Abstract
Since its first appearance, the SARS-CoV-2 has spread rapidly in the human population, reaching the pandemic scale with >280 million confirmed infections and more than 5 million deaths to date (https://covid19.who.int/). These data justify the urgent need to enhance our understanding of SARS-CoV-2 effects in the respiratory system, including those linked to co-infections.
The principal aim of our study is to investigate existing correlations in the nasopharynx between the bacterial community, potential pathogens, and SARS-CoV-2 infection.
The main aim of this study was to provide evidence pointing to possible relationships between components of the bacterial community and SARS-CoV-2 in the nasopharynx. Meta-transcriptomic profiling of the nasopharyngeal microbial community was carried out in 89 SARS-Cov-2 positive subjects from the Campania Region in Italy. To this end, RNA extracted from nasopharyngeal swabs collected at different times during the initial phases of the pandemic was analyzed by Next-Generation Sequencing (NGS). Results show a consistently high presence of members of the Proteobacteria (41.85%), Firmicutes (28.54%), and Actinobacteria (16.10%) phyla, and an inverted correlation between the host microbiome, co-infectious bacteria, and super-potential pathogens such as Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, and Neisseria gonorrhoeae.
In depth characterization of microbiota composition in the nasopharynx can provide clues to understand its potential contribution to the clinical phenotype of Covid-19, clarifying the interaction between SARS-Cov-2 and the bacterial flora of the host, and highlighting its dysbiosis and the presence of pathogens that could affect the patient's disease progression and outcome.
Keywords: Nasopharyngeal microbiome, COVID-19, Nasal swabs, Meta-transcriptomics, SARS-CoV-2
1. Introduction
Coronavirus disease 2019 (COVID-19) is caused by the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) [1]. Human coronavirus is a group of seven single-stranded RNA viruses, belonging to the Coronaviridae family, able to infect humans. Three of them, Severe Acute Respiratory Syndrome coronavirus (SARS-CoV), Middle East Respiratory Syndrome coronavirus (MERS-CoV), and Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2), belonging to the Betacoronavirus genus, can cause severe infections that can lead to deaths [2]. Like the other human coronaviruses, SARS-CoV-2 is mainly transmitted through the upper respiratory tract by aerosolized droplets carrying viral particles [3], binding to the angiotensin-converting enzyme 2 ACE-2 receptor, whose expression is particularly high in the nasal and oral cavity cells [4,5]. Generally, COVID-19 patients show a wide clinical picture after 2–14 days post-viral exposure, ranging from absence of clinical manifestation, presence of mild symptoms, such as cold, fatigue, nausea, and fever to more severe symptoms, such as respiratory distress and pneumonia [6]. Several studies indicate that multiple factors such as age, sex, lifestyle, diabetes, cigarette smoke, and use of antibiotics affect the severity of COVID-19 manifestation [7,8]. In addition, it has been recently documented a decrease in the microbiota diversity of the upper respiratory tract as well as an increase in opportunistic pathogens in SARS-CoV-2 infected patients, both with a key role in determining the clinical manifestation of the disease [9], an observation supported by the evidence that SARS-CoV-2 infections are frequently associated with an increase in secondary bacterial co-infections that leads to a clinical worsening [10,11].
The human body hosts millions of microbes, including bacteria, fungi, and viruses, most of them representing the microbiota, that can be divided into core microbiota and variable microbiota. The former represents the predominant microbial species existing in healthy conditions, while the variable one is exclusive of each individual and depends on its lifestyle and genome [12]. The human microbiota starts forming directly after birth and plays an essential role in human health and disease. Indeed, it increases the acquisition of nutrients by providing unique and specific enzymes to digest food and synthesize vitamins, provides a physical barrier that protects against external pathogens with a competitive exclusion, and stimulates the immune system of the host [13]. Inflammatory states and pathologies can alter the equilibrium of the microbiota causing microbial dysbiosis and reduction in the diversity of the microbial community, an event that promotes proliferation of pathogenic or opportunistic species. In healthy humans, the nasopharyngeal microbiome is one of the largest and is principally composed of the following genera: Bifidobacterium, Corynebacterium, Staphylococcus, Streptococcus, Dolosigranulum, and Moraxella belonging to the respective Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria phyla [14]. In recent years, meta-transcriptomic profiling of bacteria, viruses, and other microbes has emerged as a powerful tool to identify the active members of the microbiome in complex samples and explore the dynamic changes in bacterial populations over time in response to different environmental conditions, including the presence of co-infections [15]. In the present study, we investigated in-depth the nasopharyngeal microbiome composition in COVID-19 patients, evaluating with a Next-generation RNA sequencing (RNA-Seq) approach, the presence and abundance of bacterial RNAs in 89 nasopharyngeal swabs, collected in the Campania Region (Italy) during 3 different seasonal periods during COVID-19 pandemic. The principal aim of the study is to investigate the bacterial microbiota composition of the nasopharynx in SARS-CoV-2 infected patients, elucidating the presence of species belonging to the normal bacterial flora of the nasopharynx, opportunistic pathogens, and pathogen species associated with severe illness of the upper respiratory tract.
2. Materials and methods
A total of 89 patients from the Campania Region in (Italy) with confirmed SARS-CoV-2 infection were selected during the three main Covid-19 waves in Italy: I period (March–May 2020); II period (September–November 2020); III period (January–February 2021). The study was approved by the Ethics Committee of “Campania Sud” (approval code: 206/2021) and was conducted according to the Declaration of Helsinki. Patients were stratified as follows: 27 cases were from the I period, 43 from period II, and 19 from period III. The median age of patients (IQR) was 55 years (ranging from 3 to 99); regarding the gender, 46% of them were female (n = 41) and 54% were male (n = 48). The clinical outcome observed ranged from asymptomatic to severe infection and death. The demographic and basal characteristics of the participants are summarized in Table 1 .
Table 1.
Patient cohort description.
| March-Apr-May 2020 (n = 27) | Sept-Oct-Nov 2020 (n = 43) | Jan–Feb 2021 (n = 19) | |
|---|---|---|---|
| Age (years) | |||
| 0–20 | 3 (11%) | 3 (7%) | 5 (26%) |
| 21–40 | 4 (15%) | 8 (19%) | 1 (5%) |
| 41–60 | 4 (15%) | 15 (35%) | – |
| 61–80 | 10 (37%) | 13 (30%) | 6 (32%) |
| >80 | 3 (11%) | 4 (9%) | 7 (37%) |
| Unknown | 3 (11%) | – | |
| Gender | |||
| Male (%) | 16 (59%) | 27 (63%) | 5 (26%) |
| Female (%) | 8 (30%) | 16 (37%) | 14 (74%) |
| Unknown | 3 (11%) | – | – |
| Disease severity | |||
| Asymptomatic | 11 (41%) | 15 (35%) | – |
| Mild | 7 (26%) | 6 (14%) | – |
| Moderate | 2 (7%) | – | 4 (21%) |
| Severe | 5 (2 dead) (19%) | 4 (1 dead) (9%) | 1 (5%) |
| Unknown | 27%) | 18 (42%) | 14 (74%) |
Nasopharynx swab specimens from patients were collected following standardized procedures by the healthcare personnel in charge. Total RNA has been extracted from swabs and RNA amount and integrity were assessed with a Qubit 2.0 fluorometer assay (Thermo Fisher Scientific, Waltham, MA, USA) and a 4200 TapeStation instrument (Agilent Technologies, Santa Clara, CA, USA), respectively, as described earlier [16]. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR) was used to assess the presence and amount of SARS-CoV-2 RNA in each sample as follows: 2 μl RNA were converted in cDNA by using SensiFAST cDNA Synthesis Kit (Bioline) and primer sequences amplifying a region in the N gene of SARS-CoV2 (Forward Primer: GGGGAACTTCTCCTGCTAGAAT; Reverse Primer: CAGACATTTTGCTCTCAAGCTG). The observed Ct (qRT-PCR cycle threshold) values were variable, ranging between a minimum of 12,18 and a maximum of 34,96, corresponding to a viral copy number ranging from 1,5 × 107 to 8 copies/mL. Libraries were prepared using the Illumina TruSeq stranded total RNA protocol, starting from 100 ng of RNA. Eight samples were sequenced on Illumina platform NextSeq 500 (Illumina, San Diego, California) (2 × 75 nt) and eighty-one samples were sequenced on Illumina platform NovaSeq 6000 (Illumina, San Diego, California) (2 × 100 nt), yielding nearly 100 million high-quality paired-end reads per sample. Raw sequencing data were analyzed in HOME-BIO pipeline [17] to obtain bacterial taxonomy profiling of samples by querying RefSeq complete bacterial genomes/proteins database. The Quality Control module was set with default parameters, to remove low-quality sequences and filter out reads aligned to the human reference genome (GRCh38.p13 release 37). Classification data were obtained using default confidence threshold (0.5) and then imported in R software (version 3.6.3) for normalization in RPMs (reads per million) values mapped to the bacterial database. As a control, we collected meta-transcriptomic data generated from nasopharyngeal swab specimens for 25 SARS-CoV-2 negative patients with no other viral infections [18]. Fastq files were quality checked with FastQC and analyzed with the HOME-BIO pipeline. Raw read counts were normalized in RPM and differential distribution of phyla and genera was computed as a ratio between the mean values of RPM in the SARS-CoV-2 positive dataset versus the control dataset. Statistical significance was computed by applying a t-test followed by Bonferroni correction. Only comparisons with a p-value ≤ 0.05 were considered significant.
3. Results and discussion
3.1. Evaluation of the bacterial microbiota composition of the nasopharynx in SARS-CoV2 positive patients and healthy donors
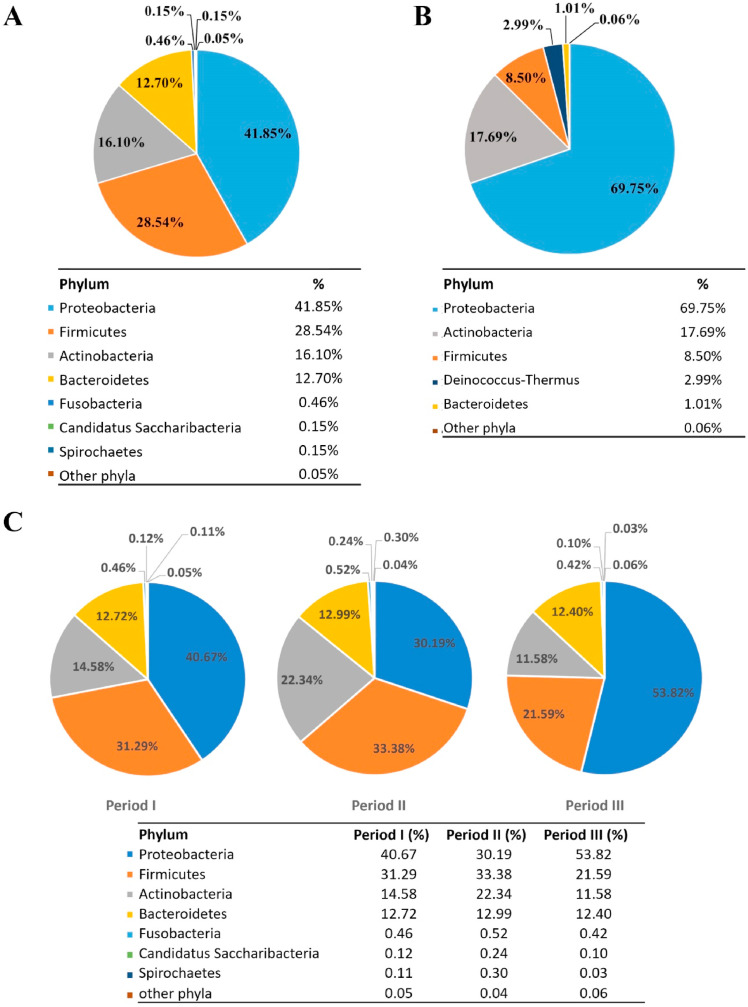
A first analysis of the bacterial microbiota composition in nasopharyngeal swabs of SARS-CoV-2 positive patients revealed the presence of three dominant phyla, also reported as highly represented in the nasopharynx of healthy humans [19]: Proteobacteria (41.85%), Firmicutes (28.54%), Actinobacteria (16.10%). Less abundant but still detectable phyla, given the great sensitivity of the method employed, were represented by Bacteroidetes (12.70%), Fusobacteria (0.46%), Candidatus Saccharibacteria (0.15%), and Spirochaetes (0.15%) (Fig. 1 A).
Fig. 1.
Bacterial microbiota composition in the nasopharynx of COVID-19 patients. Relative percentage of abundance for the main phyla identified in SARS-CoV-2 positive (panel A) and negative (panel B) patients and in SARS-CoV-2 patients across three periods of sampling (panel C).
By comparison, among the most abundant phyla detected in the control group, we found an increased amount of Proteobacteria (69.75%) and a decreased percentage of Firmicutes (8.50%), while Actinobacteria were less variable between the two groups (17.69%) (Fig. 1B). It is worth mentioning an observed variability in relative abundances of phyla within infected individuals, a result that could depend on the dynamic ecological niche of the individual nasopharynx [20] and/or biological fluctuations in response to SARS-CoV-2 infections, an interesting possibility recently suggested by Hernández-Terán et al. [21].
To go further in depth, we computed the differential distribution of phyla between infected and non-infected individuals (p-value <0.05), observing that 5 phyla were more represented in SARS-CoV-2 positive patients (Tenericutes, Candidatus Saccharibacteria, Fusobacteria, Bacteroidetes, Firmicutes), while 4 phyla were predominant in SARS-CoV-2 negative vs positive patients (Proteobacteria, Planctomycetes, Verrucomicrobia, Deinococcus-Thermus).
In addition, to evaluate whether the effect of seasonality and behavior (introduction of the widespread use of a face mask, increased attention to social distancing, etc.) could have influenced the composition of the nasopharyngeal microbiota, we divided SARS-CoV-2 patients based on the period of sampling considering the three COVID-19 waves in our Region; in detail, 27 patients were from the first period (March–May 2020), 43 patients in the second period (September–November 2020) and 19 in the third period (January–February 2021). As shown in Fig. 1C, the analyses did not reveal significant qualitative differences among phyla detected in the three sampling periods, but we noticed differences in the percentages assigned to each category. Indeed, the highest variability over time was observed for the Proteobacteria phylum. Of note, it has been reported that the overall population of Proteobacteria in the upper respiratory trait decreases upon aging or due to smoking, two factors correlated with high severity of SARS‐CoV‐2 infection and mortality rate, leading to the hypothesis that a decrease in the population of Proteobacteria could increase the susceptibility to viral infection and replication leading to severe disease [22], an observation also supported by the increased amount of Proteobacteria seen in the control group (Fig. 1B).
3.2. Effects of SARS-CoV-2 infection on the host-microbiome of the nasopharynx and possible bacterial co-infections
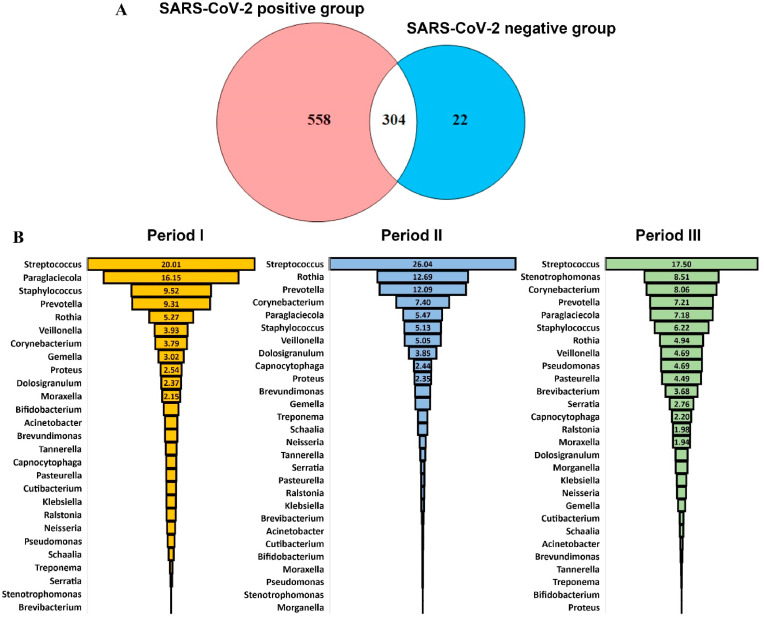
Due to the complex interaction that potential pathogens establish with the healthy bacterial flora, respiratory tract infections have been associated with changes in the microbiome [23]. In the specific case of SARS-CoV2 infection, still, there are controversial results: indeed, a recent study comparing the nasopharyngeal microbiota of positive and negative COVID-19 patients has revealed no significant differences in bacterial diversity or composition [24], while another study has identified an increase in opportunistic pathogen species and a reduction in the abundance of normal bacterial flora [25]. To evaluate whether the SARS-CoV-2 infection could have perturbed the equilibrium between commensal bacteria, we went more in-depth into the taxonomic classification by exploring the bacterial genera present in the nasopharynx of COVID-19 patients. Previous evidence demonstrated that the nasopharyngeal microbiome of healthy humans is primarily populated by microbes belonging to the phyla Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria, with a predominance of Bifidobacterium, Corynebacterium, Staphylococcus, Streptococcus, Dolosigranulum, and Moraxella genera [23,26]. By comparing the abundance of 304 common bacterial genera identified in SARS-CoV-2 and non-infected patients (Fig. 2 A), we identified 42 genera that predominated in the infected patients, among whom is worth mentioning Streptococcus, which was also the most abundant bacterial genera, followed by Prevotella, Rothia, Staphylococcus, and Veillonella, whose abundance, in addition, was similar in the three periods of sampling (Fig. 2B). In accordance with our results, Prevotella and Veillonella were identified as bacterial genera with increased abundance in COVID-19 patients compared with healthy controls [27].
Fig. 2.
Comparison between bacterial genus identified in SARS-CoV-2 positive patients and control group (A) and taxonomy profiling, expressed as a percentage of the total, in three different periods of sampling (B).
Interestingly, in SARS-CoV-2 patients compared to the control group we noticed a predominance of common potential respiratory pathogens belonging to the Streptococcus and Staphylococcus genera, accompanied by a reduced presence of commensal Bifidobacterium and Dolosigranulum. In accordance with the literature, in case of respiratory infections a dysbiosis of the microbiota occurs, with an increase in pathobionts such as Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, and Moraxella catarrhalis, with a decrease of the normal bacterial flora, mainly composed by Lactococcus, Anoxybacillus, Corynebacterium, Bifidobacterium and Dolosigranulum sp. [25,28].
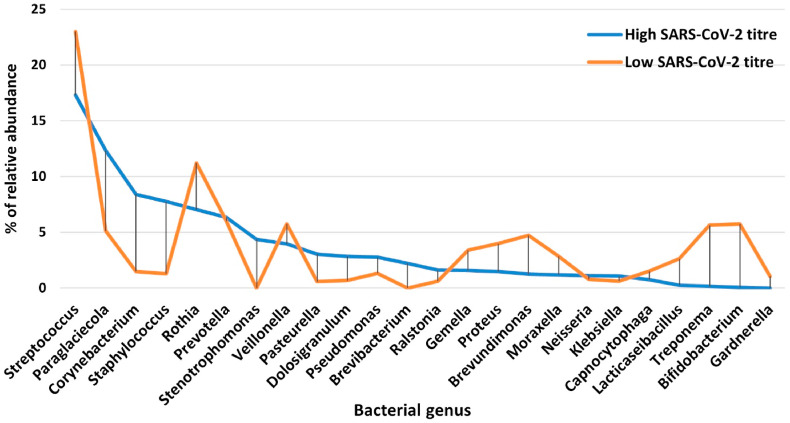
To evaluate if these changes in the microbiota composition of the nasopharynx could be directly correlated with the SARS-CoV2 viral titer, we classified infected patients into two groups, based on the amount of SARS-CoV-2 RNA detected (High and Low viral copy number amount, expressed as Ct value < 20 and Ct value > 25, respectively) and analyzed the differences at a genus level.
Interestingly, we noticed that in the High SARS-CoV-2 group, genera involved in the immunological homeostasis of nasopharyngeal microbiota decreased in favor of multidrug-resistant pathogens (Fig. 3 ). Indeed, the Streptococcus, Veillonella, Proteus, Treponema, Brevudimonas, Bifidobacterium and Lacticaseibacillus genera were generally less abundant in patients with high SARS-CoV-2 RNA detected; in addition, in accordance with a recent study comparing the nasopharyngeal microbiota in SARS-CoV-2 positive and negative patients [3], a decrease in the relative abundance of Rothia was also recorded in patients with high SARS-CoV-2 RNA. On the contrary, the genera Staphylococcus, Corynebacterium, Dolosigranulum, Pseudomonas, Pasteurella, and Stenotrophomonas were more abundant in the high than in the low SARS-CoV-2 group; these genera include pathogen species that are among the main causes of hospital infections, also with drug-resistant strains associated with a high mortality rate [29]. Despite minor differences in the relative abundance of these genera, our data are in agreement with other studies surveying the nasopharynx microbiota composition in COVID-19 patients [30]. These data suggest that SARS-CoV-2 is associated with a dysbiosis of the microbiota and a higher risk of bacterial co-infection.
Fig. 3.
Percentage of relative abundance for bacterial genera identified in nasopharyngeal swabs from patients with High or Low SARS-CoV-2 RNA amount.
3.3. Identification of super-pathogen bacteria associated with SARS-CoV-2 infection
Given its role in the clearance of pathogens from the middle ear and paranasal sinuses, the nasopharynx typically hosts both pathogenic and non-pathogenic bacteria. Viral infections, however, could alter the normal clearance patterns, allowing bacterial colonization and subsequent risk of co-infections [31,32]. Meta-transcriptomic analysis of SARS-CoV-2 patients detected 974 bacterial species with an rpm>5 in all samples. Interestingly, we noticed in a medium-high % of patients the presence of bacterial species included in the WHO global priority pathogens list (global PPL) of public health concern for antibiotic resistance [33], such as Staphylococcus aureus (98%), Klebsiella pneumoniae (80%), Streptococcus pneumoniae (52%) and Neisseria gonorrhoeae (79%); less abundant but still detectable, we also found Pseudomonas aeruginosa (19%) and Acinetobacter baumannii (16%). The most abundant, among the above mentioned, was Staphylococcus aureus; this bacterial strain, indeed, accounted for more than the 10% of all reads associated with the species in 21 out of 89 patients. Klebsiella pneumoniae was the second most frequent, representing 2% of all reads in 12 patients, followed by Neisseria gonorrhoeae (5 patients), Streptococcus pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii (1 patient) (Table 2 ). Klebsiella pneumoniae and Streptococcus pneumoniae are among the most common causes of community-acquired pneumonia, while Staphylococcus aureus and Pseudomonas aeruginosa have been frequently associated with hospitalization; furthermore, Streptococcus pneumoniae has been also associated with intubation for mechanical ventilation [34]. Neisseria gonorrhoeae, the etiologic agent of gonorrhea, a sexually transmitted infection, mainly colonizes the genital mucosa, but it can also colonize the nasopharyngeal cavity giving rise to infections in this atypical anatomic site [35,36]. However, colonization of the nasopharynx by these pathogens is a multifactorial process involving virulence factors of the pathogen, possible interactions with the microbiota, and genetic factors of the host [37].
Table 2.
Relative abundance of super-pathogen bacteria in nasopharyngeal samples from COVID-19 patients.
| Bacterial Species | OMS priority | Patients in which the pathogen has been identified | n° patients in which the pathogen represents >10% of total reads | n° patients in which the pathogen represents >5% of total reads | n° patients in which the pathogen represents >2% of total reads |
|---|---|---|---|---|---|
| Staphylococcus aureus | High | 98% | 21 | 30 | 34 |
| Klebsiella pneumoniae | Critical | 80% | 0 | 2 | 12 |
| Neisseria gonorrhoeae | High | 79% | 0 | 0 | 5 |
| Streptococcus pneumoniae | Medium | 52% | 0 | 0 | 1 |
| Pseudomonas aeruginosa | Critical | 19% | 0 | 0 | 1 |
| Acinetobacter baumannii | Critical | 16% | 0 | 0 | 1 |
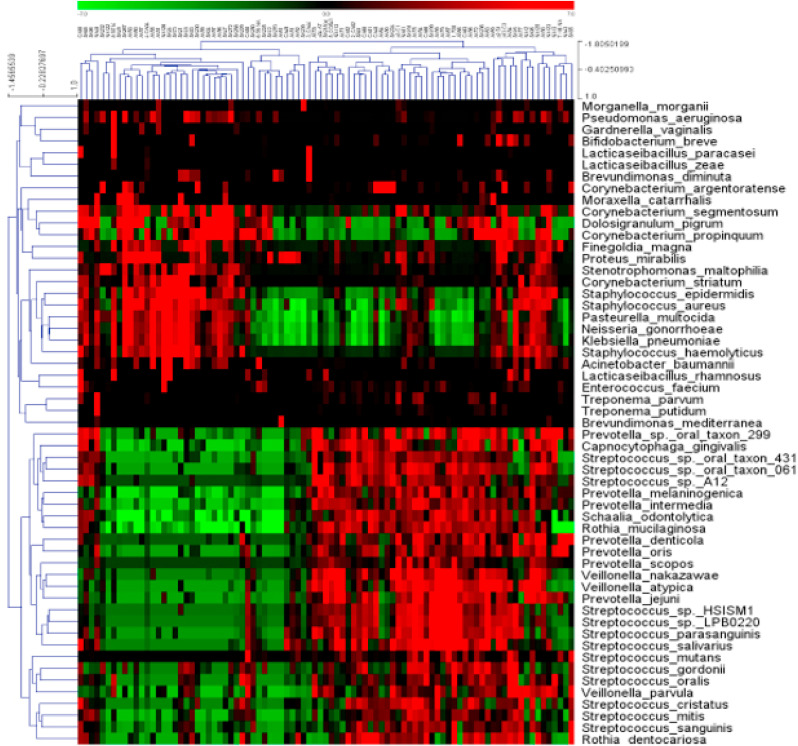
To go further in-depth in describing the complexity of the bacterial microbiota in COVID-19 patients, we then focused our attention on the most abundant bacterial species, identified in at least 25% of the whole cohort of 89 patients. These included pathogens and opportunistic pathogens that might take advantage of a microbial dysbiosis of the upper respiratory tract, as well as bacterial species known to be a part of the normal bacterial flora of the nasopharynx. Of note, unsupervised hierarchical clustering revealed a discrete segregation of 55 abundant bacterial species into two main groups (Fig. 4 ).
Fig. 4.
Heatmap showing median centered expression values of 55 bacterial species across 89 samples from SARS-CoV-2 positive patients. Unsupervised hierarchical clustering, generated with Multi-Experiment Viewer (MeV 4.5.1) using default parameters revealed clear segregation of samples in two major clusters, characterized by a different abundance of commensal, opportunistic pathogens, and super-pathogen bacteria.
As main representative species of the first group, we noticed the presence of Staphylococcus aureus, Klebsiella pneumoniae, Pseudomonas aeruginosa typically associated with pneumonia, opportunistic pathogens such as Moraxella catarrhalis [38], Finegoldia magna [39], Acinetobacter baumannii [40], and Enterococcus faecium [41], as well as normal commensal highly abundant in the human nasopharynx such as several Lactobacilli [42], Corynebacterium propinquum, Dolosigranulum pigrum [43], etc.
The second big cluster was dominated by Streptococci such as Streptococcus salivarius, Streptococcus oralis, and other bacteria predominating in periodontal diseases such as members of the Prevotella genus, Schaalia odontolitica, Veillonella parvula, Capnocytophaga gingivalis, Rothia dentocariosa, etc [44], in line with other studies even showing an increase in periodontal bacteria generally linked to poor oral hygiene and confirming their association with SARS-CoV-2 infection [27,45].
In agreement with our data, recent studies reported the presence of co-infections and superinfections associated with poor outcomes and increased mortality in more than 20% of patients with SARS-CoV-2 infection, with Klebsiella pneumoniae, Streptococcus pneumoniae, and Staphylococcus aureus as the most frequently identified bacteria [9]. Notably, these species can produce serious respiratory illnesses on their own. From this evidence, virus-host microbiota interplay seems to be a common feature of COVID-19 pathogenesis, with a disturbance of resident bacterial community, an event linked to disease severity and spread, as recently reported [3]. In agreement with this, our results indicate the presence of a dis-biotic and pro-inflammatory microbiota in COVID-19 patients, with a significant diminution in species richness of the nasopharynx; importantly, among bacteria identified as particularly abundant in a relevant number of patients, we also detected species reported as potential counteractors of COVID-19, such as Rothia mucilaginosa and Streptococcus oralis [46]. Another important observation is the identification of oral pathogens involved in periodontal diseases, including, among others, species belonging to the Prevotella and Veillonella genera, that might take advantage of oral and nasopharyngeal dysbiosis, and can propagate in distant organs such as heart [47] and lung [48]. In a consistent number of patients, potential pathogens and commensal bacteria belonging to these two clusters displayed an opposite abundance, leading to the observation that, a reduction in the normal bacterial flora, as reported in other COVID-19 patients, is frequently associated with increased susceptibility to pathogen bacteria of the respiratory system and over-proliferation of opportunistic pathogens of the oral cavity. In this regard, the nasopharynx and the oral cavity could act as a potential reservoir for a dysbiotic microbiota with pathogenic potential [49,50], thus underlying the importance of characterizing the microbiota during SARS-CoV-2 infections, as this approach might identify the presence of bacterial species associated with an increased risk of disease severity and guide the therapeutic strategy.
4. Conclusions
In this study, we applied meta-transcriptomic analysis to obtain a comprehensive picture of the whole microbiota, using a minimally invasive procedure from a very small amount of starting sample, thereby implementing a diagnostic approach for the diagnosis of the upper respiratory tract co-infections, useful to guide clinical management of the SARS-CoV-2 positive patients.
Here, we underlie how SARS-CoV-2 infection disturbs the symbiosis between commensal bacteria and opportunistic pathogens that colonize the nasopharyngeal cavity, resulting in an over-proliferation of pathogenic bacteria that predispose to the development of co-morbidities. Indeed, although the balance between microbes populating the nasopharynx is very complex, as it is subjected to variables such as sex, age, pathology, seasons, etc., our study has shown a significant difference between the microbiota composition of SARS-CoV-2 positive and negative control patients, and a clear correlation between high SARS-CoV-2 burden and proliferation of super-pathogenic bacterial species such as Staphylococcus aureus, Klebsiella pneumoniae, Streptococcus pneumoniae, Pseudomonas aeruginosa, and Acinetobacter baumannii, accompanied by a reduced abundance and bacterial diversity in the nasopharynx.
Taken together, our findings suggest that the nasopharyngeal microbiota could be a factor potentially associated with the clinical manifestation of COVID-19, as highlighted by the observed dysbiosis and the presence of pathogens that could affect disease progression and outcome. These results indicate that it is worth deepening this kind of analysis on much larger cohorts of patients from different geographical areas.
Funding statement
Work supported by Regione Campania (grant ‘Monitoring the spread and genomic variability of the COVID 19 virus in Campania using NGS technology’, POR Campania FESR 2014/2020, CUP: B14I20001980006 and grant ‘GENOMAeSALUTE’, POR Campania FESR 2014/2020, azione 1.5, CUP: B41C17000080007). AS is recipient of a post-doctoral research fellowship (Assegno di Ricerca) from University of Salerno, TR and YDA are fellows of Rete Oncologica Campana and EA is a fellow of Fondazione U. Veronesi. CF and JL are Ph.D. students of the Research Doctorate in Veterinary Sciences of the University of Napoli ‘Federico II’ and in ‘Molecular and Translational Oncology and Innovative Medical-Surgical Technologies', University of Catanzaro “Magna Graecia”.
Authors contribution
Study concept and design: AW, FR and GF. Sample preparation and sequencing: EA, TR, JL and YDA. Bioinformatics analysis: CF, DP, GG. Statistical analysis and interpretation of the data: RG, AS, GG, AW, FR. Writing of the manuscript: RG, AS, AW, FR. Writing - Review and Editing: MG, EV e PP.
Data availability
The data that support the findings described here are available from the corresponding authors upon request.
Supporting information
N/A.
CRediT authorship contribution statement
Rosa Giugliano: Formal analysis, Writing – original draft. Assunta Sellitto: Formal analysis, Writing – original draft. Carlo Ferravante: Formal analysis. Teresa Rocco: Investigation. Ylenia D'Agostino: Formal analysis, Investigation. Elena Alexandrova: Formal analysis, Investigation. Jessica Lamberti: Investigation, Methodology. Domenico Palumbo: Formal analysis. Massimiliano Galdiero: Writing – review & editing. Emilia Vaccaro: Investigation. Pasquale Pagliano: Formal analysis. Alessandro Weisz: Conceptualization, Resources, Supervision, Writing – review & editing. Giorgio Giurato: Formal analysis, Conceptualization, Supervision. Gianluigi Franci: Conceptualization, Supervision, Writing – review & editing. Francesca Rizzo: Conceptualization, Resources, Supervision, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thanks Prof. Ivan Gentile, Dr. Nicola Schiano Moriello, PO Malattie Infettive and Prof. Giuseppe Portella, Dr. Michele Cennamo, PO Patologia Clinica of “Federico II” University Hospital, Napoli; Dr Angelo Salomone Megna - U.O.C. Malattie Infettive and Dr. Vincenzo Rocco, Dr. Maurizio Fumi - U.O.C. Patologia Clinica of AORN “San Pio” PO G. Rummo, Benevento; Dr. Gregorio Goffredi, Dr. Francesca Marciano - G.O.I. Medicina di Laboratorio e Biologia Molecolare of “Maria Santissima Addolorata” Hospital, Eboli – Salerno; Dr. Paolo Sorrentino, Dr. Carmine Sanseverino, Unità Fegato, UO Malattie infettive, Dr. Maria Landi, Dr. Maria Grazia Foti, Servizio di Microbiologia e Virologia - A.O.R.N. “San Giuseppe Moscati”, Avellino; Prof. Paolo Maggi, U.O.C. Malattie Infettive e Tropicali – Dr. Maddalena Schioppa, UOSD Genetica e Biologia Molecolare - A.O.R.N. “S. Anna e S. Sebastiano”, Caserta for providing nasopharyngeal swab RNA and other information used in this study.
References
- 1.Ventero M.P., Cuadrat R.R.C., Vidal I., Andrade B.G.N., Molina-Pardines C., Haro-Moreno J.M., et al. Nasopharyngeal microbial communities of patients infected with SARS-CoV-2 that developed COVID-19. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.637430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26:729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Engen P.A., Naqib A., Jennings C., Green S.J., Landay A., Keshavarzian A., et al. Nasopharyngeal microbiota in SARS-CoV-2 positive and negative patients. Biol. Proced. Online. 2021;23:10. doi: 10.1186/s12575-021-00148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhar D., Mohanty A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020;285 doi: 10.1016/j.virusres.2020.198018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rhoades N.S., Pinski A.N., Monsibais A.N., Jankeel A., Doratt B.M., Cinco I.R., et al. Acute SARS-CoV-2 infection is associated with an increased abundance of bacterial pathogens, including Pseudomonas aeruginosa in the nose. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tu Y.F., Chien C.S., Yarmishyn A.A., Lin Y.Y., Luo Y.H., Lin Y.T., et al. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Callender L.A., Curran M., Bates S.M., Mairesse M., Weigandt J., Betts C.J. The impact of pre-existing comorbidities and therapeutic interventions on COVID-19. Front. Immunol. 2020;11:1991. doi: 10.3389/fimmu.2020.01991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musuuza J.S., Watson L., Parmasad V., Putman-Buehler N., Christensen L., Safdar N. Prevalence and outcomes of co-infection and superinfection with SARS-CoV-2 and other pathogens: a systematic review and meta-analysis. PLoS One. 2021;16 doi: 10.1371/journal.pone.0251170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaibani P., Viciani E., Bartoletti M., Lewis R.E., Tonetti T., Lombardo D., et al. The lower respiratory tract microbiome of critically ill patients with COVID-19. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-89516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X., Ge Y., Wu T., Zhao K., Chen Y., Wu B., et al. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285 doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deo P.N., Deshmukh R. Oral microbiome: unveiling the fundamentals. J. Oral Maxillofac. Pathol. 2019;23:122–128. doi: 10.4103/jomfp.JOMFP_304_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B., Yao M., Lv L., Ling Z., Li L. The human microbiota in health and disease. Engineering. 2017;3:71–82. [Google Scholar]
- 14.Kumpitsch C., Koskinen K., Schopf V., Moissl-Eichinger C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019;17:87. doi: 10.1186/s12915-019-0703-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shakya M., Lo C.C., Chain P.S.G. Advances and challenges in metatranscriptomic analysis. Front. Genet. 2019;10:904. doi: 10.3389/fgene.2019.00904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rizzo F., Vanoli A., Sahnane N., Cerutti R., Trapani D., Rinaldi A., et al. Small-bowel carcinomas associated with celiac disease: transcriptomic profiling shows predominance of microsatellite instability-immune and mesenchymal subtypes. Virchows Arch. 2020;476:711–723. doi: 10.1007/s00428-019-02675-w. [DOI] [PubMed] [Google Scholar]
- 17.Ferravante C., Memoli D., Palumbo D., Ciaramella P., Di Loria A., D'Agostino Y., et al. HOME-BIO (sHOtgun MEtagenomic analysis of BIOlogical entities): a specific and comprehensive pipeline for metagenomic shotgun sequencing data analysis. BMC Bioinf. 2021;22:106. doi: 10.1186/s12859-021-04004-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng D.L., Granados A.C., Santos Y.A., Servellita V., Goldgof G.M., Meydan C., et al. A diagnostic host response biosignature for COVID-19 from RNA profiling of nasal swabs and blood. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen E.K., Koeppel A.F., Hendley J.O., Turner S.D., Winther B., Sale M.M. Characterization of the nasopharyngeal microbiota in health and during rhinovirus challenge. Microbiome. 2014;2:22. doi: 10.1186/2049-2618-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn M., Dooley J. The microbiome of the nasopharynx. J. Med. Microbiol. 2021;70 doi: 10.1099/jmm.0.001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernandez-Teran A., Mejia-Nepomuceno F., Herrera M.T., Barreto O., Garcia E., Castillejos M., et al. Dysbiosis and structural disruption of the respiratory microbiota in COVID-19 patients with severe and fatal outcomes. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-00851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honarmand Ebrahimi K. SARS-CoV-2 spike glycoprotein-binding proteins expressed by upper respiratory tract bacteria may prevent severe viral infection. FEBS Lett. 2020;594:1651–1660. doi: 10.1002/1873-3468.13845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Stadio A., Costantini C., Renga G., Pariano M., Ricci G., Romani L. The microbiota/host immune system interaction in the nose to protect from COVID-19. Life (Basel) 2020;10 doi: 10.3390/life10120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Maio F., Posteraro B., Ponziani F.R., Cattani P., Gasbarrini A., Sanguinetti M. Nasopharyngeal microbiota profiling of SARS-CoV-2 infected patients. Biol. Proced. Online. 2020;22:18. doi: 10.1186/s12575-020-00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mostafa H.H., Fissel J.A., Fanelli B., Bergman Y., Gniazdowski V., Dadlani M., et al. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect COVID-19 patients. mBio. 2020;11 doi: 10.1128/mBio.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Boeck I., Wittouck S., Wuyts S., Oerlemans E.F.M., van den Broek M.F.L., Vandenheuvel D., et al. Comparing the healthy nose and nasopharynx microbiota reveals continuity as well As niche-specificity. Front. Microbiol. 2017;8:2372. doi: 10.3389/fmicb.2017.02372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soffritti I., D'Accolti M., Fabbri C., Passaro A., Manfredini R., Zuliani G., et al. Oral microbiome dysbiosis is associated with symptoms severity and local immune/inflammatory response in COVID-19 patients: a cross-sectional study. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.687513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schenck L.P., Surette M.G., Bowdish D.M. Composition and immunological significance of the upper respiratory tract microbiota. FEBS Lett. 2016;590:3705–3720. doi: 10.1002/1873-3468.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Govindaraj Vaithinathan A., Vanitha A. WHO global priority pathogens list on antibiotic resistance: an urgent need for action to integrate One Health data. Perspect Public Health. 2018;138:87–88. doi: 10.1177/1757913917743881. [DOI] [PubMed] [Google Scholar]
- 30.Liu J., Liu S., Zhang Z., Lee X., Wu W., Huang Z., et al. Association between the nasopharyngeal microbiome and metabolome in patients with COVID-19. Synth Syst Biotechnol. 2021;6:135–143. doi: 10.1016/j.synbio.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rawlings B.A., Higgins T.S., Han J.K. Bacterial pathogens in the nasopharynx, nasal cavity, and osteomeatal complex during wellness and viral infection. Am J Rhinol Allergy. 2013;27:39–42. doi: 10.2500/ajra.2013.27.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki Y., Togo Y., Wagner H.N., Jr., Hornick R.B., Schwartz A.R., Proctor D.F. Mucociliary function during experimentally induced rhinovirus infection in man. Ann. Otol. Rhinol. Laryngol. 1973;82:203–211. doi: 10.1177/000348947308200219. [DOI] [PubMed] [Google Scholar]
- 33.Mancuso G., Midiri A., Gerace E., Biondo C. Bacterial antibiotic resistance: the most critical pathogens. Pathogens. 2021;10 doi: 10.3390/pathogens10101310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hickey S., Giwa A. StatPearls; 2021. Mechanical Ventilation. [PubMed] [Google Scholar]
- 35.Humbert M.V., Christodoulides M. Atypical, yet not infrequent, infections with Neisseria species. Pathogens. 2019;9 doi: 10.3390/pathogens9010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noble R.C., Cooper R.M., Miller B.R. Pharyngeal colonisation by Neisseria gonorrhoeae and Neisseria meningitidis in black and white patients attending a venereal disease clinic. Br. J. Vener. Dis. 1979;55:14–19. doi: 10.1136/sti.55.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Steenhuijsen Piters W.A., Sanders E.A., Bogaert D. The role of the local microbial ecosystem in respiratory health and disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015;370 doi: 10.1098/rstb.2014.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aebi C. Moraxella catarrhalis - pathogen or commensal? Adv. Exp. Med. Biol. 2011;697:107–116. doi: 10.1007/978-1-4419-7185-2_9. [DOI] [PubMed] [Google Scholar]
- 39.Neumann A., Bjorck L., Frick I.M. Finegoldia magna, an anaerobic gram-positive bacterium of the normal human microbiota, induces inflammation by activating neutrophils. Front. Microbiol. 2020;11:65. doi: 10.3389/fmicb.2020.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antunes L.C., Visca P., Towner K.J. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis. 2014;71:292–301. doi: 10.1111/2049-632X.12125. [DOI] [PubMed] [Google Scholar]
- 41.Gao L., Xu T., Huang G., Jiang S., Gu Y., Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9:488–500. doi: 10.1007/s13238-018-0548-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Boeck I., van den Broek M.F.L., Allonsius C.N., Spacova I., Wittouck S., Martens K., et al. Lactobacilli have a niche in the human nose. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107674. [DOI] [PubMed] [Google Scholar]
- 43.Toivonen L., Hasegawa K., Waris M., Ajami N.J., Petrosino J.F., Camargo C.A., Jr., et al. Early nasal microbiota and acute respiratory infections during the first years of life. Thorax. 2019;74:592–599. doi: 10.1136/thoraxjnl-2018-212629. [DOI] [PubMed] [Google Scholar]
- 44.Gao W., Howden B.P., Stinear T.P. Evolution of virulence in Enterococcus faecium, a hospital-adapted opportunistic pathogen. Curr. Opin. Microbiol. 2018;41:76–82. doi: 10.1016/j.mib.2017.11.030. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi Y., Watanabe N., Kamio N., Kobayashi R., Iinuma T., Imai K. Aspiration of periodontopathic bacteria due to poor oral hygiene potentially contributes to the aggravation of COVID-19. J. Oral Sci. 2020;63:1–3. doi: 10.2334/josnusd.20-0388. [DOI] [PubMed] [Google Scholar]
- 46.Iebba V., Zanotta N., Campisciano G., Zerbato V., Di Bella S., Cason C., et al. Profiling of oral microbiota and cytokines in COVID-19 patients. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.671813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leishman S.J., Do H.L., Ford P.J. Cardiovascular disease and the role of oral bacteria. J. Oral Microbiol. 2010;2 doi: 10.3402/jom.v2i0.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mammen M.J., Scannapieco F.A., Sethi S. Oral-lung microbiome interactions in lung diseases. Periodontol. 2000;83:234–241. doi: 10.1111/prd.12301. 2020. [DOI] [PubMed] [Google Scholar]
- 49.Bao L., Zhang C., Dong J., Zhao L., Li Y., Sun J. Oral microbiome and SARS-CoV-2: beware of lung Co-infection. Front. Microbiol. 2020;11:1840. doi: 10.3389/fmicb.2020.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang Z., Koo H., Chen Q., Zhou X., Liu Y., Simon-Soro A. Potential implications of SARS-CoV-2 oral infection in the host microbiota. J. Oral Microbiol. 2020;13 doi: 10.1080/20002297.2020.1853451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings described here are available from the corresponding authors upon request.