Summary
Background
Multiple myeloma (MM) is still an incurable malignancy of plasma cells. Proteasome inhibitors (PIs) work as the backbone agent and have greatly improved the outcome in majority of newly diagnosed patients with myeloma. However, drug resistance remains the major obstacle causing treatment failure in clinical practice. Here, we investigated the effects of Indirubin-3’-monoxime (I3MO), one of the derivatives of Indirubin, in the treatment of MM.
Methods
MM patient primary samples and human cell lines were examined. I3MO effects on myeloma treatment and the underling molecular mechanisms were investigated via in vivo and in vitro study.
Findings
Our results demonstrated the anti-MM activity of I3MO in both drug- sensitive and -resistance MM cells. I3MO sensitizes MM cells to bortezomib-induced apoptosis. Mechanistically, I3MO acts as a multifaceted regulator of cell death, which induced DNA damage, cell cycle arrest, and abrogates NF-κB activation. I3MO efficiently down-regulated USP7 expression, promoted NEK2 degradation, and suppressed NF-κB signaling in MM. Our study reported that I3MO directly bound with and caused the down-regulation of PA28γ (PSME3), and PA200 (PSME4), the proteasome activators. Knockdown of PSME3 or PSME4 caused the inhibition of proteasome capacity and the overload of paraprotein, which sensitizes MM cells to bortezomib-mediated growth arrest. Clinical data demonstrated that PSME3 and PSME4 are over-expressed in relapsed/refractory MM (RRMM) and associated with inferior outcome.
Interpretation
Altogether, our study indicates that I3MO is agent triggering proteasome inhibition and represents a promising therapeutic strategy to improve patient outcome in MM.
Fundings
A full list of funding can be found in the acknowledgements.
Keywords: Multiple myeloma, Proteasome inhibition, Indirubin-3’-monoxime (I3MO), PSME3 (PA28γ), PSME4 (PA200)
Research in context.
Evidence before this study
Indirubin is one of many active constituents derived from a traditional Chinese prescription, Indigo Naturalis, which was identified by the researcher in our institute in the mid of 1970s. Our colleagues demonstrated that indirubin and its analogue Meisoindigo were effective agents in the treatment of chronic myeloid leukemia in preclinical and clinical trials. Indirubin-3′-monoxime (I3MO) was one of the ideal derivatives of Indirubin, with lower IC50 and enhanced cytotoxicity compared to indirubin itself. Several studies have reported that I3MO efficiently induces malignant cell apoptosis, inhibits cyclin-dependent kinases, and triggers anti-angiogenesis effects in hematologic malignancies and solid tumors.
Added value of this study
In the present study, we demonstrated the promising cytotoxicity of I3MO in myeloma. Combination with I3MO notably enhanced myeloma cell sensitivity to bortezomib treatment. Mechanistically, we found that I3MO acts as a multifaceted regulator of cell death, including promoting cell cycle arrest, DNA damage response, and efficiently induces apoptosis of tumor cells. In addition, our results strikingly demonstrated that I3MO acts as an agent in triggering proteasome inhibition by targeting the proteasome complex subunits PSME3 and PSME4.
Implications of all the available evidence
In summary, our study demonstrates that I3MO is a proteasome inhibitor targeting proteasome activators PSME3 and PSME4. PSME3 and PSME4 are overexpressed in RRMM, which associates with bortezomib resistance and inferior outcome of MM patients. It therefore represents a promising therapeutic agent in overcoming drug-resistance and improves patient outcome in MM.
Alt-text: Unlabelled box
Introduction
Multiple myeloma (MM) is an incurable disease which characterized by proliferation of malignant plasma cells in the bone marrow.1 Although there have been remarkable treatment advances, drug resistance underlying relapse of disease is common, highlighting the continued need for novel active treatments.2 Myeloma cells typically produce a substantial amount of paraprotein and are therefore heavily dependent on the unfolded protein response (UPR) and ubiquitin proteasome system (UPS) for its degradation and to maintain the cellular homeostasis.3
The ubiquitin proteasome system (UPS) is a non-lysosomal intracellular protein degradation pathway mediated via proteasome holoenzyme, ubiquitin ligases, and deubiquitylating (DUB) enzymes.4 Specifically, the covalent attachment of ubiquitin to target substrates leads to protein degradation via the multi-catalytic 26S proteasome complex.5 Deregulation of the UPS pathway is linked to the pathogenesis of various human diseases, including MM.6 DUBs overexpression also plays pivotal roles in bortezomib resistance, which reported by our previous study.7 Inhibition of DUBs, like ubiquitin-specific protease-7 (USP7) notably causes protein overload, which promotes apoptosis and overcomes bortezomib resistance in MM cells.8
One of the most successful approaches to myeloma therapy has been through ubiquitin-proteasome pathway (UPP) inhibition.9 PIs represent the backbone treatment of MM, and have greatly improved the outcome in the majority of patients with newly diagnosed multiple myeloma (NDMM).10, 11, 12 The function was in part by disturbing the balance between proteasome load and capacity.13 However, the continuous exposure to PIs induces drug resistance in vivo and in vitro.14 The molecular mechanisms underlying drug resistance has not been fully elucidated.15 Several studies identified that a single point mutation and the persistent overexpression of PSMB5, the subunit of 20S proteasome subunit, played a pivotal role in the PIs resistance in MM cells.16 These aberrant 20S proteasomes cause conformational or steric variation of the proteasome drug-binding sites, which in turn not only impairs the binding of PIs with proteasome, but also decreases its chymotryptic catalytic function in MM cells. Recently, an alternative strategy targeting the 19S and 11S subunits of proteasome complex to induce MM cell cytotoxicity has been investigated.17 In addition, the bone marrow microenvironment plays an important role in supporting bortezomib resistance as our previous study reported.18
The proteasome complexes are mainly composed of the 20 S catalytic core proteasome particle,19 19S subunit regulators, and 11s alternative activators. Recent studies have shown that inhibition of the 19S proteasome-associated ubiquitin receptor Rpn13 efficiently induced MM cell apoptosis and overcame the bortezomib resistance.20 PA28 (11S, REG) is a family of proteasome regulators whose members are widely present in many of the eukaryotic supergroups. They are represented by three paralogs, PA28α, PA28β and PA28γ. While they share similar sequence and structural, the three isoforms significantly differ in their biochemical and biological features. PA28γ (PSME3) markedly enhances degradation of target protein by 20S proteasome.21,22 PA200 (PSME4) is another 200 kDa large, monomeric proteasome activator, which binds to the 20S proteasome and modulates degradation of the protein of acetyl-histone.23
In the present study, we evaluated the efficacy and mechanisms of action of I3MO triggers cytotoxicity against MM cells. In addition to single-agent activity against drug-naïve MM cell lines in vitro and in vivo, our results supported that I3MO is an effective agent in myeloma treatment in PIs resistance ones, which provides the rational evidence for the clinical application of I3MO in the treatment of MM acting as proteasome inhibitor.
Methods
Cell lines and primary patient cells
Human MM cell lines (ARP1: CVCL_D523, U266: CVCL_0015, RPMI8226: CVCL_0014, OCI-MY5: CVCL_E332), and stromal cell line HS5 (CVCL_3720) were maintained in RPMI-1640 medium (Gibco, C11875500BT, USA) supplemented with 10% fetal bovine serum (Gibco, 10100147, AUS), 100units/mL penicillin, 100 μg/mL streptomycin (Gibco, 15140122).24 IL-6-dependent BTZ-sensitive (ANBL6, CVCL_5425) and ANBL-6 BTZ-resistant (V10R, ANBL6-BR, CVCL_QZ43) cells were provided by Dr. Robert Orlowski (MD Anderson Cancer Center, Houston, TX)25 and maintained in 1640 medium containing IL-6 (1 ng/mL) (PeproTech, 200-06, Inc., USA). ANBL6-BR cells were selected for BTZ resistance by exposure to progressively higher concentrations of BTZ for > 6 months. BTZ concentrations were started at 1 nM and escalated by increments of 1 nM when culture densities doubled in cell number.26 NEK2 overexpression BTZ-resistance MM cell lines, ARP1 and OCI-MY5 cells were constructed in our previous study.27 All the cell lines were performed STR profiling, and grown under mycoplasma-free conditions at 37 °C and 5% CO2. Primary MM cells were obtained from NDMM and RRMM patients visiting in our hospital. This study was conducted in accordance with the Declaration of Helsinki and approved by the local institutional Ethic Committees (the reference number is KT2020010-EC-2). Informed consent to participate in the study was obtained. The data were collected and anonymized for analyses.
Reagents and antibodies
Indirubin-3′-monoxime (I3MO, HY-19807, CAS Number: 160807-49-8) was obtained from MCE. Bortezomib (PS-341, Velcade) was purchased from Selleck Chemicals. Western blot analyses were performed as described previously.28,29 The primary antibody for PARP (#9542, RRID: AB_2160739), Caspase-3 (#9662, RRID: AB_331439), cleaved Caspase-3 (#9664, RRID: AB_2070042), Phospho-IKKα/β(S176/180, # 2697), Phospho-NF-κB p65 (Ser536)(#3033, RRID: AB_331284), p53 (# 9282, RRID:AB_331476), p21 (#2947, RRID: AB_823586), BCL-2 (# 15071, RRID:AB_2744528), Phospho- Histone H2A.X (ser139), # 9718, RRID: AB_2118009), Histone H3 (# 14269, RRID: AB_2756816), GAPDH (#5174, RRID: AB_10622025), anti-rabbit IgG HRP-linked antibody (#7074, RRID: AB_2099233), and anti-mouse IgG HRP-linked antibody (#7072, RRID: AB_331144) were purchased from Cell Signaling Technologies. PA200 (#18799-1-AP), PA28γ (#14907-1-AP), and USP7 (3D7D11) were purchased from Proteintech Group (Rosemont, IL, USA). NEK2(ab227958, RRID: AB_10901715) were purchased from Abcam. Protein bands were visualized with a ChemiDocTM Imaging System (BIO-RAD, Singapore). The bands on the blot were quantified with ImageJ software.
Cell proliferation and apoptosis assay
Cell proliferation assays were performed in 12-well plates and counted once a day for 6 days, using a hemocytometer and the trypan blue exclusion assay (0.4% dye in phosphate-buffered saline, pH 7.3) (T8070, Solarbio). Cell viability at different time points after I3MO treatment was assessed using a Cell Counting Kit-8 (CCK-8) assay (CK04, Dojindo Laboratories, Kumamoto, Japan), and calculated as the ratio of live cells/total cells. Apoptosis was analyzed by using the Annexin V/PE Apoptosis Detection kits (559763, BD Bioscience, USA, RRID: AB_2869265), following the manufacturer's protocol. Fluorescence was measured on an LSR II cytometer (BD Biosciences) and data analyzed using FlowJo Software v10.0 (Tree Star, Ashland, OR, USA).
RNA sequencing and GEO dataset analyses
A panel of MM cell lines, ANBL6 (wild-type), ANBL6 BR (BTZ-resistant), ARP1, U266 and RPMI8266, was treated with I3MO for 24 h, and RNA sequencing was performed as described previously.28,29 GSEA (Gene set enrichment analyses, V2.2.4, Broad Institute) analyses was performed. The GEO datasets GSE5900, GSE2658 and GSE31161 were utilized to investigate the clinical significance of PA28γ and PA200. The TCGA data of MMRF-CoMMpass was utilized.
Proteasome activity assay
Chymotrypsin-Like and Caspase-Like proteasome activity assays were conducted using Proteasome-Glo™ Chymotrypsin-Like and Caspase-Like Cell-Based Assays (G8660, Promega Corporation, Madison, USA), respectively, according to the manufacturer's protocol.
Primers or shRNA sequences
PSME3 and PSME4 shRNAs were purchased from Beijing Genomics Institute (BGI, China). shRNA sequences, sh1-PSME3: 5′-GCATCTTATCTGGACCAGATT-3′ and sh2-PSME3: 5′-CGTGACAGAGATTGATGAGAA-3′. The shRNA sequence for PSME4 were sh3: 5′-CCCTGAGTTTACTGCTCGAAT-3′ and PSME4 sh4: 5′-CGATTGTTTCTTCA GGGCTTA-3′. The non-sense sequencing was utilized as scramble control. The PCR primer sequences used were as follows: PSME3 forward: 5’-AGG ATG GCT TTC TGT ACC GC-3’, PSME3 reverse: 5’-AGA AGA ACG CAG GGC TTT GA-3’, PSME4 forward: 5’-CCA ACA GGA AAA GAA TGC CGA-3’, PSME4 reverse: 5’-CCA GGG CAG GTT TCT TTG CT-3’, GAPDH forward: 5’-GAA GGT GAA GGT CGG AGT C-3’, and GAPDH reverse: 5’-GAA GAT GGT GAT GGG ATT TC-3’.
Plasmids and transfection
ShRNA sequences for PSME3 or PSME4 were inserted into EcoRI and AgeI sites of the doxycycline-inducible vector PLKO-Tet-on. Lentivirus was packaged in HEK293T cells (RRID: CVCL_HA71). After 48 h, HEK293T cells were harvested and concentrated 10-fold by the Lenti-X™ Concentrator (Takara Bio USA, Inc.). 1 × 106 cells were plated in a 12-well plate. Lentivirus was added to the cells with polybrene (8 μg/ml, Santa, sc-134220) and the stable cells were selected according to the manufacturer's protocol. The expression of target proteins was examined by western blots.
The PDX model of primary patient MM cells in zebrafish
To confirm the in vivo anti-myeloma activity of I3MO in primary patient samples, a zebrafish PDX model of MM was utilized according to the previous report.30 Briefly, zebrafish embryos were obtained by the natural spawning of Tubingen strain and incubated at 28 °C in embryo medium. All zebrafish work was approved by the Ethical Review Committee of State Key Laboratory of Experimental Hematology, National Clinical Research Center for Blood Diseases, Institute of Hematology & Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, China (the reference number is NSFC2021057-EC-1). The purified patient primary CD138+ MM cells were fluorescently labeled with 0.1μM Calcein-AM (Invitrogen, C3100MP) and injected into the perivitelline space of each embryo of zebrafish at 48 hpf.30 Engrafted embryos were transferred into 24-well plates and incubated at 28°C in freshly prepared E3 medium31 containing DMSO or drugs. Working stocks were 10nM BTZ, 5µM I3MO or 10nM BTZ and 5µM I3MO (vehicle was DMSO). At 24 h post injection (hpi), engrafted embryos were detected by fluorescent microscopy to visualize calcein (green) positive areas.30 Comparison between groups was performed using unpaired t-test (n≥8 embryos/ treatment arm). Totally 500 zebrafish were utilized, and 50 ones for each experiment.
Myeloma xenograft model
For Figure 3b, a total of 1 × 106 ARP1 MM cells was injected subcutaneously into the left flanks of 6- to 8-weeks old female NOD/SCID mice.32 Two weeks after injection, the mice were randomized into 6 groups that were treated with (a) I3MO (1.25 mg/kg, i.p., every other day), (b) I3MO (6 mg/kg, i.p., every other day), (c) BTZ (0.5 mg/kg, i.p., 2 times/week), (d) BTZ (1 mg/kg, i.p., 2 times/week), (e) I3MO (1.25 mg/kg, i.p.) and BTZ (0.5 mg/kg, i.p.), and (f) vehicle (control group). Tumor volume was measured every 3 days as the tumor burden according to the formula tumor volume = 1/2 × length × width2.
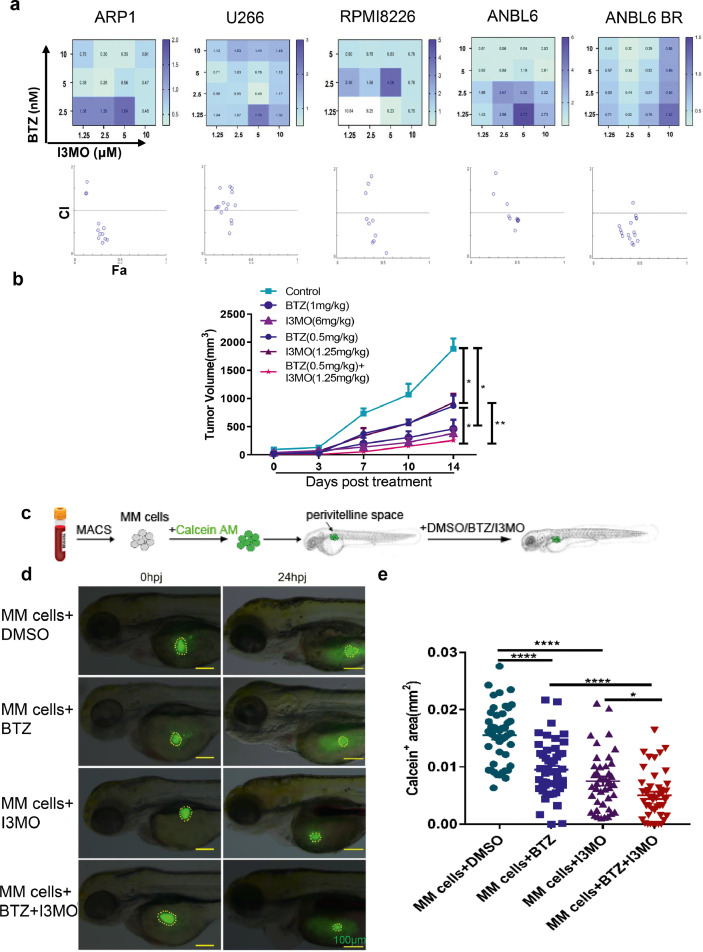
Figure 3.
I3MO sensitizes MM cells to bortezomib-induced apoptosis. (a) The apoptosis of ARP1, U266, RPMI8226, ANBL6 and ANBL6 BR cells with monotherapy or combination therapy of BTZ (1.25,2.5,5,10 nM) or I3MO (1.25,2.5,5,10 µM) were detected after the treatment for 24 h by flow cytometry, respectively. Isobologram analysis shows the synergistic cytotoxic effect of BTZ and I3MO. CI value<1 indicates synergistic interactions. The experiments were performed in triplicate. (b) Xenograft mouse model of MM was utilized to detect the I3MO effects on MM cell growth. There were six treatment groups with varying dosage of I3MO and BTZ as indicated. (P<0.05, Two-way ANOVA). (c) Primary patient samples (n=5) derived xenograft model was further utilized to detect the cytotoxicity of I3MO in zebrafish. Schematic diagram of experimental design. (d) Patient-derived MM cells were labeled with Calcein-AM and implanted into the perivitelline space of each zebrafish. The survival of engrafted cells was monitored at 0 hpi and 24 hpi. Fluorescent microscopy imaging of Calcein-AM-labeled patient MM cells in zebrafish embryos treated with DMSO, BTZ, I3MO, or BTZ and I3MO, at 0 and 24hpi, respectively. Dashed lines circle the primary MM cells. (Scale bars: 100 μm.) (e) Quantification of Calcein-AM-positive (green) areas in zebrafish embryos at 24 h post-injection (5 patients/treatment) (P<0.05, t test). Data are mean ± SEM. NS, not significant; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, unpaired t test. MACS, magnetic cell sorting; MM cells, multiple myeloma cells; hpi, hours post-injection.
For Figure 7j, PSME3, PSME4 shRNA ARP1 cells was injected subcutaneously into the left flanks of 6- to 8-weeks old female NOD/SCID mice. Mice with tumor volume of 100 mm3 (10 days after the injection) were randomized and treated for 6 weeks with doxycycline (DOXY) by oral administration (0.1 mg/ml biweekly).
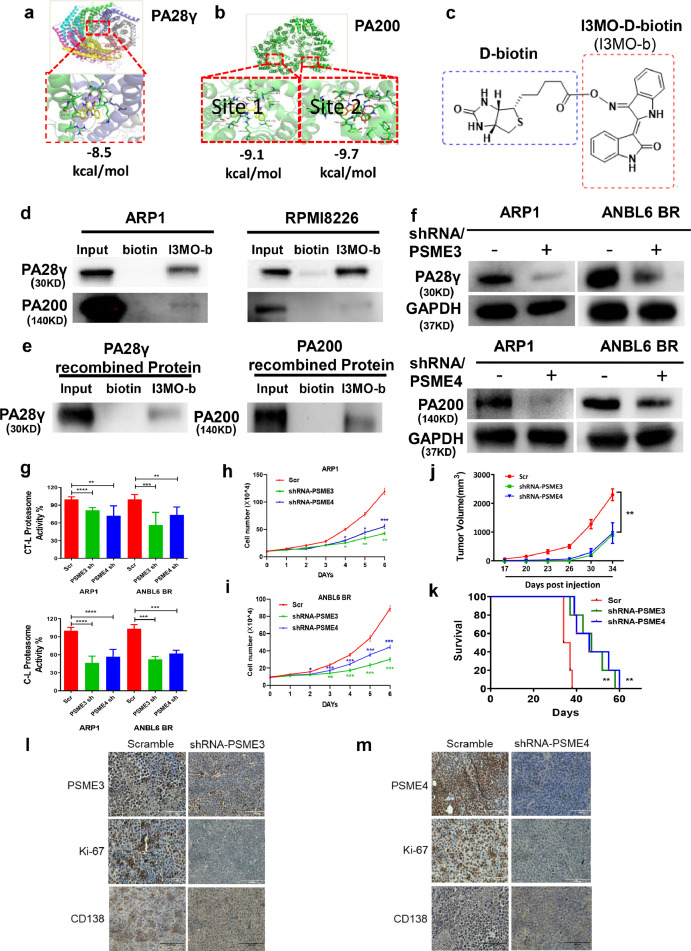
Figure 7.
Knockdown PSME3 and PSME4 inhibited proteasome activity and MM cell growth. (a-b) The binding site and the binding model between I3MO and its target proteins, PA28γ or PA200 were examined by Auto Dock Vina analyses. (c) I3MO was labeled with D-biotin. (d-e) Cell lysates from ARP1 and RPMI8226 or PSME3 and PSME4 recombinant protein were incubated overnight with either 200 µM I3MO-D-biotin or D-biotin, I3MO bound complex was separated with streptavidin MagBeads. The pull-down protein was identified by western blots with primary antibody PA28γ, PA200. (f) Knockdown PSME3 and PSME4 in ARP1 and ANBL6 BR cells were confirmed by western blots. (g) The assays of chymotrypsin- (CT-L) and caspase-like (C-L) proteasome activity were examined in PSME3 and PSME4 knocking down MM cells (P<0.05, t test). (h,i) Cell proliferation was measured by absolute cell counting in PSME3 and PSME4 knocking down MM cells. All results were presented as means ± SEM of three independent experiments. (j) Knockdown of PSME3 or PSME4 inhibits MM cell growth was investigated in vivo. Tumor volumes were monitored every other day once the tumors could be touched (n=5/group), bars represent the means ± SEM each group (P<0.01, Two-way ANOVA). (k) The survival of mice was calculated by Kaplan-Meier analyses (P<0.01, log rank test). (l,m) The levels of PSME3, PSME4, Ki-67 and CD138 in tumors were detected by immunohistochemistry. (Scale bars: 100 μm.) The experiments were performed in triplicate. *P < 0.05; **P < 0.01; ***P < 0.001.
When the tumor volume reached 2000 mm3 the mice were euthanized in a carbon dioxide chamber.26,28 Tumor formation was examined in 5 mice in each group. All procedures for in vivo studies were performed according to a protocol approved by the Animal Care and Use Committee (IACUC) of the Institute of Hematology, Chinese Academy of Medical Science, and compliance with animal use guidelines. All mouse work was approved by the Ethical Review Committee of State Key Laboratory of Experimental Hematology, National Clinical Research Center for Blood Diseases, Institute of Hematology & Blood Diseases Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, China (the reference number is IHCAMS-DWLL-2018-KT007-1).
Toxicological studies of I3MO
To determine the hematopoietic toxicity of I3MO, 6- to 8-weeks old female Balb/c mice were treated with I3MO (6 mg/kg, i.p., every other day) for a month. The amounts of white blood cell (WBC) and the body weight were monitored.
Molecular docking
The 3D structure of PA200 was taken from Protein Data Bank with code 6KWX. Since no crystal structure of PA28γ was reported previously, homology modeling method was employed to predict the PSME3 structure on Swiss Model server.33 Auto Dock Vina34 was used to determine the binding site and binding mode of I3MO and target. Both the receptors and I3MO were prepared in UCSF Chimera software with Dock Prep tool. The grid box was generated to cover the entire receptor with the receptor center as the grid center. Ten possible binding poses were obtained after docking calculations, and the one with the lowest binding affinity was selected as the most stable conformation.
Biotin-based pull-down assay
I3MO was labeled with D-biotin (#60338ES03, YEASEN).35 D-biotin worked as negative control. Cell lysates from MM cell lines (ARP1 and RPMI8226), and the recombinant proteins for PSME3 (P03968, Solarbio) or PSME4 (P05708, Solarbio) were incubated with I3MO-D-biotin (200 µM) at 4°C overnight, respectively. Then, streptavidin MagBeads (#L00424, GenScript)36 were added to cell lysates and rocked gently at room temperature for extra 3 h, followed by PBS washing three times (1 mL for each time) to thoroughly remove the non-specifically bound proteins. The beads left were re-suspended in SDS-PAGE sample buffer (90 µL), and heated at 95 °C for 5 min. The pull-down protein was identified by western blots with primary antibody of PSME3 and PSME4, respectively.
Flow cytometric assay for intracellular monoclonal light chain expression
Intracellular clonal light chain levels were determined using APC anti-human Ig light chain k antibody (#316510, RRID:AB_493615; Biolegend), and FITC anti-human Ig light chain λ antibody (#316606, RRID:AB_493625; Biolegend) staining according to the manufacturer's instructions. Data were analyzed by FlowJo software (Treestar, Ashland, OR).
Statistical analysis
Data are presented as the mean ± SD or mean ± SEM, which was indicated in the figure legends. Statistical analysis was done using GraphPad Prism (version 8.01, GraphPad software Inc.) The two-tailed Student's t-test was used to compare two groups. Two-way analysis of variance (ANOVA) was used to assess more than two groups. Kaplan-Meier was utilized in the survival analyses. Differential expressed genes (DEGs) after I3MO treatment were identified with the EdgeR package in R software with a threshold fold change (FC) >=2.0 and P < 0.05. Differences were considered significant when the P value was <0.05 (*), <0.01 (**), <0.001 (***) or <0.0001 (****).
Role of founders
This work was funded by following foundations: 82170194, 2018RC320012, 81920108006, 2018PT31006, 2018ZX09733003 and (CIFMS) 2021-1-I2M-040.The funders have no roles in study design, data collection, data analysis, interpretation or the writing of this research.
Results
I3MO presents a promising cytotoxicity in MM cells
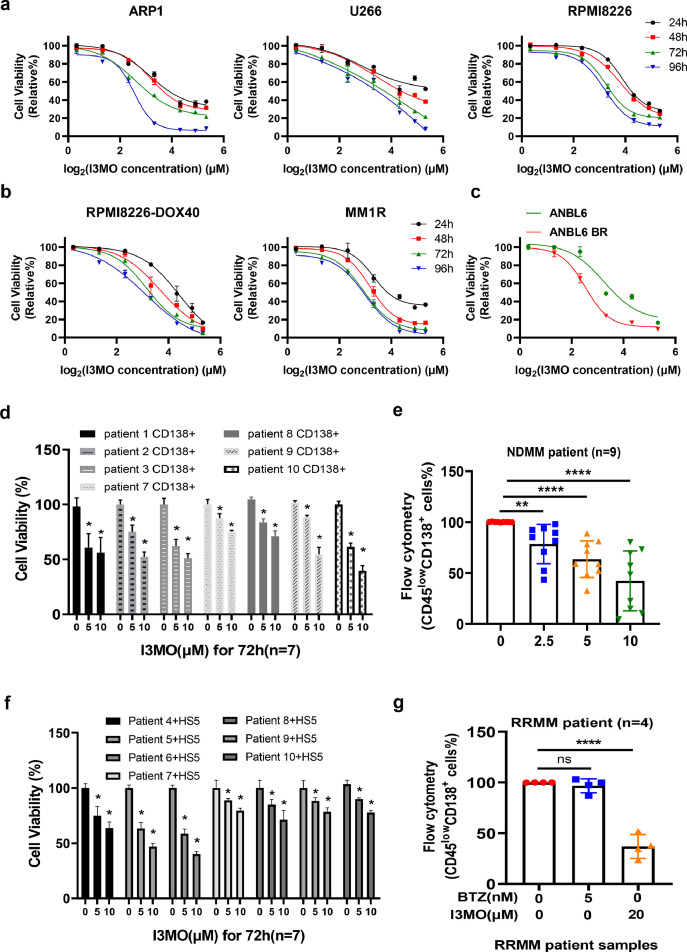
To determine the anti-MM activity of I3MO, we performed the in vitro and in vivo study with a panel of MM cell lines and patient primary samples. We found that I3MO treatment significantly suppressed the growth of MM cells. The viability of MM cell lines (ARP1, U266 and RPMI8226) were significantly decreased with I3MO treatment (Figure 1a). I3MO induced a dose-dependent decrease in viability of ARP1, U266 and RPMI8226 MM cell lines. The IC50 was (5.56±0.71) μM, (9.92±1.21) μM, and (8.88 ±0.79) μM in ARP1, U266, and RPMI8226 cells at 96 h, respectively (Figure 1a). Intriguingly, we found that I3MO also restrained the growth of drug resistance MM cells, including the resistance to conventional therapeutic agents, doxorubicin (RPMI8226-DOX40), dexamethasone (MM.1R), and the proteasome inhibitor bortezomib (ANBL6-BR) (Figure 1b and 1c). Of note, compared with the parental cells, bortezomib-resistant MM cells were more sensitive to I3MO. IC50 in ANBL6-BR cells was (5.76±0.34) μM compared to (9.47±1.37) μM in ANBL6 cells (P<0.001,t test, Figure 1c). These variable IC50 of I3MO in diverse MM cell lines probably due to their distinct cytogenetic and biological backgrounds. To further confirm the anti-MM effects of I3MO, the patient primary samples were examined. Consistently, I3MO induced a dose-dependent suppression in viability of patient primary CD138+ cells, which decreased to (75.52±15.45) % at 5 μM, and to (57.49±12.78) % at 10μM for 72 h, compared to the untreated group (n=7, P<0.05, t test, Figure 1d). In addition, I3MO treatment (5μM) did not show notably cytotoxicity on normal peripheral blood mononuclear cells (PBMCs, data not shown), which suggests a favorable therapeutic specificity and no cytotoxicity against normal cell counterparts.
Figure 1.
Cytotoxic effects of I3MO on the growth of MM cells. (a) ARP-1, U266 and RPMI8226 cells were treated with the indicated concentrations of I3MO for 96 h, followed by an assessment of cell viability by CCK-8 assays (n=3). (b) Drug-resistant MM cell lines (RPMI8226 Dox-40 and MM.1R) were treated with the indicated concentrations of I3MO for 96 h, following the cell viability detection (n=3). (c) Wild-type (ANBL6) and BTZ-resistant (ANBL6 BR) cell lines were treated with I3MO for 96 h, followed by an assessment of cell viability and the calculation of IC50. The viability experiments were performed in triplicate (P<0.001, t test). (d) Purified CD138+ cells from MM patient primary samples (n=7) were treated with the indicated concentrations of I3MO for 72 h. Cell viability was calculated by CCK-8 assays (P<0.05, t test). (e) BMNCs from newly diagnosed MM patients (NDMM) (n=9) were treated with the indicated concentrations of I3MO for 24 h, and the percentage of CD45lowCD138+ myeloma cells was determined by flow cytometry (P<0.05, t test). (f) Purified CD138+ cells from MM patients (n=7) were co-cultured with the HS-5 stromal cell line and treated with I3MO for 72 h, followed by an assessment of cell viability by CCK-8 assay (P<0.05, t test). (g) BMNCs isolated from relapsed and refractory MM patients (RRMM) were treated with BTZ or I3MO for 24 h, and the percentage of CD45lowCD138+ cells was determined by flow cytometry. Data are presented as the mean ± SEM (n=4) (P<0.0001, t test). *P < 0.05; **P < 0.01; ****P < 0.0001.
We have shown that stromal cells in the bone marrow promote MM cell survival and drug resistance.18,37 To further investigate whether the tumor associated microenvironment (TAM) inhibits the anti-MM effects of I3MO, bone marrow mononuclear cells (BMNCs) including both tumor and non-tumor cells from patient samples were isolated and treated with I3MO. The proportion of residue CD45lowCD138+ cell was then determined by flow cytometry. We also examined the effect of I3MO on CD138+ MM patient cells co-cultured with the stromal cell line HS5. Flow cytometry data showed that I3MO significantly suppressed the growth and survival of CD45lowCD138+ MM cells even in the presence of stromal cells (Figure 1e). The cell viability decreased to (78.41±12.65) % and (66.57±14.50) % after I3MO treatment at 5 μM or 10 μM for 72 h, respectively (n=7, P<0.05, t test, Figure 1f). More importantly, the survival of bone marrow mononuclear cells (BMNCs) from RRMM patients with remarkably resistance to bortezomib could be suppressed by I3MO treatment. Our data showed that the growth of CD45lowCD138+ MM cells in RRMM patients was significantly inhibited by I3MO treatment (n=4, P<0.0001, t test, Figure 1g). Altogether, these findings indicated that I3MO is an effective cytotoxicity agent against MM cells in both wide-type and drug-resistant MM cells. Moreover, I3MO could suppress MM cell survival even under the protection of tumor associated microenvironment (TAM).
I3MO sensitizes MM cells to bortezomib-induced apoptosis
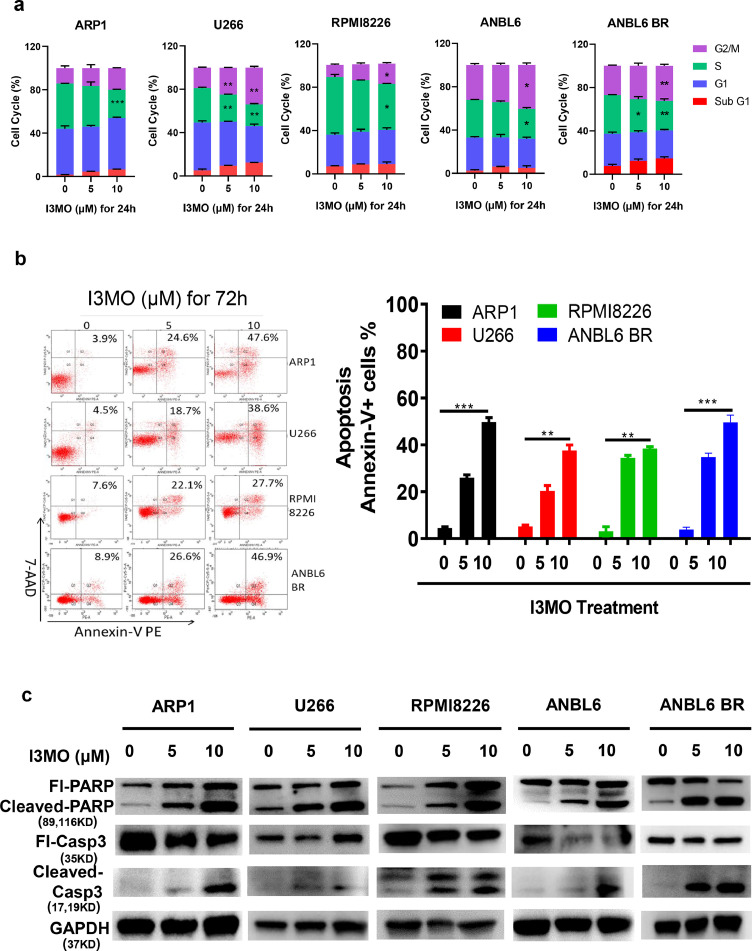
Next, we determined the effects of I3MO on cell cycle and apoptosis induction of MM cells. Flow cytometry analyses showed that I3MO treatment effectively caused cell cycle arrest of MM cells in drug-sensitive ARP1, U266, RPMI8266, ANBL6 and bortezomib-resistant ANBL6-BR cell lines (Figure 2a). Many studies have reported the cell cycle arrest induced by I3MO treatment in cancer cells, which consistently in our study. Due to the diverse background (cytogenetic and biological heterogeneity), the cell cycle arrest presents at different manner in MM cell lines. We found G2/M arrest in U266, RPMI8226, ANBL6, and ANBL6 BR MM cells. In addition, S phase significantly decreased in all five MM cell lines, especially in high dose groups. Of note, I3MO treatment triggered a remarkable apoptosis of MM cells (Figure 2b), which associated with proteins PARP and caspase 3 cleavage, as shown in Figure 2c.
Figure 2.
I3MO induces the apoptosis of MM cells. (a) The cell cycle was evaluated by flow cytometry in ARP1, U266, RPMI8226, ANBL6 and ANBL6 BR cells after the treatment with I3MO for 24 h (P<0.05, t test). (b) The cell apoptosis of MM cell lines (ARP-1, U266, RPMI8226 and ANBL6 BR) were detected with a 7AAD-Annexin V double staining assay (n=3) after the treatment of I3MO (P<0.05, t test). (c) Western blot was utilized to detect the level of apoptosis related proteins after I3MO treatment. The experiments were performed in triplicate. *P < 0.05; **P < 0.01; ***P < 0.001.
Since I3MO had a cytotoxic effect on both BTZ-sensitive and BTZ-resistant MM cells, we further asked whether I3MO and bortezomib have synergistic effects in the treatment of myeloma. The CI is considered the gold standard for the synergism of drug-drug interactions. A CI value = 1 always indicates an additive effect, while CI values <1 indicate synergistic interactions. Thus, the lower CI value, the stronger synergism. Our data showed that I3MO remarkably enhanced MM cell sensitivity to bortezomib-induced apoptosis. Annexin V positive apoptotic MM cells were significantly increased, especially in the cohort treated with combination of I3MO and bortezomib. I3MO and BTZ exhibited synergistic effects in both wild-type and BTZ-resistant cells, as shown by CI values < 1.0 (Figure 3a).
We further utilized two different in vivo strategies to address I3MO anti-MM efficacy and confirmed the combination anti-MM activity of I3MO and bortezomib. Firstly, the xenograft myeloma mouse model was performed. Thirty tumor-bearing mice were divided into six randomized treatment groups (n=5 in each group), including (a) PBS control group, (b) bortezomib treatment groups, a low dose one (0.5 mg/kg, two times/a week) or (c) regular dose one (1 mg/kg, two times/a week), (d) I3MO treatment groups, high dose, 6 mg/kg, every other day or (e) low dose, 1.25 mg/kg, every other day, and (f) I3MO and bortezomib combination group (I3MO: 1.25 mg/kg and bortezomib: 0.5 mg/kg). As Figure 3b showed, I3MO treatment (6 mg/kg) significantly reduced the tumor burden compared to that in the PBS control group (382.5±44.4 mm3 v.s. 1881.6±82.8 mm3, P<0.05, Two-way ANOVA), and the extent tumor burden was equivalent to treatment with bortezomib at regular dosage of 1 mg/kg (461.8±80.9 mm3 v.s. 1881.6±82.8 mm3, P<0.05, Two-way ANOVA). The tumor volume of the group of treatment with low dose of I3MO (1.25 mg/kg) compared to the PBS control group was (929.4±39.2 mm3 v.s. 1881.6±82.8 mm3, P<0.05, Two-way ANOVA), or with low dose of bortezomib (0.5 mg/kg) compared with the control group was (887.7±74.7 mm3 v.s. 1881.6±82.8 mm3, P<0.05, Two-way ANOVA). Strikingly, the combination treatment group with low dose of I3MO (1.25 mg/kg) and BTZ (0.5 mg/kg) prominently restrained the tumor burden compared to either treatment with regular I3MO (236.1±40.7 mm3 v.s. 929.4±39.2 mm3, P<0.01, Two-way ANOVA) or BTZ alone (236.1±40.7 mm3 v.s. 887.7±74.7 mm3, P<0.01, Figure 3b Two-way ANOVA). This data indicated that the effects of tumor suppression with lower dose combination treatment were equal to the regular treatment either with I3MO or BTZ. In addition, to determine the toxicity of I3MO treatment, we did routine examination to monitor the amounts of white blood cell (WBC) and the body weight of Balb/c mice during I3MO treatment. Our results indicated that there is not conspicuous hematological toxicity and the body weight lost in treatment with I3MO (Figure S1a), which supports the well tolerance and effective anti-MM effects of I3MO. The combination therapy with decreased dose of I3MO and BTZ might be benefit for clinical practice with alleviation side effects induced by the agents.
Zebrafish is an ideal model to investigate the cytotoxicity of compounds on primary MM patient samples, which secret growth factors, such as IL-6 and SDF1α to support primary MM cells growth.30,38 To investigate the I3MO effects on primary MM patient cells in vivo, the patient-derived xenograft (PDX) model of zebrafish was further utilized follow the protocol reported previously. Primary MM patient cells were isolated and enriched with CD138 microbeads, further labeled with Calcein-AM (0.1μM), and then transplanted into the perivitelline space of zebrafish embryos. Xenograft-positive zebrafish embryos were divided randomly and treated with (a) DMSO (control group), (b) BTZ (10 nM), (c) I3MO (5 μM), and (d) I3MO/BTZ combination (10 nM and 5 μM, respectively). The tumor load was examined by quantification of Calcein-AM (green) positive areas in zebrafish, and compared in different groups after 24 h treatment. As Figure 3c–e showed, either monotherapy or combination therapy of I3MO eliminates the engraftment of MM cells in zebrafish embryos (P<0.05, t test). Together, these in vitro and in vivo results therefore confirmed the anti-MM activity efficiently induced by I3MO induction, and strikingly enhances the cytotoxicity of bortezomib in combination therapy.
I3MO is a multifaceted regulator of myeloma cell death
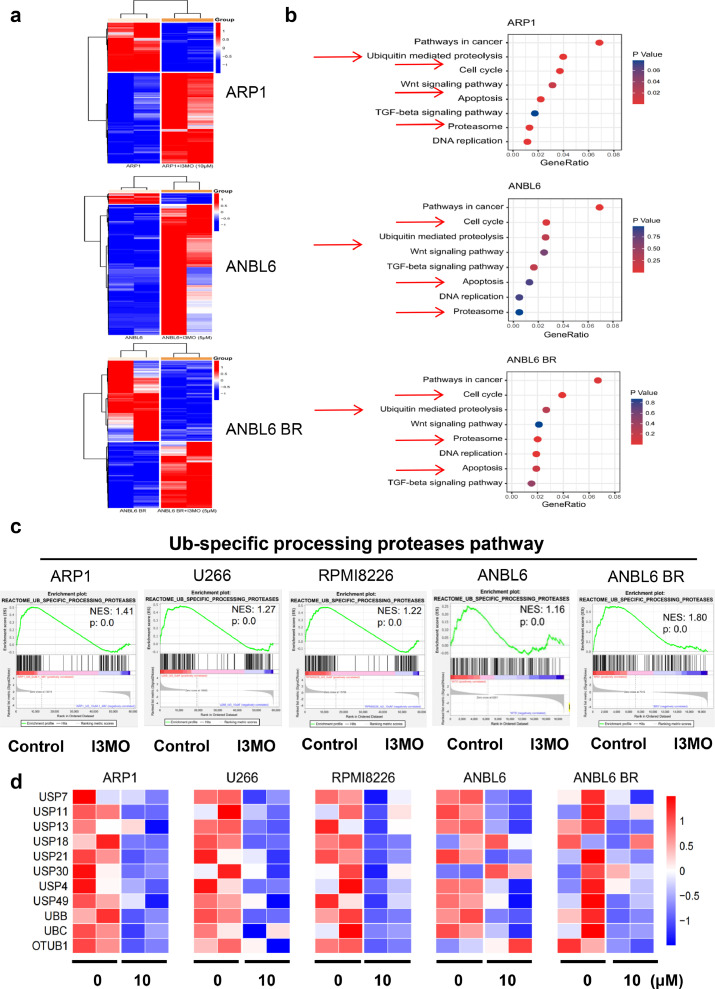
To investigate the mechanism of I3MO anti-MM ability, RNA sequencing of MM cells before and after the treatment of I3MO was performed. The heatmaps showed the differentially expressed genes (DEGs) in MM cells that induced by I3MO treatment in a panel of MM cell lines (FC>=2.0, P<0.05, edgeR). There was 3322 DEGs (2155 up and 1167 down) in ARP1, 2233 DEGs (959 up and 1274 down) in U266, 1412 DEGs (442 up, 970 down) in RPMI8226, respectively (10μM for 24 h, Figures 4a and S1b). Additionally, there was 2730 DEGs (2524 up and 206 down) in ANBL6, and 1815 DEGs (820 up and 995 down) in ANBL6BR cell lines after treatment with I3MO (5μM for 24 h). Of note, DEGs in MM cell lines treated with or without I3MO were commonly enriched in multiple pathways according to KEGG analysis, including Ubiquitin mediated proteolysis, Cell cycle, Proteasome, and Apoptosis pathways (Figure 4b). These results consistent with our data showed above that I3MO induced significantly cell cycle arrest and apoptosis in MM cell lines. Furthermore, Gene Set Enrichment Analyses (GSEA) data showed that DEGs in these MM cell lines which induced by I3MO were significantly suppressed in Ub-specific processing proteasomes pathway, as shown in Figure 4c. Furthermore, the UPP related genes were remarkably down-regulated in MM cell lines after the treatment of I3MO, which included USP7, USP11, USP13 etc., as Figure 4d heatmaps showed. These results indicated that I3MO is a multifaceted regulator of cell death. Ub-specific processing proteasomes pathway is a common mechanism that plays pivotal roles in anti-MM effect of I3MO.
Figure 4.
I3MO is a multifaceted regulator of cell death. (a) The heatmaps showed the differentially expressed genes (DEGs) in ARP1, ANBL6 and ANBL6 BR cells treated with I3MO for 24 h. (b) KEGG pathway analyses was performed after I3MO treatment. (c) GSEA analyses was performed to analyze pathways and function annotation. (d) Heatmaps showed decreased expression of genes related to UPP pathway among five MM cells after I3MO treatment.
I3MO suppresses the growth of MM cells via down-regulating USP7 expression
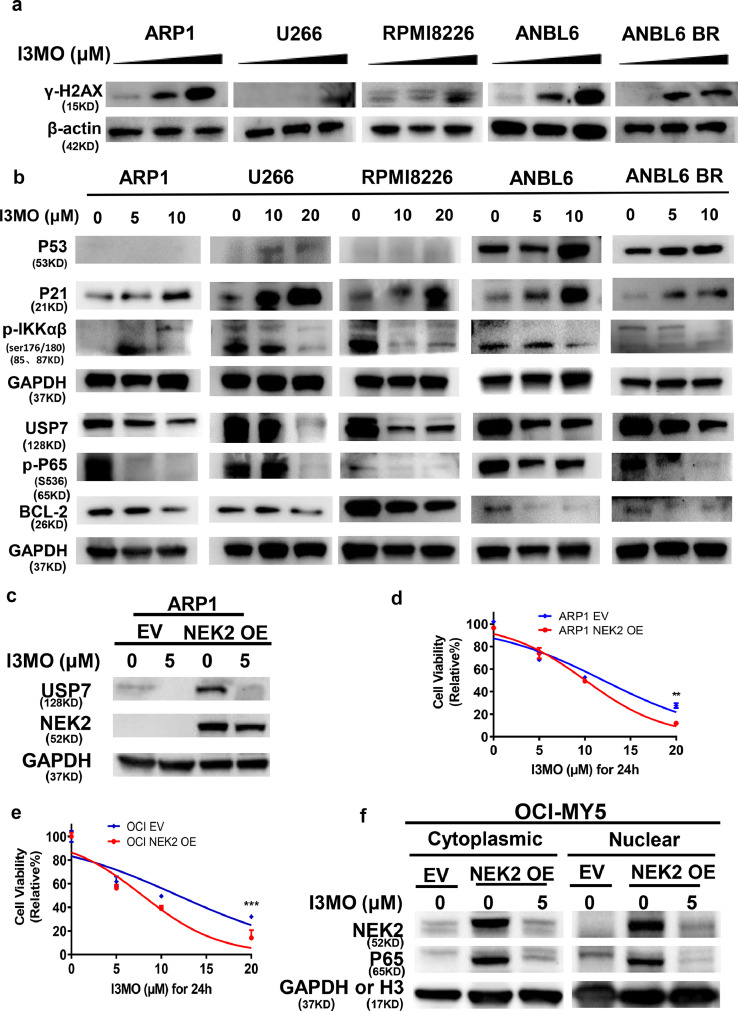
To confirm the signaling pathway mediating I3MO anti-MM activity, five MM cell lines (ARP1, U266, RPMI8266, ANBL6 and ANBL6 BR) were further analyzed. Western blots showed that γ-H2AX, a sensitive molecular marker of DNA damage and repair, was significantly enhanced in MM cell lines after I3MO treatment at dose dependent manner (Figure 5a), associated with increased p53 and p21 levels. In contrast, the anti-apoptotic protein BCL-2 was down-regulated in MM cell lines after I3MO treatment (Figure 5b). Interesting, we found that I3MO treatment upregulated p21 level even in p53 null ARP1 MM cells, suggesting that its cytotoxic activity presents a p53 independent way. Additionally, NF-kB pathway activators phosphorylated IKKαβ (s176/s180) and phosphorylated p65 (s536) were notably down-regulated after treatment of I3MO in MM cell lines. Since p53 and p65 both are the substrates of USP7, the USP7 level was examined. Western blots showed that I3MO induced a dose-dependent decrease of USP7 (Figure 5b). These findings suggested that USP7 downregulated by I3MO triggers p53 and p65 degradation and suppresses its downstream signaling transduction.
Figure 5.
I3MO suppresses the growth of bortezomib-resistant cell via down-regulating USP7 expression. (a) The level of γ-H2AX was detected by western blots in ARP1, U266, RPMI8226, ANBL6 and ANBL6 BR cells which treated with I3MO (0, 5, 10 μM) for 24 h. (b) Western blots were utilized to detect the levels of p53, p21, BCL-2, p-IKKα/β (Ser176/180), p-p65-S536, USP7 and GAPDH in ARP1, U266, RPMI8226, ANBL6 and ANBL6 BR cells which treated with I3MO for 24 h. (c) The levels of USP7 and NEK2 were evaluated by western blots in ARP1 cell line with EV and NEK2-OE. (d) ARP1 cell line with EV and NEK2-OE cells were treated with I3MO for 24 h, followed by an assessment of cell viability (P<0.01, Two-way ANOVA). (e) OCI-MY5 cell line with EV and NEK2-OE cells were treated with I3MO for 24 h, followed by an assessment of cell viability (P<0.001, Two-way ANOVA). (f) OCI-MY5 cells with EV and NEK2-OE cells were treated with or without I3MO for 24 h, and cytosolic and nuclear fractionation was carried out. The levels of NEK2 and p65 were analyzed by western blots. GAPDH or Histone H3 (H3) was used as cytosolic and nuclear markers, respectively. The experiments were performed in triplicate. **P < 0.01; ***P < 0.001.
Our previous study indicated that USP7/NEK2/NF-κB pathway activation plays pivotal roles in bortezomib resistance in MM.7,39 Since the stabilization of NEK2 by USP7 is required for the release of p65 into the nucleus, we further investigate whether downregulation of USP7 involved in I3MO efficacy via triggering NEK2 degradation and suppresses its downstream signaling transduction. The NEK2-OE BTZ-resistance ARP1 and OCI-MY5 MM cell lines were utilized. Western blots showed that I3MO triggered decreased expression of USP7 and NEK2 (Figure 5c). Cell viability significantly decreased in NEK2-OE ARP1 cells from 96.55% to 11.94% after I3MO treatment at 20 μM for 24 h compared with ARP1-EV cells from 100.00% to 27.61% (P<0.01, Two-way ANOVA, Figure 5d). To further investigate whether inhibition of NF-κB triggered by I3MO treatment could overcome bortezomib resistance, NEK2 overexpression or empty vector (EV) OCI-MY5 cells were next incubated with I3MO. We found that I3MO also caused a dose-dependent decrease in viability of NEK2-OE bortezomib resistant MM cells (from 99.99% to 14.11%, at 20μM for 24 h, compared with from 100% to 32.05% in EV OCI-MY5 cells (P<0.001, Two-way ANOVA, Figure 5e). Nuclear and cytoplasmic fractionations were performed to determine if p65 was stabilized in the nucleus of after treatment with I3MO. The protein level of p65 was increased in both cytoplasmic and nuclear fractions in NEK2-OE bortezomib resistant MM cells compared with EV control cells. Consistently, both cytoplasmic and nuclear accumulation of NEK2 and p65 was reversed after I3MO treatment (Figure 5f), suggesting that I3MO impaired the stabilization of NEK2 and p65 via down-regulation of USP7. Together, our data therefore indicate that I3MO downregulates the level of USP7 and promotes NEK2 degradation, resulting in the suppression of downstream targets and NF-κB signaling, and inducing cell death in bortezomib resistant MM cells. These findings also indicate that the anti-MM activity of I3MO is associated with decreased USP7 and its down-stream targets p53 and NF-kB pathway. These results further support that Ub-specific processing proteases pathway (UPP pathway) is the potential target of I3MO.
I3MO works as proteasome inhibitor via suppressing PSME3 and PSME4
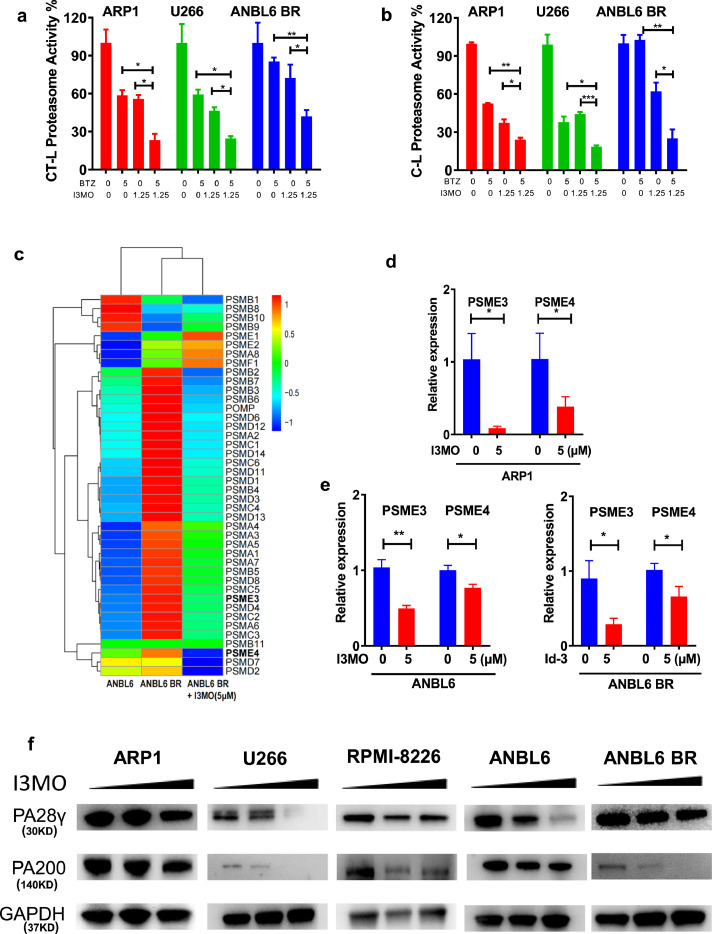
Importantly, our GSEA analyses also suggested that I3MO impacts proteasome activity (Figure 4). Therefore, to further validate the I3MO effects on proteasome activities, ARP1, U266 and ANBL6 BR MM cell lines were analyzed. Decreased proteasome activities, chymotrypsin-like (CT-L) and caspase-like (C-L), were observed in drug sensitive (ARP1, U266) and bortezomib resistant (ANBL6 BR) MM cell lines in treatment with I3MO (Figure 6a and b). Furthermore, the combination of I3MO and bortezomib further decreased the CT-L and C-L proteasome activities compared to bortezomib or I3MO alone treatment (Figure 6a and b).
Figure 6.
I3MO works as proteasome inhibitor via suppressing PSME3 and PSME4 expression. (a) The chymotrypsin- (CT-L) and (b) caspase-like (C-L) proteasome activity of ARP1, U266 and ANBL6 BR cells were examined after the treatment with I3MO monotherapy or combination therapy (P<0.05, t test). (c) The heatmap showed several genes of protease complex were down-regulated by I3MO induction. (d,e) ARP1, ANBL6 and ANBL6 BR (BTZ-resistant) cells were treated with or without I3MO (5 µM) for 24 h, and the mRNA levels of PSME3 and PSME4 were detected by real time-PCR (P<0.05, t test). (f) Western blots were utilized to detect the levels of PA28γ, PA200 before and after the I3MO treatment. The experiments were performed in triplicate. *P < 0.05; **P < 0.01; ***P < 0.001.
RNA-seq data showed that 42 protease complex genes were regulated by I3MO treatment (5 µM for 24 h) in ANBL6 BR myeloma cells, including 34 down-regulated and 7 up-regulated genes, and no difference in PSMB11 (Figure 6c). The proteasome is an intracellular protease complex including the 20S catalytic core and its associated 19S complex, PA28αβ, PA28γ, PA200, and PI31. Since alternative proteasome genes of PSME3 (PA28γ) and PSME4 (PA200) mediate substrate degradation in an ubiquitin-independent manner,41 we next evaluated the function of PSME3 and PSME4 in MM cells survival after the treatment of I3MO. Real-time PCR data confirmed that the decreased levels of PSME3 and PSME4 in ARP1, ANBL6 and ANBL6 BR MM cell lines treated with I3MO overnight at 5µM (Figure 6d and e, P<0.05, t test). Western blots confirmed that the protein levels of PA28γ and PA200 were downregulated in both drug-sensitive ARP1, U266 and bortezomib-resistance ANBL6 BR MM cell lines (Figure 6f). Together, these findings support the notion that I3MO acts as proteasome inhibitor which targets the alternative proteasome subunits PSME3 and PSME4.
To further explore whether there is a direct binding site and characterize binding between I3MO and its target proteins, PA28γ or PA200, AutoDock Vina analyses was performed.33 Both the receptors and I3MO were prepared in UCSF Chimera software with Dock Prep tool. The grid box was generated to cover the entire receptor, with the receptor center as the grid center. Ten possible binding poses were obtained after docking calculations, and the one with the lowest binding affinity was selected as the most stable conformation. As shown in Figure 7a and b, PA28γ is a doughnut-shaped homo heptamer, and the obtained top pose of I3MO locates between two monomers with a binding affinity of -8.5 kcal/mol. The interacting residues include Ala186, Ser187, Leu189, Ser137, Gly138, Lys139, Gln191, Asp190, Arg194, Ser229 and Lys225, among which three residues (Gln191 and Ser229 of one monomer, and Asp190 of another monomer) are involved in polar interactions and Lys225 creates a cation-π interaction with the aromatic ring structure of I3MO. The PA28γ heptamer is shown with each chain in a different color, and I3MO is shown in yellow carbon stick model (Figure 7a). The predicted binding of I3MO with PA200 can be classified into two sites, I3MO in site 1 and site 2 shown in yellow and orange carbon scheme, respectively. PA200 is shown in green carbon scheme (Figure 7b). Site 1 is characterized by two tyrosine residues (Tyr732 and Tyr736) as pocket bottom, and two arginine residues (Arg726 and Arg914) as pocket edge. Site 2 locates near the C-terminal of PA200. The obtained binding affinities of top poses for these two sites are -9.1 kcal/mol and -9.7 kcal/mol, respectively. Our data also showed that the two ring structures of I3MO parallel in site 1 to the phenyl ring of Tyr732 and Tyr736, respectively, creating face-to-face π-π interactions. The polar atoms of I3MO form hydrogen bonds with Arg726 and Arg914, and the guanidine group of Arg685 has the potential to produce cation-π interactions with the ring structure of I3MO. In site 2, I3MO is surrounded by Thr338, Ser339, His342, Asn1716, Ser1772, pro1774 and Tyr1775, and polar interactions exist between I3MO and Ser339 and Asn1716 (Figure 7b).
The most straight forward way to determine interaction partners of a peptide is to use the peptide as bait in affinity pull-down experiments and then by direct detection of binding proteins. The biotinylated I3MO, I3MO-D-Biotin, were synthesized (Figure 7c). Cell lysates from ARP1 and RPMI8226 were incubated with I3MO-D-Biotin, and probed with streptavidin. Protein complexes followed by streptavidin pull-down, and PA28γ and PA200 were determined with western blots (Figure 7d). Our results demonstrated that PA28γ and PA200 were the direct targets binding with I3MO. These combinations were further confirmed in recombined proteins as Figure 7e showed. These results indicate that I3MO could direct bind with, and down-regulate the expression of PA28γ and PA200, which involved in the inhibition of proteasome activity.
Since a previous study has reported that alternative proteasome complexes persisted even when cells had regained proteasome activity after pulse exposure to proteasome inhibitors,41 we hypothesized that the activation of alternative proteasome might be associated with resistance to the traditional proteasome inhibitor bortezomib. As components of the alternative proteasome complex, PSME3 and PSME4 perform pivotal roles in proteasome formation and function.19 We therefore next constructed a doxycycline-inducible PSME3/PSME4-shRNA vector that was transduced into ARP1 and ANBL6 BR myeloma cell lines (Figure 7f), and then evaluated the proteasome activity. Knockdown PSME3 or PSME4 caused a remarkable decrease of the CT-L and C-L proteasome activity in both drug-sensitive (ARP1) and drug-resistant MM cells (ANBL6 BR) (Figure 7g). The cell proliferation of ARP1 cells showed a 64% reduction after PSME3 knockdown and a 45% reduction after PSME4 knockdown. There was a 65% reduction in cell proliferation of PSME3 knockdown, and a 50% reduction in PSME4 knockdown in ANBL6 BR cell line (Figure 7h and i). An in vivo test of PSME3 and PSME4 knockdown was performed in NOD/SCID xenograft model with doxycycline (DOXY) by oral administration (0.1 mg/ml biweekly). The tumor volume was significantly inhibited in groups of PSME3 and PSME4 knockdown compared with the control groups, as Figure 7j showed (P<0.01, Two-way ANOVA). Kaplan-Meier analysis showed that knockdown PSME3 or PSME4 significantly extended the survival of tumor-bearing mouse compared with the control group (Figure 7k, P<0.01, log rank test). The CD138+ MM cell and the Ki-67 level significantly decreased in the tumor of knockdown PSME3 and PSME4 groups (Figure 7l and m).
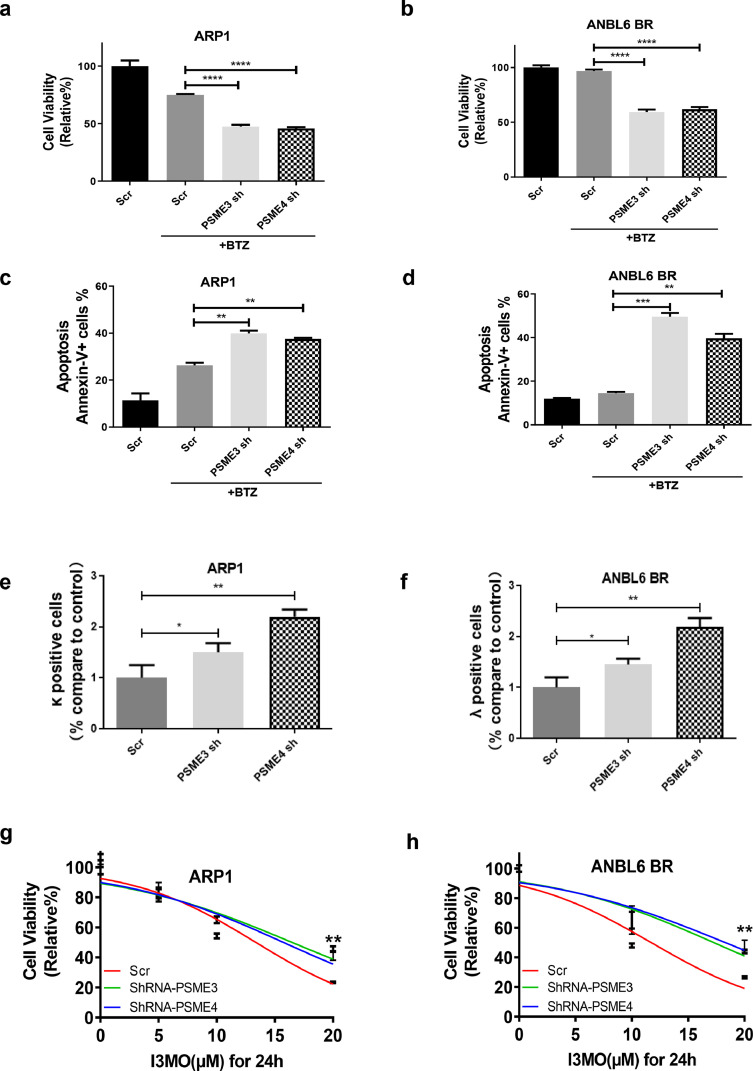
Knockdown PSME3 and PSME4 suppresses MM cell growth and sensitizes to bortezomib induction
Next, we evaluated whether PSME3 and PSME4 have effects on MM cells sensitivity to proteasome inhibitor. Cell viability analysis showed that MM cells were more sensitive to bortezomib-induced cell growth arrest when knockdown PSME3 or PSME4, especially in bortezomib-resistant cells ANBL6-BR (Figure 8a and 8b). Knockdown of PSME3 and PSME4 significantly increased MM cell apoptosis induced by bortezomib in both ARP1 and ANBL6 BR cell lines (Figure 8c and d). According to the load-versus-capacity model, cells equipped with fewer proteasomes, higher workload, or both should be more vulnerable to the toxic effects of bortezomib. To better understand the mechanism by which PSME3 and PSME4 sensitize to bortezomib-induced apoptosis in MM cells, the capability of protein synthesis in knockdown PSME3 or PSME4 ARP1 and ANBL6 BR MM cell lines was analyzed. Intracellular clonal light chain levels in ARP1 and ANBL6 BR MM cells were examined using APC anti-human Ig light chain k antibody and FITC anti-human Ig light chain λ antibody, respectively. We found that the κ light chain synthesis in ARP1 cells or λ light chain in ANBL6 BR cells was significantly increased when PSME3 or PSME4 were down-regulated (Figure 8e and f). In addition, we found that knockdown PSME3 or PSME4 significantly decreased MM cell sensitivity to I3MO (Figure 8g and h). These findings indicate that the efficacy against MM of I3MO by disturbing the balance between proteasome load and capacity. PSME3 and PSME4 in the setting of bortezomib resistance represent promising potential therapeutic targets.
Figure 8.
Knockdown PSME3 and PSME4 significantly enhances myeloma cell sensitivity to proteasome inhibitor. (a–d) The sensitivity to bortezomib treatment of MM cells with knockdown PSME3 and PSME4 was detected, including cell viability (a,b) and apoptosis (c,d) (P<0.05, t test). (e,f) Intracellular clonal light chain levels were determined in those MM cells after knockdown PSME3 and PSME4 (P<0.05, t test). (g,h) The sensitivity to I3MO treatment of MM cells with knockdown PSME3 and PSME4 was detected by CCK-8 assays (n=3) (P<0.01, log rank test). The experiments were performed in triplicate. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
High level of PSME3 or PSME4 associated with inferior outcome in MM patients
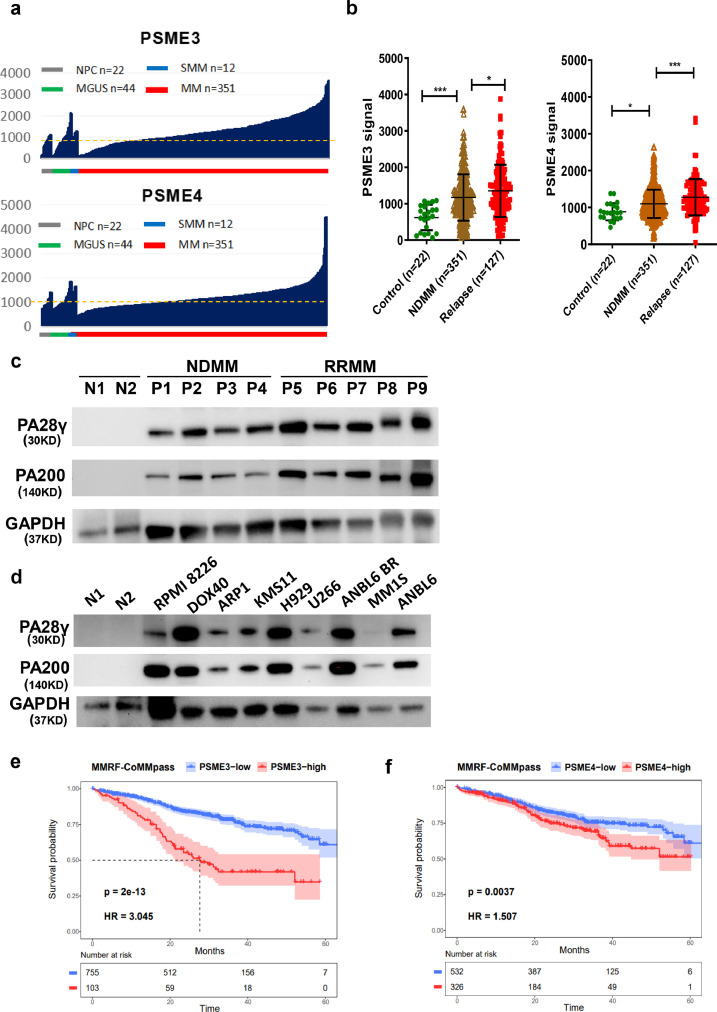
To further demonstrate the clinical significance of our finding, multiple independent clinical data sets were analyzed. Based on the public available GEO datasets (GSE5900 & GSE2658) of MM patients, we found that the levels of PSME3 (200987_x_at) and PSME4 (212219_x_at) were significantly increased in plasma cells of newly diagnosed MM patients (NDMM, n=351). When the cutoff for the affymetrix signals for PSME3 and PSME4 were set at 1,000, 134 out of 351 (38.2%, PSME3) and 123 out of 351 (35.0%, PSME4) NDMM patients exhibited significantly increased levels of PSME3 and PSME4, respectively, compared to the plasma cells from healthy donors (Figure 9a). Of note, the levels of PSME3 and PSME4 were further increased in patients with RRMM (P<0.05, t test) (Figure 9b). The Cancer Cell Line Encyclopedia (CCLE) transcriptome data set also showed that the levels of PSME3 and PSME4 were significantly higher in B-cell–derived malignancies, including MM, relative to other hematologic malignancies and solid tumors (FigureS1c).
Figure 9.
PSME3 and PSME4 are drug-resistance genes and are associated with inferior outcomes in myeloma patients. (a)The clinical significance of PSME3 and PSME4 in the GEO datasets of MM patient data was investigated. The expression of PSME3 and PSME4 was compared in plasma cells from healthy donors (NPC, n=22), individuals with monoclonal gammopathy of undetermined significance (MGUS, n=44), individuals with smoldering multiple myeloma (SMM, n=12) and newly diagnosed MM patients (n=351) from the total therapy 2 (TT2) datasets (GSE5900 & GSE2658). (b) The levels of PSME3 and PSME4 at baseline and after relapse (GSE31161) were compared (P<0.05, t test). (c) Western blots assay was utilized to detect the protein level of PA28γ (PSME3) and PA200 (PSME4) in purified CD138+ cells from new diagnosed (NDMM) (n=4) and relapsed MM patients (RRMM) (n=5). Normal plasma cells (n=2) were utilized as control. (d) PA28γ (PSME3) and PA200 (PSME4) expression in a panel of MM cell lines with normal plasma cells as control. (e and f) Kaplan-Meier analysis was performed in MM patients with varying levels of PA28γ (PSME3) (P<0.0001, log rank test). and PA200 (PSME4) in MMRF-CoMMpass clinical trial. (P=0.0037, log rank test). *P < 0.05; ***P < 0.001.
To confirm PSME3 or PSME4 upregulation at the protein level, the primary patient samples of NDMM and RRMM were examined. Western blots showed that the protein level of PA28γ (PSME3) and PA200 (PSME4) increased in NDMM patients, with further upregulation in the RRMM compared to normal plasma cells (Figure 9c). PA28γ and PA200 were significantly increased in a panel of MM cell lines relative to normal plasma cells (Figure 9d). Importantly, Kaplan-Meier analysis showed that MM patients with highly PA28γ (PSME3) or PA200 (PSME4) at the best expression signal cutoff using R package had inferior outcome (Figure 9e and f). A total of 12.0% (103/858) of MM patients with elevated PSME3 expression had inferior overall survival (OS) in MMRF-CoMMpass clinical trial (hazard ratio, HR=3.045, P<0.0001, log rank test, Figure 9e). And 37.0% (326/858) of MM patients with elevated PSME4 expression had inferior overall survival (OS) in MMRF-CoMMpass clinical trial (HR=1.507, P=0.0037, log rank test, Figure 9f). Together, these findings indicate that PA28γ (PSME3) and PA200 (PSME4) play pivotal roles in RRMM, and therefore represent promising therapeutic targets.
Discussion
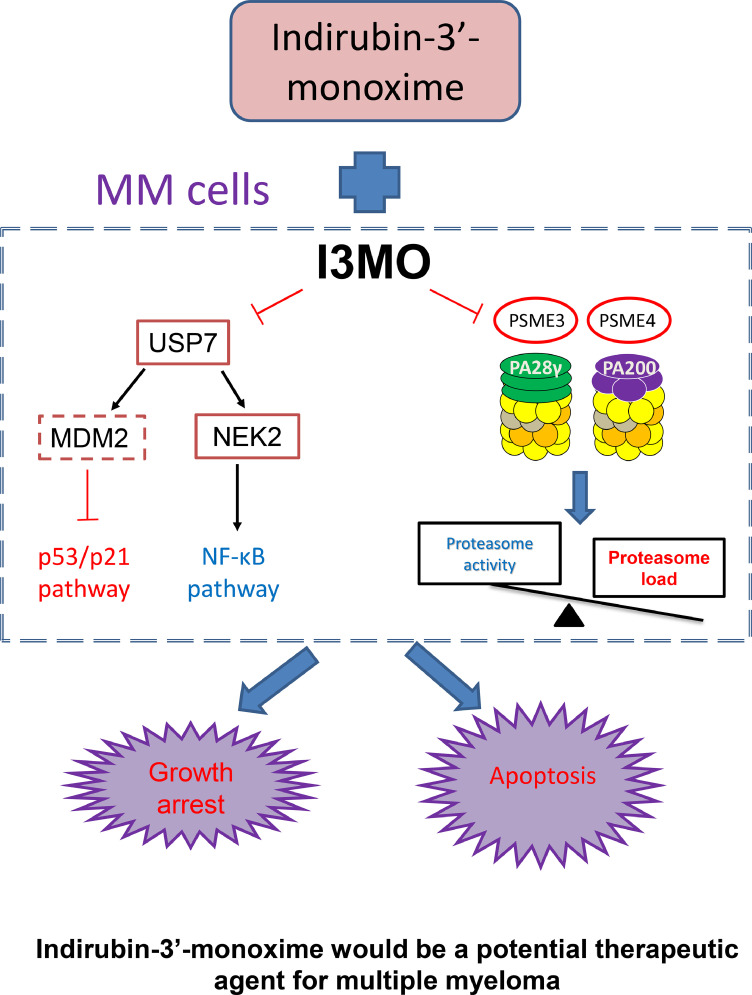
Three proteasome inhibitors have garnered regulatory approvals in MM, but drug resistance is an emerging challenge, prompting interest in potential therapies blocking regulator components of the proteasome pathway.25 Indirubin is one of many active constituents derived from a traditional Chinese prescription, Indigo Naturalis, which was identified by the researcher in our institute in the mid of 1970s.42, 43, 44 Indirubin-3′-monoxime (I3MO) was one of the ideal derivatives of Indirubin, with lower IC50 and enhanced cytotoxicity compared to indirubin itself.44 Several studies have reported that I3MO efficiently induces malignant cell apoptosis, inhibits cyclin-dependent kinases, and triggers anti-angiogenesis effects in hematologic malignancies and solid tumors.45, 46, 47 In the current report, the in vitro and in vivo study demonstrated the promising therapeutic role of I3MO on inhibiting of MM cell growth, even in the presence of the protective tumor associated microenvironment (TAM). Of note, combination of I3MO with bortezomib notably enhanced MM cell sensitivity to bortezomib-induced apoptosis. Mechanism, I3MO acts as multifaceted regulators of cell death in MM, which promotes cell cycle arrest, DNA damage response, and efficiently induces apoptosis of tumor cells. Of note, I3MO also suppresses NF-κB signaling, a critical pathway mediating growth and drug-resistance in MM cells, through downregulating USP7 expression. Our results strikingly demonstrate that I3MO act as proteasome inhibitor by targeting the proteasome activators PSME3 and PSME4. These molecular mechanisms of anti-MM activity are illustrated in Figure 10.
Figure 10.
The working model of anti-myeloma activity of I3MO.
Ubiquitination controls the stability of most cellular proteins, and its deregulation contributes to human diseases including cancer. Deubiquitinases (DUBs) remove ubiquitin from proteins, and their inhibition can induce degradation of select proteins. Cheng et al. has reported that indirubin derivatives enhanced SMAD protein degradation through targeting USP9x and USP34.48 Recent study reported by this team further demonstrated that pVHL (a component of an E3 ubiquitin ligase complex) also contributes for SMAD ubiquitination degradation induced by indirubin treatment.49 Consistent with that, our study shows that Ub-specific processing proteases pathway (UPP pathway) was notably suppressed after I3MO treatment in a panel of MM cell lines. I3MO treatment significantly suppresses the expressions of USP7, USP11, USP13, USP18, UBB, and UBC etc. These results indicated that UPP pathway is the potential target of I3MO, which involved in the anti-MM efficacy. Excessive production of abnormal immunoglobulins is an important hallmark of MM, which makes MM cells heavily dependent on the ubiquitin-proteasome system (UPS) to maintain protein homeostasis.39,50 Ubiquitin specific peptidase 7 (USP7) is a deubiquitinating enzyme (DUB) which removes ubiquitin tags from target protein substrates to alter their degradation and sub-cellular localization. USP7 was proposed to be as a therapeutic target in several cancers, including MM51 because it has many reported substrates with a role in cancer progression including NEK2,7,39 MDM2,52 N-Myc53 and PTEN.54 NEK2, a CIN gene, is the most significant gene associated with drug resistance and poor outcome in MM and other cancers. NEK2 is stabilized by USP7 and promoted the bortezomib resistance of MM cells as our previous study reported.7,39 MDM2 ubiquitylates p53 and targets it for proteasome-mediated degradation. As normal, USP7 stabilizes the levels of MDM2, which consequently drives p53 ubiquitylation and its subsequent degradation.55,56 The functional consequences of inhibiting USP7 therefore include decreased MDM2 levels with accumulation of p53, induction of growth arrest via p21, and cell death. Our results demonstrate that I3MO treatment remarkably downregulates USP7 and stabilizes p53, thereby inducing MM cell growth arrest via upregulation of p21. Kon et al.57 also highlighted a p53-dependent and -independent function of USP7. These findings have important clinical implications, since 10–15% of MM patients have p53 mutations/deletions which confer drug resistance; a therapeutic strategy using I3MO would have potential anti-MM activity even in this patient population. Andrew et al. reported the USP7 inhibition by selective small molecule inhibitors.51 However, our data showed that I3MO cannot direct bind with USP7 (data not shown). In addition, USP7 is proposed to exist in an oligomeric form like other deubiquitinases.58,59 Therefore, the related regulatory mechanisms of USP7 downregulation by I3MO are still need to be further clarified.
Additionally, our study demonstrated that I3MO treatment inhibits proteasome activity via directly binding with PA28γ and PA200. The proteasome complex is composed by chymotrypsin-like, trypsin-like and caspase-like activities that are associated with three catalytic core particles: β5, β2, β1, and its associated activators and regulators including PA28αβ, PA28γ (PSME3), PA200 (PSME4) and PI31, respectively.60,61 This study identified that I3MO direct binds with proteasome complex activator, PA28γ and PA200, and down-regulates the expression of PA28γ (PSME3) and PA200 (PSME4) in both mRNA and protein level, which causes the inhibition of proteasome activity. Therefore, these findings support I3MO as proteasome inhibitor with potential to overcome bortezomib resistance in MM cells by targeting PA28γ (PSME3) and PA200 (PSME4), the diverse subunits compared with traditional proteasome inhibitor bortezomib. Knockdown PSME3 or PSME4 by shRNA not only inhibited proteasome activity, but also increased protein synthesis, which in turn led to cell growth arrest and increased MM cell sensitivity to traditional proteasome inhibitor bortezomib. The sensitivity of MM cells to bortezomib heavily depends on the balance between proteasome load and capacity. The sensitivity of certain MM cell line to BTZ could stem from decreased proteasomal capacity, increased proteasomal workload, or both.5,13 Gu et al.62 found that downregulation of PA28α inhibits both proteasomal load and activity, resulting in protein homeostasis less dependent on the proteasome activity, which causes MM cells are resistance to proteasome inhibitors. Jagannathan and colleagues63 study also supports that upregulation the level of PA200 would increase proteasome activity and render cells therapeutically resistant to PIs. Our results indicated that downregulation PSME3 or PSME4 by I3MO decreases the proteasome capacity, and notably increases the proteasomal workload by increasing the protein synthesis in MM cells. Moreover, knockdown PSME3 or PSME4 significantly decreased MM cell sensitivity to I3MO. These findings demonstrate that I3MO targeting PSME3/4 and promotes MM cells sensitive to BTZ.
The activator PA28γ and PA200 of proteasome complex is constitutively expressed in most cell types and involved in the pathogenesis of malignancy. PA28γ is found exclusively in the nucleus and associates as a homoheptamer with the 20S complex.64 PA28γ has been implicated in the regulation of cell cycle progression and apoptosis, and negatively regulates p53.65 High levels of PA28γ promote aggressive pancreatic cancer cell growth via c-Myc-glycolysis signaling axis.66,67 Boulpicante et al. reported that overexpressing the proteasome activator PA28γ promotes tumor escape from immunosurveillance.67, 68, 69 The activator PA200 functions in a variety of processes including proteasome assembly, DNA repair, genomic stability, spermatogenesis, and mitochondrial checkpoint regulation,70 as well as stimulates the peptide-hydrolyzing activity of proteasomes.71 Welk et al. reported that catalytic proteasome inhibition resulted in the rapid (within 2–6 h) recruitment of PA28γ and PA200 to the 20S and 26S proteasomes to form alternative proteasome complexes.41 Myeloma cells are characterized by high proteasome complex activity, which is responsible for the degradation of paraprotein and maintenance of cellular homeostasis. When proteasome activity was inhibited by bortezomib, compensatory alternative proteasome formation increased.41 Most importantly, our results confirmed that targeting the components of proteasome activators PSME3 (PA28γ) and PSME4 (PA200) caused the inhibition of alternative proteasome function and efficiently induced the suppression of MM cell growth. We have shown that an increased NEK2 signature is linked to drug resistance in MM using systematic analyses of sequential MM samples.7,72 Here, we demonstrated that I3MO efficiently suppresses cell growth of NEK2 overexpression MM cells via downregulating the proteasome activator PA28γ and PA200.
Utilizing the publicly available datasets, we found that more than 30% MM patients have increased levels of PSME3 and PSME4, especially in RRMM. Moreover, high levels of PSME3 or PSME4 are associated with an inferior outcome in MM patients. All these findings support a critical role of the alternative proteasome composed with PSME3 and PSME4 in the growth and progression of MM, especially in bortezomib resistant MM. Additionally, relapse phenotype may be acquired via therapy induced selection of resistant minor clones present at diagnosis or by direct adaptation to therapy of the original malignant clone. Increased expression of PSME3 and PSME4 in RRMM patients represents the biomarkers of drug resistance clones. I3MO targets PA28γ (PSME3) and PA200 (PSME4) and leads to the inactivation of downstream signals, which results in cell death and overcomes drug resistance in MM cells. Due to the diverse targets of proteasome complex mentioned above, I3MO exerts therapeutic effects against bortezomib-resistant MM cells, and enhances the cytotoxicity of bortezomib in combination treatment. These findings highlight the potential value of PSME3 and PSME4 inhibition in sensitizing to PI or overcoming PI resistance in MM.
In summary, our study demonstrated that I3MO is an effective agent against MM, which targets UPP signal pathway and works as proteasome inhibitor. Proteasome activators PSME3 and PSME4 as the targets of I3MO are overexpressed in RRMM and associated with bortezomib resistance. It therefore represents a promising therapeutic application overcome PIs resistance and improve patient outcome in MM. Whereas, more effective inhibitors need to be further developed given the high micromolar dose of I3MO in MM treatment.
Contributors
YZ, WXJ, LLT, contributed equally to this paper and did the experiments. HM designed the study. SH, SWW, FT and HWY contributed to data collection, data analyses and interpretation. WKF, WL and LY worked on laboratory data and PDX model. AG, HY and ZYZ engaged in sample collection. MFC and HCJ performed the molecular docking analyses. YTT contributed to the data analysis. KC Anderson and CT provides insightful comments. HM, CT and QLG contributed to literature search, writing of the manuscript. All authors contributed to data acquisition, data interpretation, and all reviewed and approved the final version of the manuscript. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. YZ, WXJ, LLT and HM have verified the underlying data.
Data sharing statement
All original sequencing data sets of the cell line are deposited in the NCBI's Gene Expression Omnibus database (GSE194379).
Declaration of interests
Dr. Kenneth. C. Anderson is the consultant: Pfizer, Amgen, Astrazeneca, Janssen, Precision Biosciences, Mana and Window. Founder/Stock Shareholder: C4 Therapeutics, Oncopep, Raqia and NextRNA. Other authors declared no competing financial interests in relation to the work described.
Acknowledgments
This work was supported by following foundations: (a). the Natural Science Foundation of China (82170194), the Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (2018RC320012) hosted by M.H. (b). the Natural Science Foundation of China (81920108006), the Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences (2018PT31006), the National Program on Key Basic Research Project 2018ZX09733003 hosted by L.G.Q. CAMS Innovation Fund for Medical Sciences (CIFMS) 2021-1-I2M-040 hosted by T.C.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103950.
Contributor Information
Lugui Qiu, Email: qiulg@ihcams.ac.cn.
Mu Hao, Email: haomu@ihcams.ac.cn.
Appendix. Supplementary materials
References
- 1.Kumar S.K., Anderson K.C. Immune Therapies in Multiple Myeloma. Clin Cancer Res. 2016;22(22):5453–5460. doi: 10.1158/1078-0432.CCR-16-0868. [DOI] [PubMed] [Google Scholar]
- 2.An G., Yan Y., Xu Y., et al. Monitoring the cytogenetic architecture of minimal residual plasma cells indicates therapy-induced clonal selection in multiple myeloma. Leukemia. 2020;34(2):578–588. doi: 10.1038/s41375-019-0590-x. [DOI] [PubMed] [Google Scholar]
- 3.Wang D., Xu Q., Yuan Q., et al. Proteasome inhibition boosts autophagic degradation of ubiquitinated-AGR2 and enhances the antitumor efficiency of bevacizumab. Oncogene. 2019;38(18):3458–3474. doi: 10.1038/s41388-019-0675-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12(9):1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 5.Adams J. The proteasome: structure, function, and role in the cell. Cancer Treat Rev. 2003;29:3–9. doi: 10.1016/s0305-7372(03)00081-1. [DOI] [PubMed] [Google Scholar]
- 6.Hoeller D., Hecker C.M., Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6(10):776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 7.Franqui-Machin R., Hao M., Bai H., et al. Destabilizing NEK2 overcomes resistance to proteasome inhibition in multiple myeloma. J Clin Invest. 2018;128(7):2877–2893. doi: 10.1172/JCI98765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chauhan D., Tian Z., Nicholson B., et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22(3):345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manasanch E.E., Orlowski R.Z. Proteasome inhibitors in cancer therapy. Nat Rev Clin Oncol. 2017;14(7):417–433. doi: 10.1038/nrclinonc.2016.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avet-Loiseau H. Introduction to a review series on advances in multiple myeloma. Blood. 2019;133(7):621. doi: 10.1182/blood-2018-11-877795. [DOI] [PubMed] [Google Scholar]
- 11.Chim C.S., Kumar S.K., Orlowski R.Z., et al. Management of relapsed and refractory multiple myeloma: novel agents, antibodies, immunotherapies and beyond. Leukemia. 2018;32(2):252–262. doi: 10.1038/leu.2017.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hideshima T., Anderson K.C. Biologic impact of proteasome inhibition in multiple myeloma cells–from the aspects of preclinical studies. Semin Hematol. 2012;49(3):223–227. doi: 10.1053/j.seminhematol.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bianchi G., Oliva L., Cascio P., et al. The proteasome load versus capacity balance determines apoptotic sensitivity of multiple myeloma cells to proteasome inhibition. Blood. 2009;113(13):3040–3049. doi: 10.1182/blood-2008-08-172734. [DOI] [PubMed] [Google Scholar]
- 14.Qin Y., Zhang S., Deng S., et al. Epigenetic silencing of miR-137 induces drug resistance and chromosomal instability by targeting AURKA in multiple myeloma. Leukemia. 2017;31(5):1123–1135. doi: 10.1038/leu.2016.325. [DOI] [PubMed] [Google Scholar]
- 15.Niewerth D., Jansen G., Assaraf Y.G., Zweegman S., Kaspers G.J., Cloos J. Molecular basis of resistance to proteasome inhibitors in hematological malignancies. Drug Resist Updat. 2015;18:18–35. doi: 10.1016/j.drup.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 16.Barrio S., Stuhmer T., Da-Via M., et al. Spectrum and functional validation of PSMB5 mutations in multiple myeloma. Leukemia. 2019;33(2):447–456. doi: 10.1038/s41375-018-0216-8. [DOI] [PubMed] [Google Scholar]
- 17.Song Y., Ray A., Li S., et al. Targeting proteasome ubiquitin receptor Rpn13 in multiple myeloma. Leukemia. 2016;30(9):1877–1886. doi: 10.1038/leu.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hao M., Zhang L., An G., et al. Bone marrow stromal cells protect myeloma cells from bortezomib induced apoptosis by suppressing microRNA-15a expression. Leuk Lymphoma. 2011;52(9):1787–1794. doi: 10.3109/10428194.2011.576791. [DOI] [PubMed] [Google Scholar]
- 19.Wang X., Meul T., Meiners S. Exploring the proteasome system: A novel concept of proteasome inhibition and regulation. Pharmacol Ther. 2020;211 doi: 10.1016/j.pharmthera.2020.107526. [DOI] [PubMed] [Google Scholar]
- 20.Du T., Song Y., Ray A., Chauhan D., Anderson KC. Proteomic analysis identifies mechanism(s) of overcoming bortezomib resistance via targeting ubiquitin receptor Rpn13. Leukemia. 2021;35(2):550–561. doi: 10.1038/s41375-020-0865-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cascio P. PA28gamma: new insights on an ancient proteasome activator. Biomolecules. 2021;11(2) doi: 10.3390/biom11020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S., Jiang C., Pan J., et al. Regulation of c-Myc protein stability by proteasome activator REGgamma. Cell Death Differ. 2015;22(6):1000–1011. doi: 10.1038/cdd.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welk V., Meul T., Lukas C., et al. Proteasome activator PA200 regulates myofibroblast differentiation. Sci Rep. 2019;9(1):15224. doi: 10.1038/s41598-019-51665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao M., Li B., Sun X., et al. Preclinical activity of DCZ3301, a novel aryl-guanidino compound in the therapy of multiple myeloma. Theranostics. 2017;7(15):3690–3699. doi: 10.7150/thno.18345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhuang J., Shirazi F., Singh R.K., et al. Ubiquitin-activating enzyme inhibition induces an unfolded protein response and overcomes drug resistance in myeloma. Blood. 2019;133(14):1572–1584. doi: 10.1182/blood-2018-06-859686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zang M., Li Z., Liu L., et al. Anti-tumor activity of the proteasome inhibitor BSc2118 against human multiple myeloma. Cancer Lett. 2015;366(2):173–181. doi: 10.1016/j.canlet.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Xia J., Xu H., Zhang X., et al. Multiple myeloma tumor cells are selectively killed by pharmacologically-dosed ascorbic acid. EBioMedicine. 2017;18:41–49. doi: 10.1016/j.ebiom.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu T., Du C., Ma X., et al. Polycomb-like protein 3 induces proliferation and drug resistance in multiple myeloma and is regulated by miRNA-15a. Mol Cancer Res. 2020;18(7):1063–1073. doi: 10.1158/1541-7786.MCR-19-0852. [DOI] [PubMed] [Google Scholar]
- 29.Li Z., Liu L., Du C., et al. Therapeutic effects of oligo-single-stranded DNA mimicking of hsa-miR-15a-5p on multiple myeloma. Cancer Gene Ther. 2020 doi: 10.1038/s41417-020-0161-3. [DOI] [PubMed] [Google Scholar]
- 30.Sacco A., Roccaro A.M., Ma D., et al. Cancer cell dissemination and homing to the bone marrow in a zebrafish model. Cancer Res. 2016;76(2):463–471. doi: 10.1158/0008-5472.CAN-15-1926. [DOI] [PubMed] [Google Scholar]
- 31.Zhang C., Chen Y., Sun B., et al. m(6)A modulates haematopoietic stem and progenitor cell specification. Nature. 2017;549(7671):273–276. doi: 10.1038/nature23883. [DOI] [PubMed] [Google Scholar]
- 32.Liu L., Yu Z., Cheng H., et al. Multiple myeloma hinders erythropoiesis and causes anaemia owing to high levels of CCL3 in the bone marrow microenvironment. Sci Rep. 2020;10(1):20508. doi: 10.1038/s41598-020-77450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trott O., Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterhouse A., Bertoni M., Bienert S., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kemper B., von Groning M., Lewe V., et al. Reversible Covalent and Supramolecular Functionalization of Water-Soluble Gold(I) Complexes. Chemistry. 2017;23(25):6048–6055. doi: 10.1002/chem.201700588. [DOI] [PubMed] [Google Scholar]
- 36.Hachimi M., Grabowski C., Campanario S., et al. Smoothelin-like 2 inhibits coronin-1B to stabilize the apical actin cortex during epithelial morphogenesis. Curr Biol. 2021;31(4):696–706.e9. doi: 10.1016/j.cub.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao M., Zhang L., An G., et al. Suppressing miRNA-15a/-16 expression by interleukin-6 enhances drug-resistance in myeloma cells. J Hematol Oncol. 2011;4:37. doi: 10.1186/1756-8722-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin J., Zhang W., Zhao J.J., et al. A clinically relevant in vivo zebrafish model of human multiple myeloma to study preclinical therapeutic efficacy. Blood. 2016;128(2):249–252. doi: 10.1182/blood-2016-03-704460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao M., Franqui-Machin R., Xu H., et al. NEK2 induces osteoclast differentiation and bone destruction via heparanase in multiple myeloma. Leukemia. 2017;31(7):1648–1650. doi: 10.1038/leu.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen X., Barton L.F., Chi Y., Clurman B.E., Roberts J.M. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol Cell. 2007;26(6):843–852. doi: 10.1016/j.molcel.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welk V., Coux O., Kleene V., et al. Inhibition of proteasome activity induces formation of alternative proteasome complexes. J Biol Chem. 2016;291(25):13147–13159. doi: 10.1074/jbc.M116.717652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoessel R., Leclerc S., Endicott J.A., et al. Indirubin, the active constituent of a Chinese antileukaemia medicine, inhibits cyclin-dependent kinases. Nat Cell Biol. 1999;1(1):60–67. doi: 10.1038/9035. [DOI] [PubMed] [Google Scholar]
- 43.Xiao Z., Hao Y., Liu B., Qian L. Indirubin and meisoindigo in the treatment of chronic myelogenous leukemia in China. Leuk Lymphoma. 2002;43(9):1763–1768. doi: 10.1080/1042819021000006295. [DOI] [PubMed] [Google Scholar]
- 44.Xiao Z., Wang Y., Lu L., et al. Anti-angiogenesis effects of meisoindigo on chronic myelogenous leukemia in vitro. Leuk Res. 2006;30(1):54–59. doi: 10.1016/j.leukres.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Blazevic T., Schaible A.M., Weinhaupl K., et al. Indirubin-3′-monoxime exerts a dual mode of inhibition towards leukotriene-mediated vascular smooth muscle cell migration. Cardiovasc Res. 2014;101(3):522–532. doi: 10.1093/cvr/cvt339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee M.Y., Liu Y.W., Chen M.H., et al. Indirubin-3′-monoxime promotes autophagic and apoptotic death in JM1 human acute lymphoblastic leukemia cells and K562 human chronic myelogenous leukemia cells. Oncol Rep. 2013;29(5):2072–2078. doi: 10.3892/or.2013.2334. [DOI] [PubMed] [Google Scholar]
- 47.Cao Z., Yang F., Wang J., et al. Indirubin derivatives as dual inhibitors targeting cyclin-dependent kinase and histone deacetylase for treating cancer. J Med Chem. 2021 doi: 10.1021/acs.jmedchem.1c01311. [DOI] [PubMed] [Google Scholar]
- 48.Cheng X., Alborzinia H., Merz K.H., et al. Indirubin derivatives modulate TGFβ/BMP signaling at different levels and trigger ubiquitin-mediated depletion of nonactivated R-Smads. Chem Biol. 2012;19(11):1423–1436. doi: 10.1016/j.chembiol.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Zhou J., Dabiri Y., Gama-Brambila R.A., et al. pVHL-mediated SMAD3 degradation suppresses TGF-beta signaling. J Cell Biol. 2022;221(1) doi: 10.1083/jcb.202012097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X.D., Baladandayuthapani V., Lin H., et al. Tight junction protein 1 modulates proteasome capacity and proteasome inhibitor sensitivity in multiple myeloma via EGFR/JAK1/STAT3 signaling. Cancer Cell. 2016;29(5):639–652. doi: 10.1016/j.ccell.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turnbull A.P., Ioannidis S., Krajewski W.W., et al. Molecular basis of USP7 inhibition by selective small-molecule inhibitors. Nature. 2017;550(7677):481–486. doi: 10.1038/nature24451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheng Y., Saridakis V., Sarkari F., et al. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13(3):285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- 53.Tavana O., Li D., Dai C., et al. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat Med. 2016;22(10):1180–1186. doi: 10.1038/nm.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schauer N.J., Liu X., Magin R.S., et al. Selective USP7 inhibition elicits cancer cell killing through a p53-dependent mechanism. Sci Rep. 2020;10(1):5324. doi: 10.1038/s41598-020-62076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Itahana K., Mao H., Jin A., et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12(4):355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 56.Cummins J.M., Rago C., Kohli M., Kinzler K.W., Lengauer C., Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428(6982) doi: 10.1038/nature02501. 1 p following 486. [DOI] [PubMed] [Google Scholar]
- 57.Kon N., Kobayashi Y., Li M., Brooks C.L., Ludwig T., Gu W. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29(9):1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernández-Montalván A., Bouwmeester T., Joberty G., et al. Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. Febs J. 2007;274(16):4256–4270. doi: 10.1111/j.1742-4658.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- 59.Elliott P.R., Liu H., Pastok M.W., et al. Structural variability of the ubiquitin specific protease DUSP-UBL double domains. FEBS Lett. 2011;585(21):3385–3390. doi: 10.1016/j.febslet.2011.09.040. [DOI] [PubMed] [Google Scholar]
- 60.Collins G.A., Goldberg AL. The logic of the 26S proteasome. Cell. 2017;169(5):792–806. doi: 10.1016/j.cell.2017.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fostier K., De Becker A., Schots R. Carfilzomib: a novel treatment in relapsed and refractory multiple myeloma. Onco Targets Ther. 2012;5:237–244. doi: 10.2147/OTT.S28911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gu Y., Barwick B.G., Shanmugam M., et al. Downregulation of PA28α induces proteasome remodeling and results in resistance to proteasome inhibitors in multiple myeloma. Blood Cancer J. 2020;10(12):125. doi: 10.1038/s41408-020-00393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jagannathan S., Vad N., Vallabhapurapu S., Vallabhapurapu S., Anderson K.C., Driscoll JJ. MiR-29b replacement inhibits proteasomes and disrupts aggresome+autophagosome formation to enhance the antimyeloma benefit of bortezomib. Leukemia. 2015;29(3):727–738. doi: 10.1038/leu.2014.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cascio P. PA28γ: new insights on an ancient proteasome activator. Biomolecules. 2021;11(2) doi: 10.3390/biom11020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z., Zhang R. Proteasome activator PA28 gamma regulates p53 by enhancing its MDM2-mediated degradation. EMBO J. 2008;27(6):852–864. doi: 10.1038/emboj.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo J., Hao J., Jiang H., et al. Proteasome activator subunit 3 promotes pancreatic cancer growth via c-Myc-glycolysis signaling axis. Cancer Lett. 2017;386:161–167. doi: 10.1016/j.canlet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 67.Jovanovic K.K., Roche-Lestienne C., Ghobrial I.M., Facon T., Quesnel B., Manier S. Targeting MYC in multiple myeloma. Leukemia. 2018;32(6):1295–1306. doi: 10.1038/s41375-018-0036-x. [DOI] [PubMed] [Google Scholar]
- 68.Boulpicante M., Darrigrand R., Pierson A., et al. Tumors escape immunosurveillance by overexpressing the proteasome activator PSME3. Oncoimmunology. 2020;9(1) doi: 10.1080/2162402X.2020.1761205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yi Z., Yang D., Liao X., Guo F., Wang Y., Wang X. PSME3 induces epithelial-mesenchymal transition with inducing the expression of CSC markers and immunosuppression in breast cancer. Exp Cell Res. 2017;358(2):87–93. doi: 10.1016/j.yexcr.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 70.Sadre-Bazzaz K., Whitby F.G., Robinson H., Formosa T., Hill CP. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol Cell. 2010;37(5):728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toste Rêgo A., da Fonseca P.C.A. Characterization of fully recombinant human 20S and 20S-PA200 proteasome complexes. Mol Cell. 2019;76(1):138–147.e5. doi: 10.1016/j.molcel.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou W., Yang Y., Xia J., et al. NEK2 induces drug resistance mainly through activation of efflux drug pumps and is associated with poor prognosis in myeloma and other cancers. Cancer Cell. 2013;23(1):48–62. doi: 10.1016/j.ccr.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.