Abstract
Yes Associated Protein 1 (YAP) and Transcriptional coactivator with PDZ-Binding Motif (TAZ) are transcriptional coactivators that have been implicated in driving metastasis and progression in many cancers, mainly through their transcriptional regulation of downstream targets. Although YAP and TAZ have shown redundancy in many contexts, it is still unknown whether or not this is true in melanoma. Here we show that while both YAP and TAZ are expressed in a panel of melanoma cell lines, depletion of YAP results in decreased cell numbers, focal adhesions, and the ability to invade matrigel. Using non-biased RNA-sequencing analysis, we find that melanoma cells depleted of YAP, TAZ, or YAP/TAZ exhibit drastically different transcriptomes. We further uncover the ARP2/3 subunit ARPC5 as a specific target of YAP but not TAZ, and that ARPC5 is essential for YAP-dependent maintenance of melanoma cell focal adhesion numbers. Our findings suggest that in melanoma, YAP drives melanoma progression, survival, and invasion.
Significance
Our findings are the first to differentiate YAP from its ortholog TAZ as a unique driver of melanoma using cell dependency studies, non-biased RNA sequencing, and a downstream target-based approach. Genetic and functional data implicate the actin nucleating ARP2/3 complex in melanoma and we discovered that the ARP2/3 subunit ARPC5 is a downstream target of YAP but not TAZ. ARPC5 acts as a downstream effector factor for YAP in focal adhesion maintenance, likely contributing to melanoma migration and metastasis.
Keywords: Melanoma, yes-associated protein (YAP), ARP2/3 complex, transcriptional coactivator, cancer
1 ∣. INTRODUCTION
Melanoma is an aggressive cancer with a high degree of metastasis (Tas, 2012). Yes Associated Protein 1 (YAP) and Transcriptional coactivator with PDZ-Binding Motif (WWTR1 or TAZ) are transcriptional coactivators implicated in driving metastasis and progression in a variety of cancers, including melanoma (Lamar et al., 2012; Nallet-Staub et al., 2014; Zanconato, Cordenonsi, & Piccolo, 2016; Zhao et al., 2008). In melanoma, increased YAP and TAZ expression correlates with lower patient survival (Q. Feng, Guo, Kang, & Zhao, 2018; Menzel et al., 2014). YAP was discovered to be a key regulator of melanoma invasiveness, and can drive non-invasive melanoma cells towards an invasive phenotype (X. Zhang et al., 2020). Furthermore, exogenous overexpression of YAP in melanoma promoted invasion, while loss of YAP and TAZ led to decreased invasion, viability, and tumorigenicity (Nallet-Staub et al., 2014; X. Zhang et al., 2019; X. Zhang et al., 2020). Activation of YAP also promotes melanoma anoikis resistance, leading to increased migration and invasion (Zhao et al., 2021). TEAD factors, common co-activator partners of YAP and TAZ, have been linked to a more invasive melanoma phenotype (Verfaillie et al., 2015). YAP promotion of melanoma metastasis was linked to its interaction with TEADs (Lamar et al., 2012). In addition to cutaneous melanoma, YAP is overexpressed in uveal melanoma and works in pathways dependent on GNAQ/GNA11 mutations that are common in this cancer type (X. Feng et al., 2014; Yu et al., 2014). In other cancer types, both YAP and TAZ have been implicated in driving various aspects of metastasis. In pancreatic cancer, YAP overexpression drives metastasis by activating the AKT cascade to induce the epithelial to mesenchymal transition (Yuan et al., 2016). In hepatocarcinoma cells, reduction of YAP phosphorylation via LATS2 inhibition reduced and increased EMT markers E-cadherin and vimentin respectively (Han, Yin, & Zhang, 2018). In colorectal cancer, YAP is shown to promote the epithelial to mesenchymal transition by driving expression of MALAT1, which in turn promotes expression of various metastasis markers, including VEGFA, SLUG, and TWIST. In addition, TAZ upregulation is able to rescue migratory and invasive phenotypes exhibited with miR-125a-5p inhibition (Tang et al., 2019). Outside of metastasis, YAP and TAZ have been shown to drive several other cancer cell processes, including cell proliferation, differentiation, and drug resistance (Choe et al., 2018; Fisher, Grun, Adhikary, Xu, & Eckert, 2017; Zanconato et al., 2015; Zhao et al., 2008). Both YAP and TAZ have been implicated in many aspects of cancer progression, with evidence of a role in migration, invasion, and metastasis.
YAP and TAZ contain many similarities and potential redundancies. Structurally, YAP and TAZ share 60% protein sequence similarity (W. Hong & Guan, 2012; Plouffe et al., 2018). YAP and TAZ do not contain a DNA binding domain and require binding partners that interact directly with DNA to exert downstream transcriptional effects. Traditionally, both YAP and TAZ bind to the TEAD family of transcription factors, but either YAP, TAZ, or both have been shown to bind to the RUNX, SMAD, and hnRNP family of proteins, among others (Ferrigno et al., 2002; J. H. Hong et al., 2005; Howell, Borchers, & Milgram, 2004; Kanai et al., 2000; Yagi, Chen, Shigesada, Murakami, & Ito, 1999; Zanconato et al., 2016). Transcriptionally, both YAP and TAZ have been shown to directly control expression of CTGF and Cyr61, genes implicated in cell differentiation and cell adhesion, among others (H. Zhang, Pasolli, & Fuchs, 2011; Zhao et al., 2008). Due to past studies from D. melanogaster, H. sapiens, and others, it is clear that YAP and TAZ have overlapping functions in driving a wide variety of biological functions.
While it is clear that YAP and TAZ share many functions, there is also evidence that these factors are not just redundant paralogs. Although YAP and TAZ contain many of the same protein binding domains, both proteins also contain unique domains of their own, most notably YAP contains an SH3 binding motif and an N terminal proline rich region, and the YAP1-2 splice variants produce a protein with an extra WW domain (Gaffney et al., 2012; Varelas, 2014). Phenotypically, several studies show stark differences in functionality between the two transcriptional cofactors. YAP knockout mice are embryonic lethal, whereas TAZ mice can live until adulthood but exhibit both kidney and lung defects (Hossain et al., 2007; Makita et al., 2008; Morin-Kensicki et al., 2006; Tian et al., 2007). When differentially expressed genes from HEK293 YAP or TAZ depleted cells were compared to YAP/TAZ null cells, only 81% and 41% of the differentially expressed genes in the YAP or TAZ only inhibited groups overlapped with the YAP/TAZ group respectively (Plouffe et al., 2018). Specifically in melanoma, TAZ was not able to compensate for YAP-dependent viability in a subset of melanoma cells (X. Zhang et al., 2019). While these studies, among others, reveal many overlapping roles for YAP and TAZ in a variety of biological settings, it is still unknown what unique functions YAP and TAZ may also be performing.
In this study, we identified YAP as the predominant player in our melanoma cell lines. We showed that YAP specific inhibition led to a reduction in melanoma cell numbers, invasion, and focal adhesions. Furthermore, RNA-sequencing revealed a YAP transcriptome highly distinct from TAZ and more enriched in genes involved in cancer progression. We demonstrated direct inhibition of one such YAP target gene, ARPC5, which led to a decrease in focal adhesion numbers, melanoma cell migration, and a shift in the ARP2/3 complex subunits. Taken together, our data support a model whereby YAP drives ARPC5 expression to enhance melanoma cell migration.
2 ∣. MATERIALS AND METHODS
2.1 ∣. Cell Culture
Human melanoma cell lines A375, M14, mel537, mel624, mel888, SKMEL-28, SKMEL-23, SKMEL-5, and mouse melanoma B16 cells (ATCC, Manassas, VA and University of Chicago Comprehensive Cancer Center Core Facilities) were cultured in DMEM with 10% FBS (Sigma-Aldrich). Melanoma marker testing, morphology, and histological analysis were used to verify melanoma cell identity and lack of mycoplasma contamination.
2.2 ∣. Immunoblotting
Thirty μg total protein melanoma lysates (MPER, Thermo Fisher – 78501) were separated on 4-15% Bis-Tris gels (Bio-Rad) and transferred to nitrocellulose membranes. The membranes were incubated overnight with antibody (Dilutions - 1:1000 for YAP/TAZ antibody (Cell Signaling Technology – D24E4), 1:1000 ARPC5 (Proteintech - 16717-1-AP), 1:1000 ARPC5L (Proteintech – 22025-1-AP), 1:10000 for GAPDH (Cell Signaling – D16H11)) in nonfat 5% milk (Santa Cruz – sc-2324). The membranes were washed with 1X TBS-T four times for 15 minutes and incubated with 1:4000 anti-rabbit IgG HRP-linked antibody (Cell Signaling – 7074). Membranes were developed with Clarity Western ECL substrate according to the manufacturer’s instructions (Bio-Rad). Blots were normalized with GAPDH or total protein input (Stain Free System, Bio-Rad). All Western analysis shown are representatives of at least three independent experiments.
2.3 ∣. Densitometry
Western blot band intensities were measured using ImageJ64 (https://imagej.nih.gov/ij/download.html). All experiments were performed minimally in triplicate, with each figure showing a representative image. The data shown are represented as a percentage of controls and normalized to total protein.
2.4 ∣. siRNA treatments
Melanoma cells were seeded at 50-70% confluency in 6-well plates. siRNA transfection using Lipfectamine 3000 (Thermo Fisher) with siRNAs and methods and parameters as performed recently (Lui et al., 2019). The experiments utilized Thermo Silencer select siRNA, designed to target all known transcripts of each gene. Validation of efficiency of siRNAs was previously verified by the manufacturer (Thermo Fisher) and our prior work (Lui et al., 2019). Gene specific siRNA targeted YAP, WWTR1 (TAZ), ARPC5, and/or siScrambled (Thermo Fisher ID Number S20366 – YAP1, S24787 – WWTR1, S19362 – ARPC5, 4390844 – siScramble). Cell lysates were collected 2 days post-siRNA transfection.
2.5 ∣. Cell shape morphology
Cell pictures were taken with a Q-Color3 Olympus camera on an Olympus CKX41 microscope 2 days post transfection. Cell shape was quantified by fitting the cell shape in to an ellipse and calculating the ratio between the short and long axis utilizing a photoshop measurement tool (PS Version CS6, Adobe Inc). 200 cells were counted per experiment, performed in triplicate.
2.6 ∣. Growth Curves Analysis
Cell confluency was measured every 6 hours over a period of 72 hours using the Incucyte FLR Live-Cell Imaging System (Essen BioScience, 2011A software). All experiments were performed in triplicate and all experimental groups were normalized to siScrambled control groups at 72 hours post treatment set at 100%.
2.7 ∣. Invasion assays
All matrigel invasion assays were performed using the Corning Matrigel matrix (Cat. No. 356234) on 8.0 μm pore cell culture inserts (Fisher) according to the manufacturer’s instructions with 10% FBS in DMEM as the chemoattractant (0.5% FBS in DMEM as the base media) and visualized using the Diff-Quik staining kit (Thermo Fisher). All analysis was performed by comparing the number of stained cells in the experimental groups as fold change compared to siScrambled control. All assays shown are representatives of at least three independent experiments, where cells were counted in 4 separate random fields of the cell culture insert.
2.8 ∣. Migration assays
Melanoma cells were plated to 90-100% confluency (50,000 cells per well). Scratch wounds were made using Essen BioScience’s 96 well woundmaker according to manufacturer’s instructions. Pictures were taken immediately post wound and 24 hours post wound using the Incucyte FLR Live-Cell Imaging System (Essen BioScience, 2011A software). Cell migration was analyzed by calculating the percentage of the distance of the wound at time zero and 12 hours (A375) or 24 hours (mel537) post wound, prior to expected impact from YAP or TAZ dependent cell growth changes. All experiments were performed in triplicate and all experimental groups were normalized to siScrambled control groups set at 100%.
2.9 ∣. Immunofluorescence
Melanoma cells grown on coverslips were washed 2x in prewarmed PBS, fixed in 4% paraformaldehyde at 37 °C, washed 2x in prewarmed PBS, and then permeabilized for 15 minutes in PBS with 0.1% Triton X-100. Blocking was performed for an hour using 5% horse serum in PBS-T, followed by incubation in anti-vinculin antibody (Millipore Sigma – 05-386) diluted 1:5000 in 5% horse serum in PBS-T overnight, washed 2x PBS-T, and incubated in DyLight horse anti-mouse secondary antibody (Vector Laboratories – DI-2488) diluted 1:200 in 5% horse serum for 2 hours. Coverslips were washed 2x with PBS-T and stained for actin using Alexa Fluor 555 Phalloidin (ThermoFisher – A34055) according to the manufacturer’s instructions. Images were captured using a Nikon Eclipse E400 microscope.
2.10 ∣. RNA Seq
All RNA sequencing experiments were performed on mel537 cells in duplicate from each group (siScrambled, siYAP, siTAZ, siYAP/TAZ). RNA was collected using Direct-Zol RNA Miniprep Kit (Zymo Research – R2060) and submitted to the University of Chicago Genomics Facility (http://fgf.uchicago.edu) for library preparation and RNA sequencing on the Illumina HiSeq4000 platform (single-end 50 base pair). Subsequent bioinformatics analysis was performed by the Center for Research Informatics (http://cri.uchicago.edu) and Gene Ontology analysis was performed through the use of IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis). Gene set enrichment analysis was performed through use of the RaNA-seq tool (https://ranaseq.eu/home, GO:0005925 (Prieto & Barrios, 2019).
2.11 ∣. Quantitative Real-Time Polymerase Chain Reaction
RNA was isolated from melanoma cells using the Direct-Zol RNA extraction kit instructions (Zymo Research - R2071, R2050-1-200) according to the manufacturer’s instructions. cDNA was then generated using the iScript cDNA Synthesis kit (Bio-Rad). The expression levels of YAP, TAZ, and ARPC5 were analyzed using SYBR-Green Master Mix (Bio-Rad) in conjunction with the CFX Connect Real-Time System (Bio-Rad), and normalized to GAPDH. The primers used were as follows (5’->3’): YAP1-1F - GTG AGC CCA CAG GAG TTA GC; YAP1-1R - CTC GAG AGT GAT AGG TGC CA; YAP1-2F - TCT TCC TGA TGG ATG GGA AC; YAP1-2R - GGC TGT TTC ACT GGA GCA CT; TAZF - GTA TCC CAG CCA AAT CTC G; TAZR - TTC TGA GTG GGG TGG TTC; GAPDHF - ACA TCA TCC CTG CCT GTA CT; GAPDHR - CTC TCT TCC TCT TGT GCT CTT G; ARPC5F - AGA GCC CGT CTG ACA ATA G; ARPC5R - CAG TCA AGA CAC GAA CAA TG
2.12 ∣. Meta Analysis
Protein expression scores for normal tissue versus cancer were obtained from the web server GEPIA (http://gepia.cancer-pku.cn/index.html). Expression scores for cutaneous melanoma versus benign nevi were obtained from Oncomine (https://www.oncomine.org). Oncoprints for members of the ARP2/3 complex were obtained from cBioPortal (cbioportal.org).
2.13 ∣. Creation of ARPC5 expression rescue cell lines
Constructs that expressed high levels of ARPC5 from a CMV promoter were obtained from GenScript USA Inc. (ARPC5_pcDNA3.1(+), cat. # SC1849). Initial experiments attempting to create stable lines expressing these vectors were unsuccessful due to a loss of cell viability. To create cell lines that expressed low but stable levels of ARPC5, The CMV promoter was replaced with a partial PAX3 promoter sequence cloned from the pGL2-PAX3pm vector, which was previously shown to drive low but measurable expression levels of genes in melanoma cells (Kubic, Little, Kaiser, Young, & Lang, 2016). This newly created vector, P3-ARPC5, was transfected into A375 and mel537 cells using TransIT-X2 Dynamic Delivery System (Mirus, MIR 6004). Cells were selected with Geneticin (Thermo Fisher) 24 hrs post transfection, and maintained with drug (A375 2.5mg/ml, mel537 3.1mg/ml) as stable lines.
2.14 ∣. Proximity ligation assay
Melanoma cells were treated as described above, and then transferred to 24 hours post-treatment to 6 well plates containing flame sterilized cover slips, where they were to attach for 24 hours. The proximity ligation assay was then performed according to the manufacturer’s instructions (Sigma-Aldrich - DUO 92101) using antibodies for ACTR2 (Santa Cruz – sc 166103) and ARPC5L (Proteintech – 22025-1-AP).
2.15 ∣. Statistical analysis
Statistics were performed using student’s two-tailed T-test between control and the appropriate groups, or ANOVA for analysis of multiple groups. Sample sizes were calculated to result in at least 85% power or greater for all experiments. All experiments were performed minimally in triplicate, with a representative image shown in the figure or bar graphs compiling at least three independent experiments, and all findings stated as significant have p < 0.05 unless otherwise stated. Unless stated, all error bars indicate standard deviation.
3 ∣. RESULTS
3.1 ∣. YAP specific inhibition results in a decrease in melanoma cell number, invasive capacity, and morphological changes
To determine expression levels of both YAP and TAZ in melanoma cells, Western blot analysis was performed on eight melanoma cell lines (1 mouse, 7 human) for the presence of YAP and TAZ proteins. Utilizing a dual YAP/TAZ antibody, all 8 cell lines examined expressed varying degrees of YAP and TAZ, with 6/8 cell lines showing higher expression of TAZ than YAP (Figure 1A). Mel537 cells showed relatively equal expression of YAP and TAZ, while SKMEL-23 cells showed higher YAP expression than TAZ (Figure 1A). Higher exposure of the blots detected TAZ expression in SKMEL-23 cells, but the other cell lines were overexposed (data not shown). For YAP and/or TAZ depletion experiments, we first verified YAP and TAZ specific siRNAs in our melanoma lines to validate our prior studies testing the specificity and efficiency of these siRNA sets (Figure 1B)(Lui et al., 2019). Initial observation of the various knockdown groups (siYAP, siTAZ, siYAP/TAZ) as compared to siScrambled control revealed dynamic morphological changes in the siYAP and siYAP/TAZ, but not siTAZ, knockdown groups, resulting in more elongated cells as compared to siScrambled control (Figure 1C). As compared to the siScrambled groups that showed a length to width ratio of 1.99±1.25 (mel537), 2.54±1.33 (M14), and 1.73±0.97 (A375), siYAP and siYAP/TAZ knockdown groups have ratios of 3.51±2.58, 2.92±2.13, and 2.32±1.18 (siYAP) and 2.98±1.67, 3.07±1.87, and 2.27±1.24 (siYAP/TAZ), respectively. Cell growth is also YAP dependent, with cells significantly reduced to 75% or less of control levels (Figure 1D, p<0.05). In 3/6 of the lines (mel537, M14, UACC62), cell numbers decreased after 72 hours in a YAP specific manner (only in the YAP, or YAP/TAZ depleted cells) while TAZ knockdowns were similar in growth to siScrambled control. The remaining three lines (A375, SKMEL5, SKMEL23) showed decreased cell numbers with inhibition of either YAP or TAZ. Next, we examined whether YAP, TAZ, or YAP/TAZ inhibition had an effect on invasion through the use of Matrigel invasion assays. As we saw two distinct patterns of growth inhibition, we utilized a representative cell line that showed varying degrees of YAP versus TAZ expression from each growth pattern group (A375 and mel537). Both A375 and mel537 cell lines showed decreased invasion capacity with inhibition of YAP or YAP/TAZ, but not TAZ alone (p<0.05) (Figure 1E). Taken together, our observations suggest that YAP plays a predominant role in the morphology, cell growth, and invasion of melanoma cell lines.
Figure 1.
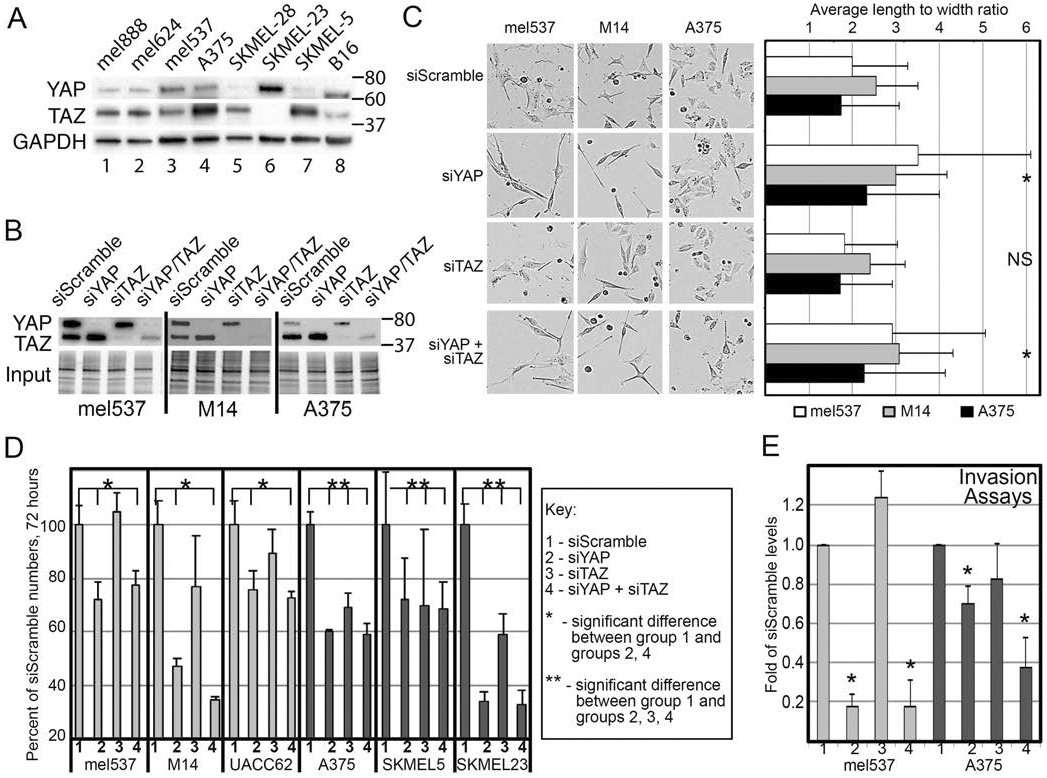
YAP inhibition results in decreased cell numbers and invasion in melanoma cells. (A) Melanoma cell lines express both YAP and TAZ at various levels. Western blot analysis for YAP and TAZ, with GAPDH as a loading control, was performed in a panel of melanoma cell lines (lanes 1-8). (B) siRNA mediated knockdowns of YAP, TAZ, and YAP/TAZ. Western blot analysis measuring levels of YAP and TAZ were performed in mel537, M14, and A375 cells transfected with siYAP, siTAZ, or both siYAP and siTAZ. (C) Morphology of mel537, M14, and A375 melanoma cells change with inhibition of YAP, TAZ, or YAP/TAZ. Cells were best fit into an ellipse, where the cellular ratio of width to length was calculated, 48 hours post siRNA transfection targeting YAP, TAZ, both YAP/TAZ, or siScrambled control. Representative photos of each cell line and knockdown group are shown. (D) Inhibition of YAP and TAZ result in decreased cell numbers. Cells were initially transfected with siRNA targeting YAP, TAZ, both YAP/TAZ, or siScrambled control and subsequently plated at 5-10% confluency. The confluency levels of the cells were then measured at 72 hours and compared to siScrambled control using the Essen BioScience IncuCyte Live-Cell imaging system. (E) Inhibition of YAP and TAZ result in decreased invasion. Matrigel invasion assays were performed on mel537 and A375 cells that were transfected with siRNA targeting YAP, TAZ, both YAP/TAZ, or siScrambled control. The values are expressed as the average of three independent experiments 12 (A375) or 24 (mel537) hours post seeding into transwell chambers, prior to any expected differences due to YAP/TAZ dependent cell growth changes. Values are mean ± SD (n = >200 (morphology and growth) or => 60 (invasion) for each experiment, three experiments normalized to controls) (* indicates p < 0.05 2-tailed ANOVA or t-tests, NS indicates not significant).
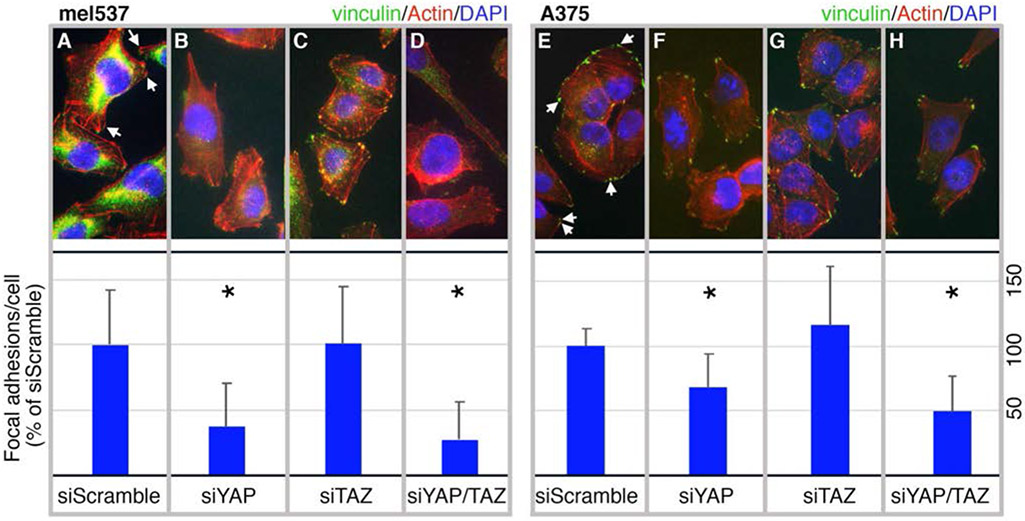
3.2 ∣. Inhibition of YAP decreases the number of focal adhesions in melanoma cells
Since we saw several melanoma cell functions (morphology, cell numbers, invasion) specific for inhibition of YAP, but not TAZ, we predicted that inhibition of YAP may deregulate the ability of melanoma cells to migrate through regulation of focal adhesion numbers, as focal adhesion dynamics have been shown to be important for cell migration and cancer metastasis (Mitra & Schlaepfer, 2006; Nagano, Hoshino, Koshikawa, Akizawa, & Seiki, 2012). To visualize and quantify focal adhesions, we performed a series of immunofluorescent stains for vinculin (a key focal adhesion protein) and actin on mel537 (Figure 2A-D) and A375 (Figure 2E-H) cells treated with siRNA targeting YAP, TAZ, or both YAP/TAZ, and compared numbers to siScrambled control. Focal adhesions were visualized as vinculin clusters at the end of, or co-stained with, the tips of F-actin polymers. Quantification of the focal adhesions revealed that both cell lines demonstrated a significant decrease per cell in a YAP specific manner (p < 0.05, n = 200 cells/group). YAP knockdowns had only 71±36% and 70±44% of the siScrambled control levels of focal adhesions in mel537 and A375 cells respectively. Similarly, YAP/TAZ knockdowns had 69±52% and 71±31% focal adhesion levels as compared to siScrambled group in mel537 and A375 cells. This indicates that YAP in mel537 and A375 melanoma cells either directly or indirectly control the number of focal adhesions.
Figure 2.
Inhibition of YAP decreases the number of focal adhesions in melanoma cells. Mel537 (A-D) and A375 (E-H) melanoma cells were transfected with siRNA targeting YAP (B, F), TAZ (C, G), YAP/TAZ (D, H), or siScrambled control (A, E). Representative cell images are shown (top row) as well as graphs of overall focal adhesions per cell from three independent experiments (bottom row). Examples of focal adhesions are indicated by arrows in the control group images. At 72 hours post siRNA treatment, cells were fixed and stained with antibodies against vinculin (Alexa 488, green), phalloidin (Alexa 555, red) for actin, and DAPI (blue) for nuclei. The number of vinculin bundles at the end of F-actin fibers, representing focal adhesions, were then counted for each cell (n = 50 cells/experiment). For each experiment, average control focal adhesion numbers per cell in siScramble is set at 100% and the Y axis of the graphs are percentage of controls. Values are mean ± SD and p values are calculated with 2-tailed t-test comparison to siScramble control groups. An asterisk indicates a significant difference from controls (p < 0.05).
3.3 ∣. YAP and TAZ have both overlapping and unique transcriptomes
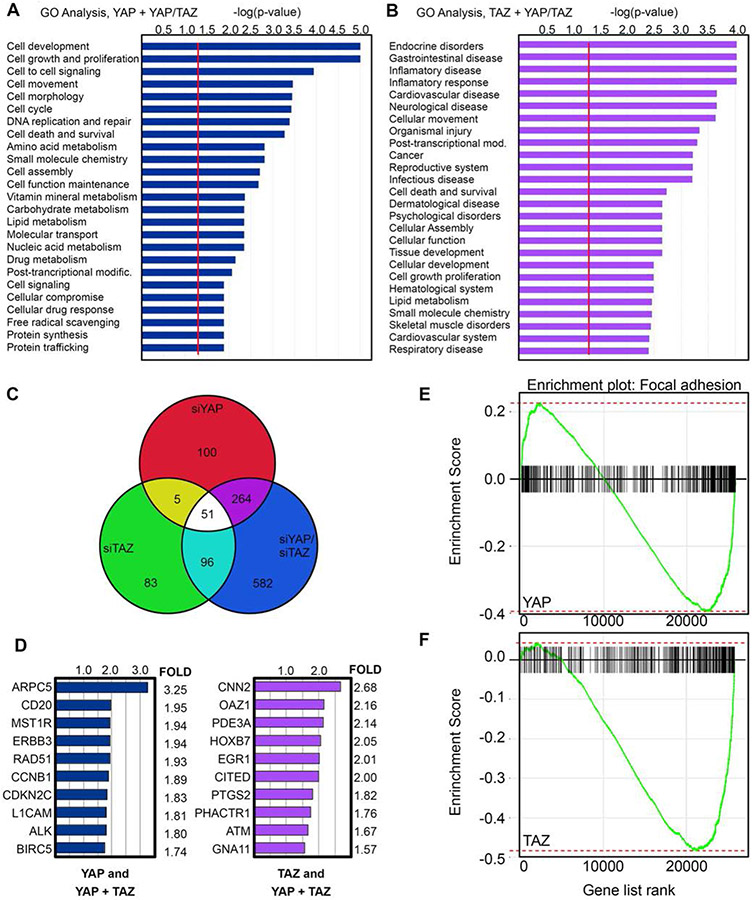
Both YAP and TAZ are transcriptional co-regulators and exert their downstream effects through transcriptional regulation of target genes. Based on our initial results, we theorized that YAP plays a unique role from TAZ in melanoma and that the differences between YAP and TAZ physiology in our experiments were due to differences in downstream transcriptional regulation. As our earlier experiments supported the idea of YAP specific roles in melanoma, we performed RNA-sequencing analysis on YAP, TAZ, and YAP/TAZ inhibited mel537 cells to look for unique transcriptome signatures in our various knockdown groups. To reduce false positives, we used a stringent criterion (FDR = p<0.05, LogFC>1.5) for analysis of our RNA-sequencing data. We grouped differentially expressed genes that were common between YAP and YAP/TAZ knockdowns (termed “YAP specific”) and those between TAZ and YAP/TAZ knockdowns (termed “TAZ specific). There were 264 YAP specific and 96 TAZ specific differentially expressed genes (Figure 3C). Taking these two groups, we performed pathway analysis using Qiagen’s Ingenuity Pathway Analysis software. Pathway analysis revealed stark differences in the molecular function and biological processes between the two groups, with the YAP specific genes more highly enriched in cellular movement, growth, and development groups, whereas the majority of genes for the TAZ specific group were more oriented towards endocrine and inflammatory processes (Figure 3A, 3B). Several of the top differentially expressed genes for the YAP and TAZ specific groups are shown in Figure 3D. Taken together, YAP and TAZ have both overlapping and unique transcriptomes in melanoma.
Figure 3.
YAP and TAZ have both overlapping and unique transcriptomes. (A,B) Gene Ontology (GO) analysis of differentially expressed genes in YAP and YAP/TAZ (A), and to TAZ and YAP/TAZ (B) depleted RNA-seq analysis in mel537 melanoma cells. Data represented as -log(p-value), where significance is determined as being above the threshold as indicated by the red line. For the RNA-seq analysis, the false discovery rate was set at p<0.05 and a fold change of > 1.5 as compared to siScrambled control. (C) Venn Diagram schematic representing the total numbers of differentially expressed genes of the three groups: YAP knockdown, TAZ knockdown, YAP/TAZ knockdown. (D) Representative list of differentially expressed genes from the RNA-Sequencing. Data represented as the mean of the fold changes from the biological duplicates. (E-F) Gene Set Enrichment Analysis (GSEA) of siYAP (E) and siTAZ (F) RNA-sequencing samples (GO:0005925). Statistics for GSEA for siYAP (E) padj = 0.004, NES = −1.657, ES = −0.395, and siTAZ (F) padj = 0.008, NES = −2.35, ES = −0.484 (both sets p<0.05).
3.4 ∣. ARPC5 drives a pro-migratory phenotype in melanoma cells
To look for potential YAP specific genes that controlled focal adhesion numbers, we performed gene set enrichment analysis (GSEA) for the siYAP and siTAZ knockdown groups. GSEA revealed focal adhesion enrichment for both of the groups but due to our previous results that demonstrated a loss of focal adhesion numbers only in our melanoma cells specific to YAP knockdowns, we hypothesized that genes enriched in the focal adhesion gene set for YAP but not for TAZ must be essential for the phenotype examined (Figure 3E, 3F). A closer examination of the differentially expressed genes for the YAP specific group revealed ARP2/3 complex member ARPC5 as a downregulated gene with YAP inhibition (Figure 3D). Inhibition of YAP alone led to a 3.25 fold decrease in ARPC5 transcript (p = 5.4x10−19, FDR = 1.9x1015). As our earlier results showed decreased cellular migration, invasion, and a loss of focal adhesion numbers in YAP inhibited melanoma cells (Figure 1, 2), we speculated that ARPC5, as a member of the actin nucleating ARP2/3 complex, could be driving melanoma cell migration as a downstream target of YAP activity. To recapitulate our RNA-sequencing results, we examined ARPC5 protein and transcript expression in YAP, TAZ, and YAP/TAZ inhibited conditions in different cell lines. 5/5 melanoma cell lines examined showed reduced ARPC5 transcript under YAP, but not TAZ, inhibition (Figure 4A). To determine changes in protein levels, we performed western blots and subsequent densitometry analysis for ARPC5 expression in 3 melanoma cell lines under YAP, TAZ, and YAP/TAZ knockdown conditions. Inhibition of YAP expression led to decreased ARPC5 protein levels in mel537 (27.8±16.9% of controls, Figure 4 B,C), M14 (59.7±9.1%, Figure 4 E,F), and A375 (26.1±12.3%, Figure 4 H, I). Taken together, inhibition of YAP but not TAZ results in loss of ARPC5 expression in melanoma cell lines.
Figure 4.
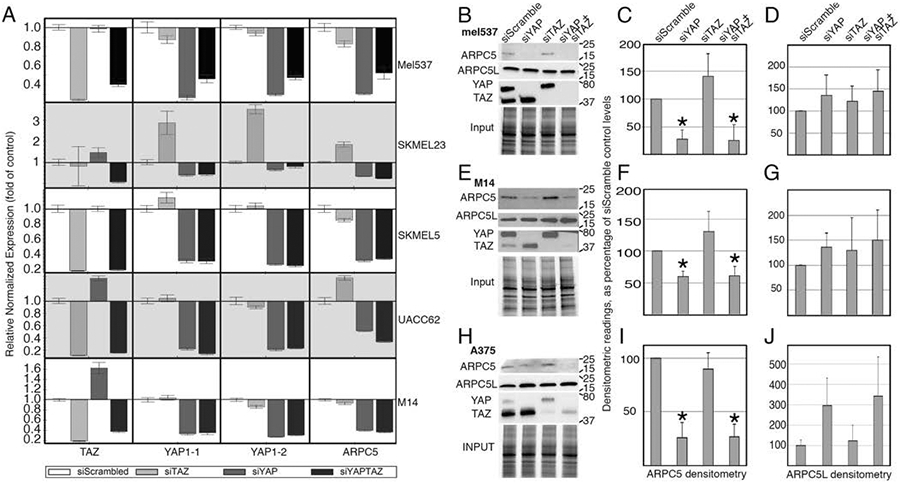
Inhibition of YAP results in a decrease in ARPC5 transcript and protein in melanoma cells. (A) Inhibition of YAP results in decrease of ARPC5 transcript in five melanoma cell lines (mel537, SKMEL23, SKMEL5, UACC62, and M14). Quantitative Real Time Polymerase Chain Reaction analysis examined expression levels of ARPC5, YAP1-1, YAP1-2, and TAZ in melanoma cells treated with siRNA targeting YAP, TAZ, YAP/TAZ, or siScrambled control. The data presented were normalized to control expression levels (GAPDH). Successful gene expression inhibition was indicated by a greater than two fold decrease or undetectable Delta Ct levels when compared to control (siScramble). Inhibition of YAP in all cell lines lead to significant decrease in ARPC5 (p<0.05, 2 tailed Student t-test) in comparison to levels in siScramble controls. (B-J) Inhibition of YAP results in decrease of ARPC5 protein. Western blot analysis for YAP, TAZ, ARPC5, and ARPC5L for the knockdown groups (siScrambled, siYAP, siTAZ, siYAP/TAZ) for the cell lines mel537 (B), M14 (E), and A375 (H). Densitometry quantification of the Western blot analysis for ARPC5 (C, F, I) and ARPC5L (D, G, J) are shown on the right for mel537, M14, and A375, respectively. All data represented as the mean of the fold changes from the biological triplicates and were normalized to a percentage of siScrambled control. Values are mean ± SD. (*, p < 0.05, 2-tailed Student t-test comparisons to siScramble controls).
To determine if inhibition of ARPC5 will phenocopy YAP inhibition, we directly targeted ARPC5 in mel537 and A375 melanoma cells using siRNA specific for ARPC5. Inhibition of ARPC5 in mel537 and A375 cells did not produce significant changes in cell length, cell population size, or ability to invade into matrigel (Figure 5A-C). However, we did observe that both YAP and ARPC5 inhibition reduced migration in a wound healing assay for both mel537 and A375 cells. Compared to the siScrambled control groups, both YAP and ARPC5 knock down groups exhibited decreased migration in both mel537 (73.6± 19.2% and 73±15.33% of siScrambled controls, respectively) and A375 cells (75.0±16.0% and 59.9±16.9% respectively) (p<0.05, 2-tailed t-tests compared to control groups, Figure 5D). Furthermore, inhibition of ARPC5 also led to a reduction of focal adhesions in the cell lines examined. Similar to numbers when YAP expression was blocked, direct ARPC5 inhibition resulted in focal adhesion levels of 70.5%±32.6 (mel537) and 85.1±40% (A375) when compared to siScrambled controls (Figure 5E, p<0.05). To summarize, direct ARPC5 inhibition led to decreased numbers of focal adhesions and migration, but no differences in cell numbers or capacity to invade into matrigel (Figure 5F).
Figure 5.
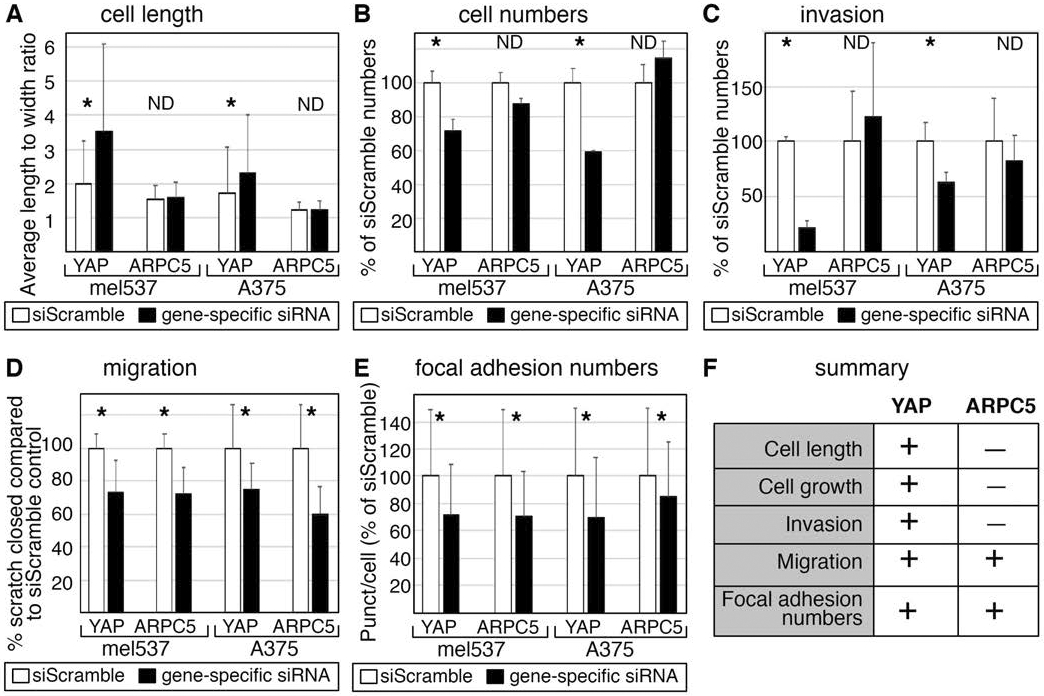
Direct inhibition of ARPC5 results in decrease of melanoma cell migration and focal adhesion numbers. (A,B,C) Direct ARPC5 inhibition does not phenocopy YAP inhibition in A375 and mel537 cells with cell morphology (A), cell numbers (B), or invasion (C). Methods follow procedures outlined in Figure 1. (D,E) ARPC5 inhibition phenocopies YAP depletion for cell migration (D) and focal adhesions numbers in melanoma cells (E). For cell migration, the percent of wound closure after 24 hours was calculated. For focal adhesion numbers, methods outlined in Figure 2 are followed. For A-E, white bars indicate siScrambled control and black bars indicate the gene specific siRNA knockdown group.Each assay was performed at least in triplicate, with a minimum of 200 (A,E) or 60 (B,C) cells per experiment. Asterisk (*) indicates a p < 0.05 and ND (not detected) a p > 0.05 as determined by 2-tailed t-tests in comparison to siScrambled controls. (F) Summary chart of phenotypes induced by direct ARPC5 or YAP inhibition.
In our experiments, ARPC5 is necessary to maintain focal adhesions numbers and migration ability (Figure 5D-F). To determine if ARPC5 is sufficient to rescue YAP-dependent reduction of migration and focal adhesions, exogenous ARPC5 was introduced into YAP-depleted cells. Initial experiments utilized vectors producing high levels of ARPC5 protein under a strong promoter (CMV). This led to a complete loss of viability in both cell lines tested, suggesting that significant increases in ARPC5 protein levels were lethal. To produce physiological levels of ARPC5, a new system made use of a weak and melanocyte native promoter from the PAX3 gene driving expression of ARPC5 (Figure 6A). This partial 1.6 proximal PAX3 promoter region expresses at low but detectable levels in melanoma cells (Kubic et al., 2016). This new model, P3-ARPC5, was able to maintain low levels of ARPC5 transcript even when YAP expression was significantly reduced after YAP siRNA treatment (Figure 6B, first column black bars 2,4). The exogenous vector maintained lower but significant ARPC5 levels even after YAP inhibition (32%±13% mel537, 60%±14% A375) in comparison to non-detectable levels in untransfected controls (Figure 6B, second column black bars 6,8). Exogenous ARPC5 partially rescued YAP-dependent decrease in migration in A375 cells but not in mel537 cells (Figure 6C). All groups had a significant decrease in migration after YAP inhibition (p<0.05, marked with *) except A375 cells, with a partial rescue (p=0.67, marked with ND, with levels of 83.4%±28.8% of control). Exogenous ARPC5 was able to rescue YAP-dependent focal adhesion reduction partially (mel537) or fully (A375) to control levels (Figure 6D). In P3-ARPC5 cells, focal adhesion numbers were maintained after YAP depletion (97.2%±33.6% p=0.62 MEL537, 106.5%±27.7% p=0.60 A375). While loss of YAP led to a number of phenotypic changes, the downstream factor ARPC5 was able to at least partially rescue migration (in A375 cells) and focal adhesion numbers (in mel537 and A375 cells) in the absence of YAP.
Figure 6.
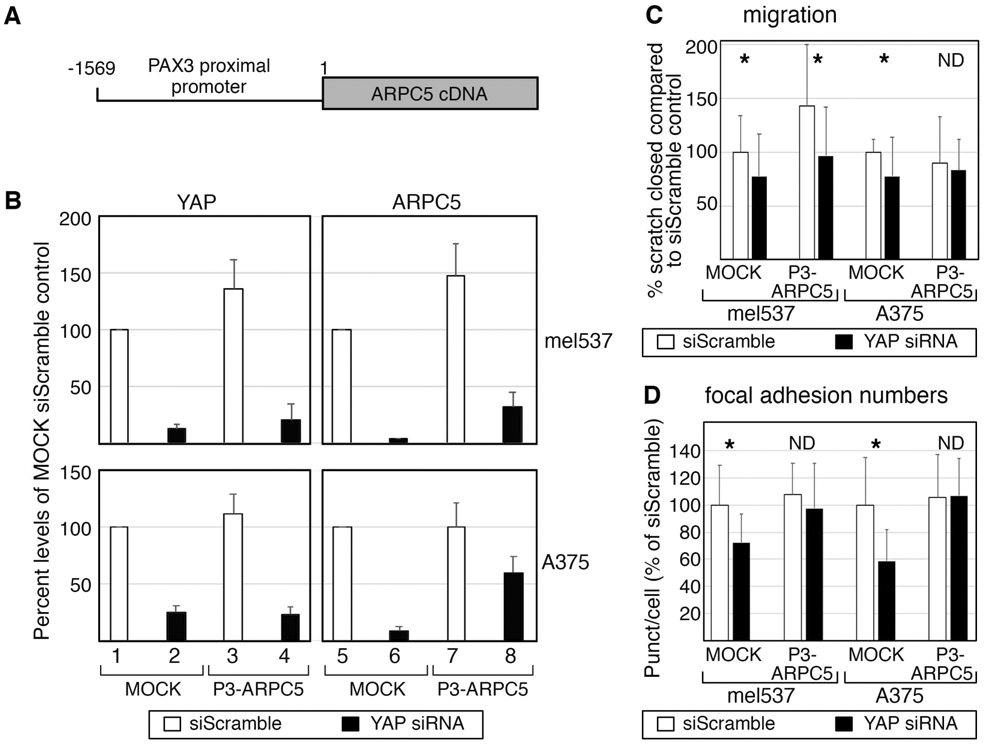
Exogenous ARPC5 expression partially protects against migration or focal adhesion reduction after YAP loss. (A,B) The P3-ARPC5 model utilizes an expression vector expressing physiological levels of ARPC5. The expression vector contains a partial 1569 bp PAX3 promoter driving the expression of an ARPC5 expression cassette. The PAX3 promoter (P3) is active in melanoma cells with low but measurable expression levels (Kubic et al., 2016). Inhibition of YAP in both MOCK and P3-ARPC5 led to undetectable levels (left column graphs in B, siScramble controls (white bars 1,3) versus siYAP (black bars 2,4)) in both mel537 (top row) and A375 (bottom row) cells. However, while ARPC5 levels dropped to undetectable levels in MOCK cells (B, black bar 6) in compared to siScramble control cells (white bars 5,7), the P3-ARPC5 vector partially rescued ARPC5 expression in YAP depleted cells (black bar 8, 32%±13% of control levels in mel537, 60%±14% A375). (C) Exogenous ARPC5 expression partially rescues YAP-dependent reduction in migration in A375 but not mel537 cells. All groups had a significant decrease in migration after YAP inhibition except for A375 cells expressing the P3-ARPC5 vector (insignificant difference from control, p=0.67 marked with ND, with levels of 83.4%±28.8% of control). For cell migration, the percent of wound closure after 24 hours was calculated. (D) Exogenous ARPC5 was able to rescue YAP-dependent focal adhesion reduction partially (mel537) or fully (A375) to control levels. In P3-ARPC5 cells, blocking of YAP did not lead to a significant decrease in focal adhesion numbers, with insignificant differences in comparison to controls (97.2%±33.6% p=0.62 mel537, and 106.5%±27.7% p=0.60 A375). For focal adhesion numbers, methods outlined in Figure 2 are followed. For C and D, white bars indicate siScrambled control and black bars indicate YAP specific siRNA knockdown. Each assay was performed at least in triplicate. Asterisk (*) indicates a p < 0.05 and ND (not detected) a p > 0.05 as determined by 2-tailed t-tests in comparison to siScrambled controls.
3.5 ∣. ARPC5 and ARPC5L have an inverse relationship in melanoma
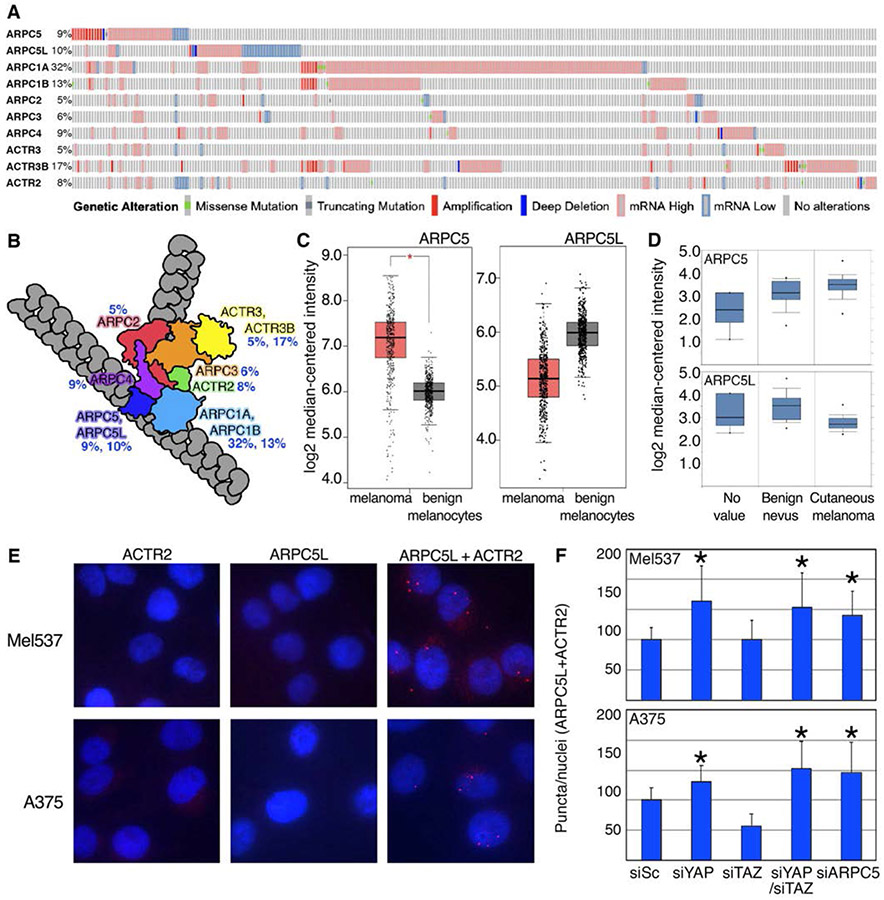
ARPC5 is a member of the 7 subunit ARP2/3 complex, a crucial regulator of actin nucleation, with subunits listed and shown schematically in Figure 7 A,B (Weaver et al., 2001). Meta-analysis of the melanoma TCGA dataset reveals that expression levels of each of the ARP2/3 family members are often altered in melanoma patients (percentage shown next to each subunit in Figure 7 A,B). Taken together, alterations to any of the ARP2/3 subunits occurs in 61% of melanoma patients from the TCGA dataset, and that the majority of these differences are due to increased expression, as opposed to mutations or gene duplications (Figure 7A).
Figure 7.
The relationship between ARPC5, ARPC5L in melanoma cells. (A,B) ARP2/3 subunits, except ARPC5L, are frequently overexpressed in human melanoma patients. Meta-analysis of the melanoma TCGA dataset for expression levels of ARP2/3 complex subunits from cBioportal (287 with ARP2/3 abnormalities of 471 total patients shown) represented as an oncoprint. (B). Representation of ARP2/3 complex subunits on actin filaments are grouped by color and designated with percent of cases altered in human melanoma TCGA dataset. (C,D) Expression of ARPC5 and ARPC5L is increased and decreased respectively in human melanoma. (C) ARPC5 GEPIA expression analysis of ARPC5 and ARPC5L transcripts comparing tumor to normal tissue from GTex and TCGA human melanoma datasets. (D) Oncomine expression analysis comparing benign nevus to cutaneous melanoma samples for ARPC5 and ARPC5L (E-F) Proximity ligation assay (PLA) analysis on cells transfected with siRNA targeting YAP, TAZ, YAP/TAZ, and ARPC5. (E) Representative images of PLA control samples, with mel537 (top row) and A375 (bottom) cells, with ACTR2 (ARP2 subunit, first column) or ARPC5L (second column) alone (negative controls), and with combined ACTR2 and ARPC5L antibodies together (third column, positive control). Interactions of ACTR2 and ARPC5L were visualized with red fluorescent puncta. Nuclei shown with DAPI (blue). (F) Quantified PLA puncta counts of ACTR2 and ARPC5L for each siRNA treated group, normalized to control (first column, siScramble) in mel537 (top graph) and A375 (bottom graph) cells. Average puncta per nuclei per group were counted (n=200) and values were normalized to percentage of control levels of puncta per nuclei. Values are mean ± SD. (*, p < 0.05, 2 tailed t-test comparison to siScramble control groups).
In addition to ARPC5, there is a separate isoform, ARPC5L, which can substitute for ARPC5 in the ARP2/3 complex. Previous studies have shown that these two subunits result in unique ARP2/3 complexes, which drive different actin dynamics depending on which isoform is present (Abella et al., 2016). In contrast to the other ARP2/3 family members, ARPC5L expression in the TCGA melanoma dataset is more commonly under expressed than overexpressed (23/36 altered cases, Figure 7A). A comparison of the two isoforms reveal an approximately 2-fold increase in ARPC5 expression and a subsequent 2-fold decrease in ARPC5L expression in tumor versus normal tissue samples (Figure 7C). Furthermore, examination of ARPC5 and ARPC5L expression in benign nevus versus cutaneous melanoma in a separate melanoma dataset shows the same pattern (Figure 7D (Talantov et al., 2005)). In summary, ARP2/3 complex members are often overexpressed in melanoma, but ARPC5L and ARPC5 have an inverse relationship with regards to expression levels.
3.6 ∣. Inhibition of YAP results in higher numbers of ARP2/3 complexes containing ARPC5L
ARPC5 levels are dependent on YAP expression in melanoma cells (Figure 3,4). In terms of ARPC5L, expression post YAP inhibition is not altered in the majority of lines examined, with the exception of a trend toward an increase in A375 cells (Figure 4). To investigate whether or not this decrease in ARPC5 expression could physiologically change the number of ARP2/3 complexes with ARPC5 as opposed to ARPC5L, we performed proximity ligation assays (PLA) with ARPC5L and ACTR2 (a complex member with no substitute, also known as ARP2) in mel537 and A375 cells treated with siYAP, siTAZ, siYAP/TAZ, or siARPC5. Quantification of the results revealed an increase in complexes with ARPC5L in YAP, YAP/TAZ, and ARPC5 inhibited cells but not in cells with expression of TAZ blocked (p<0.05, n = 200 cells/group). Collectively, this suggests that YAP predominantly drives expression of ARPC5, resulting in higher number of ARP2/3 complexes containing ARPC5 in melanoma (Figure 7E,F).
In summary, YAP, but not TAZ, inhibition results in decreased invasion, migration, and focal adhesion numbers in melanoma cells, most likely due to YAP-specific downstream targets. One YAP target, ARPC5, is essential to maintain YAP-dependent focal adhesion numbers. ARPC5 is a member of the ARP2/3 complex, a part of the actin assembly machinery that is frequently misexpressed in melanoma.
4 ∣. DISCUSSION
YAP and TAZ are often dysregulated in many different types of cancers, where they drive expression of genes crucial for cancer initiation, progression, and metastasis (Q. Feng et al., 2018; Liu, Yu, Huang, Cui, & Hong, 2017; Martinez et al., 2019; Nallet-Staub et al., 2014; Zanconato et al., 2018; Zhao et al., 2008). Recent studies have linked YAP overexpression, as well as canonical TEAD transcription factor partners, to melanoma invasion and metastasis (Lamar et al., 2012; Verfaillie et al., 2015; X. Zhang et al., 2020). Exogenous overexpression of YAP promotes invasion, metastasis, and anoikis resistance, while inhibition led to decreases in invasion and lung colonies in nude mice tail vein injections (Nallet-Staub et al., 2014; X. Zhang et al., 2020; Zhao et al., 2021). In the context of drug resistance, PLX4032 resistant melanoma cell lines exhibited a gene signature similar to that of increased YAP activity, while YAP/TAZ inhibition led to lower viability in the presence of PLX4032 (Kim et al., 2016). Furthermore, increased YAP activity led to increased PLX4032 resistance (Fisher et al., 2017; Kim et al., 2016). While these studies support the role of YAP in melanoma, the role that TAZ plays is not always directly addressed experimentally. It is possible that the two cofactors play unique roles specific for certain cancer processes. Here, we examine each factor and discover YAP-specific roles for melanoma migration and focal adhesion numbers.
Although genetic data indicates functional redundancy in some contexts, it should not be unexpected that these two proteins could have evolved unique functions, particularly given that YAP and TAZ contain unique moieties (Varelas, 2014). Indeed, recent reports have started to delineate differential roles between these two transcriptional coactivators. YAP was found to be elevated in benign nevi and primary cutaneous melanomas as compared to normal melanocytes (X. Zhang et al., 2019). In our studies, we discovered variable responses to YAP loss, as well as sensitivity to TAZ inhibition (Figure 1). This variable viability and TAZ loss sensitivity has parallels to a prior study (X. Zhang et al., 2019) that also found no obvious correlation to BRAF/NRAS mutation status or melanoma stage. The cause for this variable response to YAP and/or TAZ is, at present, unclear. However, there is some evidence that melanoma phenotype may play a role. Melanoma cells have the ability to phenotype switch between a more invasive or proliferative cell type based on environmental signals and gene expression (Hoek et al., 2008; Hoek et al., 2006; Tirosh et al., 2016; Widmer et al., 2012). Prior studies support a stronger tie of YAP to an invasive phenotype (X. Zhang et al., 2020); therefore, a cell line's dominant base phenotype may impact on the YAP and TAZ inhibition responses. We found that YAP and TAZ have different gene expression profiles after inhibition (Figure 3), and that TAZ was not able to compensate for YAP loss and reduced cell invasion and focal adhesion numbers (Figure 1,2). The observation that TAZ was unable to compensate for YAP loss are limited but not without precedence. For example, TAZ was unable to rescue cell survival after YAP loss for a subset of melanoma cells (X. Zhang et al., 2019), and in HEK293 cells, YAP was found to have greater impact than TAZ on cell proliferation and migration, among other cell functions (Plouffe et al., 2018). We find that YAP, but not TAZ, is the predominant player in our melanoma cell lines. Functional experiments (Figure 1,2) and RNA-sequencing (Figure 3) all reveal a more substantial role of YAP than TAZ in driving human melanoma. It is especially interesting that different cancers have unique and redundant roles for YAP and TAZ. A clue to the differential roles YAP and TAZ play may lay in the protein domains unique to YAP or TAZ. Due to the lack of DNA binding domain in YAP and TAZ, it is possible that YAP and TAZ have different binding partners specific to their unique binding domains to help facilitate their downstream effects. Future studies into protein domains specific to YAP, such as the SH3 and the YAP1-2 specific second WW domain, will be needed to fully understand how YAP drives migration and survival.
Here we find ARPC5 is down-regulated with inhibition of YAP, but not TAZ. We show that direct inhibition of ARPC5 or YAP led to decreased cell migration and focal adhesion numbers. In head and neck squamous cell carcinoma, inhibition of ARPC5 decreased cancer cell migration and invasion (Kinoshita et al., 2012). In multiple myeloma, ARPC5 has been implicated in its potential use as a biomarker for the disease (Xiong & Luo, 2018). Indirectly, microRNA-141 has anti-metastatic properties in prostate cancer, with at least one mechanism of tumor suppression through the targeting and inhibition of ARPC5 (C. Liu et al., 2017). Our studies complement these prior reports by implicating a role of ARPC5 in driving melanoma migration and invasion. Direct knockdown of ARPC5 phenocopies YAP inhibition in terms of decreased melanoma cell migration and focal adhesion numbers (Figure 5). While exogeneous expression of ARPC5 in YAP-depleted cells had some rescue of YAP-dependent migration, a more significant impact was seen in protecting overall focal adhesion numbers (Figure 6). As focal adhesion dynamics and cell migration are closely linked, it is possible that YAP controls focal adhesion turnover, dynamics, and stability through its regulation of ARPC5 (Hoock et al., 2019; Nagano et al., 2012).
We discovered that overall ARPC5 levels were critical for melanoma cells, where high overexpression driven from a CMV promoter led to cell death but low physiological expression levels were able to have significant functional consequences (Figure 6). Further, a prior study overexpressing YAP did not find an abundance of ARPC5 levels (X. Zhang et al., 2020). This may be due, at least in part, to the overall function of the ARP2/3 complex, which has different properties with respect to the stability and turnover when it contains either ARPC5 or ARPC5L (Abella et al., 2016). The ratio of ARPC5 to ARPC5L has direct impact on complex activity, where ARPC5L supports more nucleation (the first step of actin polymerization prior to elongation) and assembly, but is 2 fold slower than ARPC5-containing complexes in terms of turnover (Abella et al., 2016; von Loeffelholz et al., 2020). We find that ARPC5L does not change with either YAP or TAZ inhibition (Figure 4). Furthermore, melanoma cells shift to larger numbers of ARPC5L-ARP2/3 complexes as compared to control than with inhibition of YAP or ARPC5, but not TAZ (Figure 7). This sets up a possible model in melanoma, where YAP promotes an increase in ARPC5-containing ARP2/3 complexes leading to decreased cytoskeletal stability but increased turnover, resulting in an overall more dynamic system promoting cell migration and invasion. The dynamic control of actin has been shown to control melanoma drug resistance, tumorigenesis, and even YAP/TAZ activity, among other functions (Kang et al., 2018; Kim et al., 2016; Panciera, Azzolin, Cordenonsi, & Piccolo, 2017). Further studies into how ARPC5 are needed to elucidate the mechanism of ARPC5-ARP2/3 driven metastasis.
In conclusion, this study provides evidence that YAP plays a more crucial role than TAZ in driving pro-tumorigenic phenotypes in melanoma cells, suggesting that YAP is a driver of melanoma progression, migration, and invasion. We provide support that YAP drives melanoma migration through YAP specific regulation of ARPC5, shifting ARP2/3 dynamics towards a pro-migratory phenotype. Lastly, we postulate that melanoma is a great model to study differences between YAP and TAZ, as both biased and unbiased screens show a higher reliance on YAP in melanoma. While future studies will be needed to understand fully how YAP and TAZ differ, it is clear that these two genes have both overlapping and unique targets in driving their downstream effects.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health, grant numbers NIH R01CA130202, R01AR062547, 1UL1TR001430, 1TL1TR001410, T32GM007183 and R01CA184001, the American Cancer Society, grant number RSG-CSM-121505, the Friends of Dermatology-Chicago, the American Skin Association Daneen and Charles Stiefel Investigative Scientist Award, the Harry J. Lloyd Charitable Trust, the Falanga Scholar endowment (Boston University), and the Department of Dermatology at Boston University.
Footnotes
CONFLICT OF INTEREST – The authors declare that they have no conflicts of interests with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
DATA AVAILABILITY STATEMENT
The raw reads produced in this study were deposited in the NCBI GEO database with accession number GSE146918.
REFERENCES
- Abella JV, Galloni C, Pernier J, Barry DJ, Kjaer S, Carlier MF, & Way M (2016). Isoform diversity in the Arp2/3 complex determines actin filament dynamics. Nat Cell Biol, 18(1), 76–86. doi: 10.1038/ncb3286 [DOI] [PubMed] [Google Scholar]
- Choe MH, Yoon Y, Kim J, Hwang SG, Han YH, & Kim JS (2018). miR-550a-3-5p acts as a tumor suppressor and reverses BRAF inhibitor resistance through the direct targeting of YAP. Cell Death Dis, 9(6), 640. doi: 10.1038/s41419-018-0698-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Guo P, Kang S, & Zhao F (2018). High expression of TAZ/YAP promotes the progression of malignant melanoma and affects the postoperative survival of patients. Pharmazie, 73(11), 662–665. doi: 10.1691/ph.2018.8499 [DOI] [PubMed] [Google Scholar]
- Feng X, Degese MS, Iglesias-Bartolome R, Vaque JP, Molinolo AA, Rodrigues M, … Gutkind JS (2014). Hippo-independent activation of YAP by the GNAQ uveal melanoma oncogene through a trio-regulated rho GTPase signaling circuitry. Cancer Cell, 25(6), 831–845. doi: 10.1016/j.ccr.2014.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrigno O, Lallemand F, Verrecchia F, L'Hoste S, Camonis J, Atfi A, & Mauviel A (2002). Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene, 21(32), 4879–4884. doi: 10.1038/sj.onc.1205623 [DOI] [PubMed] [Google Scholar]
- Fisher ML, Grun D, Adhikary G, Xu W, & Eckert RL (2017). Inhibition of YAP function overcomes BRAF inhibitor resistance in melanoma cancer stem cells. Oncotarget, 8(66), 110257–110272. doi: 10.18632/oncotarget.22628 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gaffney CJ, Oka T, Mazack V, Hilman D, Gat U, Muramatsu T, … Sudol M (2012). Identification, basic characterization and evolutionary analysis of differentially spliced mRNA isoforms of human YAP1 gene. Gene, 509(2), 215–222. doi: 10.1016/j.gene.2012.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han LL, Yin XR, & Zhang SQ (2018). miR-103 promotes the metastasis and EMT of hepatocellular carcinoma by directly inhibiting LATS2. Int J Oncol, 53(6), 2433–2444. doi: 10.3892/ijo.2018.4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, … Dummer R (2008). In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res, 68(3), 650–656. doi: 10.1158/0008-5472.CAN-07-2491 [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, … Dummer R (2006). Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res, 19(4), 290–302. doi: 10.1111/j.1600-0749.2006.00322.x [DOI] [PubMed] [Google Scholar]
- Hong JH, Hwang ES, McManus MT, Amsterdam A, Tian Y, Kalmukova R, … Yaffe MB (2005). TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science, 309(5737), 1074–1078. doi: 10.1126/science.1110955 [DOI] [PubMed] [Google Scholar]
- Hong W, & Guan KL (2012). The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol, 23(7), 785–793. doi: 10.1016/j.semcdb.2012.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoock SC, Ritter A, Steinhauser K, Roth S, Behrends C, Oswald F, … Yuan J (2019). RITA modulates cell migration and invasion by affecting focal adhesion dynamics. Mol Oncol, 13(10), 2121–2141. doi: 10.1002/1878-0261.12551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain Z, Ali SM, Ko HL, Xu J, Ng CP, Guo K, … Hunziker W (2007). Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1. Proc Natl Acad Sci U S A, 104(5), 1631–1636. doi: 10.1073/pnas.0605266104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M, Borchers C, & Milgram SL (2004). Heterogeneous nuclear ribonuclear protein U associates with YAP and regulates its co-activation of Bax transcription. J Biol Chem, 279(25), 26300–26306. doi: 10.1074/jbc.M401070200 [DOI] [PubMed] [Google Scholar]
- Kanai F, Marignani PA, Sarbassova D, Yagi R, Hall RA, Donowitz M, … Yaffe MB (2000). TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. Embo J, 19(24), 6778–6791. doi: 10.1093/emboj/19.24.6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Wang J, Yao Z, Hu Y, Ma S, Fan Q, … Sun J (2018). Fascin induces melanoma tumorigenesis and stemness through regulating the Hippo pathway. Cell Commun Signal, 76(1), 37. doi: 10.1186/s12964-018-0250-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Kim J, Hong H, Lee SH, Lee JK, Jung E, & Kim J (2016). Actin remodeling confers BRAF inhibitor resistance to melanoma cells through YAP/TAZ activation. Embo J, 35(5), 462–478. doi: 10.15252/embj.201592081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Nohata N, Watanabe-Takano H, Yoshino H, Hidaka H, Fujimura L, … Seki N (2012). Actin-related protein 2/3 complex subunit 5 (ARPC5) contributes to cell migration and invasion and is directly regulated by tumor-suppressive microRNA-133a in head and neck squamous cell carcinoma. Int J Oncol, 40(6), 1770–1778. doi: 10.3892/ijo.2012.1390 [DOI] [PubMed] [Google Scholar]
- Kubic JD, Little EC, Kaiser RS, Young KP, & Lang D (2016). FOXD3 Promotes PAX3 Expression in Melanoma Cells. J Cell Biochem, 117(2), 533–541. doi: 10.1002/jcb.25306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamar JM, Stern P, Liu H, Schindler JW, Jiang ZG, & Hynes RO (2012). The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci U S A, 109(37), E2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Liu R, Zhang D, Deng Q, Liu B, Chao HP, … Tang DG (2017). MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat Commun, 8, 14270. doi: 10.1038/ncomms14270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Yu T, Huang Y, Cui L, & Hong W (2017). ETS (E26 transformation-specific) upregulation of the transcriptional co-activator TAZ promotes cell migration and metastasis in prostate cancer. J Biol Chem, 292(22), 9420–9430. doi: 10.1074/jbc.M117.783787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JW, Xiao S, Ogomori K, Hammarstedt JE, Little EC, & Lang D (2019). The Efficiency of Verteporfin as a Therapeutic Option in Pre-Clinical Models of Melanoma. J Cancer, 10(1), 1–10. doi: 10.7150/jca.27472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita R, Uchijima Y, Nishiyama K, Amano T, Chen Q, Takeuchi T, … Kurihara H (2008). Multiple renal cysts, urinary concentration defects, and pulmonary emphysematous changes in mice lacking TAZ. Am J Physiol Renal Physiol, 294(3), F542–553. doi: 10.1152/ajprenal.00201.2007 [DOI] [PubMed] [Google Scholar]
- Martinez B, Yang Y, Harker DMR, Farrar C, Mukundan H, Nath P, & Mascarenas D (2019). YAP/TAZ Related BioMechano Signal Transduction and Cancer Metastasis. Front Cell Dev Biol, 7, 199. doi: 10.3389/fcell.2019.00199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel M, Meckbach D, Weide B, Toussaint NC, Schilbach K, Noor S, … Bauer J (2014). In melanoma, Hippo signaling is affected by copy number alterations and YAP1 overexpression impairs patient survival. Pigment Cell Melanoma Res, 27(4), 671–673. doi: 10.1111/pcmr.12249 [DOI] [PubMed] [Google Scholar]
- Mitra SK, & Schlaepfer DD (2006). Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr Opin Cell Biol, 18(5), 516–523. doi: 10.1016/j.ceb.2006.08.011 [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, … Milgram SL (2006). Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol, 26(1), 77–87. doi: 10.1128/MCB.26.1.77-87.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M, Hoshino D, Koshikawa N, Akizawa T, & Seiki M (2012). Turnover of focal adhesions and cancer cell migration. Int J Cell Biol, 2012, 310616. doi: 10.1155/2012/310616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallet-Staub F, Marsaud V, Li L, Gilbert C, Dodier S, Bataille V, … Mauviel A (2014). Pro-invasive activity of the Hippo pathway effectors YAP and TAZ in cutaneous melanoma. J Invest Dermatol, 134(1), 123–132. doi: 10.1038/jid.2013.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panciera T, Azzolin L, Cordenonsi M, & Piccolo S (2017). Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol, 18(12), 758–770. doi: 10.1038/nrm.2017.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plouffe SW, Lin KC, Moore JL 3rd, Tan FE, Ma S, Ye Z, … Guan KL (2018). The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell. J Biol Chem, 293(28), 11230–11240. doi: 10.1074/jbc.RA118.002715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto C, & Barrios D (2019). RaNA-Seq: Interactive RNA-Seq analysis from FASTQ files to functional analysis. Bioinformatics. doi: 10.1093/bioinformatics/btz854 [DOI] [PubMed] [Google Scholar]
- Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, … Wang Y (2005). Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res, 11(20), 7234–7242. doi: 10.1158/1078-0432.CCR-05-0683 [DOI] [PubMed] [Google Scholar]
- Tang L, Zhou L, Wu S, Shi X, Jiang G, Niu S, & Ding D (2019). miR-125a-5p inhibits colorectal cancer cell epithelial-mesenchymal transition, invasion and migration by targeting TAZ. Onco Targets Ther, 12, 3481–3489. doi: 10.2147/OTT.S191247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tas F (2012). Metastatic behavior in melanoma: timing, pattern, survival, and influencing factors. J Oncol, 2012, 647684. doi: 10.1155/2012/647684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Kolb R, Hong JH, Carroll J, Li D, You J, … Benjamin T (2007). TAZ promotes PC2 degradation through a SCFbeta-Trcp E3 ligase complex. Mol Cell Biol, 27(18), 6383–6395. doi: 10.1128/MCB.00254-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, … Garraway LA (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science, 352(6282), 189–196. doi: 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X (2014). The Hippo pathway effectors TAZ and YAP in development, homeostasis and disease. Development, 141(8), 1614–1626. doi: 10.1242/dev.102376 [DOI] [PubMed] [Google Scholar]
- Verfaillie A, Imrichova H, Atak ZK, Dewaele M, Rambow F, Hulselmans G, … Aerts S (2015). Decoding the regulatory landscape of melanoma reveals TEADS as regulators of the invasive cell state. Nat Commun, 6, 6683. doi: 10.1038/ncomms7683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Loeffelholz O, Purkiss A, Cao L, Kjaer S, Kogata N, Romet-Lemonne G, … Moores CA (2020). Cryo-EM of human Arp2/3 complexes provides structural insights into actin nucleation modulation by ARPC5 isoforms. Biol Open, 9(7). doi: 10.1242/bio.054304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer DS, Cheng PF, Eichhoff OM, Belloni BC, Zipser MC, Schlegel NC, … Hoek KS (2012). Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res, 25(3), 343–353. doi: 10.1111/j.1755-148X.2012.00986.x [DOI] [PubMed] [Google Scholar]
- Xiong T, & Luo Z (2018). The Expression of Actin-Related Protein 2/3 Complex Subunit 5 (ARPC5) Expression in Multiple Myeloma and its Prognostic Significance. Med Sci Monit, 24, 6340–6348. doi: 10.12659/MSM.908944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, & Ito Y (1999). A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. Embo J, 18(9), 2551–2562. doi: 10.1093/emboj/18.9.2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, … Guan KL (2014). Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell, 25(6), 822–830. doi: 10.1016/j.ccr.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Li D, Li H, Wang L, Tian G, & Dong Y (2016). YAP overexpression promotes the epithelial-mesenchymal transition and chemoresistance in pancreatic cancer cells. Mol Med Rep, 13(1), 237–242. doi: 10.3892/mmr.2015.4550 [DOI] [PubMed] [Google Scholar]
- Zanconato F, Battilana G, Forcato M, Filippi L, Azzolin L, Manfrin A, … Piccolo S (2018). Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nat Med, 24(10), 1599–1610. doi: 10.1038/s41591-018-0158-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Cordenonsi M, & Piccolo S (2016). YAP/TAZ at the Roots of Cancer. Cancer Cell, 29(6), 783–803. doi: 10.1016/j.ccell.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, … Piccolo S (2015). Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol, 17(9), 1218–1227. doi: 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Pasolli HA, & Fuchs E (2011). Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci U S A, 108(6), 2270–2275. doi: 10.1073/pnas.1019603108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tang JZ, Vergara IA, Zhang Y, Szeto P, Yang L, … Shackleton M (2019). Somatic hypermutation of the YAP oncogene in a human cutaneous melanoma. Mol Cancer Res. doi: 10.1158/1541-7786.MCR-18-0407 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yang L, Szeto P, Abali GK, Zhang Y, Kulkarni A, … Harvey KF (2020). The Hippo pathway oncoprotein YAP promotes melanoma cell invasion and spontaneous metastasis. Oncogene, 39(30), 5267–5281. doi: 10.1038/s41388-020-1362-9 [DOI] [PubMed] [Google Scholar]
- Zhao B, Xie J, Zhou X, Zhang L, Cheng X, & Liang C (2021). YAP activation in melanoma contributes to anoikis resistance and metastasis. Exp Biol Med (Maywood), 246(8), 888–896. doi: 10.1177/1535370220977101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, … Guan KL (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev, 22(14), 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw reads produced in this study were deposited in the NCBI GEO database with accession number GSE146918.