids and small molecules, which takes into account the effect of lipids on membrane protein allostery. Alfayate et al. [28] established a torsional network model to analyze dynamic couplings between amino acids in a protein. The dynamic couplings dissect a protein into different domains with correlated motion and the couplings between these domains can be applied to identify potential allosteric sites [28]. The Berezovsky group developed a structure-based statistical mechanical model of allostery to study allosteric communication between regulatory and functional sites [29]. Based on this model, they created the AlloMAPS database that can be used to analyze the causality and energetics of allosteric signaling, access the allosteric effects of perturbations, and evaluate the modulatory effects of new allosteric mutations and sites [29]. Recently, Schupfner et al. [30] developed an ancestral sequence reconstruction method that resurrects the highly probable sequences of extinct proteins based on a phylogenetic tree and multiple sequence alignment of modern proteins. Using this method and further sequence comparison, they identified four residues that contributed to the allosteric signal transduction within the tryptophan synthase complex.
Using experimental and computational methods, integrative methods have been developed that combine the two to illustrate allosteric communications. Subramanian et al. [31] revealed critical residues in the allosteric signal transduction pathway by deep mutagenesis and MD simulations. Ni et al. [32] applied MD simulations, Markov state models, and site-directed mutagenesis to identify a novel cryptic allosteric site of nicotinamide dinucleotide (NAD+)-dependent protein lysine deacetylase sirtuin 6. Sztain et al. [33] combined solution NMR spectroscopy and MD to decode the communication mechanism between acyl carrier protein and its corresponding partner enzyme. They elucidated an allosteric communication network in which the binding of the substrate within the four-helical bundle of the acyl carrier protein causes conformational changes to the exterior, and thus determined its interaction with a specific partner enzyme [33]. Using solution NMR and Gaussian-accelerated MD simulations, East et al. [34] discovered a signal transduction pathway within the CRISPR-Cas9 HNH domain that propagates from the region interacting with the RuvC nuclease to the DNA recognition site.
Construction of hybrid and long-range allosteric networks
The allosteric pathways identified in two different proteins can be rationally linked to form a long-range allosteric pathway. In this hybrid system, perturbation introduced (signal detected) at the ligand-binding site of protein A can be propagated to the functional site of protein B and then affect its activity. By engineering the hybrid allosteric network, we can generate diverse combinations of signal detection and functional outputs, where various signals can be used to manipulate different functions. This application is significant as it provides more options for synthetic biologists and makes regulation more sophisticated.
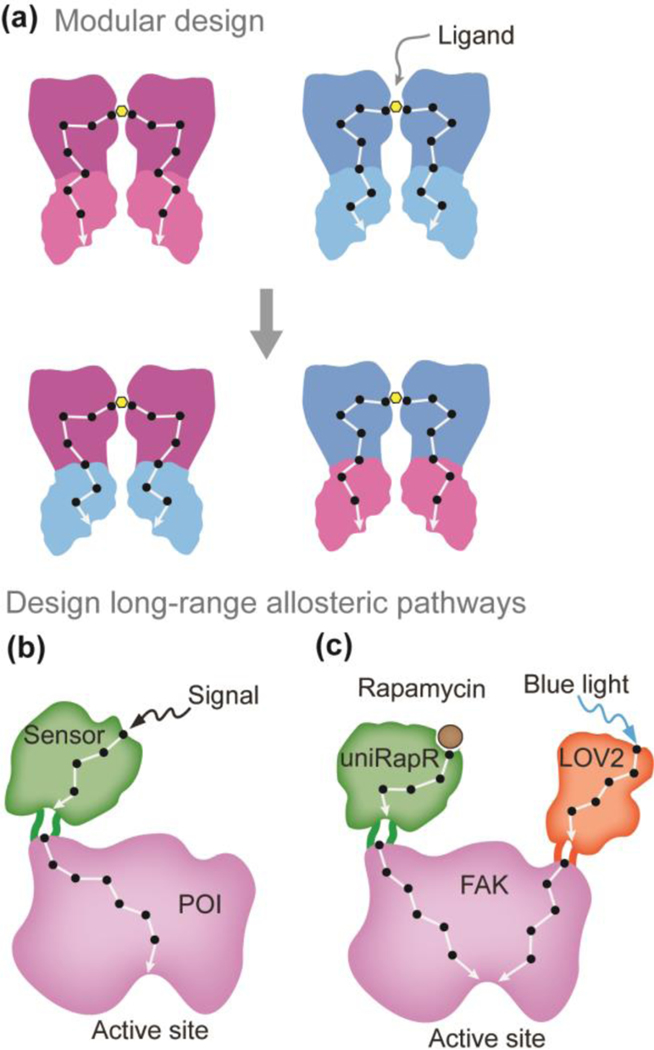
Modular design is a general strategy to create hybrid proteins and allosteric communication pathways [14]. It divides a system into smaller intact parts called modules that can be exchanged and fused with other modules to create a hybrid system. By coupling various allosteric modules and connecting different allosteric communication pathways, we can establish a synthetic allosteric communication network (Figure 1a). A candidate that has been exploited for modular design is the transcriptional repressor LacI, which consists of a DNA binding domain that interacts with specific promoters and a ligand sensing module that detects environmental signals. The ligand interaction at the environmental sensing module allosterically regulates the DNA binding with the promoter. However, not all possible combinations of DNA and ligand-binding domains will generate hybrid repressors that are functional. To address this problem and facilitate the rewiring of allosteric networks, Dimas et al. [35] developed a computational model based on coevolution theory. They propose that a mutation in the ligand binding region is coupled with a specific mutation in the DNA binding domain if these two domains are structurally or functionally related. Using direct coupling analysis, this model identifies strong coupling pairs that can result in functional hybrid repressors [35]. The generalizability of this modular design strategy has been validated by several studies and described in this review [14]. Recently, Chen et al. [36] designed de novo protein modules that can be fused to create a heterodimer (e.g. A-B) endowed with interaction cooperativity. Specifically, the interaction of subunit A (within the heterodimer A-B) with monomer A’ induces the binding between subunit B and monomer B’, which might be attributed to the hybrid allosteric network constructed by fusing A and B. This ground-breaking research demonstrated the possibility to engineer de novo allosteric communication pathways in proteins.
Figure 1.
Construction of hybrid and long-range allosteric pathways. (a) Schematic representation of the modular design strategy. Synthetic allosteric communications are established by connecting signaling pathways from different modules. (b) Design long-range allosteric networks to control the functions of proteins of interest (POI) using chemical or light responsive sensor domains. Black dots represent critical residues in the allosteric pathway and white lines indicate covalent or non-covalent interactions.
Modular design is limited to proteins with multiple distinct domains (building blocks) that can be separated and functional. Over the last decade, a more universal approach has been developed to engineer long-range allosteric networks in most proteins of interest [37–40]. In this method, a chemical or light-sensitive sensor domain is inserted at an allosteric site of the target protein using molecular cloning techniques [41,42]. Through the connected allosteric pathway, the signal detected by the sensor domain can be propagated to the functional site of the target protein and thus influence its function (Figure 1b). Gil et al. [41] designed opto-nanobodies by inserting a photoswitchable light-oxygen-voltage (LOV) domain at a solvent-exposed loop of the nanobodies. The LOV domain recognizes blue light and leads to structural perturbations that are further transmitted to the binding surface of nanobodies and then interfere with the recognition of a target protein. The reversible control of binding between nanobody and its target has many applications such as modulating intracellular signaling and controlling protein binding in cells. Recently, the Dokholyan’s group [43] engineered a complex allosteric communication network by inserting two sensor domains, a rapamycin responsive uniRapR domain and a blue light sensitive LOV2 domain in the focal adhesion kinase (FAK) (Figure 1c). By connecting allosteric pathways between the sensor domains and FAK, they built a ‘two-input logic OR gate’, where the inputs rapamycin and blue light independently control the kinase activity of FAK. This innovative work demonstrated that two allosteric communication networks can exist in one protein design to control its activity without crosstalk.
Engineering of alternative allosteric communication pathways
Allosteric communications are highly adaptable during evolution as one mutation that interrupts the original signaling pathway can be functionally compensated by another or a few mutations that create an alternative allosteric communication [15]. The cutting-edge methods used to confer potential allosteric networks include directed evolution and rational design.
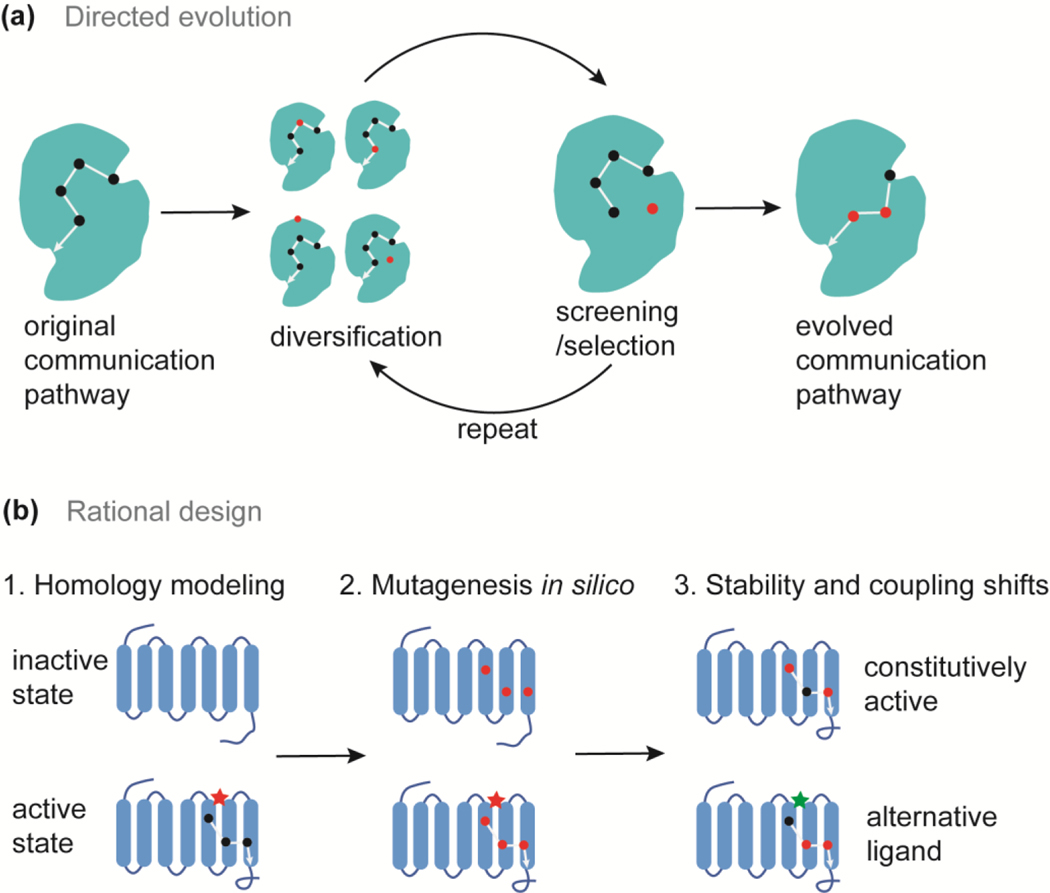
Directed evolution utilizes the principle of natural selection by imposing stringent rules to screen targets with desired functions from a library of randomly mutated proteins (Figure 2a) [44]. Directed evolution does not require structural information of proteins of interest. However, a high-throughput screening methodology is necessary for efficient protein design. Using a cell growth-based assay, D’Amico et al. [45] identified a mutant involved in the allosteric network that enhances tryptophan synthase function. Substitution of this residue in the alpha subunit of tryptophan synthase allosterically affects the opening of the indole channel and stimulates the activity of the beta subunit [45]. The Wilson lab extensively engineered the allosteric communications in the LacI system based on directed evolution [14,46]. For example, to confer alternate ligand binding in the PurR scaffold, Rondon et al. [47] blocked the original allosteric communication pathway and introduced additional mutations through error-prone PCR in order to reconstruct alternate allosteric routes with alternative ligand recognition [47].
Figure 2.
Directed evolution and rational design of alternative allosteric communication pathways. (a) The process of directed evolution in engineering alternative allosteric communications. (b) The rational design steps in designing alternate allosteric communication pathways in GPCRs. Black dots and white lines represent essential residues and interactions within the allosteric network. Red dots indicate mutated residues.
The rational design of allosteric communication requires a thorough understanding of the target protein in terms of structure, function, and the signal transition pathway. To preserve allosteric regulation during evolution, allosteric networks are expected to be conserved and residues on these networks have coevolved [48]. Based on statistical coupling analysis, Jiao et al. [48] identified coevolved residues that may form important communication networks in the enzyme, 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase. Through site-directed mutagenesis of a critical node within this network, they successfully blocked allosteric communication and constitutively activated the enzyme function [48]. To rationally engineer allosteric communications, Campitelli et al. [49] calculated the effects of residue substitution on flexibilities of the functional surface (the DNA-binding domain in LacI repressor), and the dynamic coupling between the mutation site and the DNA-binding domain. Based on the change in flexibility and dynamic coupling, they successfully rewired the allosteric signaling pathways to adjust DNA binding affinity by substituting non-conserved residues in the linker region of the LacI repressor [49]. To engineer allosteric functions, Chen et al. [50] developed a general approach to reprogram allosteric communications in membrane receptors, such as GPCRs (Figure 2b). First, this method predicts GPCR conformations in the active (ligand-bound) and inactive (ligand-free) states using homology modeling. Allosteric sites and communication pathways are identified through MD simulations. Then residues that mediate allosteric signaling in the transmembrane region are mutated in silico to all possible 20 amino acids. Mutations that shift conformational stability or structural coupling between allosteric sites are selected. For instance, proteins with enhanced constitutive activity can be designed by mutating residues that stabilize the active state rather than the inactive ligand-free state [50].
Conclusions
Genetic variants that disrupt allosteric communication can be detrimental, while rationally designed or directed evolved allosteric networks provide many opportunities for protein engineering and synthetic biology. Over the past two years, we have seen great strides in the identification of allosteric communications in various biological systems and the development of new approaches to the design and engineering of allosteric networks. However, a more general and effective design method, as well as the de novo design of allosteric communications, are still challenges to be addressed in the future. We highlight computationally-guided methods as they are effective means of designing and engineering allosteric communications, surpassing sequence-based deep mutagenesis in terms of efficiency, speed, and success rate. One major limitation to consider when designing allosteric communications is preserving the structural integrity of the target protein. Evolutionary conserved residues can be important in maintaining structural integrity, while non-conserved residues in the allosteric network may be targeted to reprogram allosteric communications.
Acknowledgements
We acknowledge the support from the National Institutes of Health (1R35 GM134864) and the Passan Foundation.
Footnotes
Conflict of interest statement
Nothing declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dokholyan NV: Controlling allosteric networks in proteins. Chem Rev 2016, 116:6463–6487. [DOI] [PubMed] [Google Scholar]
- 2.Proctor EA, Kota P, Aleksandrov AA, He L, Riordan JR, Dokholyan NV: Rational coupled dynamics network manipulation rescues disease-relevant mutant cystic fibrosis transmembrane regulator. Chem Sci 2015, 6:1237–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cournia Z, Chatzigoulas A: Allostery in membrane proteins. Curr Opin Struct Biol 2020, 62:197–204. [DOI] [PubMed] [Google Scholar]
- 4.Wensien M, von Pappenheim FR, Funk LM, Kloskowski P, Curth U, Diederichsen U, Uranga J, Ye J, Fang P, Pan KT, et al. : A lysine–cysteine redox switch with an NOS bridge regulates enzyme function. Nature 2021, 593:460–464. [DOI] [PubMed] [Google Scholar]
- 5.Buddingh’ BC, Elzinga J, van Hest JCM: Intercellular communication between artificial cells by allosteric amplification of a molecular signal. Nat Commun 2020, 11:1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee CH, MacKinnon R: Voltage sensor movements during hyperpolarization in the HCN channel. Cell 2019, 179:1582–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper A, Dryden DT: Allostery without conformational change. A plausible model. Eur Biophys J 1984, 11:103–109. [DOI] [PubMed] [Google Scholar]
- 8.Hilser VJ, Wrabl JO, Motlagh HN: Structural and energetic basis of allostery. Annu Rev Biophys 2012, 41: 585–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatzigoulas A, Cournia Z: Rational design of allosteric modulators: challenges and successes. Wiley Interdiscip Rev Comput Mol Sci 2021, 11: e1529. [Google Scholar]
- 10.Banerjee-Ghosh K, Ghosh S, Mazal H, Riven I, Haran G, Naaman R: Long-range charge reorganization as an allosteric control signal in proteins. J Am Chem Soc 2020, 142: 20456–20462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wodak SJ, Paci E, Dokholyan NV, Berezovsky IN, Horovitz A, Li J, Hilser VJ, Bahar I, Karanicolas J, Stock G, et al. : Allostery in its many disguises: from theory to applications. Structure 2019, 27:566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingle BL, Shrestha B, De Jesus MC, Conrad-Webb HM, Anderson ME, Cundari TR: Genetic mutations in the S-loop of human glutathione synthetase: links between substrate binding, active site structure and allostery. Comput Struct Biotechnol J 2019, 17:31–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chávez-García C, Aguayo-Ortiz R, Dominguez L: Quantifying correlations between mutational sites in the catalytic subunit of γ-secretase. J Mol Graph Model 2019, 88:221–227. [DOI] [PubMed] [Google Scholar]
- 14.Herde ZD, Short AE, Kay VE, Huang BD, Realff MJ, Wilson CJ: Engineering allosteric communication. Curr Opin Struct Biol 2020, 63:115–122. [DOI] [PubMed] [Google Scholar]
- 15. Leander M, Yuan Y, Meger A, Cui Q, Raman S: Functional plasticity and evolutionary adaptation of allosteric regulation. Proc Natl Acad Sci U S A 2020, 117:25445–25454. *This work applied deep mutational scanning to study the molecular determinants of allostery using tetracycline repressor as the model system. They found that allosteric communications are highly flexible and redundant. Residues essential for allosteric communications are poorly conserved in evolution, while residues critical to maintain protein structures are highly conserved. This interesting discovery could be harnessed to guide future engineering of allosteric networks in proteins. Non-conserved residues may be targeted instead of conserved ones.
- 16.Rennella E, Huang R, Yu Z, Kay LE: Exploring long-range cooperativity in the 20S proteasome core particle from Thermoplasma acidophilum using methyl-TROSY–based NMR. Proc Natl Acad Sci U S A 2020, 117:5298–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köhler C, Carlström G, Gunnarsson A, Weininger U, Tångefjord S, Ullah V, Lepistö M, Karlsson U, Papavoine T, Edman K, et al. : Dynamic allosteric communication pathway directing differential activation of the glucocorticoid receptor. Sci Adv 2020, 6:eabb5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bingöl EN, Serçinoǧlu O, Ozbek P: Unraveling the allosteric communication mechanisms in T-cell receptor-peptide-loaded major histocompatibility complex dynamics using molecular dynamics simulations: an approach based on dynamic cross correlation maps and residue interaction energy calcula. J Chem Inf Model 2021, 61:2444–2453. [DOI] [PubMed] [Google Scholar]
- 19.Nierzwicki Ł, Arantes PR, Saha A, Palermo G: Establishing the allosteric mechanism in CRISPR-Cas9. Wiley Interdiscip Rev Comput Mol Sci 2021, 11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crean RM, Biler M, Van Der Kamp MW, Hengge AC, Kamerlin SCL: Loop dynamics and enzyme catalysis in protein tyrosine phosphatases. J Am Chem Soc 2021, 143:3830–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pérez-Segura C, Goh BC, Hadden-Perilla JA: All-atom md simulations of the hbv capsid complexed with at130 reveal secondary and tertiary structural changes and mechanisms of allostery. Viruses 2021, 13:564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galdadas I, Qu S, Oliveira ASF, Olehnovics E, Mack AR, Mojica MF, Agarwal PK, Tooke CL, Gervasio FL, Spencer J, et al. : Allosteric communication in class a β-lactamases occurs via cooperative coupling of loop dynamics. Elife 2021, 10:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Zhang S, Han Z, Fan H, Li C: An investigation into the allosteric mechanism of GPCR A2A adenosine receptor with trajectory-based information theory and complex network model. J Biomol Struct Dyn 2021, 39:6431–6439. [DOI] [PubMed] [Google Scholar]
- 24.Vishweshwaraiah YL, Chen J, Dokholyan NV: Engineering an allosteric control of protein function. J Phys Chem B 2021, 125:1806–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mersmann SF, Strömich L, Song FJ, Wu N, Vianello F, Barahona M, Yaliraki SN: ProteinLens: A web-based application for the analysis of allosteric signalling on atomistic graphs of biomolecules. Nucleic Acids Res 2021, 49:W551–W558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang J, Jain A, McDonald LR, Gambogi C, Lee AL, Dokholyan NV: Mapping allosteric communications within individual proteins. Nat Commun 2020, 11:1–13. ** This research developed a conceptually different computational approach to analyze allosteric communications within individual proteins based on the knowledge of physics and perturbation propagation algorithm. They built a user-friendly platform, Ohm (Ohm.dokhlab.org), to promote the usage of this method. Using the protein structure as the only input, Ohm predicts the allosteric sites, allosteric communication networks, key residues, and allosteric coupling between residues. This comprehensive web server could be used to assist in designing alternate allosteric networks.
- 27.Westerlund AM, Fleetwood O, Pérez-Conesa S, Delemotte L: Network analysis reveals how lipids and other cofactors influence membrane protein allostery. J Chem Phys 2020, 153:1–10. [DOI] [PubMed] [Google Scholar]
- 28.Alfayate A, Caceres CR, Santos HGD, Bastolla U: Predicted dynamical couplings of protein residues characterize catalysis, transport and allostery. Bioinformatics 2019, 35: 4971–4978. [DOI] [PubMed] [Google Scholar]
- 29.Tan ZW, Tee WV, Guarnera E, Booth L, Berezovsky IN: AlloMAPS: allosteric mutation analysis and polymorphism of signaling database. Nucleic Acids Res 2019, 47: D265–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schupfner M, Straub K, Busch F, Merkl R, Sterner R: Analysis of allosteric communication in a multienzyme complex by ancestral sequence reconstruction. Proc Natl Acad Sci U S A 2020, 117:346–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Subramanian S, Gorday K, Marcus K, Orellana MR, Ren P, Luo XR, O’donnell ME, Kuriyan J: Allosteric communication in DNA polymerase clamp loaders relies on a critical hydrogen-bonded junction. Elife 2021, 10:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni D, Wei J, He X, Rehman AU, Li X, Qiu Y, Pu J, Lu S, Zhang J: Discovery of cryptic allosteric sites using reversed allosteric communication by a combined computational and experimental strategy. Chem Sci 2021, 12:464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sztain T, Bartholow TG, Lee DJ, Casalino L, Mitchell A, Young MA, Wang J, McCammon JA, Burkart MD: Decoding allosteric regulation by the acyl carrier protein. Proc Natl Acad Sci U S A 2021, 118:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.East KW, Newton JC, Morzan UN, Narkhede YB, Acharya A, Skeens E, Jogl G, Batista VS, Palermo G, Lisi GP: Allosteric motions of the CRISPR-Cas9 HNH nuclease probed by NMR and molecular dynamics. J Am Chem Soc 2020, 142:1348–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimas RP, Jiang XL, De La Paz JA, Morcos F, Chan CTY: Engineering repressors with coevolutionary cues facilitates toggle switches with a master reset. Nucleic Acids Res 2019, 47:5449–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Z, Kibler RD, Hunt A, Busch F, Pearl J, Jia M, VanAernum ZL, Wicky BIM, Dods G, Liao H, et al. : De novo design of protein logic gates. Science 2020, 368: 78–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dagliyan O, Shirvanyants D, Karginov AV, Ding F, Fee L, Chandrasekaran SN, Freisinger CM, Smolen GA, Huttenlocher A, Hahn KM, et al. : Rational design of a ligand-controlled protein conformational switch. Proc Natl Acad Sci U S A 2013, 110:6800–6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dagliyan O, Karginov AV, Yagishita S, Gale ME, Wang H, Dermardirossian C, Wells CM, Dokholyan NV, Kasai H, Hahn KM: Engineering Pak1 allosteric switches. ACS Synth Biol 2017, 6:1257–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dagliyan O, Tarnawski M, Chu PH, Shirvanyants D, Schlichting I, Dokholyan NV, Hahn KM: Engineering extrinsic disorder to control protein activity in living cells. Science 2016, 354:1441–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dagliyan O, Dokholyan NV, Hahn KM: Engineering proteins for allosteric control by light or ligands. Nat Protoc 2019, 14:1863–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gil AA, Carrasco-López C, Zhu L, Zhao EM, Ravindran PT, Wilson MZ, Goglia AG, Avalos JL, Toettcher JE: Optogenetic control of protein binding using light-switchable nanobodies. Nat Commun 2020, 11:4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaaya M, Fauser J, Zhurikhina A, Conage-Pough JE, Huyot V, Brennan M, Flower CT, Matsche J, Khan S, Natarajan V, et al. : Light-regulated allosteric switch enables temporal and subcellular control of enzyme activity. Elife 2020, 9:1–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vishweshwaraiah YL, Chen J, Chirasani VR, Tabdanov ED, Dokholyan NV: Two-input protein logic gate for computation. Nat Commun 2021, 12:6615. ** Using Focal Adhesion Kinase as a candidate protein, the authors engineered a modular, two-input logic OR gate for biological computation using ‘allosteric wiring’. Using this protein design, authors showed the direct regulation of cellular phenotype at the single-protein level. This study provides proof-of-principle for multimodal control of protein function as demonstrated by allosterically wiring two orthogonal regulators to an active site of a protein to program it as a logic gate.
- 44.Packer MS, Liu DR: Methods for the directed evolution of proteins. Nat Rev Genet 2015, 16:379–394. [DOI] [PubMed] [Google Scholar]
- 45.D’Amico RN, Bosken YK, O’Rourke KF, Murray AM, Admasu W, Chang C en A, Boehr DD: Substitution of a surface-exposed residue involved in an allosteric network enhances tryptophan synthase function in cells. Front Mol Biosci 2021, 8:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Groseclose TM, Rondon RE, Hersey AN, Milner PT, Kim D, Zhang F, Realff MJ, Wilson CJ: Biomolecular systems engineering: unlocking the potential of engineered allostery via the lactose repressor topology. Annu Rev Biophys 2021, 50:303–321. [DOI] [PubMed] [Google Scholar]
- 47. Rondon R, Wilson CJ: Engineering alternate ligand recognition in the PurR topology: a system of novel caffeine biosensing transcriptional antirepressors. ACS Synth Biol 2021, 10:552–565. * In this paper, the authors engineered alternative ligand binding in the PurR repressor based on directed evolution. They successfully abolished the recognition of the native ligand hypoxanthine and simultaneously, engineered PurR to interact with caffeine. To achieve this design, first they disrupt the original allosteric pathway that is responsive to hypoxanthine and then, build an alternate communication pathway to confer caffeine binding. This paper demonstrated the methodology to engineer alternate ligand binding, which could be applied to design novel biosensors.
- 48.Jiao W, Fan Y, Blackmore NJ, Parker EJ: A single amino acid substitution uncouples catalysis and allostery in an essential biosynthetic enzyme in Mycobacterium tuberculosis. J Biol Chem 2020, 295:6252–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Campitelli P, Swint-Kruse L, Ozkan SB: Substitutions at nonconserved rheostat positions modulate function by rewiring long-range, dynamic interactions. Mol Biol Evol 2021, 38:201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen KYM, Keri D, Barth P: Computational design of G Protein-Coupled Receptor allosteric signal transductions. Nat Chem Biol 2020, 16:77–86. ** The authors developed a computational approach to engineer allosteric communications in membrane receptor GPCRs through the integration of homology modeling, molecular dynamics simulations, and design calculations. Judged by conformational stability and long-range coupling analysis, they engineered constitutive dopamine D2 receptors and repurposed the receptor to recognize non-native ligand serotonin. This work developed a rational design framework that can be readily applied to the other membrane proteins, which can further be used to redesign cell signaling in synthetic biology.