Abstract
Vaccine-induced immunity is expected to target the native antigens expressed by the pathogens. Therefore, it is highly important to generate vaccine antigens that are immunologically indistinguishable from the native antigens. Nucleic acid vaccines, comprised of DNA, mRNA, or recombinant viral vector vaccines, introduce the genetic material encoding the antigenic protein for the host to express. Because these proteins will undergo host posttranslational modifications, host glycosylation can potentially alter the structure and immunological efficacy of the antigen. In this review, we discuss the potential impact of host protein glycosylation on the immune responses generated by nucleic acid vaccines against bacterial and viral pathogens.
Keywords: Nucleic acid vaccines, viral glycoproteins, host glycosylation, immune responses, antibodies
I. Introduction
Unlike traditional vaccines that are comprised of antigens in a purified subunit vaccine form or a whole-cell vaccine (live-attenuated or inactivated), nucleic acid vaccines use genetic materials (DNA or mRNA) that encode the antigenic proteins. The manufacturing of these vaccines can be robust and cost-effective [1]. While nucleic acid vaccines are a relatively new vaccine technology, they have been evaluated in clinical trials for humans [2,3] and used in animals [4] but have not been approved for human use until recently. The arrival of COVID-19 as a global pandemic facilitated the swift approval of several nucleic acid vaccines for clinical use globally. Based on the genetic material used, nucleic acid vaccines can be categorized under three main classes [5].
DNA vaccines.
Genes of antigens are cloned into plasmid DNAs under the control of a strong eukaryotic promoter. After their introduction into host cells, these plasmids initiate the expression of the antigens they encode. DNA vaccines have high stability that does not require a cold chain transportation [6]. Several DNA vaccines were approved by FDA for veterinary use [7]. Currently, there are no FDA-approved human DNA vaccines.
mRNA vaccines.
mRNA is the intermediate genetic material that is produced (transcribed) from protein-coding DNA in the nucleus. While plasmids used in DNA vaccines must translocate to the nucleus and be transcribed into mRNAs, mRNAs introduced in mRNA vaccines can be directly used by ribosomes for protein production. However, mRNA vaccines are not as stable as DNA vaccines, and they require cold chain transportation. BNT162b2 (Pfizer-BioNTech) and mRNA-1273 (Moderna) COVID-19 vaccines are the two mRNA vaccines currently administered globally. mRNA vaccine technologies are rapidly improving. One such example is self-amplifying RNA (saRNA) vaccines, which can be administered at lower doses compared to conventional mRNA vaccines [8].
Viral vector-based vaccines.
In viral vector-based vaccines, genes of antigens are incorporated into a viral genome. Although viruses used as vectors in vaccinations can infect cells and deliver the genes of antigens, they do not cause disease. Upon infection, host cells express the target antigens. There are several licensed viral vector-based vaccines in the veterinary use [9]. Sputnik V and AstraZeneca COVID-19 vaccines are two viral vector-based vaccines in use globally for humans and more are in ongoing clinical trials [2,3]. Like mRNA vaccines, viral vector-based vaccines require a cold chain transportation [10]. Another potential limitation of this class of nucleic acid vaccines is that viral vectors may elicit viral vector-specific neutralizing antibody responses that may prevent their re-administration [11].
In this review, we discuss glycosylation as a key parameter in the design of nucleic acid vaccines utilizing two distinct types of pathogenic proteins. First, we describe nucleic acid vaccines encoding bacterial surface protein immunogens that are not glycosylated in their native form. As the newly biosynthesized antigens are secreted by host cells, the design of nucleic acid vaccines leads the antigens to a secretory pathway, where they are subject to mammalian cellular glycosylation. In the second part, we discuss nucleic acid vaccines encoding a surface glycoprotein of a membrane enveloped virus. Contrary to native bacterial proteins, surface glycoproteins of enveloped viruses are products of the mammalian host cell glycosylation machinery.
II. Impact of host protein glycosylation in nucleic acid vaccines for bacterial pathogens
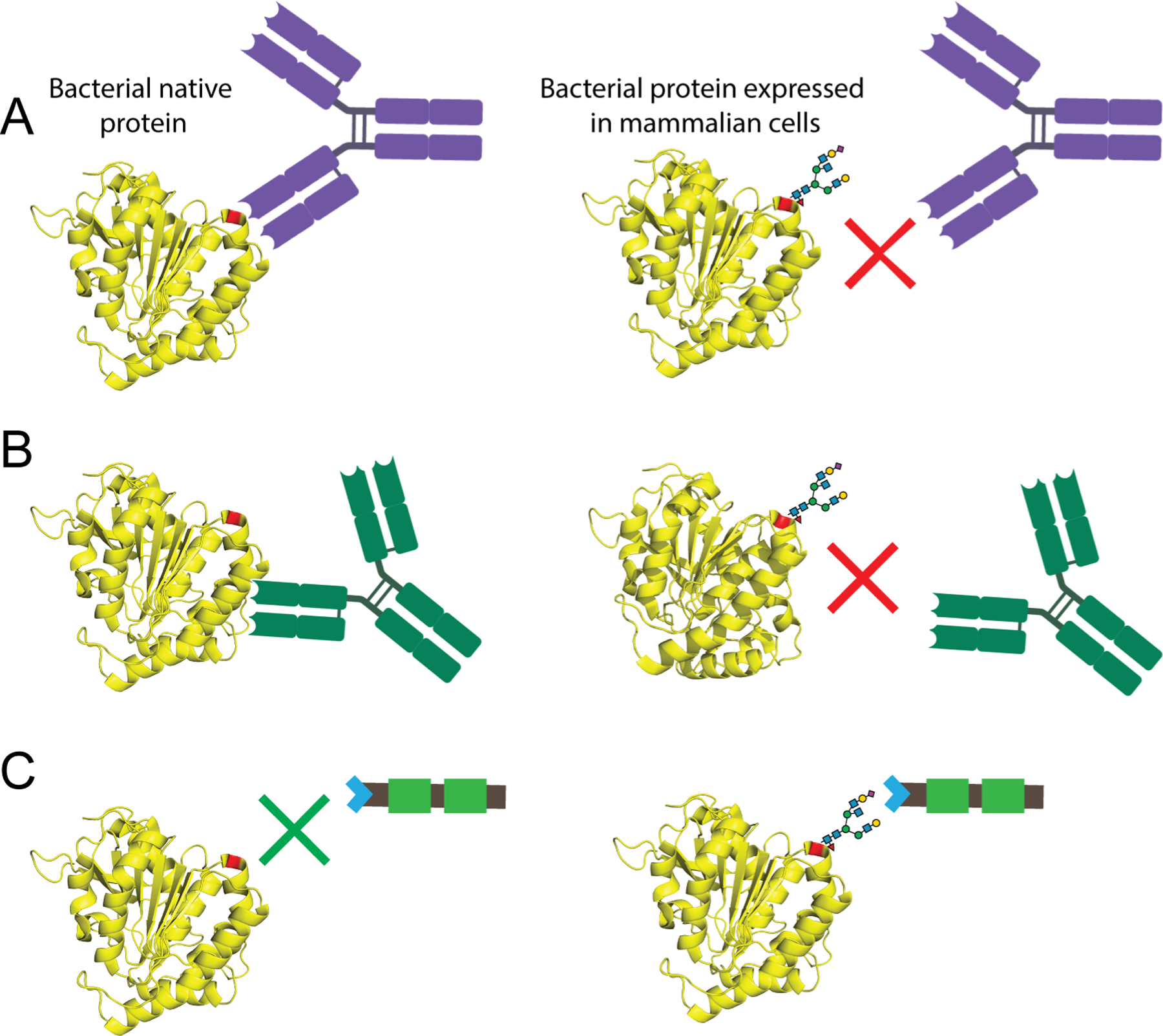
Host protein glycosylation may result in impaired immune responses when nucleic acid vaccines are used against bacterial pathogens [12]. While the bacterial pathogens express their native antigens, the mammalian host cells express the same antigens with nucleic acid vaccine administration and those antigens can be glycosylated by host cells. (Fig. 1A). As a result of host protein glycosylation, naturally non-existing antigenic determinants can potentially be produced, consequentially inducing an impaired immune response against the native antigen-bearing bacteria.
Figure 1.
Glycosylation of bacterial proteins by mammalian host cells can impair immune responses. A. Bacterial proteins can be glycosylated when expressed by mammalian host cells (right panel), while its native counterpart is not (left panel). The crystal structure of the Mycobacterium tuberculosis Antigen 85A protein was used for the depiction (PBD 1SFR). The N-glycosylation site is colored as red. Glycans can shield protein surface of the proteins and prevent generation of antibodies to those sites. B. Glycans can result in a different protein folding and distort the epitopes that native protein has. C. Sialic acids on glycoproteins expressed by mammalian host cells can induce inhibitory siglecs.
With every glycan added by the host, the protein becomes less like the native protein. The space occupied by the glycans around the proteins can vary, depending on the degree of glycosylation. One obstacle associated with glycans occupying a space around the protein is that glycans prevent the access of B cell receptors to some antigenic determinants on the protein, and therefore no antibody response is generated against those sterically hindered regions (Fig. 1A). Steric hindrance of epitopes by glycans is exploited by many pathogens to evade from host immune responses, such as the human immunodeficiency virus (HIV) that is decorated with a heavily glycosylated envelope glycoprotein [13], and many pathogenic bacteria that are encapsulated with capsular polysaccharides [14–17]. Another potential mechanism through which mammalian host glycosylation may prevent an effective immune response is altered folding of the protein, which then leads to distortion of epitopes on the protein (Fig. 1B).
In nucleic acid vaccination, the degree of glycosylation by the host largely depends on the amino acid sequence of the antigen. The proteins that enter the secretory pathway are subjected to the glycosylation machinery, and if they possess one or more N-glycosylation consensus sequons, they can be N-glycosylated. Those proteins may also undergo O-glycosylation [18].
Although not common, some bacterial proteins are glycosylated in their native form [19]. Because the glycosylation mechanisms of prokaryotes and eukaryotes are often different, the protein antigen would be glycosylated by the host and the bacterium distinctly. Adding new glycans that the native protein does not have, and/or replacing or removing the authentic glycans on the native glycoprotein may result in impaired immune responses as the epitopes are distorted. Previously, it was shown that native and recombinant (E. coli-expressed) forms of a Mycobacterium tuberculosis (Mtb) protein, alanine-proline-rich antigen (Apa), which have different glycosylation motif in each form, differ in their antigenicity and immunogenicity [20]. Another highly immunogenic Mtb surface protein, Ag85A, has been used as a component in several nucleic acid vaccine candidates [3]. In three completed clinical trials, two viral vector-based vaccine candidates carrying the gene of Ag85A were evaluated and found not to be protective or immunogenic among the participants of the trials [21–23]. In our recently published work, we investigated the effect of host protein glycosylation on the immune responses generated by nucleic acid vaccines [12]. In the study, Ag85A was expressed in human embryonic kidney (HEK) cells and Chinese hamster ovarian (CHO) cells through transient transfection with plasmids or viral infection using Adenovirus 5 as a carrier [12]. In all expression conditions, Ag85A expressed by mammalian cells was shown to be N-glycosylated, while native Ag85A expressed by Mtb did not show any evidence of being glycosylated. N-glycan analysis of Ag85A expressed by HEK 293-F cells (293-F Ag85A) by mass spectrometry revealed several distinct glycan structures many of which contained sialic acids. Humoral immune responses generated to native or mammalian Ag85As were compared in mouse immunizations, where 293-F Ag85A raised significantly lower antibody titers than native Ag85A. Similarly, in an in vitro T cell stimulation assay, native Ag85A outperformed HEK 293-F expressed Ag85A. In the same assay, nonglycosylated 293-F Ag85A that has a mutation at the glycosylation site stimulated T cells better than wild type 293-F Ag85A. Also, in a dot blot assay, antibodies raised against native Ag85A reacted to mutant 293-F Ag85A more than wild type 293-F Ag85A, indicating that some epitopes on the protein are blocked by the glycans for antibody recognition.
Another potential problem associated with the glycosylation of proteins with nonviral origin by host cells is the immune inhibitory receptors, whose ligands are glycans (Fig.1C). These receptors function in the self or non-self discrimination [24]. Sialicacid-binding immunoglobulin-like lectins (Siglecs) are a major immunoregulatory receptor family, consisting of 15 cell surface receptors in humans. Sialic acids are considered as self-associated molecular patterns [25] and most of siglecs are inhibitory as they have immunoreceptor tyrosine-based inhibitory motifs (ITIMs) and ITIM-like motifs in their cytoplasmic domains [26].
The downregulatory effects of siglecs on immune responses are exploited by many pathogens, which display sialic acids on their surfaces to pretend as “self” and evade immune recognition [27]. The sialylated capsular polysaccharide of group B Streptococcus (GBS), for example, activates neutrophil Siglec-9, which in turn suppresses the innate immune response against this pathogen, leading to an increased bacterial survival [28]. While some of these pathogens, utilizing molecular mimicry, synthesize sialic acids de novo, some of them utilize sialic acids of hosts [29]. An opportunistic pathogen Pseudomonas aeruginosa adsorbs sialoglycoproteins from serum and reduces neutrophil activity [30]. A protozoan parasite Trypanosoma cruzi transfers sialic acids from host glycoconjugates to themselves by using trans-sialidases [31]. Siglecs are utilized by not only pathogens but also cancer cells to evade immune responses [32]. Cancer cells hypersialylate their surface proteins to activate inhibitory siglecs on immune cells to downregulate anti-tumor immunity. Hypersialylation activity of cancer cells is correlated with metastatic phenotype and poor prognosis in cancer patients [33,34]. Upon using nucleic acid vaccines and generating glycosylated antigens that possess sialic acids, it is not unlikely to have an impaired immune response to target antigens. In addition to playing roles in immune regulation, sialic acids on antigen/pathogen surfaces can also potentially dampen the uptake and presentation of antigens by antigen-presenting cells for the T cell recognition [35–37]. On the other hand, it is also possible that sialic acids on the antigen can help increase the uptake of the antigen by antigen-presenting cells [38].
III. Nucleic acid vaccines employing glycoproteins of enveloped viruses
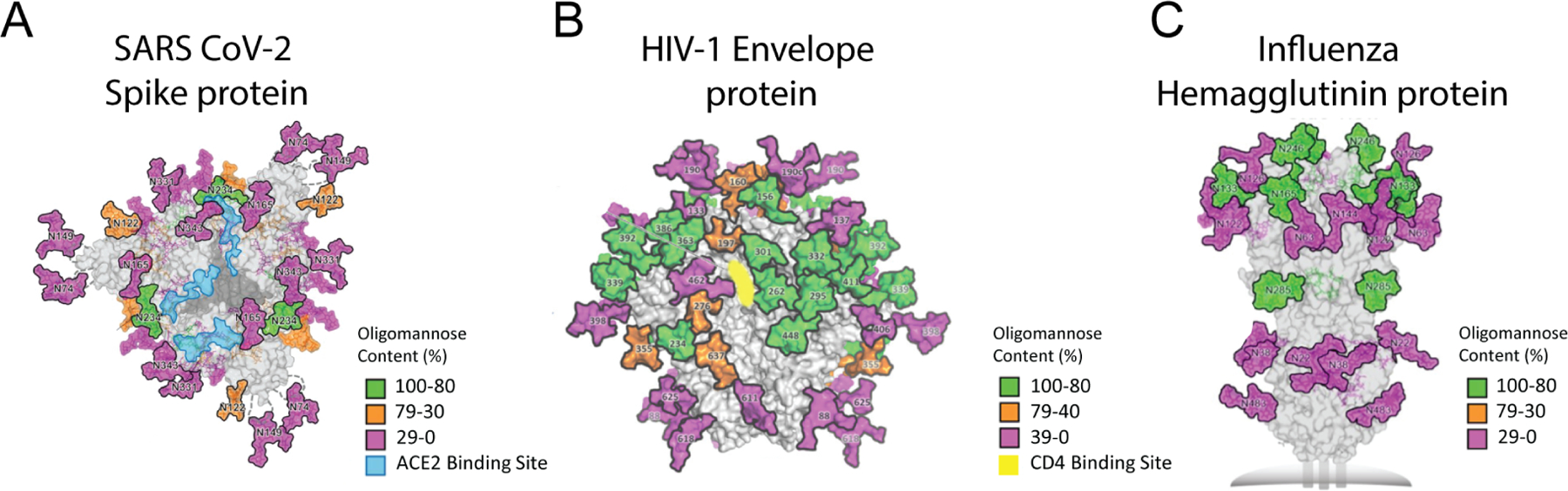
Glycans decorate the surface proteins of many pathogenic viruses, including human immunodeficiency virus-1 (HIV-1), influenza virus, Ebola virus and coronavirus. These glycans on viral surfaces play many critical roles in the life cycle of the viruses, one of which is to prevent immune recognition through molecular mimicry and shielding their surface proteins [39]. Another important role of viral surface glycans is to infect host cells through binding to their surface lectins [40]. A recent study analyzed the glycosylation of the SARS-CoV-2 spike protein to understand its role in the host-cell entry of the virion by binding to angiotensin-converting enzyme 2 (ACE2) [41]. In the study, recombinant spike proteins expressed in HEK 293-F cells were proteolyzed and the formed glycopeptides were analyzed by mass spectrometry to determine the composition of glycan structures at the 22 N-linked glycan sequons. The N-glycans proximal to the receptor-binding domain shield this conserved and thus vulnerable domain to immune attack [41] (Fig. 2A). The roles of glycans in sterically masking the polypeptide epitopes were also revealed through molecular dynamics simulations of the SARS-CoV-2 spike protein [42]. HIV envelope protein (Env), which is responsible for viral attachment to the host cells through CD4 receptors, is another heavily glycosylated viral glycoprotein. Similar to the roles of glycans on SARS-CoV-2 spike protein, glycans on Env shield the polypeptide epitopes including the CD4 binding site (Fig. 2B) [43].
Figure 2.
The oligomannose content and distribution of N-glycans on the surface of SARS CoV-2spike protein (A), HIV-1 envelope glycoprotein (B), and influenza (H3N2) hemagglutinin protein (C). Different percentages of observed oligomannose structures are represented in green, magenta and orange, and unoccupied sites are represented in gray. This figure is reproduced with permission from: American Association for the Advancement of Science [41] (Figure 2A); Annual Reviews [43] (Figure 2B); and PLOS [48] (Figure 2C).
Although viral glycans serve as a shield that protects the viral surface proteins from immune recognition, they can be the target of the immune system as well. A potently neutralizing antibody against SARS-CoV-1, for example, was shown to bind an epitope containing a glycan structure [44]. There are also several broadly neutralizing antibodies isolated from HIV infected individuals that recognize the glycan structures on HIV envelop glycoprotein [45–47]. Importantly, a cross-neutralizing glycan-specific antibody between HIV-1 and influenza virus was previously observed [48]. In addition to antibody binding, glycans on viral glycoproteins contribute to recruiting T cell help to induce adaptive humoral immune responses [35,37]. Therefore, upon administration of nucleic acid vaccines, host glycosylation of viral surface glycoproteins can be critical for inducing robust B and T cell responses. On the other hand, glycans on viral surfaces could shield the antigenic determinants on the protein backbone -particularly the conserved epitopes- preventing/inhibiting the induction of a strong immune response targeting these shielded epitopes. Conserved epitopes on viral protein immunogens have been the major targets in the generation of universal vaccines. Therefore, removal of glycosylation sites on and/or around those conserved and thus vulnerable epitopes in nucleic acid vaccine design can potentially serve as an effective strategy to develop protective and conserved/universal vaccines against viral pathogens.
Nucleic acid vaccines generated against SARS-CoV-2 and administered worldwide have shown to be highly effective in controlling COVID-19 pandemic [49]. Nucleic acid vaccines against other viral pathogens are under development [50–52]. Contrary to native bacterial surface proteins, surface glycoproteins of enveloped viruses are the products of the mammalian host glycosylation machinery. Thus, the glycosylation of viral proteins expressed through nucleic acid vaccine immunization are expected to be identical or highly similar to the glycosylation of the protein on the surface of the virion. While this is largely true, for a proper glycosylation match, nucleic acid vaccines may need to be expressed in the same cell types as the cells infected during viral infection, since glycosylation of a protein may differ when it is expressed by different cell types of the same species [53]. This may be due to the highly intricate glycosylation pathways enrolling many distinct glycosyltransferases, donor sugars, and chaperone proteins that can all be variably expressed and/or available at different concentrations in different cell types and cellular states [53,54]. In addition, the inflammation taking place during an infection can alter the glycosylation process [55–57] yielding glycoproteins that differ from the ones expressed through nucleic acid vaccination. In addition, the expression levels of enzymes participating in glycosylation can be changed when the cells are infected, leading to differentiated glycosylation. It was recently shown that transcription of HIV genes in infected T cells alters the glycosylation machinery of the cells [58]. In the study, CD4+ T cells isolated from HIV-negative donors were infected with the virus and infected cells that are active or inactive in HIV transcription were separated by cell sorting. Membrane proteins were isolated and used in a lectin microarray to illustrate the cell surface glycomic signature of each group. Demonstrating the influence of HIV transcription on glycosylation, the results revealed that cell surface proteins of cells that are active or inactive in HIV transcription have different glycosylation profiles.
IV. Conclusions
Proteins with bacterial origin can be glycosylated when expressed in mammalian host cells through nucleic acid vaccine administration and the host glycans incorporated onto these proteins can impair the immune responses generated against the target pathogen.
Viral surface glycoproteins are decorated with a glycan layer produced by mammalian host’s glycosylation machinery. This glycan layer serves as a virulence factor shielding the protein backbone to prevent immune targeting. On the other hand, the glycans can be part of the antigenic determinants for antibody recognition as well as T cell epitopes in the form of glycopeptide epitopes.
Nucleic acid vaccines against viral pathogens induce immune responses to viral glycoproteins since native viral glycoproteins on the virion surfaces are also expressed in host cells. However, nucleic acid vaccines against viral pathogens may potentially produce differentially glycosylated proteins than native viral proteins due to different cell types that express the proteins, and the potentially altered glycosylation machinery in infected cells.
Acknowledgments
This work was supported by National Institutes of Health grants R01AI123383 (FYA), R01AI152766 (FYA). The authors thank Cathryn Quinn for editing the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
Authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published with the period of review, are highlighted as:
*of special interest
**of outstanding interest
- 1.Delany I, Rappuoli R, De Gregorio E: Vaccines for the 21st century. EMBO Mol Med 2014, 6:708–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajao DS, Perez DR: Universal Vaccines and Vaccine Platforms to Protect against Influenza Viruses in Humans and Agriculture. Front Microbiol 2018, 9:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voss G, Casimiro D, Neyrolles O, Williams A, Kaufmann SHE, McShane H, Hatherill M, Fletcher HA: Progress and challenges in TB vaccine development. F1000Res 2018, 7:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Draper SJ, Heeney JL: Viruses as vaccine vectors for infectious diseases and cancer. Nature Reviews Microbiology 2009, 8:62. [DOI] [PubMed] [Google Scholar]
- 5.Ellis WR, Rappuoli R, Ahmed S: Technologies for making new vaccines. In Vaccines, edn 6th. Edited by Plotkin SA, Orenstein WA, Offit PA: Saunders; 2013:1182–1199. [Google Scholar]
- 6.Lee J, Arun Kumar S, Jhan YY, Bishop CJ: Engineering DNA vaccines against infectious diseases. Acta Biomater 2018, 80:31–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira VB, Zurita-Turk M, Saraiva TlDL, Castro CPsD, Souza BM, Agresti PM, Lima FA, Pfeiffer VN, Azevedo MSP, Rocha CS, et al. : DNA Vaccines Approach: From Concepts to Applications. World Journal of Vaccines 2014, Vol.04 No.02:22. [Google Scholar]
- 8.Bloom K, van den Berg F, Arbuthnot P: Self-amplifying RNA vaccines for infectious diseases. Gene Ther 2021, 28:117–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Draper SJ, Heeney JL: Viruses as vaccine vectors for infectious diseases and cancer. Nat Rev Microbiol 2010, 8:62–73. [DOI] [PubMed] [Google Scholar]
- 10.Crommelin DJA, Volkin DB, Hoogendoorn KH, Lubiniecki AS, Jiskoot W: The Science is There: Key Considerations for Stabilizing Viral Vector-Based Covid-19 Vaccines. J Pharm Sci 2021, 110:627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirley JL, de Jong YP, Terhorst C, Herzog RW: Immune Responses to Viral Gene Therapy Vectors. Mol Ther 2020, 28:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozdilek A, Paschall AV, Dookwah M, Tiemeyer M, Avci FY: Host protein glycosylation in nucleic acid vaccines as a potential hurdle in vaccine design for nonviral pathogens. Proc Natl Acad Sci U S A 2020, 117:1280–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Revealing the importance of considering glycosylation in nucleic acid vaccines, this study shows that bacterial proteins are glycosylated when expressed in mammalian cells and the presence of glycans can impair the pathogen-specific immune responses.
- 13.Berndsen ZT, Chakraborty S, Wang X, Cottrell CA, Torres JL, Diedrich JK, Lopez CA, Yates JR 3rd, van Gils MJ, Paulson JC, et al. : Visualization of the HIV-1 Env glycan shield across scales. Proc Natl Acad Sci U S A 2020, 117:28014–28025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerry P, Poly F, Riddle M, Maue AC, Chen YH, Monteiro MA: Campylobacter polysaccharide capsules: virulence and vaccines. Front Cell Infect Microbiol 2012, 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yother J: Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol 2011, 65:563–581. [DOI] [PubMed] [Google Scholar]
- 16.Avci F: Novel Strategies for Development of Next-Generation Glycoconjugate Vaccines. Current Topics in Medicinal Chemistry 2013, 13:2535–2540. [DOI] [PubMed] [Google Scholar]
- 17.Avci F, Berti F, Dull P, Hennessey J, Pavliak V, Prasad AK, Vann W, Wacker M, Marcq O: Glycoconjugates: What It Would Take To Master These Well-Known yet Little-Understood Immunogens for Vaccine Development. mSphere 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varki A: Biological roles of glycans. Glycobiology 2017, 27:3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Upreti RK, Kumar M, Shankar V: Bacterial glycoproteins: functions, biosynthesis and applications. Proteomics 2003, 3:363–379. [DOI] [PubMed] [Google Scholar]
- 20.Nandakumar S, Kannanganat S, Dobos KM, Lucas M, Spencer JS, Fang S, McDonald MA, Pohl J, Birkness K, Chamcha V, et al. : O-mannosylation of the Mycobacterium tuberculosis adhesin Apa is crucial for T cell antigenicity during infection but is expendable for protection. PLoS Pathog 2013, 9:e1003705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tameris MD, Hatherill M, Landry BS, Scriba TJ, Snowden MA, Lockhart S, Shea JE, McClain JB, Hussey GD, Hanekom WA, et al. : Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet 2013, 381:1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ndiaye BP, Thienemann F, Ota M, Landry BS, Camara M, Dieye S, Dieye TN, Esmail H, Goliath R, Huygen K, et al. : Safety, immunogenicity, and efficacy of the candidate tuberculosis vaccine MVA85A in healthy adults infected with HIV-1: a randomised, placebo-controlled, phase 2 trial. Lancet Respir Med 2015, 3:190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tameris M, Hokey DA, Nduba V, Sacarlal J, Laher F, Kiringa G, Gondo K, Lazarus EM, Gray GE, Nachman S, et al. : A double-blind, randomised, placebo-controlled, dose-finding trial of the novel tuberculosis vaccine AERAS-402, an adenovirus-vectored fusion protein, in healthy, BCG-vaccinated infants. Vaccine 2015, 33:2944–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pereira MS, Alves I, Vicente M, Campar A, Silva MC, Padrao NA, Pinto V, Fernandes A, Dias AM, Pinho SS: Glycans as Key Checkpoints of T Cell Activity and Function. Front Immunol 2018, 9:2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varki A: Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology 2011, 21:1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulson JC, Macauley MS, Kawasaki N: Siglecs as sensors of self in innate and adaptive immune responses. Ann N Y Acad Sci 2012, 1253:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang YC, Nizet V: The interplay between Siglecs and sialylated pathogens. Glycobiology 2014, 24:818–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlin AF, Uchiyama S, Chang YC, Lewis AL, Nizet V, Varki A: Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood 2009, 113:3333–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vimr E, Lichtensteiger C: To sialylate, or not to sialylate: that is the question. Trends Microbiol 2002, 10:254–257. [DOI] [PubMed] [Google Scholar]
- 30.Khatua B, Bhattacharya K, Mandal C: Sialoglycoproteins adsorbed by Pseudomonas aeruginosa facilitate their survival by impeding neutrophil extracellular trap through siglec-9. J Leukoc Biol 2012, 91:641–655. [DOI] [PubMed] [Google Scholar]
- 31.Freire-de-Lima L, Fonseca LM, Oeltmann T, Mendonca-Previato L, Previato JO: The trans-sialidase, the major Trypanosoma cruzi virulence factor: Three decades of studies. Glycobiology 2015, 25:1142–1149. [DOI] [PubMed] [Google Scholar]
- 32.Adams OJ, Stanczak MA, von Gunten S, Laubli H: Targeting sialic acid-Siglec interactions to reverse immune suppression in cancer. Glycobiology 2018, 28:640–647. [DOI] [PubMed] [Google Scholar]
- 33.Hudak JE, Canham SM, Bertozzi CR: Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat Chem Biol 2014, 10:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldus SE, Zirbes TK, Monig SP, Engel S, Monaca E, Rafiqpoor K, Hanisch FG, Hanski C, Thiele J, Pichlmaier H, et al. : Histopathological subtypes and prognosis of gastric cancer are correlated with the expression of mucin-associated sialylated antigens: Sialosyl-Lewis(a), Sialosyl-Lewis(x) and sialosyl-Tn. Tumour Biol 1998, 19:445–453. [DOI] [PubMed] [Google Scholar]
- 35.Sun L, Middleton DR, Wantuch PL, Ozdilek A, Avci FY: Carbohydrates as T-cell antigens with implications in health and disease. Glycobiology 2016, 26:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Paschall AV, Middleton DR, Ishihara M, Ozdilek A, Wantuch PL, Aceil J, Duke JA, LaBranche CC, Tiemeyer M, et al. : Glycopeptide epitope facilitates HIV-1 envelope specific humoral immune responses by eliciting T cell help. Nat Commun 2020, 11:2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun L, Ishihara M, Middleton DR, Tiemeyer M, Avci FY: Metabolic labeling of HIV-1 envelope glycoprotein gp120 to elucidate the effect of gp120 glycosylation on antigen uptake. J Biol Chem 2018, 293:15178–15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delputte PL, Nauwynck HJ: Porcine arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J Virol 2004, 78:8094–8101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe Y, Bowden TA, Wilson IA, Crispin M: Exploitation of glycosylation in enveloped virus pathobiology. Biochim Biophys Acta Gen Subj 2019, 1863:1480–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This review article discusses the envelope glycans, their roles during viral life-cycles and their recognition by the immune system.
- 40.Raman R, Tharakaraman K, Sasisekharan V, Sasisekharan R: Glycan-protein interactions in viral pathogenesis. Curr Opin Struct Biol 2016, 40:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M: Site-specific glycan analysis of the SARS-CoV-2 spike. Science 2020, 369:330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]; *In this study the glycan composition of SARS CoV-2 spike protein was analyzed and how glycans are shielding the ACE2 binding domain, which is a conserved and vulnrable site to immune responses, was depicted.
- 42.Zhao P, Praissman JL, Grant OC, Cai Y, Xiao T, Rosenbalm KE, Aoki K, Kellman BP, Bridger R, Barouch DH, et al. : Virus-Receptor Interactions of Glycosylated SARS-CoV-2 Spike and Human ACE2 Receptor. Cell Host Microbe 2020, 28:586–601 e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crispin M, Ward AB, Wilson IA: Structure and Immune Recognition of the HIV Glycan Shield. Annu Rev Biophys 2018, 47:499–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pinto D, Park YJ, Beltramello M, Walls AC, Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, et al. : Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 2020, 583:290–295. [DOI] [PubMed] [Google Scholar]
- 45.McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. : Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 2011, 480:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, et al. : A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 2011, 334:1097–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calarese DA, Lee HK, Huang CY, Best MD, Astronomo RD, Stanfield RL, Katinger H, Burton DR, Wong CH, Wilson IA: Dissection of the carbohydrate specificity of the broadly neutralizing anti-HIV-1 antibody 2G12. Proc Natl Acad Sci U S A 2005, 102:13372–13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee CD, Watanabe Y, Wu NC, Han J, Kumar S, Pholcharee T, Seabright GE, Allen JD, Lin CW, Yang JR, et al. : A cross-neutralizing antibody between HIV-1 and influenza virus. PLoS Pathog 2021, 17:e1009407. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study demonstrates that a glycan specific HIV Env antibody is broadly neutrilizing. MAb 2G12 isolated from an HIV infected individual is specific for mannose patches also neutrilized a strain of influenza viruses.
- 49.Tenforde MW, Olson SM, Self WH, Talbot HK, Lindsell CJ, Steingrub JS, Shapiro NI, Ginde AA, Douin DJ, Prekker ME, et al. : Effectiveness of Pfizer-BioNTech and Moderna Vaccines Against COVID-19 Among Hospitalized Adults Aged >/=65 Years - United States, January-March 2021. MMWR Morb Mortal Wkly Rep 2021, 70:674–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khalid K, Padda J, Khedr A, Ismail D, Zubair U, Al-Ewaidat OA, Padda S, Cooper AC, Jean-Charles G: HIV and Messenger RNA (mRNA) Vaccine. Cureus 2021, 13:e16197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou K, Li C, Shi W, Hu X, Nandakumar KS, Jiang S, Zhang N: Current Progress in the Development of Zika Virus Vaccines. Vaccines (Basel) 2021, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma AR, Lee YH, Nath S, Lee SS: Recent developments and strategies of Ebola virus vaccines. Curr Opin Pharmacol 2021, 60:46–53. [DOI] [PubMed] [Google Scholar]
- 53.May LT, Shaw JE, Khanna AK, Zabriskie JB, Sehgal PB: Marked cell-type-specific differences in glycosylation of human interleukin-6. Cytokine 1991, 3:204–211. [DOI] [PubMed] [Google Scholar]
- 54.Moremen KW, Tiemeyer M, Nairn AV: Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol 2012, 13:448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Groux-Degroote S, Cavdarli S, Uchimura K, Allain F, Delannoy P: Glycosylation changes in inflammatory diseases. Adv Protein Chem Struct Biol 2020, 119:111–156. [DOI] [PubMed] [Google Scholar]
- 56.Gornik O, Lauc G: Glycosylation of serum proteins in inflammatory diseases. Dis Markers 2008, 25:267–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giron LB, Colomb F, Papasavvas E, Azzoni L, Yin X, Fair M, Anzurez A, Damra M, Mounzer K, Kostman JR, et al. : Interferon-alpha alters host glycosylation machinery during treated HIV infection. EBioMedicine 2020, 59:102945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Colomb F, Giron LB, Kuri-Cervantes L, Adeniji OS, Ma T, Dweep H, Battivelli E, Verdin E, Palmer CS, Tateno H, et al. : Sialyl-Lewis(X) Glycoantigen Is Enriched on Cells with Persistent HIV Transcription during Therapy. Cell Rep 2020, 32:107991. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This study demonstrates that when cells are infected with a virus, the glycosylation machinery is altered. In the study, the transcription of HIV genes altered the surface glycomic signature of T cells.


