Abstract
Introduction
In late 2019 and after the COVID-19 pandemic in the world, many researchers and scholars tried to provide methods for detecting COVID-19 cases. Accordingly, this study focused on identifying patients with COVID-19 from chest X-ray images.
Methods
In this paper, a method for diagnosing coronavirus disease from X-ray images was developed. In this method, DenseNet169 Deep Neural Network (DNN) was used to extract the features of X-ray images taken from the patients’ chests. The extracted features were then given as input to the Extreme Gradient Boosting (XGBoost) algorithm to perform the classification task.
Results
Evaluation of the proposed approach and its comparison with the methods presented in recent years revealed that this method was more accurate and faster than the existing ones and had an acceptable performance for detecting COVID-19 cases from X-ray images. The experiments showed 98.23% and 89.70% accuracy, 99.78% and 100% specificity, 92.08% and 95.20% sensitivity in two and three-class problems, respectively.
Conclusion
This study aimed to detect people with COVID-19, focusing on non-clinical approaches. The developed method could be employed as an initial detection tool to assist the radiologists in more accurate and faster diagnosing the disease.
Implication for practice
The proposed method's simple implementation, along with its acceptable accuracy, allows it to be used in COVID-19 diagnosis. Moreover, the gradient-based class activation mapping (Grad-CAM) can be used to represent the deep neural network's decision area on a heatmap. Radiologists might use this heatmap to evaluate the chest area more accurately.
Keywords: XGBoost, Deep neural network (DNN), DenseNet169, COVID-19, Chest X-ray images
Introduction
COVID-19 virus was reported in Wuhan, China, in late December 2019 with unknown causes, after which it spread rapidly throughout the world.1, 2, 3 The virus prevailed in most parts of China within 30 days.4 The infectious disease caused by this type of virus was named COVID-19 by the World Health Organization (WHO) on February 11, 2020.2 COVID-19 was reported in Iran on February 21, 2020. About 3.5 and 192 million confirmed cases were identified in Iran and worldwide until July 20, 2021, respectively. Most types of coronavirus affect animals, but they can also be transmitted to humans due to their common nature. Severe Acute Respiratory Syndrome (SARS-CoV)-associated coronavirus causes humans’ severe respiratory disease and death.5 The well-known signs and symptoms of COVID-19 include fever, cough, sore throat, headache, fatigue, muscle pain, and shortness of breath.6 Since the prevalence of this pandemic, the COVID-19 virus has directly impacted the lifestyles of most communities, including human health, social welfare, businesses, and social relationships. It has also put its indirect effects, such as reducing the quality of education in schools and universities, weakening family relationships, decreasing sports activities, and so on.
The most common method of COVID-19 diagnosis in individuals is the Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR) assay. However, identification through this approach is time-consuming, and the results may have a high level of false-negative errors.7 , 8 Alternatively, chest radiographic imaging methods, such as Computed Tomography scan (CT-scan) and X-ray, can have a vital and effective role in the timely diagnosis and treatment of this disease,7 especially in pregnant women and children.9 , 10 Chest X-ray Radiograph (CXR) images are mostly utilized to diagnose chest pathology and have been rarely applied to detect COVID-19.
This research was conducted on these types of images due to the availability of radiographic imaging devices in most hospitals and specialized clinics.9 , 11 According to the previous studies, radiological images of patients with COVID-19 bear important and useful information for identifying the virus in the body.12 However, one of the disadvantages of using CXR images is that they cannot detect soft tissues with poor contrast and are not thus capable of determining the degree of a patient's lung involvement.13 , 14 To compensate for this shortcoming, Computer-Aided Diagnosis (CAD) systems can be employed.15 , 16 Most CAD systems depend on the development of Graphics Processing Units (GPUs), which are applied to implement medical image processing algorithms, such as image enhancement and limb or tumor segmentation.17 , 18 The development of artificial intelligence, especially some of its branches, such as Machine Learning and Deep Learning (DL), has contributed to greater intelligence in this process compared to human intelligence. Artificial intelligence has also significantly impacted the speed of the processes involved in such fields as medical sciences for performing diagnosis or even treatment. For instance, in areas like diagnosing lung19 , 20 and cardiovascular21 , 22 diseases and performing brain surgery,23 , 24 it has so effectively contributed to the medical community and patients.
Advances in DL have shown promising results in medical image analysis and radiology.25 , 26 DL has got various architectures, each of which involves a variety of applications in the related fields. A type of DL architecture is Deep Convolutional Neural Network (DCNN), which is employed specifically in image processing. Among its varied applications, pattern recognition and image classification can be mentioned.27
Depending on the problem involved, DCNNs can be used in many different ways. One of the existing methods is to use pre-trained neural networks, which were utilized in this research. Based on this approach, pre-trained models that are freely available are employed, and image features are extracted using DNNs. The second step following the extraction of image features is to utilize classification methods for conducting the classification task. Among the various classification methods, such as Support Vector Machines (SVMs), Decision Trees, etc., the XGBoost classifier was applied in this paper.
Apostolopoulos et al.28 carried out a study on a set of X-ray images from patients with pneumonia, COVID-19, and healthy individuals to assess the Convolutional Neural Network (CNN) performance. In this research, transfer learning was utilized, and the research process was done in 3 stages. The results demonstrated that the use of DL could lead to the extraction of significant features from COVID-19. In another study, Wang et al.29 presented the COVID-Net network (a DCNN for COVID-19 detection), implemented on X-ray images. Their proposed network could help physicians during the screening phase. Sethy et al.30 utilized DL and SVM to detect coronavirus-involved patients using X-ray images. Since SVM provides a powerful approach, it was applied in their classification process.
Hemdan et al.31 proposed COVIDX-Net, which consisted of VGG16 and Google MobileNet. Mishra et al.32 proposed a decision fusion approach, which combined predictions from varied DCNNs to identify COVID-19 from chest CT images. In their study, Narin et al.33 proposed five models for diagnosing patients with pneumonia and coronavirus via X-ray images. These models were based on pre-trained CNNs, such as ResNet 152, ResNet 101, ResNet 50, Inception-ResNetV2, and InceptionV3. Pandit et al.34 employed the pre-trained VGG-16 to detect COVID-19 from chest radiographs and achieved 96% accuracy. Sung et al.35 developed a system to identify patients with COVID-19 using the CT images collected from hospitals in two provinces of China. Javadi Moghaddam and Gholamalinejad36 developed a novel DL structure for COVID-19 detection. The pooling layer of their proposed structure was a combination of pooling and the Squeeze Excitation Block layer. They also used the Mish function for convergence optimization.
In a study, Ozturk et al.12 proposed a new model for detecting COVID-19 by using X-ray images. Their proposed model was presented based on the two problems of binary classification (for distinguishing COVID-19 from the “no-finding” class) and multi-class classification (for distinguishing COVID-19, pneumonia, and the “no-finding” classes). They proposed a DCNN called DarkCovidNet, which included 17 convolutional layers, and achieved 98.08% and 87.02% accuracies in their binary and multi-class classifications, respectively. The rest of the paper is organized as follows: Section: Methodology will describe the methodology. In Section: Proposed Method, results and discussion will be presented. Finally, the conclusion will be presented in Section: Results & Discussion.
Methodology
In this section, first, the dataset used in this study is presented. Then the XGBoost algorithm employed for classification in our proposed method is introduced, and finally, the proposed method is presented.
Dataset
Same as12 X-ray images from two different sources were used as the dataset in this paper: 1-Covid-19 X-ray images dataset, which was collected by Cohen,37 2-ChestX-ray 8 dataset collected by Wang et al.38 Cohen collected images from public sources and through indirect collection from hospitals and physicians. This project was approved by the University of Montreal's Ethics Committee. Fig. 1 depicts the sample images in this dataset. There are 43 female and 82 male cases in the dataset, and the subjects' average age is approximately 55 years.12
Figure 1.
Sample images in Cohen's dataset.
The ChestX-ray 838 dataset was employed for normal and pneumonia images. This dataset consists of 108,948 frontal view X-ray images of 32,717 unique patients, from which 500 no-findings and 500 pneumonia chest X-ray images were selected randomly. So overall, the dataset used in this study contained 1125 X-ray images of the studied individuals’ chests, including 125 images labeled as COVID-19, 500 images labeled as pneumonia, and 500 images labeled as no findings. Fig. 2 shows the sample images in the ChestX-ray8 dataset.
Figure 2.
Sample images in the ChestX-ray 8 dataset.
XGBoost
XGBoost is an efficient and scalable algorithm based on tree boosting proposed by Chen & Guestrin in 2016.39 , 40 It is an improved version of the Gradient Boosted Decision Tree (GBDT) method. It has proven not to have its computational limitations41, 42, 43 and thus differs from the GBDT method. GBDT uses the first-order Taylor expansion, while the second-order Taylor expansion is utilized in the XGBoost's loss function.44, 45, 46 In addition, the objective function is normalized in XGBoost to alleviate the model's complexity and prevent it from overfitting.44 , 47
Proposed method
Considering the past similar research activities and common methods of using artificial intelligence in image processing, especially for medical images, our proposed method aimed to utilize the extracted features of the images by using pre-trained networks. One of the applications of artificial intelligence is the use of transfer learning techniques. In this technique, various networks are designed and trained with a huge set of available data, and the weights of the network layers are calculated. For example, in image processing, the ImageNet dataset contains millions of images in 1000 different classes. Several methods employ pre-trained networks, as follows:
-
1
Using the structures of pre-designed networks to train one's model, remove the last layer of the presented network, and finally add layers to perform classification.
-
2
Extracting image features by using pre-trained models and using the extracted features to perform classification via other algorithms.
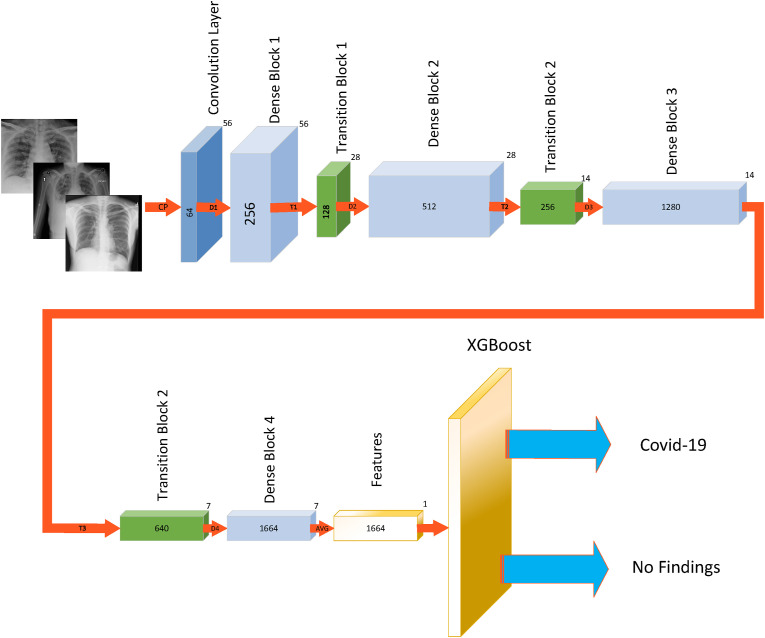
In this paper, the features were extracted using the second method, and the XGBoost classifier was employed for classification. In this way, the images were first given as input to the DenseNet169 DNN so that the network could extract image features. The extracted features were then given as input to the XGBoost algorithm to perform the classification operation. The framework of the proposed method can be seen in Fig. 3 . The proposed method was implemented using Python 3.8 and Keras 2.4 (i.e., the Python deep learning API).
Figure 3.
Framework of the proposed method.
Results
This research was done in two phases. In the first phase, the best (pre-trained) DNN was selected to extract the features, and in the second phase, the XGBoost classifier parameters were set by trial-and-error. Also, the ChestX-ray 8 dataset was used for two cases: 2-class problem, including COVID-19 and no findings (625 images), and 3-class problem, consisting of COVID-19, pneumonia, and no findings (1125 images).
In the first phase, 17 pre-trained neural networks were assessed, and the XGBoost classifier was employed along with the default parameters for classification. Table 1 shows the average accuracy of the DNNs for each of the 2-class and 3-class problems. It should be noted that a 5-fold cross-validation method was applied to obtain the average accuracies in this experiment.
Table 1.
Comparison of the average accuracies of the different DNNs.
| DNN | Average Accuracy (%) |
|
|---|---|---|
| Three-class Problem | Two-class Problem | |
| Xception | 78.84 | 93.59 |
| VGG16 | 81.68 | 96.48 |
| VGG19 | 80.08 | 95.36 |
| ResNet 50 | 80.71 | 95.51 |
| ResNet 152 | 79.55 | 95.68 |
| ResNet50V2 | 80.53 | 94.71 |
| ResNet101V2 | 76.88 | 93.95 |
| ResNet152V2 | 77.60 | 93.59 |
| InceptionV3 | 79.02 | 92.79 |
| InceptionResNetV2 | 68.44 | 90.72 |
| MobileNet | 79.55 | 95.51 |
| MobileNetV2 | 82.57 | 96.16 |
| DenseNet121 | 82.51 | 96.32 |
| DenseNet169 | 83.02 | 97.43 |
| DenseNet201 | 82.31 | 96.63 |
| NASNetMobile | 74.57 | 93.11 |
| EfficientNetB0 | 80.00 | 97.28 |
As can be seen, the DenseNet169 network has the best accuracy in both cases. As a result, this network was selected to extract the features of the proposed model in the second phase. The input to this network included images with the dimensions of 224 × 224 × 3, and its output consisted of 1664 features, which the network extracted from the given images. After determining the network type, parameters of the XGBoost classifier were set. Table 2 shows the parameters used in the XGBoost algorithm.
Table 2.
The XGBoost parameter settings.
| Parameter | Value |
|---|---|
| Base Learner | Gradient boosted tree |
| Tree construction algorithm | Exact greedy |
| Number of gradients boosted trees | 100 |
| Learning rate | 0.44 |
| Lagrange multiplier | 0 |
| Maximum depth of trees | 6 |
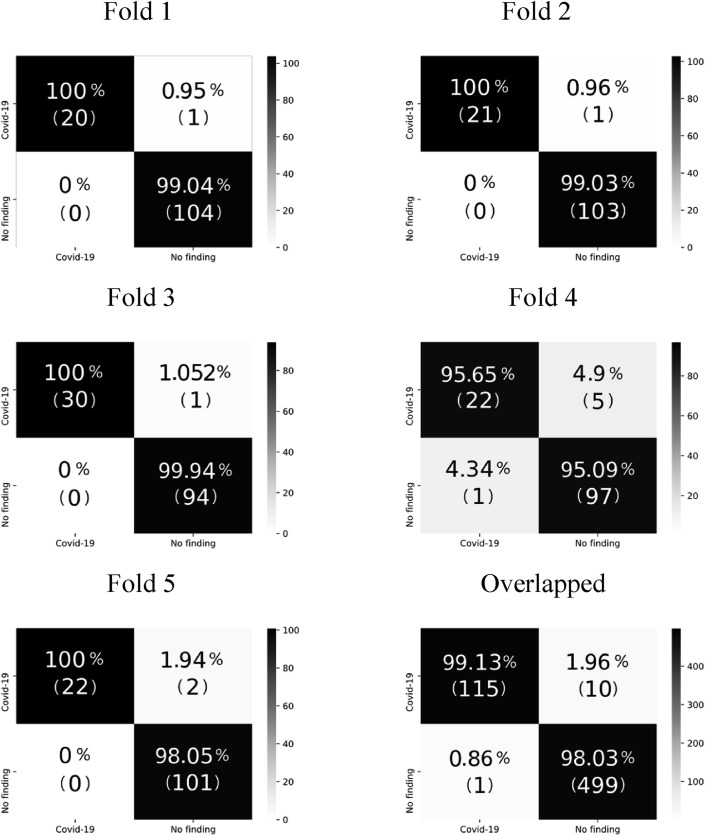
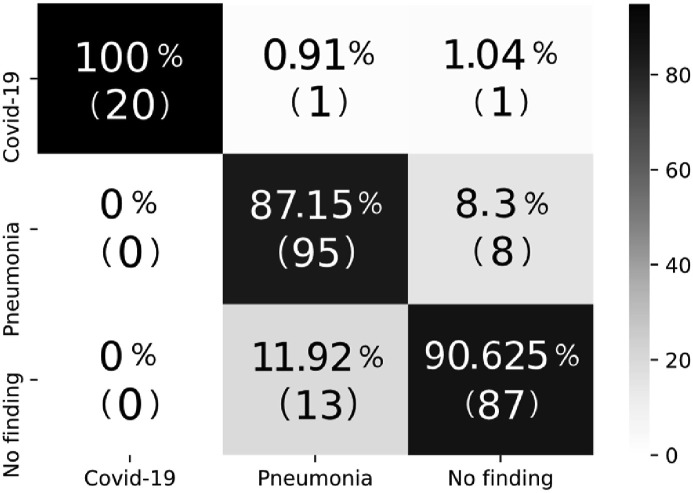
A 5-fold cross-validation was used for the 2-class problem, 80% of the dataset was utilized for training in the 3-class problem, and the remaining 20% was applied as the test set. The average accuracy for the 2-class problem was 98.23%, and the test accuracy for the 3-class problem was 89.70%. The confusion matrices for each of the five folds in the 2-class problem are shown in Fig. 4 , and the confusion matrix for the 3-class problem is shown in Fig. 5 .
Figure 4.
Confusion matrices for the 2-class problem.
Figure 5.
Confusion matrix for the 3-class problem.
The results of comparing the proposed approach with the method proposed by Ozturk et al.12 for the 3-class and 2-class problems can be observed in Table 3, Table 4 , respectively. Also, a comparison of the results obtained in this study with those of other proposed methods is given in Table 5 .
Table 3.
Comparison of the proposed method with DarkCovidNet (3-class problem).
| Proposed Method | DarkCovidNet | |
|---|---|---|
| Sensitivity | 95.20 | 88.17 |
| Specificity | 100 | 93.66 |
| Precision | 92.50 | 90.97 |
| F1-score | 91.20 | 89.44 |
| Accuracy | 89.70 | 89.33 |
Table 4.
Comparison of the proposed method with DarkCovidNet (2-class problem).
| Performance Metrics | Fold 1 | Fold 2 | Fold 3 | Fold 4 | Fold 5 | Average | |
|---|---|---|---|---|---|---|---|
| Sensitivity | Proposed Method | 95.20 | 95.40 | 96.70 | 81.40 | 91.40 | 92.08 |
| DarkCovidNet | 100 | 96.42 | 90.47 | 93.75 | 93.18 | 95.13 | |
| Specificity | Proposed Method | 100 | 100 | 100 | 89.90 | 100 | 99.78 |
| DarkCovidNet | 100 | 96.42 | 90.47 | 93.75 | 93.18 | 95.30 | |
| Precision | Proposed Method | 99.50 | 99.50 | 99.40 | 95.30 | 99.02 | 98.54 |
| DarkCovidNet | 100 | 94.52 | 98.14 | 98.57 | 98.58 | 98.03 | |
| F1-score | Proposed Method | 98.50 | 98.50 | 98.20 | 92.50 | 97.30 | 97.00 |
| DarkCovidNet | 100 | 95.52 | 93.79 | 95.93 | 95.62 | 96.51 | |
| Accuracy | Proposed Method | 99.20 | 99.20 | 99.20 | 95.20 | 98.40 | 98.24 |
| DarkCovidNet | 100 | 97.60 | 96.80 | 97.60 | 97.60 | 98.08 | |
Table 5.
Comparison of the proposed method with other DL-based methods.
| Study | Type of Images | Number of Samples | Method Used | Accuracy (%) |
|---|---|---|---|---|
| Apostolopoulos et al.28 | Chest X-ray | 1428 | VGG-19 | 93.48 |
| Wang et al.29 | Chest X-ray | 13,645 | COVID-Net | 92.40 |
| Sethy et al.30 | Chest X-ray | 50 | ResNet 50 + SVM | 95.38 |
| Hemdan et al.31 | Chest X-ray | 50 | COVIDX-Net | 90.00 |
| Narin et al.33 | Chest X-ray | 100 | Deep CNN ResNet-50 | 98.00 |
| Song et al.35 | Chest CT | 1485 | DRE-Net | 86.00 |
| Wang et al.48 | Chest CT | 453 | M-Inception | 82.90 |
| Zheng et al.49 | Chest CT | 542 | UNet + 3D Deep Network | 90.80 |
| Xu et al.50 | Chest CT | 443 | ResNet + Location Attention | 86.60 |
| Ozturk et al.12 | Chest X-ray | 625 | DarkCovidNet | 98.08 |
| 1125 | 89.33 | |||
| Proposed Method | Chest X-ray | 625 | DenseNet169 + XGBoost | 98.24 |
| 1125 | 89.70 |
Discussion
As can be seen in Table 3, Table 4, the proposed method has better performance than the DarkCovidNet network in both 3-class and 2-class problems. Noteworthy, the proposed approach had a higher speed and lower computational complexity than the method presented by Ozturk et al. because it did not require training of the DNN. Note that the proposed method has just trained the XGBoost algorithm. Table 5 demonstrates that the proposed method is more accurate than other DL-based models. However, it should be noted that the results presented in Table 5 were obtained from different datasets. This study's limitations include using an unbalanced dataset with a limited number of COVID-19 X-ray images and low sensitivity in the two-class problem.
To compare the performance of XGBoost with other machine learning algorithms, we employed Random Forest and SVM as the classifier instead of XGBoost and repeated the experiments. The linear kernel was used for SVM. Table 6 shows the result of comparing different machine learning algorithms. As can be seen, the XGBoost outperforms other machine learning algorithms in both 2-class and 3-class problems.
Table 6.
Comparison of the different machine learning algorithms.
| Accuracy (%) | ||
|---|---|---|
| Method | 2-class problem | 3-class problem |
| DenseNet169 + XGBoost | 98.24 | 89.70 |
| DenseNet169 + Random Forest | 95.85 | 80.15 |
| DenseNet169 + SVM | 96.96 | 79.20 |
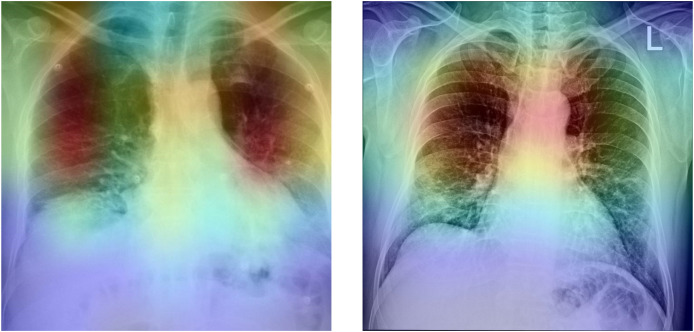
To further analyze the proposed method, the gradient-based class activation mapping (Grad-CAM)51 was used to represent the decision area on a heatmap. Fig. 6 depicts the heatmaps for two confirmed COVID-19 cases. As can be seen, the developed method extracted correct features, and the model is mainly concentrated on the lung area. Radiologists can employ the heatmap to evaluate the chest area more accurately.
Figure 6.
Heatmap of two confirmed COVID-19 cases.
Conclusion
This study aimed to detect and identify people with COVID-19, focusing on non-clinical approaches and artificial intelligence techniques. In the proposed method, DenseNet169 was employed to extract image features, and the XGBoost algorithm was used for classification. The obtained results revealed that the detection accuracy of the proposed method in the 2-class problem was 98.24%, which was higher than other proposed methods. Also, 89.70% accuracy was reached in the 3-class problem, thus indicating better performance compared to the DarkCovidNet network. Besides being highly accurate, the proposed approach had a higher speed and lower computational complexity than the other proposed methods since it did not require the training of DNN.
Funding statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
References
- 1.Wu F., Zhao S., Yu B., Chen Y., Wang W., Song Z., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sohrabi C., Alsafi Z., O’neill N., Khan M., Kerwan A., Al-Jabir A., et al. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int J Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jha N., Prashar D., Rashid M., Shafiq M., Khan R., Pruncu C., et al. Deep learning approach for discovery of in silico drugs for combating COVID-19. J Healthc Eng. 2021:2021. doi: 10.1155/2021/6668985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 5.Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection. Radiol Cardiothorac Imag. 2020;2 doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zu Z.Y., Jiang M.D., Xu P.P., Chen W., Ni Q.Q., Lu G.M., et al. Coronavirus disease 2019 (COVID-19): a perspective from China. Radiology. 2020;296:E15–E25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang P., Liu T., Huang L., Liu H., Lei M., Xu W., et al. Use of chest CT in combination with negative RT-PCR assay for the 2019 novel coronavirus but high clinical suspicion. Radiology. 2020;295:22–23. doi: 10.1148/radiol.2020200330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng M.Y., Lee E.Y., Yang J., Yang F., Li X., Wang H., et al. Imaging profile of the COVID-19 infection: radiologic findings and literature review. Radiol Cardiothorac Imag. 2020;2 doi: 10.1148/ryct.2020200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu H., Liu F., Li J., Zhang T., Wang D., Lan W. Clinical and CT imaging features of the COVID-19 pneumonia: focus on pregnant women and children. J Infect. 2020;80:e7–e13. doi: 10.1016/j.jinf.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kroft L.J., van der Velden L., Girón I.H., Roelofs J.J., de Roos A., Geleijns J. Added value of ultra--low-dose computed tomography, dose equivalent to chest x-ray radiography, for diagnosing chest pathology. J Thorac Imag. 2019;34:179. doi: 10.1097/RTI.0000000000000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karar M.E., Merk D.R., Falk V., Burgert O. A simple and accurate method for computer-aided transapical aortic valve replacement. Comput Med Imag Graph. 2016;50:31–41. doi: 10.1016/j.compmedimag.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Merk D.R., Karar M.E., Chalopin C., Holzhey D., Falk V., Mohr F.W., et al. Image-guided transapical aortic valve implantation sensorless tracking of stenotic valve landmarks in live fluoroscopic images. Innovations. 2011;6:231–236. doi: 10.1097/IMI.0b013e31822c6a77. [DOI] [PubMed] [Google Scholar]
- 16.Messerli M., Kluckert T., Knitel M., Rengier F., Warschkow R., Alkadhi H., et al. Computer-aided detection (CAD) of solid pulmonary nodules in chest x-ray equivalent ultralow dose chest CT-first in-vivo results at dose levels of 0.13 mSv. Eur J Radiol. 2016;85:2217–2224. doi: 10.1016/j.ejrad.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Tian X., Wang J., Du D., Li S., Han C., Zhu G., et al. Medical imaging and diagnosis of subpatellar vertebrae based on improved Laplacian image enhancement algorithm. Comput Methods Progr Biomed. 2020;187:105082. doi: 10.1016/j.cmpb.2019.105082. [DOI] [PubMed] [Google Scholar]
- 18.Hannan R., Free M., Arora V., Harle R., Harvie P. Accuracy of computer navigation in total knee arthroplasty: a prospective computed tomography-based study. Med Eng Phys. 2020;79:52–59. doi: 10.1016/j.medengphy.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Liang C., Liu Y., Wu M., Garcia-Castro F., Alberich-Bayarri A., Wu F. Identifying pulmonary nodules or masses on chest radiography using deep learning: external validation and strategies to improve clinical practice. Clin Radiol. 2020;75:38–45. doi: 10.1016/j.crad.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Hussein S., Kandel P., Bolan C.W., Wallace M.B., Bagci U. Lung and pancreatic tumor characterization in the deep learning era: novel supervised and unsupervised learning approaches. IEEE Trans Med Imag. 2019;38:1777–1787. doi: 10.1109/TMI.2019.2894349. [DOI] [PubMed] [Google Scholar]
- 21.Karar M.E., El-Khafif S.H., El-Brawany M.A. Automated diagnosis of heart sounds using rule-based classification tree. J Med Syst. 2017;41:60. doi: 10.1007/s10916-017-0704-9. [DOI] [PubMed] [Google Scholar]
- 22.Karar M.E., Merk D.R., Chalopin C., Walther T., Falk V., Burgert O. Aortic valve prosthesis tracking for transapical aortic valve implantation. Int J Comput Assist Radiol Surg. 2011;6:583–590. doi: 10.1007/s11548-010-0533-5. [DOI] [PubMed] [Google Scholar]
- 23.Ghassemi N., Shoeibi A., Rouhani M. Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed Signal Process Control. 2020;57:101678. [Google Scholar]
- 24.Rathore H., Al-Ali A.K., Mohamed A., Du X., Guizani M. A novel deep learning strategy for classifying different attack patterns for deep brain implants. IEEE Access. 2019;7:24154–24164. [Google Scholar]
- 25.Chen L., Bentley P., Mori K., Misawa K., Fujiwara M., Rueckert D. Self-supervised learning for medical image analysis using image context restoration. Med Image Anal. 2019;58:101539. doi: 10.1016/j.media.2019.101539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim M., Yan C., Yang D., Wang Q., Ma J., Wu G. Biomedical information technology. Elsevier; 2020. Deep learning in biomedical image analysis; pp. 239–263. [Google Scholar]
- 27.Zhou D.X. Theory of deep convolutional neural networks: Downsampling. Neural Network. 2020;124:319–327. doi: 10.1016/j.neunet.2020.01.018. [DOI] [PubMed] [Google Scholar]
- 28.Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Lin Z.Q., Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sethy P.K., Behera S.K., Ratha P.K., Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. Int J Math Eng Manag Sci. 2020;5:643–651. [Google Scholar]
- 31.Hemdan E.E.D., Shouman M.A., Karar M.E. Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv Prepr arXiv200311055. 2020 [Google Scholar]
- 32.Mishra A.K., Das S.K., Roy P., Bandyopadhyay S. Identifying COVID19 from chest CT images: a deep convolutional neural networks based approach. J Healthc Eng. 2020:2020. doi: 10.1155/2020/8843664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal Appl. 2021:1–14. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandit M.K., Banday S.A., Naaz R., Chishti M.A. Automatic detection of COVID-19 from chest radiographs using deep learning. Radiography. 2021;27:483–489. doi: 10.1016/j.radi.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., et al. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE ACM Trans Comput Biol Bioinf. 2021;18:2775–2780. doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.JavadiMoghaddam S., Gholamalinejad H. A novel deep learning based method for COVID-19 detection from CT image. Biomed Signal Process Control. 2021;70:102987. doi: 10.1016/j.bspc.2021.102987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen J.P., Morrison P., Dao L. 2020. COVID-19 image data collection. arXiv Prepr arXiv200311597. [Google Scholar]
- 38.Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings of the IEEE conference on computer vision and pattern recognition. IEEE; 2017. Chestx-ray 8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 2097–2106. [Google Scholar]
- 39.Chen T., Guestrin C. Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining. 2016. Xgboost: a scalable tree boosting system; pp. 785–794. [Google Scholar]
- 40.Chehreh Chelgani S., Nasiri H., Tohry A. Modeling of particle sizes for industrial HPGR products by a unique explainable AI tool- A “Conscious Lab” development. Adv Powder Technol. 2021;32:4141–4148. [Google Scholar]
- 41.Xu Y., Zhao X., Chen Y., Yang Z. Research on a mixed gas classification algorithm based on Extreme random tree. Appl Sci. 2019;9:1728. [Google Scholar]
- 42.Ogunleye A., Wang Q.G. XGBoost model for chronic kidney disease diagnosis. IEEE ACM Trans Comput Biol Bioinf. 2019;17:2131–2140. doi: 10.1109/TCBB.2019.2911071. [DOI] [PubMed] [Google Scholar]
- 43.Valarmathi R., Sheela T. Heart disease prediction using hyper parameter optimization (HPO) tuning. Biomed Signal Process Control. 2021;70:103033. [Google Scholar]
- 44.Nasiri H., Alavi S.A. A novel framework based on deep learning and ANOVA feature selection method for diagnosis of COVID-19 cases from chest X-ray images. Comput Intell Neurosci. 2022;2022:4694567. doi: 10.1155/2022/4694567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nasiri H., Homafar A., Chehreh Chelgani S. Prediction of uniaxial compressive strength and modulus of elasticity for Travertine samples using an explainable artificial intelligence. Results Geophys Sci. 2021;8:100034. [Google Scholar]
- 46.Chehreh Chelgani S., Nasiri H., Alidokht M. Interpretable modeling of metallurgical responses for an industrial coal column flotation circuit by XGBoost and SHAP-A “conscious-lab” development. Int J Min Sci Technol. 2021;31:1135–1144. [Google Scholar]
- 47.Song K., Yan F., Ding T., Gao L., Lu S. A steel property optimization model based on the XGBoost algorithm and improved PSO. Comput Mater Sci. 2020;174:109472. [Google Scholar]
- 48.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) Eur Radiol. 2021:1–9. doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., et al. MedRxiv; 2020. Deep learning-based detection for COVID-19 from chest CT using weak label. [Google Scholar]
- 50.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6:1122–1129. doi: 10.1016/j.eng.2020.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selvaraju R.R., Cogswell M., Das A., Vedantam R., Parikh D., Batra D. Proceedings of the IEEE international conference on computer vision. IEEE; 2017. Grad-cam: visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]