Abstract
Lipid-soluble tanshinone is one of the main bioactive substances in the medicinal plant Salvia miltiorrhiza, and its medicinal demand is growing rapidly. Yeast extract (YE) modulates the tanshinone biosynthesis, but the underlying regulatory network remains obscure. In this study, a YE-responsive transcriptional factor Scarecrow1 (SCR1) was identified in S. miltiorrhiza from the YE-induced transcriptome dataset. SmSCR1 is located in the nucleus. Overexpression of SmSCR1 in S. miltiorrhiza roots resulted in a significantly higher accumulation of tanshinone than the control, with the highest 1.49-fold increase. We also detected upregulation of tanshinone biosynthetic genes, SmSCR1 and SmHMGR1, and distinct alteration of growth and development of the hairy roots in the overexpression lines compared to the control. An inverse phenotype was observed in SmSCR1-SRDX suppression expression lines. We found that SmSCR1 can bind to the promoter of SmCPS1 to induce its expression. This study provides new insight into the regulatory mechanism on the growth and development of hairy roots, tanshinone accumulation, and the metabolic engineering of bioactive compounds in S. miltiorrhiza.
Keywords: Salvia miltiorrhiza, GRAS transcription factor, tanshinone accumulation, hairy root growth, regulatory mechanism
Introduction
Salvia miltiorrhiza Bunge is a Chinese herbal medicine that belongs to the Salvia genus in the Lamiaceae family (Jia et al., 2019; Jung et al., 2020). In Asian countries, dried roots and stems of S. miltiorrhiza are a common treatment for cardiovascular system-related diseases (Hao et al., 2015). The bioactivity of S. miltiorrhiza is associated with lipid-soluble tanshinones, including tanshinone I (TA-I), cryptotanshinone (CT), dihydrotanshinone (DT), and tanshinone IIA (TA-II) (Fu et al., 2020). Increasing attention has been given to the abovementioned compounds in recent years, mainly focusing on improving the yield of tanshinone in S. miltiorrhiza.
Tanshinones are a type of diterpenoids produced in two phases in S. miltiorrhiza roots (Supplementary Figure 1). First, common terpenoid precursors [e.g., isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP)] are generated by two distinct processes (e.g., mevalonate (MVA) pathway in cytosol and 2-C-Methyl-D-erythritol-4-phosphate (MEP) pathway in plastids) (Ma et al., 2012). The universal diterpenoid precursor, geranylgeranyl diphosphate (GGPP), is then biosynthesized GGPP synthase (GGPPS) (Shi et al., 2016a,b). Then, three known synthases, namely, copalyl diphosphate synthase 1 (CPS1), kaurene synthase-like 1 (KSL1), and miltiradiene oxidase (CYP76AH1), and a still-unidentified enzyme(s) produce tanshinones (Gao et al., 2009; Xing et al., 2018). Overexpression or downregulation of one or two synthases significantly impacts tanshinone synthesis (Kai et al., 2011; Wang et al., 2015; Ma et al., 2016). Plant transcription factors (TFs) play important regulatory roles in resistance to stress and metabolic engineering. TFs can regulate one or multiple biosynthetic genes from a single or multiple pathways (Xu et al., 2016; Wang et al., 2017; Yang et al., 2017). TFs are a diverse group of genes including many families and subfamilies, such as bHLH, MYB, ERF, bZIP, and GRAS (Deng et al., 2020a,b; Hao et al., 2020; Shi et al., 2021a,b; Zhou et al., 2021a,b). Less is known about the regulatory actions or target genes of the GRAS TFs, which play a critical role in the tanshinone biosynthetic pathway in S. miltiorrhiza.
The GRAS TF family has been discovered to be unique in plants. GRAS TFs consist of three gene members, namely, gibberellic acid insensitive (GAI), repressor of GA1-3 mutant (RGA), and Scarecrow (SCR). A whole-genome analysis from Arabidopsis thaliana, rice (Oryza sativa), and musk lily (Lilium longiflorum) grouped the GRAS families into ten distinctive subfamilies, namely, SCR, short-root (SHR), L. longiflorum SCR-like (LISCL), DELLA, SCL-like 3 (SCL3), SCL-like 4 and SCL-like 7 (SCL4/7), A. thaliana Lateral suppressor (AtLAS), hairy meristem (HAM), A. thaliana phytochrome A signal transduction 1 (AtPAT1), and dwarf and low tillering (DLT) (Sun et al., 2011). The SCR and SHR subfamilies were validated to act as positive regulators to promote the formation of root radial morphology and root monolayer cells in Arabidopsis (Koizumi et al., 2012; Rich et al., 2017). SCR is mainly expressed in root epidermal cells regulating the division of daughter cells differentiation in root, whereas SHR not only affects the polar differentiation of root endothelial cells but also activates the SCR promoter in specific tissues to regulate its function (Slewinski et al., 2012). In addition, SHR proteins are expressed explicitly in the mid-column sheath cells. They can transfer from external monolayers to endothelial cells to form a dimer with SCR, which jointly activates the transcription of the SCR gene and other downstream target genes. Therefore, it can be inferred that SCR and SHR are transcriptionally dependent on each other, and the resulting SCR-SHR complex positively modulates the transcription of SCR (Koizumi et al., 2012; Slewinski et al., 2012). Yeast extract (YE), also known as yeast flavoring, plays a vital role in promoting the accumulation of secondary metabolites in plants. In S. miltiorrhiza, after treating hairy root culture with YE, the content of tanshinone was significantly increased (Chen et al., 2001). However, the transcriptional mechanism involved in YE-triggered tanshinone biosynthesis remains obscure.
In this study, we isolated and identified a new GRAS TF named SmSCR1. Our studies uncover the regulatory role of SmSCR1 gene in response to YE-induced regulation of tanshinone biosynthesis and hairy root growth and development. Our data discover a novel TF family, GRAS, as a candidate for improving the biosynthesis potential of tanshinone through metabolic engineering strategies.
Materials and Methods
Plant Materials and Growth Conditions
Salvia miltiorrhiza plants were grown in solid Murashige and Skoog (MS) medium at 25°C, in 60% humidity, and in a light rhythm of 16-h light and 8-h dark (Shi et al., 2021a,b). S. miltiorrhiza hairy roots were subcultured in 250 ml Erlenmeyer flasks with 100 ml 1/2 MS liquid medium and placed in a shaker at 110 rpm min–1, 25°C in the dark. The hairy roots were collected after 60 days for gene expression analysis (Shi et al., 2016a,2020). Nicotiana benthamiana was directly sown in soil and cultivated in pots for further experimental needs (Huang et al., 2019). RNA was extracted from different tissues of annual plants of S. miltiorrhiza and stored at −80°C using liquid nitrogen flash freezing.
Gene Isolation and Sequence Analysis
All the GRAS families in S. miltiorrihiza were identified against local datasets. SmSCR1, which belongs to the GRAS family and intensively responds to YE induction, was cloned using a homology-based cloning method, as described previously (Zhou et al., 2016, 2017). The cloned SmSCR1 gene was analyzed using the BlastP tool in the NCBI database. The reported GRAS members, i.e., SmGRAS1 (accession number: KY435886) and SmGRAS2 (KY435887) in S. miltiorrihiza, together with O. sativa OsGRAS32 (Os06g0127800), O. sativa OsSCR1 (XP_015615402.1), O. sativa OsSCR2 (XP_015620600.1), A. thaliana AtSCR (Q9M384.1), and A. thaliana AtSCL23 (NM_123557), which are highly homologous to SmSCR1 (OM032820), were subjected to amino acid sequence alignment using Vector NTI software (Invitrogen, Carlsbad, CA, United States). The full-length amino acid sequences of the GRAS were aligned using the neighbor-joining (NJ) method in the Clustal X computer, and the MEGA 6.0 program was used to generate a phylogenetic tree. Using Clustal X computing tools, the amino acid sequences of candidate GRAS family members were aligned based on the NJ method, and MEGA 6.0 software was used to generate phylogenetic trees. Subfamily members of SCR and SCL from Arabidopsis and rice were compared with SmSCR1 using Vector NTI 10.0 software.
Subcellular Localization Analysis of SmSCR1
To verify the subcellular localization profile of SmSCR1, pHB-SmSCR1-YFP, and pHB-YFP recombinant plasmids were introduced into Agrobacterium rhizogenes strain EHA105 for transient transformation, respectively. pHB-YFP was used as the positive control. After 48 h of infection by Agrobacterium tumefaciens, YFP signals from infected N. benthamiana leaves were visualized using a high-resolution microscope observation system. The nuclei of epidermal cells of infected N. benthamiana leaves were stained with 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) solution for 3 h before observation (Zhou et al., 2020).
Quantitative Real-Time PCR
According to the procedure reported before (Shi et al., 2016b), the quantitative real-time PCR (qRT-PCR) analysis was conducted. Different tissues (e.g., taproot, lateral root, fibrous root, phloem, xylem, stem, petiole, young leaves, and mature leaves) were taken from an adult plant, as well as hairy roots with the treatment of YE, and then frozen in liquid nitrogen for RNA isolation (Zhang et al., 2011). Actin gene of S. miltiorrhiza was used as the internal reference. The sequences of all primer pairs used in qRT-PCR studies are summarized in Supplementary Table 1. The gene expression was quantified by the comparative Ct method (Hao et al., 2015).
Transformation of SmSCR1 in Salvia miltiorrhiza
The open reading frame (ORF) of SmSCR1 was introduced into the double restriction insertion site of BamHI and SpeI of the pHB vector under the CaMV 35S promoter and NOS terminator (Supplementary Figure 2A). The DNA sequence encoding the SRDX structural domain (LDLDLELRLGFA) was inserted into the ORF of SmSCR1 just before the stop codon (TAA) using the method described previously (Hiratsu et al., 2003; Deng et al., 2020a,b). The SmSCR1-SRDX was then subcloned into the pHB to create the pHB-SmSCR1-SRDX suppressive expression construct (Supplementary Figure 2B). To obtain the transgenic hairy roots, all plasmids were transformed into A. tumefaciens C58C1 and then transinfected into S. miltiorrhiza (Zhou et al., 2016).
Determination of Tanshinones by High-Performance Liquid Chromatography
After 60 days of continuous culture in 1/2 MS liquid medium, each hairy root line was harvested and dried in a freeze dryer for 24 h, and then extracted with 16 ml methanol/dichloromethane (3:1, v/v). Notably, 100 mg of dried powder of S. miltiorrhiza, sonicated for 1 h, then soaked overnight in the dark and centrifuged for 10 min at 12,000 × g. After centrifugation, the supernatant was poured into a distillation flask. After centrifugation, the supernatant was poured into a distillation flask and spun dry at 50°C. We then added 2 ml of methanol to dissolve the material in the distillation flask. After further centrifugation at 6,500 × g for 10 min, the samples were filtered separately through 0.22 μM organic membranes (Jinten, China) using a 1 ml sterile syringe (Huang et al., 2019). All metabolites were quantified by high-performance liquid chromatography (HPLC), as described previously (Agilent Technologies, Santa Clara, CA, United States) (Zhou et al., 2016; Sun et al., 2019; Yang et al., 2021).
Dual-Luciferase Assay
The dual-luciferase (Dual-LUC) assays were carried out as previously reported (Deng et al., 2019). The recombinants of pHB-YFP and pHB-SmSCR1-YFP were introduced into A. tumefaciens GV3301,respectively. Gene promoters from the tanshinone biosynthetic pathway were cloned and inserted into pGREEN0800 vector, respectively, which were co-transformed into the GV3301 strain with pSoup19. The test was carried out as previously reported (Cao et al., 2018). The pHB-YFP construct was employed as the negative control, and the Renilla gene was used as the internal control.
Yeast One-Hybrid Assay
Similar to a previous study (Deng et al., 2019), the ORF of SmSCR1 was inserted into the pB42AD vector. Three fragments of SmCPS1 promoter sequence (i.e., from −1 to −700, −701 to −1,400, and −1,401 to −2,100 bp, relative to translation start site, respectively) were inserted into a pLacZ plasmid. The resulting recombinants were co-transformed into yeast cell EGY48a. After cultivation on SD/-Ura/-Leu medium for 48 h, the positive binding activity was examined on SD/-Ura/-Leu medium with X-gal for 24 h. PB42AD and pLacZ empty carriers were set as negative controls.
Morphological Observations of SmSCR1
To investigate the effect of the SmSCR1 gene on plant growth and development, we examined different phenotypes of the transgenic hairy roots on 1/2 MS medium. In addition, three different transgenic hairy roots of SmSCR1 with good growth and similar developmental time were selected for developmental microstructure observation, as reported previously (Wu et al., 2020). The pith (Pi) and posterior xylem (Mx) of the hairy roots were observed under a light microscope (200 ×).
Statistical Analysis
Each data represent the average of three independent experimental calculations, and the data are expressed as the mean ± SD. SPSS 16.0 software was used to perform a single-sample t-test and one-way ANOVA, and p-values < 0.05 were considered statistically significant.
Results
Isolation and Sequence Analysis of SmSCR1 Gene
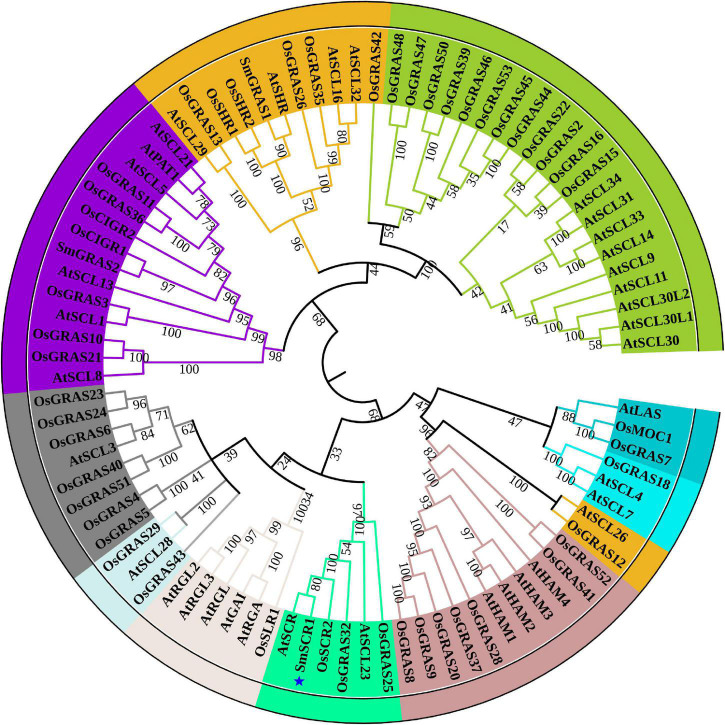
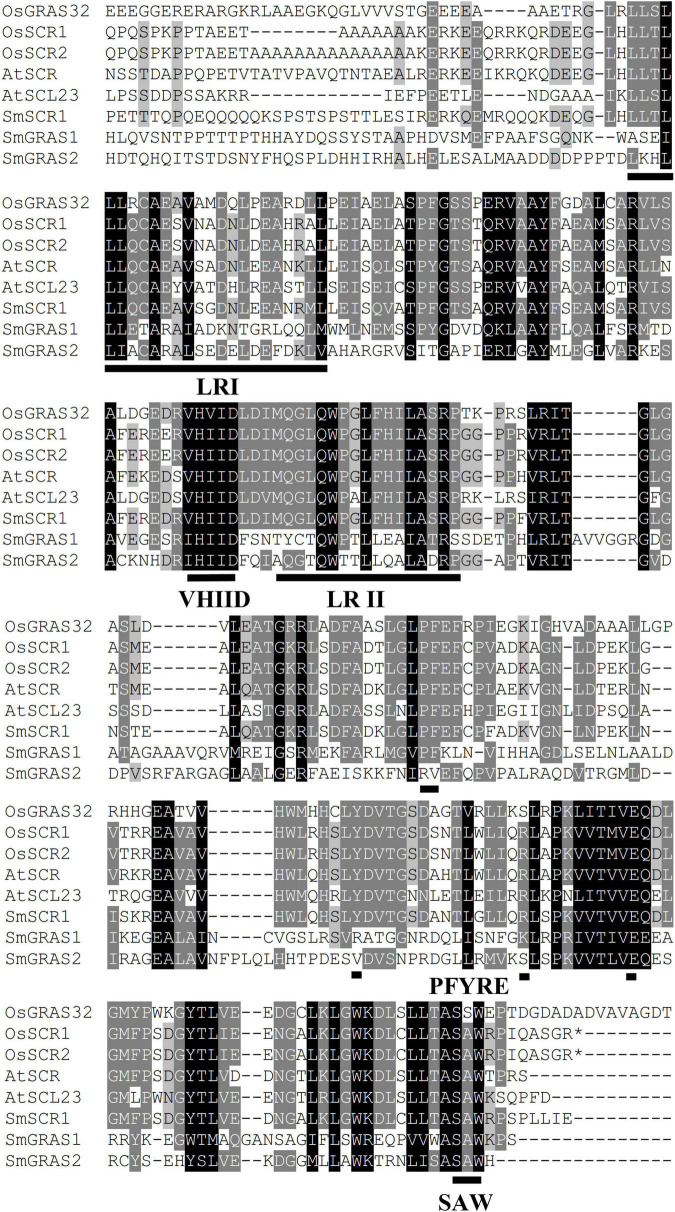
First, we identified a GRAS TF (unigene number: SmiContig9465) and SmSCR1 (NCBI accession number: OM032820), responding to YE-induction (fold change > 2) from the S. miltiorrhiza transcriptome (Zhou et al., 2017). SmSCR1 length is 1,944 bp long and encodes 647 amino acids, with a molecular weight size of 70.515 kDa coupled with a theoretical isoelectric point of 5.78. GRAS family members from Arabidopsis and rice were analyzed with SmSCR1 to construct a phylogenetic tree. As shown in Figure 1, SmSCR1 has the highest relationship with AtSCR in Arabidopsis and OsSCR2 in rice, implying that SmSCR1 belongs to the SCR subgroup of the GRAS family. SmSCR1 consists of several conserved motifs, including VHIID, PFYRE, SAW motifs, and the leucine-rich regions (LRs), i.e., LR I and LR II (Figure 2).
FIGURE 1.
Bioinformatics analysis of SmSCR1. Phylogenic tree analysis of SmSCR1 and GRAS transcription factors from different species. The MEGA 6.05 software was employed to construct the phylogenetic tree with the amino acid sequences of SmSCR1 in Salvia miltiorrhiza and 89 GRAS family members in Arabidopsis and rice using the neighbor-joining approach. The bootstrap values after 1,000 replicates are indicated by the numbers on the nodes.
FIGURE 2.
Multiple alignments of SmSCR1 with related GRAS proteins from other plants. SmGRAS1 (accession number: KY435886), SmGRAS2 (KY435887), OsGRAS32 (Os06g0127800), OsSCR1 (XP_015615402.1), OsSCR2 (XP_015620600.1), AtSCR (Q9M384.1), AtSCL23 (NM_123557), and SmSCR1 (OM032820). Black line indicates the conserved amino acid sites. ILR I, leucine-rich region I; LR II, leucine-rich region II; VHIID, the conserved region consisting of valine, histidine, isoleucine, and D-aspartic acid; PFYRE, the conserved amino acid sites consisting of proline, phenylalanine, tyrosine, arginine, and glutamic acid; SAW, the conserved region consisting of serine, alanine, and tryptophan.
Expression Analysis of SmSCR1
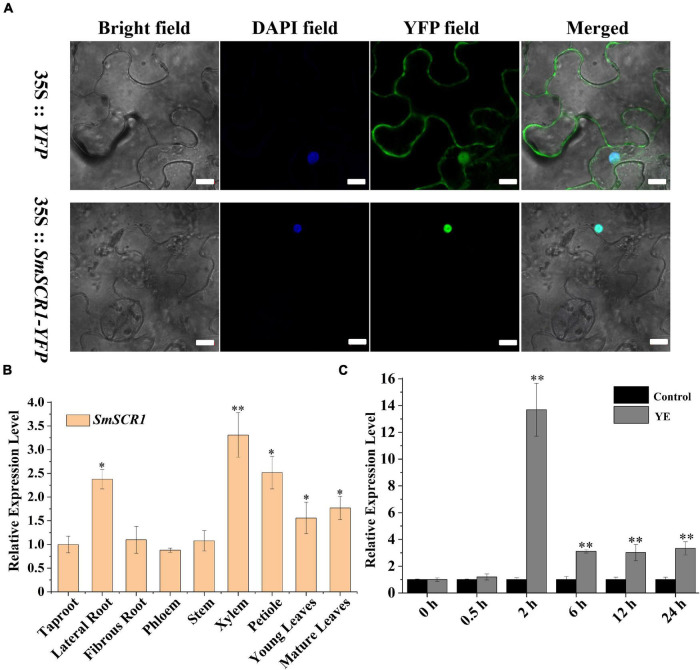
Transient expression of SmSCR1 in epidermal cells from 45-day-old N. benthamiana leaves exhibited that SmSCR1 located in the nucleus (Figure 3A). The gene expression of SmSCR1 was characterized from nine different tissues of S. miltiorrhiza by qRT-PCR. SmSCR1 was expressed in all tissues, namely, taproot, lateral root, fibrous root, phloem, xylem, stem, petiole, young leaves, and mature leaves, with the highest expression in xylem and petiole, and the lowest expression in taproot (Figure 3B). The expression of SmSCR1 after YE-induction was examined, reaching the highest levels for 2 h after post-induction with 13.6-fold compared to the control (Figure 3C).
FIGURE 3.
Expression profile of SmSCR1. (A) Transient expression of the SmSCR1 in Nicotiana benthamiana leaf epidermal cells. Scale bars represent 10 μm. (B) The expression patterns of SmSCR1 in different tissues. Fold changes of the relative expression level of SmSCR1 gene in other tissues are all normalized to the taproot. (C) Induced effect of YE on the expression of SmSCR1. Fold changes of the gene relative expression levels are all normalized to the control without induction by YE at 0 h. Asterisks indicate significant differences between the taproot and other tissues in (B) and between treatment and control in (C) at one significant level of t-test (*p < 0.05; **p < 0.01). Data are means of three replicates with SDs.
SmSCR1 Activates the Transcription of Copalyl Diphosphate Synthase 1 Gene
The role of SmSCR1 in regulating tanshinone biosynthetic gene expressions was examined. The Dual-LUC assay showed that SmSCR1 uniquely activates the transcription of the SmCPS1 promoter, leading to a 2.19-fold increase compared with the control, but not to other promoters (e.g., HMGR1 promoter) (Figure 4). Y1H assay showed that self-activating of pB42AD vector binding to the promoter of the CPS1 gene is visible (Supplementary Figure 3). Those results indicated that SmSCR1 activated the transcription of SmCPS1 to increase the production of tanshinone in S. miltiorrhiza.
FIGURE 4.
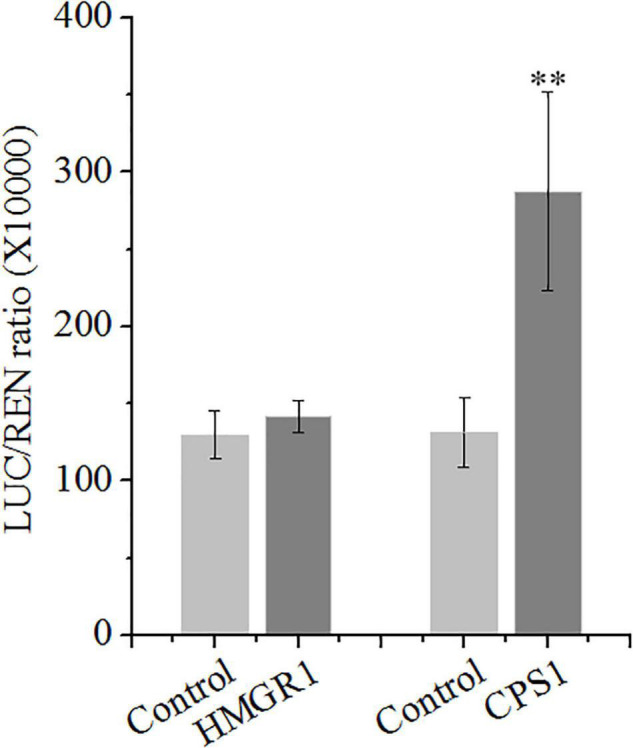
SmSCR1 activates the transcription of SmCPS1. SmSCR1 activates the promoter of SmCPS1 by the dual-luciferase (Dual-LUC) assay. The SmCPS1 promoter was fused to the LUC reporter, and its activity was determined by a transient Dual-LUC assay in Nicotiana benthamiana. The value of LUC/REN was recorded as the activating activity. Asterisk indicates significant differences at one significant level of t-test (**p < 0.01). Data are means of three replicates with SDs.
SmSCR1 Promotes Tanshinone Biosynthesis in the Transgenic Hairy Root of Salvia miltiorrhiza
The OE and SRDX suppressive hairy root lines were first examined by genomic PCR detection (Supplementary Figure 4). Then, the transcript profiles of SmSCR1 in transgenic hairy roots were quantified by qRT-PCR. Three independent OE lines (i.e., OE-5, OE-8, and OE-11) and three SRDX lines (i.e., SRDX-2, SRDX-4, and SRDX-6) of SmSCR1 with the highest expression levels in transgenic lines were subsequently chosen for further analysis (Supplementary Figure 5). After 60-day-old subculture, the hairy root lines were collected independently and were subjected to examine the content of tanshinones by HPLC (Figure 5A). The concentrations of DT, TA-1, CT, and TTA were significantly increased compared to the control in the OE lines, with the highest total tanshinone (TTA) content in SmSCR1-OE-11, reaching 1.49-fold than that of the control. In contrast, in the SRDX repression lines, DT, TA-1, CT, and TTA were significantly decreased compared to the control, with the lowest TTA content of 2.76 mg/g DW. The above results suggest that the SmSCR1 TF can promote the tanshinone accumulation in S. miltiorrhiza.
FIGURE 5.
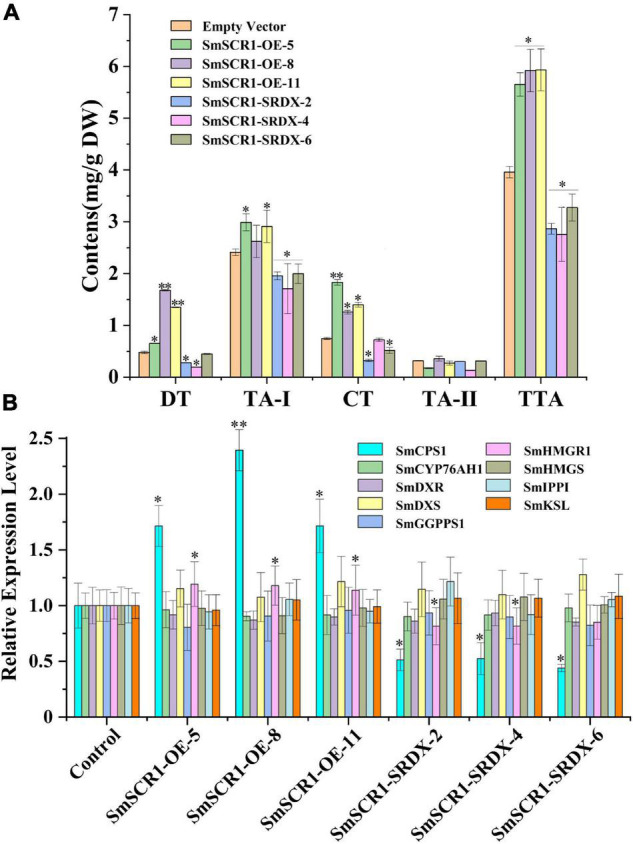
Tanshinone content and gene expression in the tanshinone biosynthetic pathway in Salvia miltiorrhiza transgenic hairy root lines. (A) High-performance liquid chromatography analysis of tanshinone content in hairy root lines of SmSCR1-OE (overexpression) and SmSCR1-SRDX (suppression). DT, dihydrotanshinone; CT, cryptotanshinone; TA-I, tanshinone I; TA-II, tanshinone IIA; TTA, total tanshinone. (B) Gene expression in the SmSCR1-OE and SmSCR1-SRDX lines for the underlying tanshinone biosynthesis. The blank vector without SmSCR1 gene is used as the control to standardize fold differences in relative gene expression levels in transgenic hairy roots. The standard errors of the mean are represented by error bars. The asterisks on the bar indicate significant differences by t-test compared to the control at two significant levels (*p < 0.05; **p < 0.01).
SmSCR1 Upregulates Copalyl Diphosphate Synthase 1 and HMGR1 in Transgenic Hairy Roots of Salvia miltiorrhiza
Copalyl diphosphate synthase 1 and HMGR1 were significantly upregulated in OE lines (e.g., OE-5, OE-8, and OE-11), with the SmSCR1-OE-11 line, showing the highest expression. In contrast, the two genes were drastically downregulated in the three SRDX lines (i.e., 2, 4, and 6), with the SmSCR1-SRDX-6 line exhibiting the lowest decrease compared to the control (Figure 5B). These findings showed that SmSCR1 stimulated CPS1 and HMGR1 together, resulting in increased production in transgenic hairy roots of S. miltiorrhiza.
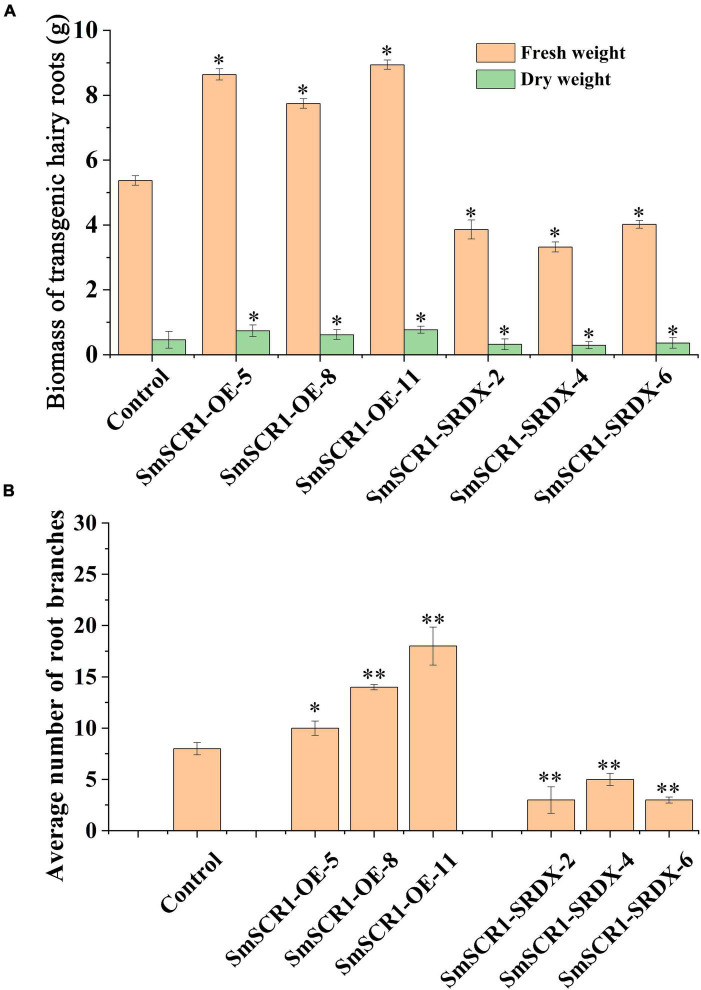
SmSCR1 Promotes the Growth of Transgenic Hairy Roots of Salvia miltiorrhiza
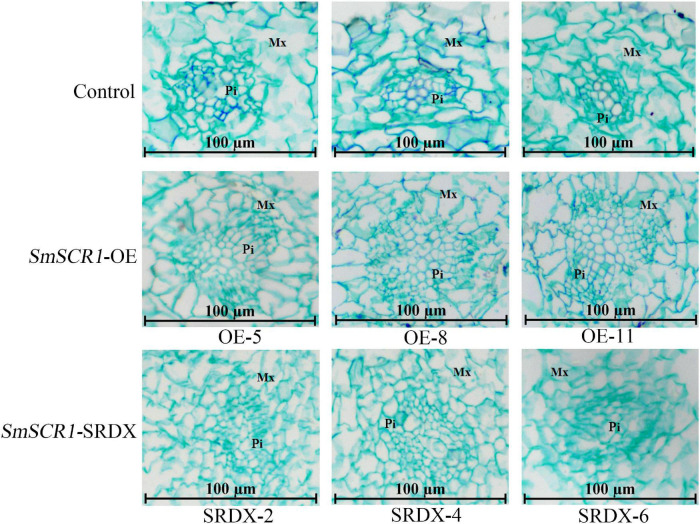
The biomass of the transgenic hairy root lines significantly increased in the SmSCR1 OE lines and significantly reduced in the SRDX lines compared to the control (Figure 6A). Moreover, the SmSCR1 overexpression hairy root lines had a better growth status than the control, with more branches in the SmSCR1 OE lines and fewer in the SRDX suppressive hair roots (Figure 6B). Roots are formed by a series of asymmetrical divisions of early differentiated cells to produce the inner and outer cortex of the root, which then develops into xylem and phloem. The development of transgenic hairy roots by paraffin sectioning demonstrated that the SmSCR1 gene promotes the formation of hairy root radiation morphology of the Mx cell and Pi cell differentiation in SmSCR1-OE hairy roots when compared with the control and the SRDX suppressive hairy roots (Figure 6). Those results suggest that SmSCR1 can modulate the growth and development of hairy roots in S. miltiorrhiza.
FIGURE 6.
Phenotype of the SmSCR1 transgenic hairy root. (A) Biomass of transgenic hairy root. (B) An average number of root branches. Asterisks indicate significant differences between the transgenic hairy root lines and control at one significant level of t-test (*p < 0.05; **p < 0.01). Data are means of three replicates with SDs.
Discussion
The GRAS TF family is an important TF class involved in regulating plant growth, development, environmental stress response, and growth signal transduction (Sun et al., 2011). With the development of genome sequencing technology, a few GRAS family members have been identified in plants, such as plum (Armeniaca mume), lotus (Nelumbo nucifera), and pine blue (Isatis indigotica) (Kamiya et al., 2003; Hou et al., 2010). However, only a few reports systematically characterized GRAS TFs in medicinal plants. We identified a novel YE-responsive GRAS TF gene, SmSCR1 gene, encoding 647 amino acids with several key GRAS conserved structural domains (e.g., VHIID, LR I, LR II, PFYRE, and SAW motifs) (Koizumi et al., 2011; Zhou et al., 2017). In a previous study, SCR and SHR families were thought to co-regulate the plant growth, development, and radial structures in roots (Koizumi et al., 2011, 2012; Fan et al., 2017). We also identified that SmSCR1 was very sensitive to YE-induction (Figure 3C), regulating tanshinone accumulation in S. miltiorrhiza (Zhou et al., 2017, 2021a).
We revealed that SmSCR1 upregulates tanshinone biosynthesis in SmSCR1 suppressive hairy root lines (Figure 5A). Recently, two GRAS families, including SmGRAS1 (GenBank accession number: KY435886) and SmGRAS2 (GenBank accession number: KY435887), clustered into SHR and PAT1 subgroups, respectively (Figure 1), were characterized in S. miltiorrhiza. SmGRAS1 and SmGRAS2 were verified by transgenic validation to upregulate tanshinone accumulation in hairy roots. Therefore, many GRAS families might play a more prominent role in regulating tanshinone accumulation in S. miltiorrhiza and require further investigation.
Both CPS1 and HMGR1 positively correlated to the expression of SmSCR1 in the transgenic lines, while SmSCR1 can only bind to the CPS1 promoter to activate its expression as examined. Due to the limited data about the specific cis-element in promoter binding by SCR1 in other plants (Li et al., 2019), further examining the direct binding of SmSCR1 to the specific promoter elements by EMSA assay in vitro would be unfeasible. In this study, overexpressing SmSCR1 upregulated not only the expression of its target gene CPS1 but also the HMGR1 gene in the tanshinone biosynthetic pathway. Overexpressing SmERF1L1 promotes the expression of five genes in the tanshinone biosynthetic pathway, but only DXR is a target (Huang et al., 2019). In SmERF128 transgenic hairy roots, the expression of six genes in the tanshinone biosynthetic pathway was activated, and only three of the six genes were validated to be targets of SmERF128 (Zhang et al., 2019). Thus, it can be inferred that the ectopic expression of certain TF genes upregulates the expression of not only their target genes but also other genes in the same biosynthetic pathway.
The root formation is a series of asymmetric divisions of stem cells to generate the inner and outer cortex of the root and then differentiate into different forms of tissue. In A. thaliana, SCR is mainly expressed in root epidermal cells. As a positive regulator, SCR regulates root radiation morphology and plays a vital role in root monolayer cell formation (Koizumi et al., 2012; Rich et al., 2017). When SCR function is lost and mutated, the normal development of the cell layer will be affected. In addition, SCR can also regulate the division of progenitor cells and promote the growth of biomass (Koizumi et al., 2011; Slewinski et al., 2012). In rice, OsSCR was thought to affect the development of plant leaf guard cells (Hou et al., 2010). In the three SmSCR1 OE lines, we found that SmSCR1 significantly promoted the biomass increase of the three transgenic hairy root lines, whereas a visible decrease was observed in SmSCR1 SRDX lines.
Moreover, SmSCR1 can regulate the development of Pi and the Mx. In SmSCR1 OE lines, the cell division rate of progeny and the root radial structure growth rate were significantly higher than that of control. In contrast, the three SRDX suppressive hairy root lines have an opposite phenotype (Figure 7). The function of SmSCR1 to regulate the growth and development of hairy roots in S. miltiorrhiza is similar to the model plants, e.g., Arabidopsis and rice.
FIGURE 7.
Paraffin sectioning analysis of the transgenic hairy roots. The control means the transgenic hairy roots with the transformation of pHB-X-YFP blank vector. pHB-SmSCR1 represents the overexpression lines, and pHB-SmSCR1-SRDX represents the suppression lines. Pi represents the pith. Mx represents the posterior xylem.
Based on the present findings, a deductive model of SmSCR1 as an activator in response to YE-induction promotes tanshinone accumulation, Pi cell division, radial structure formation, and biomass increase of hairy roots in S. miltiorrhiza was outlined (Figure 8).
FIGURE 8.
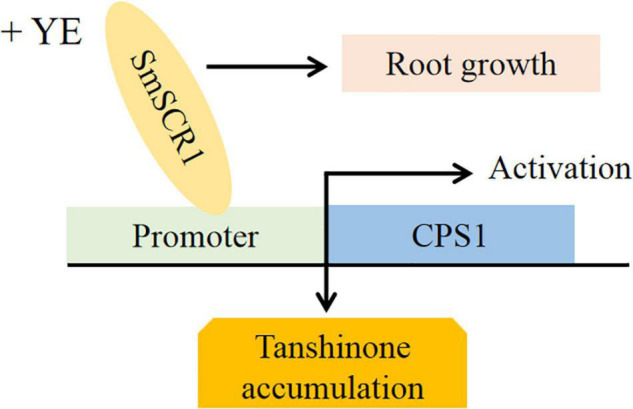
A proposed model for the role of SmSCR1 in modulating tanshinone biosynthesis and root growth in Salvia miltiorrhiza.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
GK and WZ conceived and designed the project. SW, YS, YL, XG, CC, and TL prepared materials and performed the experiments. WZ, SW, YS, IM, YL, and CW performed the bioinformatics analysis and analyzed the data. GK, WZ, SW, and YS wrote the manuscript. IM, WZ, and GK revised the manuscript. All authors read and approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We gratefully acknowledge the great experimental support of the Public Platform of Medical Research Center, Academy of Chinese Medical Science, and Zhejiang Chinese Medical University.
Abbreviations
- CPS1
copalyl diphosphate synthase I
- HMGR1
3-hydroxy-3-methylglutaryl-CoA-reductase I
- GGPPS1
geranylgeranyl diphosphate synthase I
- HMGS
hydroxymethylglutaryl-CoA synthase
- IPPI
isopentenyl diphosphate isomerase
- KSL
ent-kaurene synthase like
- CPS1
copalyl diphosphate synthase I
- CYP76AH1
miltiradiene oxidase
- DXR
1-deoxy-D-xylulose 5-phosphate reductoisomerase
- DXS
deoxy-D-xylulose 5-phosphate synthase II
- Dual-LUC
dual-luciferase
- YE
yeast extract
- GRAS1
GRAS transcriptional factor 1
- DT
dihydrotanshinone
- CT
cryptotanshinone
- TA-I
tanshinone I
- TA-II
tanshinone IIA
- TTA
total tanshinone.
Funding
We acknowledge the support extended toward this research by the bodies listed: Zhejiang Provincial Natural Science Foundation of China (LY20H280008), National Key R&D Program of China (2018YFC1706203), the Major Science and Technology Projects of Breeding New Varieties of Agriculture in Zhejiang Province (2021C02074-3-4), Pre-research Projects of Zhejiang Chinese Medical University (2021JKZKTS011A), and Sannongliufang Research Joint Project of Zhejiang Province (2021SNLF019), and the Opening Project of Zhejiang Provincial Preponderant and Characteristic Subject of Key University (Traditional Chinese Pharmacology), Zhejiang Chinese Medical University (ZYAOX2018003 and ZYAOX2018009).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.860033/full#supplementary-material
The skeleton of tanshinone biosynthetic pathway in Salvia miltiorrhiza.
Recombinants of SmSCR1-overexpressive and suppressed vectors. (A) Recombinant vector of pHB -SmSCR1-YFP. (B) Recombinant vector of pHB-SmSCR1-SRDX.
Yeast one-hybrid assay of SmSCR1 binding to the SmCPS1 promoter. P1, P2, and P3 represent the three fragments of SmCPS1 promoter sequence from −1 to −700, −701 to −1,400, and −1,401 to −2,100 bp, relative to translation start site, respectively.
Detection of the expression level of SmSCR1 gene in transgenic hairy root lines by qRT-PCR.
Identification of transgenic hairy root lines by PCR. The asterisks on the bar indicate significant differences by t-test compared to the control (**P < 0.01).
References
- Cao W., Wang Y., Shi M., Hao X., Zhao W., Wang Y., et al. (2018). Transcription factor SmWRKY1 positively promotes the biosynthesis of tanshinones in Salvia miltiorrhiza. Front. Plant Sci. 9:554. 10.3389/fpls.2018.00554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Chena F., Chiu F. C., Lo C. M. (2001). The effect of yeast elicitor on the growth and secondary metabolism of hairy root cultures of Salvia miltiorrhiza. Enzyme Microb. Tech. 28 100–105. 10.1016/s0141-0229(00)00284-2 [DOI] [PubMed] [Google Scholar]
- Deng C., Hao X., Shi M., Fu R., Wang Y., Zhang Y., et al. (2019). Tanshinone production could be increased by the expression of SmWRKY2 in Salvia miltiorrhiza hairy roots. Plant Sci. 284 1–8. 10.1016/j.plantsci.2019.03.007 [DOI] [PubMed] [Google Scholar]
- Deng C., Shi M., Fu R., Zhang Y., Wang Q., Zhou Y., et al. (2020a). ABA-responsive transcription factor bZIP1 is involved in modulating biosynthesis of phenolic acids and tanshinones in Salvia miltiorrhiza. J. Exp. Bot. 71 5948–5962. 10.1093/jxb/eraa295 [DOI] [PubMed] [Google Scholar]
- Deng C., Wang Y., Huang F., Lu S., Zhao L., Ma X., et al. (2020b). SmMYB2 promotes salvianolic acid biosynthesis in the medicinal herb Salvia miltiorrhiza. J. Integr. Plant Biol. 62 1688–1702. 10.1111/jipb.12943 [DOI] [PubMed] [Google Scholar]
- Fan S., Zhang D., Gao C., Zhao M., Wu H., Li Y., et al. (2017). Identification, classification, and expression analysis of GRAS gene family in Malus domestica. Front. Physiol. 8:253. 10.3389/fphys.2017.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Han B., Zhou Y., Ren J., Cao W., Patel G., et al. (2020). The anticancer properties of tanshinones and the pharmacological effects of their active ingredients. Front. Pharmacol. 11:193. 10.3389/fphar.2020.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Hillwig M. L., Huang L., Cui G., Wang X., Kong J., et al. (2009). A functional genomics approach to tanshinone biosynthesis provides stereochemical insights. Org. Lett. 11 5170–5173. 10.1021/ol902051v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X., Pu Z., Cao G., You D., Zhou Y., Deng C., et al. (2020). Tanshinone and salvianolic acid biosynthesis are regulated by SmMYB98 in Salvia miltiorrhiza hairy roots. J. Adv. Res. 23 1–12. 10.1016/j.jare.2020.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao X., Shi M., Cui L., Xu C., Zhang Y., Kai G. (2015). Effects of methyl jasmonate and salicylic acid on tanshinone production and biosynthetic gene expression in transgenic Salvia miltiorrhiza hairy roots. Biotechnol. Appl. Bioc. 62 24–31. 10.1002/bab.1236 [DOI] [PubMed] [Google Scholar]
- Hiratsu K., Matsui K., Koyama T., Ohme-Takagi M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34 733–739. 10.1046/j.1365-313x.2003.01759.x [DOI] [PubMed] [Google Scholar]
- Hou X., Lee L., Xia K., Yan Y., Yu H. (2010). DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dec. Cell 19 884–894. 10.1016/j.devcel.2010.10.024 [DOI] [PubMed] [Google Scholar]
- Huang Q., Sun M., Yuan T., Wang Y., Shi M., Lu S., et al. (2019). The AP2/ERF transcription factor SmERF1L1 regulates the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Food Chem. 274 368–375. 10.1016/j.foodchem.2018.08.119 [DOI] [PubMed] [Google Scholar]
- Jia Q., Zhu R., Tian Y., Chen B., Li R., Li L., et al. (2019). Salvia miltiorrhiza in diabetes: a review of its pharmacology, phytochemistry, and safety. Phytomedicine 58:152871. 10.1016/j.phymed.2019.152871 [DOI] [PubMed] [Google Scholar]
- Jung I., Kim H., Moon S., Lee H., Kim B. (2020). Overview of Salvia miltiorrhiza as a potential therapeutic agent for various diseases: an update on efficacy and mechanisms of action. Antioxidants 9:857. 10.3390/antiox9090857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai G., Xu H., Zhou C., Liao P., Xiao J., Luo X., et al. (2011). Metabolic engineering tanshinone biosynthetic pathway in Salvia miltiorrhiza hairy root cultures. Metab. Eng. 13 319–327. 10.1016/j.ymben.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Kamiya N., Itoh J., Morikami A., Nagato Y., Matsuoka M. (2003). The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J. 36 45–54. 10.1046/j.1365-313x.2003.01856.x [DOI] [PubMed] [Google Scholar]
- Koizumi K., Hayashi T., Wu S., Gallagher K. L. (2012). The SHORT-ROOT protein acts as a mobile, dose-dependent signal in patterning the ground tissue. P. Natl. Acad. Sci. U S A. 109 13010–13015. 10.1073/pnas.1205579109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi K., Wu S., MacRae-Crerar A., Gallagher K. L. (2011). An essential protein that interacts with endosomes and promotes movement of the SHORT-ROOT transcription factor. Curr. Biol. 21 1559–1564. 10.1016/j.cub.2011.08.013 [DOI] [PubMed] [Google Scholar]
- Li W., Bai Z., Pei T., Yang D., Mao R., Zhang B., et al. (2019). SmGRAS1 and SmGRAS2 regulate the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Front. Plant Sci. 10:1367. 10.3389/fpls.2019.01367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Ma X. H., Meng F. Y., Zhan Z. L., Guo J., Huang L. Q. (2016). RNA interference targeting CYP76AH1 in hairy roots of Salvia miltiorrhiza reveals its key role in the biosynthetic pathway of tanshinones. Biochem. Bioph. Res. Co 477 155–160. 10.1016/j.bbrc.2016.06.036 [DOI] [PubMed] [Google Scholar]
- Ma Y., Yuan L., Wu B., Li X., Chen S., Lu S. (2012). Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J. Exp. Bot. 63 2809–2823. 10.1093/jxb/err466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich M. K., Courty P. E., Roux C., Reinhardt D. (2017). Role of the GRAS transcription factor ATA/RAM1 in the transcriptional reprogramming of arbuscular mycorrhiza in Petunia hybrida. BMC Genomics 18:589. 10.1186/s12864-017-3988-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Gong H., Cui L., Wang Q., Wang C., Wang Y., et al. (2020). Targeted metabolic engineering of committed steps improves anti-cancer drug camptothecin production in Ophiorrhiza pumila hairy roots. Ind. Crop. Prod. 148:112277. 10.1016/j.indcrop.2020.112277 [DOI] [Google Scholar]
- Shi M., Du Z., Hua Q., Kai G. (2021a). CRISPR/Cas9-mediated targeted mutagenesis of bZIP2 in Salvia miltiorrhiza leads to promoted phenolic acid biosynthesis. Ind. Crop. Prod. 167:113560. 10.1016/j.indcrop.2021.113560 [DOI] [Google Scholar]
- Shi M., Liao P., Nile S. H., Georgiev M. I., Kai G. (2021b). Biotechnological exploration of transformed root culture for value-added products. Trends Biotechnol. 39 137–149. 10.1016/j.tibtech.2020.06.012 [DOI] [PubMed] [Google Scholar]
- Shi M., Luo X., Ju G., Li L., Huang S., Zhang T., et al. (2016a). Enhanced diterpene tanshinone accumulation and bioactivity of transgenic Salvia miltiorrhiza hairy roots by pathway engineering. J. Agr. Food Chem. 64 2523–2530. 10.1021/acs.jafc.5b04697 [DOI] [PubMed] [Google Scholar]
- Shi M., Zhou W., Zhang J., Huang S., Wang H., Kai G. (2016b). Methyl jasmonate induction of tanshinone biosynthesis in Salvia miltiorrhiza hairy roots is mediated by JASMONATE ZIM-DOMAIN repressor proteins. Sci. Rep. 6:20919. 10.1038/srep20919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slewinski T. L., Anderson A. A., Zhang C., Turgeon R. (2012). Scarecrow plays a role in establishing Kranz anatomy in maize leaves. Plant Cell Physiol. 53 2030–2037. 10.1093/pcp/pcs147 [DOI] [PubMed] [Google Scholar]
- Sun M., Shi M., Wang Y., Huang Q., Yuan T., Wang Q., et al. (2019). The biosynthesis of phenolic acids is positively regulated by the JA-responsive transcription factor ERF115 in Salvia miltiorrhiza. J. Exp. Bot. 70 243–254. 10.1093/jxb/ery349 [DOI] [PubMed] [Google Scholar]
- Sun X., Xue B., Jones W. T., Rikkerink E., Dunker A. K., Uversky V. N. (2011). A functionally required unfoldome from the plant kingdom: intrinsically disordered N-terminal domains of GRAS proteins are involved in molecular recognition during plant development. Plant Mol. Biol. 77 205–223. 10.1007/s11103-011-9803-z [DOI] [PubMed] [Google Scholar]
- Wang B., Sun W., Li Q., Li Y., Luo H., Song J., et al. (2015). Genome-wide identification of phenolic acid biosynthetic genes in Salvia miltiorrhiza. Planta 241 711–725. 10.1007/s00425-014-2212-1 [DOI] [PubMed] [Google Scholar]
- Wang N., Xu H., Jiang S., Zhang Z., Lu N., Qiu H., et al. (2017). MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90 276–292. 10.1111/tpj.13487 [DOI] [PubMed] [Google Scholar]
- Wu X., Riaz M., Yan L., Zhang Z., Jiang C. (2020). How the cells were injured and the secondary metabolites in the shikimate pathway were changed by boron deficiency in trifoliate orange root. Plant Physiol. Biochem. 151 630–639. 10.1016/j.plaphy.2020.04.009 [DOI] [PubMed] [Google Scholar]
- Xing B., Liang L., Liu L., Hou Z., Yang D., Yan K., et al. (2018). Overexpression of SmbHLH148 induced biosynthesis of tanshinones as well as phenolic acids in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 37 1681–1692. 10.1007/s00299-018-2339-9 [DOI] [PubMed] [Google Scholar]
- Xu H., Song J., Luo H., Zhang Y., Li Q., Zhu Y., et al. (2016). Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol. Plant. 9 949–952. 10.1016/j.molp.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Wang Q., Liu Y., Hao X., Wang C., Liang Y., et al. (2021). Divergent camptothecin biosynthetic pathway in Ophiorrhiza pumila. BMC Biol. 19:122. 10.1186/s12915-021-01051-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Zhou W., Su J., Wang X., Li L., Wang L., et al. (2017). Overexpression of SmMYC2 increases the production of phenolic acids in Salvia miltiorrhiza. Front. Plant Sci. 8:1804. 10.3389/fpls.2017.01804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yan X. M., Wang J., Li S. S., Liao P., Kai G. Y. (2011). Molecular cloning and expression analysis of a new putative gene encoding 3-hydroxy-3-methylglutaryl-CoA synthase from Salvia miltiorrhiza. Acta Physiol. Plant. 33 953–961. 10.1007/s11738-010-0627-2 [DOI] [Google Scholar]
- Zhang Y., Ji A., Xu Z., Luo H., Song J. (2019). The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza. Plant Mol. Biol. 100 83–93. 10.1007/s11103-019-00845-7 [DOI] [PubMed] [Google Scholar]
- Zhou W., Huang F. F., Li S., Wang Y., Zhou C. C., Shi M., et al. (2016). Molecular cloning and characterization of two 1-deoxy-D-xylulose-5-phosphate synthase genes involved in tanshinone biosynthesis in Salvia miltiorrhiza. Mol. Breed. 36:124. 10.1007/s11032-016-0550-3 [DOI] [Google Scholar]
- Zhou W., Huang Q., Wu X., Zhou Z., Ding M., Shi M., et al. (2017). Comprehensive transcriptome profiling of Salvia miltiorrhiza for discovery of genes associated with the biosynthesis of tanshinones and phenolic acids. Sci. Rep. 7:10554. 10.1038/s41598-017-10215-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Li S., Maoz I., Wang Q., Xu M., Feng Y., et al. (2021a). SmJRB1 positively regulates the accumulation of phenolic acid in Salvia miltiorrhiza. Ind. Crop. Prod. 164:113417. 10.1016/j.indcrop.2021.113417 [DOI] [Google Scholar]
- Zhou W., Shi M., Deng C., Lu S., Huang F., Wang Y., et al. (2021b). The methyl jasmonate-responsive transcription factor SmMYB1 promotes phenolic acid biosynthesis in Salvia miltiorrhiza. Hortic. Res. 8:10. 10.1038/s41438-020-00443-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Sun W., Chen J., Tan H., Xiao Y., Li Q., et al. (2020). Author correction: SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci. Rep. 10:7201. 10.1038/s41598-020-62994-w [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The skeleton of tanshinone biosynthetic pathway in Salvia miltiorrhiza.
Recombinants of SmSCR1-overexpressive and suppressed vectors. (A) Recombinant vector of pHB -SmSCR1-YFP. (B) Recombinant vector of pHB-SmSCR1-SRDX.
Yeast one-hybrid assay of SmSCR1 binding to the SmCPS1 promoter. P1, P2, and P3 represent the three fragments of SmCPS1 promoter sequence from −1 to −700, −701 to −1,400, and −1,401 to −2,100 bp, relative to translation start site, respectively.
Detection of the expression level of SmSCR1 gene in transgenic hairy root lines by qRT-PCR.
Identification of transgenic hairy root lines by PCR. The asterisks on the bar indicate significant differences by t-test compared to the control (**P < 0.01).
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.