Graphical Abstract
A palindrome reads the same forward as backward (compare top left to right). A regulatory palindrome is a typically imperfect inverted sequence repeat that is bound by a transcription factor homodimer, e.g. in mammalian steroid receptor signaling (bottom left) and Drosophila photoreceptor differentiation (bottom right). The underlying mechanisms remain incompletely understood.
Keywords: gene regulation, cis-regulatory motif, palindrome, enhancer, transcription factor, steroid hormone receptor, glucocorticoid, rhodopsin
Summary
In human languages, a palindrome reads the same forward as backward (e.g. ‘madam’). In regulatory DNA, a palindrome is an inverted sequence repeat that allows a transcription factor to bind as a homodimer or as a heterodimer with another type of transcription factor. Regulatory palindromes are typically imperfect, i.e. the repeated sequences differ in at least one base pair, but the functional significance of this asymmetry remains poorly understood. Here, we review the use of imperfect palindromes in Drosophila photoreceptor differentiation and mammalian steroid receptor signaling. Moreover, we discuss mechanistic explanations for the predominance of imperfect palindromes over perfect palindromes in these two gene regulatory contexts. Lastly, we propose to elucidate whether specific imperfectly palindromic variants have specific regulatory functions in steroid receptor signaling and whether such variants can help predict transcriptional outcomes as well as the response of individual patients to drug treatments.
Introduction
Genomic regulatory regions such as enhancers and promoters contain short DNA sequences that are called cis-regulatory motifs [1,2]. Cis-regulatory motifs are recognized by sequence-specific transcription factors that direct where, when, and the levels at which genes are expressed [3] (Fig. 1a). A special type of cis-regulatory motif is the palindrome, which resembles a palindromic word that, by definition, reads the same forward as backward (e.g. ‘racecar’; see also Fig. 1b). In cis-regulatory palindromes, the first half of the palindromic DNA sequence (which we will call left ‘half-site’) is repeated as its reverse complement (the right ‘half-site’) on the same DNA strand and thus follows in reverse orientation on the opposite strand (e.g. 5’ TAATTGAATTA 3’ and 3’ ATTAACTTAAT 5’; Fig. 1c). In this essay, we thus consider cis-regulatory palindromes to be composed of a left half-site, central sequences, and a right half-site.
Figure 1.
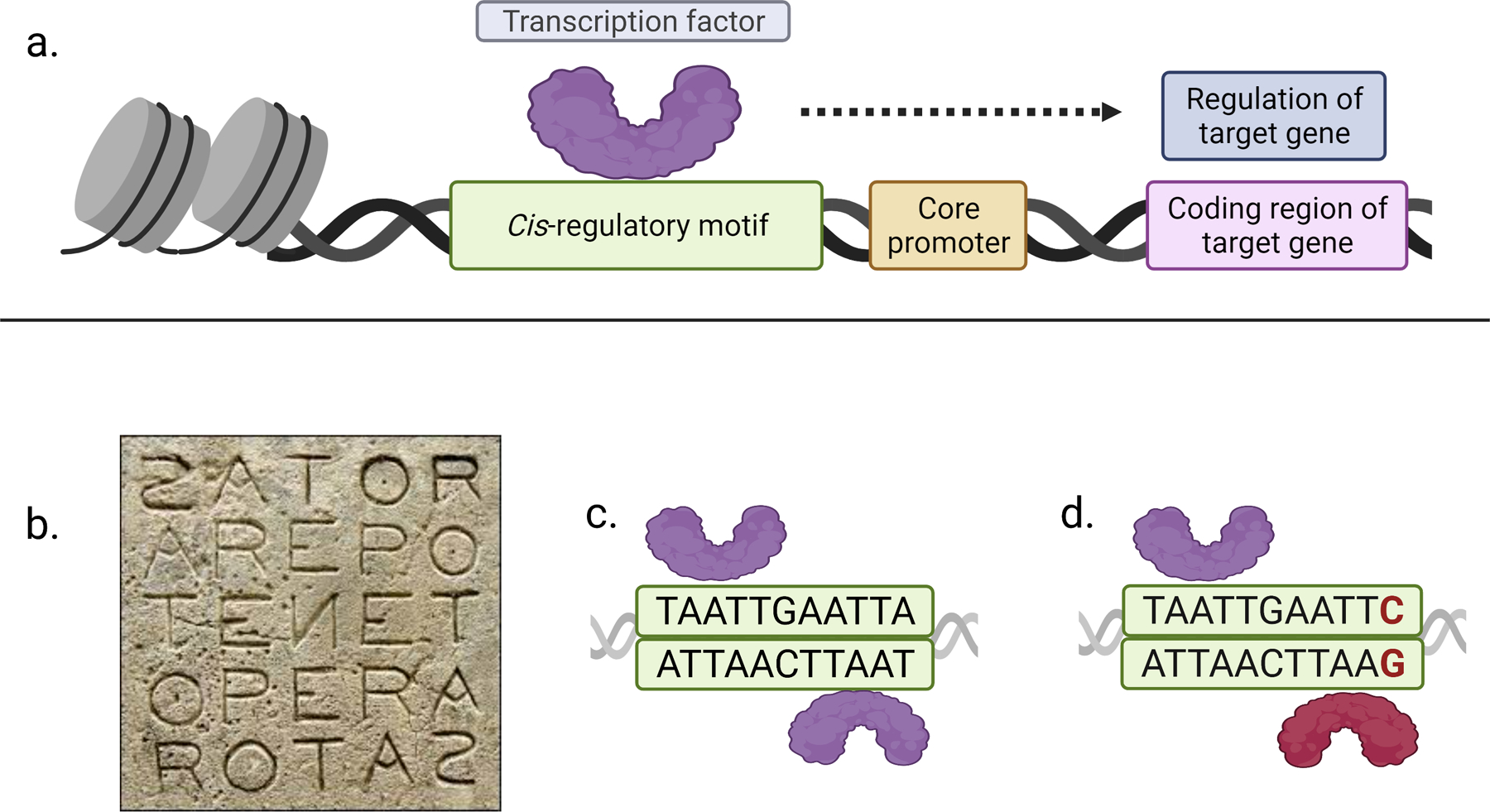
A: The schematic represents the binding of a sequence-specific transcription factor to a cis-regulatory motif in the regulatory DNA of a target gene. The transcription factor controls where, when, and at which levels the gene is expressed.
B: The ‘Sator square’ is a complex square palindrome that consists of five Latin words (‘sator arepo tenet opera rotas’) that can be read top-to-bottom, bottom-to-top, left-to-right, and right-to-left (image from wikipedia.com).
C: A homeodomain transcription factor (purple) binds as a homodimer to a palindromic cis-regulatory motif. Note the TAAT homeodomain core motif repeat on the opposite DNA strand (5’ TAAT…ATTA 3’ and 3’ ATTA…TAAT 5’).
D: The right TAAT core motif repeat is disrupted in an imperfectly palindromic motif variant (TAAT…ATTC instead of TAAT…ATTA), which recruits a different transcription factor type (red) and results in the binding of a heterodimer.
Palindromic words can have an even (e.g. ‘noon’) or an odd number of letters (e.g. ‘rotator’). Cis-regulatory palindromes can also consist of an odd number of base pairs, which can make it difficult to judge from sequence alone whether the central bases are unbound spacer sequences or belong to the bound left and right half-sites (e.g. the central G in TAATTGAATTA; Fig. 1c). In human languages, the inverted repeats in single-word palindromes have to be perfect in order to mean the same forward as backward (e.g. ‘noon’ and not ‘noun’). However, cis-regulatory palindromes rarely have such perfectly inverted repeats and the sequences of the left and right half-site therefore differ in at least one base pair. Here, we use the word ‘palindromic’ to imply ‘palindrome-like’ and to include both perfect and imperfect cis-regulatory palindrome types.
The cis-regulatory palindrome allows a sequence-specific transcription factor to cooperatively bind as a homodimer to the left and the right half-sites on opposite DNA strands such that each half-site is occupied by one DNA-binding domain [4] (Fig. 1c); the central sequences may (see Drosophila retina example below) or may not be bound. This cooperative interaction of the two DNA-binding domains can result in stronger dimeric binding compared to the binding of a monomeric DNA-binding domain to the left or right half-site [4,5].
Palindromic motifs can also be bound by heterodimers of two different transcription factor types (Fig. 1d) and thereby mediate cell type-specific combinatorial gene regulation. For instance, Jun family proteins can form homodimers but also heterodimers with Fos family proteins on the palindromic AP-1 motif (TGAG/CTCA) [6,7]. Moreover, the bHLHZ protein Max can homodimerize, but also heterodimerize with the closely related bHLHZ protein Myc on the palindromic E box motif (CACGTG) [8,9]. Max homodimers bind the E box with lower affinity than Myc-Max heterodimers [10–12]. Notably, different heterodimer combinations can drive different transcriptional outcomes: while the Myc-Max heterodimer activates gene expression, the Mad-Max heterodimer mediates transcriptional repression [13].
While the cooperative binding of transcription factor homodimers or heterodimers to palindromic motifs appears to resemble the cooperative binding of transcription factors to nonpalindromic motifs, structural studies of homodimeric glucocorticoid receptor (GR) binding to palindromic glucocorticoid response elements (GREs) suggest a unique feature of palindromic binding sites: each GRE half-site acts as a sequence-specific allosteric ligand for the GR monomer that binds to it and subtle sequence variations in the half-site affect the binding affinity as well as the conformation of the bound GR monomer [14]. Remarkably, the alteration of the sequence of one half-site can also modify the conformation of the second GR monomer that binds to the other half site [15] and the two monomers thus appear to be able to communicate with each other across the half-sites [14]. This complex interplay between the variations in both palindromic half-sites could allow the GR dimer to interpret a longer (palindromic) sequence and communicate this information to the transcription machinery to affect target gene expression [15].
To gain insights into the roles of palindromic motifs in gene expression, we review two regulatory contexts that rely on them: Drosophila terminal photoreceptor differentiation and mammalian steroid hormone signaling. We discuss mechanisms that explain the predominance of imperfect palindromes over perfect palindromes in regulatory DNA and propose that asymmetric inverted repeats are an evolutionarily conserved mechanism for differential gene expression.
1. The role of palindromic motifs in photoreceptor terminal differentiation in Drosophila
Sequence-specific terminal selector transcription factors co-regulate sets of functionally related genes through shared cis-regulatory motifs; terminal selectors thereby control the terminal differentiation of post-mitotic neurons and define their identity and function [16–18]. The role of homeodomain transcription factors [19] that act as terminal selectors through binding to cis-regulatory palindromes (Fig. 2a) in terminally differentiating Drosophila photoreceptor neurons has been studied in detail [20,21]. As we discuss below, the differential expression of phototransduction genes in all photoreceptor neurons and rhodopsin genes in a subset of photoreceptor neurons is based on specific imperfectly palindromic motifs with different degrees of half-site symmetry (Fig. 2a).
Figure 2.
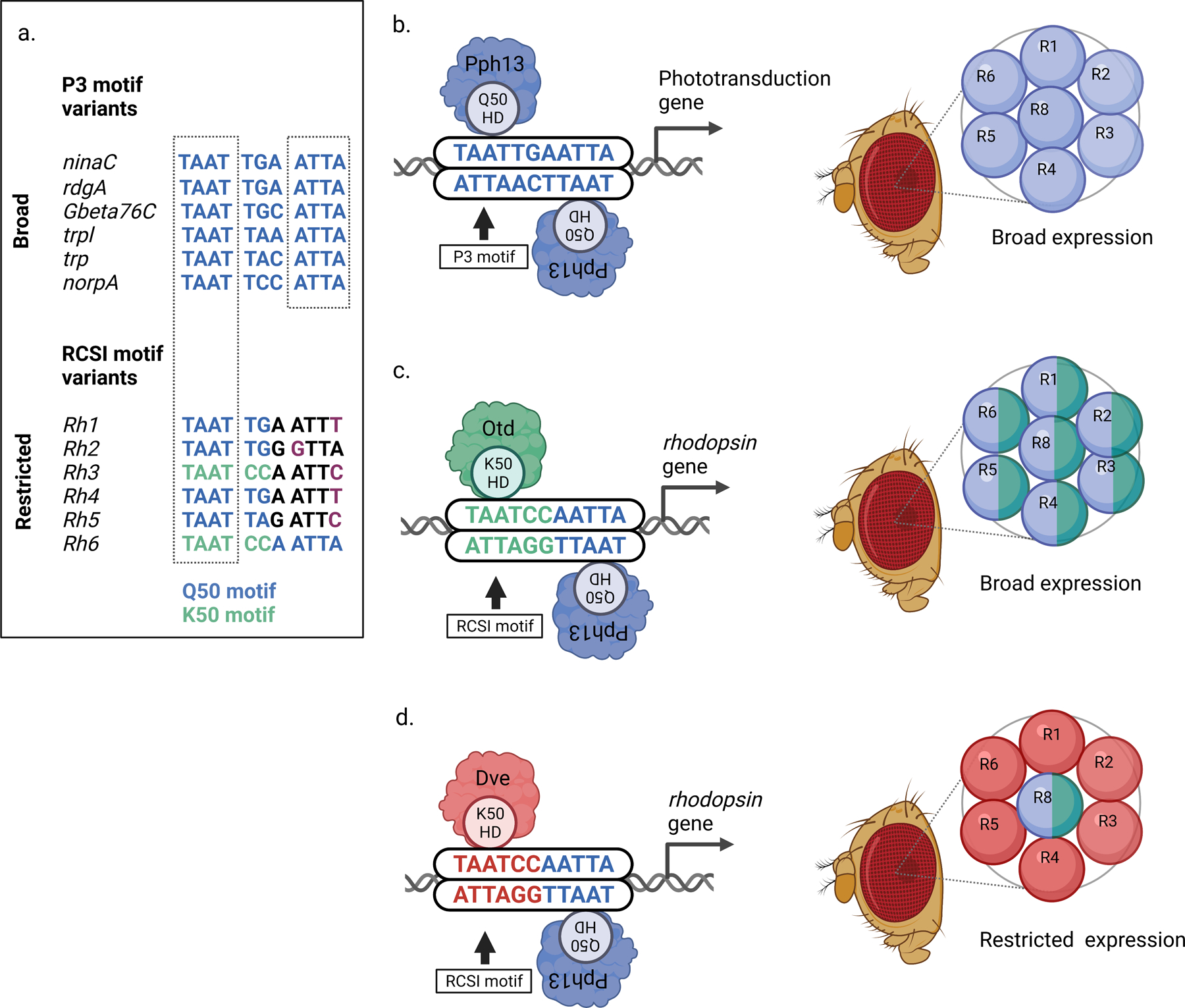
A: Comparison of palindromic P3 motif variants (top) in phototransduction genes that are broadly expressed in all Drosophila photoreceptors with RCSI motif variants (Rhodopsin Core Sequence I, bottom) in rhodopsin genes (Rh1-Rh6) whose expression is restricted to different subsets of photoreceptors.
Top, P3 motif variants in broadly expressed phototransduction genes contain Q50 motifs (TAATTA/G, blue) and have a palindromic repeat of the homeodomain core motif (TAAT…ATTA).
Bottom, each rhodopsin gene ‘prefers’ a specific imperfectly palindromic RCSI motif variant. Note that Rh3 and Rh6 contain K50 motifs (TAATCC, green). Only the 5’ TAAT core motif of rhodopsins matches the palindromic repeat in broadly expressed phototransduction genes (dashed box), while the inverted 3’ repeat is imperfect in Rh1-Rh5 (indicated by purple letters: ATTT, GTTA, or ATTC instead of ATTA).
B: Left, a homodimer of the Q50 homeodomain transcription factor Pph13 binds to a palindromic P3 motif (that contains a Q50 motif, TAATTG) in broadly expressed phototransduction genes. Right, the activator Pph13 (blue) is expressed in all photoreceptors (R) of the unit eye and drives broad expression of phototransduction genes.
C: Left, the central base pairs of the RCSI motif differ in some rhodopsin genes, for instance in Rh6. This generates a K50 motif (TAATCC) that recruits the K50 homeodomain transcription factor Otd. Right, like Pph13 (blue), the activator Otd (green) is expressed in all photoreceptors of the unit eye and drives broad expression of its target genes.
D: Left, the same K50 motif (TAATCC) recruits the K50 homeodomain transcription factor Dve, which is a repressor. Right, the repressor Dve (red) is expressed in ‘outer’ photoreceptors (R1-R6) where it prevents activation of Rh6 by Pph13 and Otd. However, the ‘inner’ photoreceptor R8 does not express Dve and therefore Pph13 and Otd can activate Rh6 in this subset of photoreceptor neurons.
1.1. Drosophila photoreceptor neurons express two types of homeodomain transcription factors that prefer different palindromic motif variants
The homeodomain transcription factors PvuII-PstI homology 13 (Pph13) and Orthodenticle (Otd) drive the broad expression of phototransduction proteins in all photoreceptor neurons (Figs. 2b and 2c), but also – in combination with repressors – the restricted expression of specific color-sensing Rhodopsin proteins in different photoreceptor subtypes [21–25]. Like most homeodomain transcription factors [26–28], Pph13 and Otd bind TAAT homeodomain core motifs with high affinity [4,19,27–31]. However, their homeodomains differ in the amino acid residue at position 50 – Pph13 has a glutamine/Q50 while Otd has a lysine/K50 – that is part of the recognition helix and binds to specific base pairs that follow the TAAT core motif [4,22,32–34]. These central base pairs are a critical part of the transcription factor type-specific left and right half-sites (Fig. 2a): the homeodomain transcription factor Pph13/Q50 [30] prefers TA or TG to follow the TAAT core (TAATTA/G; Fig. 2a) [27,33] and binds as a homodimer to palindromic ‘P3’ motifs (consensus: TAATYNRATTA; Y = C or T and R = A or G) [27,33]. The term ‘P3’ refers to palindromic TAAT core motifs that are separated by three central base pairs (TAAT…ATTA) [22,33] (Figs. 2a and 2b, left). Specific variants of these central base pairs (TAATYNRATTA) can increase the binding affinity of the Q50 homodimer [4,35], likely due to a sequence-specific allosteric effect that mediates cooperativity [35]. In this model, the binding of the first Q50 homeodomain to the left half-site of the palindromic motif affects the conformation of the DNA-homeodomain complex and thereby promotes the binding of the second Q50 homeodomain to the right half-site (Fig. 2b, left).
The binding of a Pph13/Q50 homodimer to P3 motifs drives broad expression of photoreceptor genes in all photoreceptors through a shared palindromic P3 motif in their proximal promoter [21,22,36,37] (Fig. 2b). This ensures that all photoreceptor neurons express the same set of phototransduction proteins that transduce and amplify the visual stimulus. Strikingly, Q50-type P3 motifs with perfectly palindromic TAAT…ATTA core repeats and central base pairs that promote strong homodimer binding (albeit with slightly variable central symmetry, e.g., TGA vs. TGC or TAA vs. TAC; Fig. 2a) are only found in the promoters of broadly expressed phototransduction genes but not in the photoreceptor subtype-restricted rhodopsin genes Rh1-Rh5 (Fig. 2a, compare top to bottom).
In contrast to Pph13/Q50’s binding to palindromic P3 motifs as a homodimer, the homeodomain transcription factor Otd/K50 prefers to bind as a monomer to the TAATCC (K50) motif [27,33,34] (Fig. 2a), which is not present in the P3 motifs of broadly expressed phototransduction genes but is found in the motif variants of the rhodopsin genes Rh3 and Rh6 (Figs. 2a and 2c, left). While K50 homeodomain transcription factors are able to bind as dimers to a subset of P3 motifs (e.g. TAATCCGATTA) [34], they do so with only 25-fold cooperativity of the two K50 homeodomains [4]. As we describe below, these distinct cis-regulatory motif preferences play important roles in the differential expression of Drosophila photoreceptor genes.
1.2. Drosophila terminal photoreceptor differentiation uses imperfectly palindromic motifs for the combinatorial control of complex expression patterns
The P3 motif consensus TAATYNRATTA shows the most frequent bases for each position [33,35] and reflects the palindromic nature of the half-sites that are bound by the Pph13/Q50 homodimer. However, the consensus does not convey that specific sequence variants distinguish the two major types of spatial expression patterns, broad and restricted: while general phototransduction factors are broadly expressed in all photoreceptors, six color-sensing Rhodopsins (Rh1-Rh6) are restricted to non-overlapping subsets of photoreceptors for wavelength discrimination and color vision [20,21] (Fig. 2a). As we discuss below, deviations from the palindromic P3 consensus are critical for the differential expression of rhodopsins in specific subsets of photoreceptors.
1.2.1. Variation of central base pairs of the palindromic motif affects the spatial rhodopsin expression pattern
The generation of the six non-overlapping Rhodopsin expression patterns by the six proximal rhodopsin promoters [38] involves highly conserved and imperfectly palindromic P3 motif variants that are called Rhodopsin Core Sequence I (RCSI) motifs [39–41] (Fig. 2a, bottom). There are two main types of RCSI motif variants [21]. The first type involves specific configurations of the central base pairs. The central bases ‘CC’ generate a K50 motif (TAATCC) in Rh3 and Rh6 (TAATCCAATTC and TAATCCAATTA; Fig. 2a), as opposed to the Pph13/Q50 (TAATTG) motifs in Rh1, Rh2, and Rh4 (TAATTGAATTT and TAATTGGGTTA) (Figs. 2a–2c). Since the K50 motif is bound by the broadly expressed activator Otd (see above and Fig. 2c, right), the introduction of a high affinity Otd/K50 motif instead of a high affinity Pph13/Q50 motif in the Rh3 and Rh6 RCSI would merely maintain activation in all photoreceptors [23,27,38] (Fig. 2c, right). However, this switch to a K50 motif adds repression because the same K50 motif recruits the homeodomain repressor Dve that is expressed in a subset of photoreceptors (Fig. 2d) [21,42]. In combination with additional repressors [20], Dve/K50 restricts Rh3 and Rh6 to their ‘correct’ photoreceptor subtypes (Fig. 2d). This elegant switch from a Pph13/Q50 activator motif to a dual Otd activator/Dve repressor K50 motif only requires the replacement of two central base pairs of the palindromic motif. Consistent with the requirement of these K50 motifs for restricting rhodopsins, mutating the Otd/Dve K50 motif to a Pph13/Q50 motif causes a Pph13-dependent derepression of Rh3 and Rh6 reporters in other photoreceptor subsets in vivo and thus a loss of photoreceptor subtype-specificity, which is indispensable for color vision.
The molecular mechanism for how repression by Dve prevents Otd-mediated activation through binding to the same high affinity K50 motif is incompletely understood. Differences in the relative expression levels of Dve and Otd could provide the difference in specificity (rather than affinity [43]) that allows the repressor to outcompete the activator. Indeed, Dve’s cell type-specific expression levels are tightly controlled by combinatorial transcription factor input, enhancer redundancy, and autoregulation [44]. Moreover, insufficient Dve levels in dve hypomorphs cause an expansion of Rh3 and Rh6 into other photoreceptor subsets [42,45]. Dve could also prevail over Otd due to special properties of its DNA binding domains [46]: while Dve’s recognition helix closely resembles the one of Otd, its first and second helix are more homologous to POU domain transcription factors and thus could mediate interactions with yet to be identified sequences flanking the K50 motifs. Moreover, a second homeodomain has been identified in Dve that, together with the first homeodomain, generates higher order protein-DNA interactions with the rhodopsin promoters in vitro, which are neither shown by a single Dve homeodomain nor by Otd’s homeodomain [42].
The replacement of a high affinity Pph13/Q50 activator motif with a slightly different high affinity dual function Otd activator/Dve repressor K50 motif in rhodopsins to generate restricted expression patterns, i.e. the switch from high affinity binding of one transcription factor type to high affinity binding of another transcription factor type, differs from how Hox transcription factors direct restricted expression patterns in the Drosophila embryo: in this context, low affinity homeodomain binding sites that substantially differ from the consensus motif recruit specific Hox transcription factors in distinct regions of the embryo to generate spatially restricted expression patterns, while the conversion to high affinity motifs generates broad (ectopic) expression patterns [47].
1.2.2. Variations of the right palindromic half-site affect the spatial rhodopsin expression pattern
The second type of RCSI variation breaks the symmetry of the palindromic TAAT…ATTA repeat due to specific and highly conserved replacements of the last base pair in the right half-site’s ATTA motif (Fig. 2a). For instance, the inverted repeat is ATTC (in Rh3 or Rh5) or ATTT (in Rh1 or Rh4) instead of ATTA in the more symmetrical P3 motifs of broadly expressed phototransduction genes (Fig. 2a). The RCSI consensus is thus TAATYNRATTN rather than the P3 consensus TAATYNRATTA [39]. However, the RCSI consensus is misleading because it suggests that the last base pair could be any base and thus might be functionally irrelevant. Yet, each rhodopsin prefers a specific base pair that disrupts the 3’ ATTA core repeat (Fig. 2a). The functional relevance of these single base pair differences has been demonstrated with reporter constructs in vivo: for instance, mutating a single base pair to convert the 3’ ATTC motif of Rh3 or Rh5 to a perfect inverse ATTA repeat (TAAT…ATTA) causes a derepression of the Rh3 and Rh5 reporters in other photoreceptor subsets [21]. Therefore, the specific RCSI motif variant in each rhodopsin gene is fine-tuned to recruit specific combinations of transcription factors to generate a specific spatial expression pattern. The ‘imperfections’ of the palindrome are critical for photoreceptor subtype-specificity: the RCSI motif of a given rhodopsin can neither be replaced with a palindromic P3 motif nor another RCSI sequence variant without compromising the spatial expression pattern of this rhodopsin [21,38].
In summary, the cis-regulatory logic of Drosophila terminal photoreceptor differentiation involves subtle single base pair differences that generate imperfectly palindromic variants and introduce novel motifs for specific combinations of transcriptional activators and repressors. Asymmetries in a shared palindromic motif thus allow combinatorial regulation that facilitates the generation of complex gene expression patterns, which form the basis for color vision [38].
2. Steroid receptors regulate mammalian gene expression through palindromic hormone response elements
Mammalian steroid hormone signaling uses palindromic cis-regulatory motifs to control a variety of essential processes such as immune responses, inflammation, metabolism, and the development of sexual characteristics. Steroid hormones are ligands that bind to sequence-specific transcription factors called steroid receptors [48–52] (Fig. 3), which include the glucocorticoid, mineralocorticoid, androgen, estrogen, and progesterone receptors [53]. In the absence of a steroid hormone ligand, steroid receptors remain associated with chaperones in the cytoplasm [48]. The binding of a steroid hormone activates and releases the steroid receptors from the chaperone, allows them to homodimerize, and exposes their nuclear localization sequence [54]. After translocation to the nucleus, steroid receptors bind to palindromic cis-regulatory motifs called Hormone Response Elements (HREs, Fig. 3). HREs typically consist of two hexameric half-sites that are separated by a central three base pair sequence which acts as a spacer [48,51]. Similar to specific P3 and RCSI motif variants that recruit different transcription factors as monomers or dimers (see above), the specific HRE sequence determines whether a steroid receptor binds as a monomer or a dimer [53–56] (Fig. 3).
Figure 3.
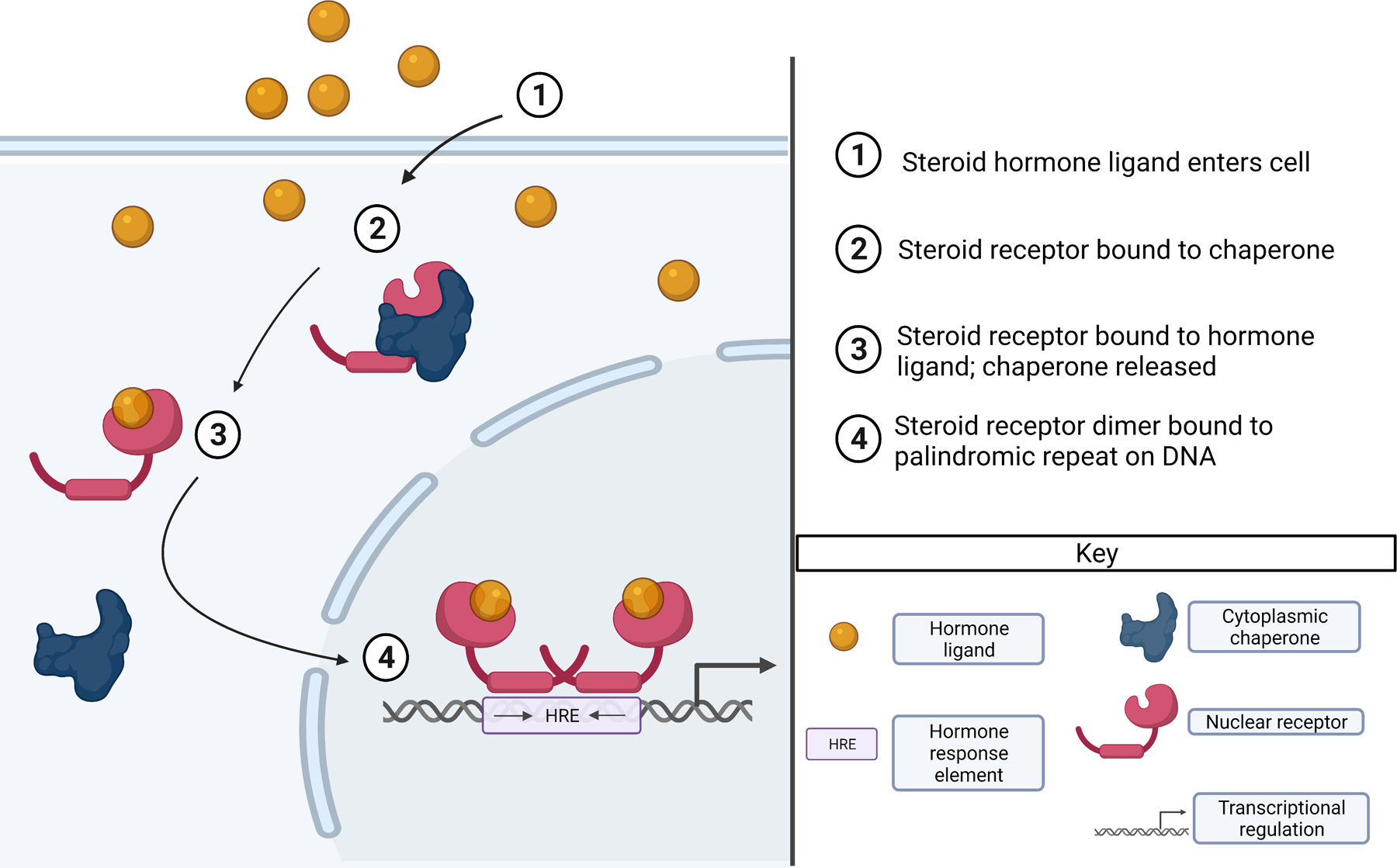
The schematic shows how a steroid hormone conveys a signal through binding to a steroid receptor. The steroid hormone enters the cell (1) and binds to the steroid receptor, which is associated with a chaperone (2). Binding the steroid hormone results in the release of the steroid receptor from the chaperone (3). After translocation to the nucleus, the steroid receptor binds as a dimer to a palindromic motif called Hormone Response Element (HRE) in regulatory DNA (4).
The corticosteroid hormone-induced binding of the glucocorticoid receptor (GR) to the glucocorticoid response element (GRE), which controls large gene regulatory networks and diverse processes in a tissue-specific manner [57], is a powerful model for steroid receptor-HRE interactions [58]. In the classical model, GR binds cooperatively as a homodimer to a palindromic GRE (Fig. 4a) that consists of hexameric left and right half-sites that are separated by a central three base pair spacer (consensus: AGAACANNNTGTTCT) [5,49,59–62]. In this essay, we focus exclusively on direct effects of GR on target gene expression through GREs rather than indirect effects through protein-protein interactions [63].
Figure 4.
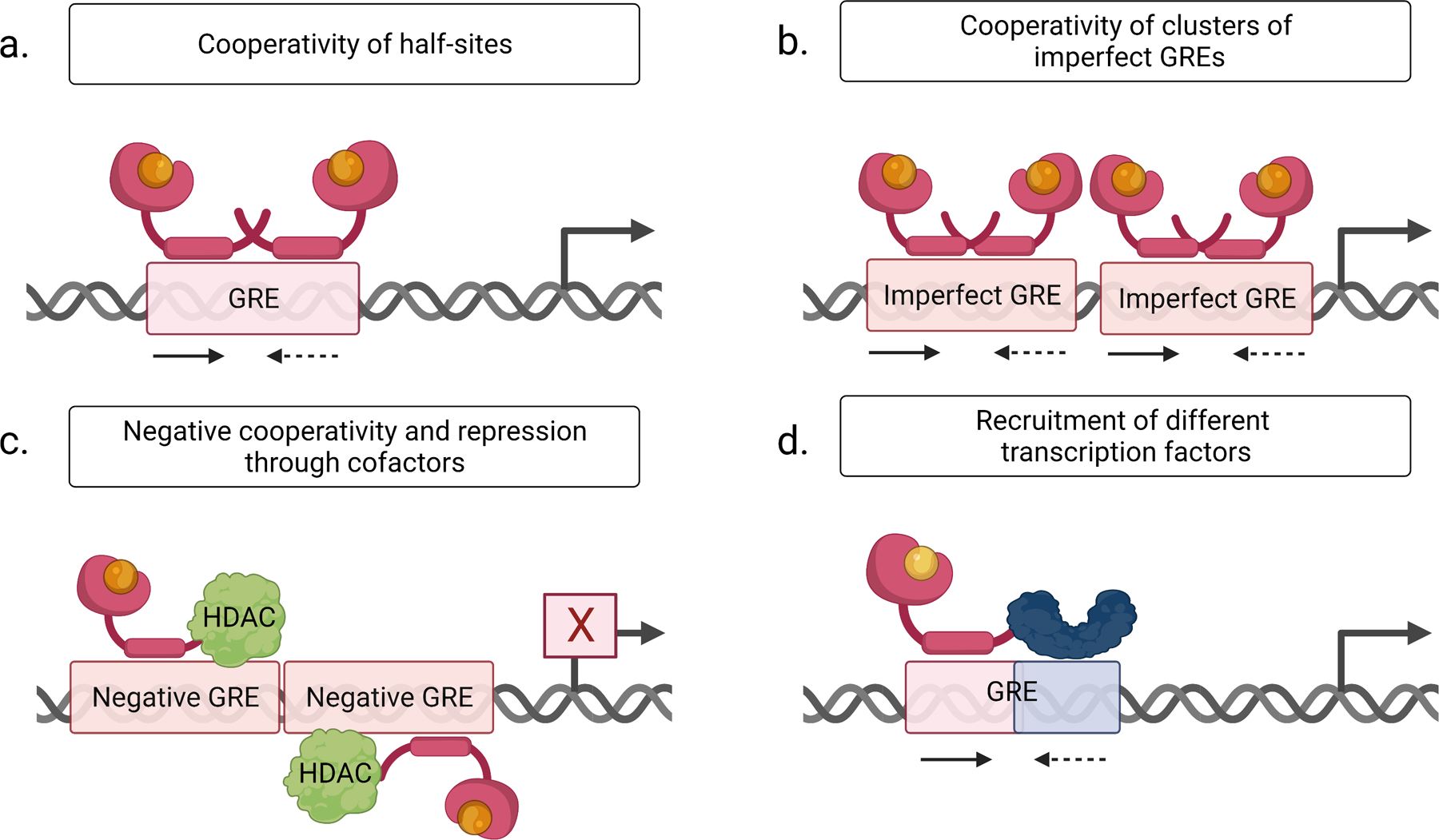
Mechanistic explanations for the predominance of imperfectly palindromic Glucocorticoid Response Elements (GREs) in glucocorticoid receptor signaling. For details, see text.
A: The left, high affinity half-site (arrow) facilitates glucocorticoid receptor binding to the right, imperfect half-site (dashed arrow) of the GRE.
B: Cooperativity of clusters of imperfectly (dashed arrow) palindromic GREs.
C: ‘Negative GREs’ have been proposed to be bound by glucocorticoid receptor monomers with negative cooperativity and reversed binding polarity, which results in new cofactor interactions (e.g. with a histone deacetylase, HDAC) and target gene repression.
D: A modified right half-site (blue, dashed arrow) of the GRE could recruit a different transcription factor (blue) and result in the formation of a heterodimer for combinatorial control of gene expression.
2.1. Imperfectly palindromic motifs in glucocorticoid receptor signaling: sequence variants and compensatory mechanisms
In this section, we discuss explanations for the predominance of imperfect GREs in the regulatory genome without the necessity of a specific functional role of specific sequence variants, which is covered in the section that follows. GREs that are bound by GR in vivo are usually imperfectly palindromic [58] (Fig. 4a): some base pair positions of the GRE are more variable than others [64] and structural studies have revealed that each DNA binding domain of the GR homodimer contacts only three base pairs of each hexameric GRE half-site [58]. The symmetry of the half-sites in the GRE consensus sequence (AGAACANNNTGTTCT), which can be read as an absolute requirement for a specific base pair at each half-site position, is thus misleading. Instead, there are few GRE variants with perfectly symmetrical half-sites, while there is a larger pool of GRE variants with minor sequence differences between the half-sites that are imperfect matches to the consensus but still generate physiologically sufficient transcriptional outcomes. This rationale also applies to the larger repertoire of lower affinity transcription factor binding sites as compared to the much fewer options for ‘perfect’ highest affinity binding sites (see discussion for Hox binding sites [65]).
When individual imperfect palindromes drive suboptimal gene expression levels, their insufficiency could be counterbalanced by compensatory mechanisms. Three examples for such compensatory mechanisms are allosteric effects, motif clustering, or cooperative interactions with other types of transcription factors. Palindromic motifs are particularly suited for mediating allosteric interactions (see also above): the binding of a steroid receptor to a higher affinity half-site can facilitate binding of the second steroid receptor to an imperfect lower affinity half site and thereby promote homodimer formation in a cooperative manner [66] (Fig. 4a). To achieve physiologically optimal hormone-responsiveness and expression levels, perfectly symmetrical half-sites might thus be unnecessary, as long as the spacing and orientation of the second, imperfect half-site provide sufficient stability for the transcription factor dimer [67].
A second compensatory mechanism is the cooperativity of clusters of imperfectly palindromic (low affinity) GREs [68] (Fig. 4b), as has also been reported for clusters of estrogen receptor (ER) response elements (EREs) [69,70] and non-palindromic low affinity binding sites in other contexts [71–73]. EREs can interact in both positive and negative ways [69,70]; for instance, two neighboring and imperfectly palindromic EREs of the vit B1 gene [74] that are inactive on their own become functional when combined [75]. Similarly, two imperfectly palindromic GREs of the tyrosine aminotransferase gene, which are individually insufficient to stimulate transcription, drive target gene expression when combined [76]. Moreover, pairs of GREs can bioinformatically predict GR-responsiveness better than single GREs [68]. However, it remains unclear whether specific sequence variations of imperfect palindromes are required for these cooperative interactions in motif clusters.
A third compensatory mechanism is the increase of the affinity of imperfect palindromes through cooperative interactions with other transcription factors that bind the same regulatory region and provide a high affinity binding environment. An example is the weak binding of GR to non-consensus (imperfect) GREs in the PEPCK gene that is enhanced by accessory motifs for other types of transcription factors (COUP-TF/HNF4 and HNF3) that generate a high affinity binding environment [77]. Taken together, allosteric effects of GRE half-sites, the cooperativity of clusters of imperfect GREs, or interactions with other types of transcription factors allow imperfectly palindromic motifs to achieve robust levels of gene expression.
2.2. Conserved imperfectly palindromic motif variants in glucocorticoid receptor signaling: potential functional implications
While the roles of imperfect palindromes in Drosophila terminal photoreceptor differentiation are well understood, the much larger repertoire of imperfectly palindromic GRE variants remains understudied. Genome-wide binding analyses revealed that conserved GR-bound GREs vary substantially around the consensus sequence in vivo [78] and include half-sites that are bound by GR monomers [79–81]. Similar findings have been reported for EREs in estrogen-stimulated cultured human cells [82]: only 8% of the bound EREs matched the perfectly palindromic consensus sequence [83]. The evolutionary conservation of specific GRE variants strongly suggests that not all variations of individual base pairs are redundant but are functionally relevant, as is the case for RCSI variants (see above). In this section, we will consider functional roles of specific GRE variants, which could also apply to other contexts that employ palindromic motifs [6–8].
One function of imperfectly palindromic GRE variants could be to generate specific target gene expression levels. Rather than directing gene expression in a binary on/off manner, imperfectly palindromic GRE variants can generate a range of GR affinities that drive a range of target gene expression levels in vitro [66,84]. While GR’s affinity to a specific GRE variant is an important parameter that influences target gene expression in vivo, its relative contribution needs further research [84]: other parameters of the cellular context like the levels of the steroid hormone, cofactor interactions, allosteric effects, posttranslational modifications, flanking sequences [85], and the chromatin architecture also influence the steroid receptor-GRE interaction and the transcriptional response.
Like P3/RCSI half-sites, a GRE half-site acts as an allosteric ligand [14] that influences the conformation of the bound GR monomer. Subtle sequence differences in the GRE sequence of one half-site thus alter the conformation of the GR bound to that half-site, but also of the GR bound to the other half-site [15]. In addition, the allosteric influence of the specific GRE half-site sequence on the conformation of the bound GR dimer [14] can also modify the interaction with other nuclear receptors or cofactors that modify chromatin and thereby cause different transcriptional outcomes [14,15,61,86].
Moreover, specific deviations from the palindromic consensus can result in different modes of GR-mediated transcriptional regulation. GR can repress transcription and it has been proposed that this involves ‘negative GREs’ [87] (consensus: CTCCN0–2GGAGA) that are bound by GR monomers on opposite sides of the DNA with negative cooperativity (Fig. 4c). This involves the formation of a complex of the GR with corepressors and histone deacetylases [55,88]. However, such interactions of GR and negative GREs have not been reproducibly enriched on a genome-wide level in vivo and alternative competition-based mechanisms have been proposed [89].
Lastly, specific GRE half-site variants could recruit heterodimers for combinatorial gene regulation, like specific RCSI motif variants recruit different transcription factors for combinatorial control of rhodopsin expression. The heterodimer could involve specific isoforms of the same steroid receptor, different types of steroid receptors, or other types of transcription factors (Fig. 4d). For instance, the symmetric replacement of only one or two base pairs in each half-site is sufficient to convert an estrogen-responsive ERE [74,75] into a GR-responsive GRE [75,90–93]. Such similar binding site preferences allow for competition of different steroid receptor types for the same motif and other modes of regulation. Like Otd and Dve bind the same K50 motif but promote opposite transcriptional outcomes, ER and thyroid receptor (TR) bind the same half-sites [94] but TR decreases ER-mediated activation in vitro [95]. Conversely, ER stimulates the expression of the human glycoprotein hormone α subunit gene by antagonizing inhibitory TR binding at a TRE in the promoter [96]. It has been suggested that the TR binds to the imperfectly palindromic TRE as a monomer rather than a dimer [96,97]. Moreover, GR and the androgen receptor (AR) can bind to the same motifs, but different sequence preferences within and outside the half-sites affect the binding specificity [98]. With increasing expression in some castration-resistant prostate cancers, the more promiscuous GR can bind to AREs and thus functionally substitute AR [98].
Taken together, genomic GREs could be divided into distinct subsets of functionally specialized variants [79] that direct different transcriptional outcomes by inducing particular GR conformations [14], cofactor interactions or competition, and/or heterodimer formation.
3. The use of imperfectly palindromic motifs in two gene regulatory contexts
The fact that two very different gene regulatory contexts use imperfectly palindromic motifs for various functions suggests that these motifs are versatile tools to fine-tune gene expression. In the Drosophila eye, stably maintained homeodomain transcription factors bind palindromic motifs with different degrees of symmetry to control the terminal differentiation of different color photoreceptor fates that need to be maintained throughout the entire life of the photoreceptor neurons. In mammalian steroid hormone signaling, steroid hormones, whose levels are variable due to circadian regulation, bind imperfectly palindromic motifs to control the tissue-specific differentiation, growth, and metabolism of a large number of cell types in a transient or a long-lasting manner. Moreover, GR signaling is very complex, because it has different effects in different organs and target genes can be activated by glucocorticoids in one tissue but repressed in another. It is conceivable that these differences between the two contexts affect the usage and diversity of the imperfectly palindromic motif variants: the pool of GRE variants might be more diverse and mediate a larger range of affinities because variable levels of steroid hormones need to control a variety of tissue-specific target gene responses, while P3/RCSI motif sequences might be more constrained and require higher affinity binding sites because rhodopsins and phototransduction genes need to be expressed and maintained at very high levels.
However, there are mechanistic similarities between the two regulatory contexts. First, in both, imperfectly palindromic motifs consist of two asymmetric half-sites that promote the cooperativity of two transcription factors and allosterically control their conformations, which affects the expression of the target gene. Second, the DNA binding domains of different types of homeodomain transcription factors or different types of steroid receptors can have a common evolutionary origin, which means that specific single base pair changes in the palindromic motif can recruit different (but related) transcription factors to change the mode of transcriptional regulation, for instance from activation to repression. Just like single base pair changes in palindromic P3/RCSI motifs recruit a different type of homeodomain transcription factor (K50 instead of Q50), single base pair differences in palindromic HRE motifs recruit a different type of steroid receptor (GR instead of ER). Third, the target gene-specific evolutionary conservation of specific imperfectly palindromic variants strongly suggests that they are functionally relevant. Like different rhodopsin genes ‘prefer’ specific evolutionarily conserved RCSI variants that deviate from the more symmetrical P3 consensus and have been shown to be critical for the spatial expression pattern, GR-regulated genes ‘prefer’ specific evolutionarily conserved deviations from the GRE consensus sequence in vivo [78]. Future studies will elucidate whether sets of conserved GRE variants similarly correlate with specific gene expression patterns.
4. Conclusions and future directions
Since the functional significance of evolutionarily conserved imperfectly palindromic GRE variants remains poorly understood, an important goal of future studies will be the identification of functional GRE variants in primary corticosteroid-responsive target genes, i.e. whose expression is modulated by the presence or absence of an exogenous glucocorticoid (e.g. dexamethasone) [99] and shows a short response time after hormone treatment. GR occupancy data alone will not suffice because they can include unspecific DNA binding events. Conversely, since motifs that match the general GRE consensus or the position weight matrix occur in any sufficiently long stretch of DNA [100], the role of specific GRE sequence variants cannot be deciphered by merely correlating the presence of a specific GRE variant with a transcriptional effect on a nearby target gene. In this respect, the evolutionary conservation of GRE sequences is a valuable indicator of functionality, since it is sufficient to predict GR occupancy at glucocorticoid-induced genes, while there is no such correlation at glucocorticoid-repressed genes [78].
The strength of the Drosophila terminal differentiation context for the analysis of cis-regulatory palindromic motifs is that the rhodopsin promoters have been dissected in detail, the binding sites for key regulators (Pph13, Otd, Dve, etc.) are known, and motif mutations can be correlated with changes of expression [21,38]. Since the GRE-controlled genes are understudied, the next critical step will be to perform functional analyses of GRE variants through mutations that either inactivate a GRE or swap a GRE variant with another conserved GRE variant (analogous to swapping RCSI motifs) in reporter assays or through CRISPR-Cas9-mediated genome editing. Complementary computational approaches will help elucidate whether specific functional GRE motif variants can be used to predict hormone responsiveness, tissue-specific effects, and/or transcriptional outcomes such as activation or repression and expression levels.
Gaining more detailed mechanistic insights into the tissue-specific regulation of target genes by specific imperfectly palindromic GRE variants has clinical relevance, since synthetic glucocorticoids are widely prescribed for various immune or inflammatory disorders [101]: a deeper understanding of the underlying mechanisms could be the foundation for personalized therapies due to better predictions of the outcomes of steroid hormone treatments as well as the patients’ sensitivity to steroid treatments. It is conceivable that specific motif variants make patients vulnerable to specific diseases and/or unwanted side effects of long-term steroid treatments such as decreased bone mass and increased fracture risk [102]. Promisingly, patient-specific GRE sequence variants (albeit a minority of the individual-specific variants that affected GR binding) have been associated with adverse effects on GR-mediated gene regulation and glucocorticoid treatment in adipocytes and hepatocytes from patient-derived stem cells [103]. Similar studies could ultimately lead to the development of personalized steroid treatments with fewer side effects.
Acknowledgements
We thank Changmeng Cai, Alexey Veraksa, Steve Small, Fareeha Syeda, Romaisa Shahid, Grace Carey, and Adaira Dumm for comments on an earlier version of this manuscript. Images for the figures were generated with Biorender.com. This work was supported by an R00/Pathway to Independence Award (R00EY023995) to J.R. from the NEI/NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wittkopp PJ, Kalay G. 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13: 59–69. [DOI] [PubMed] [Google Scholar]
- 2.Davidson EH. 2006. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution San Diego: Elsevier Science.
- 3.Yanez-Cuna JO, Kvon EZ, Stark A. 2013. Deciphering the transcriptional cis-regulatory code. Trends Genet 29: 11–22. [DOI] [PubMed] [Google Scholar]
- 4.Wilson D, Sheng G, Lecuit T, Dostatni N, et al. 1993. Cooperative dimerization of paired class homeo domains on DNA. Genes Dev 7: 2120–34. [DOI] [PubMed] [Google Scholar]
- 5.Drouin J, Sun YL, Tremblay S, Lavender P, et al. 1992. Homodimer formation is rate-limiting for high affinity DNA binding by glucocorticoid receptor. Mol Endocrinol 6: 1299–309. [DOI] [PubMed] [Google Scholar]
- 6.Chinenov Y, Kerppola TK. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20: 2438–52. [DOI] [PubMed] [Google Scholar]
- 7.Mechta-Grigoriou F, Gerald D, Yaniv M. 2001. The mammalian Jun proteins: redundancy and specificity. Oncogene 20: 2378–89. [DOI] [PubMed] [Google Scholar]
- 8.Carroll PA, Freie BW, Mathsyaraja H, Eisenman RN. 2018. The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis. Front Med 12: 412–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orian A, van Steensel B, Delrow J, Bussemaker HJ, et al. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev 17: 1101–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackwood EM, Eisenman RN. 1991. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251: 1211–7. [DOI] [PubMed] [Google Scholar]
- 11.Blackwood EM, Kretzner L, Eisenman RN. 1992. Myc and Max function as a nucleoprotein complex. Curr Opin Genet Dev 2: 227–35. [DOI] [PubMed] [Google Scholar]
- 12.Fieber W, Schneider ML, Matt T, Krautler B, et al. 2001. Structure, function, and dynamics of the dimerization and DNA-binding domain of oncogenic transcription factor v-Myc. J Mol Biol 307: 1395–410. [DOI] [PubMed] [Google Scholar]
- 13.James L, Eisenman RN. 2002. Myc and Mad bHLHZ domains possess identical DNA-binding specificities but only partially overlapping functions in vivo. Proc Natl Acad Sci U S A 99: 10429–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meijsing SH, Pufall MA, So AY, Bates DL, et al. 2009. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324: 407–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watson LC, Kuchenbecker KM, Schiller BJ, Gross JD, et al. 2013. The glucocorticoid receptor dimer interface allosterically transmits sequence-specific DNA signals. Nat Struct Mol Biol 20: 876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobert O, Kratsios P. 2019. Neuronal identity control by terminal selectors in worms, flies, and chordates. Curr Opin Neurobiol 56: 97–105. [DOI] [PubMed] [Google Scholar]
- 17.Hobert O 2011. Regulation of terminal differentiation programs in the nervous system. Annu Rev Cell Dev Biol 27: 681–96. [DOI] [PubMed] [Google Scholar]
- 18.Hobert O 2008. Regulatory logic of neuronal diversity: terminal selector genes and selector motifs. Proc Natl Acad Sci U S A 105: 20067–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gehring WJ, Affolter M, Burglin T. 1994. Homeodomain proteins. Annu Rev Biochem 63: 487–526. [DOI] [PubMed] [Google Scholar]
- 20.Rister J, Desplan C, Vasiliauskas D. 2013. Establishing and maintaining gene expression patterns: insights from sensory receptor patterning. Development 140: 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rister J, Razzaq A, Boodram P, Desai N, et al. 2015. Single-base pair differences in a shared motif determine differential Rhodopsin expression. Science 350: 1258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra M, Oke A, Lebel C, McDonald EC, et al. 2010. Pph13 and orthodenticle define a dual regulatory pathway for photoreceptor cell morphogenesis and function. Development 137: 2895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tahayato A, Sonneville R, Pichaud F, Wernet MF, et al. 2003. Otd/Crx, a dual regulator for the specification of ommatidia subtypes in the Drosophila retina. Dev Cell 5: 391–402. [DOI] [PubMed] [Google Scholar]
- 24.Jukam D, Xie B, Rister J, Terrell D, et al. 2013. Opposite feedbacks in the Hippo pathway for growth control and neural fate. Science 342: 1238016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedrich M, Cook T, Zelhof AC. 2016. Ancient default activators of terminal photoreceptor differentiation in the pancrustacean compound eye: the homeodomain transcription factors Otd and Pph13. Current opinion in insect science 13: 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Affolter M, Slattery M, Mann RS. 2008. A lexicon for homeodomain-DNA recognition. Cell 133: 1133–5. [DOI] [PubMed] [Google Scholar]
- 27.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, et al. 2008. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 133: 1277–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berger MF, Badis G, Gehrke AR, Talukder S, et al. 2008. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133: 1266–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolberger C 1996. Homeodomain interactions. Curr Opin Struct Biol 6: 62–8. [DOI] [PubMed] [Google Scholar]
- 30.Goriely A, Mollereau B, Coffinier C, Desplan C. 1999. Munster, a novel paired-class homeobox gene specifically expressed in the Drosophila larval eye. Mech Dev 88: 107–10. [DOI] [PubMed] [Google Scholar]
- 31.Finkelstein R, Smouse D, Capaci TM, Spradling AC, et al. 1990. The orthodenticle gene encodes a novel homeo domain protein involved in the development of the Drosophila nervous system and ocellar visual structures. Genes Dev 4: 1516–27. [DOI] [PubMed] [Google Scholar]
- 32.Datta RR, Ling J, Kurland J, Ren X, et al. 2018. A feed-forward relay integrates the regulatory activities of Bicoid and Orthodenticle via sequential binding to suboptimal sites. Genes Dev 32: 723–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treisman J, Gonczy P, Vashishtha M, Harris E, et al. 1989. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell 59: 553–62. [DOI] [PubMed] [Google Scholar]
- 34.Wilson DS, Sheng G, Jun S, Desplan C. 1996. Conservation and diversification in homeodomain-DNA interactions: a comparative genetic analysis. Proc Natl Acad Sci U S A 93: 6886–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson DS, Guenther B, Desplan C, Kuriyan J. 1995. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell 82: 709–19. [DOI] [PubMed] [Google Scholar]
- 36.Ranade SS, Yang-Zhou D, Kong SW, McDonald EC, et al. 2008. Analysis of the Otd-dependent transcriptome supports the evolutionary conservation of CRX/OTX/OTD functions in flies and vertebrates. Dev Biol 315: 521–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zelhof AC, Koundakjian E, Scully AL, Hardy RW, et al. 2003. Mutation of the photoreceptor specific homeodomain gene Pph13 results in defects in phototransduction and rhabdomere morphogenesis. Development 130: 4383–92. [DOI] [PubMed] [Google Scholar]
- 38.Poupault C, Choi D, Lam-Kamath K, Dewett D, et al. 2021. A combinatorial cis-regulatory logic restricts color-sensing Rhodopsins to specific photoreceptor subsets in Drosophila. PLoS Genet 17: e1009613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papatsenko D, Nazina A, Desplan C. 2001. A conserved regulatory element present in all Drosophila rhodopsin genes mediates Pax6 functions and participates in the fine-tuning of cell-specific expression. Mech Dev 101: 143–53. [DOI] [PubMed] [Google Scholar]
- 40.Fortini ME, Rubin GM. 1990. Analysis of cis-acting requirements of the Rh3 and Rh4 genes reveals a bipartite organization to rhodopsin promoters in Drosophila melanogaster. Genes Dev 4: 444–63. [DOI] [PubMed] [Google Scholar]
- 41.Mismer D, Rubin GM. 1989. Definition of cis-acting elements regulating expression of the Drosophila melanogaster ninaE opsin gene by oligonucleotide-directed mutagenesis. Genetics 121: 77–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnston RJ Jr., Otake Y, Sood P, Vogt N, et al. 2011. Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye. Cell 145: 956–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Ho TD, Buchler NE, Gordan R. 2021. Competition for DNA binding between paralogous transcription factors determines their genomic occupancy and regulatory functions. Genome Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan J, Anderson C, Viets K, Tran S, et al. 2017. Regulatory logic driving stable levels of defective proventriculus expression during terminal photoreceptor specification in flies. Development 144: 844–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thanawala SU, Rister J, Goldberg GW, Zuskov A, et al. 2013. Regional modulation of a stochastically expressed factor determines photoreceptor subtypes in the Drosophila retina. Dev Cell 25: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakagoshi H, Hoshi M, Nabeshima Y, Matsuzaki F. 1998. A novel homeobox gene mediates the Dpp signal to establish functional specificity within target cells. Genes Dev 12: 2724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crocker J, Abe N, Rinaldi L, McGregor AP, et al. 2015. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 160: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beato M 1991. Transcriptional control by nuclear receptors. FASEB J 5: 2044–51. [DOI] [PubMed] [Google Scholar]
- 49.Evans RM. 1988. The steroid and thyroid hormone receptor superfamily. Science 240: 889–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans RM, Mangelsdorf DJ. 2014. Nuclear Receptors, RXR, and the Big Bang. Cell 157: 255–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Green S 1991. Nuclear hormone receptors and rational drug discovery. Biochem Soc Trans 19: 894–7. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto KR. 1985. Steroid receptor regulated transcription of specific genes and gene networks. Annu Rev Genet 19: 209–52. [DOI] [PubMed] [Google Scholar]
- 53.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83: 835–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sever R, Glass CK. 2013. Signaling by nuclear receptors. Cold Spring Harb Perspect Biol 5: a016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hudson WH, Youn C, Ortlund EA. 2013. The structural basis of direct glucocorticoid-mediated transrepression. Nat Struct Mol Biol 20: 53–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cotnoir-White D, Laperriere D, Mader S. 2011. Evolution of the repertoire of nuclear receptor binding sites in genomes. Mol Cell Endocrinol 334: 76–82. [DOI] [PubMed] [Google Scholar]
- 57.Kadmiel M, Cidlowski JA. 2013. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci 34: 518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weikum ER, Knuesel MT, Ortlund EA, Yamamoto KR. 2017. Glucocorticoid receptor control of transcription: precision and plasticity via allostery. Nat Rev Mol Cell Biol 18: 159–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strahle U, Klock G, Schutz G. 1987. A DNA sequence of 15 base pairs is sufficient to mediate both glucocorticoid and progesterone induction of gene expression. Proc Natl Acad Sci U S A 84: 7871–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wrange O, Eriksson P, Perlmann T. 1989. The purified activated glucocorticoid receptor is a homodimer. J Biol Chem 264: 5253–9. [PubMed] [Google Scholar]
- 61.Luisi BF, Xu WX, Otwinowski Z, Freedman LP, et al. 1991. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature 352: 497–505. [DOI] [PubMed] [Google Scholar]
- 62.Tsai SY, Carlstedt-Duke J, Weigel NL, Dahlman K, et al. 1988. Molecular interactions of steroid hormone receptor with its enhancer element: evidence for receptor dimer formation. Cell 55: 361–9. [DOI] [PubMed] [Google Scholar]
- 63.Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, et al. 1998. DNA binding of the glucocorticoid receptor is not essential for survival. Cell 93: 531–41. [DOI] [PubMed] [Google Scholar]
- 64.Timmermans S, Souffriau J, Libert C. 2019. A General Introduction to Glucocorticoid Biology. Front Immunol 10: 1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Crocker J, Noon EP, Stern DL. 2016. The Soft Touch: Low-Affinity Transcription Factor Binding Sites in Development and Evolution. Curr Top Dev Biol 117: 455–69. [DOI] [PubMed] [Google Scholar]
- 66.La Baer J, Yamamoto KR. 1994. Analysis of the DNA-binding affinity, sequence specificity and context dependence of the glucocorticoid receptor zinc finger region. J Mol Biol 239: 664–88. [DOI] [PubMed] [Google Scholar]
- 67.Dahlman-Wright K, Wright A, Gustafsson JA, Carlstedt-Duke J. 1991. Interaction of the glucocorticoid receptor DNA-binding domain with DNA as a dimer is mediated by a short segment of five amino acids. J Biol Chem 266: 3107–12. [PubMed] [Google Scholar]
- 68.van Batenburg MF, Li H, Polman JA, Lachize S, et al. 2010. Paired hormone response elements predict caveolin-1 as a glucocorticoid target gene. PLoS One 5: e8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carleton JB, Ginley-Hidinger M, Berrett KC, Layer RM, et al. 2020. Regulatory sharing between estrogen receptor alpha bound enhancers. Nucleic Acids Res 48: 6597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carleton JB, Berrett KC, Gertz J. 2017. Multiplex Enhancer Interference Reveals Collaborative Control of Gene Regulation by Estrogen Receptor alpha-Bound Enhancers. Cell Syst 5: 333–44 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kratsios P, Stolfi A, Levine M, Hobert O. 2011. Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat Neurosci 15: 205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Serrano-Saiz E, Gulez B, Pereira L, Gendrel M, et al. 2020. Modular Organization of Cis-regulatory Control Information of Neurotransmitter Pathway Genes in Caenorhabditis elegans. Genetics 215: 665–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ludwig MZ, Manu, Kittler R, White KP, et al. 2011. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS Genet 7: e1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walker P, Germond JE, Brown-Luedi M, Givel F, et al. 1984. Sequence homologies in the region preceding the transcription initiation site of the liver estrogen-responsive vitellogenin and apo-VLDLII genes. Nucleic Acids Res 12: 8611–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Martinez E, Givel F, Wahli W. 1987. The estrogen-responsive element as an inducible enhancer: DNA sequence requirements and conversion to a glucocorticoid-responsive element. EMBO J 6: 3719–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jantzen HM, Strahle U, Gloss B, Stewart F, et al. 1987. Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell 49: 29–38. [DOI] [PubMed] [Google Scholar]
- 77.Stafford JM, Wilkinson JC, Beechem JM, Granner DK. 2001. Accessory factors facilitate the binding of glucocorticoid receptor to the phosphoenolpyruvate carboxykinase gene promoter. J Biol Chem 276: 39885–91. [DOI] [PubMed] [Google Scholar]
- 78.So AY, Chaivorapol C, Bolton EC, Li H, et al. 2007. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet 3: e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schiller BJ, Chodankar R, Watson LC, Stallcup MR, et al. 2014. Glucocorticoid receptor binds half sites as a monomer and regulates specific target genes. Genome Biol 15: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.John S, Sabo PJ, Thurman RE, Sung MH, et al. 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43: 264–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miranda TB, Morris SA, Hager GL. 2013. Complex genomic interactions in the dynamic regulation of transcription by the glucocorticoid receptor. Mol Cell Endocrinol 380: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mason CE, Shu FJ, Wang C, Session RM, et al. 2010. Location analysis for the estrogen receptor-alpha reveals binding to diverse ERE sequences and widespread binding within repetitive DNA elements. Nucleic Acids Res 38: 2355–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klein-Hitpass L, Schorpp M, Wagner U, Ryffel GU. 1986. An estrogen-responsive element derived from the 5’ flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell 46: 1053–61. [DOI] [PubMed] [Google Scholar]
- 84.Bain DL, Yang Q, Connaghan KD, Robblee JP, et al. 2012. Glucocorticoid receptor-DNA interactions: binding energetics are the primary determinant of sequence-specific transcriptional activity. J Mol Biol 422: 18–32. [DOI] [PubMed] [Google Scholar]
- 85.Schone S, Jurk M, Helabad MB, Dror I, et al. 2016. Sequences flanking the core-binding site modulate glucocorticoid receptor structure and activity. Nature communications 7: 12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hard T, Kellenbach E, Boelens R, Maler BA, et al. 1990. Solution structure of the glucocorticoid receptor DNA-binding domain. Science 249: 157–60. [DOI] [PubMed] [Google Scholar]
- 87.Dostert A, Heinzel T. 2004. Negative glucocorticoid receptor response elements and their role in glucocorticoid action. Curr Pharm Des 10: 2807–16. [DOI] [PubMed] [Google Scholar]
- 88.Surjit M, Ganti KP, Mukherji A, Ye T, et al. 2011. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell 145: 224–41. [DOI] [PubMed] [Google Scholar]
- 89.Gerber AN, Newton R, Sasse SK. 2021. Repression of transcription by the glucocorticoid receptor: A parsimonious model for the genomics era. J Biol Chem 296: 100687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Klock G, Strahle U, Schutz G. 1987. Oestrogen and glucocorticoid responsive elements are closely related but distinct. Nature 329: 734–6. [DOI] [PubMed] [Google Scholar]
- 91.Danielsen M, Hinck L, Ringold GM. 1989. Two amino acids within the knuckle of the first zinc finger specify DNA response element activation by the glucocorticoid receptor. Cell 57: 1131–8. [DOI] [PubMed] [Google Scholar]
- 92.Danielsen M, Northrop JP, Jonklaas J, Ringold GM. 1987. Domains of the glucocorticoid receptor involved in specific and nonspecific deoxyribonucleic acid binding, hormone activation, and transcriptional enhancement. Mol Endocrinol 1: 816–22. [DOI] [PubMed] [Google Scholar]
- 93.Green S, Chambon P. 1988. Nuclear receptors enhance our understanding of transcription regulation. Trends Genet 4: 309–14. [DOI] [PubMed] [Google Scholar]
- 94.Vasudevan N, Koibuchi N, Chin WW, Pfaff DW. 2001. Differential crosstalk between estrogen receptor (ER)alpha and ERbeta and the thyroid hormone receptor isoforms results in flexible regulation of the consensus ERE. Brain Res Mol Brain Res 95: 9–17. [DOI] [PubMed] [Google Scholar]
- 95.Glass CK, Holloway JM, Devary OV, Rosenfeld MG. 1988. The thyroid hormone receptor binds with opposite transcriptional effects to a common sequence motif in thyroid hormone and estrogen response elements. Cell 54: 313–23. [DOI] [PubMed] [Google Scholar]
- 96.Yarwood NJ, Gurr JA, Sheppard MC, Franklyn JA. 1993. Estradiol modulates thyroid hormone regulation of the human glycoprotein hormone alpha subunit gene. J Biol Chem 268: 21984–9. [PubMed] [Google Scholar]
- 97.Brent GA, Williams GR, Harney JW, Forman BM, et al. 1992. Capacity for cooperative binding of thyroid hormone (T3) receptor dimers defines wild type T3 response elements. Mol Endocrinol 6: 502–14. [DOI] [PubMed] [Google Scholar]
- 98.Zhang L, Martini GD, Rube HT, Kribelbauer JF, et al. 2018. SelexGLM differentiates androgen and glucocorticoid receptor DNA-binding preference over an extended binding site. Genome Res 28: 111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Phuc Le P, Friedman JR, Schug J, Brestelli JE, et al. 2005. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet 1: e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sasse SK, Zuo Z, Kadiyala V, Zhang L, et al. 2015. Response Element Composition Governs Correlations between Binding Site Affinity and Transcription in Glucocorticoid Receptor Feed-forward Loops. J Biol Chem 290: 19756–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vandevyver S, Dejager L, Libert C. 2014. Comprehensive overview of the structure and regulation of the glucocorticoid receptor. Endocr Rev 35: 671–93. [DOI] [PubMed] [Google Scholar]
- 102.Boling EP. 2004. Secondary osteoporosis: underlying disease and the risk for glucocorticoid-induced osteoporosis. Clin Ther 26: 1–14. [DOI] [PubMed] [Google Scholar]
- 103.Hu W, Jiang C, Kim M, Yang W, et al. 2021. Individual-specific functional epigenomics reveals genetic determinants of adverse metabolic effects of glucocorticoids. Cell metabolism 33: 1592–609 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]


