Abstract
Background
Survival heterogeneity and limited trial follow-up present challenges for estimating lifetime benefits of oncology therapies. This study used CheckMate 067 (NCT01844505) extended follow-up data to assess the predictive accuracy of standard parametric and flexible models in estimating the long-term overall survival benefit of nivolumab plus ipilimumab (an immune checkpoint inhibitor combination) in advanced melanoma.
Methods
Six sets of survival models (standard parametric, piecewise, cubic spline, mixture cure, parametric mixture, and landmark response models) were independently fitted to overall survival data for treatments in CheckMate 067 (nivolumab plus ipilimumab, nivolumab, and ipilimumab) using successive data cuts (28, 40, 52, and 60 mo). Standard parametric models allow survival extrapolation in the absence of a complex hazard. Piecewise and cubic spline models allow additional flexibility in fitting the hazard function. Mixture cure, parametric mixture, and landmark response models provide flexibility by explicitly incorporating survival heterogeneity. Sixty-month follow-up data, external ipilimumab data, and clinical expert opinion were used to evaluate model estimation accuracy. Lifetime survival projections were compared using a 5% discount rate.
Results
Standard parametric, piecewise, and cubic spline models underestimated overall survival at 60 mo for the 28-mo data cut. Compared with other models, mixture cure, parametric mixture, and landmark response models provided more accurate long-term overall survival estimates versus external data, higher mean survival benefit over 20 y for the 28-mo data cut, and more consistent 20-y mean overall survival estimates across data cuts.
Conclusion
This case study demonstrates that survival models explicitly incorporating survival heterogeneity showed greater accuracy for early data cuts than standard parametric models did, consistent with similar immune checkpoint inhibitor survival validation studies in advanced melanoma. Research is required to assess generalizability to other tumors and disease stages.
Highlights
Given that short clinical trial follow-up periods and survival heterogeneity introduce uncertainty in the health technology assessment of oncology therapies, this study evaluated the suitability of conventional parametric survival modeling approaches as compared with more flexible models in the context of immune checkpoint inhibitors that have the potential to provide lasting survival benefits.
This study used extended follow-up data from the phase III CheckMate 067 trial (NCT01844505) to assess the predictive accuracy of standard parametric models in comparison with more flexible methods for estimating the long-term survival benefit of the immune checkpoint inhibitor combination of nivolumab plus ipilimumab in advanced melanoma.
Mixture cure, parametric mixture, and landmark response models provided more accurate estimates of long-term overall survival versus external data than other models tested.
In this case study with immune checkpoint inhibitor therapies in advanced melanoma, extrapolation models that explicitly incorporate differences in cancer survival between observed or latent subgroups showed greater accuracy with both early and later data cuts than other approaches did.
Keywords: advanced melanoma, immune checkpoint inhibitor, immuno-oncology, ipilimumab, nivolumab, survival heterogeneity, survival modeling
Introduction
An increasing number of health technology assessment agencies use cost-effectiveness analysis to inform technology appraisal decisions for new cancer treatments. This type of economic evaluation produces an estimate of the cost-effect tradeoffs between 2 or more competing interventions, often over a lifetime.
Most randomized controlled trials of oncology agents have relatively short follow-up at the time of submission to health technology assessment agencies, with heavy right-censoring often observed in the Kaplan-Meier survivor function. Consequently, appropriate survival extrapolation methods are required to derive reliable estimates of the long-term effects of these treatments. The conventional approach has been to use common standard parametric models to extrapolate time-to-event data collected in the trial, using a selection algorithm developed by Latimer. 1 In the interest of parsimony, some researchers have suggested that the exponential distribution should be considered the default, unless contrary evidence is provided that others provide more accurate projections. 2
The mechanisms of action for immuno-oncology therapies such as immune checkpoint inhibitors can lead to delayed clinical effects (i.e., response) and the potential for long-term survivorship in a subgroup of patients who achieve an immunological response.3,4 The shape of the Kaplan-Meier curves for immune checkpoint inhibitors is typically characterized by an initial period of nonseparation of the curves when compared with conventional or targeted therapies due to delayed response, a subsequent period of separation as responses occur, and a final period with a plateau driven by a persistent treatment effect among those who respond.3,5 This variation in cancer survival may be indicative of latent prognostic factors or differences in treatment efficacy in observed or latent patient subgroups and result in a hazard function that standard parametric models may be unable to accurately capture.
A number of alternative, more flexible models have been described in the National Institute for Health and Care Excellence Technical Support Document 21. 6 This document provides a critique of the more sophisticated extrapolation methods that have been applied in technology appraisals of immuno-oncology treatments, including flexible parametric (piecewise and cubic spline), mixture cure, parametric mixture, and landmark response models. A simulation study was presented by the National Institute for Health and Care Excellence to highlight the strengths and weaknesses of a subset of these different approaches and their potential impact on the estimates of lifetime survival. Technical challenges and lack of access to suitable individual patient data precluded an assessment of the performance of several flexible models identified by the researchers.
The design of this study was informed by previous research that investigated the predictive accuracy of standard versus more flexible extrapolation methods used to evaluate the long-term benefit of immune checkpoint inhibitors.7–10 The main aim of this study was to retrospectively assess the performance of 6 survival extrapolation methods fitted to extended follow-up data from CheckMate 067, a phase III randomized controlled trial that compared the use of the immune checkpoint inhibitor combination nivolumab (a programmed death 1 checkpoint inhibitor) plus ipilimumab (an anti–cytotoxic T-cell lymphocyte-antigen 4 checkpoint inhibitor) or nivolumab alone with ipilimumab alone in previously untreated patients with advanced melanoma. 11 Successive artificial data cuts with varying lengths of follow-up were created to assess how the predictive accuracy of the methods may vary based on the maturity of the data.
Methods
Study Design
This study estimated and compared the following 6 survival modeling techniques that have been used to estimate the long-term survival of patients with cancer treated with immune checkpoint inhibitors: standard parametric, piecewise, cubic spline, mixture cure, parametric mixture, and landmark response models (Figure 1).7,8,10,12 Standard parametric models are more widely used and established for health technology assessments than the other methods and are typically considered the starting point for survival extrapolation in the absence of a complex hazard. Because standard hazard functions are less complex, they are generally easier to interpret and more parsimonious. 1 The other modeling approaches are increasingly considered as alternatives to standard parametric models for modeling survival data for immune checkpoint inhibitors because they allow for varying degrees of flexibility to fit complex hazards caused by delayed response and long-term survivorship.1,6 Piecewise and cubic spline models allow for additional flexibility mechanistically in fitting the hazard function.5–8,12 Mixture cure, parametric mixture, and landmark response models provide flexibility by explicitly assuming survival heterogeneity.5–8,12 In fact, parametric mixture models subsume standard parametric and mixture cure models as special cases. Detailed descriptions of the parameterization for each modeling approach are provided in the Supplemental Material.
Figure 1.
Study design. Blue boxes indicate methods that allowed for flexibility mechanistically. Purple boxes indicate methods that allowed for flexibility by accounting for survival heterogeneity. CSM, cubic spline model, MCM, mixture cure model, PMM, parametric mixture model, PSM, standard parametric model, PWM, piecewise model, RBLM, landmark response model.
Database
In the CheckMate 067 trial (NCT01844505), nivolumab either as a monotherapy (n = 316) or in combination with ipilimumab (n = 314) was compared with ipilimumab monotherapy (n = 315) in patients with previously untreated, unresectable, or advanced stage III or IV melanoma. Details on the study design and patient population have been presented previously. 13 Results from this trial showed significant improvement in long-term survival outcomes with the nivolumab-containing treatments, with sustained overall survival and progression-free survival at 4 and 5 y of follow-up. Overall survival rates at 4 and 5 y, respectively, were 53% and 52% in the nivolumab plus ipilimumab arm and 46% and 44% in the nivolumab arm, as compared with 30% and 26% in the ipilimumab arm. 11
In this case study, overall survival data from the most recent data available at the time of analysis were analyzed (minimum follow-up, 60 mo; Figure 2A). Three artificial data cuts were created based on this data set.
Figure 2.
Overall survival by data cut. (A) Kaplan-Meier curves; (B) smoothed hazard functions. Shaded areas represent 95% CIs. CI, confidence interval; WHO, World Health Organization.
The earliest data cut corresponded with 28 mo of follow-up and data that were available at the time of the initial health technology assessment review for nivolumab plus ipilimumab in Canada (Canadian Agency for Drugs and Technologies in Health). In that submission, standard parametric models were fitted to the available data for each arm and extrapolated out to 20 y using a log-normal distribution.14,15 The results from the models used in this health technology assessment review are used as a point of reference for the other models under consideration here. The other 2 data cuts were set to be 12 and 24 mo after this initial data cut (up to 40 and 52 mo, respectively) to reflect varying degrees of data maturity. Results for these 2 intermediate data cuts are presented in the Supplemental Material.
Model Estimation and Selection
Standard survival analysis involves generating Kaplan-Meier curves, testing of proportional hazards and accelerated failure time assumptions, and fitting parametric survival models (either standard or more complex). Aligned with the National Institute for Health and Care Excellence Technical Support Document 14, 1 different parametric survival functions—including exponential, Weibull, Gompertz, log-logistic, log-normal, gamma, and generalized gamma—were evaluated for standard parametric, piecewise, mixture cure, and landmark response model approaches. For cubic spline models, 1- and 2-knot hazards, odds, and probit models were evaluated. For parametric mixture models, 15 potential combinations of exponential, Weibull, log-logistic, log-normal, and gamma distributions were tested, assuming that the trial population consisted of 2 mutually exclusive and exhaustive latent subgroups in each arm. Detailed descriptions of all modeling techniques and distributional assumptions are provided in the Supplemental Material.
Model selection was informed by 3 criteria: goodness-of-fit statistics, visual inspection of the predicted survival/hazard to the observed survival/hazard, and alignment with external long-term data (available for ipilimumab only). Given the similar mechanism of action across the treatments, the same distribution was selected for all 3 arms within a given method unless there was strong statistical counterindication from the models to do otherwise (i.e., goodness-of-fit assessments strongly indicated one distribution for one treatment and a different distribution for another treatment).
Akaike and Bayesian information criteria statistics were generated separately for each arm and for each model to inform model selection. Specifically, when the Akaike and Bayesian information criteria statistics were similar, they were summed across the 3 treatment arms, and the aggregate Akaike and Bayesian information criteria statistics were compared across candidate models to identify the best fit.
Visual inspection of predicted survival curves overlaid with the Kaplan-Meier curves (including the 95% confidence interval [CI]) was used to assess the quality of within-trial fit to the observed data. Similarly, smoothed hazard plots comparing the empirical hazard and the predicted hazard from the fitted models were used to assess how close the models’ predictions were to the observed data. In addition, for ipilimumab, in which the visual fit and statistical fit to the observed data were not decisive, external data from the pooled analysis of long-term survival data from 12 studies of ipilimumab in unresectable or advanced melanoma were leveraged (based on reconstructed survival data from Schadendorf et al. to inform model selection). 16
For the piecewise models, a combination of the Kaplan-Meier survivor functions, followed by a standard parametric distribution, was evaluated. A truncated Kaplan-Meier curve at prespecified landmark time points combined with an exponential distribution is not only a commonly used piecewise model in health technology assessment but also the most preferred by certain evidence review groups.2,17,18 Therefore, this approach was used as an initial starting point for comparison, but all distributions were tested. Two time periods (corresponding to 1 cut point) for the piecewise models were estimated for all data cuts based on visual inspection of the cumulative hazards plot. A point of change in slope of the cumulative hazard plot was identified visually and set as the cut point in which independent parametric models were fitted to the data beyond this time point.
For the parametric mixture models, 2 additional selection criteria were applied to identify the best-fitting models. The potential for a local versus global solution was evaluated by testing extreme starting values for the expectation-maximization algorithm. In addition, models that estimate the weight of one class to be <5% represent the cases in which the fits were driven by one predominant class. Because the fits in such cases can be approximated fairly well with a single parametric survival model, they were omitted from consideration.
For landmark response models, survival was conditioned on patients’ response status (i.e., complete or partial response v. nonresponders) at a landmark time point. Landmark time point selection requires a balance that allows enough time for patients to respond while providing sufficient postlandmark data to model survival. After assessment of these data, a 6-mo landmark was selected. Patients alive at 6 mo were then included in the analysis, and standard parametric survival models were then fitted to the 2 response groups. The separate response-specific curves were then weighted by response rate at the landmark time point to form a single overall survival curve. Survival prior to the landmark point was based on the Kaplan-Meier curves from the unstratified population.
Analyses were conducted using the statistical packages in R and Stata. Mixture cure models were fitted using the flexsurvcure package in R. All other analyses were performed using the flexsurv package in R, with the exception of the parametric mixture models, which were fitted using the fmm package in Stata (v15; https://www.stata.com/support/faqs/resources/citing-software-documentation-faqs/).
Model Assessment and Validation
To evaluate internal validity, we compared survival extrapolations obtained by each approach for each treatment arm against the observed overall survival data from CheckMate 067 at 60 mo.
Mean estimates of survival over a 20-y time horizon were also generated for each selected model. Incremental survival benefits were calculated for nivolumab plus ipilimumab in comparison with either monotherapy and for nivolumab in comparison with ipilimumab. All mean survival times were adjusted to account for general population mortality as follows. For all extrapolation methods, except for the mixture cure models in which background mortality was explicitly accounted for in a relative survival framework, the instantaneous hazards from the fitted curves were compared with the general population hazard over time. When the general population hazard exceeded the modeled disease-related hazard, the modeled hazard was replaced with the general population hazard. General population rates were based on World Health Organization life tables for Canada, assuming a starting age of 60 y of age, which was the mean age of patients who participated in CheckMate 067. A discount rate of 5% was also applied to represent the time value of health outcomes from a Canadian health technology assessment perspective and to align with the rate used in the original submission based on the 28-mo data cut from CheckMate 067.14,15
A cross-validation against independent external data was performed to ascertain whether the survival projections are clinically plausible. As described above, the pooled overall survival data from 1861 patients with advanced melanoma across 10 prospective and 2 retrospective studies of ipilimumab were used to inform and validate survival projections across all methods. 16 The pooled data, representative of various dosing regimens of ipilimumab, comprise a mixture of treatment-naïve and previously treated patients. The extrapolated long-term survival estimates for each model were compared against the 95% CIs around the pooled overall survival rates for ipilimumab at year 7 among previously untreated patients (95% CI: 17.4%–27.0%) and at year 10 for the full pooled population (95% CI: 16.5%–20.8%), because data beyond 7 y were not available for the previously untreated group. Considering that CheckMate 067 included only previously untreated patients, a higher survival rate was expected than in the pooled population. Therefore, for the purposes of validation, the 10-y survival 95% CI upper limit (20.8%) for the pooled population was increased by 3% to 23.8%. It was not possible to repeat a similar exercise for nivolumab or nivolumab plus ipilimumab because of the lack of external data with longer follow-up currently available from CheckMate 067.
In addition, 2 clinical experts in the treatment of advanced melanoma were consulted to validate the appropriateness of the survival extrapolations and comment on the clinical plausibility (face validity) of the extrapolation estimates. These clinicians were first presented with the patient characteristics from CheckMate 067 and the observed results for overall survival, response, and subsequent therapy based on the 60-mo data cut. They were not presented the results of any of the models before they provided their opinions. The experts were requested to give their input regarding the true value, the lower limit, and the upper limit of landmark survival at 10 and 20 y for patients treated with each of the 3 treatment arms, assuming a mean age of 60 y of age at diagnosis, based on a framework that was developed by Sheffield University in the United Kingdom (Sheffield Elicitation Framework). 19 Example text of the instructions and questions from this exercise are provided in the Supplemental Material. With these outcomes, boundaries were set for what could be considered as “clinically valid,” based on the oucomes of the CheckMate 067 trial.
Results
Model Estimation and Selection
The proportional hazards and accelerated failure time assumptions were violated for all data cuts (see Supplemental Material for results supporting this assessment). As such, all models were fitted independently for the 3 treatment arms. 1 Smoothed hazard plots based on the observed data for each data cut are provided in the Supplemental Material. These showed an initial increase in the hazard followed by a steady decline across the successive data cuts, with the hazards for nivolumab plus ipilimumab and nivolumab trending toward the general population hazard at 60 mo (see Figure 2B).
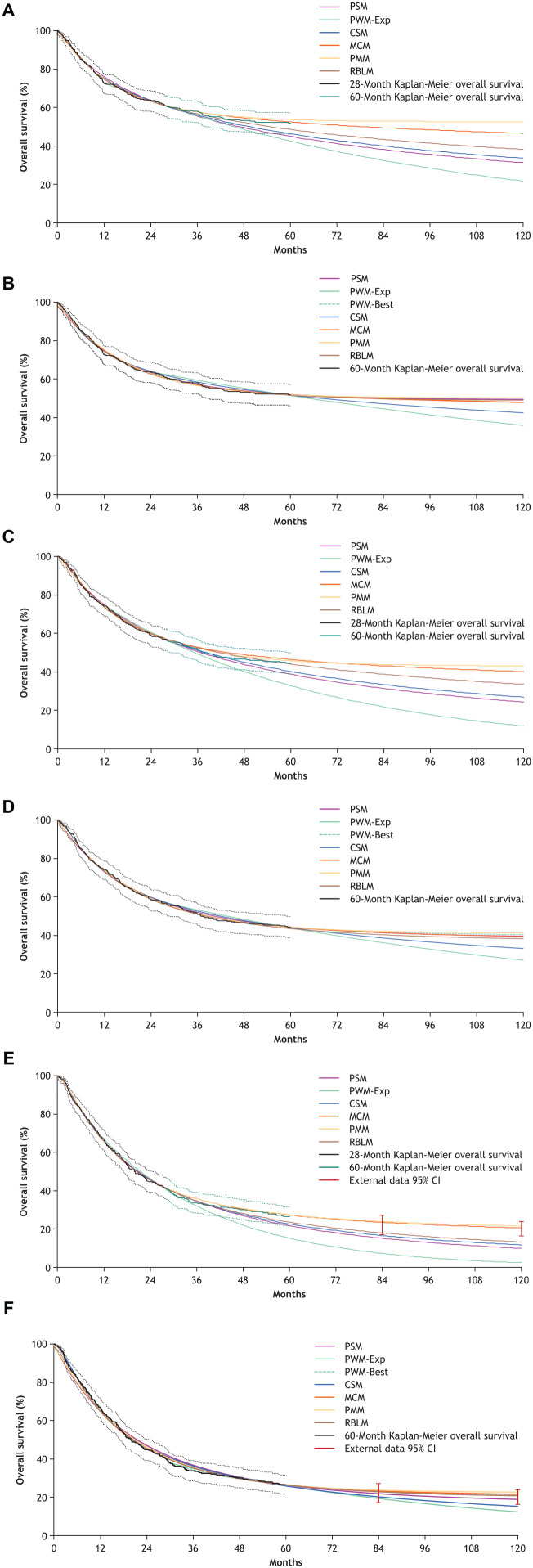
The best-fitting models within each modeling approach for the 28-mo and 60-mo data cuts are presented in Table 1 and are plotted against the Kaplan-Meier curves in Figure 3. All selected models have a close fit to the observed data for that data cut. In the 28-mo data, there was much more variation in the extrapolated portions of the curves, with the piecewise model (exponential) being the most conservative, followed by the standard parametric model (log-normal). The log-normal standard parametric model was the selected model used in the Canadian Agency for Drugs and Technologies in Health review with the 28-mo data. The parametric mixture (exp-exp [exponential-exponential]) and mixture cure (log-logistic) models were the least conservative. There was more alignment in the extrapolations for the best-fitting models when based on the 60-mo data, with the exception of the piecewise (exponential) and cubic spline models. The best-fitting standard parametric model in the 60-mo data shifted to a Gompertz distribution. Another version of the piecewise model using a Gompertz distribution was also added for comparison, as this was the best-fitting piecewise model for that data cut.
Table 1.
Selected Models for Each Modeling Approach and Data Cut
| Model | Nivolumab Plus Ipilimumab | Nivolumab | Ipilimumab |
|---|---|---|---|
| 28-mo data cut | |||
| PSM | Log-normal a | Log-normal a | Log-normal a |
| CSM | Odds, 1 knot | Odds, 1 knot | Odds, 1 knot |
| PWM b | Exponential | Exponential | Exponential |
| MCM | Log-logistic | Log-logistic | Log-logistic |
| PMM | Exp-Exp | Exp-Exp | Exp-LL |
| RBLM (6-mo landmark) | Generalized Gamma | Log-logistic | Log-logistic |
| 60-mo data cut | |||
| PSM | Gompertz | Gompertz | Gompertz |
| CSM | Odds, 1 knot | Odds, 1 knot | Odds, 1 knot |
| PWM (best fit) c | Gompertz | Gompertz | Gompertz |
| PWM (exponential) c | Exponential | Exponential | Exponential |
| MCM | Exponential | Exponential | Exponential |
| PMM | Exp-Exp | Exp-Exp | Exp-Exp |
| RBLM (6-mo landmark) | Gompertz | Gompertz | Gompertz |
CSM, cubic spline model; Exp, exponential; LL, log-logistic; MCM, mixture cure model; PMM, parametric mixture model; PSM, standard parametric model; PWM, piecewise model; RBLM, landmark response model.
Selected distribution for Canadian Agency for Drugs and Technologies in Health submission.
Best-fitting PWM in the 28-mo data cut was the exponential distribution. Kaplan-Meier data used up to 12 mo, followed by parametric model.
Kaplan-Meier data used up to 24 mo for nivolumab plus ipilimumab and nivolumab and 30 mo for ipilimumab, followed by exponential model.
Figure 3.
Overall survival curves for each modeling approach and data cut. Dashed lines represent 95% CIs for 28-month and 60-month Kaplan-Meier overall survival. (A) Nivolumab plus ipilimumab, 28-month data cut; (B) nivolumab plus ipilimumab, 60-month data cut; (C) nivolumab, 28-month data cut; (D) nivolumab, 60-month data cut; (E) ipilimumab, 28-month data cut; (F) ipilimumab, 60-month data cut. External data presented in Figures 3E and 3F were pooled from 1861 patients with advanced melanoma across 10 prospective and 2 retrospective studies of ipilimumab. 16 CI, confidence interval; CSM, cubic spline model; Exp, exponential; MCM, mixture cure model; PMM, parametric mixture model; PSM, standard parametric model; PWM, piecewise model; RBLM, landmark response model.
Model Assessment and Validation
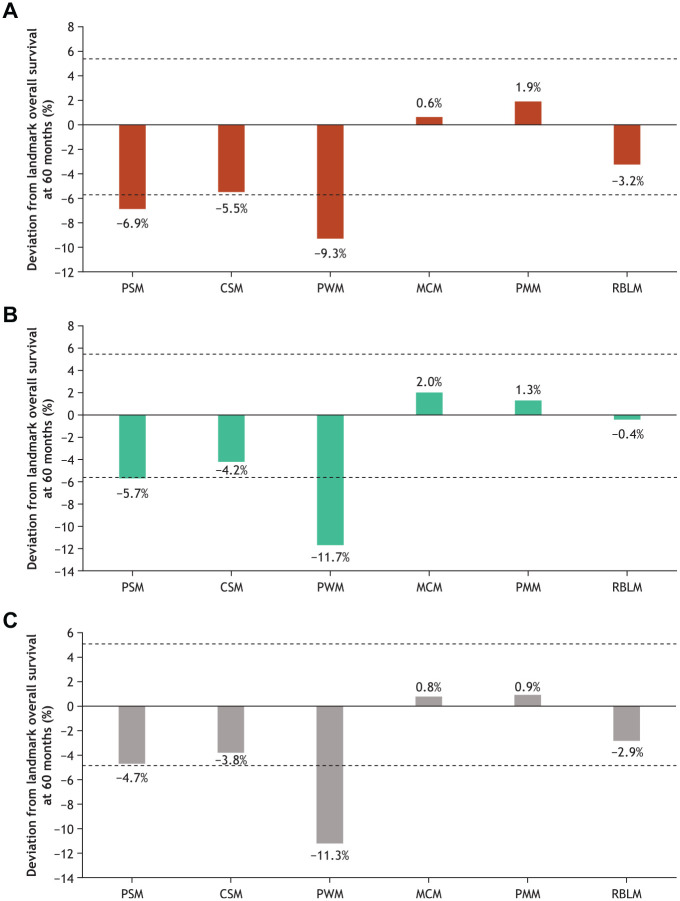
At the 60-mo landmark, standard parametric, piecewise, and cubic spline models all exhibited a common underestimating behavior for overall survival when based on the 28-mo data cut. The mixture cure, parametric mixture, and landmark response models performed consistently better than the other models across all data cuts, with all estimates falling within the 95% CI of the observed Kaplan-Meier overall survival rates (Figure 4 and Supplemental Material).
Figure 4.
Absolute deviation from landmark survival at 60 months based on the 28-month data cut. Dashed lines represent Kaplan-Meier 95% CI. (A) Nivolumab plus ipilimumab; (B) nivolumab; (C) ipilimumab. CI, confidence interval; CSM, cubic spline model; MCM, mixture cure model; PMM, parametric mixture model; PSM, standard parametric model; PWM, piecewise model; RBLM, landmark response model.
Restricted mean survival time estimates for a 20-y time horizon are presented in Table 2. In general, the models that account for survival heterogeneity (mixture cure, parametric mixture, and landmark response models) produced higher estimates of mean survival (nivolumab plus ipilimumab, 6.0–7.0 y based on the 28-mo data cut) than did those for other models (nivolumab plus ipilimumab, 4.6–5.6 y). These same modeling approaches predicted consistent estimates of survival benefit when based on the 60-mo data compared with the 28-mo estimates. This consistency was evident for all treatment arms and across the interim data cuts as well (Supplemental Material). By contrast, for the methods that had projected lower survival times with the 28-mo data (standard parametric model, cubic spline model, piecewise model + best fitting), survival times increased to align more closely with the models accounting for survival heterogeneity, with the exception of the piecewise model plus exponential.
Table 2.
Mean Survival Benefit (Years) at 20 Years a
| 20-y Mean Survival Time | |||
|---|---|---|---|
| Method | Nivolumab Plus Ipilimumab | Nivolumab | Ipilimumab |
| 28-mo data cut | |||
| PSM | 5.4 | 4.7 | 3.0 |
| CSM | 5.6 | 4.9 | 3.1 |
| PWM-Exp | 4.6 | 3.6 | 2.4 |
| MCM | 6.7 | 6.0 | 3.9 |
| PMM | 7.0 | 6.2 | 4.0 |
| RBLM | 6.0 | 5.5 | 3.3 |
| 60-mo data cut b | |||
| PSM | 6.8 | 5.9 | 3.8 |
| CSM | 6.4 | 5.5 | 3.5 |
| PWM-best | 6.8 | 6.0 | 3.9 |
| PWM-Exp | 5.8 | 4.9 | 3.2 |
| MCM | 6.8 | 5.9 | 4.0 |
| PMM | 6.8 | 6.0 | 4.0 |
| RBLM | 6.8 | 5.8 | 3.9 |
CSM, cubic spline model; Exp, exponential; MCM, mixture cure model; PMM, parametric mixture model; PSM, standard parametric model; PWM, piecewise model; RBLM, landmark response model.
All mean survival times account for Canadian-specific general population mortality, assuming a starting age of 60 y, and are discounted at a 5% rate.
Bold indicates a difference of 15% or greater as compared with the 28-mo data cut PSM shown in bold in the first row.
These findings indicate that survival estimates based on the 28-mo standard parametric model log-normal used in the Canadian Agency for Drugs and Technologies in Health analysis likely underestimated long-term survival. For nivolumab plus ipilimumab, the original model estimated 5.4 y of survival benefit in comparison with a range of 6.4–6.8 y as estimated by all other models (when based on the 60-mo data, excluding piecewise model + exponential; Table 2). This difference translates to an underestimation of 1.0–1.4 y (or a 19%–26% higher survival benefit). This underestimation was also noted for nivolumab (0.8–1.3 y difference v. 28-mo standard parametric model; 17%–29% increase) and ipilimumab (0.5–1.0 y and 16%–35% increase).
The disparities observed in the mean survival estimates between different survival models were also observed in the incremental benefit when comparing treatments, although the patterns were less pronounced (Supplemental Material). Standard parametric model, cubic spline model, and piecewise model + exponential generated lower estimates of survival benefit for comparisons versus ipilimumab when based on the 28-mo data as compared with the other methods.
External Validation
As illustrated in Figures 3E and 3F for ipilimumab, extrapolations based on the 28-mo data cut were highly variable, differing substantially depending on the survival modeling technique. Only the mixture cure and parametric mixture models generated predictions of survival estimates of 7-y and 10-y overall survival that were strongly aligned (within both CIs) with external data for ipilimumab. All other models—most significantly the piecewise model + exponential—underestimated long-term overall survival. With the availability of more mature data at 60 mo, multiple modeling approaches, including the standard parametric model (Gompertz) and piecewise model (Gompertz), appeared to show better alignment with the external data. Importantly, mixture cure, parametric mixture, and landmark response models showed alignment with the long-term ipilimumab data across all data cuts. Piecewise model + exponential and cubic spline models underestimated survival at the 10-y landmark across all data cuts.
For nearly all of the modeling approaches considered, the predicted survival estimates at 10 and 20 y lie within the plausible range provided by the clinicians (see Supplemental Material). Notable exceptions in the 28-mo data cut analysis included underestimation of survival with the piecewise model + exponential at 10 and 20 y and the standard parametric model at 20 y for all 3 arms. In addition, when based on the 60-mo data, piecewise model + exponential still underestimated survival at 20 y.
Discussion
This study evaluated the predictive accuracy of standard parametric survival models versus more flexible models for an immune checkpoint inhibitor combination that has the potential to provide durable survival for patients with advanced melanoma. A comprehensive range of extrapolation models was tested using data from CheckMate 067, 11 including standard parametric, piecewise, cubic spline, mixture cure, parametric mixture, and landmark response models.
In general, all of the selected models provided a reasonable fit to the observed data, irrespective of the level of data maturity. Extrapolations based on early data cuts were associated with a higher degree of uncertainty when predicting long-term survival, as compared with more mature data cuts, in which there was more agreement across the various modeling approaches. This is in line with what would be expected and indicates that the selection of the “right” model is especially important when dealing with immature data.
In this analysis, the methods that explicitly assumed survival heterogeneity in the patient population, namely, the mixture cure, parametric mixture, and landmark response models, outperformed other approaches across all 3 treatment arms. These 3 modeling techniques were all generally aligned with each other and provided accurate and consistent estimates of overall survival across the range of follow-up periods. In particular, the results of the internal validation showed that these models closely matched landmark survival at 60 mo for all data cuts when compared with the most mature observed data from CheckMate 067, whereas the standard parametric, cubic spline, and piecewise models consistently underestimated survival for the earlier data cuts.
Similar findings were observed in the external validation exercises. When fitted to early data cuts, mixture cure, parametric mixture, and landmark response models produced 7-y and 10-y overall survival estimates that were more aligned with external long-term data for ipilimumab than the other methods. Piecewise model + exponential, cubic spline model, and standard parametric model underestimated survival when compared with either the long-term ipilimumab data or the clinically plausible ranges provided by expert clinicians.
The original Canadian Agency for Drugs and Technologies in Health review used the 28-mo data from CheckMate 067 in the economic evaluation of these immune checkpoint inhibitors. 14 A mean survival time of 5.4 y over a 20-y time horizon was estimated for nivolumab plus ipilimumab, with a standard parametric model (log-normal) used for extrapolation (see Table 2). This estimate is well below the range estimated by the mixture cure, parametric mixture, and landmark response models for the 28-mo data cut and lower than estimates for all models based on the 60-mo data cut. When compared with the average mean survival benefit across all 60-mo models, the 28-mo log-normal standard parametric model underestimated survival by 1.2 y, 1 y, and 0.8 y for nivolumab plus ipilimumab, nivolumab, and ipilimumab, respectively. The degree of underestimation would have been greater if the piecewise model + exponential models had been used.
These findings are in general agreement with other recent studies assessing the accuracy of similar survival extrapolation techniques in immuno-oncologic therapy settings. A similar case study conducted by Bullement et al. in advanced melanoma found that mixture cure models generated more accurate survival estimates for ipilimumab plus dacarbazine than other methods when validated with external registry data. 7 In this study, a 3-part piecewise model (Kaplan-Meier data + log-normal fitted to trial data + Weibull fitted to registry data) performed well. The commonality across these 2 approaches was the incorporation of external information (background mortality for the mixture cure model and long-term registry data for the piecewise model), highlighting the importance of including external data when available. 7
A study by Ouwens et al. used data from ATLANTIC (NCT02087423), a phase II single-arm trial of durvalumab in previously treated patients with advanced lung cancer, to examine different methods of extrapolating overall survival. 8 The authors reported that the cure models provided the best fit for longer-term data, whereas the standard parametric model based on a log-normal distribution generally underestimated long-term observed overall survival. 8 This study did find, however, that the long-term mean survival estimates from these methods differed from each other. By contrast, our study showed general consistency in the mean survival estimates for these methods. This difference may be attributable to an early plateauing of the tail of the Kaplan-Meier overall survival curves in CheckMate 067, 11 whereas this was not as evident in the survival data from ATLANTIC.
The underperformance of the piecewise model + exponential in our analysis is also noteworthy and is consistent with a similar recent case study in previously treated patients with advanced renal cell carcinoma. 10 In this analysis, however, the flexible modeling approaches that accounted for survival heterogeneity (parametric mixture and landmark response models) did not perform as well for earlier data cuts as they did in our analysis. 10
Other studies have focused on more limited comparisons of standard parametric models to either cubic spline or mixture cure models in the context of immune checkpoint inhibitors. An analysis conducted by Gibson et al. also compared standard parametric and cubic spline models using 28-mo progression-free survival data from CheckMate 067 and found cubic spline models to provide a better fit to the observed data than did standard parametric models. 20 Our analysis of overall survival from the same data cut did show that the cubic spline models performed marginally better when looking at 60-mo landmark survival (both underestimated, but the cubic spline model underestimated less than the log-normal standard parametric model). An analysis by Othus et al. demonstrated that a Weibull mixture cure model was superior to a Weibull standard parametric model in estimating long-term data for ipilimumab from a different trial in advanced melanoma. 21
The survival modeling techniques considered in our study are broadly aligned with those considered in the National Institute for Health and Care Excellence Technical Support Document 21. 6 An exception to this is the implementation of relative survival models, which were not included in our analysis; however, Technical Support Document 21 does note that these have not previously been used in technology appraisals to date.
Technical Support Document 21 recommends leveraging external data and incorporating background mortality in survival extrapolations, and the importance of those recommendations has been borne out in this study and in the existing literature.7,8,22 Although long-term external data for ipilimumab were leveraged in our study, no such data were available specific to nivolumab plus ipilimumab or nivolumab alone. Ranges of clinically plausible long-term survival rates were solicited from 2 expert clinicians to fill this gap. Extending this exercise to additional clinicians could have produced a more narrow and robust range of plausible outcomes. Although we did leverage the Sheffield Elicitation Framework, 19 best practice for structural expert elicitation is an emerging area of discussion, and there are currently no standard guidelines in this setting.23,24 Furthermore, these external data were used as reference points only for external validation and were not explicitly used to inform model parameterization or extrapolation (e.g., via a Bayesian framework). Because the aim of this case study was to explore the performance of commonly used methodologies in health technology assessment in the context of heterogeneity of survival, applying these external data for the purposes of validation felt most appropriate. Advanced methods for formally integrating external data—either observed or elicited via clinical expert—have recently been explored, and this topic is an area of ongoing research and development.6,25–29
As part of Technical Support Document 21, a simulation exercise was conducted to evaluate the performance of a subset of the flexible modeling techniques under different scenarios representing various “true” complex hazard shapes (based on Weibull distributions). One such scenario assumed that there was a signal indicating a cure fraction exists, and the presented survival and hazard plots most closely aligned with the shapes seen in CheckMate 067. In this scenario, Technical Support Document 21 reported that cubic spline models (accounting for background mortality) had a large underestimation of lifetime survival. In our analysis, based on the 60-mo data, cubic spline models were not found to underestimate long-term survival at 10 y when compared with data from Schadendorf et al. 16 In addition, the Technical Support Document 21 simulation revealed minimal bias with the mixture cure model (Weibull) in lifetime survival estimates. A close alignment of the mixture cure model (exponential) with long-term data for ipilimumab was also demonstrated. 16
A further limitation of this case study is that our findings may not be generalizable to other trial data of immune checkpoint inhibitors in which the comparators and cancer type may differ. Although some of our key conclusions are supported across other studies in other disease sites, as described above, there were some differences noted in the performance of certain models when based on earlier data. Our analysis used Canadian general population mortality, matched to the CheckMate 067 trial population, to inform the long-term survival extrapolations. Applying these methods to another locality would necessitate use of country-specific, non–disease-related mortality rates. In addition, although estimated survival benefit is an important element of cost-effectiveness modeling for health technology assessment, this analysis did not account for progression-free survival, quality-of-life adjustment, or costs, all of which are also important components in economic evaluation. Therefore, it is difficult to make a clear determination on the net impact of the different modeling approaches on the estimation of incremental cost-effectiveness ratios.
For the purposes of this case study, all methodologic approaches were tested for all treatment arms for all data cuts. In a real-world analysis, it is likely that not all methodologic approaches would be necessary or appropriate to test in each setting. 6 For example, piecewise models are generally indicated when there is a clear turning point in the observed data (although there are some who recommend their use regardless because of their explicit use of observed Kaplan-Meier data). In this analysis, particularly for the early data cuts, there was not a clear turning point to select the cut point for the piecewise models; thus, their application here may be somewhat artificial in the interest of testing all methods.
Lastly, in this analysis, the “best” single model was selected within each method, and the data cut was based on a number of criteria. However, it is possible, if not likely, that secondary or alternative distribution selections could have been made and tested. Frequently in health technology assessment submissions, multiple distributions would be evaluated in sensitivity analysis. Furthermore, the “best” distribution within a methodologic approach also changed over time (e.g., log-normal standard parametric model at 28 mo v. Gompertz standard parametric model at 60 mo, exponential piecewise model at 28 mo v. Gompertz piecewise model at 60 mo). The improved performance of certain approaches that do not account for survival heterogeneity (e.g., standard parametric and piecewise models) over the successive data cuts is likely, in part, because of the selected distribution, in addition to data maturity, rather than the underlying method specifically.
Conclusions
In summary, this case study in advanced melanoma found that survival modeling techniques that explicitly assume heterogeneity showed greater accuracy than other modeling approaches with both early and later data cuts. Extrapolated survival outcomes from the earliest data cut from CheckMate 067 displayed a wide range of outcomes, with greater agreement across the methods as data matured. This study highlights the importance of considering flexible modeling approaches in earlier data cuts when estimating the long-term survival of immune checkpoint inhibitors as well as the key role of external data to validate and support model selection.
Supplemental Material
Supplemental material, sj-docx-1-mpp-10.1177_23814683221089659 for Heterogeneity in Survival with Immune Checkpoint Inhibitors and Its Implications for Survival Extrapolations: A Case Study in Advanced Melanoma by Victoria Federico Paly, Murat Kurt, Lirong Zhang, Marcus O. Butler, Olivier Michielin, Adenike Amadi, Emma Hernlund, Helen M. Johnson, Srividya Kotapati, Andriy Moshyk and John Borrill in MDM Policy & Practice
Acknowledgments
Editorial assistance was provided by Mark Palangio and Michele Salernitano at Ashfield MedComms, an Ashfield Health company, funded by Bristol Myers Squibb.
Footnotes
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: VFP is an employee of ICON plc, which received consulting service fees from Bristol Myers Squibb related to the current work. MK is an employee of Bristol Myers Squibb. LZ is an employee of ICON plc, which received consulting service fees from Bristol Myers Squibb related to the current work. MB reports receiving grants from Merck and Takara Bio for clinical trial support; personal fees for advisory roles from Bristol Myers Squibb, EMD Serono, GlaxoSmithKline, Immunocore, Merck, Novartis, Pfizer, Sanofi, and Sun Pharma; and personal fees for safety review board roles from Adaptimmune and GlaxoSmithKline. OM reports receiving personal fees for consulting/advisory roles from Amgen, Bristol Myers Squibb, Merck Sharp & Dohme, Novartis, and Roche; and research funding from NeraCare GmbH. AA is an employee of Bristol Myers Squibb. EH is an employee of ICON plc, which received consulting service fees from Bristol Myers Squibb related to the current work. HMJ reports receiving consulting fees from Bristol Myers Squibb related to the current work. SK is an employee of Bristol Myers Squibb. AM is an employee of and holds shares in Bristol Myers Squibb. JB is an employee of Bristol Myers Squibb. The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Financial support for this study was provided by Bristol Myers Squibb, Princeton, NJ, USA. The funding agreement ensured the authors’ independence in designing the study, interpreting the data, and writing and publishing the report.
ORCID iDs: Victoria Federico Paly  https://orcid.org/0000-0003-3508-1296
https://orcid.org/0000-0003-3508-1296
John Borrill  https://orcid.org/0000-0002-1047-8556
https://orcid.org/0000-0002-1047-8556
Availability of data and materials: Bristol Myers Squibb’s policy on data sharing is available at https://www.bms.com/researchers-and-partners/independent-research/data-sharing-request-process.html.
Supplemental Material: Supplementary material for this article is available on the MDM Policy & Practice website at https://journals.sagepub.com/home/mpp.
Contributor Information
Victoria Federico Paly, ICON plc, Global Health Economics and Outcomes Research, North Wales, PA, USA.
Murat Kurt, Bristol Myers Squibb, Health Economics and Outcomes Research, Princeton, NJ, USA.
Lirong Zhang, ICON plc, Global Health Economics and Outcomes Research, London, UK.
Marcus O. Butler, Department of Medical Oncology and Hematology, Princess Margaret Cancer Centre, University of Toronto, Toronto, ON, Canada
Olivier Michielin, Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland.
Adenike Amadi, Bristol Myers Squibb, Health Economics and Outcomes Research, Uxbridge, UK.
Emma Hernlund, ICON plc, Global Health Economics and Outcomes Research, Stockholm, Sweden.
Helen M. Johnson, Bristol Myers Squibb, Health Economics and Outcomes Research, Uxbridge, UK
Srividya Kotapati, Bristol Myers Squibb, Worldwide Medical Affairs, Princeton, NJ, USA.
Andriy Moshyk, Bristol Myers Squibb, Health Economics and Outcomes Research, Princeton, NJ, USA.
John Borrill, Bristol Myers Squibb, Health Economics and Outcomes Research, Uxbridge, UK.
References
- 1. Latimer N. NICE DSU Technical Support Document 14: survival analysis for economic evaluations alongside clinical trials-extrapolation with patient-level data. Report by the Decision Support Unit. Available from: http://nicedsu .org.uk/wp-content/uploads/2016/03/NICE-DSU-TSD-Survival-analysis.updated-March-2013.v2.pdf [PubMed]
- 2. Bagust A, Beale S. Survival analysis and extrapolation modeling of time-to-event clinical trial data for economic evaluation: an alternative approach. Med Decis Making. 2014;34(3):343–51. [DOI] [PubMed] [Google Scholar]
- 3. Kaufman HL, Atkins MB, Subedi P, et al. The promise of immuno-oncology: implications for defining the value of cancer treatment. J Immunother Cancer. 2019;7(1):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Michielin O, Atkins MB, Koon HB, Dummer R, Ascierto PA. Evolving impact of long-term survival results on metastatic melanoma treatment. J Immunother Cancer. 2020;8(2):e000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quinn C, Garrison LP, Pownell AK, et al. Current challenges for assessing the long-term clinical benefit of cancer immunotherapy: a multi-stakeholder perspective. J Immunother Cancer. 2020;8(2):e000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rutherford M, Lambert P, Sweeting M, et al. NICE DSU Technical Support Document 21. Flexible methods for survival analysis. Available from: http://nicedsu.org.uk/wp-content/uploads/2020/11/NICE-DSU-Flex-Surv-TSD-21_Final_alt_text.pdf
- 7. Bullement A, Latimer NR, Bell Gorrod H. Survival extrapolation in cancer immunotherapy: a validation-based case study. Value Health. 2019;22(3):276–83. [DOI] [PubMed] [Google Scholar]
- 8. Ouwens M, Mukhopadhyay P, Zhang Y, Huang M, Latimer N, Briggs A. Estimating lifetime benefits associated with immuno-oncology therapies: challenges and approaches for overall survival extrapolations. Pharmacoeconomics. 2019;37(9):1129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lanitis T, Proskorovsky I, Ambavane A, et al. Survival analysis in patients with metastatic merkel cell carcinoma treated with avelumab. Adv Ther. 2019;36(9):2327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klijn SL, Fenwick E, Kroep S, et al. What did time tell us? A comparison and retrospective validation of different survival extrapolation methods for immuno-oncologic therapy in advanced or metastatic renal cell carcinoma. Pharmacoeconomics. 2021;39(3):345–56. [DOI] [PubMed] [Google Scholar]
- 11. Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med. 2019;381(16):1535–46. [DOI] [PubMed] [Google Scholar]
- 12. Kroep S, Kiff C, Kraan C, et al. Modeling the survival benefit of immuno-oncologic therapy: a review of methods used in NICE single technology appraisals. Value Health. 2019;22(S3):S523–4. (Abstract PCN451) [Google Scholar]
- 13. Larkin J, Hodi FS, Wolchok JD. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373(13):1270–1. [DOI] [PubMed] [Google Scholar]
- 14. Pan-Canadian Oncology Drug Review. Nivolumab (Opdivo) with ipilimumab (Yervoy) for metastatic melanoma. Final economic guidance report. Available from: https://www.cadth.ca/sites/default/files/pcodr/pcodr_opdivo_yervoy_metmela_fn_egr.pdf
- 15. Quon PL, Xiao Y, Sorensen S, Monfared AAT. Economic evaluation of nivolumab plus ipilimumab combination as first-line treatment for patients with advanced melanoma in Canada. Pharmacoecon Open. 2019;3(3):321–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schadendorf D, Hodi FS, Robert C, et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J Clin Oncol. 2015;33(17):1889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. National Institute for Health and Care Excellence. Pembrolizumab for untreated PD-L1-positive metastatic non-small-cell lung cancer. Technology appraisal guidance [TA531]. Available from: https://www.nice.org.uk/guidance/ta531
- 18. National Institute for Health and Care Excellence. Atezolizumab for treating locally advanced or metastatic non-small-cell lung cancer after chemotherapy. Technology appraisal guidance [TA520]. Available from: https://www.nice.org.uk/guidance/ta520
- 19. Oakley J, O’Hagan T. SHELF: the Sheffield Elicitation Framework (version 4). Available from: http://tonyohagan.co.uk/shelf
- 20. Gibson E, Koblbauer I, Begum N, et al. Modelling the survival outcomes of immuno-oncology drugs in economic evaluations: a systematic approach to data analysis and extrapolation. Pharmacoeconomics. 2017;35(12):1257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Othus M, Bansal A, Koepl L, Wagner S, Ramsey S. Accounting for cured patients in cost-effectiveness analysis. Value Health. 2017;20(4):705–9. [DOI] [PubMed] [Google Scholar]
- 22. Kearns B, Stevens J, Ren S, Brennan A. How uncertain is the survival extrapolation? A study of the impact of different parametric survival models on extrapolated uncertainty about hazard functions, lifetime mean survival and cost effectiveness. Pharmacoeconomics. 2020;38(2):193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grigore B, Peters J, Hyde C, Stein K. A comparison of two methods for expert elicitation in health technology assessments. BMC Med Res Methodol. 2016;16:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bojke L, Soares M, Claxton K, et al. Developing a reference protocol for structured expert elicitation in health-care decision-making: a mixed-methods study. Health Technol Assess. 2021;25(37):1–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jackson C, Stevens J, Ren S, et al. Extrapolating survival from randomized trials using external data: a review of methods. Med Decis Making. 2017;37(4):377–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pennington M, Grieve R, der Meulen JV, Hawkins N. Value of external data in the extrapolation of survival data: a study using the NJR data set. Value Health. 2018;21(7):822–29. [DOI] [PubMed] [Google Scholar]
- 27. Cope S, Ayers D, Zhang J, Batt K, Jansen JP. Integrating expert opinion with clinical trial data to extrapolate long-term survival: a case study of CAR-T therapy for children and young adults with relapsed or refractory acute lymphoblastic leukemia. BMC Med Res Methodol. 2019;19(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soikkeli F, Hashim M, Ouwens M, Postma M, Heeg B. Extrapolating survival data using historical trial-based a priori distributions. Value Health. 2019;22(9):1012–7. [DOI] [PubMed] [Google Scholar]
- 29. van Oostrum I, Ouwens M, Remiro-Azócar A, et al. Comparison of parametric survival extrapolation approaches incorporating general population mortality for adequate health technology assessment of new oncology drugs. Value Health. 2021;24(9):1294–301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-mpp-10.1177_23814683221089659 for Heterogeneity in Survival with Immune Checkpoint Inhibitors and Its Implications for Survival Extrapolations: A Case Study in Advanced Melanoma by Victoria Federico Paly, Murat Kurt, Lirong Zhang, Marcus O. Butler, Olivier Michielin, Adenike Amadi, Emma Hernlund, Helen M. Johnson, Srividya Kotapati, Andriy Moshyk and John Borrill in MDM Policy & Practice