Abstract
Background:
Cannabis remains one of the most widely abused drugs during pregnancy. In utero exposure to its principal psychoactive component, Δ9-tetrahydrocannabinol (THC), can result in long-term neuropsychiatric risk for the progeny. The current study investigated epigenetic signatures underlying these enduring consequences.
Methods:
Rat dams were exposed daily to THC (0.15mg/kg) during pregnancy and adult male offspring were examined for reward and depressive-like behavioral endophenotypes. Using unbiased sequencing approaches, we explored transcriptional and epigenetic profiles in the nucleus accumbens (NAc), a brain area central to reward and emotional processing. An in vitro CRISPRa model coupled with RNA-sequencing was also applied to study specific consequences of epigenetic dysregulation and altered molecular signatures were compared to human major depressive disorder (MDD) transcriptome datasets.
Results:
Prenatal THC-exposure induced increased motivation for food, heightened learned helplessness and anhedonia, and altered stress sensitivity. We identified a robust increase specific to males in the expression of Histone-Lysine N-Methyltransferase 2A (Kmt2a) that targets lysine 4 on histone H3 (H3K4me) in cellular chromatin. Normalizing Kmt2a in the NAc restored the motivational phenotype of prenatally THC-exposed animals. Comparison of RNA and H3K4me3 sequencing datasets from the NAc of rat offspring with the in vitro model of Kmt2a upregulation revealed overlapping, significant disturbances in pathways that mediate synaptic plasticity. Similar epigenetic alterations were detected in human MDD.
Conclusions:
These studies provide direct evidence for the persistent effects of prenatal cannabis exposure on transcriptional and epigenetic deviations in the NAc via Kmt2a dysregulation and associated psychiatric vulnerability.
Keywords: cannabis, pregnancy, motivation, depression, epigenetics, chromatin
Introduction
Cannabis remains the most abused illicit drug during pregnancy, with an estimated 4% of pregnant women reporting past-month use, and 7% in the past 2 to 12 months. Among pregnant women reporting exposure the past year, 16.2% used cannabis daily (1–3). These women cited nausea, pain, and low appetite as reasons for their use, with the erroneous perception that cannabis is innocuous to the developing fetus (1,4,5). However, the main psychoactive component, Δ9-tetrahydrocannabinol (THC), crosses the placental barrier and can affect the developing fetus, with enduring neurobiological repercussions (6–8). Epidemiological evidence indicates that prenatal cannabis exposure is associated with an elevated risk of neuropsychiatric disorders later in life (9–12). Multiple studies report that progeny exhibit increased impulsivity (13,14) and risk for depression, anxiety, and substance use disorders (10,11,15), particularly when exposed to traumatic experiences (16). While many factors (e.g., environment and genetics) may account for some of the negative consequences ascribed to prenatal cannabis exposure in humans (17–19), controlled animal experiments demonstrate a direct link between cannabinoid exposure and neuropsychiatric related pathologies (20,21,22).
THC directly targets the cannabinoid CB1 receptor within the endocannabinoid system, which mediates cell fate decisions, axonal guidance, and synaptic wiring under highly specific temporal and spatial conditions during prenatal development (23–27). Given the long-term molecular and behavioral effects seen particularly in males with prenatal cannabis/THC exposure (28), we hypothesized underlying epigenetic disturbances as the cellular context by which environmental factors influence gene expression and related behaviors. To that end, we leveraged several behavioral paradigms and identified phenotypic differences in adult male offspring with prenatal THC exposure (PTE) as well as gene expression and epigenetic abnormalities in systems related to synaptic function within the nucleus accumbens (NAc), a brain region mediating reward and emotional processing (29–31). Importantly, we demonstrated a direct relationship to motivational behavior via Kmt2a (histonelysine N-methyltransferase 2A), an epigenetic regulator, as well as novel overlapping transcriptomic and epigenomic signatures common to PTE and human major depressive disorder (MDD).
Methods and Materials
Animals
Long-Evans female rats (44–50 days old; Charles River Laboratories) were carried through PTE as described previously (32) (Figure 1A). All experiments and procedures were approved by the Institutional Animal Care and Use Committee at the Icahn School of Medicine at Mount Sinai.
Figure 1. THC treatment in utero leads to enduring behavioral alterations in adult offspring.
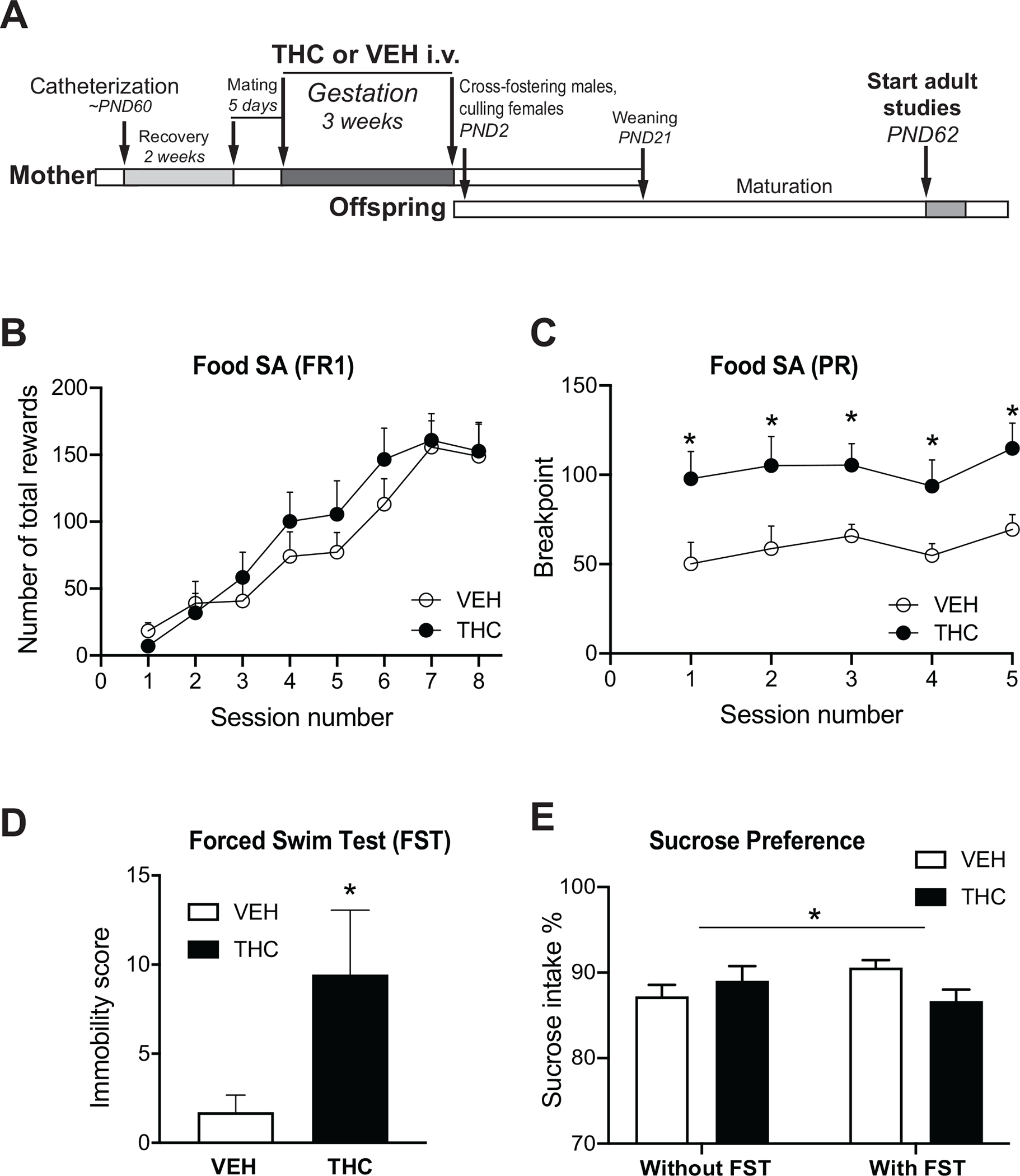
(A) Prenatal THC Paradigm. Pregnant females were treated with low dose THC or VEH and offspring were studied at adulthood. Food Self-administration under (B) Fixed Ratio 1 (FR1) and (C) Progressive Ratio (PR) schedules. (D) Immobility behavior in the Forced Swim Test (FST). (E) Preference of 1% sucrose solution over water. Horizontal line and asterisk indicate a significant interaction effect between PTE and FST. N=7–9/group in all behavioral tests. Data are expressed as mean ± SEM. *:p<0.05.
THC exposure paradigm
Descriptions of drug preparations and animal models are provided in Supplement 1. THC (0.15 mg/kg; in 0.3% tween) was intravenously injected into pregnant dams (via jugular catheter) from gestational day 5 to postnatal day (PND) 2, equivalent to the early-to-midgestational period in humans (33). At PND2, male offspring were cross-fostered to vehicle-exposed dams and THC-treated birth mothers removed from the study. No dams raised their own pups. Approximately 12 pups were randomly assigned to each dam until pups were weaned at PND21 and housed in groups of 3 or 4.
Behavioral Testing
Palatable Food Self-administration.
To assess hedonic food reinforcement, animals were placed in operant chambers where lever pressing resulted in the release of a chocolate pellet. Rats were food restricted (10–18 g/day) 1 day before and throughout testing. Self-administration behavior was studied at fixed ratio 1 (FR1) (one food pellet in response to one active lever press) to monitor reward sensitivity, or progressive ratio (PR) reinforcement schedules (34,35) to assess motivational effort. Animals that never acquired the FR1 task were excluded (1 saline and 1 THC animal).
Forced Swim Test.
Behavioral despair paradigm was used to determine learned helplessness behavior (36). Immobility score reflects time spent floating versus active.
Sucrose Preference.
The sucrose preference test was used to evaluate hedonic state in the preference for sucrose over water. Anhedonia-like behavior is operationalized as a reduction in sucrose intake.
Data Analysis
Groups of 7 to 9 rats were used for behavioral tasks to achieve 80% statistical power with an alpha level of 0.05. Using SAS, JMP, and Prism 8 (GraphPad Software Inc.) statistical software, independent t tests were used when treatment was the sole group variable. When multiple variables existed, analysis of variance statistical models were applied, including appropriate covariates (e.g., treatment, body weight, litter) and/or repeated behavioral measures, to evaluate the influence of PTE on behavioral outcomes. Outliers were detected using Grubbs’ test and removed from the analysis. All data were tested for normality and transformed if not.
Gene and Protein Expression Analysis
Quantitative reverse transcription polymerase chain reaction, NanoString, and Western blotting are described in Supplemental Methods and Materials in Supplement 1. Assay details are shown in Tables S1 and S2 in Supplement 2.
Kmt2a knockdown
One day prior to the behavioral test, 1 μL of a 20-pmol small interfering RNA ON-TARGETplus SMART pool (rat Kmt2a, L-100995-02-0005; Horizon Discovery) or nontargeting pool was injected into the NAc via infusion needles through indwelling bilateral cannulae (37–39) as described in Supplemental Methods and Materials in Supplement 1. Knockdown efficiency is shown in Figure S1 in Supplement 1.
RNA Sequencing, Chromatin Immunoprecipitation Sequencing, and In Vitro CRISPR-Mediated Kmt2a Activation
Sequencing strategies were used as discovery tools for genes and ontology networks associated with PTE. Technical and analytic procedures, as well as data comparisons, are provided in Supplemental Methods and Materials in Supplement 1.
Gene Ontology
All gene ontology enrichment analyses were conducted using GOrilla (40): http://cbl-gorilla.cs.technion.ac.il.
Gene Network Analysis
Networks were constructed using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database (41) and analyzed with NetworkAnalyzer via Cytoscape (42,43) to quantify node degrees. More details are provided in Supplemental Methods and Materials in Supplement 1.
Differential Gene Correlation Analysis
Differential gene correlation analysis (44) was conducted on each dataset to identify gene-gene regulatory relationships that were increased or decreased between experimental groups in our sequencing datasets (RNA sequencing [RNA-seq] and chromatin immunoprecipitation sequencing [ChIP-seq] from the rat NAc and RNA-seq from the CRISPR [clustered regularly interspaced short palindromic repeats] model).
Comparison of Rat NAc and CRISPR Models With Human MDD Datasets
Gene expression datasets from human studies of MDD (45,46) in the NAc; anterior insula; Brodmann areas 11, 25, and 89; and subiculum were used for comparison with our data from rat NAc after PTE and CRISPR activation models.
Results
PTE Increases Motivation for Reward and Depressive-like Behaviors in Adult Offspring
We first explored the direct contributions of PTE to rewardrelated behaviors in adult male progeny because we had previously demonstrated such long-term increase on drug reward (47,48). To assess effects on natural reward, we investigated the response of the offspring to palatable reward in a food selfadministration paradigm. There was no THC-related difference in food self-administration behavior when examined in an FR1 schedule (Figure 1B). In contrast, there was a significant effect in the PR motivational model, as evident with repeatedmeasures analysis of variance on the breakpoint threshold (Figure 1C) (F1,12 = 8.682, p = .0122). Post hoc analysis indicated group differences in the breakpoint in each session, suggesting that PTE consistently increased the motivation to obtain reward.
Depression and altered stress response are reported in the progeny of women who use cannabis while pregnant (10,11). To assess potential depressive-like behaviors in the animal model, rats underwent the forced swim test (FST) to measure learned helplessness (49,50). THC-exposed rats showed significant increases in immobility (F1,15 = 3.8372; p = .0490) (covaried for body weight) (Figure 1D), reflective of a depressive-like endophenotype. These results were replicated in a separate cohort of rats (Figure S2 in Supplement 1). The FST also presents an unexpected, highly stressful situation, and rats with or without this stress experience subsequently showed different sucrose intake behavior, a measure of hedonic state. A significant interaction was evident between PTE and postnatal FST experience for sucrose preference (F1,28 = 4.511; p = .0426) (Figure 1E). PTE animals experiencing FST had reduced sucrose intake, suggesting an anhedonic response, although the stressful event had occurred 4 weeks prior to the sucrose test.
PTE Increases Expression of Kmt2a in the NAc, Linked to Reward-Related Motivation
To gain insight into underlying epigenetic mechanisms relevant to PTE, we evaluated the expression of 80 genes (Table S1 in Supplement 2) encoding various epigenetic regulators involved in chromatin and DNA modification in the NAc, which plays a critical role in reward, motivation, and emotion processing (29–31). This analysis identified significant increases in the messenger RNA (mRNA) expression of Setdb1 (set domain, bifurcated 1) (p = .0114), Kat2b (lysine acetyltransferase 2B) (p = .0274), Kmt2a (p = .0298), and Ncoa6 (nuclear receptor coactivator 6) (p = .0358) (Table S3 in Supplement 2; Figure 2A). Of these genes, Kmt2a upregulation was the most reproducible across male cohorts and detected by quantitative reverse transcription polymerase chain reaction even at a later developmental time point (PND117, p = .007) (Figure 2B). Kmt2a in the NAc of PTE-exposed adult female rats was unchanged (Figure S3 in Supplement 1). An increase in Kmt2a was also observed at the protein level in adult male offspring at PND62 (p = .03) (Figure 2C). Kmt2a is a histone-H3-lysine-4 (H3K4) methyltransferase known to mediate chromatin modifications often associated with transcriptional activation (51–54) and has a documented role in the regulation of genes that are critical for brain development (55).
Figure 2. Increased NAc Kmt2a expression due to PTE is associated with impaired motivational behavior and dysregulation of genes functionally related to neuronal development and plasticity.
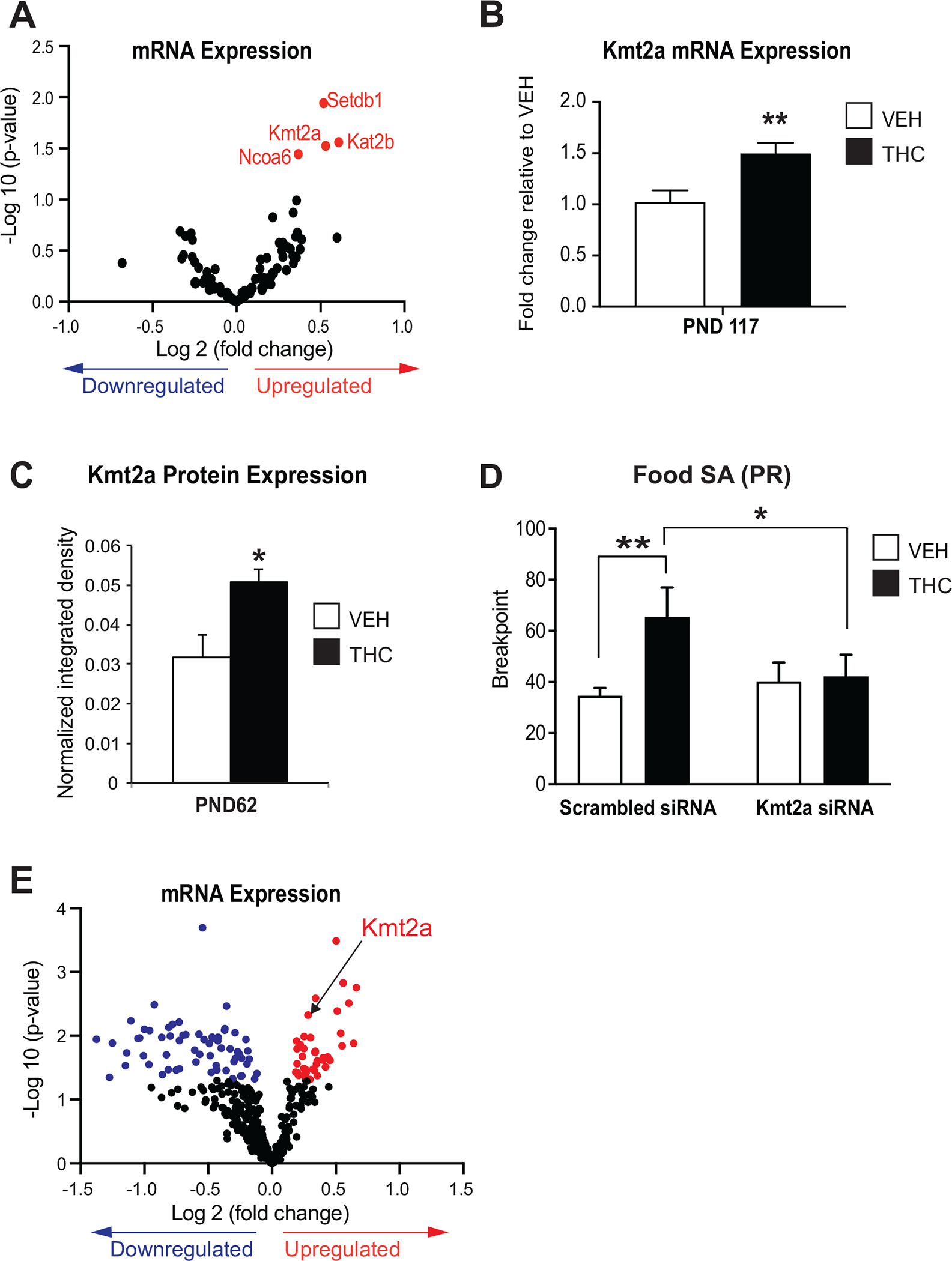
(A) Changes in the mRNA levels of epigenetic regulators. Red dots: upregulated genes at p<0.05. N=5–6/group, PND62. (B) Kmt2a mRNA expression throughout development and (C) protein level in young adult offspring. (D) PR food SA breakpoint in siRNA-infused animals, 24h post-infusion. N=4–6/group. Data represented as mean ± SEM. *:p <0.05. (E) Gene expression alterations in adult rats with prenatal THC. Blue dots: downregulated genes, red dots: upregulated genes at p<0.05. N=6–10/group, PND62.
To investigate the functional consequences of Kmt2a dysregulation in the NAc, we used a small interfering RNA–mediated knockdown strategy to alter Kmt2a levels (Figure S1 in Supplement 1). Four months after the initial PR food self-administration behavioral testing (Figure 1B, C), rats with intra-NAc infusions of the negative control small interfering RNA retained their prior PR behavior, with those exposed to prenatal THC showing higher PR breakpoint (F1,9 = 9.8756, p < .01) (Figure 2D). Interestingly, in the THC group, Kmt2a knockdown resulted in a significant reduction in PR breakpoint (F1,10 = 5.6382, p < .05) (Figure 2D). Kmt2a manipulation modulated motivational behavior and did not affect general locomotion (data not shown). In addition, in a separate study, NAc Kmt2a knockout mice showed altered FR5 responding in males (Figure S4 in Supplement 1). These results suggest that abnormal Kmt2a levels induced by PTE are directly related to the observed motivational disturbances.
PTE Is Associated With Enduring Transcriptomic Alterations Relevant to Synaptic Plasticity and Neuropsychiatric Disorders
In view of the pivotal role of NAc in emotional processing and the known role of Kmt2a, we conducted a larger screen, using the NanoString nCounter technology, to measure mRNA levels of 400 genes relevant to various neuropsychiatric phenotypes and neurobiological processes (Table S2 in Supplement 2), yielding 42 upregulated and 63 downregulated genes altered by PTE (Figure 2E; Table S4 in Supplement 2). Functionally, the affected genes related to neurotransmitter systems including glutamate and GABAergic (gamma-aminobutyric acidergic) receptors, multiple ion channels, zinc transporters, calcium sensors, and genes encoding cytoarchitectural and other cellular components related to structural integrity. This screen also confirmed, in another independent animal cohort, the significant upregulation of Kmt2a.
To gain an unbiased insight into the long-term dysregulation of NAc due to PTE, RNA-seq was used to explore the transcriptome (Figure 3). Principal component analysis (Figure S5A in Supplement 1) was performed on the RNA-seq counts, and some separation by treatment was observed. As expected, clustering based on treatment was more pronounced when only differentially expressed genes (DEGs) were included in a heatmap (Figure 3A). To ensure rigor, we used DEGs shared at p < .05 by both DESeq2 (Figure 3B; Table S5 in Supplement 2) and Voom-limma (Figure 3C; Table S6 in Supplement 2) in all downstream analyses, given that the two approaches showed good agreement (Figure S5B, C in Supplement 1). This combined approach identified 600 upregulated and 808 downregulated genes. Fisher’s exact test showed statistically significant overlap between the RNA-seq and NanoString datasets, both for upregulated (odds ratio [OR] = 6.47, p = .0002) and downregulated (OR = 4.73, p = 3.95 3 1027 ) genes.
Figure 3. Consequences of PTE on the NAc transcriptome and affected biological pathways.
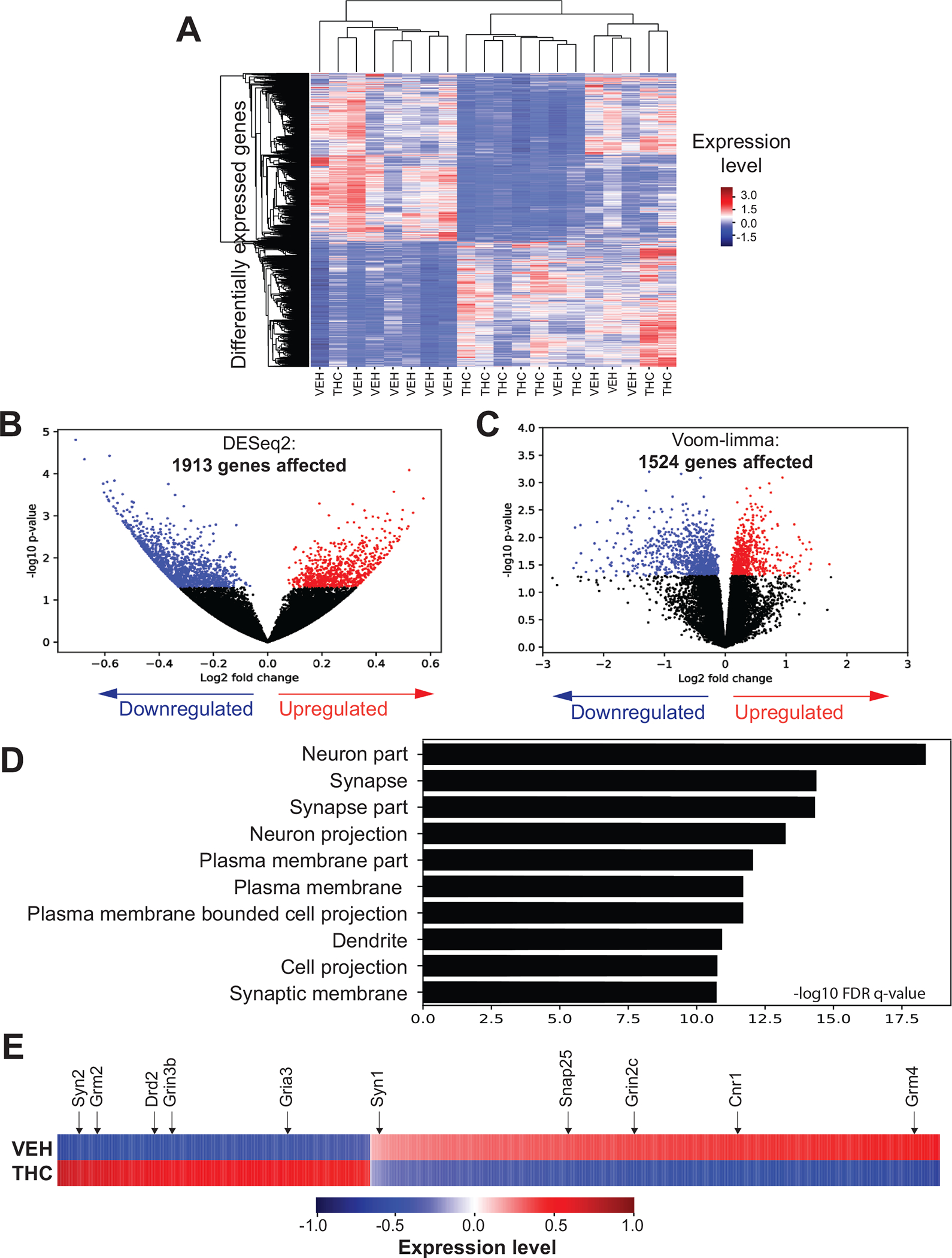
(A) Hierarchical clustering of RNA-seq reads for all DEGs (vertical dimension) and samples (horizontal dimension), PND62. (B, C) Distribution of DEGs obtained using the DESeq2 and Voom-limma analysis tools. Each dot corresponds to a single gene; colors indicate significant up-(red) or down-(blue) regulation. (D) Gene ontology analysis using GOrilla on DEGs shared by the DESeq2 and Voom-limma analyses; graph represents the top 10 categories within “cellular component”. (E) Heatmap showing changes in the RNA expression level of DEGs within the above top 10 categories. Arrows point to several interesting genes involved in synaptic regulation.
To address the biological significance of the observed transcriptome alterations, gene ontology analysis was performed on all 1408 DEGs, revealing an impact of PTE on synaptic function and neuronal communication (Figure 3D; Table S7 in Supplement 2). Genes in the top 10 enriched ontologies included cannabinoid receptor 1 (Cnr1), glutamate receptors (Grin3b, Grin2c, Gria3, Grm2, Grm4), dopamine receptor 2 (Drd2), and proteins crucial for neurotransmitter release (Snap25, Syn1, Syn2) (Figure 3E). The NanoStringbased assay in a different cohort of animals similarly showed a tendency for THC-related decrease of gene expression (Figure 2E), suggesting repressive effects of PTE on the NAc transcriptome.
PTE Is Associated With Complementary Epigenomic Alterations Relevant to Synaptic Plasticity and Psychiatric Disorders
To identify genomic loci directly relevant to Kmt2a, we conducted a ChIP-seq assay in the NAc of adult rats with PTE for H3K4me3 (H3K4-trimethylation), the methylation target of Kmt2a present at active promoters contributing to the recruitment of transcriptional machinery. Significant differences in H3K4me3 enrichment were identified at 2813 loci (Table S8 in Supplement 2). The majority of affected loci fell within proximal promoter regions of protein coding genes located <1 kb from the transcription start site (Figure 4A). No major shift in the location of H3K4me3 peaks was observed (Figure 4B), indicating that the THC-mediated chromatin modification likely affects the density of histones within a particular region rather than changing their specific positions. Figure 4C provides examples of differential ChIP-seq peaks within several genes related to cannabis and synaptic neurobiology (Cnr1, Camk2d, Gabbr2, Shank1, Kcnj3) in individual animals. The specificity of H3K4me3 enrichment in proximal promoter regions of Cnr1 and Camk2d is illustrated in Figure S6 in Supplement 1.
Figure 4. PTE changes the epigenomic profile of the NAc at gene loci functionally relevant to the regulation of synaptic plasticity.
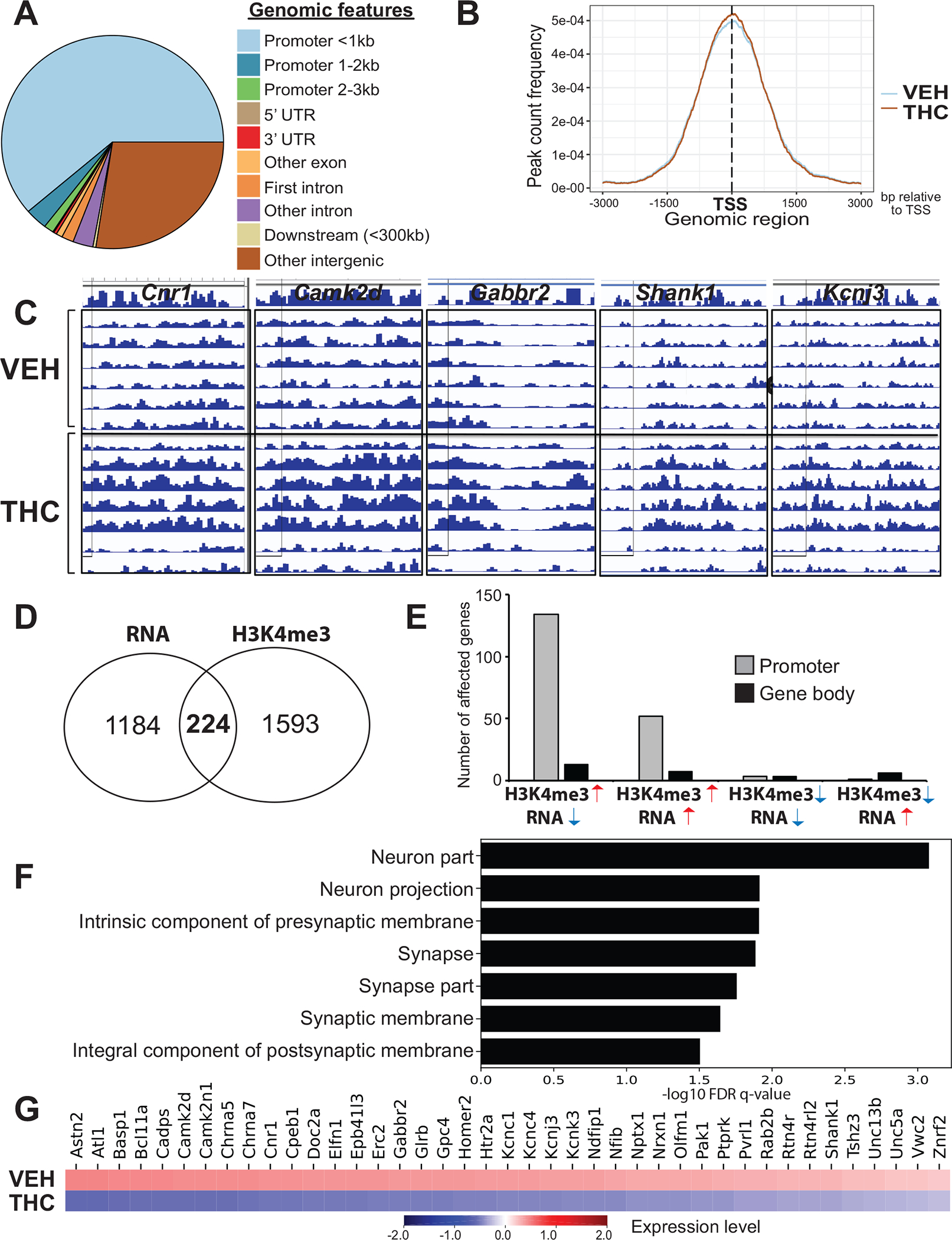
(A) Enrichment of different genomic features within chromatin domains affected by THC. N=6–7/group. (B) No significant THC-related shift in the distribution of H3K4me3 peaks relative to the transcriptional start sites of affected genes. (C) ChIP-seq peaks for five genes related to cannabis and synaptic neurobiology. Individual rows correspond to samples within the VEH and THC groups. (D) Comparison between genes affected on the mRNA (RNA-seq) and H3K4me3 (ChIP-seq) level. Numbers inside the Venn diagram indicate altered expression regardless of direction of change. (E) Relationship between mRNA transcript and H3K4me3 level changes at different gene loci. Note the higher abundance of affected promoters vs. coding regions. (F) Gene ontology analysis on “H3K4me3 up/RNA down”; graph represents the top 10 “cellular component” categories and heatmap (G) showing the affected genes.
To examine genes where mRNA expression and H3K4me3 enrichment are affected by PTE, we identified genes that contained a differential H3K4me3 enrichment site and were differentially expressed, yielding 224 genes (Figure 4D). We stratified these overlapping genes by direction in either dataset (H3K4me3-up/RNA-up, H3K4me3-down/RNA-up, H3K4me3- up/RNA-down, H3K4me3-down/RNA-down), with counts of genes whose differential H3K4me3 sites were contained in the gene promoter or gene body (Figure 4E; Table S9 in Supplement 2). Interestingly, the highest number of overlapping genes were associated with increased H3K4me3 enrichment and decreased mRNA expression (i.e., H3K4me3-up/RNA-down) at promoter regions. Fisher’s exact test further confirmed that out of the four overlaps, only H3K4me3-up/RNA-down showed statistical significance (OR = 2.004, p = 5.53 3 10212). Gene ontology analysis of this particular subset of genes identified strong enrichment of synaptic regulatory mechanisms (Figure 4F; Table S10 in Supplement 2). This extends our prior findings from the NanoString and RNA-seq assays showing reduced expression by PTE of genes involved in regulation of synaptic plasticity by demonstrating that these same genes are also differentially enriched for H3K4me3 binding (Figure 4G). We further investigated this H3K4me3-up/RNA-down set of genes by conducting a gene ontology analysis of all genes with H3K4me3 enrichment (Table S16 in Supplement 2) and identified enriched ontologies (false discovery rate-corrected) such as negative regulation of transcription by RNA polymerase II (p = .0441), negative regulation of nucleic acid–templated transcription (p = .0447), and negative regulation of transcription, DNA templated (p = .0457). These ontologies included 118 unique genes, 14 of which were also downregulated in the CRISPR model (Supplement). This demonstrates that H3K4me3 is enriched in genes that repress transcription, giving a functional explanation for H3K4me3-up/RNA-down being the largest and only statistically significant overlapping gene set. There were no significant ontologies identified in the H3K4me3-up/RNA-up list, which was the next most abundant category, but many genes falling in this group relate to functions such as skeletal muscle satellite cell activation, extracellular matrix, splicing, and transcription.
Specific Kmt2a Upregulation in Neuronal Cells Results in Molecular Disturbances Similar to PTE Effects
The neurobiological consequences of PTE develop over the period between conception and adulthood. As such, molecular mechanisms can be challenging to untangle, even using wellcontrolled in vivo animal models. To explore specific associations to Kmt2a dysregulation strictly in a neuronal context without the influence of environmental variables, we developed a CRISPR activation–based overexpression model of endogenous Kmt2a upregulation in human neuronal progenitor cells from male patients (Figure 5A) and identified specific changes in the cellular transcriptome (Figure S7A, B in Supplement 1; Tables S11 and S12 in Supplement 2), reflected in strong hierarchical clustering by treatment (Figure 5B). Comparison of our PTE-related RNA-seq dataset with the consequences of specific Kmt2a upregulation in the CRISPR model revealed, again, robust alterations of pathways related to synaptic mechanisms and neurotransmission (Figure S7C, D in Supplement 1; Table S13 in Supplement 2), demonstrating that Kmt2a indeed plays a direct role in these functional impairments. On a global level, Fisher’s exact test showed that for a given gene, differential expression in the rat NAc due to PTE is associated with 38% higher odds of also being a DEG in the CRISPR activation RNA-seq data (OR = 1.38, p < .0001).
Figure 5. Specific KMT2A upregulation is associated with similar complex alterations to PTE.
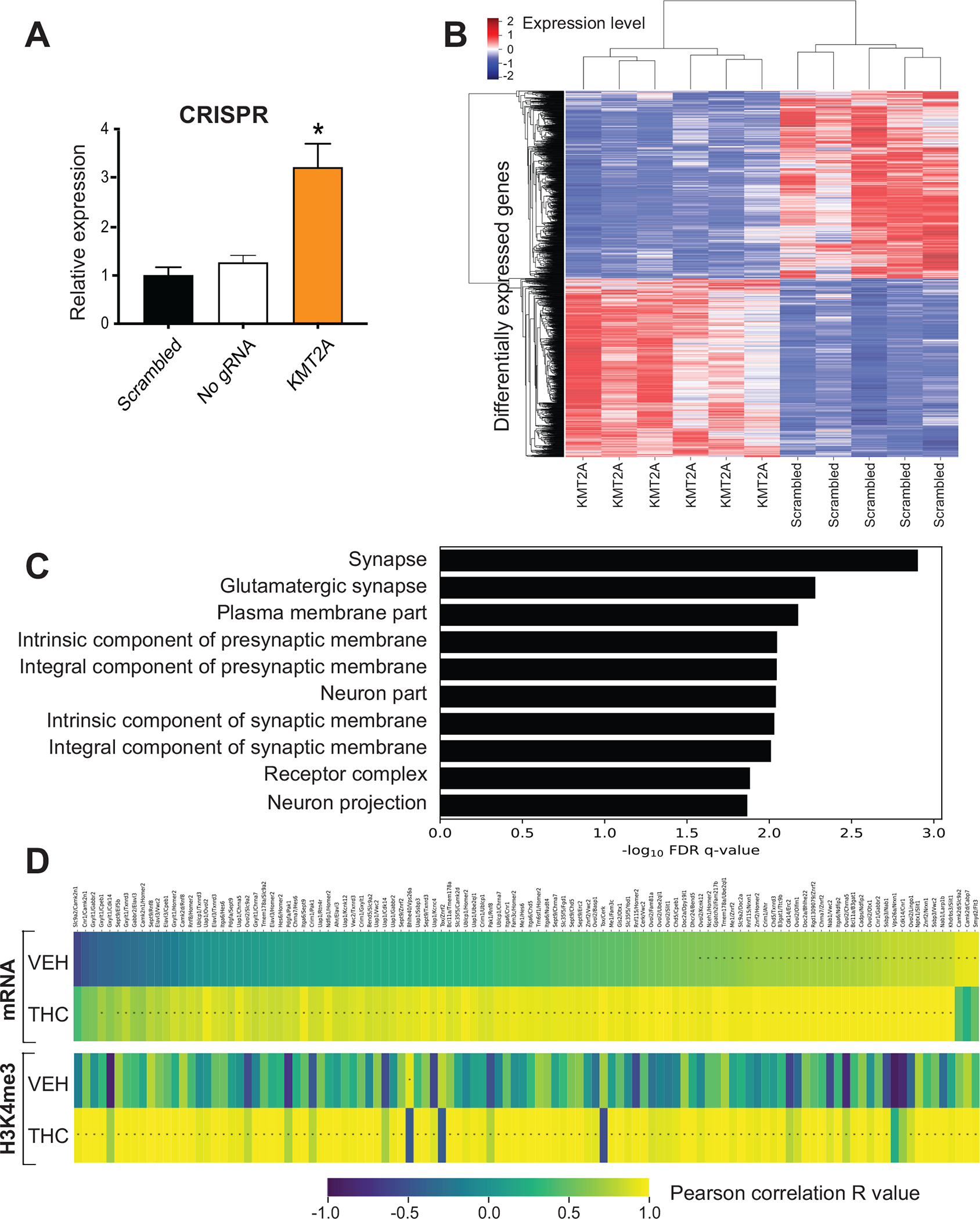
(A) KMT2A upregulation achieved using CRISPR in Neuronal Progenitor Cells. Normalized relative mRNA levels of Kmt2a (orange) compared to Scrambled gRNA control (black) following transduction of dCas9VPR in NPCs. (B) Hierarchical clustering of RNA-seq reads for all DEGs (vertical dimension) and CRISPR samples (horizontal dimension). N=5–6/group. (C) Ontological categories affected in gene-gene relationships observed in our different datasets. (D) Altered pairwise correlations on the mRNA and H3K4me3 levels in the prenatally THC-exposed NAc. Colors correspond to Pearson R values for each gene pair.
PTE and Kmt2a Upregulation Are Associated With Common Differential Gene Correlations and Hubs Within Gene Regulatory Networks
To address more complex gene-gene regulatory relationships in relation to Kmt2a and PTE, differential gene correlation analysis was conducted separately on the PTE RNA-seq, ChIP-seq, and CRISPR RNA-seq datasets (Table S14 in Supplement 2). To highlight overlapping regulatory relationships perturbed in these three datasets, we identified genegene pairs that were differentially correlated in more than one dataset. There were 1213 differential correlations common to two datasets, representing 591 unique genes. Gene ontology analysis on these 591 genes showed enrichments associated with glutamatergic synapse, postsynaptic density, synaptic membranes, and neuron part (Figure 5C). Of the 1213 differential correlations, 829 overlapped between the PTE ChIP-seq and CRISPR RNA-seq datasets, 146 between the PTE RNA-seq and CRISPR RNA-seq datasets, and 238 between the PTE ChIP-seq and PTE RNA-seq datasets. Of the 238 overlapping differential correlations between the PTE ChIP-seq and PTE RNA-seq datasets, 116 were between pairs of genes that were both part of the H3K4me3-up/RNA-down subset previously mentioned. These 116 gene pairs show robust THC-induced increases in correlation magnitude in both the RNA-seq and ChIP-seq datasets, demonstrating common, multiscale gene regulatory alterations by THC on both the gene expression and H3K4me3 enrichment levels (Figure 5D). These analyses demonstrate significant overlap of gene-gene regulatory relationships between experimental contexts (i.e., PTE and Kmt2a upregulation) and scales of analysis (i.e., gene expression, H3K4me3 enrichment).
To gain a deeper understanding of the molecular dynamics of Kmt2a in the contexts of PTE and Kmt2a upregulation, we conducted network analyses using the STRING protein-protein interaction database (Figure 6A). First, a Kmt2a reference network was built based on known protein-protein interactions between Kmt2a and its interaction partners. Second, we constructed networks based on DEGs from the CRISPR Kmt2a overexpression dataset composing the top 10 enriched gene ontology categories for Biological Process and Cellular Component. Third, we constructed a network based on the DEGs from the PTE RNA-seq dataset in the NAc. These three data-driven networks collectively included 24 proteins overlapping with the Kmt2a reference network (Table S15 in Supplement 2), demonstrating that both the PTE and Kmt2a upregulation contexts recruit many known interaction partners of Kmt2a that are part of its molecular complex. The overlapping nodes as they appear in the Kmt2a reference network are shown in Figure 6B (for full network, see Figure S8 in Supplement 1). Several of the 24 overlapping proteins between the Kmt2a and two data-driven networks have well-known functions in the regulation of chromatin modification (e.g., Kmt2b, Hdac1, Ncoa3, Kat2b, Wdr5, histone variants) or regulate different steps of the transcriptional process in concert with Kmt2a (e.g., Mta2, Tp53, Ruvbl2, Cited2). Many genes altered by PTE and Kmt2a overexpression showed comparable and consistent direction of change (Figure 6C), emphasizing the central role of Kmt2a in the network of developmental epigenetic processes influenced by PTE. This analysis also highlighted several genes with consistent expression alterations involved in well-characterized repressive transcriptional processes (H2afx, Ncoa3, Ago2, Mef2a, Cited2, Mta2), which might explain the prevalence of transcriptional downregulation by PTE (Figure 6C). To identify potential mechanisms of how these epigenetic and transcriptional regulators affect synaptic regulation and plasticity, we examined their interaction partners in our data-driven networks in relation to the gene ontology categories enriched for the overlapping DEGs between the PTE and CRISPR datasets (Figure S7C, D in Supplement 1). Several genes related to synaptic plasticity were interaction partners of the epigenetic/transcriptional regulators: Bcl11a, Dhcr24, Insig1, Klf5, Mef2c, Nup210, Smad3, Stat6, and Thbs1. This analysis demonstrates thatknown interaction partners of Kmt2a are recruited in both the PTE and Kmt2a upregulation contexts and interact with genes with known involvement in the regulation of synaptic plasticity.
Figure 6. Affected Kmt2a gene network based on specific Kmt2a dysregulation and PTE.
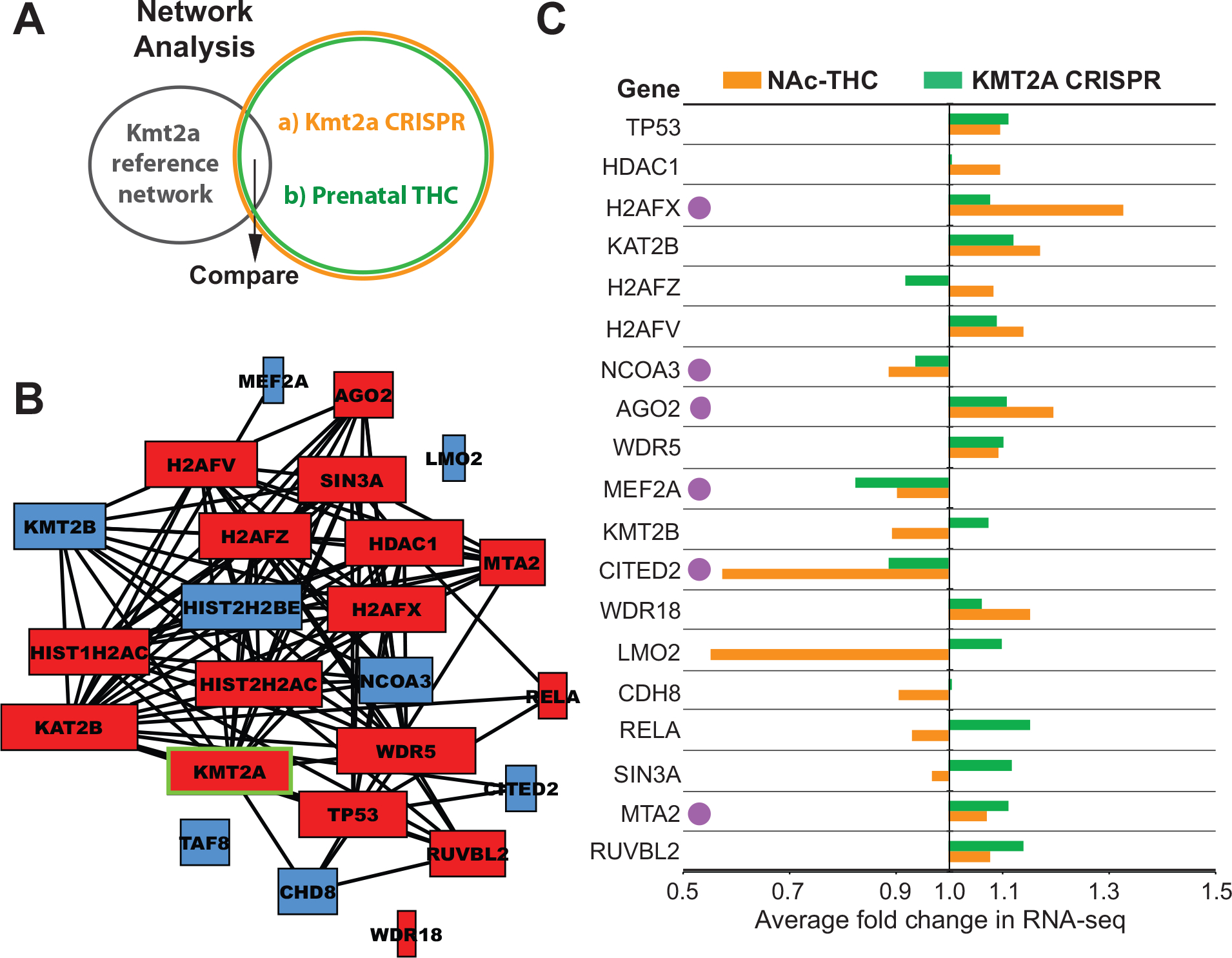
(A) Network analysis approach. (B) Kmt2a network identified. Colors indicate genes dysregulated either in our prenatal THC or CRISPR Kmt2a overexpression studies. Red: upregulation, blue: downregulation. (C) mRNA level changes of genes within the affected Kmt2a network due to prenatal THC (green bars) and specific Kmt2a upregulation by CRISPR (orange bars). Purple dots indicate potentially interesting repressors.
Gene Dysregulation Patterns Associated With PTE and Kmt2a Overexpression Are Present in Human MDD
We explored whether transcriptional networks induced by PTE or Kmt2a overexpression were also evident in human MDD because key behavioral effects observed in PTE animals included increased depressive-like phenotypes and altered hedonic state. Using publicly available RNA-seq and microarray datasets from human patients with MDD (the NAc, anterior insula, subiculum, and Brodmann areas 11, 25, and 89), we measured the overlap of DEGs in these datasets with the 591 genes mentioned previously that composed 1213 differential correlations that occurred in two of our sequencing datasets (i.e., PTE ChIP-seq and RNA-seq, Kmt2a upregulation RNA-seq). We filtered DEGs from the human datasets and calculated the percentage of DEGs overlapping with our set of 591 genes. In the PTE and Kmt2a upregulation RNA-seq datasets, 19.8% and 12.2% of DEGs overlapped with our set of 591 genes, respectively. Thus, there appears to be a core subset of genes related to synaptic plasticity that are differentially expressed and differentially correlated in PTE, Kmt2a upregulation, and MDD. Moreover, in relation to genes potentially involved in transcriptional repressive mechanisms (Figure 6C), there was consistency in the direction of their expression in the NAc between our PTE and Kmt2a conditions and the human MDD datasets (Figure S9 in Supplement 1), along with some cortical regions.
Discussion
We demonstrated a relationship between PTE and long-term perturbations in natural reward–related motivation and anhedonia, endophenotypes that can be present in depression and other psychiatric disorders, and stress sensitivity concomitant with disturbances in the epigenomic regulation of the NAc. Our findings revealed that PTE-induced epigenomic dysregulation relates to histone methyltransferase Kmt2a and downstream synaptic plasticity–related transcriptional impairments that altogether converge on a network of repressive transcriptional processes implicated in MDD.
The observations that PTE leads to increased motivation for natural reward in adulthood are consistent with the literature that emphasizes long-term influence of developmental THC on reinforcement and motivation for drug and natural rewards (56–58). Our study now provides sex-specific molecular evidence that PTE increases expression of Kmt2a in the NAc (Figure S3 in Supplement 1) that persists into adulthood and that normalization of Kmt2a levels can normalize motivation for natural reward in adult male offspring. NAc Kmt2a knockout mice showed sex-specific altered FR5 responding (Figure S4 in Supplement 1). Similar to previous findings (10,59–61), we observed that PTE induced learned helplessness and anhedonic phenotypes in males lasting into adulthood. Moreover, stress is known to negatively affect individuals depending on compromised physiological and emotional states, and our findings suggest that PTE, in combination with stressful experiences, shifts the normal functioning toward distress, resulting in increased vulnerability to endophenotypes such as anhedonia present in depression and other psychiatric disorders. Thus, consistent with a “double-hit” hypothesis (62), PTE (first hit) and postnatal stressful conditions (second hit) may exacerbate negative affect and psychiatric vulnerability. Important for future studies is in-depth interrogation of sex and brain region specificity of Kmt2a regulation.
Various lines of evidence emphasize synaptic dysregulation in psychiatric disorders, and our study revealed robust longterm effects of PTE on the regulation of synaptic plasticity that directly related to the Kmt2a network. Kmt2a is an epigenetic regulator that affects multiple forms of histone methylation and is required for both neurogenesis (54) and normal synaptic plasticity in the adult brain (63). Moreover, the CB1 receptor, which mediates the initial cascade of events induced by PTE, is well known to regulate synaptic transmission by inhibiting calcium channels and activating potassium channels (64–67). Accumbal expression of these ion channels were altered not only after PTE but also by Kmt2a upregulation, implicating this epigenetic factor in the long-term synaptic plasticity alterations evident with PTE. Interestingly, the subset of genes strongly associated with synaptic plasticity within the differential gene correlation analysis for the PTE, and Kmt2a upregulation conditions also constituted a significant portion of the DEGs in subjects with MDD. These findings suggest common synaptic disturbances in PTE and MDD that are regulated by Kmt2a.
The unbiased data-driven networks also identified consistent alterations of genes (e.g., Ago2 and Cited2 in the NAc) involved in repressive transcriptional processes, which is intriguing, given the predominant decrease in gene expression associated with PTE and Kmt2a upregulation. Our data suggest that transcriptional repressors (e.g., H2afx, Mta2; increased), transcriptional activators (e.g., Cited2, Ncoa3; reduced), and posttranscriptional repressive mechanisms involving small RNA processing by Ago2 are perturbed by PTE. Importantly, these have important roles in synaptic regulation. For example, Mta2 (metastasis-associated 1 family member 2) is a subunit of NuRD histone deacetylase complex involved in synaptic development connectivity (68). Cited2 (Cbp/P300 interacting transactivator with Glu/Asp-rich carboxy-terminal domain 2) is a transactivator involved in neuronal activity–dependent long-term potentiation (69). Ncoa3 (nuclear receptor coactivator-3) is a histone acetyltransferase that influences neuroplasticity in a microRNAdependent manner via regulating Ago2 (70). Importantly, many key Kmt2a network components were also dysregulated in human MDD, emphasizing a translational connection between PTE and depressive-like phenotypes.
Overall, our findings support a direct role of PTE to enhance neuropsychiatric risk as identified in human longitudinal studies (16) including motivational disturbances, stress sensitivity, and depressive-like phenotypes. We emphasize a role of Kmt2a dysregulation to induce behavioral changes underlying long-term neuropsychiatric vulnerability and demonstrate an epigenetic signature linked to molecular disturbances of synaptic plasticity.
Supplementary Material
KEY RESOURCES TABLE.
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Antibody | anti-Kmt2a primary antibody | Bethyl | 374A | |
| Antibody | anti-Cas9 | Millipore | 7A9 | |
| Antibody | β-Actin | Ambion | 1406030 | |
| Cell line | human neural progenitor cells | Donor from male control patient | NSB2607 | HiPSC-derived cell line. Collected as fibroblasts, reprogrammed into NPCs. More information here: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM2843554 |
| Chemical Compound or Drug | Trizol | ThermoFisher | 15596026 | |
| Chemical Compound or Drug | 200 mg/ml THC stock | NIDA | N/A | |
| Commercial Assay Or Kit | RNAqueous-micro kit | Ambion | AM1931 | |
| Commercial Assay Or Kit | TaqMan assay | ThermoFisher | Rn01444748_m1 | |
| Commercial Assay Or Kit | nCounter array | Nanostring | N/A | |
| Commercial Assay Or Kit | DNAse treatment | ThermoFisher | EN0525 | |
| Commercial Assay Or Kit | Bioanalyzer | Agilent | G2939BA | |
| Commercial Assay Or Kit | uQuant plate reader | Biotek Instruments | UNK446 | |
| Commercial Assay Or Kit | NovaSeq 6000 S1 flowcell PE50 | Illumina | N/A | |
| Commercial Assay Or Kit | Qubit instrument | ThermoFisher | Q33239 | |
| Deposited Data; Public Database | Gene Expression Omnibus | NCBI | GSE87610, GSE102556 | |
| Genetic reagent | ON-TARGETplus SMART pool, rat Kmt2a, | Horizon Discovery | L-100995–02-0005 | |
| Genetic reagent | ON-TARGET nontargeting pool | Horizon Discovery | D-001810–10-05 | |
| Genetic reagent | lentiCRISPR v2 | Addgene | plasmid # 52961 | |
| Genetic reagent | Guide RNAs | Benchling | N/A | |
| Genetic reagent | AAV8-hSyn-Cre-GFP | https://www.med.unc.edu/genetherapy/vectorcore/in-stock-aav-vectors/reporter-vectors/ | N/A | |
| Transfection Reagent | JetSI transfection reagent | Polyplus Transfection | N/A | |
| Organism/Strain | Long-Evans female dams | Charles River Laboratories | N/A | |
| Organism/Strain | Mice (Kmt2a2lox/2lox) | Bred in-house | ||
| Software; Algorithm | Gen5 | Biotek Instruments | RRID:SCR_017317 | |
| Software; Algorithm | Subread | PMID:23558742 | RRID:SCR_009803 | |
| Software; Algorithm | DESeq2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html | RRID:SCR_015687 | |
| Software; Algorithm | Voom-limma | http://bioinf.wehi.edu.au/limma/ | RRID:SCR_010943 | |
| Software; Algorithm | Bowtie | PMID:19261174 | RRID:SCR_005476 | |
| Software; Algorithm | SAMtools | PMID:21903627 | RRID:SCR_002105 | |
| Software; Algorithm | MACS | PMID:18798982 | RRID:SCR_013291 | |
| Software; Algorithm | FastQC | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | RRID:SCR_014583 | |
| Software; Algorithm | diffReps | https://doi.org/10.1371/journal.pone.0065598 | RRID:SCR_010873 | |
| Software; Algorithm | ChIPseeker | http://bioconductor.org/packages/devel/bioc/vignettes/ChIPseeker/inst/doc/ChIPseeker.html#:~:text=ChIPseeker%20is%20an%20R%20package,peaks%20binding%20to%20TSS%20regions.&text=Currently%2C%20ChIPseeker%20contains%2017%2C000%20bed%20file%20information%20from%20GEO%20database. | N/A | |
| Software; Algorithm | HISAT2 | PMID:25751142 | RRID:SCR_015530 | |
| Software; Algorithm | STRING | PMID:21045058 | RRID:SCR_005223 |
Acknowledgements
We thank Nayana Patel and Lyla Parvez for technical assistance. Our research was supported by NIH grant DA030359 for Y.L.H. and a NIDA-Inserm Postdoctoral Fellowship for A.B.
Footnotes
Financial Disclosures
The authors reported no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ko JY, Farr SL, Tong VT, Creanga AA, Callaghan WM (2015): Prevalence and patterns of marijuana use among pregnant and nonpregnant women of reproductive age. Am J Obstet Gynecol. 213:201.e201–201.e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volkow ND, Han B, Compton WM, McCance-Katz EF (2019): Self-reported Medical and Nonmedical Cannabis Use Among Pregnant Women in the United States. JAMA. 322:167–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young-Wolff KC, Sarovar V, Tucker LY, Conway A, Alexeeff S, Weisner C, et al. (2019): Self-reported Daily, Weekly, and Monthly Cannabis Use Among Women Before and During Pregnancy. JAMA Netw Open. 2:e196471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westfall RE, Janssen PA, Lucas P, Capler R (2009): Reprint of: survey of medicinal cannabis use among childbearing women: patterns of its use in pregnancy and retroactive self-assessment of its efficacy against ‘morning sickness’. Complement Ther Clin Pract. 15:242–246. [DOI] [PubMed] [Google Scholar]
- 5.Roberson EK, Patrick WK, Hurwitz EL (2014): Marijuana use and maternal experiences of severe nausea during pregnancy in Hawai’i. Hawaii J Med Public Health. 73:283–287. [PMC free article] [PubMed] [Google Scholar]
- 6.Blackard C, Tennes K (1984): Human placental transfer of cannabinoids. N Engl J Med. 311:797. [DOI] [PubMed] [Google Scholar]
- 7.Schou J, Prockop LD, Dahlstrom G, Rohde C (1977): Penetration of delta-9-tetrahydrocannabinol and 11-OH-delta-9-tetrahydrocannabinol through the blood-brain barrier. Acta Pharmacol Toxicol (Copenh). 41:33–38. [DOI] [PubMed] [Google Scholar]
- 8.Pertwee RG (2008): The diverse CB1 and CB2 receptor pharmacology of three plant cannabinoids: delta9-tetrahydrocannabinol, cannabidiol and delta9-tetrahydrocannabivarin. Br J Pharmacol. 153:199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abel E (1986): Publication trends for alcohol, tobacco, and narcotics in MEDLARS. Ann N Y Acad Sci. 477:103–104. [DOI] [PubMed] [Google Scholar]
- 10.Gray KA, Day NL, Leech S, Richardson GA (2005): Prenatal marijuana exposure: effect on child depressive symptoms at ten years of age. Neurotoxicol Teratol. 27:439–448. [DOI] [PubMed] [Google Scholar]
- 11.Leech SL, Larkby CA, Day R, Day NL (2006): Predictors and correlates of high levels of depression and anxiety symptoms among children at age 10. J Am Acad Child Adolesc Psychiatry. 45:223–230. [DOI] [PubMed] [Google Scholar]
- 12.Porath AJ, Fried PA (2005): Effects of prenatal cigarette and marijuana exposure on drug use among offspring. Neurotoxicol Teratol. 27:267–277. [DOI] [PubMed] [Google Scholar]
- 13.Goldschmidt L, Richardson GA, Willford JA, Severtson SG, Day NL (2012): School achievement in 14-year-old youths prenatally exposed to marijuana. Neurotoxicol Teratol 34:161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kooijman MN, Kruithof CJ, van Duijn CM, Duijts L, Franco OH, van IJzendoorn MH, et al. (2016): The Generation R Study: Design and cohort update 2017. Eur J Epidemiol 31:1243–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Genna NM, Richardson GA, Goldschmidt L, Day NL, Cornelius MD (2018): Prenatal exposures to tobacco and cannabis: Associations with adult electronic cigarette use. Drug Alcohol Depend 188:209–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank DA, Rose-Jacobs R, Crooks D, Cabral HJ, Gerteis J, Hacker KA, et al. (2011): Adolescent initiation of licit and illicit substance use: Impact of intrauterine exposures and post-natal exposure to violence. Neurotoxicol Teratol 33:100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loeber R, Farrington DP (2000): Young children who commit crime: Epidemiology, developmental origins, risk factors, early interventions, and policy implications. Dev Psychopathol 12:737–762. [DOI] [PubMed] [Google Scholar]
- 18.Patrick ME, Maggs JL, Greene KM, Morgan NR, Schulenberg JE (2014): The link between mother and adolescent substance use: Intergenerational findings from the British cohort study. Longit Life Course Stud 5:56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heron J, Barker ED, Joinson C, Lewis G, Hickman M, Munafò M, Macleod J (2013): Childhood conduct disorder trajectories, prior risk factors and cannabis use at age 16: Birth cohort study. Addiction 108:2129–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grant KS, Conover E, Chambers CD (2020): Update on the developmental consequences of cannabis use during pregnancy and lactation. Birth Defects Res 112:1126–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jutras-Aswad D, DiNieri JA, Harkany T, Hurd YL (2009): Neurobiological consequences of maternal cannabis on human fetal development and its neuropsychiatric outcome. Eur Arch Psychiatry Clin Neurosci 259:395–412. [DOI] [PubMed] [Google Scholar]
- 22.Morris CV, DiNieri JA, Szutorisz H, Hurd YL (2011): Molecular mechanisms of maternal cannabis and cigarette use on human neurodevelopment. Eur J Neurosci 34:1574–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howlett AC (2002): The cannabinoid receptors. Prostaglandins Other Lipid Mediat 68–69:619–631. [DOI] [PubMed] [Google Scholar]
- 24.Galve-Roperh I, Chiurchiù V, Díaz-Alonso J, Bari M, Guzmán M, Maccarrone M (2013): Cannabinoid receptor signaling in progenitor/ stem cell proliferation and differentiation. Prog Lipid Res 52: 633–650. [DOI] [PubMed] [Google Scholar]
- 25.Compagnucci C, Di Siena S, Bustamante MB, Di Giacomo D, Di Tommaso M, Maccarrone M, et al. (2013): Type-1 (CB1) cannabinoid receptor promotes neuronal differentiation and maturation of neural stem cells. PLOS ONE 8:e54271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keimpema E, Mackie K, Harkany T (2011): Molecular model of cannabis sensitivity in developing neuronal circuits. Trends Pharmacol Sci 32:551–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tortoriello G, Morris CV, Alpar A, Fuzik J, Shirran SL, Calvigioni D, et al. (2014): Miswiring the brain: D9-tetrahydrocannabinol disrupts cortical development by inducing an SCG10/stathmin-2 degradation pathway. EMBO J 33:668–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bara A, Ferland JN, Rompala G, Szutorisz H, Hurd YL (2021): Cannabis and synaptic reprogramming of the developing brain. Nat Rev Neurosci 22:423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Everitt BJ, Robbins TW (2005): Neural systems of reinforcement for drug addiction: From actions to habits to compulsion. Nat Neurosci 8:1481–1489. [DOI] [PubMed] [Google Scholar]
- 30.Girault JA (2012): Integrating neurotransmission in striatal medium spiny neurons. Adv Exp Med Biol 970:407–429. [DOI] [PubMed] [Google Scholar]
- 31.Koob GF, Volkow ND (2010): Neurocircuitry of addiction. Neuropsychopharmacology 35:217–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakovcevski M, Ruan H, Shen EY, Dincer A, Javidfar B, Ma Q, et al. (2015): Neuronal Kmt2a/Mll1 histone methyltransferase is essential for prefrontal synaptic plasticity and working memory. J Neurosci 35:5097–5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayer SA, Altman J, Russo RJ, Zhang X (1993): Timetables of neurogenesis in the human brain based on experimentally determined patterns in the rat. Neurotoxicology 14:83–144. [PubMed] [Google Scholar]
- 34.Paterson NE, Froestl W, Markou A (2004): The GABAB receptor agonists baclofen and CGP44532 decreased nicotine self-administration in the rat. Psychopharmacol (Berl) 172:179–186. [DOI] [PubMed] [Google Scholar]
- 35.Hodos W, Kalman G (1963): Effects of increment size and reinforcer volume on progressive ratio performance. J Exp Anal Behav 6:387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slattery DA, Cryan JF (2012): Using the rat forced swim test to assess antidepressant-like activity in rodents. Nat Protoc 7:1009–1014. [DOI] [PubMed] [Google Scholar]
- 37.McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, et al. (2011): HDAC3 is a critical negative regulator of longterm memory formation. J Neurosci 31:764–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aguilar-Valles A, Vaissière T, Griggs EM, Mikaelsson MA, Takács IF, Young EJ, et al. (2014): Methamphetamine-associated memory is regulated by a writer and an eraser of permissive histone methylation. Biol Psychiatry 76:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griggs EM, Young EJ, Rumbaugh G, Miller CA (2013): MicroRNA-182 regulates amygdala-dependent memory formation. J Neurosci 33:1734–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z (2009): GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. (2019): STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M (2008): Computing topological parameters of biological networks. Bioinformatics 24:282–284. [DOI] [PubMed] [Google Scholar]
- 43.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. (2003): Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenzie AT, Katsyv I, Song WM, Wang M, Zhang B (2016): DGCA: A comprehensive R package for Differential Gene Correlation Analysis. BMC Syst Biol 10:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arion D, Huo Z, Enwright JF, Corradi JP, Tseng G, Lewis DA (2017): Transcriptome alterations in prefrontal pyramidal cells distinguish schizophrenia from bipolar and major depressive disorders. Biol Psychiatry 82:594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Labonté B, Engmann O, Purushothaman I, Menard C, Wang J, Tan C, et al. (2017): Sex-specific transcriptional signatures in human depression. Nat Med 23:1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spano MS, Ellgren M, Wang X, Hurd YL (2007): Prenatal cannabis exposure increases heroin seeking with allostatic changes in limbic enkephalin systems in adulthood. Biol Psychiatry 61:554–563. [DOI] [PubMed] [Google Scholar]
- 48.DiNieri JA, Wang X, Szutorisz H, Spano SM, Kaur J, Casaccia P, et al. (2011): Maternal cannabis use alters ventral striatal dopamine D2 gene regulation in the offspring. Biol Psychiatry 70:763–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Porsolt RD, Anton G, Blavet N, Jalfre M (1978): Behavioural despair in rats: A new model sensitive to antidepressant treatments. Eur J Pharmacol 47:379–391. [DOI] [PubMed] [Google Scholar]
- 50.Porsolt RD, Bertin A, Jalfre M (1978): “Behavioural despair” in rats and mice: strain differences and the effects of imipramine. Eur J Pharmacol 51:291–294. [DOI] [PubMed] [Google Scholar]
- 51.Ford DJ, Dingwall AK (2015): The cancer COMPASS: Navigating the functions of MLL complexes in cancer. Cancer Genet 208:178–191. [DOI] [PubMed] [Google Scholar]
- 52.Cosgrove MS, Patel A (2010): Mixed lineage leukemia: A structure-function perspective of the MLL1 protein. FEBS Journal 277:1832–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang P, Bergamin E, Couture JF (2013): The many facets of MLL1 regulation. Biopolymers 99:136–145. [DOI] [PubMed] [Google Scholar]
- 54.Huang YC, Shih HY, Lin SJ, Chiu CC, Ma TL, Yeh TH, Cheng YC (2015): The epigenetic factor Kmt2a/Mll1 regulates neural progenitor proliferation and neuronal and glial differentiation. Dev Neurobiol 75:452–462. [DOI] [PubMed] [Google Scholar]
- 55.Akbarian S, Huang HS (2009): Epigenetic regulation in human brainfocus on histone lysine methylation. Biol Psychiatry 65:198–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Levin ED, Hawkey AB, Hall BJ, Cauley M, Slade S, Yazdani E, et al. (2019): Paternal THC exposure in rats causes long-lasting neurobehavioral effects in the offspring. Neurotoxicol Teratol 74:106806. [DOI] [PubMed] [Google Scholar]
- 57.Silva L, Zhao N, Popp S, Dow-Edwards D (2012): Prenatal tetrahydrocannabinol (THC) alters cognitive function and amphetamine response from weaning to adulthood in the rat. Neurotoxicol Teratol 34:63–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szutorisz H, Dinieri JA, Sweet E, Egervari G, Michaelides M, Carter JM, et al. (2014): Parental THC exposure leads to compulsive heroinseeking and altered striatal synaptic plasticity in the subsequent generation. Neuropsychopharmacology 39:1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fried PA, Watkinson B (1990): 36- and 48-month neurobehavioral follow-up of children prenatally exposed to marijuana, cigarettes, and alcohol. J Dev Behav Pediatr 11:49–58. [PubMed] [Google Scholar]
- 60.Day NL, Leech SL, Goldschmidt L (2011): The effects of prenatal marijuana exposure on delinquent behaviors are mediated by measures of neurocognitive functioning. Neurotoxicol Teratol 33: 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.United States Department of Health and Human Services (2014): The health consequences of smoking–50 years of progress: a report of the surgeon general. Public Health Service, Office of the Surgeon General. Rockville, MD: United States Department of Health and Human Services, Public Health Service, Office of the Surgeon General. [Google Scholar]
- 62.Richardson KA, Hester AK, McLemore GL (2016): Prenatal cannabis exposure—The “first hit” to the endocannabinoid system. Neurotoxicol Teratol 58:5–14. [DOI] [PubMed] [Google Scholar]
- 63.Kerimoglu C, Sakib MS, Jain G, Benito E, Burkhardt S, Capece V, et al. (2017): KMT2A and KMT2B mediate memory function by affecting distinct genomic regions. Cell Rep 20:538–548. [DOI] [PubMed] [Google Scholar]
- 64.Ohno-Shosaku T, Maejima T, Kano M (2001): Endogenous cannabinoids mediate retrograde signals from depolarized postsynaptic neurons to presynaptic terminals. Neuron 29:729–738. [DOI] [PubMed] [Google Scholar]
- 65.Wilson RI, Nicoll RA (2001): Endogenous cannabinoids mediate retrograde signalling at hippocampal synapses. Nature 410:588–592. [DOI] [PubMed] [Google Scholar]
- 66.Guo J, Ikeda SR (2004): Endocannabinoids modulate N-type calcium channels and G-protein-coupled inwardly rectifying potassium channels via CB1 cannabinoid receptors heterologously expressed in mammalian neurons. Mol Pharmacol 65:665–674. [DOI] [PubMed] [Google Scholar]
- 67.Zou Z, Lu Y, Zha Y, Yang H (2016): Endocannabinoid 2- arachidonoylglycerol suppresses LPS-induced inhibition of A-type potassium channel currents in caudate nucleus neurons through CB1 receptor. J Mol Neurosci 59:493–503. [DOI] [PubMed] [Google Scholar]
- 68.Yamada T, Yang Y, Hemberg M, Yoshida T, Cho HY, Murphy JP, et al. (2014): Promoter decommissioning by the NuRD chromatin remodeling complex triggers synaptic connectivity in the mammalian brain. Neuron 83:122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Esvald EE, Tuvikene J, Sirp A, Patil S, Bramham CR, Timmusk T (2020): CREB family transcription factors are major mediators of BDNF transcriptional autoregulation in cortical neurons. J Neurosci 40:1405– 1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sananbenesi F, Fischer A (2015): New friends for Ago2 in neuronal plasticity. EMBO J 34:2213–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


