Abstract
Regulatory T cells (Tregs) utilize multiple mechanisms to attenuate inflammation and prevent autoimmunity. Tregs residing in peripheral (i.e. nonlymphoid) tissues have specialized functions, specifically skin Tregs promote wound healing, suppress dermal fibrosis, facilitate epidermal regeneration and augment hair follicle cycling. Here, we demonstrated that skin Tregs were transcriptionally attuned to interact with their tissue environment through increased expression of integrin and TGF-β pathway genes that influence epithelial cell biology. We identified a molecular pathway where skin Tregs license keratinocytes to promote innate inflammation following skin barrier breach. Using a single cell discovery approach, we identified preferential expression of the integrin αvβ8 on skin Tregs. Upon skin injury, Tregs utilized this integrin to activate latent TGF-β which acted directly on epithelial cells to promote CXCL5 production and neutrophil recruitment. Induction of this circuit delayed epidermal regeneration but provided protection from Staphylococcus aureus infection across a compromised barrier. Thus, αvβ8 expressing Tregs in skin, somewhat paradoxical to their canonical immunosuppressive functions, facilitated inflammation acutely after loss of barrier integrity to promote host defense against infection.
One Sentence Summary:
Regulatory T cells in skin license pro-inflammatory signaling from keratinocytes acutely following epidermal injury.
Introduction
Barrier tissues such as the gastrointestinal tract, lungs and skin are home to relatively large proportions of Tregs in both rodents and humans (1, 2). Tregs seed these organs early in life and receive local cues that facilitate their survival and function (1, 2). Skin is in constant contact with the outside environment and highly prone to barrier breach in the contexts of both superficial abrasion and full thickness wounding. Thus, regulating immune responses in this organ are important to avoid chronic inflammation incited by environmental stimuli. However, these mechanisms must be precisely balanced in order to control and eliminate potential pathogens. The cellular and molecular mechanisms utilized by Tregs that establish and maintain this balance are largely unknown.
Given the specialized function of tissue Tregs (1–3), we hypothesized that these cells might exhibit a transcriptional and proteomic profile that is imprinted by their environment. Single cell RNA-sequencing studies give credence to this hypothesis by revealing heterogeneity among Tregs present in various lymphoid and non-lymphoid tissues and by identifying transcriptionally related cell subsets primed for a non-lymphoid environment (4, 5). Nonetheless, whether Tregs in skin are specifically adapted for tissue localized, cell-cell communication remains unclear.
Transforming growth factor β (TGF-β) signaling is a compelling candidate pathway to mediate tissue specific Treg functions, as this cytokine plays a major role in tissue repair/remodeling after injury (6–9). Furthermore, there is an intimate association between TGF-β biology and Tregs. This cytokine acts on naive CD4+ T cells to help differentiate them towards Tregs, leading to regulation of various immune responses, especially Th17 inflammation (10–13). In addition, Tregs express the receptors for TGF-β, they can secrete large amounts of TGF-β and they can express integrins capable of activating latent TGF-β (14–16). Despite numerous studies linking TGF-β and Tregs and the role these play in tissue homeostasis, it is unclear if and how Tregs utilize the TGF-β pathway in the context of tissue repair.
Here, we discovered that Tregs residing in skin possessed a tissue specific transcriptional profile that potentiates cell-cell communication with skin epithelial cells. Notably, Tregs in close proximity to keratinocytes were enriched for interacting receptor or ligand gene partners, including Notch1/Jag1, Egfr/Areg, and Tgfb1/Tgfbr2. TGF-β pathway genes, especially the TGF-β activating integrin αvβ8, were among the most preferentially expressed in skin Tregs. Expression of αvβ8, but not TGF-β itself or TGF-β receptor, endowed these cells with an ability to directly promote innate inflammation following epidermal injury. Our results suggest that Tregs in close proximity to skin epithelium maintain tissue integrity by balancing reparative and pro-inflammatory functions.
Results
Regulatory T cells in skin were transcriptionally enriched for TGF-β and integrin pathways
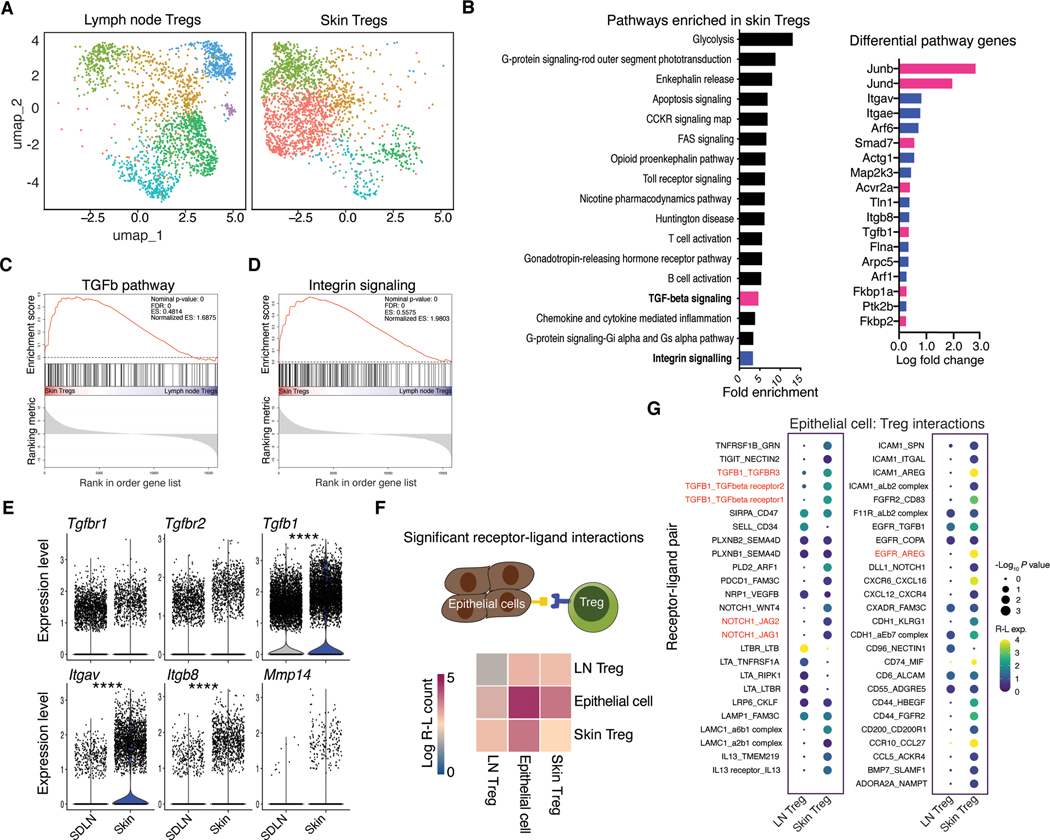
To begin to understand the transcriptional heterogeneity of skin resident Tregs, we performed single cell RNA-sequencing (scRNA-seq) on cells isolated from the skin and skin draining lymph nodes (SDLNs) of Foxp3-GFP reporter mice (17) (Fig. 1a and S1a). Comparative analysis with PANTHER (18) revealed a set of transcriptional pathways preferentially enriched in skin Tregs over Tregs from SDLNs. Interestingly, both TGF-β and integrin signaling were preferentially enriched in skin Tregs, with greater than a 3-fold enrichment in both (Fig. 1B). We performed gene set enrichment analysis (GSEA) (19) to verify increased expression of these pathways in skin Tregs. Both TGF-β and integrin associated genes were significantly elevated in these cells (Fig. 1C, D). At an individual gene level, expression of the TGF-β receptor constituents, Tgfbr1 and Tgfbr2 were not increased in skin Tregs while Tgfb1 was significantly elevated (Fig. 1E). Additionally, both Itgav and Itgb8, which form the TGF-β activating integrin avβ8 (9), were highly enriched in skin Tregs (Fig. 1E). Mmp14, which can aid αvβ8-mediated activation of TGF-β, trended towards significant enrichment (20) (Fig. 1E). Itgb6, another TGF-β activating integrin, was expressed at very low levels on skin Tregs (Fig. S1B). Therefore, comparative transcriptional profiling of Tregs revealed a distinctive skin Treg signature defined by enrichment of TGF-β and TGF-β activating genes.
Fig. 1. Regulatory T cells in skin were transcriptionally attuned for their local tissue environment.
(A) UMAP of unsupervised clustering of single cell RNA-sequencing data comparing skin and lymph node Foxp3+ T cells isolated from Foxp3-GFP mice. Each tissue represents cells pooled from 3–4 mice (B) Significantly enriched (FDR <0.05, Fisher exact test) PANTHER Gene Ontology pathways identified from genes differentially expressed (non-parametric Wilcoxon rank sum test) between lymph node and skin Tregs in single cell sequencing data. (C, D) Gene set enrichment analysis (GSEA) showing enrichment of genes for skin versus lymph node Tregs probing TGF-β (C) or integrin (D) gene sets. (E) Differential gene expression analysis for select TGF-β pathway genes. (F), CellPhoneDB receptor-ligand interaction repository analysis identifying significant (P < 0.05) interactions between lymph node or skin Tregs from single cell RNA-seq data (A) and epithelial cells, reference (21). Heatmap indicates number of significant interactions between two cell populations. (G) Overview of statistically significant interactions between epithelial cells and Tregs using the CellPhoneDB pipeline. Dot size indicates interaction p values and color indicates expression means of the receptor-ligands pairs between 2 cell populations. Single cell RNA-seq data are derived from a single experiment where either skin or lymph node Tregs were pooled from 3–4 mice. For differential gene expression Seurat was used to perform a non-parametric Wilcoxon rank sum test. ****P < 0.0001.
Regulatory T cells in skin were preferentially poised to interact with epithelium
Given the unique transcriptional signature of skin Tregs and their previously described tissue specific functions (1–3), we hypothesized that enriched pathways would include genes functionally relevant to skin epithelium. To directly test this we screened our skin and lymph node Treg scRNA-seq data against a published scRNA-seq dataset of murine epithelial cells using the CellPhoneDB repository of receptor-ligand interactions (21, 22). This analysis revealed that skin derived Tregs had more than double the number of bidirectionally significant receptor-ligand interaction pairs than Tregs from the SDLNs (Fig. 1F, G). Among significant Treg-epithelial cell interactions, 31 were specific to skin Tregs. Notably, significant interactions included gene pathways identified to be involved in tissue specific Treg function, including Notch-Jag and Egfr-Areg (23, 24). Tgfb1-Tgfbr interactions were also preferentially enriched in skin Tregs (Fig. 1G).
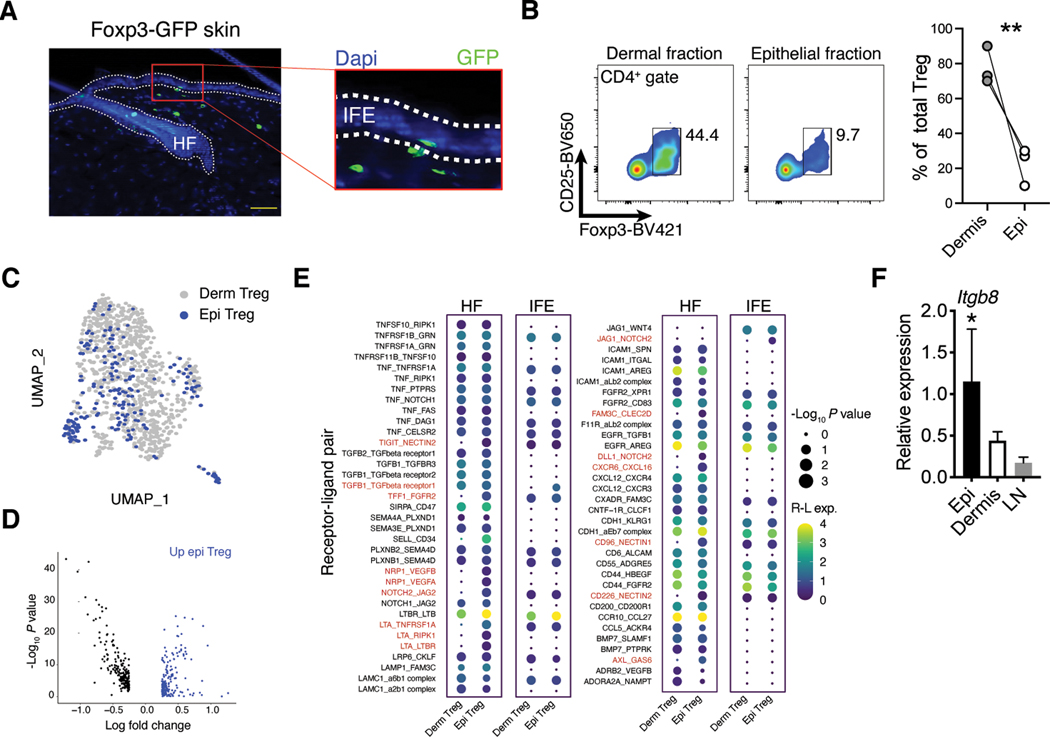
We have previously observed that Tregs in skin primarily localize to hair follicles where they facilitate epithelial stem cell differentiation through Notch-Jag1 interactions (23, 25). Nonetheless, specific analysis of skin from Foxp3-GFP mice revealed that a small fraction of Foxp3+ cells localize to other anatomically distinct portions of skin, mainly the interfollicular epidermis (IFE) (Fig. 2A). This region forms the functional skin barrier and is preferentially inhabited by dendritic epidermal T cells (DETCs) expressing a restricted γδTCR (26). To further elucidate skin-Treg epithelial interactions, we utilized an established enzymatic approach to separate skin epithelium from dermal layers (27). This method successfully isolated epithelial localized cells as demonstrated by enrichment of DETCs (Fig. S1C). Tregs were readily identified in the epithelial fraction by flow cytometry, albeit at significantly reduced frequencies when compared to the dermis (Fig 2B).
Fig. 2. Both dermal and epidermal Tregs were poised to interact with skin epithelium.
(A) Representative immunofluorescence imaging of skin sections from Foxp3-GFP mice and counterstained with DAPI. HF = hair follicle; IFE = interfollicular epidermis; scale bar is 200 μm. (B) Flow plots and quantification of Foxp3+ cells in the epithelial and dermal skin fractions. Lines indicated matched samples from the same skin sample (C) UMAP of unsupervised clustering and integration of scRNA-seq data from dermal and epithelial derived Tregs isolated from Foxp3-GFP mice. Each sample is pooled from 4 mice. (D) Identified differentially expressed genes between dermal and epithelial Tregs calculated from nonintegrated data scRNA-seq data. (E) CellPhoneDB identified significant (P < 0.05) receptor-ligand interactions between dermal node or epithelial Tregs from scRNA-seq data in (C) and epithelial cells from reference (21). Epithelial cells were further stratified into hair follicle or interfollicular epidermis cells based on Krt14, Mt2, Cd34, Ptn, and Postn expression (see supplemental figure 1). Dot size indicates interaction p values and color indicates expression means of the receptor-ligands pairs between 2 cell populations. (F) Quantitative RT-PCR for Itgb8 expression on Tregs isolated from epidermis, dermis, or SDLN and normalized to Hprt. All data are representative of at least three independent experiments and error bars represent SD, n = 3–4 mice per group. For comparison between two groups, Student’s two-tailed t test was used. For comparison between multiple groups, one-way ANOVA was used. *P<0.05, **P < 0.01.
We hypothesized that Tregs associated with the epithelial skin niche represent a distinct transcriptional subset. To test this, we isolated Tregs directly from dermal or epithelial fractions and performed scRNA-seq on both groups. Contrary to our hypothesis, unsupervised clustering analysis and sample integration revealed little difference between the two cell populations, with the epithelial fraction of Tregs homogenously overlapping with dermal Treg clusters (Fig. 2C) (28). However, despite overall similarities, many genes were differentially expressed between these fractions (Fig. 2D). While this divergence could in part be due to small differences in tissue processing (i.e., the dermal fraction is exposed to an additional enzymatic digestion step), receptor-ligand pairing analysis identified 16 receptor-ligand interactions unique to epithelial Tregs (Fig. 2E). We further stratified the epithelial compartment into hair follicle and IFE cells based on Krt14, Mt2, Cd34, Ptn, and Postn expression and examined significant interaction pairs (Fig. S1D). Most interactions unique to epithelial Tregs were with hair follicle partner cells, as Tregs closely associate with the hair follicle epithelium (23, 25). Interestingly, interactions between Tgfb1 and Tgfbr1 were enriched with the IFE (Fig. 2E). Consistent with this finding, quantitative RT-PCR analysis of purified epithelial Tregs revealed these cells preferential expressed Itgb8 when compared to Tregs purified from the dermis or SDLNs (Fig. 2F). While, TGF-β stimulation of dermal and lymph node Tregs induced similar phosphorylation levels of the downstream signaling protein SMAD2/3, epithelial associated Tregs exhibited slightly increased SMAD2/3 phosphorylation (Fig. S1E). Taken together, these data demonstrate that dermal and epidermal Tregs are largely similar with both populations preferentially expressing transcriptional pathways relevant to epithelial cell biology.
Activation of latent TGF-β by Tregs delayed epidermal repair
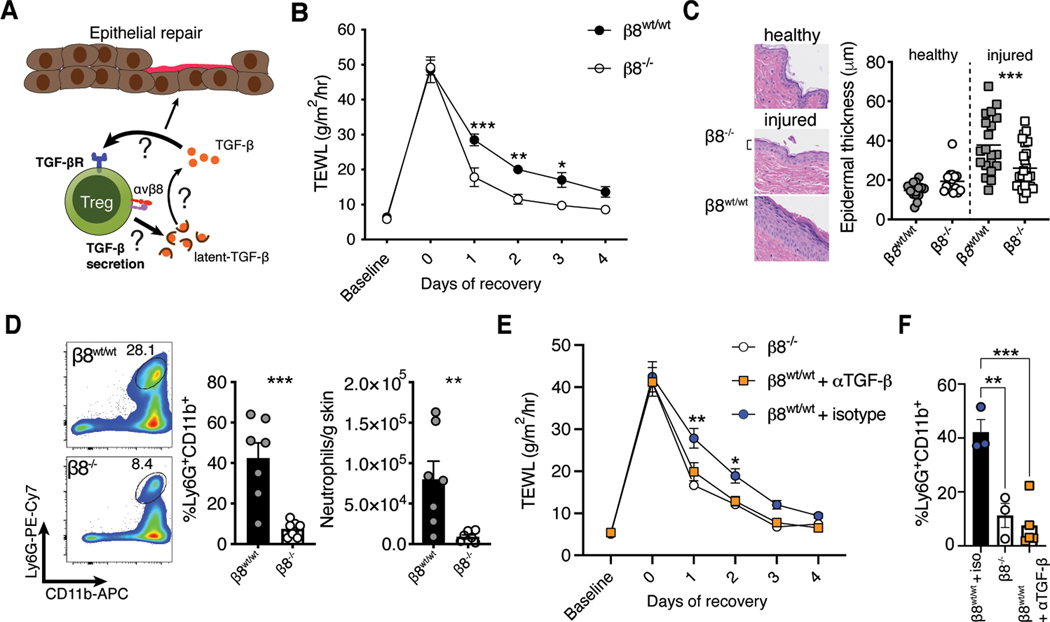
We have previously observed that Tregs facilitate epidermal barrier repair following superficial skin abrasion (29). In this model, repeated application of adhesive tape disrupts the stratum corneum while leaving the underlying tissue layers intact. Following this form of injury, Tregs attenuate the CXCL5/IL-17/neutrophil axis, and in doing so, enable hair follicle stem proliferation and differentiation to help regenerate the epithelium (29). Given these findings and the important role for TGF-β signaling in wound repair (6), we hypothesized that skin Tregs harness the TGF-β pathway to modulate epithelial responses to injury (Fig. 3A). The specific enrichment of Itgb8 on epithelial associated Tregs led us to postulate that Treg activation of latent-TGF-β could regulate epithelial cell activity following injury. Unlike other integrins, αvβ8 does not transmit tensile force through the actin cytoskeleton and its only known function is activation of latent-TGF-β via conformational exposure of TGF-β to TGF-βR (30–32). Interestingly, murine Tregs in secondary lymphoid organs have been reported to increase expression of αvβ8 after activation, consistent with the activated signature of skin Tregs (Fig. 1B) (14, 15).
Fig. 3. Activation of latent TGF-β by Tregs delayed epithelial repair.
(A) Schematic depicting our hypothesis that skin Tregs harness the TGF-β pathway to modulate epithelial responses to injury. scRNA-seq analysis identified skin Tregs as poised to respond to TGF-β, secrete TGF-β, and activate latent TGF-β. All of these functions could regulate epithelial cell biology. (B) Kinetic transepidermal water loss (TEWL) measurements taken before and after tape stripping Foxp3Cre-ERT2Itgb8f/f mice treated with either vehicle (β8wt/wt) or tamoxifen (β8−/−). (C) Representative histology and quantification of epidermal thickness comparing β8wt/wt and β8−/− mice before or two days after injury. Symbols indicate individual measurements. (D) Flow cytometric quantification of neutrophils in skin following tape stripping. (E, F) Treg β8wt/wt mice were treated i.d. with 100 ug of TGF-β neutralizing or isotype control antibody immediately prior to tape stripping. TEWL (E) and neutrophil accumulation (F) were monitored as before. All data are representative of at least two independent experiments, n = 3–7 mice per group; error bars are SEM. Statistical analysis was performed using two-way ANOVA and Sidak’s multiple comparisons test (B, E, F) or a unpaired Student’s two-tailed t test (C, D). ***P<0.001.
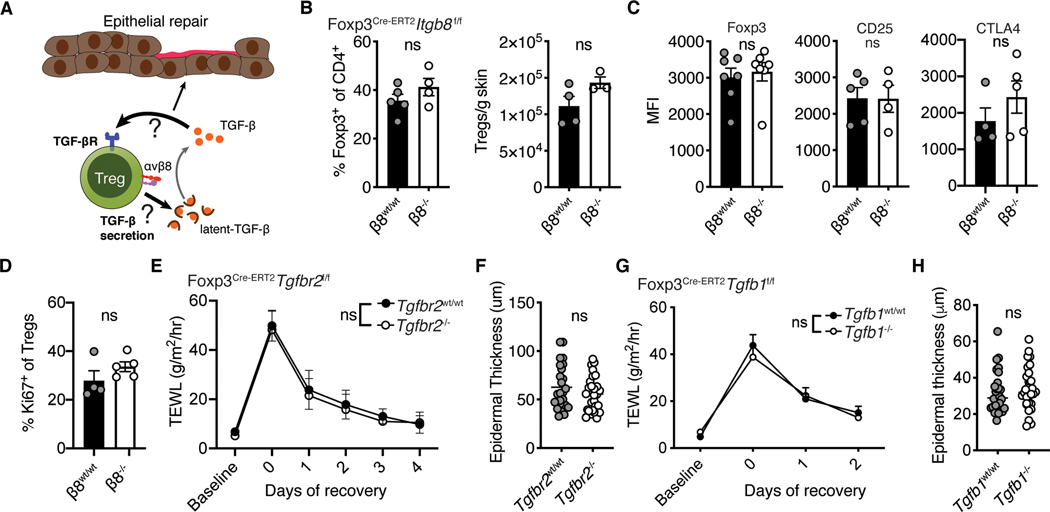
To test if Itgb8 was involved in Treg responses to epithelial injury, we bred Foxp3ERT2-Cre mice (33) to Itgb8flox/flox animals (34) to generate an inducible and lineage specific model for deleting αvβ8 in Tregs. Sex matched Foxp3ERT2-CreItgb8flox/flox littermates were treated with tamoxifen or vehicle control and subjected to superficial skin injury by tape stripping the stratum corneum (Fig. S2A). Administration of tamoxifen reduced Treg expression of Itgb8 although deletion was incomplete (Fig. S2B). Skin barrier recovery was quantified by measuring transepidermal water loss and epidermal thickness. Surprisingly, deletion of αvβ8 on Tregs resulted in significantly faster epithelial barrier repair, with reduced transepidermal water loss early in the recovery process (Fig. 3B). Consistent with this finding, tamoxifen treated mice also exhibited significantly reduced epidermal thickening (Fig. 3C). These results were recapitulated in Foxp3CreItgb8flox/flox animals whose Tregs are constitutively deficient in αvβ8 (Fig. S2C, D).
Epidermal injury induces an inflammatory response that recruits neutrophils into skin. Excessive influx of these cells hinders epithelial differentiation and delays restoration of the barrier (29). We therefore surveyed immune cell populations in our Foxp3Cre-ERT2Itgb8flox/flox animals (Fig. S3A). Following tape stripping, tamoxifen treated animals exhibited strikingly reduced accumulation of neutrophils in the skin (Fig. 3D). To directly link deletion of αvβ8 on Tregs with TGF-β activity, we treated wild type mice with TGF-β neutralizing antibody immediately prior to tape stripping. In contrast to isotype treatment, TGF-β blockade closely mimicked epidermal repair kinetics and impaired neutrophil recruitment observed with Treg deletion of Itgb8 (Fig. 3E, F). These results support the hypothesis that activation of latent TGF-β by Tregs is a relevant pathway in epithelial repair. Surprisingly, our data point to a mechanism whereby Treg utilization of this pathway impairs barrier repair capacity instead of facilitating it.
αvβ8 expressing Tregs promoted innate skin inflammation
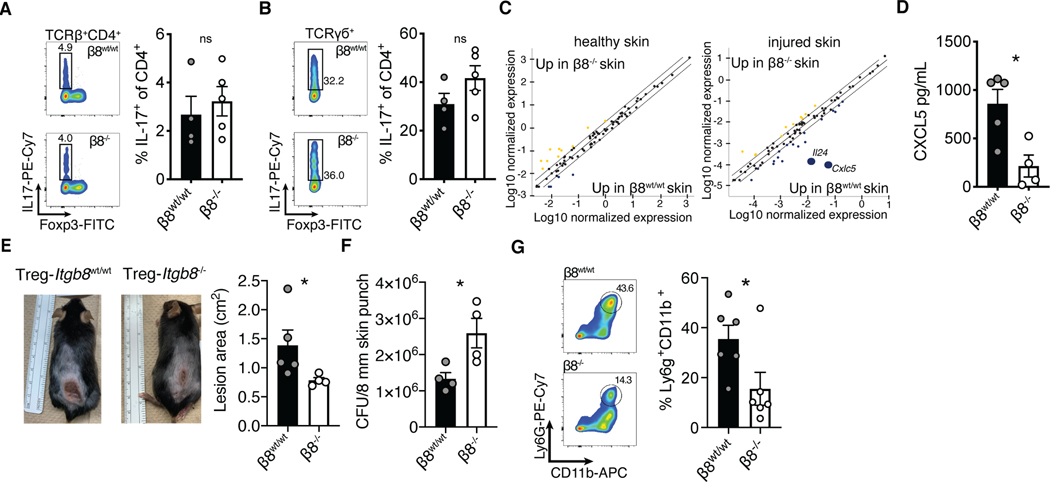
Delayed barrier repair, epidermal thickening, and increased neutrophil recruitment are markers of skin inflammation (29). Thus, the delayed epidermal regeneration observed in control animals suggested that the skin of mice with functional integrin αvβ8 on Tregs (i.e., wild type Tregs) was more inflamed (Fig. 3). Because Tregs efficiently restrain IL-17 mediated inflammation and Th17 cell differentiation can be induced by TGF-β (29, 35), we examined whether IL-17 production was enhanced in skin of mice that express functional integrin αvβ8 on Tregs. Following barrier injury, there was no difference between wildtype and Foxp3ERT2-CreItgb8flox/flox animals in IL-17 production from either αβ or γδ T cells, indicating that integrin αvβ8 expression on Tregs played no discernable role in regulating the IL-17 pathway in skin (Fig. 4A, B). Similarly, we did not observe any difference in accumulation of Th1 or Th2 cells (Fig. S3B, C).
Fig. 4. αvβ8+ Tregs promoted innate skin inflammation.
(A, B) Tregs from Foxp3Cre-ERT2Itgb8f/f mice treated with either vehicle or tamoxifen were phenotyped by flow cytometry for IL-17 expression in skin CD4+ αβT cells (A), or γσT cells (B) after epithelial injury. (C) Whole skin quantitative PCR array analysis comparing cytokine and chemokine gene expression levels between β8wt/wt and β8−/− healthy, or tape strip injured mice. (D) Whole skin biopsy lysate ELISA for CXCL5. (E, F) Treg β8wt/wt and β8−/− mice intradermally infected with 5×107 CFU Staphylococcus aureus (USA300) were monitored for six days before an 8 mm punch biopsies surrounding lesions were excised for bacterial colony forming unit analysis. (G) Representative flow plots and quantification of neutrophil accumulation in skin following Staphylococcus aureus infection. Data are representative of at least three independent experiments, n = 4–7 mice per group; error bars are SEM. Statistical analysis was performed with a unpaired Student’s two tailed t test. *P<0.05.
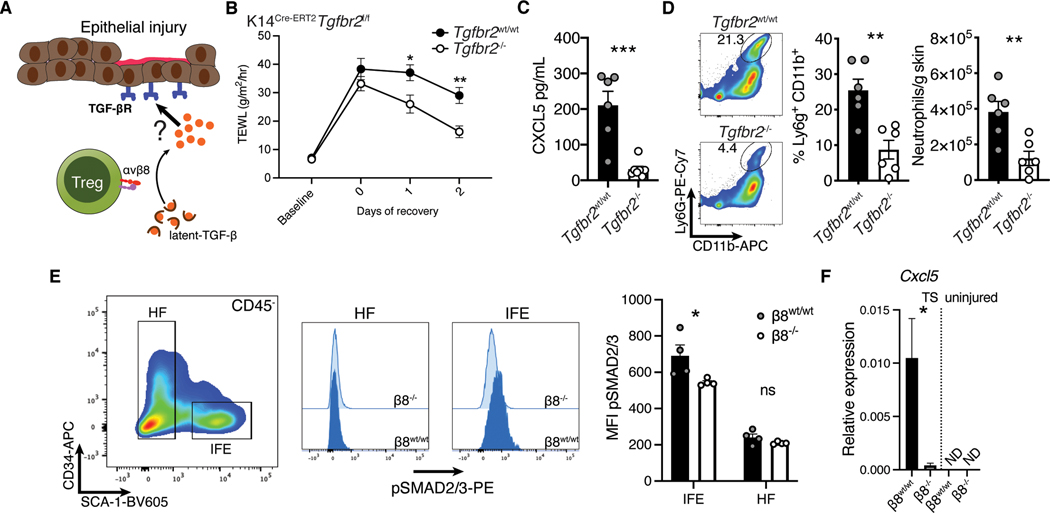
To screen for candidate inflammatory mediators induced by αvβ8 expressing Tregs, we quantified cytokine and chemokine expression in tape stripped and uninjured whole skin tissue by qPCR array. While there were no overt differences at steady-state, two days following barrier injury the expression of both Il24 and Cxcl5 were dramatically reduced in Treg Itgb8 deleted mice (Fig. 4C). Cxcl5 codes for the chemokine CXCL5 and is known to be a potent driver of neutrophil accumulation in tissues (36, 37). We confirmed the reduction in CXCL5 protein levels by ELISA on whole skin lysates (Fig. 4D).
Given the proinflammatory function of Tregs in promoting neutrophil recruitment, we sought to determine whether the presence or absence of αvβ8 on Tregs affects their lineage identity and/or stability as regulatory cells. To this end, we quantified transcript levels of Treg lineage defining genes (Foxp3, Il2ra, Ctla4, Icos and Ikzf2) between Itgb8+ and Itgb8− Tregs in our skin single cell RNAseq dataset. This analysis revealed no significance differences in Treg defining genes between these populations (Fig. S3D). In addition, we tested whether deletion of Itgb8 on Tregs results in loss of Treg identity and/or stability during steady-state. Foxp3ERT2-CreItgb8flox/flox mice treated with tamoxifen proliferated normally (Fig. S3E). There was also normal Treg frequencies and absolute numbers in skin (Fig. S3F). Tregs in these mice exhibited equivalent protein levels of Foxp3 and CD25 when compared to vehicle treated controls (Fig. S3G). Taken together, these results demonstrate that bona fide Tregs mediated activation of latent TGF-β, which acted to enhance CXCL5 production and neutrophil accumulation acutely following skin barrier injury. While this comes at a cost of delayed epidermal regeneration, this effect is only transient, as barrier recovery in mice with functional integrin αvβ8 on Tregs is equivalent to that of αvβ8-deficient mice 4 days after injury (Fig. 3B).
Activation of latent TGF-β by Tregs protected against bacterial skin infection
A defining function of skin epithelium is to form a tissue barrier to protect against pathogens. Following injury, tissue repair processes act to restore barrier function but do so in the context of immediate and ongoing anti-microbial immune responses. We hypothesized that Tregs in skin utilize the αvβ8/TGF-β pathway to facilitate anti-microbial immunity immediately following epithelial injury. This concept is not without precedent, as Tregs in the vaginal mucosa help facilitate tissue entry of specific immune cell populations in order to augment early protective immune responses to viral infections (38). To test this hypothesis, we infected the skin of Foxp3CreItgb8flox/flox or tamoxifen treated Foxp3Cre-ERT2Itgb8flox/flox mice with Staphylococcus aureus and quantified skin infection area and bacterial burden relative to control animals. Mice with Tregs deficient in Itgb8 exhibited significantly smaller lesions surrounding the injection site consistent with reduced skin inflammation previously observed in these animals (Fig. 4E). However, these mice maintained approximately double the bacterial burden, indicating an inability to control the infection (Fig. 4F). Neutrophil infiltration was also reduced (Fig. 4G). Taken together, these data indicate that while expression of αvβ8 on skin Tregs slightly reduced the efficiency of epithelial repair, it simultaneously served to enhance innate immunity against bacterial threats occurring immediately following skin barrier breach.
TGF-β signaling did not influence Tregs in a cell intrinsic fashion during epidermal repair
We next set out to elucidate the mechanisms by which Treg mediated activation of latent TGF-β leads to enhanced innate inflammation following epidermal injury. As skin Tregs are highly enriched in TGF-β pathway genes, including Tgbr2 and Tgfb1, we hypothesized that integrin αvβ8-mediated activation of latent TGF-β by skin Tregs works in an autocrine fashion to control their homeostasis and suppressive capacity in skin (Fig. 5A). Following epidermal injury, there were no differences in the frequency or absolute numbers of Tregs in either the skin or SDLNs of Treg Itgb8 deleted mice (Fig. 5B and Fig. S3H). In addition, flow cytometric quantification of critical Treg effector molecules, such as Foxp3, CD25 and CTLA-4, revealed no differences between these groups (Fig. 5C). Levels of Ki67 were also unchanged, indicating that αvβ8 deficient Tregs proliferate normally in vivo after skin barrier disruption (Fig. 5D). These results suggest that activation of latent TGF-β by integrin αvβ8 does not markedly alter Treg numbers or activation in the context of skin barrier breach.
Fig. 5. Cell intrinsic TGF-β signaling did not drive Treg responses to epidermal injury.
(A) Schematic depicting the possibility that skin Tregs either respond to, or release, TGF-β to regulate epithelial repair. TGF-β secretion could alter tissue bioavailability of this cytokine while TGF-β sensing by Tregs has the potential to alter their functionality. (B-D) Tregs from Foxp3Cre-ERT2Itgb8f/f mice treated with either vehicle or tamoxifen were phenotyped by flow cytometry for (B) their frequency and absolute abundance in skin, (C) their expression of lineage defining proteins, (D) and the frequency of proliferating cells. (E) Kinetic transepidermal water loss measurements taken before and after tape stripping Foxp3Cre-ERT2Tgfbr2f/f mice treated with either vehicle or tamoxifen. (F) Epidermal thickness two days following injury, as measured from histology. (G) Kinetic transepidermal water loss measurements taken before and after tape stripping Foxp3Cre-ERT2Tgfb1f/f mice treated with either vehicle or tamoxifen. (H) Epidermal thickness two days following injury, as measured from histology. Data are representative of at least two independent experiments, n = 4–7 per group; error bars are SEM. Statistical analysis was performed using two-way ANOVA and Sidak’s multiple comparisons test (E, G) or a unpaired Student’s two-tailed t test (B-D, F, H).
To definitively test whether TGF-β signaling acts on Tregs in an autocrine fashion to influence their function during epidermal repair, we generated Foxp3Cre-ERT2Tgfbr2f/f mice (39). These animals are inducibly refractory to TGF-β signaling due to tamoxifen sensitive deletion of TGF-βRii (which is required for productive signaling through all TGF-β receptors (39, 40)), enabling us to determine if TGF-β acts directly on Tregs. Tamoxifen treatment reduced Tgfrb2 transcript, protein levels and impaired pSMAD2/3 induction after TGF-β stimulation (Fig. S4A–C). Deletion of TGF-β signaling in Tregs had no effect on skin barrier regeneration, as Foxp3Cre-ERT2Tgfbr2flox/flox animals treated with tamoxifen showed no difference from sex-matched littermate vehicle treated controls with respect to transepidermal water loss or epidermal thickness (Fig. 5E, F). Th17 cells also accumulated normally (Fig. S4D).
In addition to activating latent TGF-β and responding to this cytokine, Tregs can synthesize latent TGF-β (41) and this molecule was highly enriched in skin Tregs (Fig. 1E). Thus, we set out to determine if Treg production of this cytokine plays a role in their early response to epithelial injury (Fig. 5A). To do so, we generated Foxp3Cre-ERT2Tgfb1flox/flox animals in which latent TGF-β production can be inducibly depleted in Tregs (Fig. S4E, F). Upon barrier disruption, mice deficient in Treg production of latent TGF-β had no discernable differences in transepidermal water loss or epidermal thickness when compared to age- and sex-matched littermate controls (Fig. 5G, H). Taken together, these results suggest that activation of latent TGF-β and not secretion or cell intrinsic sensing of this cytokine was the primary mechanism active in skin Tregs to promote inflammation after epithelial injury. This raised the possibility that Treg activation of latent-TGFβ acts extrinsically on other cell populations in skin.
Epithelial cell TGF-β sensing promoted neutrophil recruitment during injury
Given that Tregs were primed to interact with keratinocytes (Fig. 1F,G and 2E), we hypothesized that Treg mediated activation of TGF-β could act directly on these cells (Fig. 6A). To test this, we generated K14Cre-ERT2Tgfbr2f/f animals which allows lineage specific inducible deletion of Tgfbr2 in keratinocytes (Fig. S4G) (42). We treated age and sex-matched littermates with tamoxifen or vehicle control, disrupted the epithelial barrier and quantified epidermal regeneration over time. We observed a significant improvement in transepidermal water loss in mice with a keratinocyte specific deletion of Tgfbr2, similar to that observed with Foxp3CreItgb8flox/flox animals (Fig. 6B). ELISA quantification of CXCL5 on whole skin lysates revealed significantly less of this chemokine in mice with keratinocyte-specific deletion of Tgfbr2 (Fig. 6C). Concordantly, both the frequency and absolute number of skin infiltrating neutrophils was decreased in Tgfbr2 deficient animals (Fig. 6D). To demonstrate that these effects were due to Treg αvβ8 expression, we examined epithelial TGF-β signaling in Treg Itgb8 deleted mice. In Itgb8 deficient animals IFE cells, but not hair follicle associated cells, had reduced SMAD2/3 phosphorylation. (Fig. 6E). Moreover, IFE cells exhibited significantly less upregulation of Cxcl5 expression following tape strip injury (Fig. 6F). Thus, TGF-β signaling in keratinocytes following epidermal injury led to increased CXCL5 production and neutrophil recruitment. Taken together, our data suggests that Tregs are uniquely positioned and possess the molecular machinery to help mediate this effect.
Fig. 6. TGF-β signaling in keratinocytes promoted neutrophil recruitment acutely following epidermal injury.
(A) Schematic representation of our hypothesis that during epithelial injury keratinocytes sense TGF-β activated by Tregs. While Treg intrinsic sensing of TGF-β was not important during epithelial repair, a paracrine response through a secondary cell type was possible. (B) Kinetic transepidermal water loss measurements taken before and after tape stripping K14Cre-ERT2Tgfbr2f/f mice treated with either vehicle (Tgfbr2wt/wt) or tamoxifen (Tgfbr2f/f). (C) Whole skin biopsy lysate ELISA for CXCL5. (D) Flow cytometric quantification of neutrophils in skin two days after injury. (E) Keratinocytes from Foxp3Cre-ERT2Itgb8f/f mice treated with either vehicle or tamoxifen were phenotyped by flow cytometry for SMAD2/3 phosphorylation after injury. (F) CD45−Sca-1+ epidermal cells from Treg β8wt/wt and β8−/− mice were sorted from tape stripped, or uninjured mice and analyzed for Ccxl5 expression by qPCR. Data are representative of three independent experiments, n = 4–6 mice per group; error bars are SEM. Statistical analysis was performed using two-way ANOVA and Sidak’s multiple comparisons test (B) or a unpaired two-tailed t test (C-F).
Discussion
Regulatory T cells have classically been defined by their ability to robustly control inflammation and promote immune tolerance. Nonetheless, it is increasingly recognized that tissue resident Tregs influence their local environment in many ways that are seemingly independent of their role in suppressing inflammation (2, 3, 23, 29, 43). Our data is consistent with others (38) and supports a model whereby Tregs in barrier tissues enable specific inflammatory pathways acutely after barrier breach. Implicit in this model is that subsets of these cells maintain molecular pathways for instructing tissue biology beyond dampening inflammation. These pathways appear to be driven by tissue specificity and heterogeneity within the Treg pool.
Here, we provide evidence of a transcriptional profile unique to skin Tregs and tailored for specific interaction with cell types comprising their tissue niche. This module was defined by enrichment of both TGF-β and integrin pathway genes. Notably, the genes coding for integrins αv and β8, which belong to both gene sets, were preferentially enriched on skin Tregs and even further on Tregs closely associated with skin epithelium. By examining the transcriptional states of both Tregs and their epithelial counterparts, we were able to find a significant co-association of these pathways. These interactions were relatively absent in lymph node derived Tregs. In functionally dissecting the TGF-β molecular circuit in skin Tregs, we revealed that αvβ8 expressing cells activate latent TGF-β to work in a cell extrinsic fashion to induce keratinocytes to increase expression of the neutrophil chemoattractant CXCL5. While this process slightly delayed epithelial regeneration after injury, it provided protection from bacterial infection.
Activation of TGF-β by αvβ8 expressed on Tregs suppresses inflammatory T cell responses and attenuates T cell mediated colitis (14). Our results are consistent with this activity as we have uncovered a locally regulated circuit that underlines the importance of tissue context in the integration of barrier repair and functional immunity. Epithelial cells were the dominant recipient of Treg activated TGF-β in the context of barrier breach, but we have not ruled out the possibility that effector T cells are also suppressed by this activity. TGF-β simultaneously mediating opposed functions is not unprecedented, as this cytokine is able to exert both pro- and anti-inflammatory responses in tissues (7–9). The TGF-β pathway can augment neutrophil accumulation and activation (including through induction of CXCL5) and Tregs themselves are capable of producing neutrophil recruiting chemokines (44–47). Notably, expression of αvβ8 on skin Tregs delayed epithelial regeneration but did not prevent it. In addition, activation of latent TGF-β and not secretion of this cytokine by Tregs predominated in this mechanism, suggesting that already available TGF-β was necessary to mediate this acute response. Expression of another TGF-β activating integrin, αvβ6, is functionally important in the IFE (48). Interestingly, skin Tregs did not express appreciable amounts of αvβ6. However, it is possible αvβ6 expression on non-immune cells synergizes with Tregs to activate latent TGF-β during epidermal injury. A limitation of our study is that we have not dissected how and if αvβ8 and αvβ6 expression on other cell types contribute to epidermal barrier repair. Although we demonstrated deletion of Tgfbr2 and Tgfb1 Tregs at both mRNA and protein levels, we were unable to verify protein knockout of αvβ8 with currently available reagents. In addition, deletion of all three genes was not completely efficient in Tregs. Given that tamoxifen induced deletion of Itgb8 was incomplete, it is likely that our results underreported the magnitude and physiological significance of this pathway.
Seemingly paradoxical to the current study, our previous work demonstrated that Tregs promote skin healing by suppressing the CXCL5-neutrophil axis (29). However, several notable features of our collective data indicate that these results are not inconsistent. Rather, together they reveal a critical role for skin Tregs in maintaining tissue integrity by establishing a balance between the opposing need for protective immunity and tissue repair. Ablation of all Tregs leads to robust skin inflammation and hinders epithelial regeneration, but the potency of this approach obscures more nuanced activity of individual molecular pathways. Data presented herein does not suggest that Tregs lose their suppressive capabilities when expressing αvβ8. Rather, acquisition of this molecular pathway in the context of epithelial injury allowed rapid promotion of neutrophilic infiltration through additional independent rather than opposed mechanisms. This is supported by our observations that skin Tregs maintain normal suppression of IL-17 production from effector T cell populations. Thus, deletion of αvβ8 on Tregs uncoupled Th17 immunity and neutrophil recruitment in the context of epidermal injury. We postulate that co-option of inflammatory pathways by epithelial associated Tregs may have arisen from evolutionary pressure to balance the kinetics of tissue healing with defense against pathogens. In this context, tissue Tregs may serve as a fulcrum balancing the need for epithelial regeneration with that of acute host defense.
It is becoming increasingly accepted that Tregs residing in non-lymphoid organs are functionally heterogenous across tissues and distinct from their lymphoid counterparts. Our data supports this notion. The fact that these cells promote optimal skin function by both suppressing aberrant inflammation and selectively recruiting inflammatory cells raises a number of interesting avenues for further investigation. In particular, several skin diseases, such as atopic dermatitis, are characterized by uncontrolled inflammation, barrier disruption, and recurrent bacterial or viral infection (49). Interestingly, patients with congenital Treg defects develop this skin disease (50). Thus, functional manipulation of specific pathways active in subsets of Tregs may be an attractive approach to more precisely restore the balance between inflammation and barrier integrity in patients with atopic dermatitis and other conditions characterized by compromised barrier integrity and chronic inflammation.
Materials and Methods
Study design:
The objective of this study was to define the transcriptional identity and functional activity of regulatory T cells present in skin. The TGF-β pathway was of specific interest. To investigate this we surveyed the transcriptional landscape of Tregs and epithelial cells isolated from murine skin with scRNA-seq. Functional investigations were carried out with inducible Cre/Lox mouse model systems. Replicate information and animal numbers are indicated in appropriate figure legends. Quantification of histology was performed blinded.
Mice:
Foxp3GFP (B6.Cg-Foxp3tm2(EGFP)Tch/J), Foxp3Cre (B6.129(CG)-Foxp3tm4(YFP/icre)Ayr/J), Foxp3Cre-ERT2 (Foxp3tm9(EGFP/cre/ERT2)Ayr/J), and K14Cre-ERT2 (Tg(KRT14-cre/ERT)20Efu/J) mice were purchased from The Jackson Laboratory. Itgb8flox/flox (B6.129-Itgb8tm2Lfr) animals were a kind gift of Dr. Dean Sheppard (UCSF), Tgfbriiflox/flox (B6;129-Tgfbr2tm1Karl/J) animals were gifted by Dr. Qizhi Tang (UCSF), and Tgfb1flox/flox (C57BL/6J-Tgfb1em2Lutzy /Mmjax) mice were gifted by Dr. Max Krummel (UCSF). Cre lines were crossed with LoxP strains until homozygosity to generate animals conducive for inducible and lineage specific gene deletion. Cre-ERT2 animals were treated for 5 consecutive days with 2.5 mg tamoxifen dissolved in corn oil i.p. (Sigma-Aldrich) to induce Cre recombinase activity. All animal experiments were performed on littermate age and sex-matched 6–16 week old mice maintained through routine breeding at the UCSF School of Medicine in a specific pathogen free facility. Experimental procedures were approved by IACUC and performed in accordance with guidelines established by the Laboratory Animal Resource Center (LARC) at UCSF.
Skin injury, bacterial infection, and TGF-β blocking
Mechanical injury of mouse skin and transepidermal water loss were measured as previously described (29). Briefly, the entire back skin was shaved and tape stripped 5–10 times using cellophane tape for three consecutive days. Transepidermal water loss was measured with a Tewameter TM 300 probe (Courage+Khazaka electronic GmbH) according to the manufacturer’s protocols. Baseline (prior to tape stripping), day 0 (immediately following last tape strip), and recovery (every 24 hours thereafter) measurements were collected on each quadrant of the back skin to monitor epidermal healing. For bacterial injection mice were challenged intradermally (i.d.) with 5×107 CFU of Staphylococcus aureus strain USA300. Bacterial burden was measured on day 6 by excising an 8 mm punch of skin surrounding the lesion injection site, homogenizing tissue with a TissuRuptor (Qiagen), and plating supernatants in a dilution series on chloramphenicol containing agar petri dishes. Colonies were manually counted after an overnight incubation. TGF-β blocking experiments were conducted by a single i.d. injection of 100 ug/mouse of clone 1D11.16.8 (ThermoFisher), or isotype control MOPC-21 (BioXCell) immediately prior to the first round of tape stripping. Mice were sedated with isoflurane during shaving, mechanical injury, and TEWL measurements.
Tissue processing:
Single cell suspensions of skin were prepared by finely mincing tissues and digesting 45 minutes in an enzyme cocktail containing collagenase XI, DNase, and hyaluronidase in complete RPMI at 37°C. For separation of epithelial and dermal fractions skin sections were floated on thermolysin containing buffer, as previously described (27). Lymphoid tissues we mashed through a 40 μm filter to release leukocytes. ELISA analysis was performed by homogenizing whole skin punch biopsies and lysing cells with Cell Lysis Buffer II (R&D Systems). CXCL5 was detected with the Mouse LIX DuoSet ELISA kit (R&D Systems). For quantitative PCR analysis of whole skin RNA was extracted using the RNeasy Fibrous Tissue Kit (Qiagen) and the RT2 Profiler PCR Array Kit (PAMM-150ZC, Qiagen) used for cDNA synthesis and amplification. Itgb8, Tgfbr2, Tgfb1, and Cxcl5 expression on sorted cells was quantified using the TaqMan Gene Expression Cells-to-CT Kit (Thermo Fisher Scientific) and TaqMan assays Mm00623991_m1, Mn03024075_m1, Mm03024091_m1, Mm00436451_g1, and Mm01178820_m1.
For histological analysis skin tissue was fixed in 10% formalin and paraffin-embedded. Tissue was stained with hematoxylin and eosin (H&E) by the University of California San Francisco Mouse Pathology Core. Epidermal thickness was quantified from H&E slides in a blinded manner by measuring 5 randomly selected points per section using Zen 2 (Zeiss) software analysis tools. Immuofluorescent tissue staining was performed by fixing skin from FoxpGFP reporter mice with 2% PFA for 4 hours, washed with PBS and left in 30% sucrose overnight before embedding in OCT and freezing. Slides were mounted with DAPI containing medium before imaging on a standard fluorescent microscope.
Flow cytometry and cell sorting:
Cells prepared as described in the above tissue processing section were stained with Ghost 510 viability dye (Tonbo Biosciences) in PBS. Surface makers were stained in PBS with 2% FCS. Cells were fixed and permeablized with the Foxp3/Transcription Factor Staining Buffer Set (eBiosciences) prior to staining intracellular antigens. For cytokine analysis, cells were stimulated 4 hours with phorbol ester and ionomycin (Cell Stimulation Cocktail, Tonbo Biosciences) prior to staining. For flow cytometric analysis, samples were run on a LSRFortessa analyzer (355; 405; 488; 532; 561; 640 nm laser configuration; BD Biosciences) in the UCSF flow cytometry core and collected using FACS Diva software (BD Biosciences). Compensation was performed using UltraComp eBeads as single color controls (ThermoFisher Scientific). Data was analyzed using FlowJo software (Tree Star Inc.). FACS isolation of Tregs was performed using a FACSaria Fusion sorter (BD Biosciences). Foxp3GFP reporter mice were used to allow identification of CD45+TCR-β+CD4+Foxp3+ cells.
Fluorophore conjugated antibodies specific for mouse and human antigens were purchased from eBioscience, BD Biosciences, R&D Systems, and Biolegend. Antibodies for staining mouse cells: α-CD8α (clone 53–6.7); α-TCR-β (clone H57–597); α-CD4 (clone GK1.5); TCR-γ (clone GL3); α-CD45 (clone 30-F-11); α-Ki67 (clone B56); α-Ly6G (clone 1A8); α-CD11b (clone M1/70); α-Foxp3 (clone FJK-16s); α-CTLA-4 (clone UC10–4F10–11); α-CD25 (clone PC61.5); α-CD34 (clone MEC14.7); α-IL-17 (TC11–18H10.1), α-IL-5 (TRFK5), α-IFN-γ (XMG1.2), α-SCA-1 (clone D7); α-pSMAD2/3 (clone 072–670); α-TGF-βR2 (polyclonal); α-LAP (TW7–20B9).
RNA-sequencing:
Single-cell RNA-sequencing was performed by the UCSF Functional Genomics core facility using the 10X Chromium Single Cell 3’ Gene Expression kit, according to the manufacturer’s instructions (10X Genomics, Pleasanton, CA). 150 paired-end sequencing was performed on a Novaseq 6000 instrument. The Cell Ranger analysis pipelines (version 3.0.2, 10X Genomics) were then used to process the generated sequencing data. The R package Seurat (version 3.0) was used for gene expression analysis. Filtered gene-barcode matrices were loaded and quality-control steps were performed (low quality or dying cells and cell doublets/multiplets were excluded from subsequent analysis). Data was normalized and scaled, and then linear dimensional reduction using principle component analysis (PCA) was performed. Highly variable genes were used to perform unsupervised clustering, and non-linear dimensional reduction with Uniform Maniford Approximation and Projection (UMAP) was used to visualize the data. Differential genes were calculated with Seurat on non-integrated data. CellPhoneDB analysis was performed as previously described (22). Bulk RNA-sequencing was performed by Expression Analysis, Quintiles (Morrisville, NC). Normalized RNA counts were determined using the R package DESeq2.
Statistical analysis:
Statistical analyses were performed with Prism software (GraphPad). A two-tailed unpaired Student’s t-test or two way ANOVA with Sidak’s multiple comparisons test were applied to calculate P values assuming a normal sample distribution, as indicated. The test used for each analysis are indicated in the figure legends. All experiments were performed with at least 2 independent trials, as indicated. Statistical analysis of single cell RNA sequencing data was handled with either Seurat (28), GSEA (19), PANTHER (18), or CellPhoneDB (22) as indicated. P values correlate with symbols as follows: ns = not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Supplementary Material
Acknowledgements:
We thank the staff of the UCSF Laboratory Animal Resource Center and Parnassus Flow Cytometry Core and for research support.
Funding:
The Parnassus Flow Cytometry Core is funded by a Diabetes Research Center (DRC) grant, NIH P30 DK063720. This work was primarily supported by NIAMS NIH grants: (R01AR071944 and DP2-AR068130) to M.D.R.. J.M.M is supported by the Human Frontier Science Program Long-term Fellowship (LT000183/2018-L).
Footnotes
Competing interests:
MDR is a consultant and co-founder of TRex Bio, Inc. and Sitryx Bio, Inc. He also is a consultant for Mozart Bio, Inc. The authors declare that they have no other competing interests.
References and Notes:
- 1.Lui PP, Cho I, Ali N, Tissue regulatory T cells. Immunology. 161, 4–17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Panduro M, Benoist C, Mathis D, Tissue Tregs. Annu. Rev. Immunol. 34, 609–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito M. et al. , Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature. 565, 246–250 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Miragaia RJ et al. , Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity. 50, 493–504.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delacher M. et al. , Precursors for Nonlymphoid-Tissue Treg Cells Reside in Secondary Lymphoid Organs and Are Programmed by the Transcription Factor BATF. Immunity. 52, 295–312.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pakyari M, Farrokhi A, Maharlooei MK, Ghahary A, Critical Role of Transforming Growth Factor Beta in Different Phases of Wound Healing. Adv. Wound Care. 2, 215–224 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han G. et al. , A role for TGFΒ signaling in the pathogenesis of psoriasis. J. Invest. Dermatol. 130, 371–377 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodyga M, Hinz B, TGF-β1 - A truly transforming growth factor in fibrosis and immunity. Semin. Cell Dev. Biol. 101, 123–139 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Travis MA, Sheppard D, TGF-β activation and function in immunity. Annu. Rev. Immunol. 32, 51–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Konkel JE et al. , Transforming Growth Factor-β Signaling in Regulatory T Cells Controls T Helper-17 Cells and Tissue-Specific Immune Responses. Immunity. 46, 660–674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li MO, Wan YY, Flavell RA, T Cell-Produced Transforming Growth Factor-β1 Controls T Cell Tolerance and Regulates Th1- and Th17-Cell Differentiation. Immunity. 26, 579–591 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Ouyang W, Beckett O, Ma Q, Li MO, Transforming growth factor-β signaling curbs thymic negative selection promoting regulatory T cell development. Immunity. 32, 642–653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Josefowicz SZ, Lu L-F, Rudensky AY, Regulatory T Cells: Mechanisms of Differentiation and Function. Annu. Rev. Immunol. 30, 531–564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Worthington JJ et al. , Integrin avB8-Mediated TGF-B Activation by Effector Regulatory T Cells Is Essential for Suppression of T-Cell-Mediated Inflammation. Immunity. 42, 903–915 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards JP, Thornton AM, Shevach EM, Release of Active TGF-β1 from the Latent TGF-β1/GARP Complex on T Regulatory Cells Is Mediated by Integrin β8. J. Immunol. 193, 2843–2849 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira C. et al. , Type 1 Treg cells promote the generation of CD8+ tissue-resident memory T cells. Nat. Immunol. 21, 766–776 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Haribhai D. et al. , Regulatory T Cells Dynamically Control the Primary Immune Response to Foreign Antigen. J. Immunol. 178, 2961–2972 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Thomas PD et al. , PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 13, 2129–2141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A. et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mu D. et al. , The integrin ανβ8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-β1. J. Cell Biol. 157, 493–507 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joost S. et al. , The Molecular Anatomy of Mouse Skin during Hair Growth and Rest. Cell Stem Cell. 26, 441–457.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Efremova M, Vento-Tormo M, Teichmann SA, Vento-Tormo R, CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 15, 1484–1506 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Ali N. et al. , Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell. 169, 1119–1129.e11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burzyn D. et al. , A Special Population of Regulatory T Cells Potentiates Muscle Repair. Cell. 155, 1282–1295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gratz IK et al. , Cutting Edge: Memory Regulatory T Cells Require IL-7 and Not IL-2 for Their Maintenance in Peripheral Tissues. J. Immunol. 190, 4483–4487 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thelen F, Witherden DA, Get in Touch With Dendritic Epithelial T Cells! Front. Immunol. 11, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walzer C, Benathan M, Frenk E, Thermolysin treatment: A new method for dermo-epidermal separation. J. Invest. Dermatol. 92, 78–81 (1989). [DOI] [PubMed] [Google Scholar]
- 28.Stuart T. et al. , Comprehensive Integration of Single-Cell Data. Cell. 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathur AN et al. , Treg-Cell Control of a CXCL5-IL-17 Inflammatory Axis Promotes Hair-Follicle-Stem-Cell Differentiation During Skin-Barrier Repair. Immunity. 50, 655–667.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell MG et al. , Cryo-EM Reveals Integrin-Mediated TGF-β Activation without Release from Latent TGF-β. Cell. 180, 490–501.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Su Y, Iacob RE, Engen JR, Springer TA, General structural features that regulate integrin affinity revealed by atypical αVβ8. Nat. Commun. 10, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ozawa A. et al. , Molecular basis of the ligand binding specificity of αvβ8 integrin. J. Biol. Chem. 291, 11551–11565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubtsov YP et al. , Stability of the Regulatory T Cell Lineage in Vivo. Science.. 329, 1667–1671 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proctor JM, Zang K, Wang D, Wang K, Reichardt LF, Vascular development of the brain requires β8 integrin expression in the neuroepithelium. J. Neurosci. 25, 9940–9948 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawlak M, Ho AW, Kuchroo VK, Cytokines and transcription factors in the differentiation of CD4+ T helper cell subsets and induction of tissue inflammation and autoimmunity. Curr. Opin. Immunol. 67, 57–67 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mei J. et al. , CXCL5 regulates chemokine scavenging and pulmonary host defense to bacterial infection. Immunity. 33, 106–117 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mei J. et al. , Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. J. Clin. Invest. 122, 974–986 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lund JM, Hsing L, Pham TT, Rudensky AY, Coordination of early protective immunity to viral infection by regulatory T cells. Science. 320, 1220–1224 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levéen P. et al. , Induced disruption of the transforming growth factor beta type II receptor gene in mice causes a lethal inflammatory disorder that is transplantable. Blood. 100, 560–568 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Derynck R, Budi EH, Specificity, versatility, and control of TGF-b family signaling. Sci. Signal. 12 (2019), doi: 10.1126/scisignal.aav5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura K, Kitani A, Strober W, Cell contact-dependent immunosuppression by CD4+CD25+ regulatory T cells is mediated by cell surface-bound transforming growth factor β. J. Exp. Med. 194, 629–644 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasioukhin V, Degenstein L, Wise B, Fuchs E, The magical touch: Genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. U. S. A. 96, 8551–8556 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalekar LA et al. , Regulatory T cells in skin are uniquely poised to suppress profibrotic immune responses. Sci. Immunol 4 (2019), doi: 10.1126/sciimmunol.aaw2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parekh T, Saxena B, Reibman J, Cronstein BN, Gold LI, Neutrophil chemotaxis in response to TGF-beta isoforms (TGF-beta 1, TGF-beta 2, TGF-beta 3) is mediated by fibronectin. J. Immunol. 152, 2456–66 (1994). [PubMed] [Google Scholar]
- 45.Haider C. et al. , Transforming Growth Factor-β and Axl Induce CXCL5 and Neutrophil Recruitment in Hepatocellular Carcinoma. Hepatology. 69, 222–236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Himmel ME et al. , Human CD4+FOXP3+ regulatory T cells produce CXCL8 and recruit neutrophils. Eur. J. Immunol. 41, 306–312 (2011). [DOI] [PubMed] [Google Scholar]
- 47.Fava RA et al. , Transforming growth factor β1 (TGF-β1) induced neutrophil recruitment to synovial tissues: Implications for TGF-β-driven synovial inflammation and hyperplasia. J. Exp. Med. 173, 1121–1132 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohammed J. et al. , Stromal cells control the epithelial residence of DCs and memory T cells by regulated activation of TGF-β. Nat. Immunol. 17, 414–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ong PY, Leung DYM, Bacterial and Viral Infections in Atopic Dermatitis: a Comprehensive Review. Clin. Rev. Allergy Immunol. 51, 329–337 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Barzaghi F, Passerini L, Bacchetta R, Immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome: A paradigm of immunodeficiency with autoimmunity. Front. Immunol. 3, 1–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.