Abstract
The RidA subfamily of the Rid (YjgF/YER057c/UK114) superfamily of proteins is broadly distributed and found in all domains of life. RidA proteins are enamine/imine deaminases. In the organisms that have been investigated, lack of RidA results in accumulation of the reactive enamine species 2-aminoacrylate (2AA) and/or its derivative imine 2-iminopropanoate (2IP). The accumulated enamine/imine species can damage specific pyridoxal phosphate (PLP)-dependent target enzymes. The metabolic imbalance resulting from the damaged enzymes is organism specific and based on metabolic network configuration. Saccharomyces cerevisiae encodes two RidA homologs, one localized to the cytosol and one to the mitochondria. The mitochondrial RidA homolog, Mmf1p, prevents enamine/imine stress and is important for normal growth and maintenance of mitochondrial DNA. Here we show that Mmf1p is necessary for optimal heme biosynthesis. Biochemical and/or genetic data herein support a model in which accumulation of 2AA and or 2IP, in the absence of Mmf1p, inactivates Hem1p, a mitochondrially located PLP-dependent enzyme required for heme biosynthesis.
Keywords: RidA, 2-aminoacrylate, heme, Hem1, Mmf1
INTRODUCTION
The Rid protein family (YjgF/YER057c/UK114) has been split into nine subfamilies in the NCBI conserved domain database (cd00448) (Marchler-Bauer et al., 2013; Niehaus et al., 2015). Members of the RidA subfamily have enamine/imine deaminase activity and are found in all domains of life (Digiovanni et al., 2020; ElRamlawy et al., 2016; D. C. Ernst & Downs, 2018; J. Irons, Hodge-Hanson, & Downs, 2018; J. Irons, Sacher, Szymanski, & Downs, 2019; Lambrecht, Schmitz, & Downs, 2013; Martínez-Chavarría et al., 2020; Niehaus et al., 2015). 2-aminoacrylate (2AA) is a reactive enamine that is generated by pyridoxal 5’-phosphate (PLP)-dependent enzymes as an obligatory intermediate in some reactions, specifically the breakdown of serine. Like other enamines, 2AA rapidly tautomerizes to its imine, 2-iminopropanoate (2IP). The imine is subsequently hydrolyzed to form a stable keto acid, which in the case of 2IP is pyruvate. 2AA can react with and irreversibly damage enzymes by forming an adduct with the PLP-enzyme complex (Borchert, Ernst, & Downs, 2019; Esaki & Walsh, 1986; Walsh, 1982). In strains lacking RidA, the accumulation of 2AA/2IP generates endogenous metabolic stress that has diverse phenotypic manifestations characteristic of the organism involved (J. L. Irons, Hodge-Hanson, & Downs, 2020). The inability to easily distinguish between enamine/imine molecules, or control their tautomerization, makes it difficult to definitively assign a role for one over the other in vivo. However, in vitro mechanistic studies support the assumption that the 2AA enamine is the agent of direct damage in ridA mutants (Borchert et al., 2019; Lambrecht et al., 2013).
Saccharomyces cerevisiae encodes two RidA homologs, one localized to the cytoplasm (Hmf1p) and one to the mitochondrion (Mmf1p). There are no phenotypes reported for the lack of Hmf1p, but loss of Mmf1p results in growth defects and the loss of mitochondrial DNA (D. C. Ernst & Downs, 2018; Kim, Yoshikawa, & Shirahige, 2001; Oxelmark et al., 2000). Analysis of mmf1-Δ::KanMX (mmf1Δ) strains showed that the generation and accumulation of 2AA/2IP was responsible for the phenotypes of the mutant (D. C. Ernst & Downs, 2018), closely mirroring the paradigm identified and rigorously characterized in Salmonella enterica (Borchert et al., 2019; D. C. Ernst & Downs, 2018; J. L. Irons et al., 2020; Lambrecht et al., 2013). In the case of S. enterica, serine/threonine dehydratase (E.C. 4.3.1.19, IlvA) is the primary generator of 2AA/2IP from endogenous L-serine. While 2AA/2IP can be hydrolyzed nonenzymatically to form pyruvate, low availability of free water is presumed to limit this reaction in vivo, and RidA is thus required to expedite the hydrolysis (Lambrecht et al., 2013). Phenotypes of ridA mutant strains of S. enterica are due to damage of specific PLP-dependent enzymes caused by 2AA (Downs & Ernst, 2015; Lambrecht et al., 2013).
The growth defects and loss of mtDNA in S. cerevisiae strains lacking MMF1 are dependent on the activity of one or both mitochondrial serine/threonine dehydratases (Ilv1p/Cha1p)(D. C. Ernst & Downs, 2018) (Figure 1). The differential roles of these two enzymes allowed modulation of 2AA/2IP formation by changing growth conditions. CHA1 encodes the catabolic serine dehydratase and is transcribed only in the presence of exogenous L-serine or L-threonine (Bornaes, Ignjatovic, Schjerling, Kielland-Brandt, & Holmberg, 1993). In contrast, the product of ILV1 is active when no exogenous L-serine is provided and is allosterically inhibited by L-isoleucine (Ahmed, Bollon, Rogers, & Magee, 1976; D. C. Ernst & Downs, 2018). When mmf1Δ yeast cells are grown under conditions in which 2AA/2IP accumulates, they grow poorly and lose mitochondrial DNA (mtDNA) at high frequency making these cells unable to respire (cytoplasmic petite) (D. C. Ernst & Downs, 2018). While the phenotypes displayed by the mmf1Δ mutants are definitively caused by accumulation or 2AA and/or 2IP, the enzyme(s) targeted by 2AA to cause the metabolic perturbations are not currently known.
Figure 1. Enamine/imine production and the resulting stress in mitochondria.
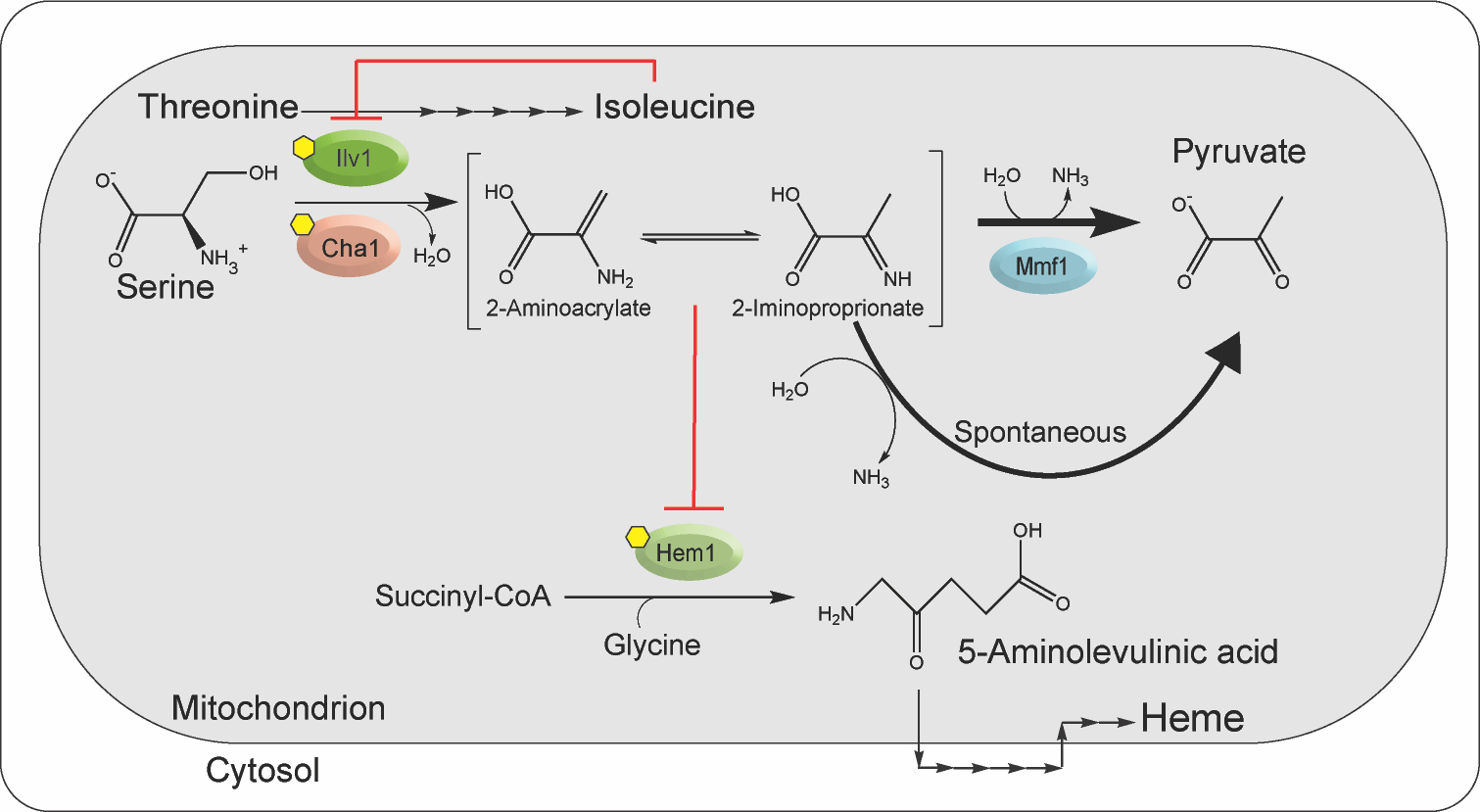
2-aminoacrylate (2AA) is generated in yeast mitochondria by PLP-dependent serine/threonine dehydratases Ilv1p and Cha1p (D. C. Ernst & Downs, 2018). CHA1 is transcribed in the presence of exogenous L-serine or L-threonine and has a role in catabolism of those amino acids. Ilv1p is constitutively synthesized, involved in biosynthesis of L-isoleucine and allosterically inhibited by L-isoleucine. Once formed, 2AA tautomerizes to 2-iminoproprionate (2IP) and is ultimately deaminated by solution water, or accelerated by Mmf1p, to pyruvate. In the absence of Mmf1p, 2AA/2IP accumulate in vivo to levels sufficient to damage target PLP enzymes. Work herein shows that Hem1p is such a target. Hem1p catalyzes the first committed step in heme biosynthesis, generating 5-aminolevulinic acid in the mitochondria. Subsequent steps in heme synthesis take place in the cytoplasm before the final steps are accomplished in the mitochondrion, as schematically represented. Past work demonstrated that branched chain amino acid transaminase is also a target for damage by 2AA/2IP (D. C. Ernst & Downs, 2018). The relevant pathways are schematically shown in the figure; red lines indicate points of post translational inhibition, yellow hexagons depict PLP cofactors and the biosynthetic steps to heme are depicted where they occur, either internal, or external to the mitochondrion. Both Ilv1p and Cha1p act on L-threonine and via enamine/imine formation generate the α-ketobutyrate that is an intermediate in the biosynthesis of L-isoleucine.
This study was initiated to extend our understanding of the 2AA/2IP mediated phenotypes of a mmf1Δ mutant and identify targeted enzyme(s) beyond those previously reported. Results herein identified a heme deficiency in mmf1Δ yeast strain that contributed to the growth defect of the mutant. Aminolevulinic acid synthase (ALAS, Hem1p, E.C. 2.3.1.37) catalyzes the condensation of succinyl-CoA and L-glycine to form 5-aminolevulinic acid (ALA) (Hunter et al., 2012). Hem1p is a PLP-dependent enzyme located in the mitochondrion that is responsible for the first step in heme biosynthesis. Here we show that 2AA/2IP caused damage to Hem1p, which contributed to the growth defects and heme deficiency of mmf1Δ yeast.
MATERIALS AND METHODS
Strains, media, and chemicals.
Saccharomyces cerevisiae strain YJF153 (MATa ho::dsdAMX4) was derived from an oak tree isolate (YPS163) and provided by Justin Fay (Washington University) (X. C. Li & Fay, 2017). S. cerevisiae strain S288c (MATα) (Mortimer & Johnston, 1986) was a gift from David Garfinkel (University of Georgia). Derivative strains are listed in Table S1.
Rich medium (YP) contained 20 g/l peptone (Fisher Scientific) and 10 g/l yeast extract. Minimal medium (S) contained 1.71 g/l yeast nitrogen base without amino acids or nitrogen (Sunrise Science; #1500–100) and ammonium sulfate (5 g/l). Either dextrose (D; 20 g/l) or glycerol (G; 30 g/l) was provided as the sole carbon source. Solid medium included 20 g/l agar (Difco). Antibiotics used for deletion marker selection were added at the following final concentrations: 400 μg/ml Geneticin (G418; Gold Biotechnology), 100 μg/ml nourseothricin sulfate (cloNAT; Gold Biotechnology). Supplements added to SD medium were: L-isoleucine (1 mM), ALA (0.24 mM) and L-glycine (1 mM).
Escherichia coli strain BL21-AI, which contains T7 polymerase under control of the araBAD promoter, was used for recombinant protein overproduction. Standard E. coli growth medium (LB broth) consisted of 10 g/l tryptone, 5 g/l yeast extract, and 10 g/l NaCl. Superbroth containing tryptone (32 g/l), yeast extract (20 g/l), sodium chloride (5 g/l), and sodium hydroxide (0.2 g/l) was used when high cell densities were desired for protein overproduction. Ampicillin (150 μg/ml) was added to the growth medium as needed. Reagents and chemicals were purchased from Sigma-Aldrich unless otherwise specified.
Genetic techniques and growth methods.
Gene disruptions in S. cerevisiae were made following the standard gene replacement method described by Hegemann and Heick (Hegemann & Heick, 2011). Disruption cassettes were amplified using the appropriate primers and plasmid templates listed in Table S2. Purified DNA (1–5 μg) was transformed into S. cerevisiae by incubating cells suspended in a mixture of 33% polyethylene glycol 3350 (PEG 3350), lithium acetate (100 mM), and salmon sperm DNA (0.28 mg/ml) at 30°C for 30 min followed by 30 min of heat shock at 42°C. The transformed cells were recovered in rich medium containing dextrose (YPD) or glycerol (YPG) for 1 h at 30°C and were subsequently plated on solid YPD or YPG containing the relevant selective agent. Colonies that arose after 2 to 3 days of incubation were transferred to selective medium, and individual colonies were screened via PCR to identify the appropriate recombinants.
For growth analyses, yeast strains were revived from glycerol stocks stored at −80°C and streaked for isolation on YPG. Selection and propagation of mmf1Δ mutants on YPG prevented the loss of the mitochondrial genome that results after growth on dextrose (D. C. Ernst & Downs, 2018). Single colonies were inoculated into 5 ml cultures of SD + L-isoleucine (1 mM) and incubated at 30°C with shaking (200 rpm) overnight. Two μl of overnight cultures were placed into wells of a 96-well microplate. Ninety-eight μl of medium was added to each well and OD650 measurements are used to monitor growth using a microplate reader (Biotek). Growth curves were plotted as averages and standard deviations of results from three independent cultures using GraphPad Prism 7.0.
Molecular techniques.
Plasmids were constructed using standard molecular techniques. DNA was amplified using Q5 DNA polymerase (New England Biolabs) with primers purchased from Eton Bioscience Inc. (Research Triangle Park, NC). Plasmids were isolated using PureYield plasmid miniprep system (Promega), and PCR products were purified using QIAquick PCR purification kit (Qiagen). Restriction endonucleases used for molecular cloning were purchased from New England Biolabs. T4 ligase (Thermo Scientific) was used to ligate inserts to vectors. The plasmids and primers used are listed in Table S2. Plasmids pDM1390 and pDM1672 were provided by David Garfinkel (University of Georgia). pDM1479 was constructed by the following method: primers Hem1_NcoI_F and Hem1_XbaI_R were used to amplify HEM1 from YJF153, omitting the mitochondrial localization sequence (9 amino acids) and introducing Nco1 and Xba1 restriction sites. The insert was then ligated into pET20b following restriction digest of the insert and vector and transformed into DH5α.
Protein purification.
Hem1- His6 lacking the N-terminal mitochondrial localization sequence was purified from an E. coli strain containing pDM1479. IlvA-His6 was purified from E. coli containing pDM1578 (pET20b-IlvA). Both proteins were purified with a similar protocol. An overnight culture of BL21 grown in 50 ml of superbroth (SB) containing ampicillin was inoculated into 6 liters of SB with ampicillin distributed in four 2.8 liter baffled Fernbach flasks. Cultures were grown at 37°C to an OD650 of ~ 0.7. Arabinose (0.2%) was added to induce expression of the inserted gene, and cultures were grown at 30°C overnight (16 hours). Cells were harvested by centrifugation (15 min at 8k × g) and resuspended in binding buffer containing potassium phosphate pH 8 (100 mM), sodium chloride (100 mM), imidazole (20 mM), PLP (10 μM), TCEP (tris(2-carboxyethyl)phosphine; 1 mM), and glycerol (10% w/v). Lysozyme (1 mg/ml), phenylmethylsulfonyl fluoride (1 mM), and DNase (25 μg/ml) were added, and the cell suspension was placed on ice for 1 h. Cells were mechanically lysed using a One-shot cell disruptor (once at 124 MPa). The resulting lysate was clarified by centrifugation (45 min at 48k × g) and filtered through a membrane (0.45 μm pore size). Filtered lysate was loaded onto HisTrap HP Ni-Sepharose columns (5 ml) and washed with five column volumes of binding buffer. Protein was eluted by increasing the concentration of imidazole in the elution buffer from 20 to 300 mM over 10 column volumes. Purified protein was concentrated by centrifugation with a 10,000-molecular-weight-cutoff filter unit (Millipore), and the buffer was replaced with potassium phosphate pH 8 (100 mM), containing PLP (10 μM), NaCl (100 mM), and glycerol (10% w/v) using a PD-10 desalting column (GE Healthcare). Protein concentration was determined by BCA assay (Pierce) and a recovery from a typical purification was ~100mg. Protein aliquots were frozen in liquid nitrogen and stored at 80°C. Densitometry estimated purity at >97% for Hem1p and IlvA. Mmf1p of similar purity (D. C. Ernst & Downs, 2018) was available in the laboratory.
Purification of Hem1p expressed in S. enterica ridA mutants.
Hem1-His6 was purified from each of two S. enterica strains carrying pDM1479, an isogenic pair with (DM13509) or without (DM17050) a functional RidA. Both strains contain T7 polymerase in the chromosome under control of the araBAD promoter. Cultures of each strain were grown overnight in LB (50 ml) containing ampicillin. For each strain, 15 ml of the overnight culture was used to inoculate each of three baffled Fernbach flasks (2.8 L) containing 1.5 liters minimal glucose medium with ampicillin (15 μg/ml), L-serine (5 mM) and L-glycine (1 mM). The resulting cultures were grown, with shaking, to an OD650 of 0.3 before arabinose (0.08%) was added to induce expression. Cells were harvested by centrifugation (15 min at 7k × g) after 18 hours of growth at 30°C. The cell pellet was resuspended in 2 ml binding buffer / g cell wet weight and subjected to the lysis method described above. Binding buffer contains potassium phosphate pH 8 (50 mM), NaCl (100 mM), imidazole (40 mM), glycerol (10% w/v). Filtered lysates were loaded onto Histrap HP Ni-Sepharose columns (1 ml), washed with 10 column volumes binding buffer, and eluted with 80 mM imidazole in binding buffer. Densitometry estimated purity at 70% for the protein samples from both WT and ridA mutant strains (Figure S1).
Characterization of cofactor content.
Hem1-bound cofactors were release from the protein as described previously (Flynn & Downs, 2013). KOH (30 mM final concentration) was added to purified Hem1p (50 nmol protein) and incubated at room temperature for 10 min. Protein was precipitated by addition of 10% trifluoroacetic acid to generate a visible precipitate. The precipitate was removed by centrifugation (3 min at 16k × g). The supernatant was filtered used a 0.45 μm centrifugal tube filter (Costar 8170) and the cofactors separated by high-performance liquid chromatography. Separation was performed on a Shimadzu HPLC equipped with a Luna C18 column (250 by 4.60 mm) (Phenomenex) using a 2-step isocratic method with a flow rate of 0.8 ml/min as follows: 0 to 5 min with 100% buffer A (0.06% [vol/vol] trifluoroacetic acid) and 5 to 18 min with methanol- buffer A (3:97). Between each run, the column was washed for 10 min with methanol-buffer A (60:40). The eluant was monitored at 305 nm using a photodiode array detector (Shimadzu SPD-M20A). Authentic pyridoxal 5’-phosphate (>98% pure; Sigma-Aldrich) and pyruvate/PLP served as standards. Pyruvate/PLP was synthesized as described previously (Schnackerz, Ehrlich, Giesemann, & Reed, 1979) purified by HPLC, and concentrated.
Heme measurements.
Intracellular heme levels were measured using a protocol adapted from Hans et. al. (Hans, Heinzle, & Wittmann, 2001). Overnight cultures of S. cerevisiae (2.5 ml) were used to inoculate 50 ml of SD media in a 500 ml flask and incubated with shaking (200 rpm) until an OD650 of 0.4 was reached. Cells were harvested by centrifugation (3k × g for 5 min), washed with 50 ml of distilled H2O, resuspended in 1 ml of deionized H2O, and placed in a microcentrifuge tube. Samples were pelleted (8k × g for 5 min), resuspended in 500 μl of oxalic acid (20 mM), and stored at 4°C for 16 hours. After incubation, 500 μl of warm oxalic acid (2 M) was added and 500 μl of each sample was transferred to a new amber microfuge tube. The original tubes were transferred to a heat block at 95°C for 30 min, while the new tubes were left at room temperature. Samples were then centrifuged at 16k × g for 2 min. 200 μl of each sample was transferred to a black 96-well microwell plate and top-read fluorescence measurements were obtained using a Spectramax Gemini EM, with 400 nm excitation light and recording emission intensity at 620 nm. Values of each sample before boiling were subtracted from their boiled counterparts. The resulting values were plotted on a standard curve generated with hemin and are presented in nmols heme/OD650. It was formally possible that normalizing heme concentration to OD650 could alter the ratios between various strains. Comparison of CFUs/OD between strains failed to suggest that the differences in heme levels reported in Table 1 and 2 were not valid (data not shown). Further, assays of representative strains that normalized heme levels to protein detected trends between strains similar to those reported in Table 1 and 2 (data not shown).
Table 1.
An mmf1Δ mutant has decreased heme levels.
| Heme levelsa | |||||
|---|---|---|---|---|---|
| Medium | |||||
| Strain | Genotype | SD | SD + Ile | SD + ALA | YPD |
| YJF153 | YJF153 | 254 ± 29 | 238 ± 10 | 251 ± 26 | 258 ± 26 |
| Dmy41 | YJF153 mmf1Δ | 90 ± 10 | 241 ± 13 | 157 ± 13 | 80 ± 7 |
| Dmy74 | YJF153 hem1Δ | ND | ND | 124 ± 26 | ND |
| Dmy61 | S288c | 140 ± 25 | 186 ± 39 | 176 ± 12 | 217 ± 8 |
| Dmy67 | S288c mmf1Δ | 84 ± 8 | 176 ± 19 | 125 ± 4 | 65 ± 9 |
Mutants lacking MMF or HEM1 in each of two strain backgrounds (YJF153 and S288c) were grown in the indicated medium. When added, L-isoleucine and aminolevulinic acid were at 1 mM and 240 μM, respectively. Data are from a representative experiment with three biological replicates and reported as average plus or minus 1 standard deviation. Abbreviations: SD, synthetic dextrose; YPD, yeast peptone dextrose; Ile, L-isoleucine; ALA, aminolevulinic acid.; ND, not determined.
Heme levels were measured as described in Materials and Methods and are reported in pmol/OD650.
Table 2.
Cha1p contributes to the decreased heme levels of an mmf1Δ mutant on YPD.
| Strain | Genotype | Heme levelsa |
|---|---|---|
| YJF153 | YJF153 | 206 ± 26 |
| Dmy41 | YJF153 mmf1Δ | 64 ± 24 |
| Dmy16 | YJF153 cha1Δ | 216 ± 21 |
| Dmy20 | YJF153 mmf1Δ cha1Δ | 96 ± 9 |
| Dmy61 | S288c | 209 ± 4 |
| Dmy67 | S288c mmf1Δ | 69 ± 10 |
| Dmy111 | S288c mmf1Δ cha1Δ | 133 ± 16 |
Mutants lacking MMF1 and/or CHA1 in two strain backgrounds (YJF153 and S288c) were grown in YPD medium. Data are from a representative experiment with 4 biological replicates and reported as average plus or minus 1 standard deviation.
Heme levels were measured as described in Materials and Methods and are reported in pmol/OD650.
Aminolevulinic acid synthase (ALAS) activity assays.
ALAS activity of Hem1p was quantified using a protocol adapted from Whittaker et. al. (Whittaker, Penmatsa, & Whittaker, 2015). Reactions consisted of potassium phosphate pH 6.8 (50 mM), L-glycine (100 mM), PLP (10 μM) and the relevant protein (~200–1000 nM) in a total volume of 170 μl. All components were added and allowed to incubate on a heat block at 30°C for 10 min. When assayed in the presence 3-chloroalanine (3CA), the incubation was at 37°C and 3CA was at 1 mM concentration. The reaction was started with the addition of succinyl-CoA (1 mM final concentration). After incubation for either 30 min (with 3CA) or 1 hour (other assays), the reaction was stopped by addition of 10% TCA. Precipitated protein was removed by centrifugation at 17k × g, after which samples were added to 1M sodium acetate (pH 4.6) containing 7.7% v/v acetylacetone. Aminolevulinate in the samples was derivatized by heat treatment (95°C) for 10 min to form a pyrrole. Ehrlich reagent was added and the resulting pyrrole derivative was detected by absorbance at 553 nm. Aminolevulinate present in each sample was determined by interpolating absorbance values onto a standard curve and reported in μmol.
In situ generation of 2AA and assessment of damage to ALAS.
A complete reaction contained IlvA (200 nM), Hem1p (1 μM) in potassium phosphate pH 8 (50 mM) with NaCl (10 mM). Microfuge tubes containing reaction components were incubated at 30°C for 5 min before adding L-serine (to 100 mM) to start the reaction and bring the total volume to 55 μl. The reaction proceeded for 1 hr at 30°C followed by cooling on ice for 5 min. Fifty μl of reaction mix was dispensed onto a 0.025 μm MCE membrane filter (MF Millipore VSWP02500) floating on the surface of 25 ml dialysis buffer (KPO4 pH 8 (50 mM), NaCl (10 mM)) in a petri dish. Dialysis proceeded for approximately one hour in a 4°C cold room, conditions that were empirically determined to eliminate the inhibitory concentration of L-serine. Forty-five μl was removed and transferred into 108 μl of ALAS reaction buffer (50 mM potassium phosphate (pH 8), 10 mM NaCl, 170 mM L-glycine) and placed on a heat block at 30°C for 5 min. The ALAS assay was started by the addition of 17 μl 10mM succinyl-CoA, bringing the final reaction mix to a total volume of 170 μl. ALA was quantified after 60 min.
RESULTS AND DISCUSSION
ρ+ mmf1Δ mutants of S. cerevisiae are compromised in synthesis of aminolevulinic acid.
ρ+mmf1Δ mutants in both YJF153 and S288c strain backgrounds were used to better understand phenotypic consequences of 2AA/2IP accumulation. Two strain backgrounds were used to provide broader insights into enamine/imine stress by identifying if any effects differed between the strains. Genomic differences in YJF153 and S288c did not significantly impact the growth of a mmf1Δ yeast in SD. Parental strains YJF153 and S288c reached a similar final OD650 (0.6 and 0.55 respectively), and the mmf1Δ mutation caused a growth defect in both strains (Figure 2). In both cases full growth of the mmf1Δ mutant was restored when L-isoleucine was added to the medium. This result was previously reported for the YJF153 strains, where L-isoleucine restored growth by allosterically inhibiting Ilv1p, and preventing the accumulation of 2AA/2IP (D. C. Ernst & Downs, 2018). This behavior is similar to that characterized in S. enterica and based on the data in Figure 2, is assumed to extend to the S288c strains.
Figure 2. Growth of mmf1Δ strains is compromised.
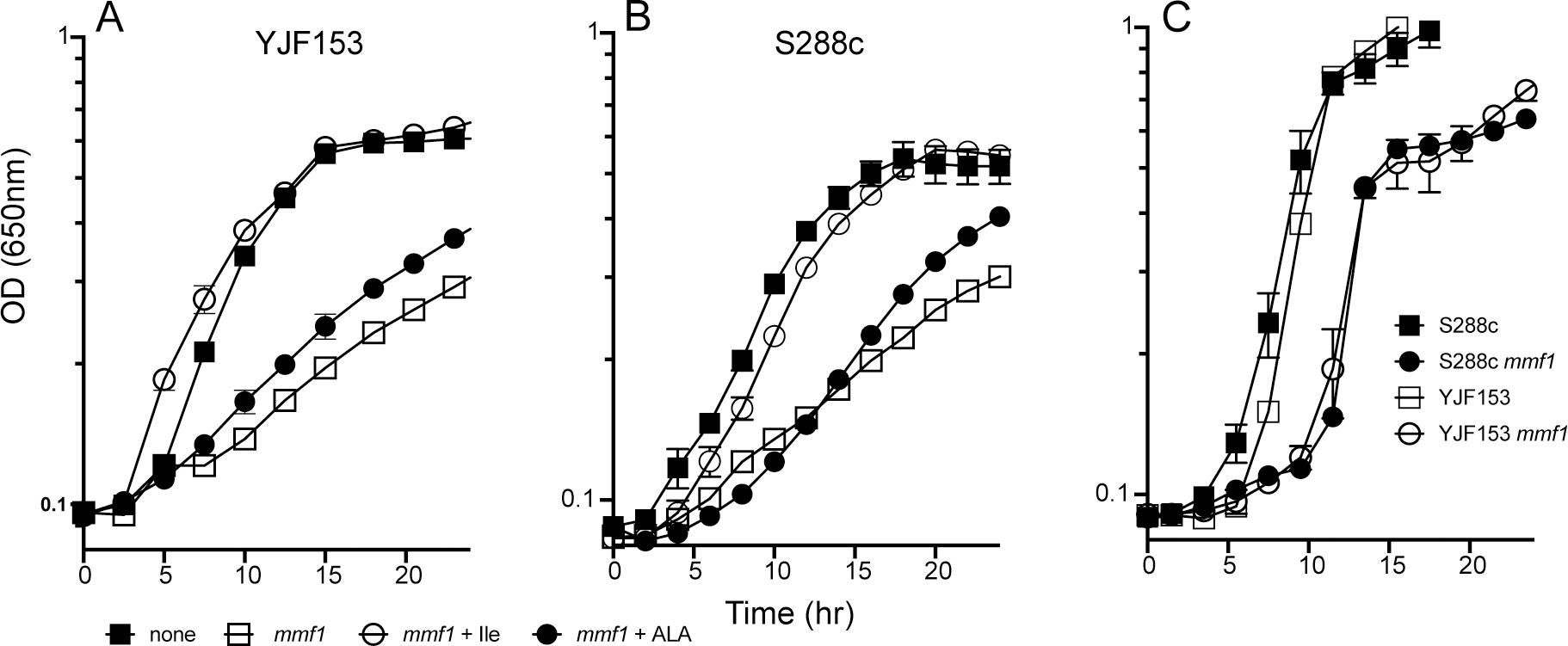
YJF153, S288c and mmf1Δ derivatives of these strains were grown on SD medium (panel A,B) or YPD (panel C). Growth was monitored as the OD650 over time. SD medium was supplemented with 5-aminolevulinic acid (ALA, 240 uM), or L-isoleucine (Ile, 1 mM) as indicated. The data represent the average and standard error of three biological replicates.
Growth on Yeast extract Peptone medium containing Dextrose (YPD) was also compromised by a mmf1Δ (Figure 2). In this case the mmf1Δ strains had a period of slower growth before reaching a growth rate that was similar to the parental strains. Due to the presence of L-isoleucine in YPD, there was little to no 2AA/2IP being generated via Ilv1p. However, the presence of L-serine in YPD induces the expression of CHA1 and would thus allow 2AA/2IP to be generated. Introduction of cha1Δ restored growth of the mmf1Δ yeast to that of the parental strains on YPD (data not shown), further supporting a role for 2AA/2IP in causing the growth phenotypes seen.
A culture of a ρ+mmf1Δ mutant of YJF153 was grown overnight in YP-Glycerol (YPG) and used to seed a soft agar overlay on minimal-glucose (SD) medium. Informed by knowledge of the RidA paradigm in bacteria, forty-nine nutritional supplements were spotted on the surface and stimulation of growth was noted (Table S3). L-Isoleucine stimulated growth likely due to its allosteric inhibition the major producer of 2AA (Ilv1p) and the iron chelator bathophenanthrolinedisulfonic acid (BPS) stimulated growth reflecting the sensitivity of mmf1Δ mutants to iron accumulation (D. C. Ernst & Downs, 2018). While a few additional nutrients showed some growth stimulation, the growth allowed by aminolevulinic acid (ALA), an intermediate in the biosynthesis in heme, was noteworthy. ALA is the product of the mitochondrially located PLP-dependent aminolevulinic acid synthase enzyme, Hem1p. Quantification of growth in liquid SD medium showed that exogenous ALA (240 μM) had a small but reproducible positive effect on the growth of a ρ+mmf1Δ mutant of both YJF153 and of S288c (Figure 2).
Heme biosynthesis is compromised by accumulated 2AA.
Partial restoration of growth by an intermediate in heme biosynthesis (ALA) suggested mmf1Δ strains might have lowered heme levels. Isogenic MMF1 and mmf1Δ strains in both the S288c and YJF153 background were grown in different media and total heme content was measured (Table 1). In SD medium, deletion of mmf1 lowered the heme levels significantly in both S288c and YJF153 strain backgrounds. Mutant derivatives of S288c and YJF153 had ~60% and ~35% of the heme found in their respective parental strains. When L-isoleucine was present in the growth medium, heme levels in the mmf1Δ mutants were restored to those in the respective wildtype. This result implicated 2AA/2IP in generating the lowered heme levels, since exogenous L-isoleucine allosterically inhibits Ilv1p and prevents the formation of these molecules (D. C. Ernst & Downs, 2018). When the growth medium contained ALA (240 μM), heme levels in the mmf1Δ mutants were restored to ~60–70% of the wild-type levels but notably to the full levels found in the parental strains. The failure of exogenous ALA to fully restore heme levels suggested there was: i) poor transport of ALA, ii) incomplete incorporation of exogenous ALA into the biosynthetic pathway, or iii) an additional bottleneck downstream of Hem1p in heme biosynthesis. A hem1Δ mutant of YJF153 was used to distinguish these possibilities. The growth of a hem1Δ mutant was restored to near that of wildtype with the addition of 240 μM ALA (Figure 3). Thus, growth occurred despite the fact that internal levels of heme reached only ~50% of the wild-type levels in this condition (Table 1). These data suggested that ALA was inefficiently transported and/or incorporated into the biosynthetic pathway. Importantly, the data also showed the level of heme allowed by this ALA supplementation could support wild-type growth. Thus, the minimal growth stimulation achieved by supplementing mmf1Δ mutants with ALA showed there were metabolic defects beyond limited heme biosynthesis that impacted growth of the mmf1Δ yeast.
Figure 3. 5-aminolevulinic acid restores growth of a hem1Δ mutant.
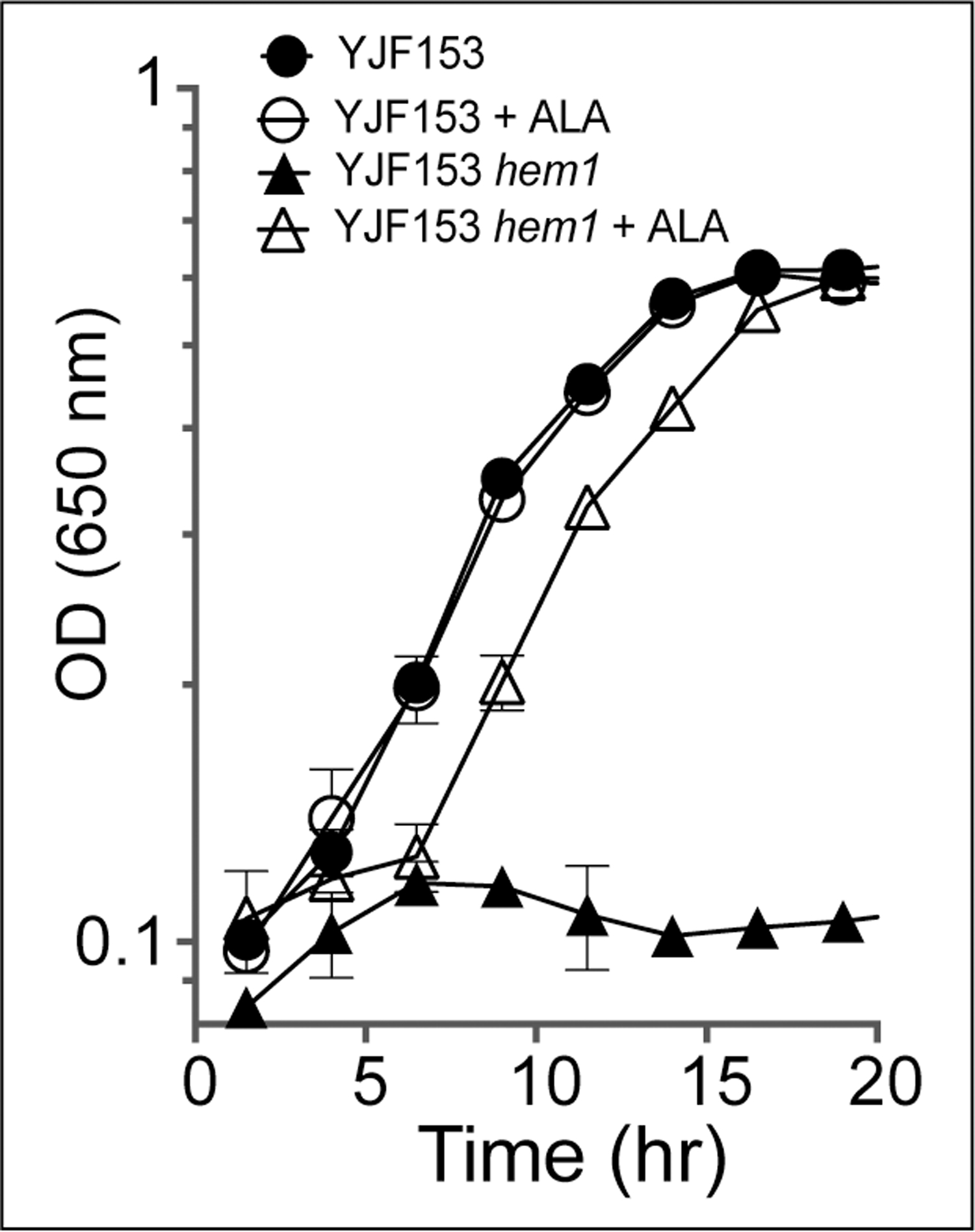
Growth of a YJF153 parent and hem1Δ mutant strain was monitored in SD media with or without the addition of 5-aminolevulinic acid (ALA) as indicated.
A mmf1Δ reduced heme levels in YPD medium in addition to SD (Table 1). The presence of L-isoleucine in YPD medium allosterically inhibits Ilv1p, while L-serine in the medium induces expression of CHA1. Thus, Cha1p was the presumed source of 2AA/2IP responsible for the reduced heme levels in YPD (D. C. Ernst & Downs, 2018). In this simple scenario, if CHA1 were eliminated, it would restore heme levels on YPD by preventing 2AA/2IP formation. However, although deleting CHA1 increased the levels of heme, they were not restored to those of the parental strains (Table 2). This result appeared to eliminate the simple model above. The formal possibility that Ilv1p was not completely inhibited by the level of L-isoleucine in YPD was eliminated since adding L-isoleucine to YPD did not increase heme levels in the mmf1Δ strain (data not shown). In total, these data suggested that if 2AA/2IP were responsible for the decreased heme levels in a mmf1Δ cha1Δ yeast in YPD, the source of these metabolites was unknown.
Specific activity of Hem1p decreases in a mmf1Δ mutant.
Hem1p is a fold type I PLP-dependent enzyme that catalyzes the condensation of succinyl-CoA and L-glycine to generate ALA, which is the first committed step of heme biosynthesis and occurs in the mitochondria (Figure 1). The data above raised the possibility that Hem1p was covalently modified by the 2AA/2IP accumulating in the mmf1Δ mutant, consistent with the paradigm established for other PLP-enzymes (Borchert et al., 2019). In this scenario and based on precedent in S. enterica (Flynn & Downs, 2013; Schmitz & Downs, 2004), the specific activity of Hem1p was predicted to be lower in an mmf1Δ yeast compared to the parental MMF1 strain. The low level of Hem1p in S. cerevisiae, in addition to its mitochondrial location and the inability to easily measure expression, complicated further analysis (Gollub, Liu, Dayan, Adlersberg, & Sprinson, 1977). To circumvent these hurdles, the well-characterized S. enterica ridA system was used to determine whether yeast Hem1p was a target of 2AA/2IP in vivo. A S. enterica ridA mutant (DM17050), and an isogenic wildtype (DM13509) were transformed with pDM1479, which contains HEM1 expressed by the T7 promoter. Expression of HEM1 was induced as the wildtype and ridA strain grew in minimal glucose medium with added L-serine and L-glycine. L-Serine was added to increase the production of 2AA/2IP, and L-glycine was present to allow growth in the presence of the elevated 2AA/2IP that is present in a ridA mutant (Christopherson, Lambrecht, Downs, & Downs, 2012; Dustin C. Ernst & Downs, 2016; Lambrecht et al., 2013). Hem1-His6 was purified from each strain and ALA synthase specific activity was determined (Figure 4). Hem1p that was purified from the S. enterica ridA mutant had 66% the specific activity of the Hem1p that was purified from cells of wildtype (8.1 ± 0.3 and 12.3 ± 0.8 μmol ALA/mg ALAS, respectively). These data were consistent with damage to Hem1p by 2AA/2IP in a ridA mutant.
Figure 4. Hem1p specific activity is affected by genetic background.
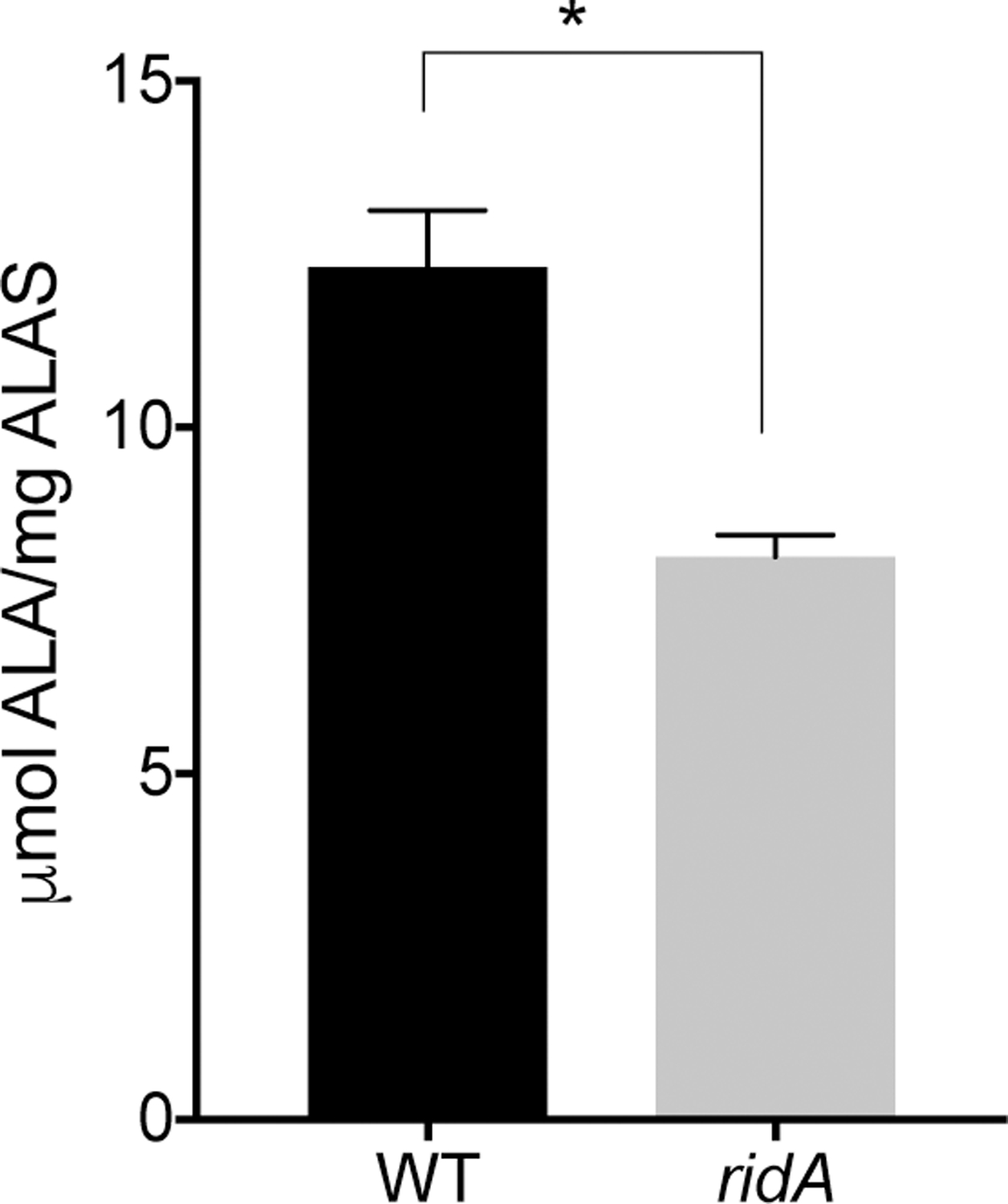
Hem1p was purified from either the wildtype (WT) or a ridA mutant strain of Salmonella enterica as indicated on the X axis. The purified protein was assayed for ALAS activity and the specific activity reported in μmol ALA/mg ALAS. The data shown are averages of triplicate technical replicates and significance (indicated by asterisk) was confirmed when a P value of 0.0042 was determined with an unpaired t test. Data are from a representative experiment that was repeated twice with two independent biological samples.
Cofactors present in the Hem1p purified from each S. enterica strain were extracted and separated by HPLC. If a PLP-dependent enzyme is attacked by 2AA, a pyruvate-PLP adduct that can be released from the enzyme after with treatment by base is generated (Flynn & Downs, 2013; Likos, Ueno, Feldhaus, & Metzler, 1982). The data in Figure 5 showed that a pyruvate-PLP adduct (in addition to PLP) was released from Hem1p purified from a ridA mutant background. Significantly, the Hem1p purified from wildtype, where 2AA/2IP does not accumulate, released only the PLP cofactor and had a barely detectable peak where the pyruvate-PLP was expected (Figure 5). Together these data supported the hypothesis that Hem1p is attacked in vivo by 2AA/2IP, generating a stably modified enzyme that is inactive.
Figure 5. Pyruvate-PLP is released from Hem1p after synthesis in a ridA mutant.
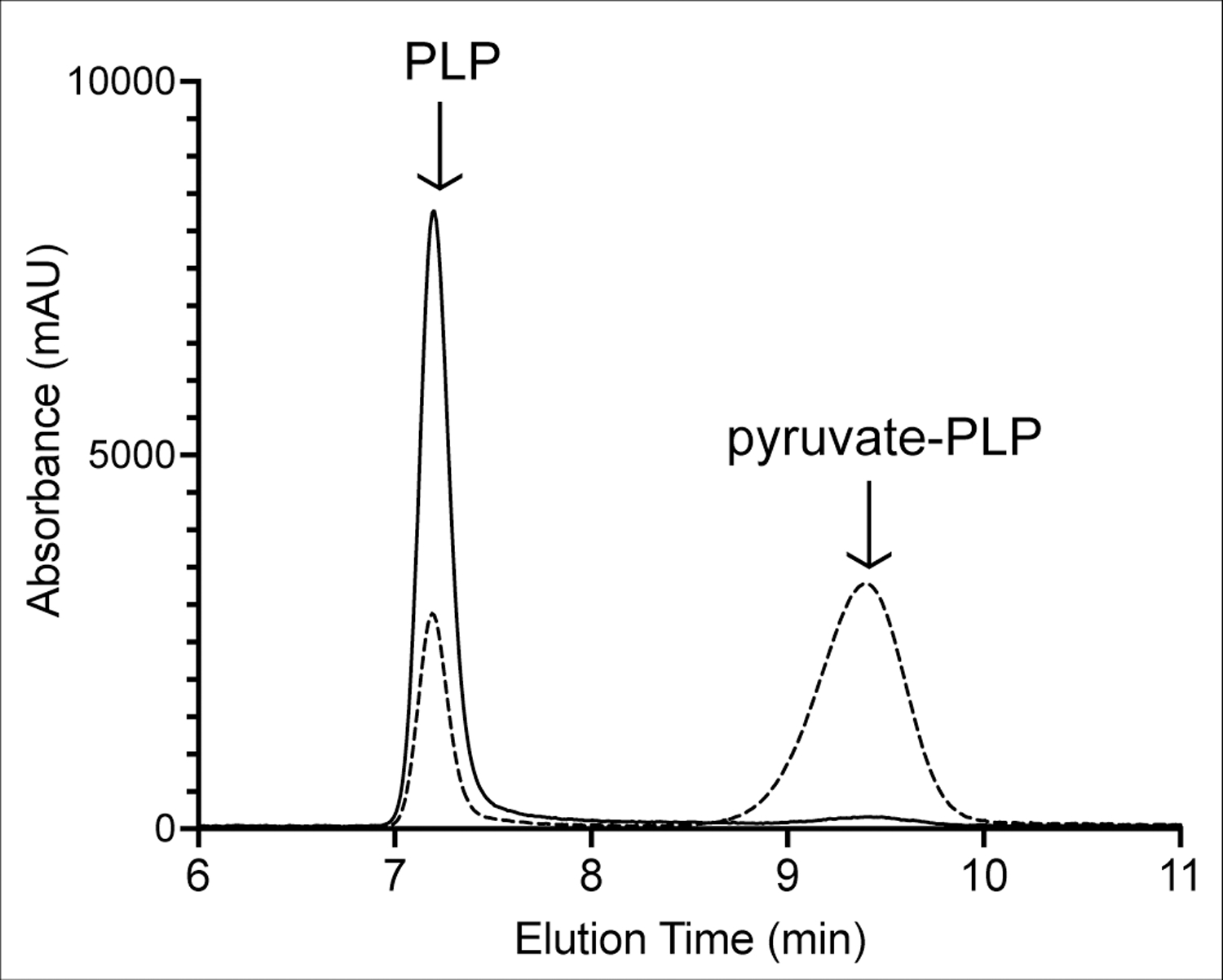
Hem1p was purified from two S. enterica strains; wildtype (solid lines) and a ridA mutant (dashed lines). Cofactors were released from the protein by treatment with base and separated with HPLC, while monitored by absorbance at 305 nm. The peak eluting with a retention time of ~7.4 min was PLP and the one at ~9.5 min was pyruvate-PLP. Peak assignment was based on retention time, UV-Vis spectra, and co-injection with authentic compounds (Figure S3).
Hem1p is damaged by 2AA in vitro.
To corroborate the in vivo data, Hem1p was purified from E. coli strain BL21-AI, which contains wild-type ridA, and exposed to 2AA/2IP in vitro. Initially, the purified protein was assayed in the presence and absence of 3-chloroalanine (3CA). 3CA reacts with PLP in the active site of some PLP-dependent enzymes, where loss of the chlorine substituent generates 2AA which can damage the active site of target enzymes (Badet, Roise, & Walsh, 1984; Henderson & Johnston, 1976; Relyea, Tate, & Meister, 1974). As such, reaction with 3CA can serve as a predictor of sensitivity to free 2AA or the 2IP tautomer. In the absence of 3CA, Hem1p generated 26.1 ± 1.1 μmol ALA/mg Hem1p after 30 min, while in the presence of 3CA (1 mM), ALA synthesis dropped to 7.8 ± 0.1 (Figure 6A).
Figure 6. Hem1p is sensitive to 2AA in vitro.
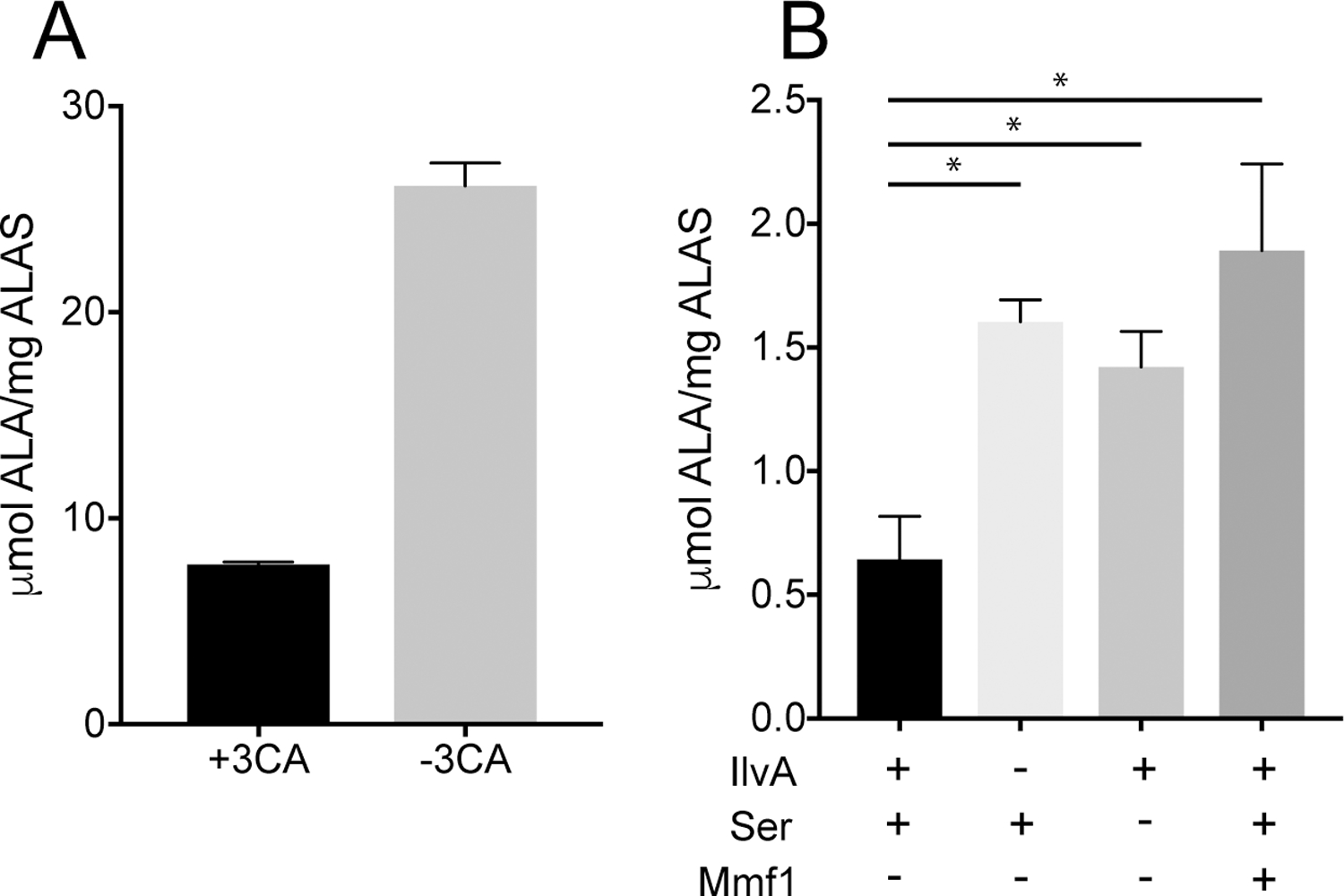
Hem1p was expressed and purified from BL21 E. coli and assayed for activity under multiple conditions. (A) Hem1p was incubated with 3-chloroalanine (3CA) prior to assaying ALA synthesis. Data are representative averages of three technical replicates. The difference is significant as determined by unpaired t test (P < .0001). (B) ALAS activity of Hem1p was determined after exposure to the 2AA/2IP generated in situ by IlvA as described in Materials and Methods. Components present in the preincubation stage of the protocol are indicated below the Y axis. Data shown are representative of at least 2 independent experiments. Each condition consisted of 4 technical replicates with significance determined by one way ANOVA. * indicates P value < .001.
Analysis of direct damage by 2AA/2IP requires that the enamine/imine is generated in situ due to the short half-life (3 sec) of these molecules in aqueous solution (Hillebrand, Dye, & Suelter, 1979). Serine dehydratase (IlvA) and cysteine desulfurase (CdsH) have been used to generate 2AA/2IP in situ from L-serine or L-cysteine, respectively (D. C. Ernst, Lambrecht, Schomer, & Downs, 2014; Lambrecht et al., 2013). Control reactions showed that Hem1p was inhibited by both L-serine and L-cysteine, complicating the use of these enzymes and requiring that a two-step assay be implemented. Hem1p was incubated with L-serine and serine dehydratase (IlvA) for 60 min to allow generation of 2AA/2IP and time for it to damage Hem1p. Excess L-serine was then removed by filter dialysis before the components needed to assay ALA formation by Hem1p were added. The data are in Figure 6B. When no 2AA was generated in the initial step (i.e., L-serine or IlvA was not present), 1.4 ± 0.1 and 1.6 +/− 0.1 μmol ALA/mg ALAS was generated, respectively. In contrast, when both L-serine and IlvA were present to generate 2AA, significantly less activity was detected, 0.6 ± 0.2 μmol ALA/mg ALAS. Finally, when Mmf1p was present in the complete assay, ALAS activity was restored to 1.9 ± 0.3 μmol ALA/mg ALAS. Mmf1p has 2AA/2IP deaminase activity (D. C. Ernst & Downs, 2018), and the restoration of ALAS activity supports the conclusion that Hem1p can be attacked by 2AA/2IP and its activity decreased.
Conclusions.
Loss of the mitochondrial ridA homolog MMF1 results in numerous phenotypes, which are collectively due to the accumulation of 2AA/2IP in the strain (D. C. Ernst & Downs, 2018). Herein we identify and characterize one target of the accumulated 2AA/2IP as Hem1p, the first enzyme in heme biosynthesis. Our work shows that Hem1p is sensitive to damage by 2AA/2IP in vivo and in vitro. Further, the data are in support of the conclusion that the reduction of heme levels found in an mmf1Δ mutant is at least partially due to damage to Hem1p mediated by 2AA/2IP.
Demonstrating that Hem1p is damaged by 2AA/2IP defines a novel target of enamine/imine damage. The inability of ALA to restore an mmf1Δ mutant to full growth on SD showed that Hem1 was not the sole growth determining target of 2AA/2IP in S. cerevisiae. Hem1p is the second identified target of 2AA in the yeast mitochondria, with branched chain amino acid transferase (BAT) being the first (D. C. Ernst & Downs, 2018; Kim et al., 2001). Neither the lack of Hem1p or of Bat1p phenocopy the respiratory defects or mtDNA loss associated with a mmf1Δ mutant (data not shown). These results suggest multiple small perturbations caused by the accumulation of 2AA/2IP exist in mmf1Δ yeast and that they act together to generate phenotypic outcomes. Mitochondrially localized PLP-dependent enzymes are the putative targets for 2AA/2IP in mmf1Δ yeast. Several such enzymes are linked directly or indirectly to mitochondrial stability: aspartate amino transferase (EC 2.6.1.1; AAT1), mitochondrial serine hydroxymethyltransferase (EC 2.1.2.1; SHM1), and cysteine desulfurase (EC 2.8.1.7; NFS1) (J. Li, Kogan, Knight, Pain, & Dancis, 1999; Luzzati, 1975; Sliwa, Dairou, Camadro, & Santos, 2012). Homologues of AAT1 and SHM1 are inhibited by 2AA/2IP in S. enterica and it is likely their eukaryotic counterparts are also targets of enamine/imines attack (Downs & Ernst, 2015). Continued genetic and biochemical studies will define components of the metabolic network in the mitochondria that are impacted by enamine/imine stress and how they combine to generate the multiple phenotypes of an mmf1Δ mutant.
Supplementary Material
TAKE AWAYS:
mmf1Δ yeast accumulate reactive species 2-aminoacrylate and/or 2-iminopropanoate.
Imine/enamine accumulation results in low levels of heme in mmf1Δ yeast.
Aminolevulinic acid synthase (Hem1p) is attacked and damaged by 2-aminoacrylate.
Defective heme biosynthesis is not the cause of all mmf1Δ phenotypes.
ACKNOWLEDGEMENTS.
The authors would like to thank David Garfinkel for plasmids, technical advice and helpful discussions. This work was supported by an award from the competitive grants program at the NIH (GM095837) to DMD. Authors contributed to; (i) the conception or design of the study (DCE, DMD,GHW), (ii) the acquisition, analysis, or interpretation of the data (GHW, DCE, DMD) and (iii) writing of the manuscript (GHW, DMD).
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
DATA AVAILABILITY
The data that supports the findings of this study are within the manuscript or the supplementary material of this article, and additional information is available upon request from the authors.
REFERENCES
- Ahmed SI, Bollon AP, Rogers SJ, & Magee PT (1976). Purification and properties of threonine deaminase from Saccharomyces cerevisiae. Biochimie, 58(1–2), 225–232. doi: 10.1016/s0300-9084(76)80374-4 [DOI] [PubMed] [Google Scholar]
- Badet B, Roise D, & Walsh CT (1984). Inactivation of the dadB Salmonella typhimurium alanine racemase by D and L isomers of b-substituted alanines: kinetics, stoichiometry, active site peptide sequencing, and reaction mechanism. Biochemistry, 23, 5188–5194. [DOI] [PubMed] [Google Scholar]
- Borchert AJ, Ernst DC, & Downs DM (2019). Reactive Enamines and Imines In Vivo: Lessons from the RidA Paradigm. Trends Biochem Sci, 44(10), 849–860. doi: 10.1016/j.tibs.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornaes C, Ignjatovic MW, Schjerling P, Kielland-Brandt MC, & Holmberg S (1993). A regulatory element in the CHA1 promoter which confers inducibility by serine and threonine on Saccharomyces cerevisiae genes. Mol Cell Biol, 13(12), 7604–7611. doi: 10.1128/mcb.13.12.7604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopherson MR, Lambrecht JA, Downs D, & Downs DM (2012). Suppressor analyses identify threonine as a modulator of ridA mutant phenotypes in Salmonella enterica. PLoS ONE, 7(8), e43082. doi: 10.1371/journal.pone.0043082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digiovanni S, Visentin C, Degani G, Barbiroli A, Chiara M, Regazzoni L, . . . Popolo L (2020). Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage. Sci Rep, 10(1), 10135. doi: 10.1038/s41598-020-66663-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs DM, & Ernst DC (2015). From microbiology to cancer biology: the Rid protein family prevents cellular damage caused by endogenously generated reactive nitrogen species. Mol Microbiol, 96(2), 211–219. doi: 10.1111/mmi.12945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElRamlawy KG, Fujimura T, Baba K, Kim JW, Kawamoto C, Isobe T, . . . Kawamoto S (2016). Der f 34, a Novel Major House Dust Mite Allergen Belonging to a Highly Conserved Rid/YjgF/YER057c/UK114 Family of Imine Deaminases. J Biol Chem, 291(41), 21607–21615. doi: 10.1074/jbc.M116.728006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst DC, & Downs DM (2016). 2-Aminoacrylate Stress Induces a Context-Dependent Glycine Requirement inridAStrains of Salmonella enterica. Journal of Bacteriology, 198(3), 536–543. doi: 10.1128/jb.00804-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst DC, & Downs DM (2018). Mmf1p Couples Amino Acid Metabolism to Mitochondrial DNA Maintenance in Saccharomyces cerevisiae. MBio, 9(1). doi: 10.1128/mBio.00084-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst DC, Lambrecht JA, Schomer RA, & Downs DM (2014). Endogenous Synthesis of 2-Aminoacrylate Contributes to Cysteine Sensitivity in Salmonella enterica. Journal of Bacteriology, 196(18), 3335–3342. doi: 10.1128/jb.01960-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki N, & Walsh CT (1986). Biosynthetic alanine racemase of Salmonella typhimurium: purification and characterization of the enzyme encoded by the alr gene. Biochemistry, 25(11), 3261–3267. [DOI] [PubMed] [Google Scholar]
- Flynn JM, & Downs DM (2013). In the absence of RidA, endogenous 2-aminoacrylate inactivates alanine racemases by modifying the pyridoxal 5’-phosphate cofactor. J Bacteriol, 195(16), 3603–3609. doi: 10.1128/JB.00463-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub EG, Liu KP, Dayan J, Adlersberg M, & Sprinson DB (1977). Yeast mutants deficient in heme biosynthesis and a heme mutant additionally blocked in cyclization of 2,3-oxidosqualene. J Biol Chem, 252(9), 2846–2854. [PubMed] [Google Scholar]
- Hans MA, Heinzle E, & Wittmann C (2001). Quantification of intracellular amino acids in batch cultures of Saccharomyces cerevisiae. Appl Microbiol Biotechnol, 56(5–6), 776–779. [DOI] [PubMed] [Google Scholar]
- Hegemann JH, & Heick SB (2011). Delete and repeat: a comprehensive toolkit for sequential gene knockout in the budding yeast Saccharomyces cerevisiae. Methods Mol Biol, 765, 189–206. doi: 10.1007/978-1-61779-197-0_12 [DOI] [PubMed] [Google Scholar]
- Henderson LL, & Johnston RB (1976). Inhibition studies of the enantiomers of beta chloroalanine on purified alanine racemase from B. subtilis. Biochem Biophys Res Commun, 68(3), 793–798. [DOI] [PubMed] [Google Scholar]
- Hillebrand GG, Dye JL, & Suelter CH (1979). Formation of an intermediate and its rate of conversion to pyruvate during the tryptophanase-catalyzed degradation of S-o-nitrophenyl-L-cysteine. Biochemistry, 18(9), 1751–1755. [DOI] [PubMed] [Google Scholar]
- Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, . . . Yong SY (2012). InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res, 40(Database issue), D306–312. doi: 10.1093/nar/gkr948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons J, Hodge-Hanson KM, & Downs DM (2018). PA5339, a RidA Homolog, Is Required for Full Growth in Pseudomonas aeruginosa. J Bacteriol, 200(22). doi: 10.1128/JB.00434-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons J, Sacher JC, Szymanski CM, & Downs DM (2019). Cj1388 Is a RidA Homolog and Is Required for Flagella Biosynthesis and/or Function in. Front Microbiol, 10, 2058. doi: 10.3389/fmicb.2019.02058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irons JL, Hodge-Hanson K, & Downs DM (2020). RidA Proteins Protect against Metabolic Damage by Reactive Intermediates. Microbiol Mol Biol Rev, 84(3). doi: 10.1128/MMBR.00024-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Yoshikawa H, & Shirahige K (2001). A member of the YER057c/yjgf/Uk114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae. Genes Cells, 6(6), 507–517. doi: 10.1046/j.1365-2443.2001.00443.x [DOI] [PubMed] [Google Scholar]
- Lambrecht JA, Schmitz GE, & Downs DM (2013). RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. MBio, 4(1), e00033–00013. doi: 10.1128/mBio.00033-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kogan M, Knight SA, Pain D, & Dancis A (1999). Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J Biol Chem, 274(46), 33025–33034. doi: 10.1074/jbc.274.46.33025 [DOI] [PubMed] [Google Scholar]
- Li XC, & Fay JC (2017). Cis-Regulatory Divergence in Gene Expression between Two Thermally Divergent Yeast Species. Genome Biology and Evolution, 9(5), 1120–1129. doi: 10.1093/gbe/evx072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likos JJ, Ueno H, Feldhaus RW, & Metzler DE (1982). A novel reaction of the coenzyme of glutamate decarboxylase with L-serine O-sulfate. Biochemistry, 21(18), 4377–4386. [DOI] [PubMed] [Google Scholar]
- Luzzati M (1975). Isolation and properties of a thymidylate-less mutant in Saccharomyces cerevisiae. Eur J Biochem, 56(2), 533–538. doi: 10.1111/j.1432-1033.1975.tb02259.x [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, . . . Bryant SH (2013). CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res, 41(Database issue), D348–352. doi: 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Chavarría LC, Sagawa J, Irons J, Hinz AK, Lemon A, Graça T, . . . Vadyvaloo V (2020). Putative Horizontally Acquired Genes, Highly Transcribed during Yersinia pestis Flea Infection, Are Induced by Hyperosmotic Stress and Function in Aromatic Amino Acid Metabolism. J Bacteriol, 202(11). doi: 10.1128/JB.00733-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, & Johnston JR (1986). Genealogy of principal strains of the yeast genetic stock center. Genetics, 113(1), 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehaus TD, Gerdes S, Hodge-Hanson K, Zhukov A, Cooper AJ, ElBadawi-Sidhu M, . . . Hanson AD (2015). Genomic and experimental evidence for multiple metabolic functions in the RidA/YjgF/YER057c/UK114 (Rid) protein family. BMC Genomics, 16, 382. doi: 10.1186/s12864-015-1584-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxelmark E, Marchini A, Malanchi I, Magherini F, Jaquet L, Hajibagheri MA, . . . Tommasino M (2000). Mmf1p, a novel yeast mitochondrial protein conserved throughout evolution and involved in maintenance of the mitochondrial genome. Mol Cell Biol, 20(20), 7784–7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relyea NM, Tate SS, & Meister A (1974). Affinity labeling of the active center of L-aspartate-beta-decarboxylase with beta-chloro-L-alanine. J Biol Chem, 249(5), 1519–1524. [PubMed] [Google Scholar]
- Schmitz G, & Downs DM (2004). Reduced Transaminase B (IlvE) Activity Caused by the Lack of yjgF Is Dependent on the Status of Threonine Deaminase (IlvA) in Salmonella enterica Serovar Typhimurium. Journal of Bacteriology, 186(3), 803–810. doi: 10.1128/jb.186.3.803-810.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackerz KD, Ehrlich JH, Giesemann W, & Reed TA (1979). Mechanism of action of D-serine dehydratase. Identification of a transient intermediate. Biochemistry, 18(16), 3557–3563. doi: 10.1021/bi00583a019 [DOI] [PubMed] [Google Scholar]
- Sliwa D, Dairou J, Camadro JM, & Santos R (2012). Inactivation of mitochondrial aspartate aminotransferase contributes to the respiratory deficit of yeast frataxin-deficient cells. Biochem J, 441(3), 945–953. doi: 10.1042/BJ20111574 [DOI] [PubMed] [Google Scholar]
- Walsh C (1982). Suicide substrates: mechanism-based enzyme inactivators. Tetrahedron, 38, 871–909. [DOI] [PubMed] [Google Scholar]
- Whittaker MM, Penmatsa A, & Whittaker JW (2015). The Mtm1p carrier and pyridoxal 5’-phosphate cofactor trafficking in yeast mitochondria. Arch Biochem Biophys, 568, 64–70. doi: 10.1016/j.abb.2015.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that supports the findings of this study are within the manuscript or the supplementary material of this article, and additional information is available upon request from the authors.


