Abstract
Lipid droplets (LDs) are intracellular organelles whose primary function is energy storage. Known to emerge from the endoplasmic reticulum (ER) bilayer, LDs have a unique structure with a core consisting of neutral lipids, triacylglycerol (TG) or sterol esters (SE), surrounded by a phospholipid (PL) monolayer and decorated by proteins that come and go throughout their complex lifecycle. In this Feature Article, we review recent developments in computational studies of LDs, a rapidly growing area of research. We highlight how molecular dynamics (MD) simulations have provided valuable molecular-level insight into LD targeting and LD biogenesis. Additionally, we review the physical properties of TG from different force fields compared with experimental data. Possible future directions and challenges are discussed.
Introduction
Lipid droplets (LDs) are storage organelles that play a central role in lipid metabolism. With a core composed of neutral lipids such as triacylglycerol (TG) or sterol esters (SE), they are the organelle surrounded by a phospholipid (PL) monolayer as opposed to a bilayer.1,2 They are universal intracellular organelles but are found in greater abundance in adipose cells because their principal function is to store excess energy in the form of highly reduced TG. The neutral lipids are also used as building blocks to generate new membranes. Although they were once considered to be mostly static, LDs are now recognized as highly dynamic organelles whose size, composition, and population respond to the metabolic needs of the cell. For instance, if cells have excess energy, fatty acids will be synthesized into TG and stored in growing LDs. In contrast, if cells require energy, TG will be hydrolyzed to provide energy and LDs will reduce in size. These lipogenesis and lipolysis processes are controlled by coat proteins on LD surfaces.
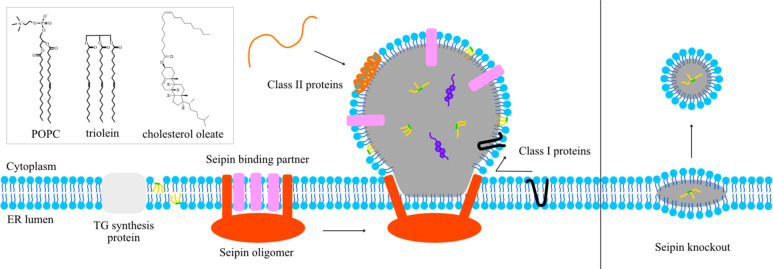
LDs are formed at the endoplasmic reticulum (ER) bilayer. Current models posit that TG is synthesized in the ER membrane by diacylglycerol acyltransferase (DGAT) enzymes and released into the bilayer (Figure 1). Once they reach a critical concentration, dispersed TG in the bilayer undergoes a phase separation and forms a distinct phase between the ER bilayer leaflets.3 Provided that cells have extra energy (e.g., in experiments, cells are incubated with oleate), TG will be continuously synthesized, resulting in the growth of the TG clusters. This process is accompanied by membrane remodeling in the ER bilayer in which the two leaflets of the bilayer are separated and the space between the leaflets is filled with TG. In the current model, the ER protein seipin maintains an ER–LD contact site via a unique neck structure, facilitating LD growth by enabling continuous transport of TG from ER to LD (Figure 1).4,5 Consistent with this perspective, seipin knockout (KO) cells have much smaller LDs accumulated at the early time of LD formation after oleate treatment because of improper LD maturation.6
Figure 1.
Illustration of lipid droplet formation and targeting. The chemical structures of three primary molecules in the system are shown in the upper left.
Although MD simulations are more advanced in their application to biomolecules such as proteins,7 they have also been extensively developed for PL bilayers because of their many critical cellular roles.8 Their thermodynamic, dynamical, mechanical, and structural properties have been calculated from the simulations and validated against the experimental data. While LD simulations are nowhere as numerous as bilayer simulations, the field is growing. In most all-atom simulations, the LD surface is modeled using a trilayer structure in which a bulk TG/SE phase is sandwiched by phospholipid monolayers (Figure 2). Since the typical diameter of a LD is greater than 500 nm, the approximation of treating LD as a trilayer structure with zero curvature is reasonable. While many important insights obtained from PL bilayers are helpful in studying LD systems via simulation, LDs should also be considered as different structures from bilayers. For instance, LDs have dynamically intercalating TG molecules within PL monolayers (Figure 2).9,10
Figure 2.
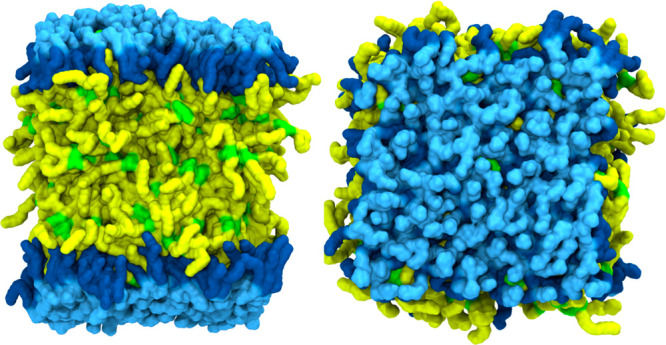
Trilayer model of LD. Side and top views are shown on the left and right, respectively. Sky blue and dark blue represent PL head/glycerol groups and PL acyl chains, respectively. Green and yellow indicate TG glycerol groups and TG acyl chains, respectively.
Lipoproteins have a similar PL-monolayer-based structure, with their core composed of various molar ratios of TG and SE.11,12 However, there are many notable differences between LDs and lipoproteins in their structures and locations. First, lipoproteins are much smaller than LDs: High-density lipoproteins, low-density lipoproteins, and very low-density lipoproteins have a typical diameter of 10, 25, and 60 nm, respectively, while LDs are usually larger than 500 nm. Second, lipoproteins have static apoproteins on their surfaces, stabilizing their structures. In contrast, LDs contain more coat proteins whose compositions dynamically change depending on the metabolic status and cellular cycles. Finally, lipoproteins are first budded to the ER lumen and then exported to blood plasma, while LDs are intracellular organelles located in the cytoplasm. However, given their comparable composition of lipids, it should be noted that knowledge obtained from one community enhances knowledge of the other. For instance, previous computational studies of lipoproteins made a good foundation for understanding PL–neutral lipid interactions and are therefore useful for LD simulators.13−16 Many experimental physical characterizations of lipoproteins are also helpful for the LD community.3,17−21
In this Feature Article, we review the recent developments in understanding LD targeting and LD biogenesis from all-atom (AA) and coarse-grained (CG) MD simulations. We also calculate the physical properties of TG with various force fields and evaluate the quality of the force fields for neutral lipids.
Physical Properties of Lipids
LD simulations have been performed with different force fields, including CHARMM,22 MARTINI,23 and Shinoda–DeVane–Klein (SDK).24 However, the original force fields do not include parameters for LD-relevant neutral lipids such as TG, diacylglycerol (DG), or SE. To circumvent these issues, the LD simulation community has developed its own parameters for neutral lipids. In this section, we summarize the recent developments in TG parameters and discuss the shared limitations.
There are key experimental quantities relevant when developing a force field for TG. The first is the interfacial tension of a bulk TG at the water interface (γTG/W), which is experimentally measured to be 32 mN/m.19 The Vanni group developed AA and CG force fields that can reproduce this experimental quantity.25,26 The second is the area per phospholipid of LDs (ALD), which is experimentally measured to be ∼15% higher than the area per phospholipid of bilayers (ABI).27,28 In other words, the surface area of the LD is approximately 15% larger than that of the bilayer if the number and composition of PLs in each leaflet are the same. Finally, the experimentally measured density of bulk TG is 0.899 g/mL.29 Accordingly, the volume occupied by a single TG molecule (VTG) is 1.64 nm3.
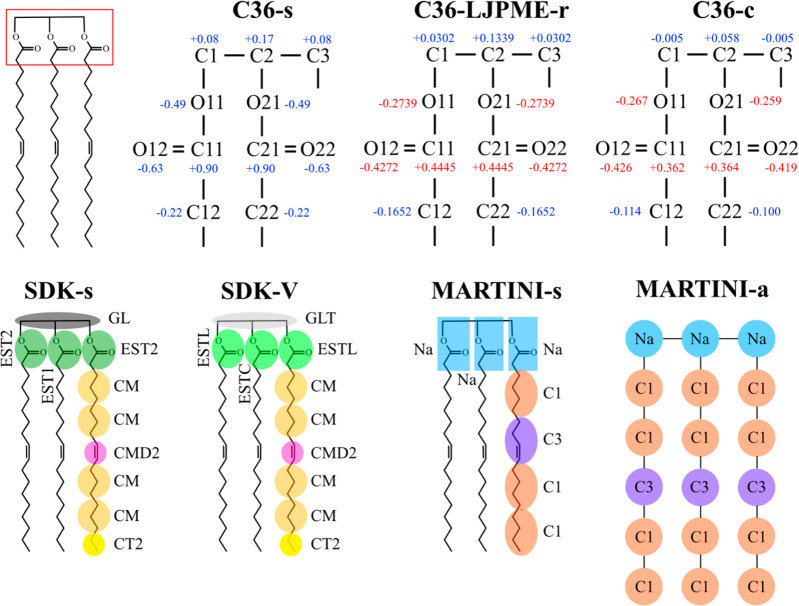
It is straightforward to obtain TG parameters from PL parameters. For instance, the TG force field can be constructed by replacing the headgroup (sn-3 position) of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) with its sn-1 chain. Such PL-derived TG force fields do not require new nonbonded parameters or partial charges, although there are new angles and dihedrals that may need to be defined. This strategy to make TG from PL without any modifications of partial charges and nonbonded parameters can be applied to various force fields. For the remainder of this article, the TG force fields obtained by modifying the reference PL force fields will be referred to as “standard force fields”. For instance, the AA force field for TG from CHARMM36 (C36) derived this way will be called C36-s (C36-standard). The same strategy can be used for MARTINI v2.2 and SDK, and the resulting force fields will be called MARTINI-s (MARTINI-standard) and SDK-s (SDK-standard), respectively.
Unfortunately, C36-s and SDK-s underestimate γTG/W (see Table 1). As the chains in these TG models are from PL force fields, the glycerol moieties of the standard force field TG are more hydrophilic than they should be when TG is in an oil phase. Previous studies also showed that water density in the core of a pure-TG LD calculated with C36-s is approximately 10 times higher than the experimentally measured value for water in bulk TG, 1.8 × 10–3 g/mL.9,30 To resolve this issue, various approaches have been taken for different force fields. The Vanni group has modified the depth of the potential well (epsilon) of the Lennard-Jones (LJ) interaction between the TG ester group (atom type GLT in SDK-V of Figure 3) and water to target the experimentally measured γTG/W.25 The resulting force field will be referred to as SDK-V (SDK-Vanni). Recent AA studies took an alternative approach of reducing the partial charges of the TG glycerol moiety. Applying this strategy, two of the present authors have developed a TG force field compatible with a LJ cutoff-free version of C36 (C36-LJPME).10,31 This force field will be referred to as C36-LJPME-r (C36-LJPME-revised), consistent with the name appearing in the original paper. Independently, the Vanni group has developed CHARMM force fields for TG with reduced partial charges of TG glycerol moiety, C36-c (C36-cutoff) and C36-p (C36-pme), compatible with C36 and C36-LJPME, respectively.26 Given the similarity between C36-c and C36-p, we will only discuss C36-c in this study.
Table 1. Calculated and Experimentally Measured Properties of TG, Bilayer, and LD.
| AA |
CG |
|||||||
|---|---|---|---|---|---|---|---|---|
| Exp | C36-s | C36-LJPME-r | C36-c | MARTINI-s | MARTINI-a | SDK-s | SDK-V | |
| VTG (nm3) | 1.64a | 1.67 | 1.64 | 1.67 | 1.60 | 1.89 | 1.63 | 1.63 |
| γTG/W (mN/m) | 32.0b | 25.3 | 34.0 | 31.5 | 31.1 | 37.6 | 17.6 | 32.9 |
| ABI (Å2) | 67.4c | 65.0 | 65.7 | 65.0 | 65.3 | 65.3 | 66.2 | 66.2 |
| ALD (Å2) | 78d | 76.9 | 67.9 | 67.3 | 66.3 | 66.0 | 68.4 | 67.3 |
| ALD/ABI | 15%d | 18.3% | 3.3% | 3.5% | 1.5% | 1.1% | 3.3% | 1.7% |
Figure 3.
TG force fields. The top panel shows the AA models and the bottom panel the CG models. The partial charges of the atoms in the glycerol moiety are shown in the AA force fields. Partial charges that deviate more than 0.2 from C36-s are indicated with red fonts in the top panel.
For MARTINI-s, no further modifications were necessary to reproduce γTG/W. However, as we will discuss later, TG does not nucleate with MARTINI-s at a critical concentration or above. Therefore, computational studies of LD formation have used MARTINI-a (MARTINI-addition of the atom) in which one more tail atom is added to each TG acyl chain (see Figure 3).32,33 This increases the hydrophobicity of TG, demonstrating TG nucleation at a critical concentration. However, the computed values of VTG and γTG/W with MARTINI-a are higher than the experimentally measured values (see Table 1). The illustrations of the discussed TG models are shown in Figure 3.
It is worth noting that C36-s is the only force field that has a value of the ratio of ALD to ABI close to the experimental data (see Table 1). In the AA simulations run with C36-s, we found that some TG molecules function as a membrane component on LD surfaces, making ALD automatically larger than ABI. Such TG molecules near the surface, referred to as SURF-TG, have PL-like conformations in which their glycerol moieties are exposed to the LD surface and their acyl chains are extended toward the LD core.9,34 This exposure of TG molecules to the LD surface would not only explain the increased area per lipid (ALD) but also the observed surface tension of LDs, experimentally measured to be 1–2 mN/m,27,28 since TG molecules are worse surfactants than PLs. Additionally, previous experiments by Miller and Small on lipoproteins confirmed the presence of SURF-TG in LD or lipoprotein surfaces.18 In contrast, the remainder of the force fields do not accurately describe LD surfaces. ALD and ABI differ by only a small amount, and LD surfaces are barely different from bilayer surfaces. Increasing the hydrophobicity of TG by reducing the hydrophilicity of the glycerol moieties clearly retains TG in the LD core. The likely explanation of the limitations of all of the developed force fields to date is lack of polarizability. As a weakly polar lipid, TG should shift its partial charge distribution depending on the environment. This would make it more polar at the PL/water interface, stabilizing some number of SURF-TG, and less polar in the LD core, decreasing the previously observed high core hydration. The reader should thus be aware of the current limitations and quality of various force fields when reading the other studies, recognizing that accurate results likely lay somewhere between those obtained to date.
Another popular neutral lipid is diacylglycerol (DG). The revised force fields for DG, consistent with C36, C36-LJPME, and SDK, have been developed by the Vanni group.26,35 The impact of DG on TG nucleation will be discussed later. Additionally, we note the other efforts to simulate realistic LDs composed of more neutral lipids other than TG. SE (in particular, cholesteryl ester) is another major component of the LD core in cellular systems, which could significantly modulate the physical properties of the LD core. In vitro and in vivo cellular imaging have confirmed that, when the SE:TG ratio is sufficiently high, the SE molecules transform to a liquid-crystalline smectic phase with layers of concentric rings that line up under the PL surface and surround an amorphous TG-rich phase in the very middle of the LD core.36−39 The smectic liquid-crystalline phase has been described as an onion-like structure with a lattice spacing of 3.4–3.7 nm.39,40 Such high SE concentrations can be induced with mitotic arrest or under starvation conditions (glucose restriction), when TG is preferentially hydrolyzed.39 It is also associated with the transformation of macrophages to foam cells in the formation of plaques in atherosclerosis.41 Recent studies have revealed that, once this phase transformation happens, the LD proteome is altered.38,42 Recent MD simulations have captured this phase transformation and indicate the PL monolayer properties are significantly altered (pending publication, Braun and Swanson).
LD Targeting
Proteins located on the LD surface regulate lipid metabolism or promote/delay lipolysis. Therefore, the LD surface proteins modulate various properties of LDs, such as size, population, and neutral lipid composition depending on the metabolic state of the cell. Additionally, proteins involved in lipid transfer and seemingly unrelated processes, such as glucose metabolism (via the transcription factor MLX),42 the innate immune system, and viral attack, have been found associated with LDs, broadening the relevance of LD biology beyond lipid metabolism.44 LD proteins are classified into two classes based on their origin. Proteins that target the LD from the ER bilayer are class I proteins and thought to contain one or more hydrophobic hairpin motifs. Proteins that target the LD from the cytosol are class II proteins and seem to rely on amphipathic helices for LD monolayer association (Figure 1). In this section, we will review joint computational–experimental and computational studies that investigate model peptides. The topic of LD targeting is an active and growing research field.
LiveDrop is the hairpin motif from class I glycerol-3-phosphate acyltransferase 4 (GPAT4) known to independently migrate from the ER to LDs.6,45 Due to its targeting abilities, LiveDrop has been used as a biomarker for LD surfaces. Using AA MD simulations, two of us with our experimental collaborators have investigated the thermodynamic process of LiveDrop targeting.45 Specifically, we calculated the average insertion depth of each amino acid of LiveDrop when LiveDrop is in the bilayer or LD. Following that, we computed the free energy profiles of single amino acids with respect to the insertion depth in the bilayer or LD. Using this information, we estimated how each amino acid of LiveDrop gains or loses thermodynamic free energy when LiveDrop is in the bilayer versus the LD. The analysis of the residue-wise LD accumulation energy showed that certain residues gain energetic stabilization by shifting locations in the LD monolayer compared to where they lay in the PL bilayer. In some cases, TG molecules facilitated more stable interactions. A comparable conclusion was made in another joint computational–experimental study.46 Furthermore, we found that two tryptophan residues of LiveDrop (W172 and W197) play a key role in the relocation process, as they had the largest energetic stabilization upon transfer to the LD. Both residues are trapped in the less favorable tail region when LiveDrop is in the bilayer. Upon LiveDrop’s migration into the LD, W172 becomes stabilized by residing at the LD surface and W197 by forming a hydrogen bond with TG glycerol moieties in the LD core. The significance of the tryptophan residues was validated via experiments: the mutated LiveDrop, in which tryptophan was substituted to valine, no longer targeted LDs in cellular systems. Interestingly, recent experiments have showed that LiveDrop moves back to the ER when the LD becomes SE-rich.38 The impact of the neutral lipid composition on the LD proteome will be an important topic for future studies.
Class II amphipathic helix-containing cytosolic proteins can target the ER bilayer or LD surface via a general targeting mechanism in which hydrophobic residues of the amphipathic helices detect exposed hydrophobic sites at the surface.47 These hydrophobic sites on the membrane surface—alternatively named packing defects—are characterized by the exposure of hydrocarbon atoms to the aqueous cytosol.48−51 Early all atom MD simulations suggested that LD surfaces have larger and more persistent packing defects than bilayer surfaces given the same PL composition.52 The difference in packing defects between the LD and bilayer can then lead to preferential targeting of amphipathic peptides of LD surfaces over bilayers. For instance, the all-atom MD simulations in the same study show that the autoinhibitory (AI) motif of CTP:phosphocholine cytidylyltransferase (CCT) binds to the LD surface with higher probability than it does to the bilayer. This finding was also validated by cell experiments in which the AI motif preferentially localized near the LD surface.
Two of the present authors have further characterized CCT association under an applied surface tension, since CCT is known to preferentially associate with expanding LD and bilayer membranes.34,53 The involved packing defects were classified into three types: PL acyl defect, TG glycerol defect, and TG acyl defect.9,34 The distinction is made based on the atomic and molecular types exposed to water at the membrane surface (Figure 4). All-atom unbiased MD simulations were then used to investigate the mechanism by which the AI motif from CCT binds to the LD surface.34 In 9 out of 13 MD trajectories, the AI motif initially binds with the TG defects. Moreover, similar to its importance in the LiveDrop targeting process, a tryptophan residue (W278) plays a central role in targeting, forming a hydrogen bond with a SURF-TG glycerol moiety. Subsequent full association is then dependent on the availability of proximal large packing defects that can accommodate a series of three phenylalanines. These large packing defects are more probable in the bilayer and monolayer surfaces when a surface tension is applied, representative of expansion conditions and explaining CCT’s preferential targeting to expanding surfaces. However, the additional W278–SURF-TG interaction likely increases CCT’s binding affinity and/or association rate with the LD surfaces. This work also revealed an interesting interplay (a direct correlation) between applied surface tension and the amount of SURF-TG. SURF-TG is a worse amphiphile than PL, yet a better amphiphile than PL or TG acyl tails. Under the LD growth conditions, the LD surface can increase the population of SURF-TG to reduce the effective surface tension. In contrast, under LD shrinkage, more SURF-TG would return to the LD core. Therefore, how much SURF-TG is presented at the LD surface can be a valuable indicator of the metabolic status of the LD and potentially indicative of the status of the cell. Accordingly, because of the different surface properties that the LD has due to altering SURF-TG concentrations, proteins that associate with the LD surface can also change depending on metabolic needs. While an enticing hypothesis, this SURF-TG-driven LD proteome awaits further validation.
Figure 4.
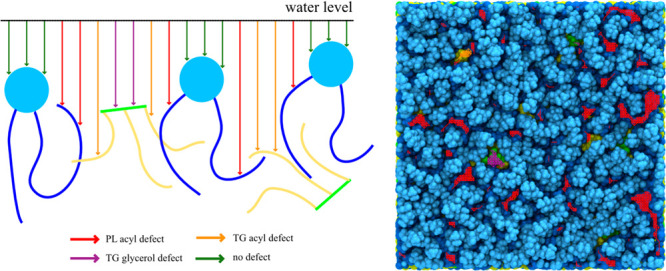
Packing defect analysis. An illustration of the analysis is shown on the left. At the water level, a grid is made, and from each grid point, a probe goes down until it meets lipid atoms. The molecular groups that probes meet determine defect types. Co-localization of the snapshot and packing defects is shown on the right. Red, purple, and orange lines indicate PL acyl defects, TG glycerol defects, and TG acyl defects, respectively. The color code for atoms is as in Figure 2.
It is valuable here to also discuss potential future research directions worth pursuing. Comparative gene identification-58 (CGI-58), a coactivator of adipose triglyceride lipase (ATGL), has a strong binding affinity to LD surfaces.54 Its targeting motif is a short amphipathic helix with three tryptophan residues. The experimental studies have shown that mutating CGI-58’s tryptophan residues to alanine inhibits LD targeting.55,56 The binding affinity calculation of the peptide and the mutated peptide (e.g., tryptophan to alanine) would support the importance of tryptophan residues in LD targeting. Another example of LD surface targeting protein is ADP-ribosylation factor GTPase-activating protein 1 (ArfGAP1). The amphipathic helix of ArfGAP1 has a small number of charged residues in the polar face and targets highly curved surfaces.57,58 Given that the LD curvature is small because of its large size, it would be intriguing to explore the biophysics of how ArfGAP1 sits on the LD surface. Finally, the perilipin family is a class of proteins that prevents the neutral lipid hydrolysis in LDs.59 Instead of bulky hydrophobic residues (e.g., tryptophan), the perilipin family has a long repetitive amphipathic helix that promotes LD targeting. An experimental study showed that shortening the helix reduces the efficiency of perilipin’s LD targeting.60 Not only that, but the perilipin family has specificity in LD targeting: some members target TG-rich LD surfaces, while some target SE-rich LD surfaces.61 How perilipin amphipathic helices achieve specificity in LD targeting remains unknown.
TG Nucleation
TG solubility in a bilayer has been measured experimentally.3 If the TG concentration is below critical, approximately 2.4 mol %, TG is dissolved in a bilayer with the same conformation as that of SURF-TG in which the glycerol moiety is exposed to water and the three acyl chains are extended toward the PL tail region. When the concentration is above the critical level, TG nucleates and forms a distinct phase in the bilayer, as shown in MD simulations.32,62,63 TG lensing can be understood using the classical nucleation theory in which the free energy ΔG is given by
| 1 |
where Δgv is the bulk free energy per unit volume, which is the difference in free energy per unit volume between the bulk TG phase and the PL phase (mostly hydrocarbon phase). Here, KC and KG are the bending modulus and Gaussian bending modulus, c0 is the spontaneous curvature, c1 and c2 describe local curvatures, A and V are the area and volume of the TG cluster, and γ is the LD surface tension.
The driving force of TG nucleation is the TG bulk free energy, proportional to V. However, γ hinders the LD formation given by the second term in eq 1 that is proportional to A. LDs have a measurable surface tension (∼1–2 mN/m) compared to PL bilayers that typically have zero surface tension.27,28 Finally, since LD formation happens in the membrane, the membrane deformation term needs to be considered. This energy term, which is approximately independent of the LD size, is proportional to the bilayer’s surface area, bending modulus, and square of curvature. Interplay between the two energy penalties, surface tension energy and membrane deformation energy, determines how the LD shape evolves during the LD growth process. For instance, when a TG cluster is small, the surface tension energy is smaller than the membrane deformation energy because the LD area is small, and the bending modulus of the bilayer (15–35 mN/m) is higher than the LD surface tension (∼1–2 mN/m). This flattens the TG cluster to minimize membrane deformation. In contrast, when the LD becomes large, the surface tension term becomes much larger than the membrane deformation one, which shapes LD to become spherical. The validity of eq 1 has been predicted in theoretical work64,65 and shown via CG MD simulations.63,66
We have tested TG nucleation in the bilayers that contain 6% mol TG using the C36-s, C36-LJPME-r, and C36-c AA force fields. However, we did not observe TG nucleation during 1–2 μs of simulation time due to the limited time scale. As a result, CG simulations are usually performed with SDK-V or MARTINI-a to study TG nucleation. For instance, the Vanni group has recently investigated the impact of lipid compositions on TG nucleation.62 With SDK-V simulations, they have shown that DG, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), and cholesterol facilitate TG nucleation while 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and dipalmitoylphosphatidylcholine (DPPC) hinder oil formation. Acyl chain saturation, chain length, and lipid shape collectively impact the mechanical properties of bilayers such as bending modulus and lateral pressure profiles, and therefore impact the TG chemical potential. This study could draw a correlation between TG nucleation and lipid chemistry because the CG model was high enough resolution to retain chemical specificity. By contrast, Kim et al. have recently developed a highly coarse-grained generic lipid model for PL and TG in which each molecule consists of linear four-site CG beads.63 This model lacks chemical specificity as a result. However, the bilayer properties such as bending modulus can be modulated via the PL angle potential. The unbiased simulations and free energy calculations of TG nucleation using the newly developed CG model suggest that an oil lens is more planar and has a lower degree of nucleation in high membrane rigidity. Moreover, due to the smaller degrees of freedom in our CG model, we were able to simulate large spherical bilayers with a diameter of 40 or 60 nm, comparable to the actual ER tubule curvature, and observed two LD growth mechanisms, oil coalescence, and Ostwald ripening.
It is worth noting that TG flip-flop free energy (the free energy profile of TG with respect to z position in a bilayer) is closely related to TG nucleation (Figure 5). We developed a generic PL and TG model, the Kim–Voth (KV) model, whose resolution is comparable to the MARTINI-s resolution but has only one glycerol moiety for both PL and TG.10 The attraction strength between PL and TG’s glycerol moieties (C in Figure 5a) can also be parametrized, resulting in various free energy barriers. When the barrier is greater than ∼1.2 kcal/mol, TG does not nucleate in a bilayer containing 6% TG (Figure 5a). The dependence of TG nucleation on TG flip-flop free energy is observed in other CG force fields. TG flip-flop has a higher barrier with SDK-s than SDK-V (Figure 5b). Accordingly, TG nucleates at the critical concentration or above with simulations performed with SDK-V, while TG remains dissolved with SDK-s. The same behavior is observed with MARTINI-s and MARTINI-a (Figure 5c). Interestingly, the free energy barrier for TG flip-flop becomes higher in POPC bilayers than DOPC bilayers in SDK-V and MARTINI-a simulations. This is consistent with the previous SDK-V simulations that show the higher TG chemical potential and lower TG clustering rate in POPC bilayers than DOPC bilayers.62 Finally, three AA force fields (C36-s, C36-LJPME-r, and C36-c) have different TG-flip flop energy profiles (Figure 5d). It is likely that C36-c and C36-LJPME-r will eventually show TG nucleation given the low free energy barriers if simulations can be extended more than 2 μs. In that regard, it would be beneficial to coarse-grain those systems with bottom-up approaches such as force matching or relative entropy minimization to study TG nucleation with CG models derived systematically from different AA force fields.67,68
Figure 5.
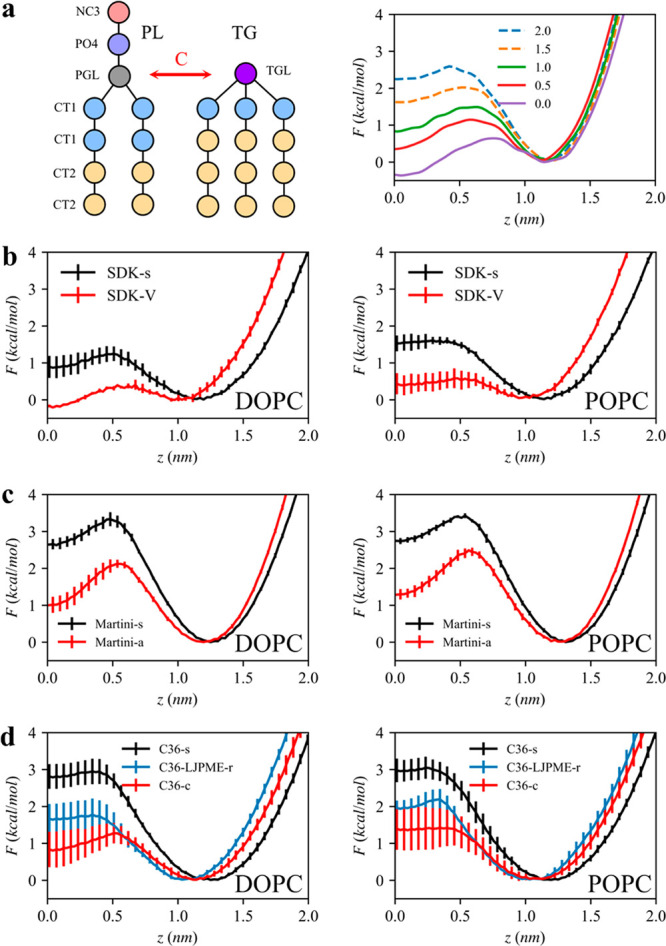
TG flip-flop in bilayers. A collective variable is the z distance between the center of mass of the phosphate atoms to the TG glycerol moiety. A depth of 0 represents the membrane center. (a) Illustration of the KV model (left). Attraction between the PL glycerol moiety (PGL) and the TG glycerol moiety (TGL) is controlled via the parameter C. The free energy profiles for TG flip-flop with various C parameters (right). TG does not nucleate with high C values, indicated with dashed lines. (b–d) TG flip-flop profiles with CG and AA force fields in DOPC bilayers (left column) or POPC bilayers (right column).
LD Formation
The ER protein seipin plays a critical role in LD biogenesis by defining LD formation sites, catalyzing TG nucleation, and providing the binding sites for its binding partner, lipid droplet assembly factor 1 (LDAF1).4 The absence of seipin results in aggregated, small LDs and/or supersized LDs as well as Bernardinelli–Seip congenital lipodystrophy type 2 (BSCL2). Seipin forms an unusual ring-like structure that consists of 10–12 subunits depending on the species.69−72 Each subunit has a highly conserved lumenal domain, flanked by two transmembrane (TM) segments and nonconserved variable cytosolic tails. Cell experiments indicate that the cytosolic tails are dispensable while other parts are critical for seipin function.73 For instance, fly and human seipin have conserved hydrophobic helices (HHs) in the lumenal domain positioned toward the lumenal leaflet. In contrast, yeast seipin does not have the HH and is not functional by itself. Removing the TM segments or replacing one of them with other ER protein FIT2 results in the LD phenotype of seipin knockout cells.4,71
Recent MD simulations have provided important insights into the molecular functioning of human seipin.33,73,74 Seipin–lipid interactions were analyzed in the seipin-containing bilayer simulations. The results show that the TG–HH and TG–TM interactions facilitate TG nucleation. Interestingly, results from four different force fields (C36-s, C36-LJPME-r, SDK-V, and MARTINI-a) all demonstrated that S166 of human seipin has the strongest interaction with TG and acts as a TG tethering site. The final structures observed in the CG simulations show a distinct TG phase accumulated in the cage-like structure of human seipin, in the form of a disk. Additionally, the simulations with SDK-V showed that DG preferentially accumulates near the TM segments, promotes TG nucleation, and potentially serves as a structural support for negative curvature at the ER–LD neck structure.74 In contrast, yeast seipin simulations with MARTINI-a show that TG preferentially clusters near the TM segments but TG does not nucleate.72 It is worth mentioning that the structure of transmembrane segments of human seipin is not fully resolved. Therefore, the findings from human seipin simulations are subject to further experimental validation.
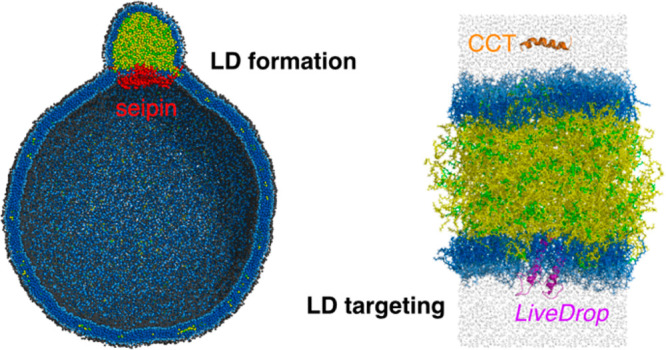
Two of us have developed a phenomenological CG model for seipin and studied TG nucleation and LD maturation with the four-site PL and TG lipid model.73 Exploiting the low resolution of these CG models, we simulated a large spherical bilayer with a diameter of 40 or 60 nm to investigate the advanced stages of LD formation. Our simulations demonstrate that a cage geometry of seipin TM segments is a key component for forming a unique ER–LD neck structure and in modulating the morphology of a forming oil lens (Figure 6). The seipin-containing system constricted the area of TG in the bilayer and transferred excess TG into LD. Therefore, seipin TM segments surrounded the oil lens, creating a unique ER–LD neck structure. The resulting shape of the oil lens then became spherical. In contrast, the pure lipid system or the system that only contains the lumenal domain of seipin had a flat oil lens to minimize membrane deformation (eq 1). Interestingly, when seipin has only five continuous subunits by removing six continuous subunits, the resulting oil lens has an elongated TG tail toward the region where no protein density presents while constricted in the seipin region. This simulation experiment emphasized the critical importance of the conserved oligomeric structure of seipin.
Figure 6.
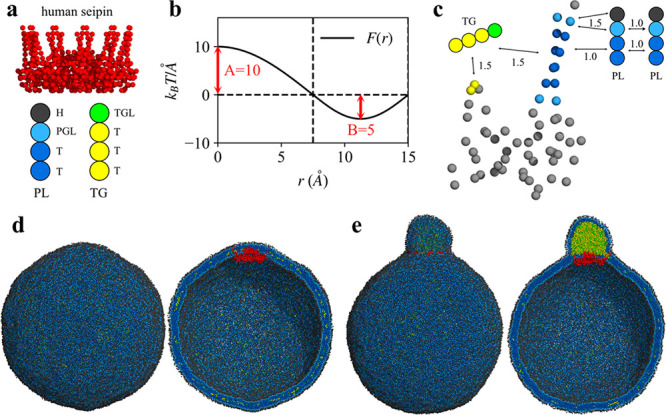
LD formation investigated with CG simulations. (a) Protein and lipid models. (b) Cosine- and sine-based forces that act between CG beads. (c) Protein–lipid attraction factors (B in part b) are indicated. The repulsion factor, A, was set to 25 in the simulations. (d) Initial structure of the spherical bilayer with a diameter of 60 nm. The exterior (left) and interior (right) views. (e) Final structure. The exterior (left) and interior (right) views. The simulation data are from ref (73).
We also note the importance of the positively charged residues (R23, R24, R26, R265, H266, and R267 of human seipin) on LD maturation located at the ends of the TM segments.73 As confirmed by the CG MD simulations, these residues anchor the TM segments in the bilayer and prevent the TM segments from entering the TG phase. The experimental collaborators in that work have mutated the charged residues to alanine and demonstrated the LD phenotypes between the seipin knockout cells and the seipin wild type cells, a result consistent with our CG results. Additionally, our CG simulations show that the rate of oil coalescence near the seipin site is slower than seipin-free sites, since these charged residues work as a zipper to maintain the bilayer thickness at the boundary of seipin. When a seipin-free oil lens approaches seipin, TG in the seipin-free lens is transported to a seipin-containing oil lens in a controlled manner, while not disrupting the ER–LD neck structure. Finally, our CG simulations show that the diameter of the ring defined by the TM segments is larger during LD maturation than during the initial stages of LD formation. Such an increase in the diameter may increase the lipid and protein flux toward the expanding LD. Although the CG models used in these simulations are simplified, the importance of the conserved oligomeric property was shown and a working model of human seipin was developed.
Computational studies of LD biogenesis are mostly in their beginning stages, and thus, there are many fundamental questions that remain unanswered. The first question is how different cell types control the neutral lipid composition of the LD core. For instance, adipose cells typically have TG-rich LDs while macrophages have SE-rich LDs. It would be valuable to study whether the differences in the neutral lipid composition of the LD core can be attributed to different activation levels of neutral lipid synthesis or a different LD biogenesis mechanism. Metabolic status and cellular cycles can also affect neutral lipid compositions.39 A second aspect concerns the binding partners of seipin. Human seipin has LDAF1, and yeast seipin has Ldb16, critical for the yeast seipin function. Including the binding partners in the MD simulations may change some of the observed behaviors. Also, one interesting potential way would be to study the correlation between desired local curvature and lipid distribution, given that the neck structure is negatively curved while the forming lipid droplet is positively curved.62,63,74,75 Finally, the way in which the lumenal domain of human seipin is involved in seipin function is largely unknown. According to the computational studies of human seipin, the hydrophobic helices and transmembrane segments are the most important aspects in the role of human seipin. However, a significant mutation is known to be N88S and S90L in a glycosylation motif facing the ER lumen, causing distal hereditary motor neuropathy and Silver syndrome.76 The molecular and functional role of the beta-sandwich domain remains unknown.
Conclusions
MD simulations have provided valuable insight into the physical and structural properties of LDs as well as the mechanisms of LD targeting and LD biogenesis at the molecular level. In this Feature Article, we summarized the relative performance of different AA (C36-s, C36-LJPME-r, and C36-c) and CG (SDK-s, SDK-V, MARTINI-s, MARTINI-a) LD models based on computed physical properties (TG/W interfacial tension, area per phospholipids of LDs, and TG flip-flop in the bilayer) compared to experimental data.
In studying the mechanisms of protein targeting to LDs, packing defect analysis has been fruitful as it was in lipid bilayers. Larger and more persistent packing defects of LDs, compared to bilayers of the identical PL composition, and LD-specific defects (due to SURF-TG molecules) offer binding sites for large hydrophobic residues of amphipathic helices from class II proteins. Furthermore, the measurable LD surface tension, which oscillates during growth and shrinkage, increases the ratio of SURF-TG to PL, enhancing TG-related defects. Thus, surface properties reflecting the LD metabolic state may play a role in life-cycle specific targeting. Class I proteins are driven by favorable conformational changes and the repositioning of residues from the ER bilayer to the LD monolayer, sometimes facilitated by interactions with TG.
LD formation is a liquid–liquid phase separation process occurring at the ER bilayer. Due to its time and length scales, multiscale simulations across the atomistic to the coarse-grained scale complement one another in order to better understand this fascinating biological process. We have reviewed here the recent MD studies focused on TG nucleation in bilayers with seipin. Different force fields (C36-s, C36-LJPME-r, SDK-V, and MARTINI-a) consistently have identified the importance of S166 of human seipin in clustering TG inside the seipin ring.33,73,74 S166 forms a hydrophilic interaction with the TG glycerol moiety in the hydrophobic phase of PL bilayers. It is worth noting that such a hydrophilic interaction has sometimes been critical in LD targeting pathways. For instance, the importance of hydrophilic interactions in Trp197 of LiveDrop and Trp278 of the AI motif of CCTα has been highlighted in recent studies.34,45
The more advanced LD formation steps have alternatively been studied with CG simulations. SDK-V and MARTINI-a simulations have demonstrated that seipin–TG interactions catalyze TG nucleation and lower the critical concentration for nucleation. Using highly CG lipid and protein models, the importance of seipin TM segments has been revealed in LD maturation. The conserved cage geometry of seipin TM segments is key to changing an oil lens into a spherical shape and forming a unique ER–LD neck structure. The same study also has demonstrated that the conserved and positively charged residues located at the borders of the seipin TM segments are key for LD growth. An effort to simulate the LD formation at a multiscale resolution will be a valuable future study.
The computational studies described here have thus addressed a broad range of different features in LD biology. This work is, however, far from finished and many intriguing questions remain unanswered. We can therefore look forward to future simulation studies contributing even more to better understanding LD biology and biophysics.
Acknowledgments
The authors thank Tobias Walther, Robert Farese, Jr., Stefano Vanni, Richard Pastor, Jeeyun Chung, and Matthew Mitsche for critical discussions. This work was supported by a grant from the National Institute of General Medical Science (NIGMS) of the National Institutes of Health (NIH), grant R01 GM063796.
Biographies
Siyoung Kim is a postdoctoral fellow at D. E. Shaw Research. He received his B.S. degree in Chemical and Biomolecular Engineering from Korea Advanced Institute of Science and Technology (KAIST). He earned his Ph.D. in Molecular Engineering from the University of Chicago. His research is focused on understanding biology and biophysics using multiscale molecular dynamics simulations.
Jessica M. J. Swanson is an Assistant Professor of Chemistry at the University of Utah. She earned her B.S. in Biochemistry from the University of California Davis and Ph.D. in Physical Chemistry from the University of California San Diego. Prior to her Assistant Professorship, she was NIH NRSA Postdoctoral Fellow at the University of Utah, a staff researcher at Argonne National Laboratory, and an Assistant to Associate Research Professor at the University of Chicago. Her areas of research include multiscale simulations and kinetic modeling of biomolecular phenomena, including membrane permeation, molecular motors, transporters, lasso peptides, and lipid droplets.
Gregory A. Voth is the Haig P. Papazian Distinguished Service Professor of Chemistry at the University of Chicago. He is also Professor of the James Franck Institute and the Institute for Biophysical Dynamics. He earned his Ph.D. in Theoretical Chemistry from the California Institute of Technology. Prior to joining the University of Chicago, he was a Distinguished Professor of Chemistry at the University of Utah. His research concerns the development and application of theoretical and computational methods to address structural and dynamical problems of complex condensed phase systems, including biomolecules and materials.
The authors declare no competing financial interest.
References
- Walther T. C.; Chung J.; Farese R. V. Jr Lipid Droplet Biogenesis. Annu. Rev. Cell Dev Biol. 2017, 33 (1), 491–510. 10.1146/annurev-cellbio-100616-060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducharme N. A.; Bickel P. E. Minireview: Lipid Droplets in Lipogenesis and Lipolysis. Endocrinology 2008, 149 (3), 942–949. 10.1210/en.2007-1713. [DOI] [PubMed] [Google Scholar]
- Hamilton J. A.; Small D. M. Solubilization and localization of triolein in phosphatidylcholine bilayers: a 13C NMR study. Proc. Natl. Acad. Sci. U. S. A. 1981, 78 (11), 6878–6882. 10.1073/pnas.78.11.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J.; Wu X.; Lambert T. J.; Lai Z. W.; Walther T. C.; Farese R. V. Jr LDAF1 and Seipin Form a Lipid Droplet Assembly Complex. Dev Cell 2019, 51 (5), 551–563. 10.1016/j.devcel.2019.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo V. T.; Li S.; Vihinen H.; Holtta-Vuori M.; Szkalisity A.; Horvath P.; Belevich I.; Peranen J.; Thiele C.; Somerharju P.; et al. Seipin Facilitates Triglyceride Flow to Lipid Droplet and Counteracts Droplet Ripening via Endoplasmic Reticulum Contact. Dev Cell 2019, 50 (4), 478–493. 10.1016/j.devcel.2019.05.016. [DOI] [PubMed] [Google Scholar]
- Wang H.; Becuwe M.; Housden B. E.; Chitraju C.; Porras A. J.; Graham M. M.; Liu X. N.; Thiam A. R.; Savage D. B.; Agarwal A. K.; et al. Seipin is required for converting nascent to mature lipid droplets. Elife 2016, 5, e16582 10.7554/eLife.16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brini E.; Simmerling C.; Dill K. Protein storytelling through physics. Science 2020, 370 (6520), eaaz3041 10.1126/science.aaz3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venable R. M.; Brown F. L. H.; Pastor R. W. Mechanical properties of lipid bilayers from molecular dynamics simulation. Chem. Phys. Lipids 2015, 192, 60–74. 10.1016/j.chemphyslip.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Swanson J. M. J. The Surface and Hydration Properties of Lipid Droplets. Biophys. J. 2020, 119 (10), 1958–1969. 10.1016/j.bpj.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Voth G. A. Physical Characterization of Triolein and Implications for Its Role in Lipid Droplet Biogenesis. J. Phys. Chem. B 2021, 125 (25), 6874–6888. 10.1021/acs.jpcb.1c03559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S.; Levy R. I.. Lipoprotein Metabolism. In Advances in Lipid Research; Paoletti R., Kritchevsky D., Eds.; Elsevier: 1975; Vol. 13, pp 1–89. [PubMed] [Google Scholar]
- Jackson R. L.; Morrisett J. D.; Gotto A. M. Lipoprotein structure and metabolism. Physiol. Rev. 1976, 56 (2), 259–316. 10.1152/physrev.1976.56.2.259. [DOI] [PubMed] [Google Scholar]
- Murtola T.; Vuorela T. A.; Hyvönen M. T.; Marrink S.-J.; Karttunen M.; Vattulainen I. Low density lipoprotein: structure, dynamics, and interactions of apoB-100 with lipids. Soft Matter 2011, 7 (18), 8135. 10.1039/c1sm05367a. [DOI] [Google Scholar]
- Ollila O. H. S.; Lamberg A.; Lehtivaara M.; Koivuniemi A.; Vattulainen I. Interfacial Tension and Surface Pressure of High Density Lipoprotein, Low Density Lipoprotein, and Related Lipid Droplets. Biophys. J. 2012, 103 (6), 1236–1244. 10.1016/j.bpj.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S. M.; Pourmousa M.; Sampson M.; Sviridov D.; Islam R.; Perrin B. S. Jr.; Kemeh G.; Pastor R. W.; Remaley A. T. Identification of a novel lipid binding motif in apolipoprotein B by the analysis of hydrophobic cluster domains. Biochim Biophys Acta Biomembr 2017, 1859 (2), 135–145. 10.1016/j.bbamem.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolska A.; Lo L.; Sviridov Denis O.; Pourmousa M.; Pryor M.; Ghosh Soumitra S.; Kakkar R.; Davidson M.; Wilson S.; Pastor Richard W.; et al. A dual apolipoprotein C-II mimetic–apolipoprotein C-III antagonist peptide lowers plasma triglycerides. Science Translational Medicine 2020, 12 (528), eaaw7905 10.1126/scitranslmed.aaw7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton J. A.; Miller K. W.; Small D. M. Solubilization of triolein and cholesteryl oleate in egg phosphatidylcholine vesicles. J. Biol. Chem. 1983, 258 (21), 12821–12826. 10.1016/S0021-9258(17)44044-0. [DOI] [PubMed] [Google Scholar]
- Miller K. W.; Small D. M. Surface-to-core and interparticle equilibrium distributions of triglyceride-rich lipoprotein lipids. J. Biol. Chem. 1983, 258 (22), 13772–13784. 10.1016/S0021-9258(17)43986-X. [DOI] [PubMed] [Google Scholar]
- Mitsche M. A.; Wang L.; Small D. M. Adsorption of egg phosphatidylcholine to an air/water and triolein/water bubble interface: use of the 2-dimensional phase rule to estimate the surface composition of a phospholipid/triolein/water surface as a function of surface pressure. J. Phys. Chem. B 2010, 114 (9), 3276–3284. 10.1021/jp908730t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsche M. A.; Small D. M. C-terminus of apolipoprotein A-I removes phospholipids from a triolein/phospholipids/water interface, but the N-terminus does not: a possible mechanism for nascent HDL assembly. Biophys. J. 2011, 101 (2), 353–361. 10.1016/j.bpj.2011.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsche M. A.; Packer L. E.; Brown J. W.; Jiang Z. G.; Small D. M.; McKnight C. J. Surface tensiometry of apolipoprotein B domains at lipid interfaces suggests a new model for the initial steps in triglyceride-rich lipoprotein assembly. J. Biol. Chem. 2014, 289 (13), 9000–9012. 10.1074/jbc.M113.540955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda J. B.; Venable R. M.; Freites J. A.; O’Connor J. W.; Tobias D. J.; Mondragon-Ramirez C.; Vorobyov I.; Mackerell A. D.; Pastor R. W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114 (23), 7830–7843. 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink S. J.; Risselada H. J.; Yefimov S.; Tieleman D. P.; de Vries A. H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111 (27), 7812–7824. 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- Shinoda W.; DeVane R.; Klein M. L. Multi-property fitting and parameterization of a coarse grained model for aqueous surfactants. Mol. Simul. 2007, 33 (1–2), 27–36. 10.1080/08927020601054050. [DOI] [Google Scholar]
- Bacle A.; Gautier R.; Jackson C. L.; Fuchs P. F. J.; Vanni S. Interdigitation between Triglycerides and Lipids Modulates Surface Properties of Lipid Droplets. Biophys. J. 2017, 112 (7), 1417–1430. 10.1016/j.bpj.2017.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campomanes P.; Prabhu J.; Zoni V.; Vanni S. Recharging your fats: CHARMM36 parameters for neutral lipids triacylglycerol and diacylglycerol. Biophysical Reports 2021, 1 (2), 100034. 10.1016/j.bpr.2021.100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorlay A.; Thiam A. R. Neutral lipids regulate amphipathic helix affinity for model lipid droplets. J. Cell Biol. 2020, 219 (4), e201907099 10.1083/jcb.201907099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorlay A.; Foret L.; Thiam A. R. Origin of gradients in lipid density and surface tension between connected lipid droplet and bilayer. Biophys. J. 2021, 120 (24), 5491–5503. 10.1016/j.bpj.2021.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouw T. H.; Vlugter J. C. Physical Properties of Triglycerides. I. Density and Refractive Index. Fette, Seifen, Anstrichmittel 1966, 68 (7), 544–549. 10.1002/lipi.19660680705. [DOI] [Google Scholar]
- Platford R. F. The octanol-water partitioning of some hydrophobic and hydrophilic compounds. Chemosphere 1983, 12 (7–8), 1107–1111. 10.1016/0045-6535(83)90264-3. [DOI] [Google Scholar]
- Yu Y.; Kramer A.; Venable R. M.; Brooks B. R.; Klauda J. B.; Pastor R. W. CHARMM36 Lipid Force Field with Explicit Treatment of Long-Range Dispersion: Parametrization and Validation for Phosphatidylethanolamine, Phosphatidylglycerol, and Ether Lipids. J. Chem. Theory Comput 2021, 17 (3), 1581–1595. 10.1021/acs.jctc.0c01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelia H.; Duelund L.; Pakkanen K. I.; Ipsen J. H. Triglyceride blisters in lipid bilayers: implications for lipid droplet biogenesis and the mobile lipid signal in cancer cell membranes. PLoS One 2010, 5 (9), e12811 10.1371/journal.pone.0012811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanna X.; Salo V. T.; Li S.; Ven K.; Vihinen H.; Jokitalo E.; Vattulainen I.; Ikonen E. Seipin traps triacylglycerols to facilitate their nanoscale clustering in the endoplasmic reticulum membrane. PLoS Biol. 2021, 19 (1), e3000998 10.1371/journal.pbio.3000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Oh M. I.; Swanson J. M. J. Stressed Lipid Droplets: How Neutral Lipids Relieve Surface Tension and Membrane Expansion Drives Protein Association. J. Phys. Chem. B 2021, 125 (21), 5572–5586. 10.1021/acs.jpcb.1c01795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campomanes P.; Zoni V.; Vanni S. Local accumulation of diacylglycerol alters membrane properties nonlinearly due to its transbilayer activity. Communications Chemistry 2019, 2 (1), 72. 10.1038/s42004-019-0175-7. [DOI] [Google Scholar]
- Shimobayashi S. F.; Ohsaki Y. Universal phase behaviors of intracellular lipid droplets. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (51), 25440–25445. 10.1073/pnas.1916248116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamme F.; Cinquin B.; Gohon Y.; Pereiro E.; Réfrégiers M.; Froissard M. Synchrotron multimodal imaging in a whole cell reveals lipid droplet core organization. Journal of Synchrotron Radiation 2020, 27 (3), 772–778. 10.1107/S1600577520003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S.; Gui L.; Kovalenko A.; Reetz E.; Nicastro D.; Henne M. W.. Liquid-crystalline lipid phase transitions in lipid droplets selectively remodel the LD proteome. 2021, 10.1101/2021.08.30.458229. https://www.biorxiv.org/content/10.1101/2021.08.30.458229v1. [DOI]
- Mahamid J.; Tegunov D.; Maiser A.; Arnold J.; Leonhardt H.; Plitzko J. M.; Baumeister W. Liquid-crystalline phase transitions in lipid droplets are related to cellular states and specific organelle association. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (34), 16866–16871. 10.1073/pnas.1903642116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabany T.; Wagner A.; Zweytick D.; Lohner K.; Leitner E.; Ingolic E.; Daum G. Structural and Biochemical Properties of Lipid Particles from the Yeast Saccharomyces cerevisiae. J. Biol. Chem. 2008, 283 (25), 17065–17074. 10.1074/jbc.M800401200. [DOI] [PubMed] [Google Scholar]
- Small D. M.; Shipley G. G. Physical-Chemical Basis of Lipid Deposition in Atherosclerosis. Science 1974, 185 (4147), 222–229. 10.1126/science.185.4147.222. [DOI] [PubMed] [Google Scholar]
- Mejhert N.; Kuruvilla L.; Gabriel K. R.; Elliott S. D.; Guie M.-A.; Wang H.; Lai Z. W.; Lane E. A.; Christiano R.; Danial N. N.; et al. Partitioning of MLX-Family Transcription Factors to Lipid Droplets Regulates Metabolic Gene Expression. Mol. Cell 2020, 77 (6), 1251–1264. 10.1016/j.molcel.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučerka N.; Nagle J. F.; Sachs J. N.; Feller S. E.; Pencer J.; Jackson A.; Katsaras J. Lipid Bilayer Structure Determined by the Simultaneous Analysis of Neutron and X-Ray Scattering Data. Biophys. J. 2008, 95 (5), 2356–2367. 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarte M. J.; Swanson J. M. J.; Walther T. C.; Farese R. V. Jr The CYTOLD and ERTOLD pathways for lipid droplet-protein targeting. Trends Biochem. Sci. 2022, 47 (1), 39–51. 10.1016/j.tibs.2021.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olarte M. J.; Kim S.; Sharp M. E.; Swanson J. M. J.; Farese R. V. Jr.; Walther T. C. Determinants of Endoplasmic Reticulum-to-Lipid Droplet Protein Targeting. Dev Cell 2020, 54 (4), 471–487. 10.1016/j.devcel.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillon L.; Nieto V.; Gehan P.; Omrane M.; Rodriguez N.; Monticelli L.; Thiam A. R. Triacylglycerols sequester monotopic membrane proteins to lipid droplets. Nat. Commun. 2020, 11 (1), 3944. 10.1038/s41467-020-17585-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J.; Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell 2012, 23 (5), 886–895. 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Cui H.; Lyman E.; Voth G. A. Mechanism of Membrane Curvature Sensing by Amphipathic Helix Containing Proteins. Biophys. J. 2011, 100 (5), 1271–1279. 10.1016/j.bpj.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanni S.; Vamparys L.; Gautier R.; Drin G.; Etchebest C.; Fuchs P. F.; Antonny B. Amphipathic lipid packing sensor motifs: probing bilayer defects with hydrophobic residues. Biophys. J. 2013, 104 (3), 575–584. 10.1016/j.bpj.2012.11.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamparys L.; Gautier R.; Vanni S.; Bennett W. F.; Tieleman D. P.; Antonny B.; Etchebest C.; Fuchs P. F. Conical lipids in flat bilayers induce packing defects similar to that induced by positive curvature. Biophys. J. 2013, 104 (3), 585–593. 10.1016/j.bpj.2012.11.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R.; Bacle A.; Tiberti M. L.; Fuchs P. F.; Vanni S.; Antonny B. PackMem: A Versatile Tool to Compute and Visualize Interfacial Packing Defects in Lipid Bilayers. Biophys. J. 2018, 115 (3), 436–444. 10.1016/j.bpj.2018.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevost C.; Sharp M. E.; Kory N.; Lin Q.; Voth G. A.; Farese R. V. Jr.; Walther T. C. Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets. Dev Cell 2018, 44 (1), 73–86. 10.1016/j.devcel.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northwood I. C.; Tong A. H.; Crawford B.; Drobnies A. E.; Cornell R. B. Shuttling of CTP: Phosphocholine cytidylyltransferase between the nucleus and endoplasmic reticulum accompanies the wave of phosphatidylcholine synthesis during the G0 → G1 transition. J. Biol. Chem. 1999, 274 (37), 26240–26248. 10.1074/jbc.274.37.26240. [DOI] [PubMed] [Google Scholar]
- Kory N.; Thiam A. R.; Farese R. V. Jr.; Walther T. C. Protein Crowding Is a Determinant of Lipid Droplet Protein Composition. Dev Cell 2015, 34 (3), 351–363. 10.1016/j.devcel.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A.; Cornaciu I.; Lass A.; Schweiger M.; Poeschl M.; Eder C.; Kumari M.; Schoiswohl G.; Wolinski H.; Kohlwein S. D.; et al. The N-terminal region of comparative gene identification-58 (CGI-58) is important for lipid droplet binding and activation of adipose triglyceride lipase. J. Biol. Chem. 2010, 285 (16), 12289–12298. 10.1074/jbc.M109.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeszoermenyi A.; Nagy H. M.; Arthanari H.; Pillip C. J.; Lindermuth H.; Luna R. E.; Wagner G.; Zechner R.; Zangger K.; Oberer M. Structure of a CGI-58 motif provides the molecular basis of lipid droplet anchoring. J. Biol. Chem. 2015, 290 (44), 26361–26372. 10.1074/jbc.M115.682203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B.; Drin G.; Levi S.; Rawet M.; Cassel D.; Bigay J.; Antonny B. Two lipid-packing sensor motifs contribute to the sensitivity of ArfGAP1 to membrane curvature. Biochemistry 2007, 46 (7), 1779–1790. 10.1021/bi062288w. [DOI] [PubMed] [Google Scholar]
- Drin G.; Casella J. F.; Gautier R.; Boehmer T.; Schwartz T. U.; Antonny B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct Mol. Biol. 2007, 14 (2), 138–146. 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- Sztalryd C.; Brasaemle D. L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim Biophys Acta Mol. Cell Biol. Lipids 2017, 1862 (10), 1221–1232. 10.1016/j.bbalip.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copic A.; Antoine-Bally S.; Gimenez-Andres M.; La Torre Garay C.; Antonny B.; Manni M. M.; Pagnotta S.; Guihot J.; Jackson C. L. A giant amphipathic helix from a perilipin that is adapted for coating lipid droplets. Nat. Commun. 2018, 9 (1), 1332. 10.1038/s41467-018-03717-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh K.; Lee Y. K.; Londos C.; Raaka B. M.; Dalen K. T.; Kimmel A. R. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. Journal of Cell Science 2012, 125 (17), 4067–4076. 10.1242/jcs.104943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoni V.; Khaddaj R.; Campomanes P.; Thiam A. R.; Schneiter R.; Vanni S. Pre-existing bilayer stresses modulate triglyceride accumulation in the ER versus lipid droplets. Elife 2021, 10, e62886 10.7554/eLife.62886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Li C.; Farese R. V. Jr.; Walther T. C.; Voth G. A. Key Factors Governing Initial Stages of Lipid Droplet Formation. J. Phys. Chem. B 2022, 126 (2), 453–462. 10.1021/acs.jpcb.1c09683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes F.; Thiam A. R.; Foret L. Lipid Droplets Can Spontaneously Bud Off from a Symmetric Bilayer. Biophys. J. 2017, 113 (1), 15–18. 10.1016/j.bpj.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Yi X. Bulging and budding of lipid droplets from symmetric and asymmetric membranes: competition between membrane elastic energy and interfacial energy. Soft Matter 2021, 17 (21), 5319–5328. 10.1039/D1SM00245G. [DOI] [PubMed] [Google Scholar]
- Zoni V.; Nieto V.; Endter L. J.; Risselada H. J.; Monticelli L.; Vanni S. To Bud or Not to Bud: A Perspective on Molecular Simulations of Lipid Droplet Budding. Frontiers in Molecular Biosciences 2019, 6, 124. 10.3389/fmolb.2019.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izvekov S.; Voth G. A. A Multiscale Coarse-Graining Method for Biomolecular Systems. J. Phys. Chem. B 2005, 109 (7), 2469–2473. 10.1021/jp044629q. [DOI] [PubMed] [Google Scholar]
- Shell M. S. The relative entropy is fundamental to multiscale and inverse thermodynamic problems. J. Chem. Phys. 2008, 129 (14), 144108. 10.1063/1.2992060. [DOI] [PubMed] [Google Scholar]
- Yan R.; Qian H.; Lukmantara I.; Gao M.; Du X.; Yan N.; Yang H. Human SEIPIN Binds Anionic Phospholipids. Dev Cell 2018, 47 (2), 248–256. 10.1016/j.devcel.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Sui X.; Arlt H.; Brock K. P.; Lai Z. W.; DiMaio F.; Marks D. S.; Liao M.; Farese R. V. Jr.; Walther T. C. Cryo-electron microscopy structure of the lipid droplet-formation protein seipin. J. Cell Biol. 2018, 217 (12), 4080–4091. 10.1083/jcb.201809067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H.; Sui X.; Folger B.; Adams C.; Chen X.; Remme R.; Hamprecht F. A.; DiMaio F.; Liao M.; Goodman J. M.; et al. Seipin forms a flexible cage at lipid droplet formation sites. 2021, 10.1101/2021.08.05.455270. https://www.biorxiv.org/content/10.1101/2021.08.05.455270v1. [DOI] [PMC free article] [PubMed]
- Klug Y. A.; Deme J. C.; Corey R. A.; Renne M. F.; Stansfeld P. J.; Lea S. M.; Carvalho P. Mechanism of lipid droplet formation by the yeast Sei1/Ldb16 Seipin complex. Nat. Commun. 2021, 12 (1), 5892. 10.1038/s41467-021-26162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Chung J.; Arlt H.; Pak A. J.; Farese R. V.; Walther T. C.; Voth G. A.. Seipin transmembrane segments critically function in triglyceride nucleation and lipid droplet budding from the membrane. 2021, 10.1101/2021.12.05.471300. https://www.biorxiv.org/content/10.1101/2021.12.05.471300v1. [DOI] [PMC free article] [PubMed]
- Zoni V.; Khaddaj R.; Lukmantara I.; Shinoda W.; Yang H.; Schneiter R.; Vanni S. Seipin accumulates and traps diacylglycerols and triglycerides in its ring-like structure. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (10), e2017205118 10.1073/pnas.2017205118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yesylevskyy S.; Khandelia H. EnCurv: Simple Technique of Maintaining Global Membrane Curvature in Molecular Dynamics Simulations. J. Chem. Theory Comput. 2021, 17 (2), 1181–1193. 10.1021/acs.jctc.0c00800. [DOI] [PubMed] [Google Scholar]
- Windpassinger C.; Auer-Grumbach M.; Irobi J.; Patel H.; Petek E.; Hörl G.; Malli R.; Reed J. A.; Dierick I.; Verpoorten N.; et al. Heterozygous missense mutations in BSCL2 are associated with distal hereditary motor neuropathy and Silver syndrome. Nat. Genet. 2004, 36 (3), 271–276. 10.1038/ng1313. [DOI] [PubMed] [Google Scholar]