Abstract
Recent research provides insight into the ability of miRNA to regulate various pathways in several cancer types. Despite their involvement in the regulation of the mRNA via targeting the 3′UTR, there are relatively few studies examining the changes in these regulatory mechanisms specific to single cancer types or shared between different cancer types.
We analyzed samples where both miRNA and mRNA expression had been measured and performed a thorough correlation analysis on 7494 experimentally validated human miRNA-mRNA target-gene pairs in both healthy and tumoral samples.
We show how more than 90% of these miRNA-mRNA interactions show a loss of regulation in the tumoral samples compared with their healthy counterparts.
As expected, we found shared miRNA-mRNA dysregulated pairs among different tumors of the same tissue. However, anatomically different cancers also share multiple dysregulated interactions, suggesting that some cancer-related mechanisms are not tumor-specific. 2865 unique miRNA-mRNA pairs were identified across 13 cancer types, ≈ 40% of these pairs showed a loss of correlation in the tumoral samples in at least 2 out of the 13 analyzed cancers. Specifically, miR-200 family, miR-155 and miR-1 were identified, based on the computational analysis described below, as the miRNAs that potentially lose the highest number of interactions across different samples (only literature-based interactions were used for this analysis).
Moreover, the miR-34a/ALDH2 and miR-9/MTHFD2 pairs show a switch in their correlation between healthy and tumor kidney samples suggesting a possible change in the regulation exerted by the miRNAs. Interestingly, the expression of these mRNAs is also associated with the overall survival. The disruption of miRNA regulation on its target, therefore, suggests the possible involvement of these pairs in cell malignant functions.
The analysis reported here shows how the regulation of miRNA-mRNA interactions strongly differs between healthy and tumoral cells, based on the strong correlation variation between miRNA and its target that we obtained by analyzing the expression data of healthy and tumor tissue in highly reliable miRNA-target pairs. Finally, a go term enrichment analysis shows that the critical pairs identified are involved in cellular adhesion, proliferation, and migration.
Keywords: Cancer, Non-coding RNA, miRNA regulation, Cancer biomarkers
1. Introduction
Micro-RNAs (miRNAs) are short molecules, usually 19-25 nucleotide-long, that work as target recognition elements of an RNA-protein complex known as RNA induced silencing complex (RISC) in post-transcriptional regulation [[1], [2], [3]].
Many biological processes, such as development, cell differentiation, and even diseases, have been associated with the activity of miRNAs [4,5].
Their major mode of action is mRNA target regulation via sequence-complementary pairing with 3′ untranslated region (3′UTR) in the cytoplasm, which leads to the target translational repression, through a temporary and reversible control, or to the target transcript degradation, in a non-reversible way, with a consequent decrease in the translation of the mRNA [1,[6], [7], [8]]. Several positive regulation mechanisms exerted by miRNAs have also been described [9,10].
Small RNA deep sequencing shows that some miRNAs are also present in the nucleus [11], and some evidence indicates that certain miRNAs exert their biological function in the nucleus [12]. Zou and collaborators suggest that the nuclear activating miRNAs (NamiRNAs) could play a crucial role in gene expression, in fact they can promote transcription by targeting enhancers in the nucleus [13]. Despite the fact that dysregulation in gene expression has been widely described in many different types of cancer [14,15], a growing number of evidences is shedding light on the role of several dysregulated miRNAs, to which an oncogenic or an onco-suppressor role can be assigned (e.g. miR-2110 [16] and miR-452 miRNA families [17]). miRNAs are also involved in more complicated relationships where different RNAs, e.g., mRNAs and lncRNAs, compete for the same endogenous miRNA (ceRNA) therefore positively regulating each other [18,19].
As a consequence of gene expression dysregulation in cancer, a loss of connectivity in a coding transcript co-expression matrix has been described [20].
Moreover, a differential regulation, dominated by a loss of regulation mechanisms, in miRNA-mediated competitive interactions between mRNA and lncRNA (ceRNA) has been observed in breast and ovarian cancers [19,21].
To date several experimental procedures are used in the detection of RNA interactions increasing the number of known validated miRNA-mRNA interactions (e.g. RAID v2.0 [22], RAIN [23], RNAcentral [24]). The resulting data increases the performance of computational predictive methods and at the same time contributes to their improvement.
Despite the great importance given to the miRNA regulation, there are relatively few studies that compared the mRNA-miRNA landscape in tumor vs healthy tissue. Andrés-León and collaborators developed a statistical approach to identify novel miRNA-target relationships in different tumor types within cancer-relevant pathways [25]. Later, another research group demonstrated that correlation between miRNAs and target genes was greatly reduced in tumors [26].
Furthermore, univariate and multivariate Cox regression analysis has been used to identify miRNAs as novel biomarkers in early diagnosis and prognosis, in order to improve outcomes in gastric cancer [27]. Here we present a thorough analysis of the miRNA-mRNA interaction landscape in different cancer types thus highlighting the differences that exist in the miRNA-mRNA regulatory networks between healthy and tumor tissue. Our results show that multiple miRNA-mRNA interactions are consistently dysregulated across different cancer types, and that the extent of these perturbations is associated with overall survival.
2. Material and methods
2.1. TCGA data collection and preprocessing
The expression data of cancer patients were downloaded from TCGA Data Portal (https://tcga-data.nci.nih.gov) using the recommended GDC data transfer tool. The processed data (level 3) were used. To date, there are 13 cancer types that are associated with data both unrestricted for publication and containing paired miRNA and mRNA expression for at least 15 matched normal and tumor samples (Table 1). RNA-seq(V2) and miRNA-seq were used for mRNA and miRNA expression data, respectively. Normal/tumor information for each sample were obtained through the Biospecimen Metadata Browser (https://tcga-data.nci.nih.gov/uuid/uuidBrowser.htm) and mapped based on the sample ID. For the sequencing data, we used the FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values for mRNA and RPM (Reads Per Million miRNA mapped) for miRNA. To ensure the homogeneity among the samples, we further transformed the FPKM and RPM in log2(TPM +1) values.
| TPM(i) = ([FPKM/RPM](i) / sum ([FPKM/RPM] all transcripts)) * 10^6 |
Table 1.
Paired expression data from TCGA.
| Cancer Symbol | Cancer Type | # matched samples |
|---|---|---|
| LUSC | lung squamous cell carcinoma | 37 |
| HNSC | head and neck squamous cell carcinoma | 42 |
| KICH | kidney chromophobe | 23 |
| KIRC | kidney renal clear cell carcinoma | 70 |
| KIRP | kidney renal papillary cell carcinoma | 31 |
| BRCA | breast invasive carcinoma | 87 |
| STAD | stomach adenocarcinoma | 27 |
| LIHC | liver hepatocellular carcinoma | 49 |
| PRAD | prostate adenocarcinoma | 51 |
| LUAD | lung adenocarcinoma | 18 |
| BLCA | bladder urothelial carcinoma | 19 |
| UCEC | uterine corpus endometrial carcinoma | 21 |
| THCA | thyroid carcinoma | 56 |
2.2. miRNA-mRNA pairs identification
The experimentally validated miRNA-mRNA interactions were collected from RAID (www.rna-society.org/raid/) [22]. This database stores more than 4 million RNA-RNA interactions collected from numerous resources.
Only human miRNA-mRNA pairs with at least one experimental evidence were selected, using a threshold score ≥0,9. A total number of 7494 miRNA-mRNA pairs were selected consisting of 557 unique miRNAs and 2678 unique mRNAs.
2.3. Correlation analysis
Pearson correlation (r) has been calculated using the miRNA and mRNA log2(TPM +1) transformed expressions. The Pearson correlation test was used to estimate the correlations between miRNAs and their mRNA targets and correlated pairs were selected using as thresholds a p-value strictly lower than 0.05 and an absolute value of r equal or higher than 0.4.
2.4. Survival curve and statistical analysis
Overall survival probability curves were plotted using the Kaplan-Meier method and comparisons between the curves were analyzed using the logrank test [28]. All tests were performed at the 0.05 level of significance.
The samples were split into 4 groups based on miRNA and mRNA median expression values: i. samples with a low expression of the miRNA and a low expression of the mRNA; ii. samples with a low expression of the miRNA and a high expression of the mRNA; iii. samples with a high expression of the miRNA and a low expression of the mRNA; iv. samples with a high expression of the miRNA and a high expression of the mRNA.
We performed differential expression analysis and overall survival curves through GEPIA 2 web server for these highlighted genes [29].
2.5. Functional enrichment
Gene Ontology (GO) classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using the online tools of Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) [30,31]. We focused on Biological Process and Molecular Function GO term categories, providing the whole genome as background.
3. Results
3.1. Identification of miRNA-mRNA target-gene pairs deregulated in tumor
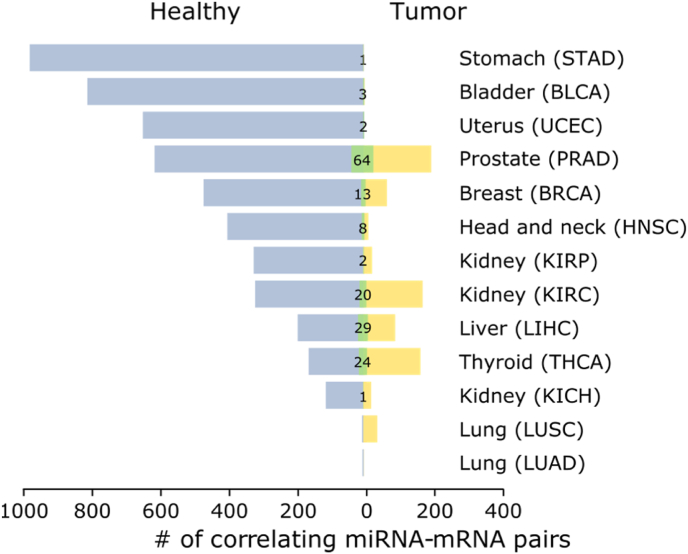
We set up to analyze the gene expression of experimentally validated mRNA and miRNA pairs in a set of 13 cancer tissues, listed in Table 1. We collected 7494 miRNA-mRNA targets interactions annotated in the RAID database [22] with strong evidence supported by at least one experimental validation. For each cancer type we calculated the Pearson correlation (r) between the miRNA and mRNA expression profiles in both healthy and tumoral samples (see the Methods section). On average we found 385 miRNA-mRNA pairs negatively correlated across the healthy samples analyzed (Pearson r < −0.4, adjusted p-value < 0.05) (Fig. 1, blue bars). A marked decrease in the number of correlated pairs was found in the tumoral samples, where, on average, only 62 miRNA-mRNA showed a negative correlation across all tumor samples within the defined thresholds (Fig. 1, yellow bars). Moreover, the comparison between healthy and tumoral samples highlighted that only a small number of miRNA-mRNA pairs preserved their correlation in both conditions (Fig. 1, green bars) (Table 2). These findings are in agreement with data published by Li and colleagues [27], who reported a decrease in miRNA-mRNA correlations in tumor tissue. As reported in Fig. 1, the number of miRNA-mRNA correlating pairs varies widely across the 13 cancer types analyzed in this study (Table 1).
Fig. 1.
For each tumor, we report the number of miRNA-mRNA correlating pairs identified in healthy and tumoral samples on the left (blu bars) and on the right (yellow bars), respectively. In green we report the number of miRNA-mRNA correlating pairs that preserved their correlation in tumoral samples.
Table 2.
miRNA-mRNA correlation comparison between healthy and tumor samples.
| Cancer Symbol | +/+ | −/− | −/+ | +/− |
|---|---|---|---|---|
| LUSC | 1 | 0 | 0 | 0 |
| HNSC | 13 | 8 | 0 | 0 |
| KICH | 7 | 1 | 0 | 0 |
| KIRC | 27 | 20 | 3 | 3 |
| KIRP | 11 | 2 | 2 | 2 |
| BRCA | 17 | 13 | 2 | 4 |
| STAD | 2 | 1 | 1 | 0 |
| LIHC | 50 | 29 | 1 | 0 |
| PRAD | 17 | 64 | 3 | 0 |
| LUAD | 0 | 0 | 0 | 0 |
| BLCA | 0 | 3 | 0 | 0 |
| UCEC | 0 | 2 | 0 | 0 |
| THCA | 46 | 24 | 0 | 0 |
Notably, thyroid carcinoma (THCA) and lung squamous cell carcinoma (LUSC) are the only tumor types where the number of correlating pairs is higher in the tumor samples (Fig. 1). On the other hand, we also found a high number of positive correlations in the healthy samples (Pearson r > 0.4, adjusted p-value < 0.05) and a drastic reduction when compared with the corresponding tumoral one (as discussed below).
Firstly, we focused our attention on the negative correlation which describes the classic translational regulation carried out by miRNAs in the cytoplasm.
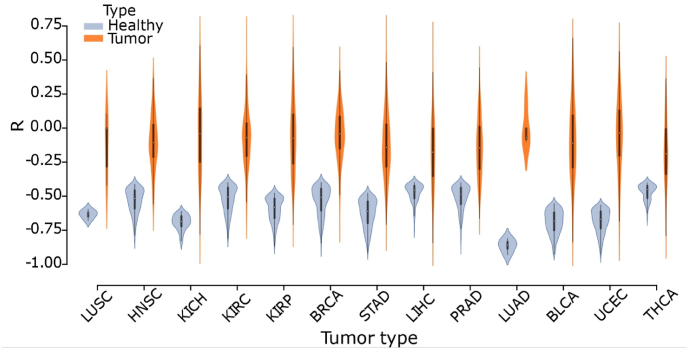
We found that only few miRNA-mRNA pairs preserve their negative correlation in the tumoral samples (1 in STAD and KICH, 2 in UCEC and KIRP, 3 in BLCA, 8 in HNSC, 13 in BRCA, 20 in KIRC, 24 in THCA, 29 in KIHC and 64 in PRAD). The Pearson coefficient distributions of the selected miRNA-mRNA pairs in healthy samples and their tumoral counterparts are shown in Fig. 2. The medians of Pearson correlation values are lower in the healthy (−0.59 on average) than in the tumoral samples (−0.09 on average) for the 13 analyzed cancers (t-test p < 0.05). Moreover, variance is much greater in the tumor samples compared with the healthy ones, showing a more tight regulation in the healthy tissue.
Fig. 2.
Distributions of correlation values of the selected miRNA-mRNA pairs in the healthy (R < −0.4, adjusted p-value < 0.05) and analogous pairs in the tumoral samples (blue and orange respectively).
Some of the miRNAs-mRNAs pairs that loose correlation in tumor samples are already known to be associated with cancer processes in several tissues. For example, ZEB1, which is targeted by the miR-200 family, regulates epithelial to mesenchymal transition in BRCA [32], and FERMT2-miR-200b pair has been associated with invasion in breast cancer [32,33]. Additionally, miRNAs like miR-182, miR-183, miR-21 are known to be associated with tumorigenesis [[34], [35], [36]].
3.2. miRNA-mRNA regulation switches
Interestingly, we found 12 miRNA-mRNA pairs showing a switch in their regulation from negative in the healthy to positive in the tumoral samples: 3 in prostate adenocarcinoma (PRAD), 1 in liver hepatocellular carcinoma (LIHC), 1 in stomach adenocarcinoma (STAD), 2 in breast invasive carcinoma (BRCA), 3 in kidney renal clear cell carcinoma (KIRC) and 2 in kidney renal papillary cell carcinoma (KIRP) (Table 2).
Table 2 The number of miRNA-mRNA pairs that showed: i. positive-positive (+/+), ii. negative-negative (−/−), iii. negative-positive (−/+), iv. positive-negative (±) correlation in healthy vs tumor samples respectively.
Out of 12 mRNA targets identified, 4 have a transcription factor activity, 1 is known to have a tumor suppressor activity and 2 are annotated as cell differentiation markers (Table 3).
Table 3.
miRNA-mRNA correlation switches between healthy and tumor samples.
| Tumor | miRNA | Target gene | Target function |
|---|---|---|---|
| KIRK | miR-22-3p | IRF5 | transcription factor |
| KIRK | miR-9-5p | MTHFD2 | Enzyme |
| KIRK | miR-483-3p | RASGRF1 | Rho guanine nucleotide exchange factor |
| KIRP | miR-34a-5p | ALDH2 | Aldehyde Dehydrogenase |
| KIRP | miR-200c-3p | ERRFI1 | EGFR family member |
| BRCA | miR-196a-5p | HOXC8 | transcription factor |
| BRCA | miR-223-3p | ABCB1 | cell differentiation marker |
| STAD | miR-126-3p | NFKBIA | transcription factor |
| LIHC | miR-194-5p | CDH2 | cell differentiation marker |
| PRAD | miR-204-5p | MEIS2 | transcription factor |
| PRAD | miR-29b-3p | TUBB2A | Tubulins |
| PRAD | miR-378a-5p | SUFU | tumor suppressor |
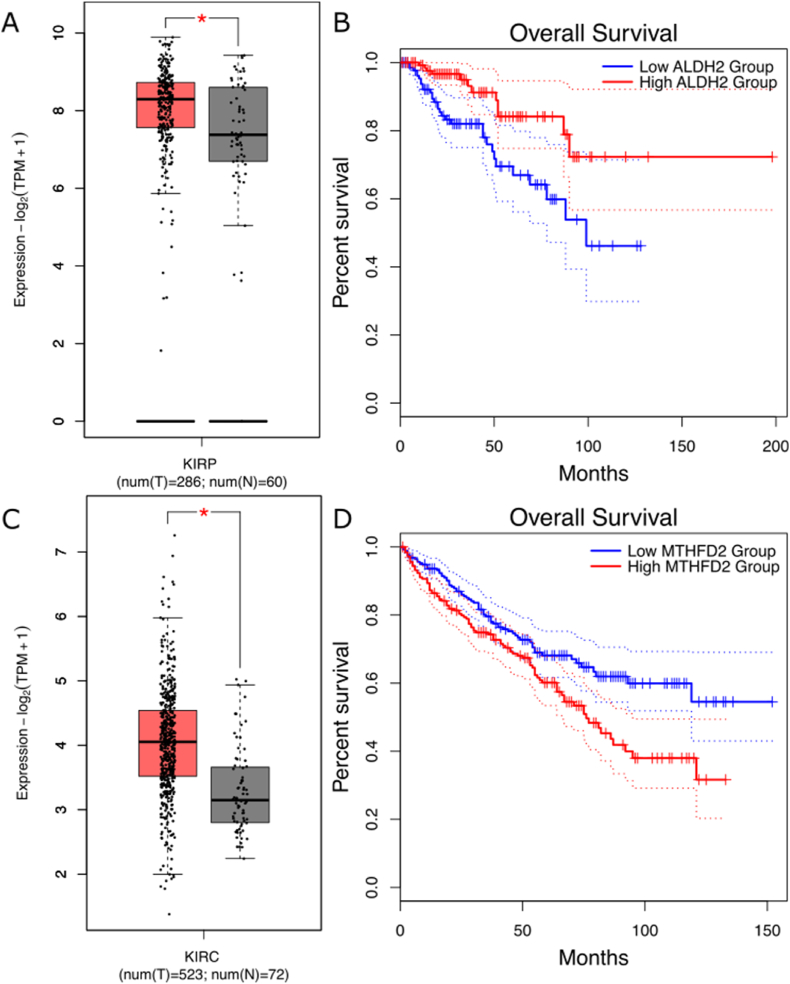
ALDH2 and MTHF2 showed a switch in the correlation with their respective miRNA regulator in kidney renal papillary cell carcinoma (KIRP) and Kidney Renal Clear Cell Carcinoma (KIRC) (Table 2). Both genes are up-regulated in tumors (TCGA data, log2FC > 0.8, p-value < 0.05) (Fig. 3a–c) and their expression is also associated with overall survival in KIRP (Kaplan-Meier log rank p < 0.05) and KIRC (Kaplan-Meier log rank p < 0.05) (Fig. 3b–d).
Fig. 3.
a) ALDH2 gene expression in KIRP (data from TCGA), the expression distributions for healthy and tumoral samples are shown respectively in grey and red. ALDH2 expression proves to be higher in the tumoral samples compared with the healthy one (log2FC > 0.8, p-value < 0.05); b) KIRP population was split into two groups based on ALDH2 expression median. Patients with overexpressed ALDH2 (red line) show an overall survival higher than the patients with a low expression of the same gene (blue line) (Kaplan-Meier log rank p < 0.05); c) MTHFD2 gene expression in KIRC (data from TCGA), the expression distributions for healthy and tumoral samples are shown respectively in red and grey. MTHFD2 expression is lower in the tumoral samples compared with the healthy one (log2FC > 0.80 and p < 0.05); d) KIRC population was split into two groups based on MTHFD2 expression median. Patients with overexpressed MTHFD2 (red line) show an overall survival lower than the patients with a low expression of the same gene (blue line) (Kaplan-Meier log rank p < 0.05).
Interestingly, the expression of MTHFD2 is already known to be associated with poor prognosis, migration and invasion in the renal cell carcinoma patients [37]. The regulation switch of these miRNA-mRNA interactions could suggest a disruption of the regulation of the miRNA on its target and subsequently the possibility to give a new malignant function to the cells.
3.3. miRNA-mRNA pairs that are consistently dysregulated across multiple cancer types
Most of the identified miRNA-mRNA dysregulated interactions are shared among different cancer types. 2865 unique miRNA-mRNA pairs were identified across 13 cancer types, ≈ 40% of these pairs showed a loss of correlation in the tumoral samples in at least 2 out of the 13 analyzed cancers.
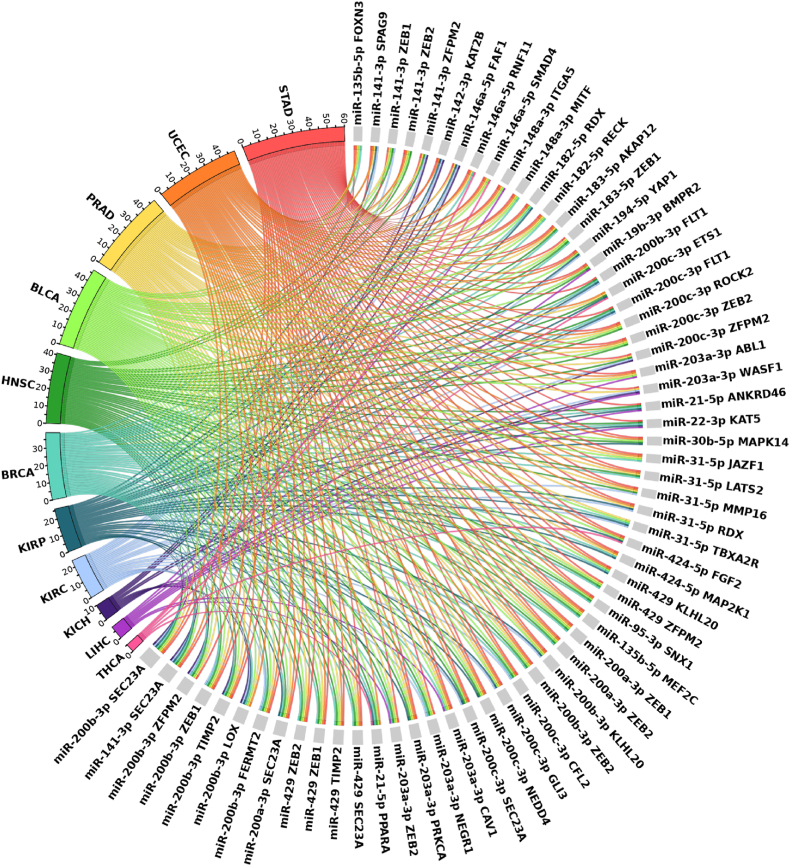
The top 64 pairs, sorted according to the descending number of tumors in which they are deregulated, with a loss of correlation in at least 5 different cancer types are shown in Fig. 4.
Fig. 4.
The circle plot displays all miRNA-mRNA interactions that are dysregulated across at least 5 different cancer types. Identified pairs are listed counterclockwise according to the descending number of tumors in which they are dysregulated. miRNA-mRNA pairs are reported in light grey. Colored chords identify the cancer in which the miRNA-mRNA relationship is lost.
miR-200b and SEC23a show a loss of correlation in 9 different cancer types (stomach, uterus, prostate, bladder, head and neck, breast, and kidney (KIRP, KIRC and KICH)). This gene and miR-200 family have been extensively studied and their expression has been associated with metastasis in different cancer types (lung, breast, and bladder) [38].
Likewise, we found a loss of correlation between ZEB1 and its regulator miR-429 in 6 different cancer types (head and neck, breast, stomach, prostate, bladder, uterus). This interaction has been associated with epithelial to mesenchymal transition in breast cancer, suggesting that the ZEB1-miR-429 axis could play a key role in promoting tumor progression [32].
3.4. Our global analysis thus extends these observations to additional tumor types
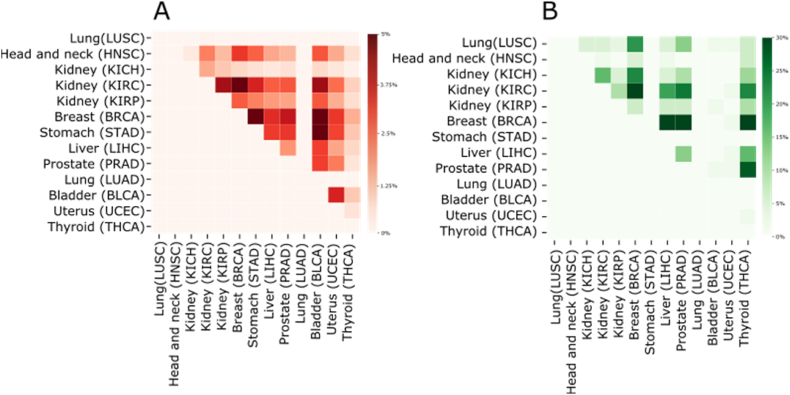
As expected, we found shared miRNA-mRNA dysregulated pairs among different tumors of the same tissue (KICH, KIRC, KIRP). However anatomically different cancers also share multiple dysregulated interactions. As shown in Fig. 5, ∼30% of miRNA-mRNA dysregulated interactions are shared between stomach (STAD) and bladder (BLCA), while ∼20% are shared between uterus (UCEC) and bladder (BLCA) (Fig. 5).
Fig. 5.
Similarity between cancer types, according to the number of dysregulated miRNA-mRNA pairs they share.
The green scale represents the magnitude of the similarity between 2 different tissues, in dark green are shown the couple of tumors that share a high number of miRNA-mRNA interaction that lost their negative correlation in the tumoral samples. The red scale represents the magnitude of the similarity between 2 different tissues, in dark red are shown the couple of tumors that share a high number of miRNA-mRNA interaction that lost their positive correlation in the tumoral samples.
3.5. Functional enrichment
We performed a gene ontology enrichment analysis to describe the function of genes that either lost or preserved their negative correlation with the cognate miRNA in tumor samples (thus possibly leading to lack of repression or over-expression in cancer). In particular we found an enrichment in positive regulation of cell division and proliferation, cell growth and migration, blood vessel development and remodeling, movement of cellular or subcellular component for the genes that lose their correlation with the cognate miRNA while genes that preserved their correlation are enriched in cell-cell adhesion, cell aging and regulation of apoptotic processes. Given the high number of genes involved in the lost of correlation with their cognate miRNA and taking in to account the biological processes in which they are involved, we can hypothesize that the loss of regulation carried out by the miRNA on its own targets may be the cause of the tumor onset or cancer progression.
3.6. miRNA-mRNA and overall survival correlation in breast cancer patients
We estimated the overall survival (OS) over time on BRCA patients considering both the high number of matched healthy and tumor biopsies available and the high number of miRNA-mRNA dysregulated pairs identified with the previous analysis. To identify the clinical relevance of miRNA-mRNA gene-target pairs we split the population in four groups according to the miRNA and the mRNA median expression values (i. miRNA under expressed - mRNA over expressed; ii. miRNA under expressed - mRNA under expressed; iii. miRNA over expressed - mRNA over expressed; iv. miRNA over expressed - mRNA under expressed). Then we performed survival analysis comparing the four identified groups in pairs. Out of 454 miRNA-mRNA dysregulated interactions in BRCA, 2 were associated with OS (p < 0.05) while using single miRNA or mRNA as discriminant no association was found.
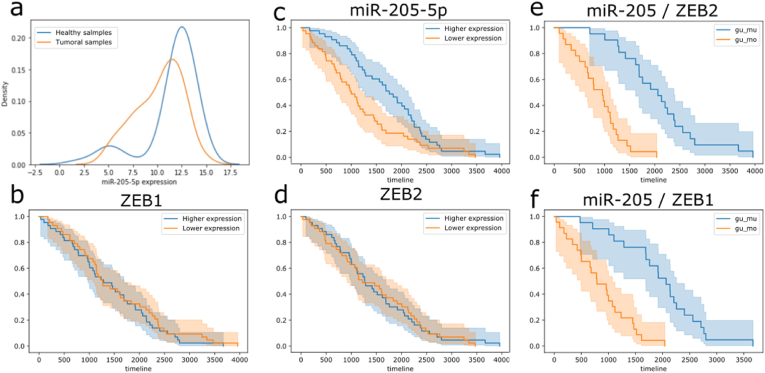
miR-205 and ZEB2 showed a negative correlation in healthy samples (R = −0.48, p-value < 0.05) while no correlation was found in tumors. Individually miR-205 and ZEB2 expression were not found to be associated with OS (Fig. 6c and d). On the other hand, when using both miRNA-mRNA expression levels to split the population in 2 cohorts, we found a significant association between miR-205 over-expression/ZEB2 under-expression and low survival rate (Kaplan-Meier log rank adjusted p-value < 0.05) (Fig. 6f). Likewise, miR-205 and ZEB1 showed a negative correlation in the healthy samples (R = −0.52, p < 0.05) while no correlation was found in tumoral samples, also in this case individually miR-205 and ZEB1 expression resulted not associated with OS (Fig. 6b and c). However, miR-205 over-expression/ZEB1 under-expression were significantly associated with low survival rate (Kaplan-Meier log rank adjusted p-value < 0.05) (Fig. 6f).
Fig. 6.
BRCA samples miR-205 expression distribution in healthy (blue) and tumoral (orange) samples (a); BRCA patients have been split into two groups based on ZEB1, ZEB2, miR-205-5p median (respectively in b, c, d). The blue line represents the group with higher expression of the analyzed feature while the orange line represents the group associated with lower expression. No significant association between expression and overall survival have been found using ZEB1, ZEB2, miR-205-5p individually (Kaplan-Meier log rank p > 0.05). The BRCA population has been split using both miR-205/ZEB1 and miR-205/ZEB2 expressions (e, f respectively). In both cases miRNA overexpression and gene under expression appears to be significantly associated with low survival rate (orange line, p-value < 0.05).
In this study we showed the prognostic power of 2 new markers for BRCA patients, the expression of these miRNA-mRNA dysregulated pairs resulted statistically associated with OS rate (Kaplan-Meier log rank p < 0.05).
3.7. Loss of miRNA-mRNA positive correlations in several cancer types
In contrast to the general assumption that miRNA-mediated downregulation is a one-way process leading to decreased mRNA stability and/or translational inhibition, several recent studies highlighted how miRNAs could upregulate gene expression in specific cell types and conditions [9,10]. Two different mechanisms underlying this process were described by Liang and collaborators: i) cytoplasmic mRNA stabilization or ii) transcription activation [39]. Accordingly, we focused our analysis on positive correlations between miRNAs and their targets.
On average we found 380 miRNA-mRNA positively correlated pairs across all healthy samples (Pearson r > 0.4, adjusted p-value < 0.05). Conversely, a drastic reduction in the number of positive correlations was found in tumor tissue (on average 63 pairs within the same thresholds).
The expression of miR-183-5p is positively correlated with that of its target PDCD6 in 8 out of 13 healthy tissues (head and neck, kidney, breast, stomach, liver, prostate, and bladder) and this relationship is completely lost in the corresponding tumor samples. PDCD6 mediates apoptosis via p53 dependent and independent pathways, and its expression is dysregulated in different cancers [40].
More generally, the genes for which a loss of positive regulation was observed are enriched in biological processes such as cell adhesion, apoptotic process and inflammatory or immune response, cell division and DNA replication, recombination, and repair (data not shown).
The positive correlation between the expression of miRNAs and their targets suggests how the miRNA could promote an increase in the expression level of the targets themselves in the physiological condition, following the molecular mechanisms described in the last years [9,10]. Given that the positively regulated genes are involved in key processes for the cell homeostasis we can hypothesize that the disruption of the positive regulation exerted by miRNA could be associated with an altered state for the cell possibly causing the cancer onset or progression.
4. Discussion
The miRNA-mRNA regulatory network has been shown to be associated with the onset and progression of several human diseases including cancer [27,[41], [42], [43], [44], [45], [46]].
In this work we investigated the relationship between the expression of miRNAs and that of their target mRNAs, first in the tumoral samples and later in the healthy biopsies simulating a physiological scenario. Comparing the two conditions we highlighted miRNA-mRNA interactions dysregulated in cancer.
From a computational point of view, regulation of gene expression by miRNA has been extensively studied in the last years using several methods [19,[47], [48], [49]]. Negative or positive regulation exerted by miRNA on its targets could result in a negative or positive correlation respectively between the expression of the miRNA and its targets. With this analysis, we shed light on the miRNA-mRNA regulation landscape comparing the healthy and tumoral states. We found a global reduction of both positive and negative miRNA-mRNA interactions in the tumor samples when compared with the corresponding healthy samples for 11 out of 13 cancers analyzed. We also highlighted some miRNA-mRNA pairs with a switch in the sign of correlation between healthy and tumoral samples in some tissues. These findings suggest possible changes in the regulation operated by a miRNA on its targets, moving from the target translational repression or degradation in the healthy condition to a putative target upregulation in the tumoral or vice versa.
Interestingly, we demonstrate how this loss of correlation for a single miRNA-mRNA pair quite often is not a tumor specific related event, highlighting how in different tissues the same changes may occur in the miRNA regulatory landscape resulting in the determination of cancer onset, stage or progression.
Since the expression changes of miRNA or mRNA regulated by various factors could cause the changes of correlation between them, the factors which result in the reduced correlation in tumor tissues were indistinct.
The loss of miRNA-target correlation could be explained by several hypotheses. Firstly, miRNAs are globally less expressed in tumor tissues, and this could influence the regulation of its target genes. Only the most abundantly expressed miRNAs could affect their target mRNAs stability by effectively binding to the available mRNA target sites [50].
Additionally, transcription factors and endogenous long noncoding RNA could also mediate mRNAs regulation exerted by miRNAs [51,52].
Moreover, we tested the clinical relevance of the dysregulated miRNA-mRNA pairs we found in BRCA patients. A significant association with overall survival was found for 2 miRNA-mRNA pairs, these findings highlight putative biomarkers which were not observed in previous studies. For both ZEB1 and ZEB2 we found no association between their expression and overall survival rate, on the contrary, samples where these 2 genes were downregulated and at the same time present an upregulation of their cognate miRNA showed a low survival rate compared with the samples where the miRNAs were downregulated.
Finally, with this analysis we highlight the changes in the miRNA-mRNA regulation that occur between the tumoral and the healthy samples. The miRNA-mRNA critical pairs, for which a loss of correlation in the tumoral samples was found, result enriched in biological processes and molecular functions such as cell proliferation and migration, regulation of cell death and metabolism, epithelial-mesenchymal transition, IL2 and MTOR signaling, inflammatory response and angiogenesis. Our analysis provides both pan-cancer and cancer specific dysregulated miRNA-mRNA interactions in cancer, a starting point to better understand the miRNA regulatory network and its relationship with cancer onset and progression. Experimentally validated miRNA-mRNA interactions, already associated with a specific cancer type, show a similar deregulation in other types of cancers suggesting a shared mechanism across several tumors. Experimental validation of these results could be useful for the identification of new critical miRNA-mRNA pairs not yet described in the literature that can be biologically and clinically meaningful.
Data availability
Datasets and scripts used in this manuscript are available upon request.
Funding
AIRC project [to MHC] (grant number IG 23539).
CRediT authorship contribution statement
Gerardo Pepe: Investigation, Writing – original draft, Supervision. Luca Parca: Conceptualization, Supervision. Gabriele Ausiello: Supervision. Manuela Helmer-Citterich: Funding acquisition, Supervision.
Declaration of competing interest
None declared.
Acknowledgment
We thank prof PF Gherardini and dr Silvia Galardi for insightful discussions.
References
- 1.Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Y. Zeng, B.R. Cullen, The biogenesis and function of MicroRNAs, Gene Expression and Regulation. (n.d.) 481–492. 10.1007/978-0-387-40049-5_29. [DOI]
- 3.Pratt A.J., MacRae I.J. The RNA-induced silencing complex: a versatile gene-silencing machine. J. Biol. Chem. 2009;284:17897–17901. doi: 10.1074/jbc.R900012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanjay S., Girish C. Role of miRNA and its potential as a novel diagnostic biomarker in drug-induced liver injury. Eur. J. Clin. Pharmacol. 2017;73:399–407. doi: 10.1007/s00228-016-2183-1. [DOI] [PubMed] [Google Scholar]
- 5.Vishnoi A., Rani S. MiRNA biogenesis and regulation of diseases: an overview. Methods Mol. Biol. 2017;1509:1–10. doi: 10.1007/978-1-4939-6524-3_1. [DOI] [PubMed] [Google Scholar]
- 6.Pasquinelli A.E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 7.Iwakawa H.-O., Tomari Y. The functions of MicroRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015;25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Wilczynska A., Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22:22–33. doi: 10.1038/cdd.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasudevan S. Posttranscriptional upregulation by MicroRNAs. Wiley Interdiscipl. Rev. RNA. 2012;3:311–330. doi: 10.1002/wrna.121. [DOI] [PubMed] [Google Scholar]
- 10.Xiao M., Li J., Li W., Wang Y., Wu F., Xi Y., Zhang L., Ding C., Luo H., Li Y., Peng L., Zhao L., Peng S., Xiao Y., Dong S., Cao J., Yu W. MicroRNAs activate gene transcription epigenetically as an enhancer trigger. RNA Biol. 2017;14:1326–1334. doi: 10.1080/15476286.2015.1112487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., Xue C., Marinov G.K., Khatun J., Williams B.A., Zaleski C., Rozowsky J., Röder M., Kokocinski F., Abdelhamid R.F., Alioto T., Antoshechkin I., Baer M.T., Bar N.S., Batut P., Bell K., Bell I., Chakrabortty S., Chen X., Chrast J., Curado J., Derrien T., Drenkow J., Dumais E., Dumais J., Duttagupta R., Falconnet E., Fastuca M., Fejes-Toth K., Ferreira P., Foissac S., Fullwood M.J., Gao H., Gonzalez D., Gordon A., Gunawardena H., Howald C., Jha S., Johnson R., Kapranov P., King B., Kingswood C., Luo O.J., Park E., Persaud K., Preall J.B., Ribeca P., Risk B., Robyr D., Sammeth M., Schaffer L., See L.-H., Shahab A., Skancke J., Suzuki A.M., Takahashi H., Tilgner H., Trout D., Walters N., Wang H., Wrobel J., Yu Y., Ruan X., Hayashizaki Y., Harrow J., Gerstein M., Hubbard T., Reymond A., Antonarakis S.E., Hannon G., Giddings M.C., Ruan Y., Wold B., Carninci P., Guigó R., Gingeras T.R. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts T.C. The MicroRNA biology of the Mammalian nucleus. Mol. Ther. Nucleic Acids. 2014;3 doi: 10.1038/mtna.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou Q., Liang Y., Luo H., Yu W. miRNA-mediated RNAa by targeting enhancers. Adv. Exp. Med. Biol. 2017;983:113–125. doi: 10.1007/978-981-10-4310-9_8. [DOI] [PubMed] [Google Scholar]
- 14.Ali H.E.A., Lung P.-Y., Sholl A.B., Gad S.A., Bustamante J.J., Ali H.I., Rhim J.S., Deep G., Zhang J., Abd Elmageed Z.Y. Dysregulated gene expression predicts tumor aggressiveness in African-American prostate cancer patients. Sci. Rep. 2018;8:16335. doi: 10.1038/s41598-018-34637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumgart S.J., Nevedomskaya E., Haendler B. Dysregulated transcriptional control in prostate cancer. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20122883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Z., Partridge V., Sousares M., Shelton S.D., Holland C.L., Pertsemlidis A., Du L. microRNA-2110 functions as an onco-suppressor in neuroblastoma by directly targeting Tsukushi. PLoS One. 2018;13 doi: 10.1371/journal.pone.0208777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W., Li G., Fan Z., Liu T. Tumor-suppressive microRNA-452 inhibits migration and invasion of breast cancer cells by directly targeting RAB11A. Oncol. Lett. 2017;14:2559–2565. doi: 10.3892/ol.2017.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J., Li Y., Lu J., Pan T., Ding N., Wang Z., Shao T., Zhang J., Wang L., Li X. The mRNA related ceRNA-ceRNA landscape and significance across 20 major cancer types. Nucleic Acids Res. 2015;43:8169–8182. doi: 10.1093/nar/gkv853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paci P., Colombo T., Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst. Biol. 2014;8:83. doi: 10.1186/1752-0509-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anglani R., Creanza T.M., Liuzzi V.C., Piepoli A., Panza A., Andriulli A., Ancona N. Loss of connectivity in cancer co-expression networks. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou M., Wang X., Shi H., Cheng L., Wang Z., Zhao H., Yang L., Sun J. Characterization of long non-coding RNA-associated ceRNA network to reveal potential prognostic lncRNA biomarkers in human ovarian cancer. Oncotarget. 2016;7:12598–12611. doi: 10.18632/oncotarget.7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yi Y., Zhao Y., Li C., Zhang L., Huang H., Li Y., Liu L., Hou P., Cui T., Tan P., Hu Y., Zhang T., Huang Y., Li X., Yu J., Wang D. RAID v2.0: an updated resource of RNA-associated interactions across organisms. Nucleic Acids Res. 2017;45:D115–D118. doi: 10.1093/nar/gkw1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Junge A., Refsgaard J.C., Garde C., Pan X., Santos A., Alkan F., Anthon C., von Mering C., Workman C.T., Jensen L.J., Gorodkin J. RAIN: RNA–protein association and interaction networks. Database. 2017 doi: 10.1093/database/baw167. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The RNAcentral Consortium, Petrov A.I., Kay S.J.E., Kalvari I., Howe K.L., Gray K.A., Bruford E.A., Kersey P.J., Cochrane G., Finn R.D., Bateman A., Kozomara A., Griffiths-Jones S., Frankish A., Zwieb C.W., Lau B.Y., Williams K.P., Chan P.P., Lowe T.M., Cannone J.J., Gutell R., Machnicka M.A., Bujnicki J.M., Yoshihama M., Kenmochi N., Chai B., Cole J.R., Szymanski M., Karlowski W.M., Wood V., Huala E., Berardini T.Z., Zhao Y., Chen R., Zhu W., Paraskevopoulou M.D., Vlachos I.S., Hatzigeorgiou A.G., Ma L., Zhang Z., Puetz J., Stadler P.F., McDonald D., Basu S., Fey P., Engel S.R., Cherry J.M., Volders P.-J., Mestdagh P., Wower J., Clark M.B., Quek X.C., Dinger M.E. RNAcentral: a comprehensive database of non-coding RNA sequences. Nucleic Acids Res. 2017;45:D128–D134. doi: 10.1093/nar/gkw1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andrés-León E., Cases I., Alonso S., Rojas A.M. Novel miRNA-mRNA interactions conserved in essential cancer pathways. Sci. Rep. 2017;7:46101. doi: 10.1038/srep46101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin X., Zhang F., Guo Z., Kong W., Wang Y. Integrative analysis of miRNA and mRNA expression profiles reveals a novel mRNA/miRNA signature to improve risk classification for patients with gastric cancer. Oncol. Lett. 2019 doi: 10.3892/ol.2019.10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., Yu X., He Y., Meng Y., Liang J., Huang L., Du H., Wang X., Liu W. Integrated analysis of MicroRNA (miRNA) and mRNA profiles reveals reduced correlation between MicroRNA and target gene in cancer. Biomed Res. Int. 2018. 2018 doi: 10.1155/2018/1972606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bland J.M., Altman D.G. Statistics notes: survival probabilities (the Kaplan-Meier method) BMJ. 1998;317:1572–1580. doi: 10.1136/bmj.317.7172.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019;47:W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 33.Yu Y., Wu J., Guan L., Qi L., Tang Y., Ma B., Zhan J., Wang Y., Fang W., Zhang H. Kindlin 2 promotes breast cancer invasion via epigenetic silencing of the microRNA200 gene family. Int. J. Cancer. 2013;133:1368–1379. doi: 10.1002/ijc.28151. [DOI] [PubMed] [Google Scholar]
- 34.Reddy K.B. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi: 10.1186/s12935-015-0185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct. Targeted Therapy. 2016;1:1–9. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge X., Liu X., Lin F., Li P., Liu K., Geng R., Dai C., Lin Y., Tang W., Wu Z., Chang J., Lu J., Li J. MicroRNA-421 regulated by HIF-1α promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget. 2016;7:24466–24482. doi: 10.18632/oncotarget.8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin H., Huang B., Wang H., Liu X., Hong Y., Qiu S., Zheng J. MTHFD2 overexpression predicts poor prognosis in renal cell carcinoma and is associated with cell proliferation and vimentin-Modulated migration and invasion. Chem. Pharm. Bull. 2018;51:991–1000. doi: 10.1159/000495402. [DOI] [PubMed] [Google Scholar]
- 38.Korpal M., Ell B.J., Buffa F.M., Ibrahim T., Blanco M.A., Celià-Terrassa T., Mercatali L., Khan Z., Goodarzi H., Hua Y., Wei Y., Hu G., Garcia B.A., Ragoussis J., Amadori D., Harris A.L., Kang Y. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat. Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Y., Xu P., Zou Q., Luo H., Yu W. An epigenetic perspective on tumorigenesis: loss of cell identity, enhancer switching, and NamiRNA network. Semin. Cancer Biol. 2019;57:1–9. doi: 10.1016/j.semcancer.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 40.Hashemi M., Bahari G., Markowski J., Małecki A., Łos M.J., Ghavami S. Association of PDCD6 polymorphisms with the risk of cancer: evidence from a meta-analysis. Oncotarget. 2018;9:24857–24868. doi: 10.18632/oncotarget.25324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng J., Zhuo H., Xu M., Wang L., Xu H., Peng J., Hou J., Lin L., Cai J. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J. Transl. Med. 2018;16:216. doi: 10.1186/s12967-018-1582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue J., Zhou D., Poulsen O., Hartley I., Imamura T., Xie E.X., Haddad G.G. Exploring miRNA-mRNA regulatory network in cardiac pathology in Na/H exchanger isoform 1 transgenic mice. Physiol. Genom. 2018;50:846–861. doi: 10.1152/physiolgenomics.00048.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J., Song H., Cao K., Song J., Zhou J. Comprehensive analysis of Helicobacter pylori infection-associated diseases based on miRNA-mRNA interaction network. Briefings Bioinf. 2019;20:1492–1501. doi: 10.1093/bib/bby018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar V., Kumar V., Chaudhary A.K., Coulter D.W., McGuire T., Mahato R.I. Impact of miRNA-mRNA profiling and their correlation on Medulloblastoma tumorigenesis. Mol. Ther. Nucleic Acids. 2018;12:490–503. doi: 10.1016/j.omtn.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li C., Hong Z., Ou M., Zhu X., Zhang L., Yang X. Integrated miRNA-mRNA expression profiles revealing key molecules in ovarian cancer based on bioinformatics analysis. Biomed Res. Int. 2021. 2021 doi: 10.1155/2021/6673655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X., Zhu X., Wei Z., Lv Q. Integrated mRNA-Seq and miRNA-Seq analysis of PLCγ2-overexpressing hepatocarcinoma cells and identification of the associated miRNA-mRNA network. J. Cell. Biochem. 2019;120:19878–19890. doi: 10.1002/jcb.29294. [DOI] [PubMed] [Google Scholar]
- 47.Lee E., Ito K., Zhao Y., Schadt E.E., Irie H.Y., Zhu J. Inferred miRNA activity identifies miRNA-mediated regulatory networks underlying multiple cancers. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.List M., Dehghani Amirabad A., Kostka D., Schulz M.H. Large-scale inference of competing endogenous RNA networks with sparse partial correlation. Bioinformatics. 2019;35:i596–i604. doi: 10.1093/bioinformatics/btz314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang S., Ning Q., Zhang G., Sun H., Wang Z., Li Y. Construction of differential mRNA-lncRNA crosstalk networks based on ceRNA hypothesis uncover key roles of lncRNAs implicated in esophageal squamous cell carcinoma. Oncotarget. 2016;7:85728–85740. doi: 10.18632/oncotarget.13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J., Le T.D., Liu L., He J., Li J. A novel framework for inferring condition-specific TF and miRNA co-regulation of protein–protein interactions. Gene. 2016;577:55–64. doi: 10.1016/j.gene.2015.11.023. [DOI] [PubMed] [Google Scholar]
- 51.Peng C., Wang M., Shen Y., Feng H., Li A. Reconstruction and analysis of transcription factor–miRNA Co-regulatory feed-forward loops in human cancers using filter-wrapper feature selection. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sui J., Li Y.-H., Zhang Y.-Q., Li C.-Y., Shen X., Yao W.-Z., Peng H., Hong W.-W., Yin L.-H., Pu Y.-P., Liang G.-Y. Integrated analysis of long non-coding RNA-associated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int. J. Oncol. 2016;49:2023–2036. doi: 10.3892/ijo.2016.3716. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets and scripts used in this manuscript are available upon request.