Abstract
The emergence of the SARS-CoV-2 Omicron (B.1.1.529) variant with a surprising number of spike mutations raises concerns about reduced sensitivity of this virus to antibody neutralization and subsequent vaccine breakthrough infections. Here, we infect Moderna mRNA-vaccinated or previously infected hamsters with the Omicron BA.1 variant. While the Moderna mRNA vaccine reduces viral loads in the respiratory tissues upon challenge with an early S-614G isolate, the vaccine efficacy is not as pronounced after infection with the Omicron variant. Previous infection with the early SARS-CoV-2 isolate prevents replication after rechallenge with either virus in the lungs of previously infected hamsters, but the Omicron variant replicates efficiently in nasal turbinate tissue. These results experimentally demonstrate in an animal model that the antigenic changes in the Omicron variant are responsible for vaccine breakthrough and re-infection.
Keywords: SARS-CoV-2, Omicron, BA.1, immunity, vaccination, previous infection
Graphical abstract
Halfmann et al. report that while the Moderna mRNA vaccine reduces BA.1 replication in vaccinated hamsters, the vaccine efficacy is not as pronounced as that against an early prototypical SARS-CoV-2 isolate. Previously infected hamsters are better protected than vaccinated hamsters against re-infection with the BA.1 variant.
Introduction
On November 26, 2021, the World Health Organization classified B.1.1.529 (Omicron) as a variant of concern. First detected in South Africa, the Omicron BA.1 variant has quickly become the dominant variant, replacing the former dominant Delta variant (B.1.617.2) in many parts of the world. Staggeringly, the Omicron variant contains over 30 amino acid substitutions in the spike protein including 15 amino acid substitutions in the receptor-binding domain. These extensive amino acid changes in the spike protein are associated with in vitro escape of neutralization to most therapeutic monoclonal antibodies (Cameroni et al., 2021; VanBlargan et al., 2021) and with reduced vaccine-induced neutralizing antibody reactivity with the Omicron variant (Ai et al., 2021; Dejnirattisai et al., 2021) leading to vaccine breakthrough infections (Helmsdal et al., 2021; Zhou et al., 2021).
Because the Omicron variant is antigenically substantially different from the previously circulating viruses (Tada et al., 2021) and the level of immunity differs among those previously infected and those vaccinated, due to the infecting strains or vaccine types and vaccination dates, it is difficult to determine whether Omicron infections in humans are due to immunity waning or antigenic changes in the infecting virus or both. Here, in the hamster model, we addressed these issues under controlled conditions using the Omicron variant and isolates that are no longer circulating in nature (i.e., a Wuhan-like isolate and an isolate with only a D614G spike mutation).
Results
Challenge of Moderna mRNA-vaccinated hamsters with the Omicron variant
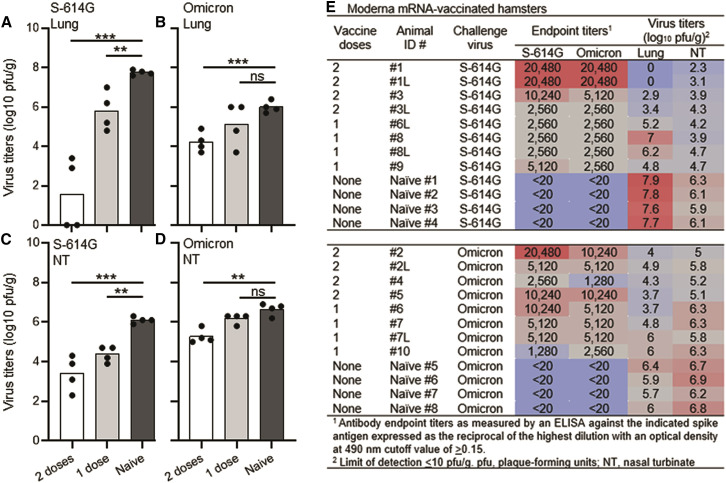
A significant decrease in the reactivity of vaccine-induced antibodies against the Omicron variant has been reported (Ai et al., 2021; Dejnirattisai et al., 2021), which may explain vaccine breakthrough infections in the human population (Helmsdal et al., 2021; Zhou et al., 2021). To experimentally investigate these findings in an animal model, we vaccinated groups of eight female Syrian golden hamsters once or twice at a 4-week interval with the Moderna mRNA vaccine (35 μg of vaccine per dose). Seven months after vaccination, serum samples were collected to evaluate binding IgG antibody titers as measured by an ELISA. All animals had measurable IgG antibody titers against the spike (S-614G) with approximately 2.6-fold higher antibody titers after two vaccinations (geometric mean titer [GMT] = 8,611, compared to one vaccination: GMT = 3,620). Antibody titers against the Omicron spike were similar with GMTs of 6,640 and 3,320 after two vaccinations and one vaccination, respectively (Figure 1E).
Figure 1.
Virus titers in Moderna mRNA-vaccinated hamsters
Virus replication of the S-614G isolate in the lungs (A) or nasal turbinates (NT) (C) or of the Omicron variant in the lungs (B) or NT (D) of naive hamsters or those vaccinated twice or once with the Moderna mRNA vaccine. Individual symbols in the figure indicate the number of hamsters in each group from one experimental study. Cumulative antibody and virology data on the Moderna mRNA-vaccinated hamsters (E). Black dots on the x axis represent virus titers below the limit of detection (≤10 pfu/g). ∗∗∗p < 0.0001, ∗∗p < 0.001, and ns; not significant.
To examine the protective efficacy of the Moderna mRNA vaccine seven months after vaccination, hamsters (8-month-old; vaccinated once or twice) were infected with 1,000 plaque-forming units (pfu) of SARS-CoV-2, specifically a S-614G isolate (n = 4 in each vaccine group) or the Omicron variant (n = 4 in each vaccine group). Age-matched, unvaccinated hamsters served as controls (n = 4 for each virus).
In the unvaccinated, naive hamsters, the S-614G isolate replicated better than the Omicron variant in the lungs with about a 2-log difference between the isolates (Figures 1A and 1B [Naive bars], Figure 1E). However, the virus titers in the nasal turbinates were similar between the two virus isolates (Figures 1C and 1D [Naive bars], Figure 1E). Two vaccinations reduced the virus titers by 6-log units in the lungs of hamsters infected with the S-614G isolate compared to the infected, unvaccinated control animals (Figures 1A and 1E). In contrast, two vaccinations only reduced the virus titers by 1.5-log units in the lungs of hamsters infected with the Omicron variant compared to the infected, unvaccinated control animals (Figures 1B and 1E). Virus loads were also significantly reduced in the lungs of hamsters vaccinated only once and infected with the S-614G isolate (2-log unit reduction compared to the naive group, p = 0.008; Figures 1A and 1E), but one vaccination did not significantly reduce Omicron virus titers in the lungs (0.9-log reduction compared to the naive group, p = 0.177; Figures 1B and 1E). Similar reductions in virus titers were observed in the nasal turbinates with the greatest reduction after two vaccinations in hamsters infected with the S-614G isolate compared to the Omicron isolate (Figures 1C–1E).
Next, we examined the relationship between the antibody levels induced by vaccination and the virus titers in different tissues. We observed a strong correlation between antibody levels and S-614G virus titers in the lungs (p < 0.0001) and nasal turbinates (p = 0.0005) (Figure S1A). In Omicron-infected, vaccinated hamsters, there was a lower but significant correlation between the antibody levels and virus titers in both tissues (lungs [p = 0.0071], nasal turbinates [p = 0.0058] [Figure S1B]).
Rechallenge of previously infected hamsters with the Omicron variant
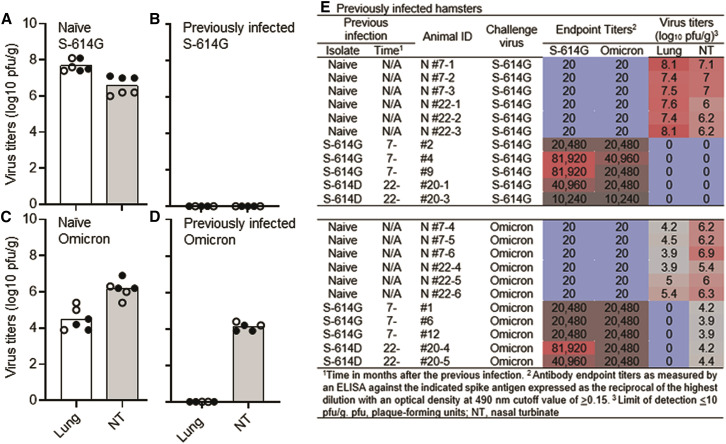
Next, we determined whether a previous infection with SARS-CoV-2 would protect hamsters against a rechallenge with the Omicron variant. Animals were first infected with a S-614G isolate (n = 6; 7 months prior) or an original Wuhan-like S-614D isolate (n = 4; 22 months prior). Prior to rechallenge, serum was analyzed for spike-specific IgG antibody titers. All animals had high IgG antibody titers against the S-614G antigen as measured by an ELISA (7-month GMT = 32,510; 22-month GMT = 34,443) with 1.4- and 3.4-fold decreases in IgG antibody titers against the Omicron spike antigen (7-month GMT = 22,988; 22-month GMT = 10,159) (Figure 2E).
Figure 2.
Virus titers in previously infected hamsters
Virus replication of the S-614G isolate or the Omicron variant in groups of naive hamsters (A and C, respectively) or groups of hamsters previously infected 7 months prior with an S-614G isolate (closed circles) or 22 months prior with an earlier S-614D isolate (open circles) and re-infected with the S-614G isolate (B) or the Omicron variant (D). Individual symbols in the figure indicate the number of hamsters in each group from one experimental study. Cumulative antibody and virology data on the previously infected hamsters (E). Black dots on the x axis represent virus titers below the limit of detection (≤10 pfu/g).
We next determined whether the immunity generated by previous infection was sufficient to prevent replication of the Omicron variant in hamsters. We re-infected hamsters with 1,000 pfu of the S-614G isolate or the Omicron variant (infected 7 months prior, n = 3 for each virus; infected 22 months prior, n = 2 for each virus). Naive, age-matched hamsters (9-month-old, n = 3 for each virus and 23-month-old, n = 3 for each virus) were also infected with either virus at the same dose. Three days after rechallenge, we did not detect any replicating virus in the lungs or nasal turbinates in either group of previously infected hamsters re-challenged with the S-614G isolate (Figures 2B and 2E), although this isolate replicated efficiently in the same tissues of naive animals of both age groups (Figures 2A and 2E). We also found no detectable replicating virus in the lungs of either group of previously infected hamsters re-challenged with the Omicron variant (Figures 2D and 2E). The lack of infectious virus (both the S-614G and the Omicron variant) in the lungs of previously infected, re-challenged hamsters was confirmed by qRT-PCR, as indicated by high Ct values (Table S1). However, the Omicron variant replicated efficiently in the nasal turbinates in both groups of hamsters infected 7 months prior with the S-614G isolate and those infected 22 months prior with the S-614D isolate (Figures 2D and 2E). In naive hamsters, the Omicron variant again grew less efficiently in the lungs, but to similar titers in the nasal turbinates compared to the S-614G isolate in naive hamsters (Figures 2A, 2C, and 2E).
Discussion
As the SARS-CoV-2 pandemic persists, we are faced with the challenges of waning antibody immunity that was either induced by vaccination or previous infection, along with antigenic changes in the viral spike protein. The latter challenge is exemplified by the Omicron BA.1 variant, which has more than 30 amino acid substitutions in its spike protein, half of which are in the receptor-binding domain. Given this change in antigenicity, a key question is whether the Omicron BA.1 variant can evade immunity induced by vaccination or a previous natural infection or both.
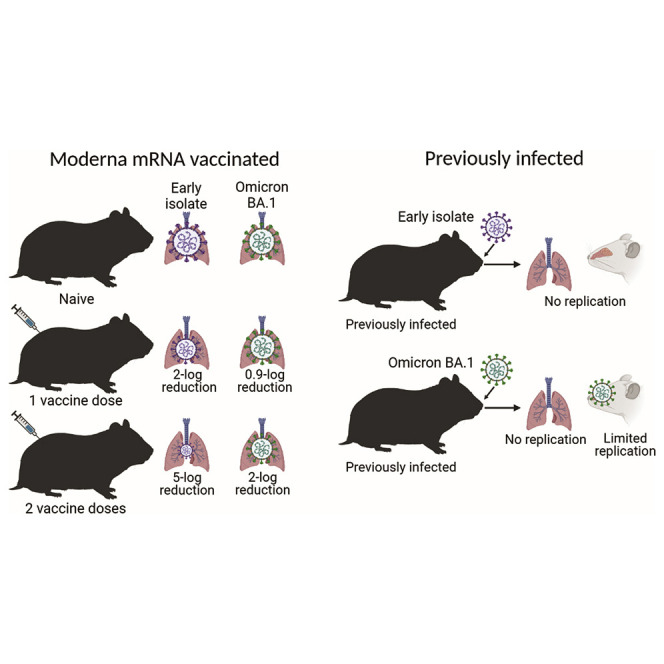
While vaccination of hamsters with the Moderna mRNA vaccine reduced viral loads in the respiratory tissues upon challenge with an early S-614G isolate, the vaccine efficacy was not as pronounced after infection with the Omicron variant. This reduction in efficacy against the Omicron variant was despite similar overall spike-specific IgG antibody titers against both viruses.
Previous infection with either early isolate (S-614D or S-614G) resulted in higher spike-specific IgG antibody titers in hamsters when compared to Moderna mRNA-vaccinated animals. This better immune response to a previous infection compared to vaccination prevented replication of both isolates (S-614G and the Omicron variant) after rechallenge in the lungs of previously infected hamsters, demonstrating that both short- and long-term immunity was protective in the lower respiratory tract. However, unlike the S-614G isolate, the Omicron variant replicated efficiently in the upper respiratory tract (nasal turbinate tissue) in previously infected hamsters. Whether this replication in the nasal turbinates of previously infected hamsters can lead to transmission to naive hamsters will be addressed in the next set of experiments.
Limitations of the study
Although the hamster model of SARS-CoV-2 infection shares many similarities with infection in humans (Imai et al., 2020), the differences in immunity induced by vaccination and the kinetics of waning immunity between hamsters and humans is not fully understood or appreciated. These potential differences are limitations in these studies of the Omicron variant. Furthermore, the Omicron variant replicates less efficiently in the lungs of infected hamsters compared to previous variants. Our group (Halfmann et al., 2022) and another group (Abdelnabi et al., 2022) have shown that the Omicron BA.1 variant is attenuated in the hamster model, resulting in minimal antiviral antigen detection and little inflammation in the lung tissue relative to previous variants. Nevertheless, our results in the hamster model do demonstrate that the Omicron variant partially evades vaccine-induced immunity, whereas a robust immune response to a first infection will protect the lower respiratory tract from a subsequent infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| hCoV-19/USA/WI-WSLH-221686/2021 | In-house; upon request | Omicron BA.1 EPI_ISL_7263803 |
| SARS-CoV-2/UT-NCGM02/Human/2020/Tokyo | In-house; upon request | S-614D |
| SARS-CoV-2/UT-HP095-1N/Human/2020/Tokyo | In-house; upon request | S-614G |
| Biological samples | ||
| Residual Moderna vaccine | In-house; upon request – limited supply; frozen at -80°C | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| S-614G purified spike protein | In-house; upon request | N/A |
| Omicron purified spike protein | AcroBiostems | SPD-C522e |
| Experimental models: Cell lines | ||
| Vero E6/TMPRSS2 cells | JCRB Cell Bank | 1819 |
| Experimental models: Organisms/strains | ||
| Syrian hamsters; females | Envigo | 8903F |
| Software and algorithms | ||
| Graphpad Prism 9 | Graphpad Prism 9 | N/A |
Resource availability
Lead contact
Requests for resources and reagents should be directed to the lead contact, Peter J. Halfmann (pjhalfma@wisc.edu).
Materials availability
All unique reagents generated in this study are available from the lead contact with completed Material Transfer Agreements.
Experimental model and subject details
Biosafety and animal experiment approvals
All experiments with SARS-CoV-2 were performed under a biosafety protocol approved by the University of Wisconsin–Madison Institutional Biosafety Committee and were performed in biosafety level-3 agriculture (BSL-3 AG) designed laboratories, which are approved for use by the Centers for Disease Control and Prevention and the US Department of Agriculture. Hamster studies were conducted under an approved animal protocol reviewed by the Institutional Animal Care and Use Committee at the University of Wisconsin-Madison (V006426). Female Syrian golden hamsters (4-weeks old at the time of purchase) were used in these studies at the ages indicated in the text. were purchased from Envigo (Indianapolis, IN) and allowed to acclimate for at least three days inside the BSL-3 AG laboratories prior to the start of studies. The age of the animals at different experiment time points in the studies is indicated in the text.
Cells
Vero E6 cells expressing TMPRSS2 (Vero E6-TMPRSS2 cells) from the Japanese Collection of Research Bioresources (JCRB) Cell Bank (1819) were propagated in DMEM containing 10% fetal bovine serum, antibiotics, and 1 mg/mL geneticin (G418) at 37°C with 5% CO2. The cell line was verified to be negative for mycoplasma contamination by monthly PCR analysis.
Viruses
The Omicron BA.1 variant (hCoV-19/USA/WI-WSLH-221686/2021; EPI_ISL_7,263,803) was isolated from a residual clinical sample from an individual who had recently traveled to South Africa. The virus was amplified once on Vero E6-TMPRSS2 cells and harvested when the cell monolayer was nearly completely dead. The cell supernatant was then clarified by low-speed centrifugation, aliquoted, and stored at −80°C until needed. Virus titration was performed on cells overlayed with 1% methylcellulose media until plaques were large enough to visualize by eye. Cells were fixed and stained with crystal violet solution to count plaques and determine the titer of the stock virus. Early contemporary S-614D and S-614G isolates (SARS-CoV-2/UT-NCGM02/Human/2020/Tokyo and SARS-CoV-2/UT-HP095-1N/Human/2020/Tokyo, respectively) were described previously (Imai et al., 2020, 2021).
Method details
Vaccination and infection of hamsters
Residual Moderna (mRNA-1273) vaccine was acquired through a university health clinic 24 h or less after the vaccine was reconstituted. Hamsters (females, 4-week-old at the time of the first vaccination) were vaccinated with 35 μg of the vaccine at injection sites in the thigh muscle. If a second dose was administered, there was a 28-day interval between vaccinations. Hamsters were infected with 1,000 pfu of SARS-CoV-2 by intranasal inoculation (50 μL total volume) while under anesthesia (isoflurane).
Titration of virus from hamster tissue samples
Three days after infection, hamsters were humanely euthanized, and respiratory tissue samples (nasal turbinate and lung) were collected. For lung samples, a piece of each lobe was collected and pooled. After the samples were frozen at −80°C for at least 24 h, tissue samples (pooled lung and nasal turbinates) were homogenized in 1 mL of media and clarified by centrifugation. Undilute or 10-fold serially diluted clarified tissue samples (100 μL per well) were used to infect a monolayer of Vero E6 TMPRSS2 cells for 30 min at 37°C. The cells were washed once to remove unbound virus and then overlayed with 1% methylcellulose media containing 1% fetal bovine serum for four days. Crystal violet solution was added directly to the wells overnight to fix and visualize plaques.
RT-qPCR assay
To detect viral RNA from the lungs, tissue homogenates were clarified by centrifugation at 10,000 rpm for 5 min, and viral RNA was isolated by using an RNeasy kit (Qiagen) and detected by using the Luna SARS-CoV-2 one-step RT-qPCR assay kit (New England Biolabs) on an ABI 7500 Fast instrument.
Enzyme-linked immunosorbent assay (ELISA)
The ELISA was performed using a recombinant S-614G protein with a C-terminal HIS-tag purified by using TALON metal affinity resin from Expi293F cells (Thermo Fisher Scientific). The Omicron spike (C-terminal HIS-tag) was obtained from AcroBiostems (Catalog # SPD-C522e). The ELISA plates were coated overnight at 4°C with 50 μL of the spike antigen at a concentration of 2 μg/mL in phosphate-buffered saline (PBS). After blocking the plates with PBS containing 0.1% Tween 20 (PBS-T) and 3% milk powder, the plates were incubated in duplicate with heat-inactivated (56°C for 30 min) serum diluted in PBS-T with 1% milk powder. After a 4-h incubation at room temperature, the plates were washed with PBS-T three times and then incubated with a hamster IgG secondary antibody conjugated with horseradish peroxidase (Invitrogen; 1:7,000 dilution in PBS-T with 1% milk powder). After a 1-h incubation with the secondary antibody, the plates were washed three times with PBS-T and then developed with SigmaFast o-phenylenediamine dihydrochloride solution. After a 10-min incubation, the reaction was stopped with the addition of 3 M hydrochloric acid. The absorbance was measured at a wavelength of 490 nm (OD490). The IgG antibody endpoint titer was defined as the highest serum dilution with an OD490 cut-off value of ≥0.15.
Quantification and statistical analysis
Statistical analysis
The sample sizes for the hamster studies were determined from previous studies that demonstrated significant differences among groups. The researchers were not blinded to the group allocations during the experiments. Virus titers from animals are expressed as scatterplots with bars and individual datapoints, obtained by using Graphpad Prism 9. Statistical analyses were performed using two-tailed unpaired Student’s t-tests. Correlations between antibody levels and virus titers were determined with the Spearman correlation test.
Acknowledgments
We thank Susan Watson for scientific editing. This work was supported, in part, by the National Institute of Allergy and Infectious Diseases Center for Research on Influenza Pathogenesis (HHSN272201400008C), the Center for Research on Influenza Pathogenesis and Transmission (CRIPT) (75N93021C00014), and the Japan Program for Infectious Diseases Research and Infrastructure (JP21wm0125002) from the Japan Agency for Medical Research and Development (AMED).
Author contributions
Conceptualization, P.J.H. and Y.K.; methodology, P.J.H. and Y.K.; investigation, P.J.H., M.K., T.M., S.C., T.A., R.W., and A.B.; writing – original draft, P.J.H., and Y.K.; funding acquisition, Y.K.; resources, K.R.F. and A.C.B.; supervision, P.J.H. and Y.K; all authors reviewed the final version of the manuscript.
Declaration of interests
Y.K. has received unrelated funding support from Daiichi Sankyo Pharmaceutical, Toyama Chemical, Tauns Laboratories, Inc., Shionogi & Co. LTD, Otsuka Pharmaceutical, KM Biologics, Kyoritsu Seiyaku, Shinya Corporation, and Fuji Rebio. The remaining authors have no competing interests.
Published: March 28, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110688.
Supplemental information
Data and code availability
The datasets supporting the current study are available from the lead contact on request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abdelnabi R., Foo C.S., Zhang X., Lemmens V., Maes P., Slechten B., Raymenants J., Andre E., Weynand B., Dallmeier K., et al. The omicron (B.1.1.529) SARS-CoV-2 variant of concern does not readily infect Syrian hamsters. Antivir. Res. 2022;198:105253. doi: 10.1016/j.antiviral.2022.105253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai J., Zhang H., Zhang Y., Lin K., Zhang Y., Wu J., Wan Y., Huang Y., Song J., Fu Z., et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2021;11:337–343. doi: 10.1080/22221751.2021.2022440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameroni E., Saliba C., Bowen J.E., Rosen L.E., Culap K., Pinto D., VanBlargan L.A., De Marco A., Zepeda S.K., Iulio J.d., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. bioRxiv. 2021 doi: 10.1101/2021.12.12.472269. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Shaw R.H., Supasa P., Liu C., Stuart A.S., Pollard A.J., Liu X., Lambe T., Crook D., Stuart D.I., et al. Reduced neutralisation of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum. Lancet. 2021;399:234–236. doi: 10.1016/S0140-6736(21)02844-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann P.J., Iida S., Iwatsuki-Horimoto K., Maemura T., Kiso M., Scheaffer S.M., Darling T.L., Joshi A., Loeber S., Singh G., et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603:687–692. doi: 10.1038/s41586-022-04441-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmsdal G., Hansen O.K., Møller L.F., Christiansen D.H., Petersen M.S., Kristiansen M.F. Omicron outbreak at a private gathering in the Faroe Islands, infecting 21 of 33 triple-vaccinated healthcare workers. medRxiv. 2021 doi: 10.1093/cid/ciac089. Preprint at. ciac089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Halfmann P.J., Yamayoshi S., Iwatsuki-Horimoto K., Chiba S., Watanabe T., Nakajima N., Ito M., Kuroda M., Kiso M., et al. Characterization of a new SARS-CoV-2 variant that emerged in Brazil. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2106535118. e2106535118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. U S A. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Chivukula V., Herati R.S., Hubbard S.R., Mulligan M.J., Landau N.R. Increased resistance of SARS-CoV-2 omicron variant to neutralization by vaccine-elicited and therapeutic antibodies. bioRxiv. 2021 doi: 10.1101/2021.12.28.474369. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by several therapeutic monoclonal antibodies. bioRxiv. 2021 doi: 10.1101/2021.12.15.472828. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., To K.K.-W., Peng Q., Chan J.M.-C., Huang H., Yang D., Lam B.H.-S., Chuang V.W.-M., Cai J.-P., Liu N., et al. Vaccine-breakthrough infection by the SARS-CoV-2 Omicron variant elicits broadly cross-reactive immune responses. bioRxiv. 2021 doi: 10.1101/2021.12.27.474218. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the current study are available from the lead contact on request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.