Abstract
Proteins, such as the Ah receptor (AHR), hold potential as sensors to detect ligands in environmental and biological samples, and may also serve as tools to regulate biosynthetic and industrial processes. The AHR is also a prototype system for the PAS superfamily that can sense and mediate adaptation to signals as diverse as light, voltage, oxygen and an array of small molecules. The yeast, S. cerevisiae, has proven to be an important model to study the signal transduction of sensors like the AHR because of its ease of use, numerous available strategies for genetic manipulation, and capacity for heterologous expression. To better understand the utility of sensor proteins as components of yeast detection systems, we characterized a chimeric AHR-LexA system that drives expression from a Lex operator (LexO) driven, beta-galactosidase (β-Gal) reporter. In this report, we demonstrate that improvements in assays sensitivity and pharmacology can arise from the careful optimization of yeast growth phase and the duration of ligand exposure. We also report that the coexpression of heterotypic modifiers from mammalian cells (e.g., the ARA9 and ARA3 proteins), can improve yeast assay performance. We propose that complementing these assay improvements with previously reported yeast mutations described by others will expand the utility of the AHR for biotechnology applications.
Keywords: Ah receptor, Yeast, Assay, Ligands, Reporter, Polycyclic aromatic hydrocarbons
Graphical Abstract
Highlights
-
•
Optimization of a yeast based chimeric assay for AHR ligands.
-
•
Description of assay conditions to enhance collaboration and harmonize the related data between many laboratories.
-
•
Sensitivity increases in the assay by reduction of ligand exposure times, andcoexpression of of AHR associated proteins.
1. Introduction
The heterologous expression of sensor proteins in the yeast Saccharomyces cerevisiae (S. cerevisiae), holds great potential as a system for the detection of a variety of biologically active molecules. This yeast has proven to be an important model to study many nuclear sensor proteins, including members of the PAS family, such as the aryl hydrocarbon receptor (AHR), as well as members of the steroid receptor family. As a eukaryotic microorganism, yeast confer certain advantages over mammalian cell culture or animal models. It has a small genome, it can exist in both a haploid and diploid state, it contains organelles similar to many mammalian systems, and its mechanisms of transcriptional initiation and regulation support many human sensor responses [1]. These characteristics of yeast have been employed to make important contributions to our understanding of nuclear sensor function. For AHR signal transduction, these include the identification of several required chaperones of the AHR, defining the importance of polymorphisms in the AHR, as well as the classification of signal transduction steps that occur in response to ligand activation [2], [3], [4], [5], [6], [7], [8], [9], [10].
The expression of the AHR in yeast is an important example of the potential of sensor systems as bioassays for ligand detection, as well as systems to regulate biosynthetic processes in industrial settings. Because yeast can grow in liquid culture and has a rapid doubling time, yeast can be propogated at low cost under common ambient conditions. These simple growth conditions allow employment of yeast systems in a variety of field and industrial settings with little supporting infrastructure. As a proof of this idea, numerous examples exist in the literature where yeast bioassays have shown initial success evaluating contaminated environmental samples for the presence of AHR ligands [11], [12], [13], [14], [15], [16], [17]. While yeast-based detection systems may never completely replace modern analytical chemistry, they may complement such approaches. For example, bioassays offer great potential when hazard identification is time sensitive or when the high cost of analysis can be reduced through sample prioritization.
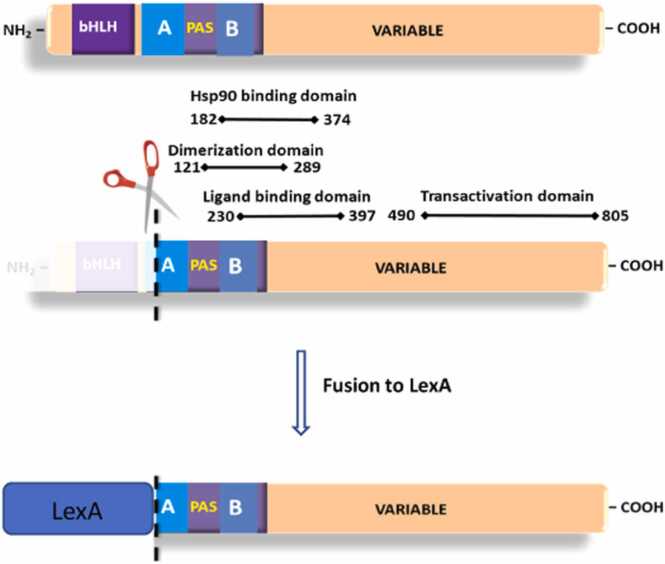
There are currently two main approaches to the yeast AHR bioassay. In the most commonly employed form, yeast is engineered through the coexpression of the pathway’s three major components: The AHR, the AHR nuclear translocator (ARNT) and a reporter gene driven by the cognate enhancer of the AHR-ARNT pair, known as a xenobiotic response element (XRE) [8], [11]. In the alternative approach pioneered in our laboratory, the system is reduced to a chimera of AHR, where its DNA binding and its primary ARNT dimerization domain is replaced by the DNA binding domain/dimerization domain of the bacterial LexA protein [2] (Fig. 1). The advantage of the complete system (i.e., plasmids expressing the AHR, ARNT and an XRE-driven reporter) is its more accurate reflection of mammalian signaling mechanism, in that it incorporates AHR, ARNT and XRE interactions. The advantages of the yeast chimeric AHR system are that it is dependent on only a single plasmid expressing an AHR-LexA chimera and a LexO driven reporter integrated into the genome. It does not require expression of ARNT, nor does it require an XRE-driven reporter. Thus, the chimeric system allows the coexpression of multiple modifiers through common expression plasmids with the available auxotrophic markers. Additionally, the use of a common integrated reporter in the chimeric system offers the potential for additional PAS or steroid receptor based chimeric sensors to be run in parallel. Finally, the replacement of the AHR’s bHLH and PAS-A domains removes a primary PAS dimerization domain and thus reduces the potential for confounding interactions in scenarios where additional sensors might be employed.
Fig. 1.
Domain map of the AHR and LexA-AHR fused protein. Schematic diagram of the murine AHRb1 structure domains and the LexA substitution made on the N- terminal side of the AHR. Specifically, the bHLH domain, which contains the DNA binding domain of AHR, was swapped for the DNA binding domain of LexA.
Given the rising application of the yeast bioassay in recent years, and its potential value for environmental analysis and ligand discovery, we set out to further characterize the chimeric system, with an eye toward documenting its pharmacology, improving its sensitivity, and better understanding its utility. In this effort, we report here a detailed description of the chimeric system, with an emphasis on identifying those variables that influence detection sensitivity and reproducibility.
2. Methods
2.1. Chemicals
Chemicals used in this study were purchased from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA) . The yeast minimal media, Synthetic-Defined (SD) with specific amino acids “dropped-out,” were purchased from TaKara Bio Inc. (Mountain View, CA USA).
2.2. Yeast strains and construction of the AHR expression systems
Competent cells of Escherichia coli strain JM109 (Promega, Madison, WI, USA) were used in all bacterial plasmid transformations in this study. Recombinant colonies were selected on Luria-Bertani (LB) agar plates supplemented with 100 μg/mL Ampicillin (Amp) and incubated for 18 h. A single colony from the transformant plate was used to inoculate 10 mL of LB broth supplemented with Amp to a final concentration of 100 μg/mL and grown overnight at 37 °C with constant shaking at 220 rpm. For plasmid propagation, 1 mL of the overnight culture was used to inoculate 500 mL of LB broth supplemented with 100 μg/mL Amp and incubated overnight at 37 °C; 220 rpm. Plasmids were prepared for transformations using ZymoPURE™ II Maxiprep Kit (Zymo Research Corp., Irvine, CA, USA).
The yeast, S. cerevisiae strain L40 (MATa HIS3∆200, trp1–901, leu2–3, 112, ade2, LYS2::(LexO)4 -HIS3 URA3::(LexO)8 -lacZ, GAL4), was used in all experiments [18]. Before transformations with any plasmid, the strain was maintained in Yeast-Peptone-Dextrose media (YPD). This strain facilitates the use of numerous auxotrophic markers and its genome contains four integrated lexA operator (LexO, also called LexAop) elements controlling the expression of HIS3 (integrated at the LYS2 locus) and eight integrated LexO elements controlling the expression of β-Gal (i.e., the LacZ gene) integrated at the URA3 locus. The plasmid, pBTM116 (pL535), is a 2-μm TRP-marked plasmid containing the 202 amino acid full length bacterial LexA protein (GenBank Accession number KX357893.1) under the control of the ADH1 promoter, followed by multiple cloning sites allowing for expression of LexA fusion proteins [18]. This plasmid was used to make all LexA-AHR chimera expressing plasmids and is available through a public repository (Plasmid #111232, Addgene, Watertown, MA). The AHR used in these constructions was obtained from the murine AHR-B1 receptor form (GenBank Accession Number M94623), with the amino acids 1–166 deleted and replaced with the LexA DNA binding sequence from the vector pBTM116. The ARA9 and ARA3 cDNAs have been described previously (GenBank Accession Numbers U78521.1 and DQ443529.1, respectively [7], [8], [19].
2.3. Yeast transformation
The yeast L40 competent cells were prepared using the Frozen-EZ Yeast Transformation II Kit™ (Zymo Research, Irvine, CA, USA). Competent cells were stored as 50 µL aliquots in sterile 1.7 mL microcentrifuge tubes and kept at − 80 °C until needed. Transformants were selected on agar plates of “drop-out media,” lacking tryptophan (SDT) (Takara, Mountain View, CA, USA). To prevent bacterial growth, media was supplemented with Amp at a final concentration of 100 μg/mL. Plates were incubated at 30 °C until visible colonies were observed (~2–4 days). After growth, a single colony was transferred onto a new SDT agar plate and incubated at 30 °C for two days. At this point, plates were kept at room temperature for up to a week or stored at 4 °C for a month.
2.4. Yeast β-gal assay
All compounds used in this study were dissolved in dimethyl sulfoxide (DMSO) at stock concentrations of 10 mM, except for FICZ and ICZ that were dissolved at a maximum concentration of 1 mM. Direct dilutions were performed in a Costar 3912, opaque white, flat bottom 96-well plates, by distributing various sub microliter volumes of each compound using the digital dispenser Tecan D300e (San Francisco, CA USA). Additional DMSO, was then distributed to each well to normalize the total volume to 5 µL per well. This was followed by addition of 95 µL of the yeast culture into each well for a total volume of 100 µL per well. This was incubated at 30 °C; 220 rpm for the designated times, and then 100 µL of Gal-Screen® solution was added to each well (Applied Biosystems Inc., Foster City, CA). The β-Gal activity was measured at 28 °C for using a CLARIOstar® microplate reader (BMG LabTech Cary, NC USA).
2.5. Influence of growth phase on the assay
Single colonies of transformed L40 strain were transferred from SDT plates into 10 mL of SDT broth. The cultures were grown overnight at 30 °C with constant shaking at 220 rpm and then diluted to an optical density at 600 nm (OD600) of 0.08 + /- 0.02. One hundred microliters of diluted cultures were transferred into individual wells on a clear, flat bottom 96 well plate. Optical density at 600 nm (OD600) was determined every 2 h up to 36 h. To determine the influence of growth phase on the β-Gal response, aliquots of yeast from 16 h (early log phase), 20 h (mid log phase), and 24 h (late log phase) were diluted to 0.5 OD and exposed to 10 μM beta-napthoflavone (BNF), in 100 μl total volume in 96 well plates for two hours at 30 °C; 220 rpm. All experiments were performed in triplicate.
2.6. Linearity experiment
The L40 yeast, transformed with the LexA-AHR fusion (Fig. 1, PL703), were grown in SDT and adjusted to an OD600 of 0.5. The culture was then split in half. One-half was transferred in 95 µL aliquots into 96 well plates containing 5 µL of BNF dilutions in DMSO. The other half was subjected to centrifugation at 2000 x g, the SDA removed, and the pellet was resuspended with the same volume of YPD. These resuspended cells were transferred to 96 well plate containing dilutions of BNF (as with the SDA cultures). Plates were incubated at 30 °C; 220 rpm for 15 min, 30 min, 1 h, 2 h, 4 h, and 16 h.
2.7. Pharmacology in mammalian cells
A modified CALUX assay emplying recombinant mouse hepatocellular carcinoma cells H1L6.1c3 [20], were cultured in high glucose DMEM medium supplemented with 10% (v/v) fetal bovine serum, 1% non-essential amino acids, 100 U/mL penicillin-streptomycin, and 1% glutamine at 37 °C with 5% CO2. Media was also supplemented with G418 (Sigma St. Louis, MO, USA) to achieve a final concentration of 0.6 mg/mL. The H1L6.1c3 cells were plated by adding 100 μL into 96-well clear bottom white wall plates to achieve a seeding concentration of 10,000 cells per well and allowed to grow overnight. Cells were treated with various compounds at logarithmic growth phase. After 4 h of treatment incubated at 37 °C with 5% CO2, cells are subjected to the Promega Luciferase Assay System (Promega, Madison, WI). Media was carefully removed from the individual wells, and 20 μL of 1x passive lysis buffer was added to each well. The plate was then placed on a plate shaker in the dark for 15 min. The ligand-induced signal was measured using the CLARIOstar® microplate reader (BMG LabTech Cary, NC USA) after the injection of 90 μL of luciferase reagent into each well. Values were expressed in relative luciferase units and assays were performed in triplicate.
2.8. Modifier experiments
To examine the influence of known interacting proteins on the dose response of the AHR chimera system, previously described cDNAs of both ARA3 and ARA9 (see accession numbers above) were employed, using the dose response conditions described above and in figure legends.
2.9. Statistics
Statistical analyses and logarithmic transformation and nonlinear regression analysis for the generation of the dose-response curve and EC50 calculations, as well as the t-test and 2-way ANOVA, Multiple comparison analysis and the calculations for the coefficient of variance were performed using PRISM program (GraphPad Software, Inc., San Diego, CA) and Microsoft Excel (Office 2016, USA).
3. Results and discussion
3.1. Influence of yeast growth phase
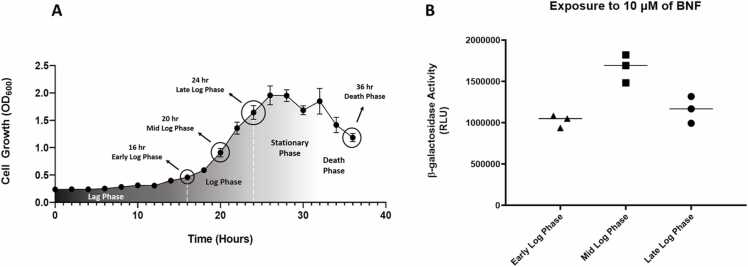
As a first step in the optimization of the yeast chimeric AHR system, we examined how the ligand response was influenced by growth phase in SD media + Amp (SDA). Using the L40 yeast transformed with the LexA-AHR chimera (L40-PL703), we observed a normal growth curve, with a lag phase between 0 and 13 h of culture seeding, a log growth phase between 13 and 25 h, and a stationary phase beginning at about 25 h (Fig. 2). The approximate doubling time observed was about 5 h, slightly slower than the 1.5 h doubling time reported in previous studies using wildtype S. cerevisiae in YPD [21].
Fig. 2.
Growth Curve of the yeast L40AHRNΔ166 SD-TRP media. A: Growth curve experiment in SD-Trp media with the yeast L40 strain harboring the plasmid pBTMAHRN∆166-B1. Cultures were grown for up to 32 h (30 °C; 220 rpm) taking OD600 measurements every two hours. All data points are average of a replica of at least 3 experiments. B: Cultures were grown for 16 h (Early Log Phase), 20 h (Mid Log Phase), and 24 h (Late Log Phase). When the cultures reached their set time they were exposed to BNF for two hours at 30 °C; 220 rpm. β-Gal activity was measured to determine AHR activation. Data points reflect 3 replicate experiments with error bars representing standard deviation.
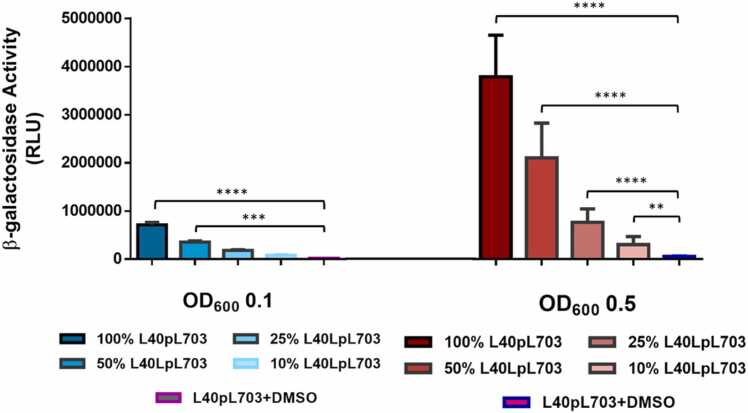
To examine the influence of growth phase and cell density of the system on the response to ligand, we performed two experiments. First, we examined the influence of growth phase on the level of reporter activation in response to 10 µm BNF after two hours. While robust responses were observed at the early, mid and late growth phases (16, 20 and 24 h, respectively), an approximately 30% greater signal was observed at mid log phase. Thus, this condition was chosen for all remaining experiments because it also provided significant flexibility with respect to timing of experimental steps (Fig. 2). To characterize the influence of cell density on the sensitivity and linearity assay, we performed an exposure-response study using yeast at various dilutions of a mid-log phase culture. Three replicate overnight cultures, grown to either 0.1 0D600 per mL, or 0.5 OD600 per mL were each diluted by multiple 2-fold serial dilutions in SD-Trp media, and the sample was assayed for β-Gal activity (Fig. 3). The, 0.5 OD was chosen as a standard condition for all following experiments as it was easy to generate and provided a greater signal intensity.
Fig. 3.
Linearity of the response after a 1:2 cell dilution. An overnight culture of the L40Δ166 was divided and diluted to an OD600 of 0.1 and 0.5. Each cell dilution was serially diluted even further to 1:2 dilutions. Each sample was exposed to 10 µM BNF for 2 h. All data points are average of a replica of 3 experiments with error bars representing standard deviation. The β-Gal units were determined on a ClarioStar plate reader.
3.2. Response optimization
Based upon the above results, we performed a time course response study using mid log cultures in SDA. In parallel, we compared the same exposure-response experiment in a richer media, YPD. We considered both media of potential value in future studies. The short time in SDA would ensure maintenance of the LexA-AHR fusion plasmid (PL703, Fig. 7) over the course of the assay. In contrast, the YPD could prove of value when using the system to characterize ligands in crude environmental samples, some of which might contain nutrients, such as Trp. Finally, although maximizing assay sensitivity was a goal, we also recognized the importance of assay time-length in future experiments. That is, we looked for the development of an assay time that would; (1) provide a high level of sensitivity to ligand, (2) produce conditions consistent with known modes of AHR signaling as defined by the 4-parameter logistic model [22], and (3) allow conception of an experiment and its completion within one working day.
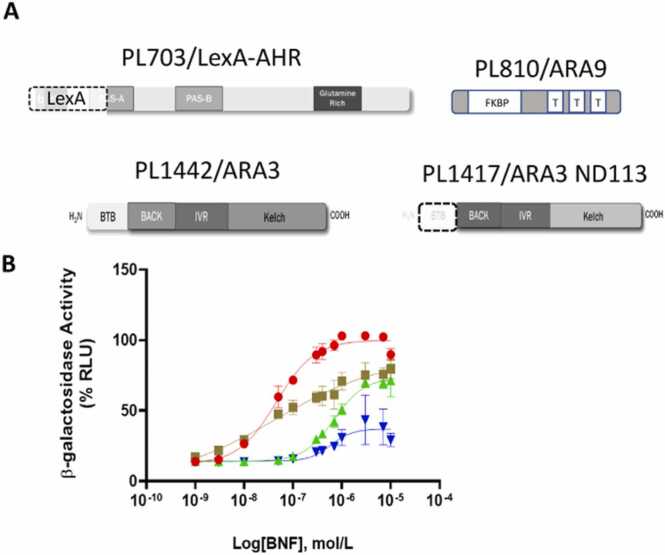
Fig. 7.
Influence of modifiers on chimeric system in yeast. The LexA-AHR chimeric system’s response to BNF (PL703, blue triangles), compared to its expression with the coexpression of the full length ARA9 modifier (PL703 + PL810, brown squares), the full length ARA3 modifier (PL703 + PL1142, green triangles) and the ARA3 modifier with its BTB domain deleted (PL703 + PL1417, red circles).
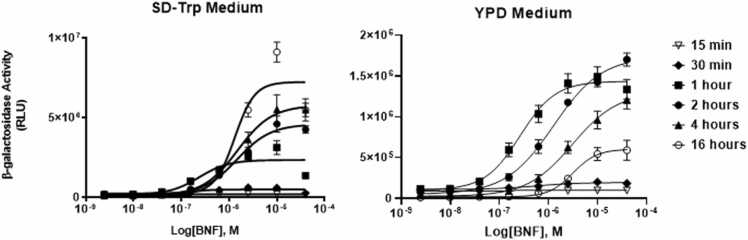
Analysis of dose-response curves under these various times and conditions revealed that both media and time of ligand exposure have a marked influence on the system’s response (Fig. 4). Visual inspection of the maximal response, slope, shape and EC50 revealed a number of interesting results. For SDA media, we did not observe a dose-response at either 15 or 30 min, but observed curves at all greater time durations. This result is in keeping with the idea that shorter times may not allow sufficient receptor movement to the nucleus, interaction with genomic elements and transcription and translation of the target reporter gene, β-Gal. In essentially all dose-response studies, we observed a maximal response at the 40 μM BNF (40 µM), consistent with ligand maximal receptor occupancy, but also possibly limited by ligand insolubility and or yeast toxicity at this dose and higher. Finally, there was a leftward shift in the slopes of the dose-response curves generated between one hour and two hours, indicating the assay is maximally sensitive under shorter incubation times. For yeast grown in YPD, the maximal response was about one fourth of the SDA media. Also of interest was the observation that shorter time points generated the most left-shifted dose-response curves and potential for higher sensitivity compared to longer incubation times. That is, in YPD, the greatest maxima and the most left shifted curve is observed at a time of one hour of exposure followed by 2 h, then 4 h and finally 16 h is the least sensitive (Fig. 4).
Fig. 4.
Time of exposure. Cultures of L40pBTMAHRN∆166-B1 were exposed to direct dilution (1:4) of BNF for 15 min, 30 min, 1 h, 2 h, 4 h and 16 h at 30 °C; 220 rpm. β-Gal activity was measured to determine AHR activation. β-Gal units are reported as relative light units (RLU). All data points are average of a replica of at least 3 experiments with error bars representing standard deviation.
3.3. Pharmacological comparison with a mammalian system
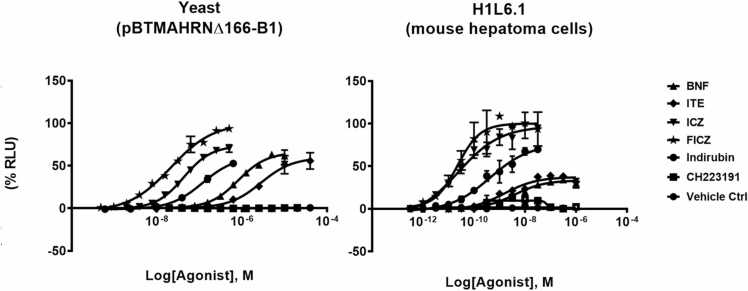
To compare the pharmacology of the yeast system to the mammalian system, we compared the structure activity of common nonhalogenated ligands to that of a popular mammalian reporter system H1L6.1c3 [20]. This mouse hepatoma cell line endogenously expresses high levels of AHR and ARNT and has an integrated set of XREs driving expression of a luciferase transgene reporter. For comparison, we used a variety of ligands (Fig. 5, Fig. 6) to study AHR activation and its pharmacology in both systems. For a pharmacological comparison between the two systems, yeast were exposed to varying concentrations of the same ligands and the AHR activation was determined by measuring β-Gal activity. The results of these experiments demonstrate that for both systems, essentially all agonists that we tested showed activity as AHR activators in both systems and the known antagonists, CH223191, did not activate the reporter in either system. In addition, the two systems displayed similar rank order potency according to EC50 based on the four-parameter model (Table 1) [23]. Interestingly, when comparing potency, the mammalian system displayed increased sensitivity of greater than an order of magnitude over the yeast system (Table 1).
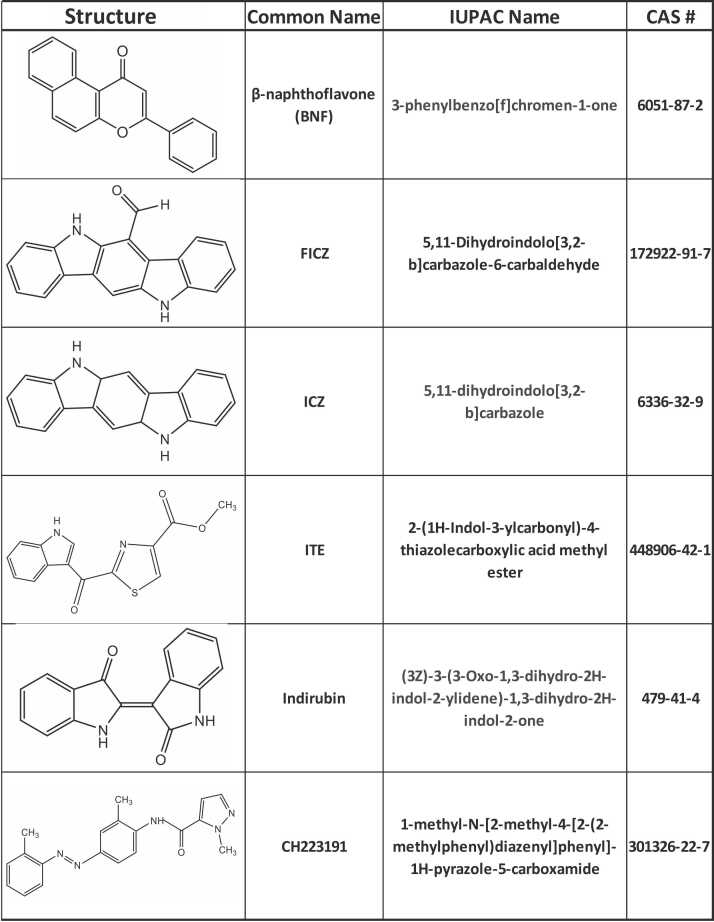
Fig. 5.
Structures of chemicals used in this study.
Fig. 6.
Pharmacology of the yeast system. Ligand-induced activity measured using the yeast system and a mouse hepatoma cell line. Dose-response curves for L40pBTMAHRN∆166-B1 yeast reporter system and for the H1L6.1c mouse hepatoma cells. Cultures were exposed to ligands for two hours at 30 °C; 220 rpm. β-Gal or luciferase activities were measured to determine AHR activation. Units were converted to percent using the maximal and minimal response from all the ligands used in the experiment. All data points are average of a replica of 3 experiments with error bars representing standard deviation.
Table 1.
Chemical potency in the yeast and mammalian expression system. Comparison of the EC50 values for the AHR agonists used in this study. (n.d. = not determined).
|
Yeast (Saccharomyces cerevisiae) |
Mammalian cell line (H1L6.1c3) |
|||
|---|---|---|---|---|
| Chemical | EC50(M) | Hill Slope | EC50(M) | Hill Slope |
| BNF | 8.22 × 10−7 | 1.29 | 3.93 × 10−9 | 0.74 |
| FICZ | 1.90 × 10−8 | 1.00 | 8.21 × 10−12 | 0.30 |
| ICZ | 4.30 × 10−8 | 1.33 | 1.23 × 10−11 | 0.29 |
| ITE | 2.78 × 10−6 | 1.16 | 1.52 × 10−9 | 0.68 |
| Indirubin | 1.19 × 10−7 | 1.14 | 2.88 × 10−10 | 0.66 |
| CH223919 | n.d. | n.d. | n.d. | n.d. |
3.4. Improving assay sensitivity
In an effort to improve the sensitivity of the yeast system, we investigated the potential of known AHR modifiers to influence the dose response curve. Specifically, we performed experiments to investigate the possibility that specific coexpressed cDNAs could left-shift the dose response curve. In our initial attempts towards this goal, we investigated two known mammalian interacting proteins we refer to as ARA9 (Ah receptor associated 9, also known as XAP2 or AIP), and ARA3 (Ah receptor associated 3, also known as NS1BP, for their capacity to left-shift the dose response curve and/or increase its maxima [8], [19], [23], [24]). Our focus on these candidates was based primarily on the fact that they were previously identified and cloned in our laboratory, so they were readily available, and they do not have yeast orthologues. For ARA9, we employed the full-length open reading frame (PL810/ARA3, Fig. 7). For ARA3, we employed both the full-length open reading frame (PL1442/ARA3) and an N-terminal deletion fragment (PL1417/ARA3 ND113, Fig. 7). The latter ARA3 deletion fragment corresponds to the original cDNA identified in our two-hybrid screen and was included as an initial test of the idea that fragments of modifiers might display differential modifier activity as compared to the full length modifiers [19]. This examination of modifier influence on the dose response curve revealed that ARA9, ARA3 and an ARA3 fragment, all increased the maximal response more than two-fold. Additionally, ARA3 and its fragment both significantly left shifted the dose response curve, with the deletion mutant having the greatest effect (approximately two orders of magnitude left shift as determined by reduction of EC50, Fig. 7).
3.5. The path forward
In our development of yeast as an environmental sensor, we began with the examination of the AHR as a model system, but experiments were designed with the future development of multiple parallel systems in mind. One future goal is to develop single tube systems that employ multiple unique sensor proteins in parallel. We propose that a spectrum of sensors can be employed on a single sample, with each sensor derived from the LexA fusion of distinct PAS family members, or steroid receptor family members. We foresee a few potential strategies to generate a sensor system that can detect a broad range of stimuli in parallel. In a “strain mixture approach,” we envision mixing multiple yeast strains, each designed to detect a specific stimulus through the reporting of a single LexO driven response. In a second, “coexpression approach,” a single yeast strain can be engineered to harbor multiple sensors within the same strain, again, all designed to drive the same LexO driven reporter. Samples that yield positive signals through either of these strategies can then be followed up in secondary screens, perhaps single sensor strains, or classical analytical approaches, that can more precisely pinpoint that class of activator being identified.
With the long-term strategy of multiple parallel sensors in mind, we began this investigation into the conditions that influence biosensor optimization for the AHR system. One early decision in this assay’s development was the choice of the LexA fusion described in Fig. 1, Fig. 7. This particular LexA fusion protein was chosen for two reasons. First, the fusion has a long history in our laboratory, responding to ligands in an ARNT independent manner, showing utility as a ligand responsive two-hybrid bait in the identification of the ARA3 and ARA9 proteins, and as a model sensor useful in understanding how the yeast genome influences AHR signal transduction [7], [19]. Second, while the replacement of the bHLH and disruption of the PAS-A domain with LexA DNA binding domain was originally chosen based on a convenient restriction-cloning site, it also removes those domains that are not required in a ligand responsive bioassay. That is, this fragment removes the primary drivers of AHR ARNT dimerization and the DNA contact region for XRE elements, neither of which is required in a LexO reporter system. Importantly, this sensor retains the AHR signaling domains within the PAS-B and C-terminal half of the protein. The retained domains include those required for ligand binding, chaperone binding, ligand dependent transformation, as well as transcriptional activation of target promoters (Fig. 1) [25], [26]. Moreover, given our interest in developing a coexpression system within a single yeast strain, reducing the burden of dimerization sequences found in the bHLH and PAS-A regions should reduce background interactions across sensor proteins of the PAS family and perhaps crossing over to steroid receptors.
The choice of the yeast, S. cerevisiae, was based upon its proven value as a model system for the study of AHR signal transduction and steroid receptor signaling. Early experiments with yeast mutants allowed proof for the essentiality of Hsp90 in receptor signaling and yeast two-hybrid strains led to the identification of additional chaperones. Later, a high throughput genetic screen provided a list of yeast genes that modified signaling and allowed the classification of yeast modifiers based upon their influence on four distinct AHR signaling steps: receptor folding, receptor translocation to the nucleus, receptor expression, and receptor activation of transcriptional targets [2]. Perhaps one of the most important lessons from these collective studies was that yeast provides a robust system that recapitulates many of the fundamental early steps in AHR signal transduction in chordates.
In addition to the mechanistic insights offered by the yeast model, proposals for more practical application of this system also arose. That is, the AHR system in yeast could serve as a tool to detect ligands in environmental and biological samples. This idea initially arose from the observation that for many halogenated environmental ligands, such as the “dioxin like compounds” (DLCs), environmental concern is related to their affinity for and capacity to activate the AHR [27]. Interestingly, early investigations into yeast as a bioassay for DLCs revealed that this system is relatively insensitive to prototype DLCs, such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) [8], [13], [28]. Early speculation was that this insensitivity was due to DLC insolubility in the aqueous systems optimal for yeast growth, due to their inability of DLCs to traverse the yeast cell wall, or their propensity to be pumped out by active transporters [29]. Recently, these issues have been partially addressed with the identification of several mutations within the yeast genome, e.g., CWP and PDR, which increase their sensitivity towards the prototype DLC, TCDD. Presumably, these intrinsic modifiers act by increasing the intracellular concentrations of this ligand.
In our laboratory, we have turned attention away from the response of yeast to DLCs and have begun to focus on enhancing the response to nonhalogenated ligands, such as the indigoid, carbazole and flavonoid classes of ligands. Our reasoning is two-fold. First, we propose that nonhalogenated classes of ligands have greater utility in agricultural, industrial and academic settings given they are not subject to many of the regulatory, disposal and environmental concerns related to DLCs. Second, we see utility in the use of the yeast system as a rapid and sensitive means to identify physiological ligands of the AHR from human and other biological tissues. Given our assumption that such physiological ligands are unlikely to be of the halogenated class of molecules, we have initiated efforts to optimize the sensitivity of the yeast system for nonhalogenated ligands. Put another way, we are interested in developing a system that responds to ligands generated by plant and animal systems endogenously.
One of the shortcomings of our early work, and the reports of others in this field, is that there has been little description of the conditions that lead to optimization of each assay system. Therefore, our experiments described here were designed to evaluate the influence of culture density, ligand exposure time, and presence of modifiers, on response parameters. Our results suggest considerable flexibility in the assay in that culture density had only a modest influence on the dose-response. What was surprising, and to our knowledge, not reported previously, is that the yeast assay can be accomplished with incredibly short incubation times that can be more easily applied in the field and that allow for more rapid completion of experiments. Our time course studies suggest that incubation of ligand for periods as short as one or two hours leads to optimal sensitivity as defined by the EC50 for BNF. This is important for two reasons. First, the majority of yeast assay results published in the literature use much longer incubation time, often 24–48 h. Thus, those systems may also be improved by this simple modification. Second, a one- or two-hour assay allows multiple experiments to be completed, from conception to analysis, in one working day. Such rapid experimentation could expedite the discovery of ligands as well as additional heterologous modifiers through high content screens of heterologously expressed cDNAs and their fragments.
To examine the idea that signaling modifiers could be coexpressed and improve sensitivity of the system, we performed an initial test with two interacting proteins that had been cloned over the past decades. Because previous studies mapped the ARA9 and ARA3 interactions to the PAS-B region (co-mapping with the known ligand and Hsp90 binding site) we predicted that increase maximal signaling would arise through an increase in receptor number resulting from improved folding and stability. To our surprise, while the ARA9 and ARA3 modifiers employed here both improved maximal response, ARA3 also left-shifted the dose response curve. Basic receptor theory suggests that both modifiers influence signaling through an increase in receptor number (thus increasing maxima), but that ARA3 may also act by increasing affinity for binding ligand (i.e., a change in KD) or perhaps by influencing receptor transformation/activation [30]. The mechanism by which modifiers left-shift the dose response curve is currently under investigation and its elucidation will require the development of novel tools and reagents over the coming years. It is also important to note that while the modifier approach holds promise to improve assay performance, our studies suggest that synthetic fragments of modifiers may be engineered to further improve assay performance. In this regard, the most potent modifier described here is an N-terminal deletion of ARA3, corresponding to the original cDNA fragment identified as an interacting protein using the two-hybrid assay almost twenty years ago [19]. This result leads us to suggest that interrogation of additional modifier mutants and fragments may further assay performance even further. Such experiments are ongoing and may also shed light on mechanism of action.
In summary, we have further characterized the yeast bioassay for ligands of the AHR and provided pathways for its improvement and optimal performance dependent on task. These results provide guidance for shorter assay times, demonstrate the pharmacological relevance of the assay, and also document the potential utility of receptor modifiers for improved assay performance. We propose the use of this LexA-chimeric system, along with its optimal assay conditions and coexpression of heterologous modifiers, may also work with approaches developed by other labs that employ deletions of specific yeast genes that allow increased concentration of xenobiotics within cells. If these parallel approaches are in fact complementary, it may be that yeast models can be improved even further, perhaps exceeding what is commonly achieved in mammalian cell models or even analytical methods such as GC/LC-mass spec. Furthermore, if we can better understand the yeast system using the AHR as a model system, we may ultimately be able to engineer bioassays that can detect a broad spectrum of biologically active molecules at low cost and with rapid turnaround times.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Institutes of Health Grants R35ES028377, T32ES007015, T32GM008692, T32CA009135, R25ES020720, and P30CA014520. The UW-Madison Science & Medicine Graduate Research Scholars Program, and the National Science Foundation Graduate Research Fellowship under the grant no. DGE-1256259. thanks also to our team of undergraduates, Camilo Mesa, Brandon Bocanegra, Carrie (Stroetz) Marcis, and A. Hoover.
Conflict of Interest
The authors have no conflicts of interest related to this manuscript.
References
- 1.Barr M.M. Super models. Physiol. Genomics. 2003;13(1):15–24. doi: 10.1152/physiolgenomics.00075.2002. [DOI] [PubMed] [Google Scholar]
- 2.Yao G., et al. Interaction networks in yeast define and enumerate the signaling steps of the vertebrate aryl hydrocarbon receptor. PLOS Biol. 2004;2(3) doi: 10.1371/journal.pbio.0020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox M.B., Miller C.A. The p23 co-chaperone facilitates dioxin receptor signaling in a yeast model system. Toxicol. Lett. 2002;129(1):13–21. doi: 10.1016/s0378-4274(01)00465-9. [DOI] [PubMed] [Google Scholar]
- 4.Cox M.B., Miller C.A., 3rd Cooperation of heat shock protein 90 and p23 in aryl hydrocarbon receptor signaling. Cell Stress Chaperones. 2004;9(1):4–20. doi: 10.1379/460.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Q., Whitlock J.P., Jr. A novel cytoplasmic protein that interacts with the ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-Tetrachlorodibenzo-<em>p</em>-dioxin *. J. Biolo. Chem. 1997;272(14):8878–8884. [PubMed] [Google Scholar]
- 6.Maier A., et al. Aromatic hydrocarbon receptor polymorphism: development of new methods to correlate genotype with phenotype. Environ. Health Perspect. 1998;106(7):421. doi: 10.1289/ehp.106-1533118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carver L.A., Bradfield C.A. Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. J. Biol. Chem. 1997;272(17):11452–11456. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- 8.Carver L.A., Jackiw V., Bradfield C.A. The 90-kDa heat shock protein is essential for Ah receptor signaling in a yeast expression system. J. Biol. Chem. 1994;269(48):30109–30112. [PubMed] [Google Scholar]
- 9.Whitelaw M.L., et al. Heat shock protein hsp90 regulates dioxin receptor function in vivo. Proc. Natl. Acad. Sci. 1995;92(10):4437–4441. doi: 10.1073/pnas.92.10.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hogenesch J.B., et al. Characterization of a Subset of the Basic-Helix-Loop-Helix-PAS Superfamily That Interacts with Components of the Dioxin Signaling Pathway. J. Biol. Chem. 1997;272(13):8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 11.Miller C.A. A human aryl hydrocarbon receptor signaling pathway constructed in yeast displays additive responses to ligand mixtures. Toxicol. Appl. Pharmacol. 1999;160(3):297–303. doi: 10.1006/taap.1999.8769. [DOI] [PubMed] [Google Scholar]
- 12.Leskinen P., et al. Detecting AhR ligands in sediments using bioluminescent reporter yeast. Biosens. Bioelectr. 2008;23(12):1850–1855. doi: 10.1016/j.bios.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 13.Kawanishi M., et al. Construction of reporter yeasts for mouse aryl hydrocarbon receptor ligand activity. Mutat. Res. 2003;540:99. doi: 10.1016/s1383-5718(03)00174-8. [DOI] [PubMed] [Google Scholar]
- 14.Noguerol T.-N., et al. Evaluating the interactions of vertebrate receptors with persistent pollutants and antifouling pesticides using recombinant yeast assays. Anal. Bioanal. Chem. 2006;385(6):1012–1019. doi: 10.1007/s00216-006-0476-4. [DOI] [PubMed] [Google Scholar]
- 15.Kamata R., et al. Mono-hydroxylated polychlorinated biphenyls are potent aryl hydrocarbon receptor ligands in recombinant yeast cells. Toxicol. in vitro: Int. J. Publ. Assoc. BIBRA. 2009;23(4):736–743. doi: 10.1016/j.tiv.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Rowlands J.C., Gustafsson J.-Å. Human dioxin receptor chimera transactivation in a yeast model system and studies on receptor agonists and antagonists. Pharmacol. Toxicol. 1995;76(5):328–333. doi: 10.1111/j.1600-0773.1995.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 17.Adachi J., et al. Indirubin and Indigo Are Potent Aryl Hydrocarbon Receptor Ligands Present in Human Urine *. J. Biol. Chem. 2001;276(34):31475–31478. doi: 10.1074/jbc.C100238200. [DOI] [PubMed] [Google Scholar]
- 18.Fields S. The two-hybrid system to detect protein-protein interactions. Methods. 1993;5(2):116–124. [Google Scholar]
- 19.Dunham E.E., et al. The aryl hydrocarbon receptor signaling pathway is modified through interactions with a kelch protein. Mol. Pharmacol. 2006;70(1):8–15. doi: 10.1124/mol.106.024380. [DOI] [PubMed] [Google Scholar]
- 20.Denison M.S., et al. Recombinant cell bioassay systems for the detection and relative quantitation of halogenated dioxins and related chemicals. Talanta. 2004;63(5):1123–1133. doi: 10.1016/j.talanta.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 21.Sherman F. In: Methods in Enzymology. Guthrie C., Fink G.R., editors. Academic Press; 2002. Getting started with yeast; pp. 3–41. [DOI] [PubMed] [Google Scholar]
- 22.Vølund A. Application of the four-parameter logistic model to bioassay: comparison with slope ratio and parallel line models. Biometrics. 1978;34(3):357–365. [PubMed] [Google Scholar]
- 23.Gadagkar S.R., Call G.B. Computational tools for fitting the Hill equation to dose–response curves. J. Pharmacol. Toxicol. Methods. 2015;71:68–76. doi: 10.1016/j.vascn.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 24.Tsai P.-L., et al. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza A virus RNA splicing. PLoS Pathogens. 2013;9(6) doi: 10.1371/journal.ppat.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vazquez-Rivera E., et al. The aryl hydrocarbon receptor as a model PAS sensor. Toxicol. Rep. 2021;9:1–11. doi: 10.1016/j.toxrep.2021.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dolwick K.M., Swanson H.I., Bradfield C.A. In vitro analysis of Ah receptor domains involved in ligand-activated DNA recognition. Proc. Natl. Acad. Sci. 1993;90(18):8566–8570. doi: 10.1073/pnas.90.18.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Safe S. Polychlorinated biphenyls (PCBs), dibenzo-p-dioxins (PCDDs), dibenzofurans (PCDFs), and related compounds: environmental and mechanistic considerations which support the development of toxic equivalency factors (TEFs) Crit. Rev. Toxicol. 1990;21(1):51–88. doi: 10.3109/10408449009089873. [DOI] [PubMed] [Google Scholar]
- 28.Miller C.A., Martinat M.A., Hyman L.E. Assessment of aryl hydrocarbon receptor complex interactions using pBEVY plasmids: Expression vectors with bi-directional promoters for use in Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26:3577. doi: 10.1093/nar/26.15.3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawanishi M., et al. Improvement of reporter gene assay for highly sensitive dioxin detection using protoplastic yeast with inactivation of CWP and PDR genes. Environ. Sci. Pollut. Res. 2020;27(9):9227–9235. doi: 10.1007/s11356-019-07484-x. [DOI] [PubMed] [Google Scholar]
- 30.Chow C.C., et al. Inferring mechanisms from dose-response curves. Methods Enzymol. 2011;487:465–483. doi: 10.1016/B978-0-12-381270-4.00016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]