Preface
SARS-CoV-2 infection is benign in most individuals but, in ~10% of cases, it triggers hypoxemic COVID-19 pneumonia, which becomes critical in ~3% of cases. The ensuing risk of death (~1%) doubles every five years from childhood onward and is ~1.5 times greater in men than in women. What are the molecular and cellular determinants of critical COVID-19 pneumonia? Inborn errors of type I IFNs, including autosomal TLR3 and X-linked TLR7 deficiencies, are found in ~1-5% of patients with critical pneumonia under 60 years old, and a lower proportion in older patients. Pre-existing autoantibodies neutralizing IFN-α, –β, and/or –ω, which are more common in men than in women, are found in ~15-20% of patients with critical pneumonia over 70 years old, and a lower proportion in younger patients. Thus, at least 15% of cases of critical COVID-19 pneumonia can apparently be explained. The TLR3- and TLR7-dependent production of type I IFNs by respiratory epithelial cells and plasmacytoid dendritic cells, respectively, is essential for host defense against SARS-CoV-2. In ways that can depend on age and sex, insufficient type I IFN immunity in the respiratory tract during the first few days of infection may account for the spread of the virus, leading to pulmonary and systemic inflammation.
Introduction
More than 5 million people have died from COVID-19, and infection fatality rates (IFR) in unvaccinated populations are ~1%[1, 2]. Indeed, infection with SARS-CoV-2 is silent in ~40% of cases, underlies a benign upper respiratory tract disease in another 40%, and causes pneumonia in ~20%[3, 4]. Non-hypoxemic, ‘moderate’ pneumonia is seen in ~10% of cases, whereas the remaining 10% of cases present hypoxemic pneumonia, typically requiring hospitalization for oxygen therapy. In ~3% of cases, the administration of O2 at a rate < 6 L/min (the cutoff for ‘severe’ pneumonia) is not sufficient to alleviate hypoxemia. In such cases, high-flow oxygen (O2 > 6 L/min), mechanical ventilation (non-invasive or by intubation), or extracorporeal membrane oxygenation (ECMO) is required (any of these three options, typically provided in intensive care units, defines ‘critical pneumonia’)[5, 6]. The IFR increases exponentially with age, doubling every five years, from 0.001% in individuals aged 5-9 years to 8.29% in those over the age of 80 years[1, 7–10]. Ancestry, social status, and several comorbid conditions have been associated with higher disease severity and death rates, but with modest odds ratios (OR, typically <1.5, rarely >2)[7–9]. Men have a 1.5 times greater risk of death than women, after adjustment for other risk factors[1, 11]. Overall, the striking epidemiological feature of life-threatening COVID-19 is its strong dependence on age, steadily increasing throughout life, with a 10,000 times greater risk at ages > 80 years than in the first decade of life[1,12,13]. A similar pattern is seen with the more transmissible viral variants[14, 15]. The same viruses are found in patients with silent and lethal infections, excluding the hypothesis that interindividual clinical variability is primarily a consequence of viral diversity.
The hypothesis that a large amount of viral inoculum is more life-threatening than a small inoculum is more plausible, in line with the findings of 100 years of experimental inoculations of animals with pathogens[16]. However, it is difficult to test this hypothesis in humans. One alternative hypothesis is that humans with life-threatening COVID-19 were particularly prone to critical illness due to an underlying and hitherto silent immunodeficiency[17, 18]. The traditional view of immunodeficiency, characterized by overt immunological abnormalities and broad vulnerability to infectious agents — as illustrated by patients with acquired immunodeficiency syndrome or severe combined immunodeficiency, who lack T cells due to HIV infection and germline mutations, respectively — has turned out to be the tip of an iceberg. Since 1996, previously healthy patients with rare or common infectious diseases but normal resistance to other infectious agents have been found to carry inborn errors of immunity (IEIs) rendering them particularly susceptible to specific microbes. Rare IEIs have been implicated in at least 20 different types of viral, bacterial, fungal, and parasitic infections[17, 18]. These rare IEIs led to the discovery of a common IEI, accounting for about 1% of cases of tuberculosis in populations of European descent[19, 20]. Based on all these findings, we launched the COVID Human Genetic Effort (CHGE, www.covidhge.com) with the aim of discovering the molecular, cellular, and immunological determinants of the various SARS-CoV-2-related manifestations by searching for causal IEIs[13]. We review here these and other studies that have clarified the human genetic and immunological determinants of life-threatening COVID-19 pneumonia[12, 13, 21–24]. We do not consider other phenotypes, such as resistance to infection[25], pernio (“COVID toes”)[26], multisystem inflammatory syndrome in children or adults (MIS-C/A)[27], neuro-COVID[28], or long COVID[10, 29], for which genetic and immunological studies have only just begun.
Inborn errors underlying critical influenza
The first breakthrough emerged from a study of candidate inborn errors of TLR3-, IRF7-, and IRF9-dependent type I IFN immunity that had previously been shown to underlie life-threatening influenza pneumonia (Figure 1)[5, 17, 18, 24, 30–32]. Predispositions to critical COVID-19 and influenza were hypothesized to be allelic because both conditions are respiratory infections caused by RNA viruses[12]. The first influenza susceptibility gene discovered encodes IRF7, the inducible transcription factor responsible for amplifying type I IFN production in virus-infected cells[33]. Plasmacytoid dendritic cells (pDCs) constitutively express high levels of IRF7 and are the most potent producers of type I IFN[34, 35]. The second encodes IRF9, the DNA-binding component of the interferon-stimulated gene factor 3 (ISGF-3) complex activated by type I and III IFNs[36]. The third encodes TLR3, an endosomal dsRNA sensor that regulates basal levels of type I IFN in various non-hematopoietic cells[37], possibly including respiratory epithelial cells (RECs)[24, 32]. Germline mutations at these three human loci are causal for critical influenza pneumonia[30–32]. We also considered 10 other genes, the products of which are biochemically and immunologically connected to these three core genes (Figure 1), and for which deleterious genotypes have been shown to underlie other severe viral diseases (suggesting incomplete penetrance for influenza)[5]. These 13 loci encode proteins for which a genetic deficiency can be considered to confer a high risk of critical influenza.
Figure 1. Inborn errors of type I IFN immunity and autoantibodies neutralizing type I IFNs underlie life-threatening COVID-19 pneumonia by interfering with type I IFN immunity in tissue-resident respiratory epithelial cells and blood plasmacytoid dendritic cells.
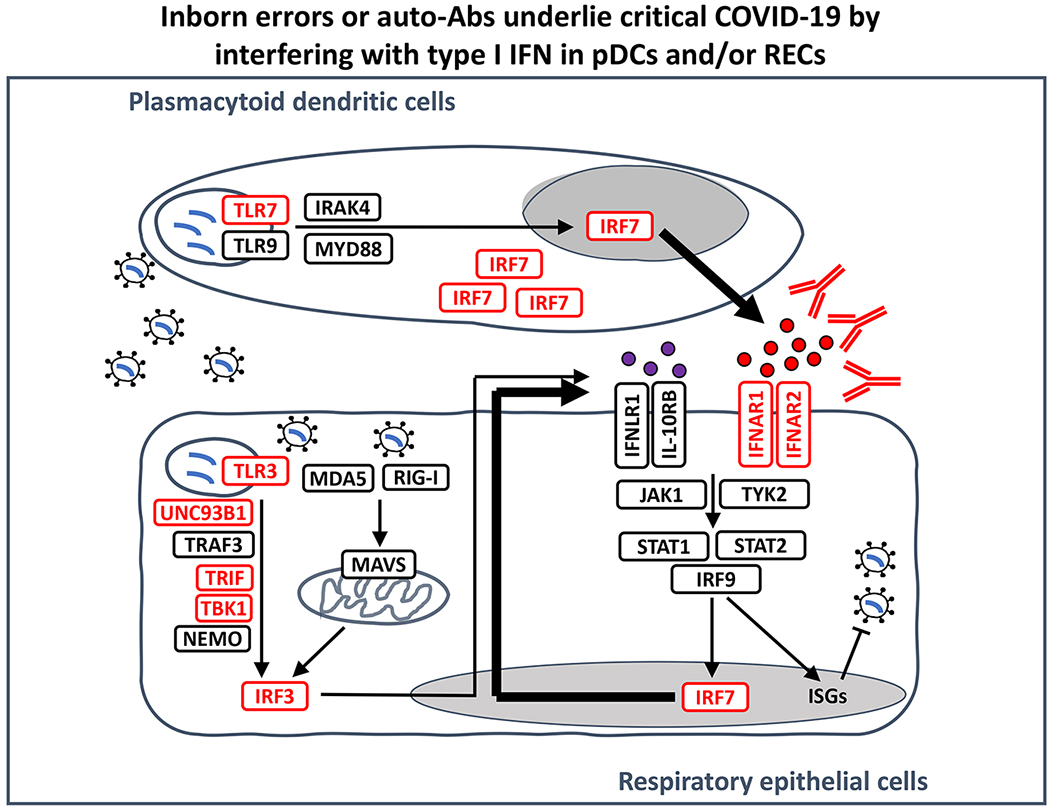
There are 17 human type I IFNs, each encoded by a specific, intron-less gene: 13 subtypes of IFN-α, IFN-β, IFN-ε, IFN-κ, and IFN-ω, and three human type III IFNs (IFN-λ1-3). Autoantibodies to IFN-α, IFN-β, and/or IFN-ω have been identified in about 15% of patients with critical COVID-19 pneumonia. Monogenic inborn errors of TLR3- and/or TLR7-dependent type I IFN immunity have been identified in about 1-5% of patients with critical COVID-19 pneumonia (genes shown in red). SARS-CoV-2 infection can induce type I IFN production in a TLR3-dependent manner in tissue-resident respiratory epithelial cells (RECs, which express TLR3 but not TLR7) and in a TLR7-dependent manner in circulating plasmacytoid dendritic cells (pDCs, which express TLR7 but not TLR3)[200]. IRF7 is constitutively expressed in pDCs, at higher levels than in other cell types, whereas it is mostly induced by viral infection in RECs[200]. IRF7 activation is required to produce type I IFNs other than IFN-β[33]. IFN: interferon; Auto-Ab: autoantibody, ISGs: interferon-stimulated genes.
Autosomal inborn errors of type I IFNs
Biochemically deleterious germline mutations of eight of the 13 genes were found in 23 of 659 patients with critical COVID-19 (3.5%) aged 17 to 77 years, including 18 patients under 60 years old (3.8%). Remarkably, four unrelated previously healthy adults, aged 26 to 50 years, had autosomal recessive (AR) complete IRF7 or IFNAR1 deficiency. The other patients had known (n=11) or previously unreported (n=8) autosomal dominant (AD), partial deficiencies. None of these patients had ever been hospitalized for other viral infections, including influenza. The penetrance of these disorders for critical COVID-19 is also probably incomplete, but higher for the AR than for the AD disorders, and for the known than for the unreported AD disorders (Table 1). A 13-year-old boy with AR IFNAR1 deficiency[38] and a three-year old girl with AR TBK1 deficiency[39] were independently reported to have critical COVID-19[40]. Fibroblasts presenting AD or AR TLR3 deficiency, AR IRF7 deficiency, or AR IFNAR1 deficiency displayed defective type I IFN-dependent control of SARS-CoV-2 in vitro[5], suggesting that RECs may display the same phenotype[32]. Moreover, pDCs from an IRF7-deficient patient were unable to induce type I IFNs upon stimulation with SARS-CoV-2 in vitro. This experimental approach provided proof-of-concept that IEIs affecting type I IFNs, including disorders of TLR3-dependent type I IFN immunity in RECs, and even AR defects that blunt type I IFN immunity across cell types, can underlie life-threatening COVID-19 pneumonia in previously healthy patients[12, 21] (Figure 1).
Table 1.
Major human genetic and immunological determinants of critical COVID-19 pneumonia§
| Risk estimatea | Frequency in the general population (%) | Frequency in patients with critical COVID (%) | References | |
|---|---|---|---|---|
| Genetic risk factors | ||||
| rs73064425/rs10490770 (3p21, intronic LZTFL1) | 1.89 - 2.14b | 8c (0.1-28) | 15d | [145–147] |
| Known AD deficiencies (TLR3, TRIF, TBK1, IRF3) | >20e | <0.1 | 1.7 | [5] |
| New AD deficiencies (UNC93B1, IRF7, IFNAR1, IFNAR2) | N.A. | 0.2 | 1.2 | [5] |
| Known AR deficiencies (IRF7, IFNAR1) | >20e | <0.1 | 0.6 | [5] |
| New XR deficiency (TLR7) | 34.4f | 0.065g | 1.3h | [41] |
| Immunological risk factors i | ||||
| anti-IFNω auto-Abs only (10 ng/mL) | 2.9j/3.6k | 0.2l | 0.8 | [109] |
| anti-IFNβ auto-Abs only (10 ng/mL) | 4.7j/4.5k | 0.3m | 1.3 | |
| anti-IFNα2 or anti-IFNω auto-Abs (100 pg/mL) | 12.7j/6.9k | 2.0n | 13.6 | |
| anti-IFNα2 or anti-IFNω auto-Abs (10 ng/mL) | 17.5j /14.9k | 0.5l | 9.8 | |
| anti-IFNα2 and anti-IFNω auto-Abs (10 ng/mL) | 67.6j/29.8k | 0.13l | 5.6 | |
We considered as major determinants only genetic or immunological abnormalities conferring an estimated OR greater than 2. Minor risk factors have been reviewed elsewhere[12]. Note that the heritability of all common SNPs (not only the chr3p21 region) was estimated at 6.5% for severe COVID-19 in [146] and < 1% in [147]. For rare variants we provide the proportion of carriers in critical COVID-19 patients.
Risk estimates are the ratio of the odds of critical COVID-19 in individuals carrying the genetic /immunological factor to those in individuals not carrying the factor. All studies compared patients with critical COVID-19 pneumonia (patients) with individuals presenting mild or asymptomatic SARS-CoV-2 infection (serving as controls), except for the GWAS of Ellinghaus et al[145], Pairo-Castineira et al[146] and the COVID-19 Host Genetics Initiative[147], which used controls from the general population.
Range of odds ratios (OR) for the risk allele under an additive model accounting for ethnicity, age and sex in the GWAS by Ellinghaus et al[145], Pairo-Castineira et al[146] and the COVID-19 Host Genetics Initiative[147].
The frequency is that of the risk allele observed in patients with critical COVID-19 pneumonia in the study by Pairo-Castineira et al[146].
The frequency is that of the risk allele in the study by Pairo-Castineira et al[146]. The range of allele frequencies observed across nine populations of gnomAD v3 is also provided in parentheses.
Based on predicted loss-of-function variants of the corresponding genes and their absence in 534 asymptomatic/paucisymptomatic infected controls. Functional tests were performed for variants from the asymptomatic/mild cases.
OR adjusted for ethnicity (PCA) and age (in years) for XR TLR7 deficiency in male patients only.
Cumulative MAF of biochemically deleterious TLR7 variants in the male gnomAD general population.
Proportion of critically ill male patients with XR TLR7 deficiency.
The types of type I IFN auto-Ab shown were selected both to cover the full range of ORs and to include all tested patients with critical COVID-19 pneumonia. The other data are available from Bastard et al[109].
OR, adjusted for age and sex, for critical COVID-19 pneumonia relative to asymptomatic or mild infection.
OR, adjusted for age and sex, for critical COVID-19 pneumonia relative to the general population.
Prevalence of auto-Ab in >34,000 samples from the general population.
Prevalence of auto-Ab in ~9,500 samples from the general population.
Prevalence of auto-Ab in >10,000 samples from the general population.
X-linked recessive TLR7 deficiency
In parallel, an X chromosome-wide approach resulted in the discovery of X-linked recessive (XR) TLR7 deficiency, a previously unknown IEI[41]. In a cohort of 1,202 unrelated male patients with critical pneumonia, 17 patients (1.4%) from 16 kindreds were hemizygous for biochemically deleterious TLR7 variants, whereas none of the 331 men with asymptomatic or mild COVID-19 carried such mutations[41]. Sixteen of the 17 patients are below the age of 60 years (1.8%). One of these patients also had ataxia-telangiectasia (AT), which was not causal for critical COVID-19 in other patients with AT infected with SARS-CoV-2[42]. TLR7 deficiency was also found in 1% of patients with severe, but not critical COVID-19 (i.e. with O2 < 6 L/min). The penetrance of XR TLR7 deficiency for severe or critical COVID-19 among relatives of index cases was high, but incomplete, especially in children (Table 1). We also found that the cumulative minor allele frequency (MAF) of deleterious alleles in men was < 6.5x10−4. Moreover, six of the 11 TLR7 variants previously reported in other patients were deleterious (carried by 9 of 16 patients)[43–46], whereas the variants in another study were not disclosed[47]. We further showed that the TLR7 genotype was deleterious in patients’ EBV-transformed B (EBV-B) cell lines. Overall, these genetic and biochemical data implicated XR TLR7 deficiency due to deleterious variants in at least 1% of critical cases of COVID-19 in male patients under the age of 60 years, with high penetrance.
Deficiency of plasmacytoid dendritic cells
TLR7-deficient pDCs did not respond to the TLR7-specific agonists tested. Moreover, when challenged with SARS-CoV-2 in vitro, they displayed severely impaired, but not entirely absent type I IFN induction[41]. TLR9 is probably responsible for the residual response, as UNC-93B- and IRAK4-deficient pDCs do not respond at all to the virus[48] (Figure 1). The discovery of XR TLR7 deficiency through an unbiased approach thus confirmed the key role of type I IFN immunity in protection against SARS-CoV-2 in the respiratory tract[41]. It also suggested that pDCs are essential for this process. It has long been known that pDCs are the most potent discernible type I IFN-producing cell type[34, 49–51]; this experiment of nature suggests that these cells are essential for antiviral immunity, as the other TLR7-expressing myeloid and lymphoid cells are poor producers of type I IFNs[52]. Human TLR7 is now firmly established as a key player in host defense. The activation of TLR7 by viral RNA was long known[53–57], with its gene shown to be subject to strong negative selection in the general population[58], but its role in host defense had remained elusive, as patients with deficiencies of MYD88 or IRAK4 displayed no severe viral illnesses and the viral infections observed in UNC-93B-deficient patients had been attributed to their TLR3 pathway defects[59]. Overall, TLR3-dependent type I immunity in RECs and TLR7-dependent type I IFN immunity in pDCs appear to be strong determinants of protection against SARS-CoV-2 in the respiratory tract.
Other inborn errors of type I IFN immunity
Nine IEIs of type I IFN immunity were thus found to underlie life-threatening COVID-19 with low (AD disorders) or high (AR, XR) penetrance. In addition, five young patients with related IEIs — MYD88[60], IRAK4[61], and GATA2 deficiencies[62, 63] — were hospitalized, for COVID-19 pneumonia, albeit of moderate severity. Severe influenza infections had been reported in patients with GATA2 deficiency, probably caused at least partly by low counts of circulating pDCs[64], which do not require TLR7 to sense influenza virus[30, 48]. Other patients with MYD88, IRAK4, or GATA2 deficiency are probably susceptible to hypoxemic COVID-19 pneumonia[48]. Defects of other genes involved in type I IFN immunity may also increase susceptibility to COVID-19 (Figure 1). Overall, the nine IEIs of type I IFN immunity identified may already account for about 1–5% of life-threatening cases of COVID-19, especially among patients under 60 years old, with XR TLR7 deficiency alone accounting for over 1% of critical cases in men. This proportion is high, exceeding the 1% of cases of tuberculosis in Europeans for which a genetic explanation has been obtained, for example[19, 20]. Other causal IEIs affecting type I IFN will probably be discovered in the future. Indeed, AR IFNAR1 and IRF7 deficiencies have already acted like a compass, pointing us in the right direction for the discovery of a more common cause of life-threatening COVID-19.
From inborn errors to their phenocopy
Auto-Abs against type I IFNs were first detected in the 1980s, in patients treated with type I IFN or with systemic lupus erythematosus (SLE)[65–67]. Their production can be genetically driven, as in patients with autoimmune polyendocrine syndrome type-1 (APS-1) due to germline mutations of AIRE, which controls the thymic expression of peripheral self-antigens and, thus, central T-cell tolerance[68–70]. They are also found in men with immunodysregulation polyendocrinopathy enteropathy X-linked (IPEX) due to mutations of FOXP3, encoding a protein that governs the development of regulatory T cells and thus, peripheral T-cell tolerance[71, 72], and in patients with combined T/B cell immunodeficiency due to hypomorphic mutations of RAG1 or RAG2[73]. Auto-Abs against type I IFN may also be produced in two overlapping conditions[74] of elusive etiology: thymoma[75] and myasthenia gravis[76, 77]. Patients with APS-1 and thymoma have thymic epithelial-intrinsic defects, whereas patients with RAG1/RAG2 and FOXP3 mutations have T cell-intrinsic defects[70, 78, 79]. These auto-Abs have been widely recognized for 40 years, and were even reported in an otherwise healthy patients with severe varicella zoster virus (VZV) infection by Ion Gresser as early as 1984[80], but they were not thought to confer a predisposition to viral diseases. By contrast, autoimmune phenocopies of IEIs disrupting type II IFN (IFN-γ), IL-6, IL-17A/F, and granulocyte-macrophage colony-stimulating factor (GM-CSF), have long been known to underlie mycobacterial disease, staphylococcal disease, mucocutaneous candidiasis, and nocardiosis, respectively[18, 81–88].
Autoantibodies neutralizing type I IFNs
We found that at least 10% of individuals with critical COVID-19 had auto-Abs neutralizing supraphysiological concentrations (10 ng/mL, in plasma diluted 1/10) of IFN-α2 and/or IFN-ω[6]. These findings were widely replicated[89–102]. In our and another study, these auto-Abs were not found in patients with silent or benign SARS-CoV-2 infections[6, 92]. Alarmingly, auto-Abs neutralizing type I IFN were found in therapeutic convalescent plasma from a few patients hospitalized for COVID-19[99]. In the few patients tested, the auto-Abs pre-existed SARS-CoV-2 infection. Moreover, APS-1 patients, who produce such auto-Abs from early childhood, were at very high risk of developing severe or critical COVID-19 pneumonia, especially in patients over 20 years old[103, 104]. An elegant unbiased study reported that a number of patients with hypoxemic COVID-19 pneumonia displayed diverse auto-Abs[92], most of which were probably triggered by SARS-CoV-2 infection and may have influenced the course of disease. This and a longitudinal study of a small group of patients suggested that SARS-CoV-2 infection might boost the levels of pre-existing type I IFN auto-Abs[105]. The auto-Abs blocked the protective effect of IFN-α2 against SARS-CoV-2 in vitro[6]. Furthermore, circulating IFN-α concentrations were low or undetectable in vivo in patients with auto-Abs against IFN-α2, which also target the 13 forms of IFN-α[6]. These auto-Abs also impair type I IFN activity in peripheral blood mononuclear cells[93]. Impaired expression of IFN-stimulated genes (ISGs) was also observed in the respiratory tract in patients with auto-Abs[96, 106] (Figure 2). Indeed, these auto-Abs were also detected in tracheal aspirates and nasal swabs[106, 107].
Figure 2. Inborn errors of type I IFN immunity and autoantibodies neutralizing type I IFNs underlie life-threatening COVID-19 pneumonia by facilitating the spread of the virus during the first few days of infection, triggering secondary leukocytic inflammation.
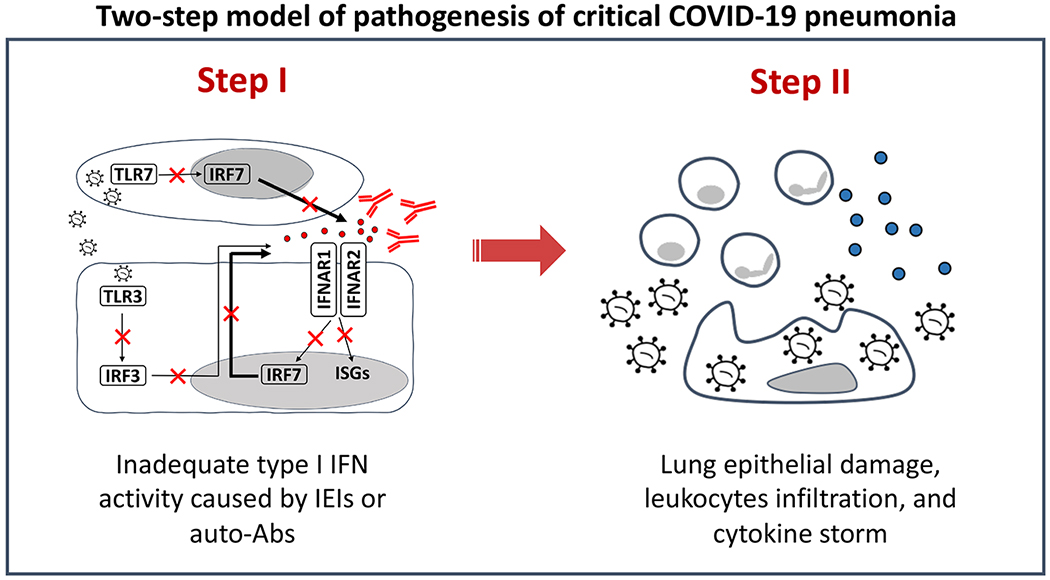
In a two-step model of pathogenesis of critical COVID-19[12], inadequate type I IFN immunity during the first few hours and days of infection results in the spread of the virus to the lungs, blood, and beyond. This results, one to two weeks later, in pulmonary and systemic hyperinflammation, largely due to the recruitment and activation of leukocytes, which produce excessive amounts of cytokines in a last-ditch attempt to eradicate the virus that should have been eradicated by type I IFN but was not. The two-step model suggests that early administration of type I IFN at the onset of SARS-CoV-2 infection, in ambulatory patients, or even before infection in exposed individuals at risk of severe disease, may halt disease progression in patients without auto-Abs to the corresponding type I IFN and without IEIs downstream from type I IFN receptors. IFN: interferon; IEI: inborn errors of immunity; Auto-Ab: autoantibody, ISGs: interferon-stimulated genes.
Neutralization of lower concentrations
The physiological concentrations of IFN-α in the blood during SARS-CoV-2 infection are much lower (between 1 and 100 pg/mL in undiluted plasma)[108] than the concentrations used in our initial experiments (10 ng/mL in plasma diluted 1/10). We found that ~14% of patients with critical COVID-19 pneumonia had auto-Abs neutralizing lower, more physiological, concentrations of IFN-α and/or IFN-ω (100 pg/mL in plasma diluted 1/10)[109]. The proportion of such patients increased after the age of 65 years and was greater in men than in women. In addition, another ~1% of patients had auto-Abs neutralizing 10 ng/mL IFN-β only. Globally, ~20% of patients with critical COVID-19 over 80 years of age, and ~20% of deceased patients across all ages, had these auto-Abs. Moreover, ~7% of patients with severe, but not critical, COVID-19 had these auto-Abs, too. We estimated ORs by comparing the prevalence of auto-Abs in patients with critical disease with that in patients with asymptomatic or mild infection[109] (Table 1). For most categories of auto-Abs to type I IFN, their prevalence was not null in patients with silent or mild infection, as previously documented for patients with APS-1[103, 104]. The highest ORs were obtained for auto-Abs neutralizing both IFN-α and IFN-ω at concentrations of 10 ng/mL or 100 pg/mL, followed by auto-Abs against IFN-α only, whereas the ORs for auto-Abs against IFN-ω only were lower. For auto-Abs against IFN-β only, the ORs for critical disease were even lower. Remarkably, however, auto-Abs neutralizing only IFN-β can underlie life-threatening COVID-19, as can auto-Abs against IFN-α only or IFN-ω only[6, 109].
Autoantibodies in the general population
We tested more than 34,000 individuals from the general population aged 18 to 100 years. We found that the prevalence of auto-Abs neutralizing 10 ng/mL (or 100 pg/mL) IFN-α or IFN-ω was not only higher in men than in women, but also increased significantly with age in the general population, with 0.17% (1.1%) of individuals positive for these antibodies before the age of 70 years, and more than 1.4% (4.4%) positive after the age of 70 years[109]. This striking distribution probably contributes to the higher risk of death from COVID-19 in the elderly population. Interestingly, auto-Abs neutralizing IFN-α and/or IFN-ω are much more prevalent in the elderly population, whereas auto-Abs neutralizing IFN-β seem to have a similar prevalence in all age groups tested. IFN-ω and the 13 forms of IFN-α are very similar biochemically, closely related phylogenetically, and found in the blood, whereas IFN-β, IFN-ε, and IFN-κ differ structurally and functionally. IFN-β is widely required to initiate the production of other type I IFNs, whereas IFN-ε and IFN-κ are predominantly expressed in reproductive and cutaneous tissues (and not tested in our studies of auto-Abs)[110–112]. Defective activity for all 13 IFN-α, or IFN-ω, or IFN-β, or a combination of these molecules may remain silent for long periods, until a virus, such as SARS-CoV-2, reveals the deficiency[112–114]. Overall, auto-Abs to type I IFNs appear to be strong determinants of critical COVID-19 pneumonia.
Clinical implications
Auto-Abs neutralizing type I IFNs apparently underlie already almost one million deaths from COVID-19 worldwide (15-20%). These studies thus have clinical implications, because (i) it is straightforward to test for these neutralizing auto-Abs before infection, (ii) individuals with these antibodies should be vaccinated early and given priority for booster injections, (iii) it is also possible to test for these antibodies during the early stages of COVID-19, (iv) specific treatments, such as IFN-β, mAbs neutralizing SARS-CoV-2, or plasma exchange could then be considered and tested in unvaccinated, and perhaps even in vaccinated individuals[115, 116]. Finally, these auto-Abs against type I IFNs also underlie severe adverse reactions to vaccination with the live attenuated virus vaccine against yellow fever and perhaps other viral infections[80, 117, 118]. Together with IEIs of type I IFN immunity, these findings may explain the pathogenesis of about 15-20% of cases of critical COVID-19 pneumonia, especially in patients over 70 years old (Table 1, Figure 3). We know from IPEX[71], RAG1/2 deficiencies[73], incontinentia pigmenti[6, 119], and APS-1[103–105, 120, 121] that some IEIs can underlie the production of auto-Abs against type I IFNs. It will be interesting to determine whether other IEIs also underlie the production of auto-Abs against type I IFN[63, 122–124]. It will also be interesting to elucidate the reasons for the sudden increase in these auto-Abs after 65 years of age, especially in men.
Figure 3. Inborn errors of type I IFN immunity and autoantibodies neutralizing type I IFNs underlie life-threatening COVID-19 pneumonia by aggravating the natural age-dependent decline of type I IFN immunity in the mucosae and blood.
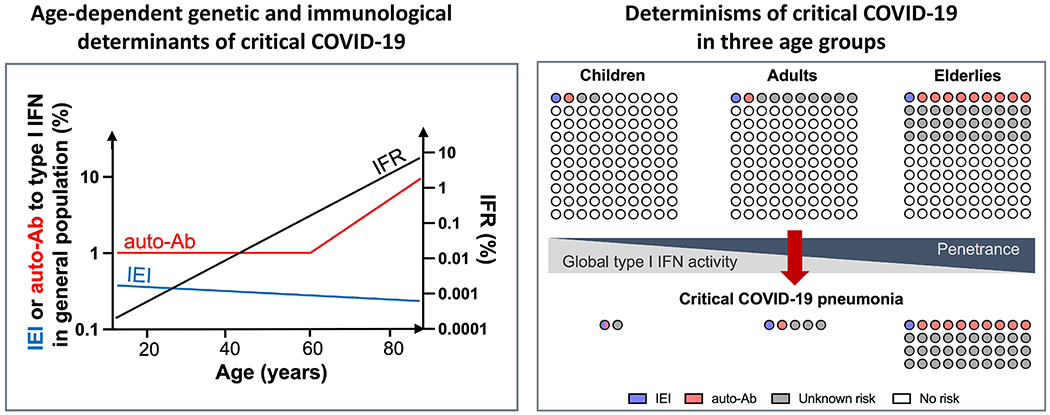
Inborn errors of type I IFN immunity conferring predisposition to critical COVID-19 pneumonia are represented in slightly declining proportion across age groups in the general population, as they may underlie critical influenza and related life-threatening viral illnesses. In contrast the frequency of auto-Abs against type I IFN increases exponentially after the age of 65 years (y axis on the left), attesting to a breakdown of tolerance in the elderly population. Global type I IFN immunity in the respiratory tract mucosae (RECs) and in the blood (pDCs) is shown to decline with age, under the influence of aging and environmental triggers[190, 191]. This decline in global type I IFN immunity over time may increase the risk of life-threatening COVID-19 (referred to as penetrance, for both IEI and autoantibodies) associated with genetic and immunological etiologies in elderly patients. All three risk factors — IEIs, auto-Abs, and tonic levels of type I IFNs — may contribute to critical COVID-19 pneumonia (right panel). IEIs and auto-Abs appear to affect different patients, while the gradual decrease in tonic levels of type I IFNs can aggravate the consequences of both IEIs and auto-Ab. Overall, the cohort of patients with life-threatening COVID-19 is enriched with IEI in young patients and with auto-Abs in elderly patients. IEI: inborn errors of immunity; IFR: infection-fatality ratio; auto-Ab: autoantibody.
Type I IFNs in unexplained COVID-19
Before the discovery that type I IFN deficiency may underlie critical COVID-19 in some patients, some observations suggested that type I IFN levels in the blood of a subset of patients with critical COVID-19 pneumonia were lower than for other forms of infection[108, 125–127]. By contrast, other studies reported enhanced type I IFN activity in a subset of patients with critical COVID-19[128–130]. Studies on patients with no known determinant of critical disease are, by nature, inconclusive. At best, the abnormalities detected can be correlated with disease severity, but it remains unclear whether they are a cause or consequence of disease. In the infinite and multidimensional matrix of causes and consequences, involving countless viruses and cell types, in individual patients, each of whom is unique, from the first day of infection to the death of the patient or viral clearance, it is difficult to establish a causal relationship. This has always been a fundamental problem in the field of infectious diseases, and in medicine at large, and has resulted in observational studies in humans gradually being replaced by experimental studies of cells in vitro and of animals in vivo, and, more recently, by the study of the human genetic determinants of infectious diseases[17, 18, 24]. The discovery of genetic lesions or pre-existing auto-Abs has provided an anchor on which observations of COVID-19 or other infections can be fixed to establish causality.
Type I IFN biology in patients with deficiencies
Only one patient with a type I IFN IEI, AR IRF9 deficiency, has been studied immunologically, early in the course of infection[131]. The impact of auto-Abs on systemic and/or mucosal immunity has been studied by scRNAseq in more patients[93, 96]. These studies showed that critically ill patients had weaker ISG responses in myeloid cells, this lack of responsiveness being particularly marked in patients with auto-Abs against type I IFN[93]. Consistently, scRNAseq on nasopharyngeal swabs showed that patients with critical COVID-19, including one patient with auto-Abs against type I IFNs, had muted ISG responses[96]. Finally, auto-Abs against type I IFN have been detected in nasal fluids, and nasal ISG responses have been shown to be correlated with nasal viral load, systematic ISG responses in leukocytes, and blood type I IFNα levels[106]. The patients with auto-Abs against type I IFN and critical COVID-19 tested also displayed increases in the levels of inflammatory cytokines in both the respiratory tract and the blood, suggesting a two-step-model for the pathogenesis of critical COVID-19, with insufficient type I IFN in the first few days of infection unleashing excessive inflammation from the second week onward[12]. Overall, these extensive studies have suggested that patients with critical COVID-19 and auto-Abs against type I IFN have insufficient systemic and nasal type I IFN activity early in the course of disease (Figure 2).
Other inborn errors of immunity
What have we learned from the study of patients with IEIs that do not impair type I IFN immunity directly or via the production of auto-Abs? In 10 retrospective cohorts of patients with various IEIs, the natural history of SARS-CoV-2 infection seemed to resemble that in the general population, albeit apparently with higher mortality in some IEI subsets[61, 63, 123, 124, 132–137]. A prospective study of IEI patients reached similar conclusions[60]. Interestingly, patients with predominant antibody deficiencies are not prone to life-threatening COVID-19 pneumonia[61, 63, 123, 124, 132–137]. This is consistent with the findings for critical influenza pneumonia, which is specifically seen in patients with IEIs of type I IFN immunity, but not in other individuals, even those lacking T and/or B cells[64]. Patients with IEIs of T and/or B cells may suffer from chronic COVID-19 infection and prolonged viral shedding[138–141], like patients with acquired adaptive immunodeficiencies[142–144]. Multi-mutated, potentially more pathogenic SARS-CoV-2 variants might arise in such cases of persistent infection[138]. No IEIs other than those impairing type I IFN immunity directly or via auto-Abs have been genetically or mechanistically associated with life-threatening COVID-19, but their vast genetic and immunological heterogeneity, and their individual rarity suggest that targeted clinical surveys are warranted. In particular, type I and III IFNs both activate ISGF-3 and induce a largely overlapping range of ISGs[64, 112] (Figure 1). It would be interesting to study the course of SARS-CoV-2 infection in patients with AR IL-10RB deficiency, whose cells respond to type I but not type III IFNs (Figure 1).
Genome-wide association studies
The key result of genome-wide association studies (GWAS) is the identification of common variants of chromosomal region 3p21.31 associated with critical COVID-19[145–148]. The risk haplotype, inherited from Neanderthals, confers an estimated OR per copy of between 1.6 and 2.1, with higher values for individuals under 60 years old[148–150]. The region encompasses six genes, including CXCR6 and LZTFL1. Five other genome-wide regions have been shown to be significantly associated with critical COVID-19[147]. Three of these regions encompass genes involved in type I IFN immunity. The first, on chr12q24.13, containing protective variants inherited from Neanderthals, includes the OAS1, OAS2, and OAS3 cluster, ISGs required for the activation of anti-viral RNaseL[151]. The second, a region on chr21q22.1, includes IFNAR2. The third, a region on chr19p13.2, includes TYK2. In these regions, one copy of the risk allele increases the risk of critical COVID-19 slightly, with ORs below 1.5. An OR of 1.5 is often presented as increasing the risk by “50%”, but, assuming that the OR does not overestimate the relative risk, the mathematical and clinical reality is that, for a COVID-19 mortality risk of 0.006% at the age of 20 years, 0.2% at the age of 50 years, and 8.3% at the age of 80 years[1], individuals carrying the at-risk genotype have risks of 0.009%, 0.3%, and 12.5%, respectively. Although modest at the individual level, the impact of these findings is significant at the population level (Table 1)[152]. These studies may not only reveal genetic modifiers of stronger determinants of disease, but also mechanisms that are type I IFN-dependent or -independent.
Genome-wide search for rare variants
In a population-based exome-wide association study[47] using a relaxed Bonferroni threshold (p<5x10−8), the authors identified eight genes, one of which, TLR7, displayed an enrichment in pLOF and in-frame variants with a MAF < 10−5 in critically ill COVID-19 patients relative to individuals of unknown or seronegative status for SARS-CoV-2 infection. By contrast, this study and a previous rare-variant candidate gene association study[153] reported no enrichment in pLOF variants of 13 type I IFN-related influenza susceptibility genes[5] in patients with critical COVID-19 pneumonia. Two possible reasons for this apparent discrepancy are of particular importance[154]. First, age, the key epidemiological factor driving COVID-19 severity was ignored. Our cohort was much younger (mean age of 52 vs. 66 years) and these IEIs are more frequent in patients under the age of 60 years[154]. Second, no tests were performed for auto-Abs against type I IFN, the most common known determinant of critical COVID-19, especially in patients over 60 years old[154]. More importantly, the proportions of patients with critical COVID-19 due to AR, XR, and AD IEIs at these (or other) loci may vary from population to population. Finally, their causal link to critical COVID-19 cannot be concluded or excluded from an enrichment analysis of untested variants: it should be based on biochemical, virological, and immunological experiments mechanistically connecting germline genotypes with clinical phenotypes[5, 40–42].
SARS-CoV-2 interference with type I IFN
The discovery that insufficient type I IFN can underlie critical COVID-19 pneumonia in vivo is remarkably convergent with various elegant virological studies conducted in human cells in vitro. Indeed, SARS-CoV-2 induces type I IFN production less strongly than seasonal influenza A viruses (IAV)[155] or Sendai virus (SeV)[156]. The ability of SARS-CoV-2 to evade type I IFN induction results not only from the non-specific inhibition of host cellular functions, such as transcription and translation[157–159], but also from the specific suppression of type I IFN induction pathways. Despite the limitations of overexpression systems, numerous studies have shown that at least 14 of the 31 products of known open reading frames (ORFs) of SARS-CoV-2 (Nsp1, Nsp5, Nsp6, Nsp13, Nsp14, Nsp15, ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF9b, M, and N) target host proteins governing type I IFN induction, including IRF3, TBK1, MAVS, RIG-I, and NEMO, or self-amplification, including IFNAR1, STAT1, STAT2, and TYK2[160–168]. Moreover, an Nsp1 mutation (ΔD500–532) frequent in viral variants is associated with even lower levels of type I IFN production[169]. It remains to be tested whether the ability of SARS-CoV-2 to resist type I IFN is also increasing in emerging variants, such as B.1, B.1.1.7 (alpha), B.1.1351 (beta), B.1.617.2 (delta), and B.1.1.529 (omicron). Current findings suggest that being able to evade type I IFN immunity is essential for viral fitness[160, 170].
Viral and human fitness depend on type I IFNs
Remarkably, three targets of the virus, IFNAR1[167], IRF3[164, 168], and TBK1[165], are encoded by COVID-19 susceptibility genes (Figure 1). We expect a greater convergence of viral targets and susceptibility genes to emerge with the genetic testing of viral targets in vivo, and the virological testing of susceptibility genes in vitro[158, 159, 171–179]. Suppression of the type I IFN response is essential for viral fitness, whereas the maintenance of type I IFN immunity is essential for human fitness. The type I IFN-blocking proteins of SARS-CoV-2 make the small amounts of type I IFN produced by infected cells in individual patients even more consequential, as attested by the catastrophic outcome of genetic or autoimmune deficiencies of type I IFN in vivo. Any further decrease in type I IFN levels due to the selection of new viral variants would tip the balance further in favor of the virus. Encouragingly, despite the ability of SARS-CoV-2 and its variants to evade type I IFN induction, these viral variants remain highly sensitive to type I IFN pretreatment in vitro[161, 180]. However, the immense numbers of viral variants worldwide raise concerns about the emergence of new variants capable of impairing type I IFN immunity to an even greater extent.
Concluding remarks
IEIs of type I IFN immunity, and pre-existing auto-Abs neutralizing type I IFNs appear to be strong determinants of critical COVID-19 pneumonia in about 15-20% of patients. This is unprecedented among common infectious diseases, this proportion being much higher than the next best example, the possible explanation of tuberculosis in only 1% of European cases[19, 20]. As these findings are consistent with those of in vitro virological studies and in vivo animal models[156, 181–187], they may reflect a general mechanism of disease. Individuals with insufficient type I IFN in the respiratory epithelium, whatever the underlying determinants, may be unable to prevent the spread of the virus to the lungs, blood, and other organs during the first few days of infection. Inflammation may then develop when activated leukocytes, including myeloid and lymphoid cells of an innate or adaptive nature are attracted to the site of infection and attempt to resolve the pulmonary and systemic infection that became established because of the lack of control by type I IFN (Figure 2)[10, 24, 188]. Understandably, at such a late inflammatory stage, therapeutic type I IFN did not help hospitalized patients[189]; clinical trials of early administration in ambulatory patients are ongoing[115]. The penetrance of known IEIs of type I IFN immunity and of auto-Abs varies, with a higher penetrance for AR and XR than for AD disorders, and for auto-Abs neutralizing high concentrations of most type I IFNs relative to those neutralizing low concentrations of a single type I IFN (Table 1). Penetrance may be influenced by the size of the viral inoculum, by prior infection with other viruses that trigger type I IFN, especially in children[190], or by human determinants, such as the age-dependent decline of pDCs[163, 191–194] and local respiratory type I IFN activity[36, 195], or common genetic variants, including those discovered by GWAS [145–147] (Figure 3).
What underlies critical COVID-19 pneumonia in the remaining 80% of cases? It would not be surprising to discover other IEIs of type I IFN immunity, including some affecting genes encoding proteins acting upstream or downstream from type I IFNs. These findings would further clarify the pathogenesis of critical COVID-19, while revealing the corresponding redundancy of these loci against other viral infections. The considerable redundancy of type I IFN in host defense against viruses is already a major surprise. Indeed, most patients with critical COVID-19 pneumonia due to an IEI or auto-Ab production had never before been hospitalized for another severe viral illness, including patients with AR (IRF7, IFNAR1) or XR (TLR7) inborn errors of type I IFN immunity. These findings suggest that there are type I IFN-independent mechanisms of cell-intrinsic immunity providing protection against a wide range of viruses[16]. Another important question is whether adaptive immunity to the vaccine can compensate for a constitutive deficiency of type I IFN. Encouragingly, mAbs neutralizing SARS-CoV-2 protected an unvaccinated but infected child with inherited IRF9 deficiency[131]. Despite their current success, it is unclear whether vaccines will remain effective in the long term and against new viral variants[196–199]. The recent spread of the omicron variant, which is not only more contagious, but also whose protein S is structurally distant from that encoded by existing vaccines, is particularly worrisome. Even prior to the emergence of omicron, an alarming increase has been reported in the number of breakthrough cases, defined as infection in fully vaccinated individuals, including cases of hypoxemic pneumonia and even death. It is tempting to hypothesize that some IEIs or auto-Abs against type I IFN may underlie some life-threatening breakthrough cases. The search for human genetic and immunological determinants of life-threatening COVID-19 pneumonia must now encompass not only various viral variants, but also both unvaccinated and vaccinated patients.
Acknowledgments
The Laboratory of Human Genetics of Infectious Diseases is supported by the Howard Hughes Medical Institute, the Rockefeller University, the St. Giles Foundation, the National Institutes of Health (NIH) (R01AI088364 and R01AI163029), the National Center for Advancing Translational Sciences (NCATS), NIH Clinical and Translational Science Award (CTSA) program (UL1 TR001866), a Fast Grant from Emergent Ventures, Mercatus Center at George Mason University, the Yale Center for Mendelian Genomics and the GSP Coordinating Center funded by the National Human Genome Research Institute (NHGRI) (UM1HG006504 and U24HG008956), the Yale High Performance Computing Center (S10OD018521), the Fisher Center for Alzheimer’s Research Foundation, the Meyer Foundation, the JPB Foundation, the French National Research Agency (ANR) under the “Investments for the Future” program (ANR-10-IAHU-01), the Integrative Biology of Emerging Infectious Diseases Laboratory of Excellence (ANR-10-LABX-62-IBEID), the French Foundation for Medical Research (FRM) (EQU201903007798), the FRM and ANR GENCOVID project (ANR-20-COVI-0003), ANRS Nord-Sud (ANRS-COV05), ANR grants GENVIR (ANR-20-CE93-003), ANR AABIFNCOV (ANR-20-CO11-0001), and ANR MIS-C (ANR 21-COVR-0039, GenMIS-C) projects, the European Union’s Horizon 2020 research and innovation program under grant agreement no. 824110 (EASI-Genomics), the Square Foundation, Grandir - Fonds de solidarité pour l’enfance, the SCOR Corporate Foundation for Science, Fondation du Souffle, Institut National de la Santé et de la Recherche Médicale (INSERM), REACTing-INSERM, and the University of Paris. P.B. was supported by the FRM (EA20170638020) and the MD-PhD program of the Imagine Institute (with the support of the Fondation Bettencourt Schueller). Giuseppe Novelli is supported by Regione Lazio (Research Group Projects 2020) No. A0375-2020-36663, GecoBiomark. Helen Su and Luigi Notarangelo are supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Appendix: List of CHGE authors
Adem Karbuz6, Adrian Gervais7, Ahmad Abou Tayoun8, Alessandro Aiuti9, Alexandre Belot10, Alexandre Bolze11, Alexandre Gaudet12, Anastasiia Bondarenko13, András N. Spaan14, Andrea Guennoun15, Andres Augusto Arias16, Anna M. Planas17, Anna Sediva18, Anna Shcherbina19, Anna-Lena Neehus7, Anne Puel20, Antoine Froidure21, Antonio Novelli22, Aslınur Özkaya Parlakay23, Aurora Pujol24, Aysun Yahşi25, Belgin Gülhan25, Benedetta Bigio26, Bertrand Boisson20, Beth A. Drolet27, Carlos Andres Arango Franco28, Carlos Flores29, Carlos Rodríguez-Gallego30, Carolina Prando31, Catherine M. Biggs32, Charles-Edouard Luyt33, Clifton L. Dalgard34, Cliona O’Farrelly35, Daniela Matuozzo7, David Dalmau36, David S. Perlin37, Davood Mansouri38, Diederik van de Beek39, Donald C. Vinh40, Elena Dominguez-Garrido41, Elena W.Y. Hsieh42, Emine Hafize Erdeniz43, Emmanuelle Jouanguy20, Esra Şevketoglu44, Estelle Talouarn7, Eugenia Quiros-Roldan45, Evangelos Andreakos46, Eystein Husebye47, Fahad Alsohime48, Filomeen Haerynck49, Giorgio Casari50, Giuseppe Novelli51, Gökhan Aytekin52, Guillaume Morelle53, Gulsum Alkan54, Gulsum Iclal Bayhan23, Hagit Baris Feldman55, Helen C. Su56, Horst von Bernuth57, Igor Resnick58, Ingrid Bustos59, Isabelle Meyts60, Isabelle Migeotte61, Ivan Tancevski62, Jacinta Bustamante20, Jacques Fellay63, Jamila El Baghdadi64, Javier Martinez-Picado65, Jean-Laurent Casanova66, Jeremie Rosain7, Jeremy Manry7, Jie Chen26, John Christodoulou67, Jonathan Bohlen7, José Luis Franco68, Juan Li26, Juan Manuel Anaya69, Julian Rojas70, Junqiang Ye71, K M Furkan Uddin72, Kadriye Kart Yasar73, Kai Kisand74, Keisuke Okamoto75, Khalil Chaïbi76, Kristina Mironska77, László Maródi78, Laurent Abel7, Laurent Renia79, Lazaro Lorenzo7, Lennart Hammarström80, Lisa F.P. Ng79, Lluis Quintana-Murci81, Lucia Victoria Erazo28, Luigi D. Notarangelo56, Luis Felipe Reyes82, Luis M Allende83, Luisa Imberti84, Majistor Raj Luxman Maglorius Renkilaraj7, Marcela Moncada-Velez26, Marie Materna7, Mark S. Anderson85, Marta Gut86, Marwa Chbihi7, Masato Ogishi26, Melike Emiroglu54, Mikko R.J. Seppänen87, Mohammed J. Uddin88, Mohammed Shahrooei89, Natalie Alexander26, Nevin Hatipoglu90, Nico Marr15, Nihal Akçay44, Oksana Boyarchuk91, Ondrej Slaby92, Ozge Metin Akcan93, Peng Zhang26, Pere Soler-Palacín94, Peter K. Gregersen95, Petter Brodin96, Pierre Garçon97, Pierre-Emmanuel Morange98, Qiang Pan-Hammarström80, Qinhua Zhou99, Quentin Philippot7, Rabih Halwani100, Rebeca Perez de Diego101, Romain Levy7, Rui Yang26, Şadiye Kübra Tüter Öz54, Saleh Al Muhsen48, Saliha Kanık-Yüksek25, Sara Espinosa-Padilla102, Sathishkumar Ramaswamy8, Satoshi Okada103, Sefika Elmas Bozdemir104, Selma Erol Aytekin93, Şemsi Nur Karabela73, Sevgi Keles105, Sevtap Senoglu73, Shen-Ying Zhang20, Sotirija Duvlis106, Stefan N. Constantinescu107, Stephanie Boisson-Dupuis20, Stuart E. Turvey108, Stuart G. Tangye109, Takaki Asano26, Tayfun Ozcelik110, Tom Le Voyer7, Tom Maniatis71, Tomohiro Morio111, Trine H. Mogensen112, Vanessa Sancho-Shimizu113, Vivien Beziat20, Xavier Solanich114, Yenan Bryceson115, Yu-Lung Lau116, Yuval Itan117
6. Cemil Taşcıoglu City Hospital, Istanbul, Turkey.
7. Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM U1163, Necker Hospital for Sick Children, Paris, France; University of Paris, Imagine Institute, Paris, France.
8. Al Jalila Children’s Hospital, Dubai, UAE.
9. San Raffaele Telethon Institute for Gene Therapy, IRCCS Ospedale San Raffaele, and Vita Salute San Raffaele University, Milan, Italy.
10. Hospices Civils de Lyon, Lyon, France.
11. Helix, San Mateo, CA, USA.
12. CHU Lille, Department of Intensive Care Medicine, Critical Care Center; Univ. Lille, CNRS, Inserm, CHU Lille, Institut Pasteur de Lille, U1019-UMR9017-CIIL-Centre d’Infection et d’Immunité de Lille, F-59000, Lille, France
13. Shupyk National Medical Academy for Postgraduate Education, Kiev, Ukraine.
14. St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY, USA. ;Department of Medical Microbiology, University Medical Center Utrecht, Utrecht, Netherlands
15. Sidra Medicine, Doha, Qatar.
16. St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY, USA; Primary Immunodeficiencies Group, Department of Microbiology and Parasitology, School of Medicine, University of Antioquia, Medellín, Colombia; School of Microbiology, University of Antioquia UdeA, Medellín, Colombia
17. IIBB-CSIC, IDIBAPS, Barcelona, Spain.
18. Department of Immunology, Second Faculty of Medicine Charles University, V Uvalu, University Hospital in Motol, Prague, Czech Republic.
19. Department of Immunology, Dmitry Rogachev National Medical Research Center of Pediatric Hematology, Oncology and Immunology, Moscow, Russia.
20. St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY, USA; Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM U1163, Necker Hospital for Sick Children, Paris, France; University of Paris, Imagine Institute, Paris, France
21. Pulmonology Department, Cliniques Universitaires Saint-Luc ; Institut de Recherche Expérimentale et Clinique (IREC), Université Catholique de Louvain, Brussels, Belgium.
22. Laboratory of Medical Genetics, IRCCS Bambino Gesù Children’s Hospital, Rome, Italy.
23. Yildirim Beyazit University, Ankara City Hospital, Ankara, Turkey.
24. Neurometabolic Diseases Laboratory, Bellvitge Biomedical Research Institute (IDIBELL), L’Hospitalet de Llobregat, Barcelona, Spain; Catalan Institution of Research and Advanced Studies (ICREA), Barcelona, Spain; Center for Biomedical Research on Rare Diseases (CIBERER), ISCIII, Barcelona, Spain.
25. Ankara City Hospital, Ankara, Turkey.
26. St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY, USA
27. School of Medicine and Public Health, University of Wisconsin, Madison, WI, USA.
28. Laboratory of Human Genetics of Infectious Diseases, Necker Branch, INSERM U1163, Necker Hospital for Sick Children, Paris, France; Grupo de Inmunodeficiencias Primarias, Departamento de Microbiología y Parasitología, Facultad de Medicina, Universidad de Antioquia UDEA, Medellín, Colombia.
29. Research Unit, Hospital Universitario Nuestra Señora de Candelaria, Santa Cruz de Tenerife; CIBER de Enfermedades Respiratorias, Instituto de Salud Carlos III, Madrid; Genomics Division, Instituto Tecnológico y de Energías. Renovables (ITER), Santa Cruz de Tenerife, Spain
30. Department of Immunology, University Hospital of Gran Canaria Dr. Negrín, Canarian Health System, Las Palmas de Gran Canaria; Department of Clinical Sciences, University Fernando Pessoa Canarias, Las Palmas de Gran Canaria, Spain
31. Faculdades Pequeno Príncipe, Instituto de Pesquisa Pelé Pequeno Príncipe, Curitiba, Brazil.
32. Department of Pediatrics, BC Children’s and St. Paul’s Hospitals, University of British Columbia, Vancouver, BC, Canada
33. Groupe Hospitalier Pitié-Salpêtrière, Paris, France.
34. Department of Anatomy, Physiology & Genetics, Uniformed Services University of the Health Sciences, Bethesda, MD, USA.
35. Comparative Immunology Group, School of Biochemistry and Immunology, Trinity Biomedical Sciences Institute, Trinity College Dublin, Ireland.
36. Hospital Universitari Mútua Terrassa; Fundació Docència i Recerca Mutua Terrassa, Terrasa; Universitat de Barcelona, Spain.
37. Center for Discovery and Innovation, Hackensack Meridian Health, Nutley, NJ, USA.
38. Department of Clinical Immunology and Infectious Diseases, National Research Institute of Tuberculosis and Lung Diseases, The Clinical Tuberculosis and Epidemiology Research Center, National Research Institute of Tuberculosis and Lung Diseases (NRITLD), Masih Daneshvari Hospital, Shahid Beheshti, University of Medical Sciences, Tehran, Iran.
39. Department of Neurology, Amsterdam Neuroscience, Amsterdam University Medical Center, University of Amsterdam, Amsterdam, The Netherlands.
40. Department of Medicine, Division of Infectious Diseases, McGill University Health Centre, Montréal, Québec, Canada; Infectious Disease Susceptibility Program, Research Institute, McGill University Health Centre, Montréal, Québec, Canada.
41. Fundacion Rioja Salud, Logroño - La Rioja, Spain.
42. Departments of Pediatrics, Immunology and Microbiology, University of Colorado, School of Medicine, Aurora, Colorado, USA.
43. Ondokuz Mayis University, Samsun, Turkey.
44. Pediatric Intensivist, University of Health Sciences, Bakirkoy Dr. Sadi Konuk Research and Training Hospital, Pediatric Intensive Care Unit, Istanbul, Turkey
45. Università degli Studi di Brescia, Brescia, Italy.
46. Laboratory of Immunobiology, Center for Clinical, Experimental Surgery and Translational Research, Biomedical Research Foundation of the Academy of Athens, Athens, Greece.
47. Department of Medicine, Haukeland University Hospital, Bergen, Norway.
48. Immunology research lab, Department of Pediatrics, College of Medicine, King Saud University, Riyadh, Saudi Arabia.
49. Department of Paediatric Immunology and Pulmonology, Centre for Primary Immunodeficiency Ghent (CPIG), PID Research Laboratory, Jeffrey Modell Diagnosis and Research Centre, Ghent University Hospital, Ghent, Belgium.
50. Clinical Genomics, IRCCS San Raffaele Scientific Institute and Vita-Salute San Raffaele University, Milan, Italy.
51. Department of Biomedicine and Prevention, Tor Vergata University of Rome, Rome, Italy.
52. Konya City, Hospital, Konya, Turkey.
53. CHU Kremlin-Bicêtre, Paris, France.
54. Selcuk University Faculty of Medicine, Department of Pediatrics, Division of Pediatric Infectious Diseases, Konya, Turkey.
55. The Genetics Institute, Tel Aviv Sourasky Medical Center and Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel.
56. National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA.
57. Department of Pediatric Pneumology, Immunology and Intensive Care, Charité Universitätsmedizin, Berlin University Hospital Center, Berlin, Germany; Labor Berlin GmbH, Department of Immunology, Berlin, Germany; Berlin Institutes of Health (BIH), Berlin-Brandenburg Center for Regenerative Therapies, Berlin, Germany.
58. University Hospital St. Marina, Varna, Bulgaria
59. Universidad de La Sabana, Chia, Colombia.
60. Department of Pediatrics, University Hospitals Leuven; KU Leuven, Department of Microbiology, Immunology and Transplantation; Laboratory for Inborn Errors of Immunity, KU Leuven, Leuven, Belgium.
61. Center of Human Genetics, Université Libre de Bruxelles (ULB), Bruxelles, Belgium.
62. Department of Internal Medicine II, Medical University of Innsbruck, Innsbruck, Austria.
63. School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland; Precision Medicine Unit, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland.
64. Genetics Unit, Military Hospital Mohamed V, Rabat, Morocco.
65. IrsiCaixa, IGTP, CIBERINFEC, UVic-UCC, ICREA, Barcelona, Spain.
66. The Rockefeller University & Howard Hughes Medical Institute, New York, NY, USA; Necker Hospital for Sick Children & INSERM, Paris, France.
67. Murdoch Children’s Research Institute and Department of Paediatrics, University of Melbourne, Australia.
68. Group of Primary Immunodeficiencies, University of Antioquia UDEA, Medellin, Colombia.
69. Center for Autoimmune Diseases Research (CREA), School of Medicine and Health Sciences, Universidad del Rosario, Bogota, Colombia.
70. Primary Immunodeficiencies Group, Department of Microbiology and Parasitology, School of Medicine, University of Antioquia, Medellín, Colombia.
71. Zukerman Mind Brain Behavior Institute, Columbia University, New York, NY, USA.
72. Head of Clinical Research, Centre for Precision Therapeutics, Genetics & Genomic Medicine Centre, NeuroGen Children’s Healthcare and Lecturer, Holy Family Red Crescent Medical College Dhaka-1000, Bangladesh.
73. Department of Infectious Diseases and Clinical Microbiology, Bakirkoy Dr. Sadi Konuk Training and Research Hospital, University of Health Sciences, Istanbul, Turkey
74. Molecular Pathology, Department of Biomedicine, Institute of Biomedicine and Translational Medicine, University of Tartu, Tartu Estonia.
75. Tokyo Medical and Dental University, Tokyo, Japan.
76. Centre Hospitalier Universitaire Avicenne, Bobigny, France.
77. University Clinic for Children’s Diseases, Department of Pediatric Immunology, Medical Faculty, University “ St. Cyril and Methodij” Skopje, North Macedonia
78. Primary Immunodeficiency Clinical Unit and Laboratory, Department of Dermatology, Venereology and Dermatooncology, Semmelweis University, Budapest, Hungary.
79. A*STAR Infectious Disease Labs, Agency for Science, Technology and Research, Singapore; Lee Kong Chian School of Medicine, Nanyang Technology University, Singapore.
80. Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm, Sweden.
81. Human Evolutionary Genetics Unit, CNRS U2000, Institut Pasteur, Paris, France; Human Genomics and Evolution, Collège de France, Paris, France.
82. Universidad de La Sabana, Bogotá, Colombia.
83. Hospital Universitario 12 de Octubre, Madrid, Spain.
84. ASST Spedali Civili di Brescia, Brescia, Italy.
85. Diabetes Center, University of California San Francisco, San Francisco, CA, USA.
86. Centre Nacional d’Anàlisi Genòmica (CNAG-CRG), Barcelona, Spain.
87. Adult Immunodeficiency Unit, Infectious Diseases, Inflammation Center, University of Helsinki and Helsinki University Hospital, Helsinki, Finland; Rare Diseases Center and Pediatric Research Center, Children’s Hospital, University of Helsinki and Helsinki University Hospital, Helsinki, Finland.
88. College of Medicine, Mohammed Bin Rashid University of Medicine and Health Sciences, Dubai, UAE; Cellular Intelligence (Ci) Lab, GenomeArc Inc. , Toronto, ON, Canada
89. Specialized Immunology Laboratory of Dr. Shahrooei, Ahvaz, Iran; Department of Microbiology and Immunology, Clinical and Diagnostic Immunology, KU Leuven, Leuven, Belgium
90. Pediatric Infectious Diseases Unit, Bakirkoy Dr Sadi Konuk Training and Research Hospital, University of Health Sciences, Istanbul, Turkey.
91. I. Horbachevsky Ternopil National Medical University, Ternopil, Ukraine
92. Central European Institute of Technology & Department of Biology, Faculty of Medicine, Masaryk University, Brno, Czech Republic.
93. Necmettin Erbakan University, Konya, Turkey.
94. Pediatric Infectious Diseases and Immunodeficiencies Unit, Vall d’Hebron Barcelona Hospital Campus, Barcelona, Catalonia, Spain.
95. Feinstein Institute for Medical Research, Northwell Health USA, Manhasset, NY, USA.
96. SciLifeLab, Department Of Women’s and Children’s Health, Karolinska Institutet, Stockholm, Sweden.
97. GHEF Marne-la-Vallée, Jossigny, France.
98. Assistance Publique des Hopitaux de Marseille, Marseille, France.
99. St. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY, USA; Clinical Allergy and Immunology, Children’s Hospital of Fudan University, Shanghai, China
100. Sharjah Institute of Medical Research, College of Medicine, University of Sharjah, Sharjah, United Arab Emirates.
101. Institute of Biomedical Research of IdiPAZ, University Hospital “La Paz”, Madrid, Spain.
102. Instituto Nacional de Pediatria (National Institute of Pediatrics), Mexico City, Mexico.
103. Department of Pediatrics, Graduate School of Biomedical and Health Sciences, Hiroshima University, Hiroshima, Japan.
104. Bursa City Hospital, Bursa, Turkey.
105. Necmettin Erbakan University Meram Medical Faculty, Konya, Turkey.
106. Faculty of Medical Sciences, University “Goce Delcev”, Republic of Northern Macedonia.
107. de Duve Institute and Ludwig Cancer Research, Brussels, Belgium.
108. BC Children’s Hospital, The University of British Columbia, Vancouver, Canada.
109. Garvan Institute of Medical Research, Darlinghurst, NSW, Australia; St Vincent’s Clinical School, Faculty of Medicine, UNSW Sydney, NSW, Australia.
110. Department of Molecular Biology and Genetics, Bilkent University, Bilkent - Ankara, Turkey.
111. Tokyo Medical & Dental University Hospital, Tokyo, Japan.
112. Department of Biomedicine, Aarhus University, Aarhus, Denmark.
113. Department of Paediatric Infectious Diseases and Virology, Imperial College London, London, UK; Centre for Paediatrics and Child Health, Faculty of Medicine, Imperial College London, London, UK.
114. Hospital Universitari de Bellvitge, L’Hospitalet de Llobregat, Spain.
115. Department of Medicine, Center for Hematology and Regenerative Medicine, Karolinska Institutet, Stockholm, Sweden.
116. Department of Paediatrics & Adolescent Medicine, The University of Hong Kong, Hong Kong, China.
117. Institute for Personalized Medicine, Icahn School of Medicine at Mount Sinai, New York, NY, USA; Department of Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Footnotes
Declaration and listing of any financial or non-financial competing interests
The authors declare that there is no financial or non-financial competing interests.
References
- 1.O’Driscoll M, Ribeiro Dos Santos G, Wang L, Cummings DAT, Azman AS, Paireau J, et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature 2021; 590(7844):140–145. [DOI] [PubMed] [Google Scholar]; Evidence that the mortality of COVID-19 doubles every 5 years from childhood onward, accounting for a 10,000-fold greater risk at 85 years of age (10%) than at 5 years of age (0.001%).
- 2.Sen P, Yamana TK, Kandula S, Galanti M, Shaman J. Burden and characteristics of COVID-19 in the United States during 2020. Nature 2021; 598(7880):338–341. [DOI] [PubMed] [Google Scholar]
- 3.Sah P, Fitzpatrick MC, Zimmer CF, Abdollahi E, Juden-Kelly L, Moghadas SM, et al. Asymptomatic SARS-CoV-2 infection: A systematic review and meta-analysis. Proc Natl Acad Sci U S A 2021; 118(34). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oran DP, Topol EJ. Prevalence of Asymptomatic SARS-CoV-2 Infection : A Narrative Review. Ann Intern Med 2020; 173(5):362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science (New York, NY) 2020; 370(6515). [DOI] [PMC free article] [PubMed] [Google Scholar]; Report of autosomal inborn errors of type I IFN, including autosomal dominant TLR3, and autosomal recessive IRF7 and IFNAR1 deficiencies, as human genetic and immunological determinants of life-threatening COVID-19 pneumonia.
- 6.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science (New York, NY) 2020; 370(6515). [DOI] [PMC free article] [PubMed] [Google Scholar]; Report of auto-Abs against type I IFNs as immunological determinants of life-threatening COVID-19 pneumonia, with auto-Abs neutralizing high concentrations of IFN-α and/or -ω.
- 7.Navaratnam AV, Gray WK, Day J, Wendon J, Briggs TWR. Patient factors and temporal trends associated with COVID-19 in-hospital mortality in England: an observational study using administrative data. Lancet Respiratory Medicine 2021; 9(4):397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett TD, Moffitt RA, Hajagos JG, Amor B, Anand A, Bissell MM, et al. Clinical Characterization and Prediction of Clinical Severity of SARS-CoV-2 Infection Among US Adults Using Data From the US National COVID Cohort Collaborative. JAMA Netw Open 2021; 4(7):e2116901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ricoca Peixoto V, Vieira A, Aguiar P, Sousa P, Carvalho C, Thomas D, et al. Determinants for hospitalisations, intensive care unit admission and death among 20,293 reported COVID-19 cases in Portugal, March to April 2020. Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin 2021; 26(33). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brodin P Immune determinants of COVID-19 disease presentation and severity. Nat Med 2021; 27(1):28–33. [DOI] [PubMed] [Google Scholar]; Review of the immunological underpinnings, correlates, and consequences of COVID-19, covering intrinsic, innate, and adaptive immunity.
- 11.Takahashi T, Ellingson MK, Wong P, Israelow B, Lucas C, Klein J, et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature 2020; 588(7837):315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Q, Bastard P, Bolze A, Jouanguy E, Zhang SY, Effort CHG, et al. Life-Threatening COVID-19: Defective Interferons Unleash Excessive Inflammation. Med (New York, NY) 2020; 1(1):14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]; Proposal of a two-step model for the pathogenesis of critical COVID-19 pneumonia.
- 13.Casanova JL, Su HC, Effort CHG. A Global Effort to Define the Human Genetics of Protective Immunity to SARS-CoV-2 Infection. Cell 2020; 181(6):1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 2021; 19(7):409–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telenti A, Arvin A, Corey L, Corti D, Diamond MS, Garcia-Sastre A, et al. After the pandemic: perspectives on the future trajectory of COVID-19. Nature 2021; 596(7873):495–504. [DOI] [PubMed] [Google Scholar]
- 16.Meyts I, Casanova JL. Viral infections in humans and mice with genetic deficiencies of the type I IFN response pathway. Eur J Immunol 2021; 51(5):1039–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casanova JL, Abel L. The human genetic determinism of life-threatening infectious diseases: genetic heterogeneity and physiological homogeneity? Hum Genet 2020; 139(6-7):681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]; Review of current concepts and approaches in the study of the human genetic determinants of life-threatening infectious diseases.
- 18.Casanova JL, Abel L. Lethal Infectious Diseases as Inborn Errors of Immunity: Toward a Synthesis of the Germ and Genetic Theories. Annu Rev Pathol 2021; 16:23–50. [DOI] [PMC free article] [PubMed] [Google Scholar]; Review of the history of concepts and findings in the field of human genetics of infectious diseases.
- 19.Kerner G, Ramirez-Alejo N, Seeleuthner Y, Yang R, Ogishi M, Cobat A, et al. Homozygosity for TYK2 P1104A underlies tuberculosis in about 1% of patients in a cohort of European ancestry. Proc Natl Acad Sci U S A 2019; 116(21):10430–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerner G, Laval G, Patin E, Boisson-Dupuis S, Abel L, Casanova JL, et al. Human ancient DNA analyses reveal the high burden of tuberculosis in Europeans over the last 2,000 years. Am J Hum Genet 2021; 108(3):517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang SY, Zhang Q, Casanova JL, Su HC, Team C. Severe COVID-19 in the young and healthy: monogenic inborn errors of immunity? Nature reviews Immunology 2020; 20(8):455–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stertz S, Hale BG. Interferon system deficiencies exacerbating severe pandemic virus infections. Trends Microbiol 2021; 29(11):973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carvalho T, Krammer F, Iwasaki A. The first 12 months of COVID-19: a timeline of immunological insights. Nature reviews Immunology 2021; 21(4):245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casanova JL, Abel L. Mechanisms of viral inflammation and disease in humans. Science (New York, NY) 2021; 374(6571):1080–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreakos E, Abel L, Vinh DC, Kaja E, Drolet BA, Zhang Q, et al. A global effort to dissect the human genetic basis of resistance to SARS-CoV-2 infection. Nat Immunol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arkin LM, Moon JJ, Tran JM, Asgari S, O’Farrelly C, Casanova J-L, et al. From your nose to your toes: A Review of SARS-CoV-2 Pandemic-associated Pernio. Journal of Investigative Dermatology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sancho-Shimizu V, Brodin P, Cobat A, Biggs CM, Toubiana J, Lucas CL, et al. SARS-CoV-2-related MIS-C: A key to the viral and genetic causes of Kawasaki disease? The Journal of experimental medicine 2021; 218(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helms J, Kremer S, Merdji H, Clere-Jehl R, Schenck M, Kummerlen C, et al. Neurologic Features in Severe SARS-CoV-2 Infection. N Engl J Med 2020; 382(23):2268–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L, Yao Q, Gu X, Wang Q, Ren L, Wang Y, et al. 1-year outcomes in hospital survivors with COVID-19: a longitudinal cohort study. Lancet 2021; 398(10302):747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciancanelli MJ, Huang SX, Luthra P, Garner H, Itan Y, Volpi S, et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science (New York, NY) 2015; 348(6233):448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]; Earliest report of an inborn error of immunity underlying life-threatening influenza pneumonia in an otherwise healthy child.
- 31.Hernandez N, Melki I, Jing H, Habib T, Huang SSY, Danielson J, et al. Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. The Journal of experimental medicine 2018; 215(10):2567–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim HK, Huang SXL, Chen J, Kerner G, Gilliaux O, Bastard P, et al. Severe influenza pneumonitis in children with inherited TLR3 deficiency. The Journal of experimental medicine 2019; 216(9):2038–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 2005; 434(7034):772–777. [DOI] [PubMed] [Google Scholar]
- 34.Reizis B Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 2019; 50(1):37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nature reviews Immunology 2006; 6(9):644–658. [DOI] [PubMed] [Google Scholar]
- 36.Stark GR, Darnell JE Jr. The JAK-STAT pathway at twenty. Immunity 2012; 36(4):503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao D, Ciancanelli MJ, Zhang P, Harschnitz O, Bondet V, Hasek M, et al. TLR3 controls constitutive IFN-beta antiviral immunity in human fibroblasts and cortical neurons. The Journal of clinical investigation 2021; 131(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hernandez N, Bucciol G, Moens L, Le Pen J, Shahrooei M, Goudouris E, et al. Inherited IFNAR1 deficiency in otherwise healthy patients with adverse reaction to measles and yellow fever live vaccines. The Journal of experimental medicine 2019; 216(9):2057–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt A, Peters S, Knaus A, Sabir H, Hamsen F, Maj C, et al. TBK1 and TNFRSF13B mutations and an autoinflammatory disease in a child with lethal COVID-19. NPJ Genom Med 2021; 6(1):55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khanmohammadi S, Rezaei N, Khazaei M, Shirkani A. A Case of Autosomal Recessive Interferon Alpha/Beta Receptor Alpha Chain (IFNAR1) Deficiency with Severe COVID-19. Journal of clinical immunology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asano T, Boisson B, Onodi F, Matuozzo D, Moncada-Velez M, Maglorius Renkilaraj MRL, et al. X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Science immunology 2021; 6(62). [DOI] [PMC free article] [PubMed] [Google Scholar]; Report of X-linked recessive TLR7 deficiency as a human genetic and immunological determinant of life-threatening COVID-19 pneumonia in male patients.
- 42.Abolhassani H, Vosughimotlagh A, Asano T, Landegren N, Boisson B, Delavari S, et al. X-Linked TLR7 Deficiency Underlies Critical COVID-19 Pneumonia in a Male Patient with Ataxia-Telangiectasia. Journal of clinical immunology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Made CI, Simons A, Schuurs-Hoeijmakers J, van den Heuvel G, Mantere T, Kersten S, et al. Presence of Genetic Variants Among Young Men With Severe COVID-19. JAMA 2020; 324(7):663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fallerini C, Daga S, Mantovani S, Benetti E, Picchiotti N, Francisci D, et al. Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: findings from a nested case-control study. Elife 2021; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pessoa NL, Bentes AA, de Carvalho AL, de Souza Silva TB, Alves PA, de Sousa Reis EV, et al. Case report: hepatitis in a child infected with SARS-CoV-2 presenting toll-like receptor 7 Gln11Leu single nucleotide polymorphism. Virol J 2021; 18(1):180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solanich X, Vargas-Parra G, van der Made CI, Simons A, Schuurs-Hoeijmakers J, Antoli A, et al. Genetic Screening for TLR7 Variants in Young and Previously Healthy Men With Severe COVID-19. Front Immunol 2021; 12:719115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kosmicki JA, Horowitz JE, Banerjee N, Lanche R, Marcketta A, Maxwell E, et al. Pan-ancestry exome-wide association analyses of COVID-19 outcomes in 586,157 individuals. Am J Hum Genet 2021; 108(7):1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Onodi F, Bonnet-Madin L, Meertens L, Karpf L, Poirot J, Zhang SY, et al. SARS-CoV-2 induces human plasmacytoid predendritic cell diversification via UNC93B and IRAK4. The Journal of experimental medicine 2021; 218(4). [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence that human plasmacytoid dendritic cells sense SARS-CoV-2 via UNC93B and IRAK4, and, by inference, via TLR7 and/or TLR9.
- 49.Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nature reviews Immunology 2015; 15(8):471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat Immunol 2004; 5(12):1219–1226. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y-J. Dendritic Cell Subsets and Lineages, and Their Functions in Innate and Adaptive Immunity. Cell 2001; 106(3):259–262. [DOI] [PubMed] [Google Scholar]
- 52.Severa M, Diotti RA, Etna MP, Rizzo F, Fiore S, Ricci D, et al. Differential plasmacytoid dendritic cell phenotype and type I Interferon response in asymptomatic and severe COVID-19 infection. PLoS Pathog 2021; 17(9):e1009878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beutler B, Inferences, questions and possibilities in Toll-like receptor signalling. Nature 2004; 430(6996):257–263. [DOI] [PubMed] [Google Scholar]
- 54.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol 2002; 3(2):196–200. [DOI] [PubMed] [Google Scholar]
- 55.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science (New York, NY) 2004; 303(5663):1529–1531. [DOI] [PubMed] [Google Scholar]
- 56.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science (New York, NY) 2004; 303(5663):1526–1529. [DOI] [PubMed] [Google Scholar]
- 57.Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, et al. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A 2004; 101(15):5598–5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barreiro LB, Ben-Ali M, Quach H, Laval G, Patin E, Pickrell JK, et al. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet 2009; 5(7):e1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang SY, Jouanguy E, Sancho-Shimizu V, von Bernuth H, Yang K, Abel L, et al. Human Toll-like receptor-dependent induction of interferons in protective immunity to viruses. Immunol Rev 2007; 220:225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deya-Martinez A, Garcia-Garcia A, Gonzalez-Navarro EA, Yiyi L, Vlagea A, Jordan I, et al. COVID-19 in children and young adults with moderate/severe inborn errors of immunity in a high burden area in pre-vaccine era. Clin Immunol 2021; 230:108821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goudouris ES, Pinto-Mariz F, Mendonca LO, Aranda CS, Guimaraes RR, Kokron C, et al. Outcome of SARS-CoV-2 Infection in 121 Patients with Inborn Errors of Immunity: A Cross-Sectional Study. Journal of clinical immunology 2021; 41(7):1479–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shields AM, Burns SO, Savic S, Richter AG, Consortium UPC-. COVID-19 in patients with primary and secondary immunodeficiency: The United Kingdom experience. J Allergy Clin Immunol 2021; 147(3):870–875 e871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meyts I, Bucciol G, Quinti I, Neven B, Fischer A, Seoane E, et al. Coronavirus disease 2019 in patients with inborn errors of immunity: An international study. J Allergy Clin Immunol 2021; 147(2):520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Q Human genetics of life-threatening influenza pneumonitis. Hum Genet 2020; 139(6-7):941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vallbracht A, Treuner J, Flehmig B, Joester KE, Niethammer D. Interferon-neutralizing antibodies in a patient treated with human fibroblast interferon. Nature 1981; 289(5797):496–497. [DOI] [PubMed] [Google Scholar]
- 66.Panem S, Check IJ, Henriksen D, Vilcek J. Antibodies to alpha-interferon in a patient with systemic lupus erythematosus. J Immunol 1982; 129(1):1–3. [PubMed] [Google Scholar]
- 67.Gupta S, Tatouli IP, Rosen LB, Hasni S, Alevizos I, Manna ZG, et al. Distinct Functions of Autoantibodies Against Interferon in Systemic Lupus Erythematosus: A Comprehensive Analysis of Anticytokine Autoantibodies in Common Rheumatic Diseases. Arthritis Rheumatol 2016; 68(7):1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levin M Anti-interferon auto-antibodies in autoimmune polyendocrinopathy syndrome type 1. PLoS Med 2006; 3(7):e292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meager A, Visvalingam K, Peterson P, Moll K, Murumagi A, Krohn K, et al. Anti-interferon autoantibodies in autoimmune polyendocrinopathy syndrome type 1. PLoS Med 2006; 3(7):e289. [DOI] [PMC free article] [PubMed] [Google Scholar]; Initial report that patients with APS-1 carry auto-antibodies against type I IFNs.
- 70.Meyer S, Woodward M, Hertel C, Vlaicu P, Haque Y, Karner J, et al. AIRE-Deficient Patients Harbor Unique High-Affinity Disease-Ameliorating Autoantibodies. Cell 2016; 166(3):582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosenberg JM, Maccari ME, Barzaghi F, Allenspach EJ, Pignata C, Weber G, et al. Neutralizing Anti-Cytokine Autoantibodies Against Interferon-alpha in Immunodysregulation Polyendocrinopathy Enteropathy X-Linked. Front Immunol 2018; 9:544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eriksson D, Bacchetta R, Gunnarsson HI, Chan A, Barzaghi F, Ehl S, et al. The autoimmune targets in IPEX are dominated by gut epithelial proteins. J Allergy Clin Immunol 2019; 144(1):327–330 e328. [DOI] [PubMed] [Google Scholar]
- 73.Walter JE, Rosen LB, Csomos K, Rosenberg JM, Mathew D, Keszei M, et al. Broad-spectrum antibodies against self-antigens and cytokines in RAG deficiency. J Clin Invest 2015; 125(11):4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Romi F Thymoma in myasthenia gravis: from diagnosis to treatment. Autoimmune Dis 2011; 2011:474512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shiono H, Wong YL, Matthews I, Liu JL, Zhang W, Sims G, et al. Spontaneous production of anti-IFN-alpha and anti-IL-12 autoantibodies by thymoma cells from myasthenia gravis patients suggests autoimmunization in the tumor. Int Immunol 2003; 15(8):903–913. [DOI] [PubMed] [Google Scholar]
- 76.Bello-Rivero I, Cervantes M, Torres Y, Ferrero J, Rodriguez E, Perez J, et al. Characterization of the immunoreactivity of anti-interferon alpha antibodies in myasthenia gravis patients. Epitope mapping. J Autoimmun 2004; 23(1):63–73. [DOI] [PubMed] [Google Scholar]
- 77.Meager A, Wadhwa M, Dilger P, Bird C, Thorpe R, Newsom-Davis J, et al. Anti-cytokine autoantibodies in autoimmunity: preponderance of neutralizing autoantibodies against interferon-alpha, interferon-omega and interleukin-12 in patients with thymoma and/or myasthenia gravis. Clin Exp Immunol 2003; 132(1):128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science (New York, NY) 2002; 298(5597):1395–1401. [DOI] [PubMed] [Google Scholar]
- 79.Cheng MH, Fan U, Grewal N, Barnes M, Mehta A, Taylor S, et al. Acquired autoimmune polyglandular syndrome, thymoma, and an AIRE defect. N Engl J Med 2010; 362(8):764–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pozzetto B, Mogensen KE, Tovey MG, Gresser I. Characteristics of autoantibodies to human interferon in a patient with varicella-zoster disease. J Infect Dis 1984; 150(5):707–713. [DOI] [PubMed] [Google Scholar]; Seminal report of a single patient with severe varicella zoster virus disease due to auto-Abs neutralizing type I IFNs, which is also the first infectious disease causally attributed to an auto-Ab directed against a cytokine.
- 81.Doffinger R, Helbert MR, Barcenas-Morales G, Yang K, Dupuis S, Ceron-Gutierrez L, et al. Autoantibodies to interferon-gamma in a patient with selective susceptibility to mycobacterial infection and organ-specific autoimmunity. Clin Infect Dis 2004; 38(1):e10–14. [DOI] [PubMed] [Google Scholar]
- 82.Hoflich C, Sabat R, Rosseau S, Temmesfeld B, Slevogt H, Docke WD, et al. Naturally occurring anti-IFN-gamma autoantibody and severe infections with Mycobacterium cheloneae and Burkholderia cocovenenans. Blood 2004; 103(2):673–675. [DOI] [PubMed] [Google Scholar]
- 83.Kampmann B, Hemingway C, Stephens A, Davidson R, Goodsall A, Anderson S, et al. Acquired predisposition to mycobacterial disease due to autoantibodies to IFN-gamma. The Journal of clinical investigation 2005; 115(9):2480–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Puel A, Picard C, Lorrot M, Pons C, Chrabieh M, Lorenzo L, et al. Recurrent staphylococcal cellulitis and subcutaneous abscesses in a child with autoantibodies against IL-6. J Immunol 2008; 180(1):647–654. [DOI] [PubMed] [Google Scholar]
- 85.Puel A, Doffinger R, Natividad A, Chrabieh M, Barcenas-Morales G, Picard C, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. The Journal of experimental medicine 2010; 207(2):291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ku CL, Chi CY, von Bernuth H, Doffinger R. Autoantibodies against cytokines: phenocopies of primary immunodeficiencies? Hum Genet 2020; 139(6-7):783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kisand K, Boe Wolff AS, Podkrajsek KT, Tserel L, Link M, Kisand KV, et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. The Journal of experimental medicine 2010; 207(2):299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosen LB, Rocha Pereira N, Figueiredo C, Fiske LC, Ressner RA, Hong JC, et al. Nocardia-induced granulocyte macrophage colony-stimulating factor is neutralized by autoantibodies in disseminated/extrapulmonary nocardiosis. Clin Infect Dis 2015; 60(7):1017–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koning R, Bastard P, Casanova JL, Brouwer MC, van de Beek D, with the Amsterdam UMCC-BI. Autoantibodies against type I interferons are associated with multi-organ failure in COVID-19 patients. Intensive Care Med 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Troya J, Bastard P, Planas-Serra L, Ryan P, Ruiz M, de Carranza M, et al. Neutralizing Autoantibodies to Type I IFNs in >10% of Patients with Severe COVID-19 Pneumonia Hospitalized in Madrid, Spain. J Clin Immunol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goncalves D, Mezidi M, Bastard P, Perret M, Saker K, Fabien N, et al. Antibodies against type I interferon: detection and association with severe clinical outcome in COVID-19 patients. Clin Transl Immunology 2021; 10(8):e1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang EY, Mao T, Klein J, Dai Y, Huck JD, Jaycox JR, et al. Diverse functional autoantibodies in patients with COVID-19. Nature 2021; 595(7866):283–288. [DOI] [PubMed] [Google Scholar]; Replication of the enrichment of auto-antibodies against type I IFNs in patients with severe COVID-19 and evidence that SARS-CoV-2 infection can trigger other auto-antibodies.
- 93.van der Wijst MGP, Vazquez SE, Hartoularos GC, Bastard P, Grant T, Bueno R, et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci Transl Med 2021; 13(612):eabh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]; Single-cell study of blood leukocytes showing that patients with auto-Abs against type I IFNs are immunologically similar to other patients with critical COVID-19 pneumonia.
- 94.Acosta-Ampudia Y, Monsalve DM, Rojas M, Rodriguez Y, Gallo JE, Salazar-Uribe JC, et al. COVID-19 convalescent plasma composition and immunological effects in severe patients. J Autoimmun 2021; 118:102598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chang SE, Feng A, Meng W, Apostolidis SA, Mack E, Artandi M, et al. New-onset IgG autoantibodies in hospitalized patients with COVID-19. Nat Commun 2021; 12(1):5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ziegler CGK, Miao VN, Owings AH, Navia AW, Tang Y, Bromley JD, et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell 2021; 184(18):4713–4733 e4722. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive and in-depth study of intrinsic immunity in the respiratory tract of patients with COVID-19.
- 97.Solanich X, Rigo-Bonnin R, Gumucio VD, Bastard P, Rosain J, Philippot Q, et al. Pre-existing Autoantibodies Neutralizing High Concentrations of Type I Interferons in Almost 10% of COVID-19 Patients Admitted to Intensive Care in Barcelona. Journal of clinical immunology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abers MS, Rosen LB, Delmonte OM, Shaw E, Bastard P, Imberti L, et al. Neutralizing type-I interferon autoantibodies are associated with delayed viral clearance and intensive care unit admission in patients with COVID-19. Immunology and cell biology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vazquez SE, Bastard P, Kelly K, Gervais A, Norris PJ, Dumont LJ, et al. Neutralizing Autoantibodies to Type I Interferons in COVID-19 Convalescent Donor Plasma. Journal of clinical immunology 2021; 41(6):1169–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chauvineau-Grenier A, Bastard P, Servajean A, Gervais A, Rosain J, Jouanguy E, et al. Autoantibodies neutralizing type I interferons in 20% of COVID-19 deaths in a French hospital. Res Sq 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carapito R, Li R, Helms J, Carapito C, Gujja S, Rolli V, et al. Identification of driver genes for critical forms of COVID-19 in a deeply phenotyped young patient cohort. Sci Transl Med 2021:eabj7521. [DOI] [PubMed] [Google Scholar]
- 102.Raadsen MP, Gharbharan A, Jordans CCE, Mykytyn AZ, Lamers MM, van den Doel PB, et al. Interferon-alpha2 Auto-antibodies in Convalescent Plasma Therapy for COVID-19. Journal of clinical immunology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bastard P, Orlova E, Sozaeva L, Levy R, James A, Schmitt MM, et al. Preexisting autoantibodies to type I IFNs underlie critical COVID-19 pneumonia in patients with APS-1. The Journal of experimental medicine 2021; 218(7). [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence that patients with autoimmune polyendocrine syndrome type 1 (APS-1) are at very high risk of critical COVID-19 due to their auto-Abs against type I IFNs.
- 104.Meisel C, Akbil B, Meyer T, Lankes E, Corman VM, Staudacher O, et al. Mild COVID-19 despite autoantibodies against type I IFNs in autoimmune polyendocrine syndrome type 1. The Journal of clinical investigation 2021; 131(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shaw ER, Rosen LB, Cheng A, Dobbs K, Delmonte OM, Ferré EMN, et al. Temporal Dynamics of Anti-Type 1 Interferon Autoantibodies in COVID-19 Patients. Clin Infect Dis 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lopez J, Mommert M, Mouton W, Pizzorno A, Brengel-Pesce K, Mezidi M, et al. Early nasal type I IFN immunity against SARS-CoV-2 is compromised in patients with autoantibodies against type I IFNs. The Journal of experimental medicine 2021; 218(10). [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence that auto-Abs against type I IFNs are found in nasal secretions and impair type I IFN immunity in the corresponding mucosa.
- 107.de Prost N, Bastard P, Arrestier R, Fourati S, Mahevas M, Burrel S, et al. Plasma Exchange to Rescue Patients with Autoantibodies Against Type I Interferons and Life-Threatening COVID-19 Pneumonia. J Clin Immunol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Galani IE, Rovina N, Lampropoulou V, Triantafyllia V, Manioudaki M, Pavlos E, et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol 2021; 22(1):32–40. [DOI] [PubMed] [Google Scholar]; Longitudinal study of antiviral immunity, including type I and III IFNs, in patients with diverse forms of COVID-19 pneumonia, in comparison with influenza pneumonia.
- 109.Bastard P, Gervais A, Le Voyer T, Rosain J, Philippot Q, Manry J, et al. Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Science immunology 2021; 6(62). [DOI] [PMC free article] [PubMed] [Google Scholar]; Discovery of auto-Abs neutralizing low concentrations of type I IFNs in the general population and as immunological determinants of life-threatening COVID-19 pneumonia, especially in the elderly population.
- 110.LaFleur DW, Nardelli B, Tsareva T, Mather D, Feng P, Semenuk M, et al. Interferon-kappa, a novel type I interferon expressed in human keratinocytes. J Biol Chem 2001; 276(43):39765–39771. [DOI] [PubMed] [Google Scholar]
- 111.Marks ZRC, Campbell N, deWeerd NA, Lim SS, Gearing LJ, Bourke NM, et al. Properties and Functions of the Novel Type I Interferon Epsilon. Semin Immunol 2019; 43:101328. [DOI] [PubMed] [Google Scholar]
- 112.Lazear HM, Schoggins JW, Diamond MS. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019; 50(4):907–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Manry J, Laval G, Patin E, Fornarino S, Itan Y, Fumagalli M, et al. Evolutionary genetic dissection of human interferons. The Journal of experimental medicine 2011; 208(13):2747–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Park A, Iwasaki A. Type I and Type III Interferons - Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020; 27(6):870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Vinh DC, Abel L, Bastard P, Cheng MP, Condino-Neto A, Gregersen PK, et al. Harnessing Type I IFN Immunity Against SARS-CoV-2 with Early Administration of IFN-beta. Journal of clinical immunology 2021:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.de Prost N, Bastard P, Arrestier R, Fourati S, Mahevas M, Burrel S, et al. Plasma Exchange to Rescue Patients with Autoantibodies Against Type I Interferons and Life-Threatening COVID-19 Pneumonia. Journal of clinical immunology 2021; 41(3):536–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bastard P, Michailidis E, Hoffmann HH, Chbihi M, Le Voyer T, Rosain J, et al. Auto-antibodies to type I IFNs can underlie adverse reactions to yellow fever live attenuated vaccine. The Journal of experimental medicine 2021; 218(4). [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence that auto-Abs neutralizing type I IFNs can underlie life-threatening adverse reactions to live-attenuated yellow fever virus vaccine in previously healthy patients.
- 118.Hetemaki I, Laakso S, Valimaa H, Kleino I, Kekalainen E, Makitie O, et al. Patients with autoimmune polyendocrine syndrome type 1 have an increased susceptibility to severe herpesvirus infections. Clin Immunol 2021; 231:108851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bastard P, Levy R, Henriquez S, Bodemer C, Szwebel TA, Casanova JL. Interferon-beta Therapy in a Patient with Incontinentia Pigmenti and Autoantibodies against Type I IFNs Infected with SARS-CoV-2. J Clin Immunol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Beccuti G, Ghizzoni L, Cambria V, Codullo V, Sacchi P, Lovati E, et al. A COVID-19 pneumonia case report of autoimmune polyendocrine syndrome type 1 in Lombardy, Italy: letter to the editor. J Endocrinol Invest 2020; 43(8):1175–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lemarquis A, Campbell T, Aranda-Guillen M, Hennings V, Brodin P, Kampe O, et al. Severe COVID-19 in an APS1 patient with interferon autoantibodies treated with plasmapheresis. J Allergy Clin Immunol 2021; 148(1):96–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Abraham RS, Marshall JM, Kuehn HS, Rueda CM, Gibbs A, Guider W, et al. Severe SARS-CoV-2 disease in the context of a NF-kappaB2 loss-of-function pathogenic variant. J Allergy Clin Immunol 2021; 147(2):532–544 e531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Drabe CH, Hansen AE, Rasmussen LD, Larsen OD, Moller A, Mogensen TH, et al. Low morbidity in Danish patients with common variable immunodeficiency disorder infected with severe acute respiratory syndrome coronavirus 2. Infect Dis (Lond) 2021:1–6. [DOI] [PubMed] [Google Scholar]
- 124.Marcus N, Frizinsky S, Hagin D, Ovadia A, Hanna S, Farkash M, et al. Minor Clinical Impact of COVID-19 Pandemic on Patients With Primary Immunodeficiency in Israel. Front Immunol 2020; 11:614086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science (New York, NY) 2020; 369(6504):718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Trouillet-Assant S, Viel S, Gaymard A, Pons S, Richard JC, Perret M, et al. Type I IFN immunoprofiling in COVID-19 patients. J Allergy Clin Immunol 2020; 146(1):206–208 e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu C, Martins AJ, Lau WW, Rachmaninoff N, Chen J, Imberti L, et al. Time-resolved systems immunology reveals a late juncture linked to fatal COVID-19. Cell 2021; 184(7):1836–1857 e1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lucas C, Wong P, Klein J, Castro TBR, Silva J, Sundaram M, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020; 584(7821):463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lee JS, Park S, Jeong HW, Ahn JY, Choi SJ, Lee H, et al. Immunophenotyping of COVID-19 and influenza highlights the role of type I interferons in development of severe COVID-19. Science immunology 2020; 5(49). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sposito B, Broggi A, Pandolfi L, Crotta S, Clementi N, Ferrarese R, et al. The interferon landscape along the respiratory tract impacts the severity of COVID-19. Cell 2021; 184(19):4953–4968 e4916. [DOI] [PMC free article] [PubMed] [Google Scholar]; Comprehensive and in-depth study of intrinsic and innate immunity along the respiratory tract in patients with severe COVID-19.
- 131.Lévy R, Zhang P, Bastard P, Dorgham K, Melki I, Hadchouel A, et al. Monoclonal antibody-mediated neutralization of SARS-CoV-2 in an IRF9-deficient child. Proc Natl Acad Sci U S A 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Evidence that mAbs neutralizing SARS-CoV-2 were able to overcome a complete deficiency of both type I and III IFNs in a child with autosomal recesive IRF9 deficiency.
- 132.Delavari S, Abolhassani H, Abolnezhadian F, Babaha F, Iranparast S, Ahanchian H, et al. Impact of SARS-CoV-2 Pandemic on Patients with Primary Immunodeficiency. Journal of clinical immunology 2021; 41(2):345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Esenboga S, Ocak M, Akarsu A, Bildik HN, Cagdas D, Iskit AT, et al. COVID-19 in Patients with Primary Immunodeficiency. Journal of clinical immunology 2021; 41(7):1515–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ho HE, Mathew S, Peluso MJ, Cunningham-Rundles C. Clinical outcomes and features of COVID-19 in patients with primary immunodeficiencies in New York City. J Allergy Clin Immunol Pract 2021; 9(1):490–493 e492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Milito C, Lougaris V, Giardino G, Punziano A, Vultaggio A, Carrabba M, et al. Clinical outcome, incidence, and SARS-CoV-2 infection-fatality rates in Italian patients with inborn errors of immunity. J Allergy Clin Immunol Pract 2021; 9(7):2904–2906 e2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Castano-Jaramillo LM, Yamazaki-Nakashimada MA, O’Farrill-Romanillos PM, Muzquiz Zermeno D, Scheffler Mendoza SC, Venegas Montoya E, et al. COVID-19 in the Context of Inborn Errors of Immunity: a Case Series of 31 Patients from Mexico. Journal of clinical immunology 2021; 41(7):1463–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Karakoc Aydiner E, Bilgic Eltan S, Babayeva R, Aydiner O, Kepenekli E, Kolukisa B, et al. Adverse COVID-19 outcomes in immune deficiencies: Inequality exists between subclasses. Allergy 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Corey L, Beyrer C, Cohen MS, Michael NL, Bedford T, Rolland M. SARS-CoV-2 Variants in Patients with Immunosuppression. N Engl J Med 2021; 385(6):562–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Giardino G, Romano R, Coppola E, Cillo F, Borzachiello C, De Luca M, et al. SARS-CoV-2 Infection in the Immunodeficient Host: Necessary and Dispensable Immune Pathways. J Allergy Clin Immunol Pract 2021; 9(9):3237–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hariharan SV, Muthusamy S, Asokan SK. Persistent Viral Shedding after SARS-CoV-2 Infection in an Infant with Severe Combined Immunodeficiency. Indian J Pediatr 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mohanty MC, Taur PD, Sawant UP, Yadav RM, Potdar V. Prolonged Fecal Shedding of SARS-CoV-2 in Asymptomatic Children with Inborn Errors of Immunity. Journal of clinical immunology 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ambrosioni J, Blanco JL, Reyes-Uruena JM, Davies MA, Sued O, Marcos MA, et al. Overview of SARS-CoV-2 infection in adults living with HIV. Lancet HIV 2021; 8(5):e294–e305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kemp SA, Collier DA, Datir RP, Ferreira I, Gayed S, Jahun A, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021; 592(7853):277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Choi B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N Engl J Med 2020; 383(23):2291–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Severe Covid GG, Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N Engl J Med 2020; 383(16):1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pairo-Castineira E, Clohisey S, Klaric L, Bretherick AD, Rawlik K, Pasko D, et al. Genetic mechanisms of critical illness in COVID-19. Nature 2021; 591(7848):92–98. [DOI] [PubMed] [Google Scholar]
- 147.C-HG Initiative. Mapping the human genetic architecture of COVID-19. Nature 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]; Genome-wide association meta-analysis of COVID-19, including genome-wide significant loci weakly associated with SARS-CoV-2 infection or severe COVID-19.
- 148.Nakanishi T, Pigazzini S, Degenhardt F, Cordioli M, Butler-Laporte G, Maya-Miles D, et al. Age-dependent impact of the major common genetic risk factor for COVID-19 on severity and mortality. The Journal of clinical investigation 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zeberg H, Paabo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 2020; 587(7835):610–612. [DOI] [PubMed] [Google Scholar]
- 150.Kerner G, Patin E, Quintana-Murci L. New insights into human immunity from ancient genomics. Curr Opin Immunol 2021; 72:116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zeberg H, Paabo S. A genomic region associated with protection against severe COVID-19 is inherited from Neandertals. Proc Natl Acad Sci U S A 2021; 118(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Colona VL, Biancolella M, Novelli A, Novelli G. Will GWAS eventually allow the identification of genomic biomarkers for COVID-19 severity and mortality? The Journal of clinical investigation 2021; 131(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Povysil G, Butler-Laporte G, Shang N, Wang C, Khan A, Alaamery M, et al. Rare loss-of-function variants in type I IFN immunity genes are not associated with severe COVID-19. The Journal of clinical investigation 2021; 131(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhang Q, Cobat A, Bastard P, Notarangelo LD, Su HC, Abel L, et al. Association of rare predicted loss-of-function variants of influenza-related type I IFN genes with critical COVID-19 pneumonia. The Journal of clinical investigation 2021; 131(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hatton CF, Botting RA, Duenas ME, Haq IJ, Verdon B, Thompson BJ, et al. Delayed induction of type I and III interferons mediates nasal epithelial cell permissiveness to SARS-CoV-2. Nat Commun 2021; 12(1):7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020; 181(5):1036–1045 e1039. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive study of the virus-host dynamics underlying COVID-19, with low levels of type I and III IFNs and high levels of other chemokines and cytokines.
- 157.Banerjee AK, Blanco MR, Bruce EA, Honson DD, Chen LM, Chow A, et al. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell 2020; 183(5):1325–1339 e1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Flynn RA, Belk JA, Qi Y, Yasumoto Y, Wei J, Alfajaro MM, et al. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell 2021; 184(9):2394–2411 e2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Schneider WM, Luna JM, Hoffmann HH, Sanchez-Rivera FJ, Leal AA, Ashbrook AW, et al. Genome-Scale Identification of SARS-CoV-2 and Pan-coronavirus Host Factor Networks. Cell 2021; 184(1):120–132 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of human cell factors that restrict SARS-CoV-2 proliferation and spreading.
- 160.Palermo E, Di Carlo D, Sgarbanti M, Hiscott J. Type I Interferons in COVID-19 Pathogenesis. Biology 2021; 10(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lowery SA, Sariol A, Perlman S. Innate immune and inflammatory responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe 2021; 29(7):1052–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]; A review of the mechanisms by which SARS-CoV-2 can induce or antagonize type I and III IFN activity.
- 162.Mariano G, Farthing RJ, Lale-Farjat SLM, Bergeron JRC. Structural Characterization of SARS-CoV-2: Where We Are, and Where We Need to Be. Front Mol Biosci 2020; 7:605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Schultze JL, Aschenbrenner AC. COVID-19 and the human innate immune system. Cell 2021; 184(7):1671–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, et al. SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep 2020; 32(12):108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sui L, Zhao Y, Wang W, Wu P, Wang Z, Yu Y, et al. SARS-CoV-2 Membrane Protein Inhibits Type I Interferon Production Through Ubiquitin-Mediated Degradation of TBK1. Front Immunol 2021; 12:662989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci U S A 2020; 117(45):28344–28354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hayn M, Hirschenberger M, Koepke L, Nchioua R, Straub JH, Klute S, et al. Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities. Cell Rep 2021; 35(7):109126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yuen CK, Lam JY, Wong WM, Mak LF, Wang X, Chu H, et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect 2020; 9(1):1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lin JW, Tang C, Wei HC, Du B, Chen C, Wang M, et al. Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe 2021; 29(3):489–502 e488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mlcochova P, Kemp S, Dhar MS, Papa G, Meng B, Ferreira I, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Daniloski Z, Jordan TX, Wessels HH, Hoagland DA, Kasela S, Legut M, et al. Identification of Required Host Factors for SARS-CoV-2 Infection in Human Cells. Cell 2021; 184(1):92–105 e116. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of human cell factors that restrict SARS-CoV-2 proliferation and spreading.
- 172.Hoffmann HH, Schneider WM, Rozen-Gagnon K, Miles LA, Schuster F, Razooky B, et al. TMEM41B Is a Pan-flavivirus Host Factor. Cell 2021; 184(1):133–148 e120. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of human cell factors that restrict SARS-CoV-2 proliferation and spreading.
- 173.Wang R, Simoneau CR, Kulsuptrakul J, Bouhaddou M, Travisano KA, Hayashi JM, et al. Genetic Screens Identify Host Factors for SARS-CoV-2 and Common Cold Coronaviruses. Cell 2021; 184(1):106–119 e114. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of human cell factors that restrict SARS-CoV-2 proliferation and spreading.
- 174.Wei J, Alfajaro MM, DeWeirdt PC, Hanna RE, Lu-Culligan WJ, Cai WL, et al. Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection. Cell 2021; 184(1):76–91 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of human cell factors that restrict SARS-CoV-2 proliferation and spreading.
- 175.Hoffmann HH, Sanchez-Rivera FJ, Schneider WM, Luna JM, Soto-Feliciano YM, Ashbrook AW, et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe 2021; 29(2):267–280 e265. [DOI] [PMC free article] [PubMed] [Google Scholar]; Identification of human cell factors that restrict SARS-CoV-2 proliferation and spreading.
- 176.Katsura H, Sontake V, Tata A, Kobayashi Y, Edwards CE, Heaton BE, et al. Human Lung Stem Cell-Based Alveolospheres Provide Insights into SARS-CoV-2-Mediated Interferon Responses and Pneumocyte Dysfunction. Cell Stem Cell 2020; 27(6):890–904 e898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Martin-Sancho L, Lewinski MK, Pache L, Stoneham CA, Yin X, Becker ME, et al. Functional landscape of SARS-CoV-2 cellular restriction. Mol Cell 2021; 81(12):2656–2668 e2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pfaender S, Mar KB, Michailidis E, Kratzel A, Boys IN, V’Kovski P, et al. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat Microbiol 2020; 5(11):1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Gordon DE, Hiatt J, Bouhaddou M, Rezelj VV, Ulferts S, Braberg H, et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science (New York, NY) 2020; 370(6521). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lokugamage KG, Hage A, de Vries M, Valero-Jimenez AM, Schindewolf C, Dittmann M, et al. Type I Interferon Susceptibility Distinguishes SARS-CoV-2 from SARS-CoV. J Virol 2020; 94(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Munoz-Fontela C, Dowling WE, Funnell SGP, Gsell PS, Riveros-Balta AX, Albrecht RA, et al. Animal models for COVID-19. Nature 2020; 586(7830):509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]; Review of animal models for SARS-CoV-2 infection and their applications.
- 182.Bessiere P, Wasniewski M, Picard-Meyer E, Servat A, Figueroa T, Foret-Lucas C, et al. Intranasal type I interferon treatment is beneficial only when administered before clinical signs onset in the SARS-CoV-2 hamster model. PLoS Pathog 2021; 17(8):e1009427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Hoagland DA, Moller R, Uhl SA, Oishi K, Frere J, Golynker I, et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity 2021; 54(3):557–570 e555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 2020; 182(3):744–753 e744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Israelow B, Song E, Mao T, Lu P, Meir A, Liu F, et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. The Journal of experimental medicine 2020; 217(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Leist SR, Dinnon KH 3rd, Schafer A, Tse LV, Okuda K, Hou YJ, et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 2020; 183(4):1070–1085 e1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dinnon KH 3rd, Leist SR, Schafer A, Edwards CE, Martinez DR, Montgomery SA, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 2020; 586(7830):560–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021; 184(4):861–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Consortium WHOST, Pan H, Peto R, Henao-Restrepo AM, Preziosi MP, Sathiyamoorthy V, et al. Repurposed Antiviral Drugs for Covid-19 - Interim WHO Solidarity Trial Results. N Engl J Med 2021; 384(6):497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Loske J, Rohmel J, Lukassen S, Stricker S, Magalhaes VG, Liebig J, et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat Biotechnol 2021. [DOI] [PubMed] [Google Scholar]; Report of stronger type I IFN activity in the respiratory tract in SARS-CoV-2-infected children than in adults.
- 191.Splunter MV, Perdijk O, Fick-Brinkhof H, Floris-Vollenbroek EG, Meijer B, Brugman S, et al. Plasmacytoid dendritic cell and myeloid dendritic cell function in ageing: A comparison between elderly and young adult women. PLoS One 2019; 14(12):e0225825. [DOI] [PMC free article] [PubMed] [Google Scholar]; Report of weaker TLR7- and TLR9-dependent responses of plasmacytoid dendritic cells in elderly than in younger adults.
- 192.Pierce CA, Preston-Hurlburt P, Dai Y, Aschner CB, Cheshenko N, Galen B, et al. Immune responses to SARS-CoV-2 infection in hospitalized pediatric and adult patients. Sci Transl Med 2020; 12(564). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Pierce CA, Sy S, Galen B, Goldstein DY, Orner E, Keller MJ, et al. Natural mucosal barriers and COVID-19 in children. JCI Insight 2021; 6(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Shaw AC, Goldstein DR, Montgomery RR. Age-dependent dysregulation of innate immunity. Nature reviews Immunology 2013; 13(12):875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bartleson JM, Radenkovic D, Covarrubias AJ, Furman D, Winer DA, Verdin E. SARS-CoV-2, COVID-19 and the aging immune system. Nature Aging 2021; 1(9):769–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Krause PR, Fleming TR, Longini IM, Peto R, Briand S, Heymann DL, et al. SARS-CoV-2 Variants and Vaccines. N Engl J Med 2021; 385(2):179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hacisuleyman E, Hale C, Saito Y, Blachere NE, Bergh M, Conlon EG, et al. Vaccine Breakthrough Infections with SARS-CoV-2 Variants. N Engl J Med 2021; 384(23):2212–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kustin T, Harel N, Finkel U, Perchik S, Harari S, Tahor M, et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat Med 2021; 27(8):1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pegu A, O’Connell SE, Schmidt SD, O’Dell S, Talana CA, Lai L, et al. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science (New York, NY) 2021; 373(6561):1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gilliet M, Cao W, Liu YJ. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nature reviews Immunology 2008; 8(8):594–606. [DOI] [PubMed] [Google Scholar]


