SUMMARY
The 12-h clock coordinates lipid homeostasis, energy metabolism, and stress rhythms via the transcriptional regulator XBP1. However, the biochemical and physiological bases for integrated control of the 12-h clock and diverse metabolic pathways remain unclear. Here, we show that steroid receptor coactivator SRC-3 coactivates XBP1 transcription and regulates hepatic 12-h cistrome and gene rhythmicity. Mice lacking SRC-3 show abnormal 12-h rhythms in hepatic transcription, metabolic functions, systemic energetics, and rate-limiting lipid metabolic processes, including triglyceride, phospholipid, and cardiolipin pathways. Notably, 12-h clock coactivation is not only preserved, with its cistromic activation priming ahead of the zeitgeber cue of light, but concomitant with rhythmic remodeling in the absence of food. These findings reveal that SRC-3 integrates the mammalian 12-h clock, energy metabolism, and membrane and lipid homeostasis and demonstrates a role for the 12-h clock machinery as an active transcriptional mechanism in anticipating physiological and metabolic energy needs and stresses.
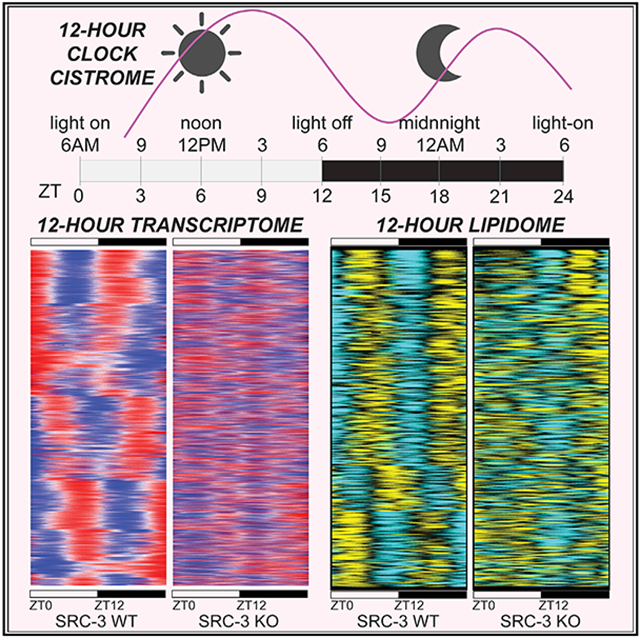
Graphical abstract
In brief
Meng et al. uncover the biochemical, physiological, and metabolic bases for rhythmic control of the 12-h clock in diverse metabolic pathways. They reveal that SRC-3 integrates the mammalian 12-h clock, lipid metabolism, and energy homeostasis as an active transcriptional mechanism in anticipating physiological and energy needs and metabolic stresses.
INTRODUCTION
Chronobiological rhythms govern biological processes and activities that are essential for maintaining physiological homeostasis, enabling organisms to anticipate environmental and nutritional changes for survival. Apart from the well-characterized 24-h circadian clock, an ultradian 12-h periodic rhythmicity is conserved in multiple species (Cretenet et al., 2010; Hughes et al., 2009; Meng et al., 2020; Pan et al., 2020; Zhang et al., 2013; Zhu et al., 2017). For example, marine organisms possess dominant ~12-h behaviors that can anticipate circatidal nutritional changes in the external environment (Zhang et al., 2013). In mammals, the 12-h periodic rhythmicity in peripheral tissues has been noted in a cluster of ER and metabolic stress genes that are regulated by X-box-binding protein-1 (XBP1) (Cretenet et al., 2010; Hughes et al., 2009; Meng et al., 2020; Pan et al., 2020; Zhu et al., 2017). With 12-h periodic rhythmicity at both the mRNA and protein levels, the spliced form of XBP1 (XBP1s) has been shown to regulate ER homeostatic and rate-limiting metabolic genes that control diverse lipid and metabolic pathways (Meng et al., 2020; Zhu et al., 2017). Notably, hepatic ablation of XBP1 disrupts the 12-h clock and promotes a progressive non-alcoholic fatty liver disease (NAFLD) phenotype via dysregulation of membrane fluidity and lipid metabolism, highlighting the importance of the 12-h clock in maintaining metabolic homeostasis (Meng et al., 2020).
The notion of the 12-h clock and the identification of XBP1s as a major 12-h cycling transcriptional factor (TF) regulator prompted the question of whether co-regulation of XBP1 contributes to 12-h clock regulation in diverse lipid and metabolic pathways. Many of the 12-h physiological cycles have been intimately connected to ER stress and unfolded protein response (UPR) pathways and systemic maintenance of metabolic homeostasis (Cretenet et al., 2010; Hughes et al., 2009; Meng et al., 2020; Zhu et al., 2017). It has been shown that perturbations in biological clocks and metabolism adversely affect metabolic homeostasis, increasing risks for metabolic diseases and cancer (Patke et al., 2020; Reinke and Asher, 2019). Thus, discovery of new 12-h clock coregulators will help determine the role of 12-h clock oscillators and further decipher their relationship with the 24-h rhythm for maintenance of metabolic homeostasis in physiology and diseases.
Here, we have extended our characterization of XBP1 coactivation by steroid receptor coactivator 3 (SRC-3; gene symbol NCOA3, Nuclear Receptor Coactivator 3) as a molecular component of the 12-h clock. We show that SRC-3 coactivation of XBP1 plays a key role in integrating the mammalian 12-h clock, lipid homeostasis, and energy metabolism. We found that SRC-3 regulates the 12-h rhythmicity of hepatic transcription, lipid metabolic processes, and systemic energetics via transcriptional regulation of triglyceride, phospholipid, and cardiolipin pathways. Notably, this coactivator component of the 12-h clock is intrinsically capable of transcriptional plasticity in hepatic physiology, offering unique opportunities for potential therapeutic intervention against metabolic diseases and cancer. This work uncovers an important module of the mammalian peripheral 12-h clock regulatory network and suggests that coactivation of the 12-h clock is an important mechanism that anticipates environmental and nutritional changes, coordinating rhythmic transcriptional and metabolic processes for whole-body energy homeostasis.
RESULTS
SRC-3 coactivates XBP1 transcription and regulates hepatic 12-h gene oscillations
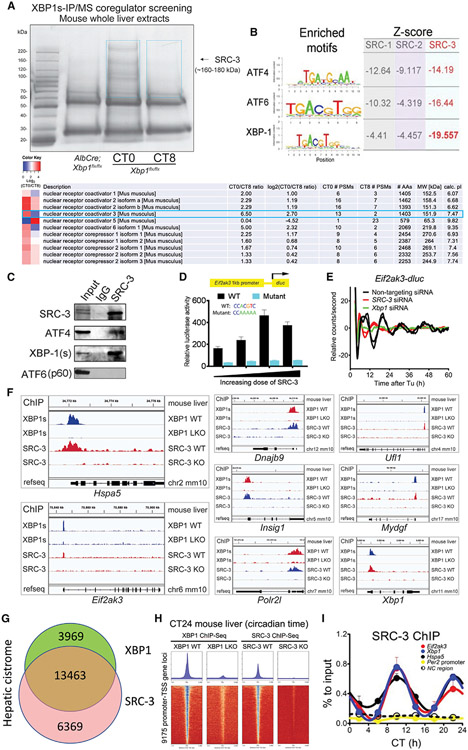
In the present study, we set out to identify and characterize novel regulatory modules of the 12-h clock. To identify proteins involved in regulating the activity and rhythmicity of the hepatic 12-h clock, XBP1s protein complexes were purified from mouse whole livers at two representative circadian times (CTs), CT0 and CT8, using an XBP1s antibody-based affinity pulldown approach (Figure 1A). We chose liver samples at CT0 and CT8 (4 before the dusk 12-h rhythmic peak) based on the temporal characteristics of 12-h hepatic cistromic binding profiles (Meng et al., 2020), which have shown that XBP1s is enriched at dawn (CT0) and at dusk (CT12). XBP1s-associated proteins were identified via quantitative liquid chromatography with tandem mass spectrometry (LC-MS/MS). Notably, several coregulatory protein components, including SRC-3, copurified with XBP1s in a rhythmic manner. We performed an unbiased motif analysis of the hepatic cistrome of all three steroid receptor coactivator (SRC) family members by chromatin immunoprecipitation sequencing (ChIP-seq) (Zhu et al., 2015), which revealed the strongest enrichment of UPR TFs motifs at SRC-3 binding sites, with the XBP1 motif exhibiting the strongest enrichment (Figure 1B). This suggested that SRC-3 is a coactivator for XBP1s. We confirmed the specific interaction between XBP1s and SRC-3 in the mouse liver in vivo using coimmunoprecipitation (Co-IP) assays (Figure 1C). Interestingly, Co-IP assays revealed a strong interaction between endogenous SRC-3 and XBP1s, while SRC-3 interaction with another ER stress regulator, activating transcription factor 6 (ATF6), was not observed, and its interaction with ATF4 was much weaker, suggesting a more prominent interaction between hepatic SRC-3 and XBP1s.
Figure 1. SRC-3 coactivates XBP1 transcription and regulates hepatic 12-h gene oscillation.
(A) Temporal XBP1s immunoprecipitation followed by mass spectrometry (IP/MS) coregulator screening in the mouse whole liver extracts identified SRC-3 as XBP1s-interacting protein candidate. Coomassie staining image of anti-XBP1s-immunoprecipitated proteins from the mouse liver extracts of Xbp1 flx/flx (WT) at circadian times CT0 and CT8, and AlbCre;Xbp1 flx/flx (LKO) mouse liver extracts are shown. SRC-3 identified by anti-XBP1s IP/MS is indicated with the black arrow. The temporal XBP1s-associated coregulators with CT0/CT8 ratio heatmaps peptide numbers, and length and molecular weight of the coregulators are shown. The two blue boxes show the gel slices submitted for MS analysis.
(B) Motif analysis of the hepatic cistrome of three major steroid receptor coactivators (SRCs) showed the strongest enrichment of XBP1 motif at SRC-3 binding sites. The enriched motifs of all three major transcriptional factors (TFs) ATF4, ATF6, and XBP1 for unfolded protein response (UPR) and the calculated Z-scores of all three major SRCs SRC-1, SRC-2, and SRC-3 are shown.
(C) Western blot (WB) verification of the specific binding of endogenous XBP1s (spliced form of XBP1) and ATF4, but not ATF6 (p60), to SRC-3 by anti-SRC-3 IP in the mouse liver. IgG was used as an IP control. The mouse whole-liver nuclei extracts prepared at CT10 were used in the IP and western blot assays. Western blotting was conducted with the antibodies specific to the indicated endogenous proteins.
(D) Transient luciferase assay using WT (XBP1s binding motif CACGTC) and XBP1s binding site mutant (a mutant promoter where CACGTC is converted to CAAAAA) Eif2ak3-dluc vector with increasing doses of SRC-3 expression vector in mouse embryonic fibroblast (MEFs) (n = 4).
(E) Real-time luminescence recording of non-targeting, SRC-3 siRNA- or Xbp1 siRNA-transfected Eif2ak3-dluc MEFs in response to tunicamycin (Tu) treatment. Detrended graphs are provided. For Tu treatment, two independent experiments are shown. Eif2ak3-dluc MEFs were transfected with different siRNAs and then shocked with a Tu treatment and real-time luciferase were recorded by Lumicycle for a total of 60 h.
(F–H) Representative binding profiles (F), Venn diagram (G), and heatmaps (H) of XBP1s and SRC-3 ChIP-seqs at the transcription start sites (TSSs) of 12-h-oscillating genes in the indicated mouse livers at CT24.
(I) ChIP-qPCR of hepatic SRC-3 on selective 12-h cycling gene promoters (Eif2ak3, Xbp1, and Hspa5), 24-h gene Per2 promoter, and noncoding (NC) region every 4 CT hours for 24 h in total (n = 3 mouse livers per time point).
Besides TFs, coactivators such as SRC family members constitute key transcriptional regulators; thus, we hypothesized that SRC-3 coactivation might be involved in regulating 12-h rhythms. This hypothesis is supported by luciferase reporter assays showing that overexpression of SRC-3 significantly increased the promoter activity of Eif2ak3 that harbors a wild-type XBP1s binding motif CACGTC, but not a mutant promoter where CACGTC is converted to CAAAAA (Figures 1D and S1). Furthermore, siRNA-mediated knockdown of SRC-3 significantly impaired tunicamycin-induced 12-h oscillation of Eif2ak3 promoter-driven luciferase activity (Figures 1E and S2), similar to that of XBP1s knockdown, substantiating that SRC-3 coactivation is involved in regulating the 12-h clock. To further test the hypothesis that SRC-3 is a coactivator for XBP1s under a constant-darkness condition, we performed ChIP-seq for XBP1s at CT24 in the mouse liver and compared its cistrome with that of SRC-3 at CT24 (Figures 1F-1H and S3). We identified over 17,432 high-confidence binding sites for XBP1s and 19,832 for SRC-3 (Figures 1F and 1G). Over 67% of the SRC-3 cistrome overlaps with that of XBP1s (Figures 1G and 1H), and this convergence is even more evident at transcription start site (TSS)-proximal regions, with overlapping XBP1s and SRC-3 peaks found in the promoter regions of key 12-h cycling genes, including Hspa5, Eif2ak3, Dnajb9, Ufl1, Insig1, Mydgf, Polr2l, and Xbp1. Furthermore, ChIP-qPCR for SRC-3 at six CT time points in the mouse liver revealed robust 12-h oscillations of SRC-3 chromatin recruitment at XBP1s binding sites, with peak occupancies found at CT10 and CT22 (Figures 1I and S4). Taken together, these data suggest that SRC-3 is a coactivator for XBP1s and for 12-h clock transcription, encouraging us to further define the role of transcriptional coactivation on the hepatic 12-h clock via SRC-3 in vivo.
SRC-3 ablation disrupts the hepatic 12-h clock transcriptome
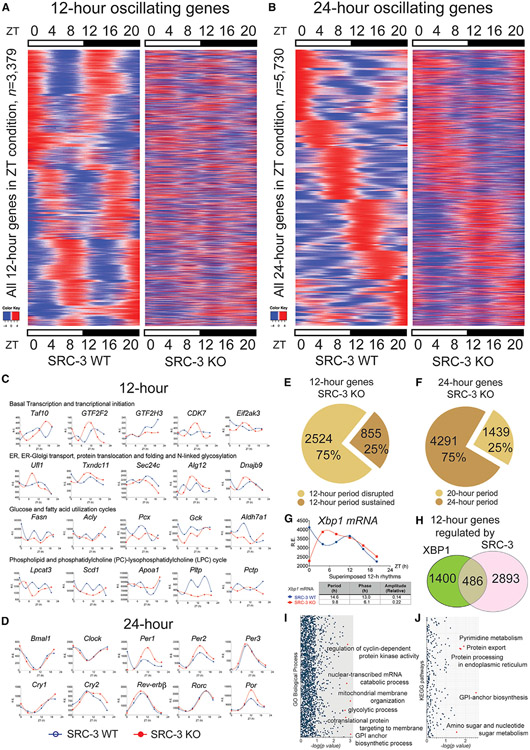
To identify the 12-h cycling transcriptome regulated by SRC-3 without bias, we profiled the temporal characteristics of oscillating transcriptomes from adult male SRC-3 wild-type (WT) and whole-body knockout (KO) littermates at 12–16 weeks of age (Figure 2). We entrained SRC-3 WT and KO mice to a 12-h light:12-h dark cycle (12h:12h LD) under constant temperature for at least 2 weeks and defined this light/dark cycle as our experimental condition in zeitgeber time (ZT). To determine whether SRC-3 ablation perturbs the hepatic 12-h clock transcriptome in vivo, we sampled two biological replicates of mouse liver tissues at a temporal resolution starting at ZT0 and proceeding every 4 h for two complete 12-h cycles (one complete circadian 24-h cycle). We pooled mouse liver RNAs from two biological replicates for each ZT time point and used RNA sequencing (RNA-seq) to profile their liver transcriptomes.
Figure 2. SRC-3 regulates the hepatic 12-h transcriptome. Rhythmic transcripts detected by RNA-seq and the RAIN method were plotted as heatmaps for 12- and 24-h cycling genes.
(A) Heatmaps of the 12-h-oscillating transcripts for the SRC-3 WT and KO mice (n = 3,379).
(B) Heatmaps of the 24-h-oscillating transcripts (n = 5,730).
(C) Representative 12-h gene oscillations plotted against zeitgeber time (ZT) from 4-h-resolution transcriptome profiling of the SRC-3 WT (blue lines) and KO (red lines) mice.
(D) Representative 24-h gene oscillations of core components of the circadian clock plotted against ZT time from 4-h resolution transcriptome profiling of the SRC-3 WT and KO mice. Relative expression (R.E.) is shown as DESeq2 normalized transcript counts.
(E) The proportion of the 12-h gene oscillations that had abolished (n = 2,524, 75%) or sustained (n = 855, 25%) 12-h periods in the SRC-3 KO mice.
(F) The proportion of the 24-h gene oscillations that had a sustained 20-h period (n = 1,439, 25%) or sustained 24-h periods (n = 4,291, 75%) in the SRC-3 KO mice. All the 24-h gene oscillations remain between 20 and 24-h periods in the SRC-3 KO mice.
(G) Xbp1 gene oscillations plotted against ZT from 4-h-resolution transcriptome profiling of the SRC-3 WT (blue lines) and KO (red lines) mice.
(H) Venn diagram of the 12-h transcriptome (n = 1,886) regulated by XBP1 and the 12-h transcriptome (n = 3,379) regulated by SRC-3. Intersection: concordant 12-h-oscillating genes regulated by both XBP1 and SRC-3 (n = 486).
(I and J) Gene enrichment analysis of the 486 concordant 12-h-oscillating genes regulated by both XBP1 and SRC-3 in GO biological processes (I) and KEGG pathways (J). Normalized p values are shown. Red dots indicate the top significant biological processes and KEGG pathways that are regulated by both XBP1 and SRC-3.
To allow unbiased characterization of all oscillations and to avoid the period pre-assignment bias in the established rhythmic algorithms such as JTK_CYCLE and ARSER (Hughes et al., 2009; Meng et al., 2020; Zhu et al., 2017), we used the RAIN method to provide unbiased detection of both 12-h and 24-h oscillations in transcript abundance (Thaben and Westermark, 2014). The key oscillating parameters, including period, phase, and the mean expression value for each gene in SRC-3 WT and KO mouse genotypes, were obtained. We used the computational criteria of 12-h and 24-h oscillations that have been previously determined before (Hughes et al., 2009); the term “12-h oscillating gene” was defined as any gene identified as cycling with a dominant 10- to 14-h period and “24-h oscillating gene” as those with a dominant 20 to 28-h period. In this context, we determined 3,379 12-h oscillating genes and 5,730 24-h oscillating genes in the mouse liver (Figures 2A-2D; Tables S1 and S2). To ascertain whether liver-specific ablation of SRC-3 globally impairs the hepatic 12-h transcriptome, we then examined the rhythmic parameters (e.g., period, phase, and curve shape) of all 3,379 12-h oscillating genes in the SRC-3 WT and KO mouse liver.
We found that 2,524 of 3,379 (75%) 12-h oscillating genes no longer possessed a 12-h periodicity in the SRC-3 KO mice (Figures 2E and 2F) and that the rhythmic profile of 12-h XBP1 transcription was also disrupted (Figure 2G), indicating that SRC-3 globally regulates XBP1 transcription and hepatic 12-h transcriptome in vivo. To make certain of the robustness of these RAIN findings, we performed a Venn analysis to compare the SRC-3 12-h transcriptome with hepatic 12-h genes regulated by XBP1 (Figure 2H). Of the 1,886 dominant 12-h oscillating genes that were identified in our previous study (Meng et al., 2020), 486 (25.8%) of the 12-h genes also were detected in the SRC-3-regulated 12-h transcriptome, again suggesting these overlapping 12-h transcriptomic oscillations are globally regulated by SRC-3 coactivation of XBP1s in vivo. By contrast, core components of the circadian clock remained largely unaffected or even enhanced, as in the case of Per1, Per2, and Por (Figure 2D), with all the hepatic 24-h oscillating genes retaining their 20- to 24 h periodicity (Figure 2F), suggesting that the disrupted 12-h gene oscillations are the consequence of SRC-3 ablation. Additionally, we found an overall reduction in the amplitude of all the 24-h oscillating genes in the SRC-3 KO mouse liver (Figure 2B), suggesting that SRC-3 may also positively regulate the amplitudes of 24-h oscillating genes. Notably, the 12-h gene oscillations regulated by SRC-3 were enriched in GO biological processes and KEGG pathways for protein export, ER protein processing, regulation of cyclin-dependent protein kinase activity and mRNA catabolic process, mitochondrial membrane organization, and several metabolic pathways including lipid, amino sugar, and nucleotide sugar metabolism (Figures 2I and 2J). Collectively, these data suggest that SRC-3 has a role in coordinating the 12-h transcriptional oscillations of ER stress, membrane organization, and metabolic processing to ensure systemic energy homeostasis.
SRC-3 regulates the hepatic 12-h lipidome
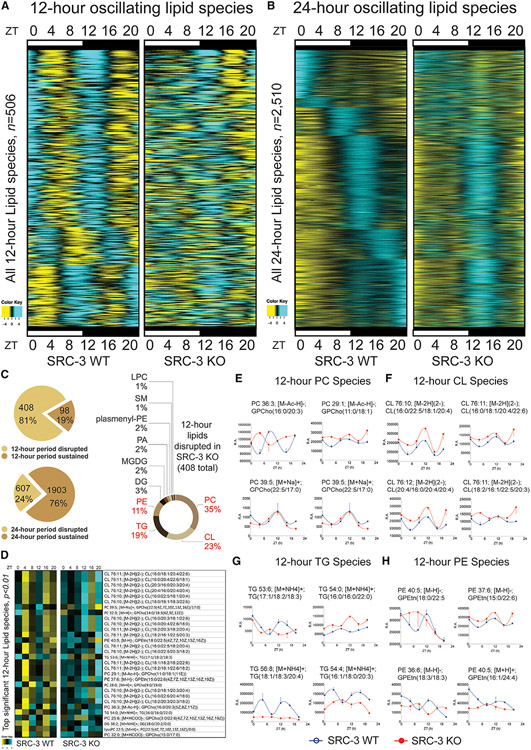
One major mechanism of 12-h clock regulation involves the cycling of genes that have important roles in lipid homeostasis (Meng et al., 2020). To further define the 12-h clock lipidome, we used a temporal-lipidomics-based approach to identify hepatic lipid species in the mouse whole liver (Figure 3). To unbiasedly characterize the 12-h cycling lipidomes regulated by SRC-3, we profiled the temporal characteristics of oscillating lipidomes from adult male SRC-3 WT and KO littermates at 12–16 weeks of age under the same ZT condition (12h:12h LD cycle). We then sampled two biological replicates of mouse liver tissues at a temporal resolution starting at ZT0 and proceeding every 4 h for two complete 12-h cycles to determine whether SRC-3 ablation perturbs the 12-h clock lipidome. Lipid species of the two biological replicates for each ZT time point were profiled using both a positive and a negative ion-switching quantitative method for untargeted lipidomics via high-resolution LC-MS/MS.
Figure 3. SRC-3 regulates the hepatic 12-h lipidome. Rhythmic hepatic lipid species detected by mouse liver lipidomics and the RAIN method were plotted as heatmaps for 12- and 24-h cycling lipids.
(A) Heatmaps of the 12-h-oscillating lipid species for the SRC-3 WT and KO mice (n = 506).
(B) Heatmaps of the 24-h-oscillating lipid species (n = 2,510).
(C) The proportion of the 12-h lipid oscillations that had abolished (n = 408, 81%) or sustained (n = 98,; 19%) 12-h periods in the SRC-3 KO mice (top left) and the proportion of the 24-h lipid oscillations that had abolished (n = 607, 24%) or sustained (n = 1,903, 76%) 24-h periods in the SRC-3 KO mice (bottom left). The hepatic 12-h lipid species that are disrupted in the SRC-3 KO mice and their specific types and proportion are shown on the right.
(D–H) Heatmap of the top significant 12-h lipid species (p < 0.01)that were disrupted in the SRC-3 KO mouse livers. The heatmap from 4-h resolution whole-liver lipidomic profiling (left) and the indicated lipid species (right) are shown.
(E–H) Representative 12-h lipid species phosphatidylcholine (PC; E), cardiolipin (CL; F), triglyceride (TG; G), and phosphatidylethanolamines (PE; H) plotted against ZT from 4-h-resolution hepatic lipidomic profiling of the SRC-3 WT (blue lines) and KO (red lines) mice. Relative abundance (R.A.) of peak spectra is shown.
We next examined the 12-h profiles in the mouse liver lipidome, including triacylglycerides (TAGs), glycerophospholipids (PLs), and sphingolipids (SLs). Of 3,840 lipid species analyzed, 3,822 lipids were detected in mouse liver tissues at all time points. Spike-in control lipid contents, including 15:0-18:1(d7) diglyceride (DAG), 15:0-18:1(d7) phosphatidylcholine (PC), 15:0-18:1(d7) phosphatidylinositol (PI), 15:0-18:1(d7) phosphatidylserine (PS), and 18:1(d7) lysophosphatidylcholine (LPC) were used to normalize the lipidomic raw data to correct for differences in the amount of starting material for each liver extract sample. We also used the RAIN method to provide unbiased detection of both 12-h and 24-h oscillations in lipidomic abundance to allow unbiased characterization of all lipid oscillations. We used our computational criteria of 12-h and 24-h oscillations and determined 506 12-h and 2,510 24-h oscillating lipid species in the mouse liver. To determine whether ablation of SRC-3 globally impairs the hepatic 12-h lipidome, we then examined key oscillating parameters including period, phase, and curve shape of all 506 12-h and 2,510 24-h oscillating lipids in SRC-3 WT and KO mouse livers (Figures 3A and 3B; Table S3).
We found that 408 of 506 (81%) 12-h oscillating lipids no longer possessed a 12-h periodicity in the SRC-3 KO mice (Figure 3C, top left). By contrast, 1,903 of 2,510 (76%) 24-h oscillating lipids had an unaffected circadian periodicity (Figure 3C, bottom left), indicating that SRC-3 globally regulates the hepatic 12-h lipidome in vivo. Four major lipid pathways, including phosphatidylcholine (PC), cardiolipin (CL), triacylglycerol (TG), and phosphatidylethanolamines (PE), showed enrichment and dramatic lipidome-wide defects upon SRC-3 ablation (Figure 3C, right). Indeed, many PC, CL, TG, and PE species were also ranked as top significant lipids in the temporal profile of the hepatic 12-h lipids in the SRC-3 KO mice (Figures 3D-3H), suggesting an association between the coactivation of 12-h clock genes and multiple lipid pathways involving PC-LPC cycle and TG pathways, and an unexpected link to CL and PE lipid species via SRC-3 ablation. It is noteworthy that 24% of the 24-h oscillating lipids also showed disrupted oscillations, consistent with the 24-h transcriptomic changes in the SRC-3 KO mouse liver (Figures 2B, 2D, and 2F), suggesting that SRC-3 may also coordinate both 12-h and 24-h lipid oscillations via coactivation of the 12-h clock and/or an unknown regulatory mechanism via 24-h gene regulation.
SRC-3 regulates hepatic lipidomic cycles, membrane fluidity and mitochondrial function
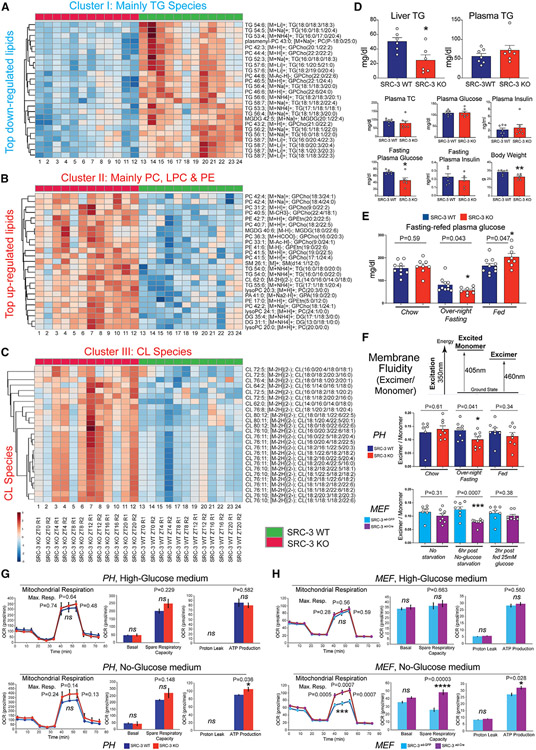
Because TG, PC, LPC, PE, and CL are critical lipid species for membrane lipid and energy homeostasis, which regulate the maintenance of plasma membrane composition and the inner mitochondrial membrane (Ackerman and Simon, 2014; Edidin, 2003; Holthuis and Menon, 2014; Ingolfsson et al., 2014; Sanders and Hutchison, 2018; van Meer et al., 2008), we next investigated whether ablation of SRC-3 leads to alterations in physiological glucose and lipid profiles, membrane fluidity, and mitochondrial function. Metabolic clustering of 12-h rhythmic lipid metabolites revealed three major interactive clusters in TG, PC, LPC, PE, and CL features (Figures 4A-4C and S5), suggesting that SRC-3 coactivation of 12-h gene transcription regulates several distinct but functionally interacting lipid metabolic pathways.
Figure 4. Coactivation of 12-h clock coordinates hepatic lipid metabolic processes.
(A) Heatmap of the cluster I 12-h TG lipid species. The indicated TG lipid species were identified as the top downregulated cluster in the SRC-3 KO mouse livers.
(B) Heatmap of the cluster II 12-h TG lipid species. The indicated PC, LPC, and PE lipid species were identified as the significantly upregulated cluster in the SRC-3 KO mouse livers.
(C) Heatmap of the cluster III 12-h CL lipid species. The indicated CL lipid species were identified as the significant upregulated cluster in the SRC-3 KO mouse livers.
(D) Liver triglycerides (TG), plasma TG, plasma total cholesterol (TC), plasma insulin, plasma glucose, fasting plasma glucose and insulin, and body weight in the indicated mouse strains (n = 6).
(E) Plasma glucose at chow, overnight fasting (16 h) and refed (3 h) in the indicated mouse strains (n = 8).
(F) Membrane fluidity of primary hepatocyte (PHs, n = 6–8 technical replicates from 1 pool of PHs combined from three mouse livers) and MEFs) (n = 6-8 replicates from the MEFs that were independently transfected with Ad-GFP or Ad-Cre for SRC-3 knockout) measured using a fluorescent lipophilic probe, pyrenedecanoic acid (PDA). The absolute excimer: monomer ratio of emission at 460 nm to emission at 405 nm is shown in the indicated SRC-3WT and KO PHs and SRC-3ad–GFP and SRC-3Ade–Cre MEFs.
(G and H) Seahorse metabolic profiling of the indicated SRC-3WT vs. KO PHs (G; n = 4 technical replicates from 1 pool of PHs combined from three mouse livers) and SRC-3ad–GFP vs. SRC-3Ade–Cre MEFs (H; n = 11–12 replicates from the MEFs that were independently transfected with Ad-GFP or Ad-Cre for SRC-3 knockout) under high-glucose medium (top) or no-glucose medium conditions (bottom). Cell Mito Stress test profiles for fundamental mitochondrial functions are shown. OCR, oxygen consumption rate. Unpaired Student’s t test was performed with p value indicated. Data are graphed as mean ± SEM. *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001.
Lipid clusters were further dissected by investigating individual lipid features (Figures 4A-4C). First, the 12-h oscillating TG species were identified as cluster I lipid features (Figure 4A), showing an overall decreased level in the SRC-3 KO mouse liver. Consistently, the SRC-3 KO mice displayed hepatic hypotriglyceridemia in the fed state, without significant functional changes in TG, cholesterol, insulin, and glucose levels in the plasma (Figure 4D, top), suggesting that SRC-3 ablation affects TG homeostasis within the mouse liver. We also observed that the SRC-3 KO mice displayed significant hypoglycemia in the plasma in the overnight (16-h) fasting state, without significant functional changes in plasma insulin levels (Figure 4D, bottom. and 4E), suggesting that SRC-3 ablation also disrupts hepatic gluconeogenesis and the fed-to-fasting transition. The 12-h disruption of TG homeostasis and the fed-to-fasting transition at least partially contributed to the reduced body weight in the SRC-3 KO mice (Figure 4D, bottom). Second, we found an enrichment of PC, LPC, and PE oscillatory lipid subgroups in the cluster II feature profile (Figure 4B), whose lipid metabolites showed upregulated levels upon SRC-3 ablation. Thus, we next investigated whether ablation of SRC-3 leads to alterations in membrane fluidity, using pyrenedecanoic acid (PDA) as a lipid analog probe to examine pyrene fluorescence-mediated fluidity (Lopez et al., 2017; Vanderkooi and Callis, 1974). We observed a significant decrease in membrane fluidity both in SRC-3 KO primary hepatocytes in the overnight fasting state and in the SRC-3ad-Cre mouse embryonic fibroblasts (MEFs) upon 6-h post-glucose deprivation (Figure 4F), suggesting that SRC-3 functionally regulates membrane fluidity-associated processes during the fed-to-fasting transition in a cell-autonomous manner. Third, we found an unexpected CL lipid profile in cluster III features (Figure 4C), displaying increased CL levels in the SRC-3 KO mouse liver. CL is an important component of the inner mitochondrial membrane, where both membrane fluidity and cardiolipin-regulated mitochondrial aggregate structures are essential for the optimal function of rate-limiting metabolic enzymes that are involved in mitochondrial energy metabolism (Claypool and Koehler, 2012). To determine how the perturbation of SRC-3-regulated membrane fluidity and cardiolipin lipids affects cellular metabolic function, we examined mitochondrial functional parameters and observed that ablation of SRC-3 promotes an upregulated respiration and ATP production under the no-glucose starvation condition (Figures 4G and 4H). Collectively, these results revealed an unexpected link among 12-h clock coactivation, TG homeostasis and fed-to-fasting transition, cellular membrane fluidity, and mitochondrial function via SRC-3 regulation.
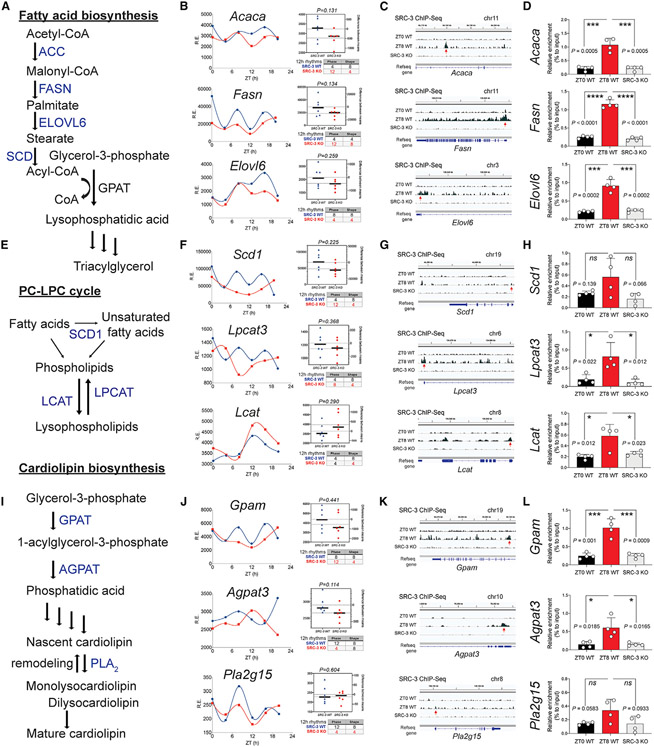
SRC-3 regulates 12-h triglyceride, phospholipid and cardiolipin biosynthesis pathways
To understand the molecular mechanisms underlying the impact of SRC-3 on regulating hepatic lipid metabolism, we next focused on three major pathways fatty acid biosynthesis, PC-LPC cycle and cardiolipin biosynthesis that are involved in 12-h lipidomic cycles, membrane fluidity, and mitochondrial function (Figure 5). We found that the 12-h rhythmicity of key genes that control critical steps of fatty acid biosynthesis (e.g., Acaca, Fasn, and Elovl6; Figures 5A and 5B), PC-LPC cycle (e.g., Scd1, Lpcat3, and Lcat; Figures 5E and 5F) and cardiolipin biosynthesis (e.g., Gpam, Agpat3, and Pla2g15; Figures 5I and 5J) were impaired upon SRC-3 ablation, suggesting that these three major pathways were affected by the absence of SRC-3. Interestingly, the mean average mRNA levels of these genes were not significantly changed in SRC-3 KO mice compared with WT mice, whereas either the phase or the curve shape of their 12-h gene oscillations was disrupted (Figures 5B, 5F, and 5J).
Figure 5. SRC-3 coactivation integrates the liver 12-h clock with lipid pathways.
(A) Schematic diagram of the fatty acid biosynthesis pathway. The key enzymes ACC, FASN, and ELOVL6 are shown.
(B) Twelve-hour gene oscillations of Acaca, Fasn, and Elovl6 plotted against ZT from 4-h-resolution transcriptome profiling of the SRC-3 WT (blue lines) and KO (red lines) mice. The respective gene expression and 12-h rhythmic RAIN parameters of each gene are shown on the right.
(C) Representative binding profiles of SRC-3 ChIP-Seqs of 12-h-oscillating genes Acaca, Fasn, and Elovl6 in SRC-3 WT at ZT0 (top) andZT8 (middle) and in the SRC-3 KO mice (bottom).
(D) ChIP-qPCR binding enrichment of SRC-3 at gene loci of Acaca, Fasn, and Elovl6 promoter regions in SRC-3 WT at ZT0 (left) and ZT8 (middle) and in the SRC-3 KO mice (right) (n = 4 per group).
(E) The schematic diagram of the PC-LPC cycle pathway. The key enzymes SCD1, LPCAT, and LCAT are shown.
(F) 12-h gene oscillations of Scd1, Lpcat3 and Lcat plotted against ZT from 4-h resolution transcriptome profiling of the SRC-3 WT (blue lines) and KO (red lines) mice. The respective gene expression and 12-h rhythmic RAIN parameters of each gene are shown in the right panels.
(G) Representative binding profiles of SRC-3 ChIP-seqs of 12-h-oscillating genes Scd1, Lpcat3, and Lcat in SRC-3 WT at ZT0 (top) and ZT8 (middle) and in the SRC-3 KO mice (bottom).
(H) ChIP-qPCR binding enrichment of SRC-3 at gene loci of Scd1, Lpcat3, and Lcat promoter regions in SRC-3 WT at ZT0 (left) and ZT8 (middle) and in the SRC-3 KO mice (right) (n = 4 per group).
(I) Schematic diagram of the cardiolipin biosynthesis pathway. The key enzymes GPAT, AGPAT, and PLA2 are shown.
(J) Twelve-hour gene oscillations of Gpam, Agpat3, and Pla2g15 plotted against ZT from 4-h-resolution transcriptome profiling of the SRC-3 WT (blue lines) and KO (red lines) mice. The respective gene expression and 12-h rhythmic RAIN parameters of each gene are shown on the right.
(K) Representative binding profiles of SRC-3 ChIP-seqs of 12-h-oscillating genes Gpam, Agpat3, and Pla2g15 in SRC-3 WT at ZT0 (top) and ZT8 (middle) and in the SRC-3 KO mice (bottom).
(L) ChIP-qPCR binding enrichment of SRC-3 at gene loci of Gpam, Agpat3, and Pla2g15 promoter regions in SRC-3 WT at ZT0 (left) and ZT8 (middle) and in the SRC-3 KO mice (right) (n = 4 per group). Unpaired Student’s t test analysis was performed with p value indicated. Data are graphed as mean ± SEM. *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001.
On the basis of the temporal characteristics of the 12-h rhythmicity enriched at dawn (ZT0) and dusk (ZT12), we chose liver samples at ZT0 and ZT8 (4 h before the dusk 12-h rhythmic peak) to interrogate the genomic binding profiles of SRC-3 in vivo. Consistently, temporal SRC-3 occupancy at the promoter regions of these gene loci showed concomitant rhythmic enrichment at ZT8, but not at ZT0, in the mouse liver (Figures 5C, 5G, and 5K), suggesting that hepatic SRC-3 directly regulates the temporal 12-h transcription of these metabolic genes in vivo. ChIP-seq analysis confirmed that hepatic SRC-3 binding sites were enriched at the promoters of their respective 12-h metabolic genes (Figures 5D, 5H, and 5L). This is of importance, since the 12-h temporal regulation of key metabolic genes via SRC-3 coactivation may integrate the liver 12-h clock with lipid metabolism. Collectively, these data suggest that SRC-3 directly regulates rate-limiting metabolic processes in fatty acid biosynthesis, PC-LPC cycle, and cardiolipin biosynthesis, at least partially, through mechanisms associated with temporal transcriptional coactivation of the key metabolic enzymes.
Ablation of SRC-3 abolishes 12-h RER oscillation
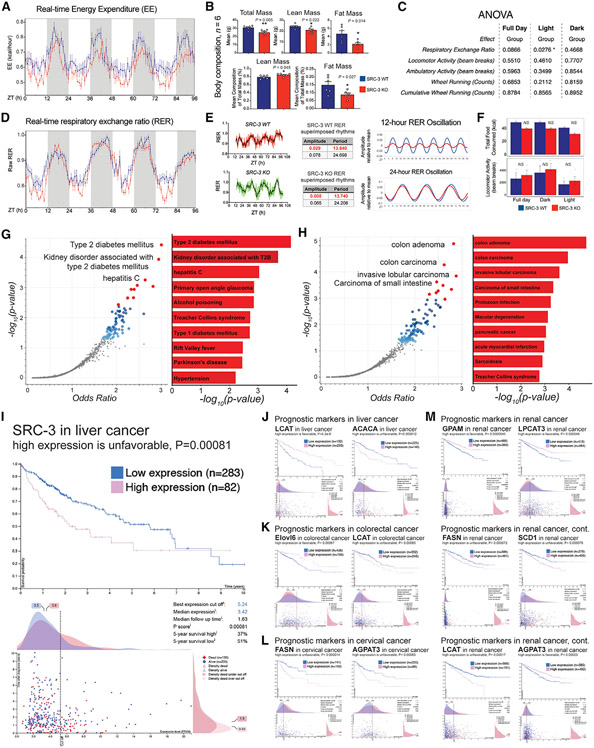
To determine the impact of metabolic dysregulation of coactivation of the 12-h clock on the synchronization of systemic energetics, we performed metabolic profiling of SRC-3 KO mice fed “regular chow” ad libitum. We identified a decrease in the real-time energy expenditure (EE) in the SRC-3 KO mice (Figure 6A), but this phenotype is accompanied by an overall reduced total mass and lean and fat body composition (Figure 6B). Thus, we performed a comprehensive statistical analysis of the EE with systemic normalization of body composition using the analytical tool CalR for interpreting the energetic balance (Mina et al., 2018). We found a statistically significant decrease in the real-time respiratory exchange ratio (RER) in the SRC-3 KO mice, with no noticeable changes in feeding behavior or activity (Figure 6C), whereas other factors, upon the normalization of body mass and lean mass in the SRC-3 KO mice, had no statistical or rhythmic changes, including EE, oxygen consumption, carbon dioxide production, and locomotor and ambulatory activities (Figure S6). Thus, we applied the unbiased eigenvalue/pencil method to the RER metabolic profiles (Antoulas et al., 2018; Zhu et al., 2017), and we observed that the superimposed 12-h rhythmicity was specifically abolished in the SRC-3 KO mice (Figures 6D-6E and S6), whereas the 24-h rhythmicity, food consumption, and locomotor activity remained largely unaffected (Figures 6E and 6F). These data revealed that ablation of SRC-3 disrupts and/or redistributes the systemic balance of whole-body energetics and leads to perturbations in 12-h RER oscillation, suggesting that coactivation of 12-h gene transcription plays an important role in coordinating 12-h oscillations with whole-body energetics.
Figure 6. SRC-3 coactivation of 12-h clock regulates energy expenditure and links to metabolic diseases and cancer.
(A) The relative real-time energy expenditure (EE) values of the indicated mice (n = 4) housed under ad libitum feeding conditions. Averaged real-time EE values of the indicated mouse strains plotted against the indicated time points.
(B) The body composition of the indicated mice (n = 6) housed under ad libitum feeding conditions. Unpaired Student’s t test was performed with p value indicated.
(C) The integrated statistical analysis is shown for the indirect calorimetry experiments of the SRC-3 WT and KO mice (n = 4) housed under ad libitum feeding conditions. One-way ANOVA analysis with Tukey’s post hoc analysis was performed with p value threshold 0.05. The ANOVA was normalized by lean mass at the indicated light, dark, and full-day conditions (n = 4).
(D) The relative real-time respiratory exchange ratio (RER) values of the indicated mice (n = 4) housed under ad libitum feeding conditions. Averaged real-time RER values of the indicated mouse strains plotted against the indicated time points.
(E) Raw, eigenvalue/pencil decompositions, and deconvolutions of the mean RER oscillations of 12-h (top, in red) and 24-h (bottom, in black) oscillations in the SRC-3 WT and KO mice.
(F) Food intake (top) and locomotor activity (bottom) in the SRC-3 WT and KO mice fed regular chow ad libitum (n = 4). Blue line,: SRC-3 WT; red line, SRC-3 KO. Mouse RER, EE, body mass, food intake, and locomotor activity data are graphed as means ± SEM. *p < 0.05, **p < 0.005, ***p < 0.001, ****p < 0.0001.
(G and H) Top enriched disease perturbations identified from the 486 concordant 12-h-oscillating genes regulated by both XBP1 and SRC-3. disease perturbations identified from downregulation (G) and upregulation (H) of the concomitant 486 12-h-oscillating genes are shown. Blue and red dots represent all the significant disease perturbations at discovery p value < 0.05. Top 10 disease perturbations are labeled as red dots.
(I–L) Survival outcome analysis of SRC-3- and SRC-3-regulated 12-h lipid metabolic genes ACACA, FASN, ELOVL6, SCD1, LPCAT3, LCAT, GPAM, and AGPAT3 in the indicated cancer types using TCGA pan-cancer clinical data resource. Clinical survival outcomes, survival scatterplot, cut-off threshold, median expression and follow-up time, p score, and 5-year survival percentage with high- and low-expression patient numbers are shown. Individual genes and their prognostic outcomes are indicated with specific cancer types.
SRC-3 coactivation of lipid metabolic genes is associated with metabolic diseases and cancers
To comprehensively uncover potential human diseases preferentially associated with the SRC-3 coactivation of 12-h genes, we performed gene enrichment analysis of the concomitant 486 12-h genes, which are regulated by both SRC-3 and XBP1 in the mouse liver, against gene expression omnibus (GEO) signatures of differentially expressed genes for diseases (Figures 6G and 6H). We found that the GEO association of 12-h clock coactivated genes links to several metabolic diseases and cancers. Of note, the downregulation of the concomitant 486 12-h oscillating genes was strongly associated with human disease perturbations connected to type 2 diabetes mellitus, kidney disorder associated with type 2 diabetes mellitus, and hepatitis C (Figure 6G). Conversely, upregulation of the 12-h genes that are regulated by 12-h clock coactivation is linked to gene expression profiles for colon adenoma and carcinoma, invasive lobular carcinoma, and carcinoma of the small intestine (Figure 6H). This is of importance, since hepatic SRC-3 coactivation integrates the liver 12-h clock with lipid metabolism and coordinates hepatic lipid metabolic processes, which implies that SRC-3 coactivation of the 12-h clock and its regulated 12-h lipid metabolic genes might be involved in a spectrum of genomic changes, not only in metabolic diseases but also in human cancers that are linked to dysfunctional lipid metabolism and/or aberrant metabolic stresses.
To specifically characterize the correlations of SRC-3 coactivation of 12-h lipid metabolic genes relating to underlying cancer types, we investigated a cohort of 365 patients with liver cancer, 597 patients with colorectal cancer, 291 patients with cervical cancer, 877 patients with renal cancer, and several other cancer types including breast, ovarian, endometrial, lung, pancreatic, prostate, skin, stomach, and head and neck cancers from the Cancer Genome Atlas (TCGA) (https://www.cancer.gov/tcga) (Figures 6I-6M). Notably, high expression of SRC-3 was associated with a worse prognostic outcome in liver cancer (5-year survival high [37%] versus 5-year survival low [51%)], p = 0.00081; Figure 6I). After correction for potentially confounding factors (e.g., expression levels, follow-up time, and 5-year survival rates) that are relevant for prognosis, we found that most of the 12-h lipid metabolic genes that are regulated by SRC-3 coactivation also were potential prognostic markers in liver, colorectal, cervical, and renal cancers, including LCAT and ACACA in liver cancer (Figure 6J), ELOVL6 and LCAT in colorectal cancer (Figure 6K), FASN and AGPAT3 in cervical cancer (Figure 6L), and GPAM, LPCAT3, FASN, SCD1, LCAT, and AGPAT3 in renal cancer (Figure 6M). Of note, high expression of LCAT in liver cancer and AGPAT3 in renal cancer appeared to be favorable for survival, whereas the high expression status of LCAT in colorectal cancer and AGPAT3 in cervical cancer correlates with poor prognosis. Additionally, ELOVL6 in colorectal cancer, GPAM in renal cancer, and LPCAT3 in renal cancer were identified as favorable prognostic markers, while a high expression of ACACA in liver cancer, SCD1 in renal cancer, and FASN in cervical and renal cancers correlated with unfavorable prognostic outcomes. Collectively, these correlative results suggest that 12-h clock coactivation of lipid metabolism could link to metabolic diseases and human cancers via regulation of several rate-limiting metabolic genes in triglyceride, phospholipid, and cardiolipin pathways.
A fasting regimen reprograms cistromic SRC-3 coactivation of the 12-h clock
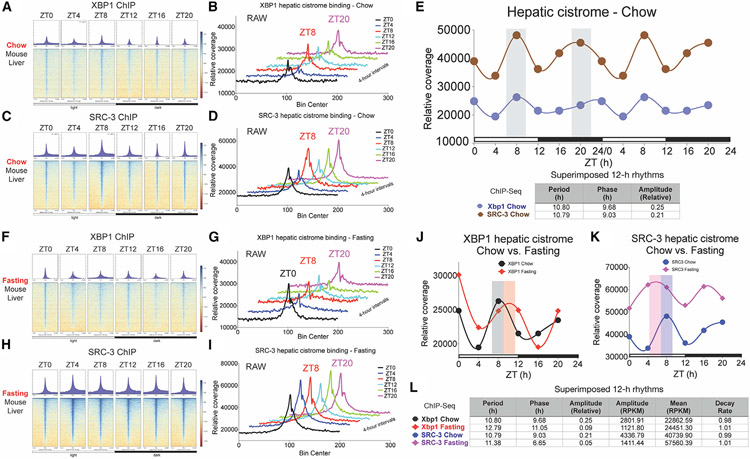
One unexpected observation of 12-h cistromic coactivation involved temporal SRC-3 occupancy enriched at ZT8 at 12h-hour metabolic gene promoters in the mouse liver under the 12h:12h LD constant-temperature condition. This is of importance, since it suggests that 12-h coactivation via SRC-3 is an active cistromic mechanism priming in anticipation of the zeitgeber cue of light. To further define the temporal SRC-3 and XBP1s cistrome in the liver under this ZT condition, we sampled two biological replicates of mouse liver tissues at a temporal resolution starting at ZT0 and proceeding every 4 h for two complete 12-h cycles. We combined the two samples at each time point and used temporal ChIP-seqs to interrogate the in vivo hepatic cistrome at a total of 13,463 concordant binding sites for both SRC-3 and XBP1s under the 12h:12h LD zeitgeber condition (Figures 7A-7E and Table S4). Indeed, the cistromic profiling revealed that hepatic SRC-3 and XBP1s concordantly regulate 12-h oscillating genes, with their cistromic activation priming ahead of the zeitgeber cue of light under constant temperature (Figure 7E).
Figure 7. Maintenance and plasticity of the cistromic 12-h clock coactivation in the mouse liver.
(A–D) Binding profiles and heatmaps of XBP1s (A and B) and SRC-3 (C and D) ChIP-seqs at 13,463 hepatic binding sites under ad libitum chow-feeding condition every 4 ZT h for 24 h in total.
(E) The relative signal of XBP1s and SRC-3 ChIP-seqs plotted as normalized binding profiles every 4 ZT h for 24 h in total (normalized uniquely mapped reads in a 50-bp bin). Gray bar indicates the time of peak in the XBP1- and SRC-3 binding profiles. The 12-h rhythmic eigenvalue parameters period, phase, and relative amplitude of the XBP1/SRC-3 genomic binding profiles are shown.
(F–I) Binding profiles and heatmaps of XBP1s (F and G) and SRC-3 (H &I) ChIP-seqs at 13,463 hepatic binding sites after 16-h overnight fasting regimen every 4 ZT h for 24 h in total.
(J and K) The relative signal of XBP1s (J) and SRC-3 (K) ChIP-seqs in chow versus fasting conditions plotted as normalized binding profiles every 4 ZT h for 24 h in total (normalized uniquely mapped reads in a 50-bp bin). The phases of XBP1 binding in chow condition (gray bar), XBP1 binding in fasting condition (orange bar), SRC-3 binding in chow condition (blue bar), and SRC-3 binding in fasting condition (purple bar) are shown.
(L) 12-h rhythmic eigenvalue parameters period, phase, amplitude (relative), amplitude (RPKM), mean (RPKM), and decay rate of the XBP1/SRC-3 genomic binding profiles in chow and fasting conditions are shown.
Food is one of the major zeitgebers for clock regulation besides light and temperature. Also, fasting has shown beneficial and therapeutic effects against various chronic over-nutritional and pathophysiological states (Wilhelmi de Toledo et al., 2013). To determine the extent to which a fasting regimen affects hepatic SRC-3 coactivation of 12-h clock, we investigated the effect of overnight fasting on SRC-3 coactivation of the XBP1s 12-h cistrome. After 16-h overnight fasting without refeeding, we harvested two biological replicates of mouse liver tissues every 4 h for two complete 12-h cycles and combined the replicates at each time point for temporal ChIP-seqs at 4-h intervals (Figures 7F-7L). We found that the 12-h rhythmicity in genomic binding of SRC-3 and XBP1s was maintained after the overnight fasting regimen, with some cistromic reprogramming occurring in their 12-h genomic occupancy (Figures 7J-7L). Specifically, a comparison in the relative coverage of temporal XBP1s hepatic cistrome under chow versus fasting conditions revealed an increased peak occupancy of XBP1s at the lights-on time and a phase delay at around ZT11 during the continuous-fasting regimen, whereas the 12-h periodicity of XBP1s hepatic occupancy was maintained in both chow and fasting states (Figure 7J and 7L). By contrast, the temporal coactivation of the 12-h clock cistrome under this continuous-fasting regimen showed a dramatically increased SRC-3 occupancy at all time-points, with the phase moving up to ZT6-7, whereas the overall 12-h rhythmicity of SRC-3 coactivation was preserved in both conditions (Figure 7K and 7L).
Collectively, these data suggest that 12-h transcriptional coactivation is not only maintained at SRC-3 and XBP1s cistromic loci, with their cistromic activation priming ahead of the zeitgeber cue of light but is also concomitant with rhythmic remodeling in the absence of food. Accordingly, the maintenance and remodeling feature of the coactivation of the 12-h clock may allow animals to anticipate and prepare for cyclical changes in their environment and their physiological and metabolic stresses.
DISCUSSION
In this study, we set out to define the role of transcriptional coactivation in regulation of 12-h clock and lipid metabolism, driven by the hypothesis that SRC-3 coactivation of XBP1 and the 12-h clock modulates hepatic 12-h transcriptional cycles and metabolic oscillations for systemic energy homeostasis. Our findings of SRC-3 integration of the mammalian 12-h clock, lipid homeostasis and energy metabolism via coactivation of the 12-h clock is of importance, since our human ancestors took advantage of favored lipid metabolism and fat storage for survival in ancient periods when food was scarce. Some lipid metabolic features may have provided unique energetic advantages that supported ancient migrations and varying exposure to environmental factors as modern humans migrated out of Africa. Although positive selection for lipid genes may have helped human ancestors survive during migration and lessened predatory selection pressures during evolution (Lingle et al., 2008; Neel, 1962; Prentice et al., 2005; Quintana-Murci et al., 1999; Sellayah et al., 2014; Speakman, 2007, 2008), these lipid-favored metabolic features may now adversely predispose humans to metabolic diseases and human cancers in industrialized societies. Accordingly, 12-h clock coactivation of lipid metabolism may have implications in identification of therapeutic targets and interventional strategies for metabolic diseases (e.g., obesity, type 2 diabetes mellitus, non-alcoholic fatty liver disease, stroke, and heart disease) and some human cancers.
While the 12-h clock and 24-h circadian clock have independent machinery and transcriptional factors (e.g., XBP1 for 12-h clock and Bmal1 for 24-h clock as the core TFs), SRC-3 coactivation of the 12-h clock appears also to affect 24-h gene and lipid oscillations. This is an interesting observation that requires future investigation because it suggests that the two independent clocks may have intimate links via common regulatory mechanisms such as transcriptional coactivation and/or repression. First, the 12-h clock and 24-h circadian clock may share some repressive machinery via circadian feedback loops, such as Per1/Per2 from this study and/or Cry1/Cry2 (Cretenet et al., 2010). Because the canonical clock machinery depends on repressive loops to maintain the circadian rhythmicity, it will be important to further characterize the unknown repressive components of the 12-h clock in future studies. Second, transcriptional coactivation may indirectly affect the BMAL1:CLOCK feedforward loop via transcriptionalcoregulation of the hepatic circadian cistrome. Thus, it is possible that other transcriptional coactivators and/or corepressors may also be involved in the feedforward loop of both the 12- and 24-h circadian clocks. Third, coactivation of the 12-h clock and lipid metabolism may affect systemic cycling machinery in the central nervous system (e.g., hypothalamus, hippocampus, and/or frontal cortex) and other peripheral organs (e.g., kidney, lung, brown fat, heart, adrenal gland, white fat, and skeletal muscle). Although human studies have highlighted many ultradian, circadian, and infradian oscillations in daily food intake, immune function, body temperature, cognitive performance, and circulating hormones, the transcriptional regulation of these cycling machineries is largely unknown and thus requires future investigations.
Our present study also reveals that cistromic binding of hepatic SRC-3 and XBP1 primes in anticipation of the zeitgeber cue of light, suggesting coactivation of the 12-h clock as an active transcriptional mechanism to anticipate and prepare for physiological and metabolic stresses. Our studies have uncovered a role for mammalian 12-h clock coactivation in lipid metabolic homeostasis and allow for potential therapeutic considerations. In summary, our findings reveal a coactivator component of the 12-h clock that coordinates the 12-h clock with diverse lipid and metabolic pathways. We show that the transcriptional coactivator SRC-3 regulates XBP1 transcription and 12-h gene oscillations and lipid metabolic cycles, highlighting the role of 12-h clock coactivation in integrating ER and metabolic stress and bioenergetics to ensure systemic energy homeostasis. SRC-3 ablation perturbs hepatic 12-h lipidomic cycles, membrane fluidity, and mitochondrial function via distinct triglyceride, phospholipid, and cardiolipin biosynthesis pathways. Clinical evidence further shows that SRC-3 coactivation of 12-h metabolic gene expression is associated with many prevailing metabolic diseases and cancers in human patients.
Limitations of the study
This study was performed using only male adult animals. Only one pool of two biological replicates was sequenced per time point in the temporal RNA-seq and ChIP-seq experiments. SRC-3 also may have non-rhythmic functions that lie outside the scope of our clock studies.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bert W. O’Malley (berto@bcm.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The RNA-Seq and ChIP-Seq data have been deposited to the Gene Expression Omnibus (GEO), the lipid metabolomics data have been deposited to the NIH Common Fund’s National Metabolomics Data Repository (NMDR), the proteomics data have been deposited to the ProteomeXchange consortium member PeptideAtlas, and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-SRC-3 | Cell Signaling Technology | Cat# 2126, RRID: AB_823642 |
| Rabbit polyclonal anti-XBP1s | Biolegend | Cat# 619501, RRID: AB_315907 |
| Rabbit monoclonal anti-ATF4 | Cell Signaling Technology | Cat# 11815S, RRID: AB_2616025 |
| Rabbit polyclonal anti-ATF6-α | Santa Cruz Biotechnology | Cat# sc-22799, RRID: AB_2242950 |
| Rabbit polyclonal anti-XBP1 | Santa Cruz Biotechnology | Cat# sc-7908, RRID: AB_2107620 |
| Rabbit polyclonal anti-BMAL1 | Abcam | Cat# ab3350, RRID: AB_303729 |
| Rabbit anti-beta-Actin Monoclonal | Cell Signaling Technology | Cat# 5125, RRID: AB_1903890 |
| Antibody, HRP Conjugated, Clone 13E5 | ||
| Bacterial and virus strains | ||
| Eif2ak3 promoter-pGreenfire-mCMV-dscGFP-EF1-Puro lentivirus | (Zhu et al., 2017) | N/A |
| XBP1s-pHAGE | A gift from Dr. Xi Chen at BCM | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Tunicamycin | EMD Millipore | Cat# 654380 |
| Eif2ak3 promoter-pGreenfire-mCMV-dscGFP-EF1-Puro lentivirus | (Zhu et al., 2017) | N/A |
| Hygromycin B | Thermo Fisher Scientific | Cat# 10687010 |
| Lipofectamine® 2000 Transfection Reagent | Thermo Fisher Scientific | Cat# 11668027 |
| Lipofectamine® RNAiMAX Transfection Reagent | Thermo Fisher Scientific | Cat# 13778030 |
| HEPES | Thermo Fisher Scientific | Cat# 15630080 |
| SuperScript® VILO™ Master Mix | Thermo Fisher Scientific | Cat# 11755050 |
| Micrococcal Nuclease | New England Biolabs | Cat# M0247S |
| Sodium Pyruvate | Thermo Fisher Scientific | Cat# 11360070 |
| PEG-it Virus Precipitation Solution | System Biosciences | Cat# LV810A-1 |
| RNase A, DNase and protease-free | Thermo Fisher Scientific | Cat# EN0531 |
| TaqMan® Universal Master Mix II, no UNG | Thermo Fisher Scientific | Cat# 4440047 |
| Power SYBR Green Master Mix | Thermo Fisher Scientific | Cat# 4367660 |
| Dialyzed Fetal Bovine Serum | Thermo Fisher Scientific | Cat# 26400036 |
| Fetal Bovine Serum | Thermo Fisher Scientific | Cat# 10437028 |
| D-glucose | Sigma | Cat# G8270 |
| Sodium Acetate | Sigma | Cat# S2889 |
| 2-Mercaptoethanol | Sigma | Cat# M3148 |
| DMSO | Sigma | Cat# D8418 |
| NP-40 | Sigma | Cat# N3516 |
| Sodium chloride | Sigma | Cat# S7653 |
| Tris/glycine/SDS buffer | Biorad | Cat# 161-0772 |
| TBST | Cell Signaling Technology | Cat# 9997 |
| 10% SDS | Gibco | Cat# 15553-035 |
| Triton X-100 | Promega | Cat# H5142 |
| Tween 20 | Fisher Scientific | Cat# BP337-100 |
| BSA | Sigma | Cat# A9647 |
| EDTA | Cell Signaling Technology | Cat# 7011S |
| Nonfat dry milk | Biorad | Cat# 170-6404 |
| BCA Protein Assay Reagent A | Thermo Fisher Scientific | Cat# 23223 |
| BCA Protein Assay Reagent B | Thermo Fisher Scientific | Cat# 23224 |
| iBlot Gel transfer stacks Nitrocellulose | Life Technologies | Cat# IB301001 |
| Sodium deoxycholate | Sigma | Cat# D6750 |
| Mouse Genomic DNA | Promega | Cat# G3091 |
| Ponceau S solution | Sigma | Cat# P7170 |
| VivoGlo™ Luciferin, In Vivo Grade | Promega | Cat# P1041 |
| Restore western blot stripping buffer | Thermo Fisher Scientific | Cat# 21063 |
| Penicillin Streptomycin | Thermo Fisher Scientific | Cat# 15070063 |
| Pellet Paint® Co-Precipitant | EMD Millipore | Cat# 69049 |
| Q5® High-Fidelity DNA Polymerase | New England Biolabs | Cat# M0491S |
| Criterion pre-cast gradient SDS-PAGE gel (4-20%) | Bio-Rad | Cat# 5671094 |
| TaKaRa Ex Taq® DNA Polymerase | Clontech | Cat# RR001A |
| T4 DNA Ligase | New England Biolabs | Cat# M0202 |
| Antarctic Phosphatase | New England Biolabs | Cat# M0289S |
| Ethanol 200 proof | Decon Labs | Cat# 2701 |
| Isopropanol | Sigma | Cat# I9516 |
| cOmplete™ Protease Inhibitor Cocktail | Roche | Cat# 11697498001 |
| PhosSTOP™ | Roche | Cat# 4906845001 |
| DMEM, high glucose | Thermo Fisher Scientific | Cat# 11965092 |
| DMEM, no glucose | Thermo Fisher Scientific | Cat# 11966025 |
| Proteinase K | Thermo Fisher Scientific | Cat# AM2544 |
| 0.45μM syringe filter | Thermo Fisher Scientific | Cat# 09-740-106 |
| Critical commercial assays | ||
| SimpleChIP® Plus Enzymatic Chromatin IP Kit | Cell Signaling Technology | Cat# 9005S |
| PureLink RNA mini kit | Life Technologies | Cat# 12183025 |
| Dual-luciferase Reporter assay system | Promega | Cat# E1960 |
| Insulin ELISA kit | EMD Millipore | Cat# EZRMI-13K |
| One touch ultra glucose meter and strips | LifeScan, Inc | N/A |
| Deposited data | ||
| GSE175598 | GEO public repository | GSE175598 |
| Study ST002079/PR001319 | NMDR public repository | datatrack_id:3065 study_id:ST002079; http://dev.metabolomicsworkbench.org:22222/data/DRCCMetadata.php?Mode=Study&StudyID=ST002079&Access=DoqF5000 |
| PASS01737 | PeptideAtlas (ProteomeXchange consortium member) | PASS01737; http://www.peptideatlas.org/PASS/PASS01737 |
| Experimental Models: Cell Lines | ||
| Immortalized mouse embryonic fibroblasts | (Zhu et al., 2017) | N/A |
| Eif2ak3-dluc or Eif2ak3-dluc with mutated XBP1s binding site stably expressing MEFs | (Zhu et al., 2017) | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J male mice | The Jackson Laboratory | JAX:000664 |
| Mouse: SRC-3 knockout male mice | (Xu et al., 2000) | N/A |
| Mouse: Xbp1 flx/flx male mice | A gift from Dr. Xi Chen at BCM (Lee et al., 2008) | N/A |
| Mouse: Xbp1 flx/flx;AlbCre male mice | (Meng et al., 2020) | N/A |
| Oligonucleotides | ||
| siRNA oligonucleotides | Dharmacon | Dharmacon, D-001206-13-05; Dharmacon, L-040825-00-0005 |
| See Table S5 for qRT-PCR and ChIP-qPCR Primer Sequences | N/A | N/A |
| Recombinant DNA | ||
| Eif2ak3-dluc | (Zhu et al., 2017) | N/A |
| Eif2ak3-dluc mutant | (Zhu et al., 2017) | N/A |
| Software and algorithms | ||
| RAIN | (Thaben and Westermark, 2014) | http://www.bioconductor.org/packages/release/bioc/html/rain.html |
| Eigenvalue/pencil | (Antoulas et al., 2018; Zhu et al., 2017) | N/A |
| Bioconductor 3.4 | R | https://www.bioconductor.org/news/bioc_3_4_release/ |
| Matlab R2019a | MathWorks | https://www.mathworks.com/products/new_products/latest_features.html |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| LumiCycle Analysis 2.54 | ActiMetrics | http://actimetrics.com/downloads/lumicycle/ |
| UCSC genome browser | UCSC | https://genome.ucsc.edu/cgi-bin/hgGateway |
| DAVID Bioinformatics Resources 6.8 | NIH | https://david.ncifcrf.gov/ |
| CLAMS data examination tool | Columbus Instruments | http://www.colinst.com/products/clams-hc-comprehensive-lab-animal-monitoring-system-for-home-cages#CLAX |
| VirtualDubMod 1.5.10 | Intel Software | https://sourceforge.net/projects/virtualdubmod/files/ |
| Venn Diagram Plotter v1.5.5228.29250 | Department of Energy and PNNL | https://omics.pnl.gov/software/venn-diagram-plotter |
| Gene Cluster 3.0 | Originally writtern by Michael Eisen at Stanford University | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm#ctv |
| Java TreeView 3.0 alpha03 | Open Source software | https://bitbucket.org/TreeView3Dev/treeview3/ |
| Other | ||
| NCBI Aceview (AceView offers a comprehensive annotation of human, mouse and nematode genes) | NIH | https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and housing conditions
All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine (BCM). All animal studies were conducted in compliance with ARRIVE guidelines. SRC-3 knockout mice were generated in house and have been previously described (Xu et al., 2000). Xbp1flx/flx mice were a gift from Dr. Xi Chen at Baylor College of Medicine (Lee et al., 2008). Xbp1flx/flx controls and Xbp1flx/flx; AlbCre transgenic mice were generated in house and have been previously described (Meng et al., 2020). Male mice at the age of 3-8 months were used for experiments unless indicated otherwise in figure legends or the main text. All offspring from this cross were maintained on a C57BL/6 background. Mice were maintained on a 12h:12h light:dark cycle and allowed free access to regular chow and water under strict temperature control. This study was conducted in male mice. All mice were bred in our animal facility and we have complied with all relevant ethical regulations at the Baylor College of Medicine.
Cell culture
Mouse embryonic fibroblasts (MEFs) isolated from the indicated mice were immortalized using SV40 Large T antigen transfection (Zhu et al., 2017). For Ade-Cre treatment, the indicated MEF strain was infected with adenovirus harboring an expression cassette for CRE recombinase (Cre-GFP as control). Each MEF replicate in the experiments was transfected with Ade-Cre or Ade-GFP independently. MEFs were cultured in DMEM (4.5g/L glucose) supplemented with 10% FBS at 37°C with 5% CO2. Primary hepatocytes isolated from the indicated littermate mice (n = 3) were used and plated on 96-wells (Stashi et al., 2014). Primary hepatocytes were cultured overnight in Williams E media (Invitrogen) containing 10% FBS before membrane fluidity or Seahorse assays.
METHOD DETAILS
Circadian time (CT) animal preparation and organ collection
Male mice between 12 and 16 weeks of age were entrained to a 12h:12h light:dark schedule for at least 2 weeks (the subjective day was beginning at 06:00 with subjective night beginning at 18:00, respectively). The entrained mice were then released into constant darkness for 24 hours. After the first 24 hours continuous darkness, mice were euthanized at selected CT times in constant darkness. Mouse liver tissues were collected and snap-frozen in liquid nitrogen at the indicated time points. Food and water were supplied ad libitum at all stages prior to euthanization.
Zeitgeber time (ZT) animal experiments and organ collection
For experimental chronology measured in ZT, the subjective day was in a 12h:12h light:dark schedule, beginning at 06:00 (ZT0) with subjective night beginning at 18:00 (ZT12), respectively. Mice were euthanized at selected ZT times in a 12h:12h light:dark schedule. Mouse liver tissues were collected and snap-frozen in liquid nitrogen from zeitgeber time 0 (ZT0) every 4 hours for 24 hours. Food and water were supplied ad libitum at all stages prior to euthanization unless otherwise indicated under fasting regimen condition.
RNA-seq data
RNA samples were isolated from two biological replicates of mouse liver tissues from ZT0 every 4 hours for 24 hours or at the indicated CT time points with the miRNeasy Mini Kit (Qiagen) per the manufacturer’s instructions. For the time course transcriptomics, the two biological replicates of RNA samples at each ZT time point (250ng of total RNA for each sample) were pooled and converted into strand-specific total mRNA-Seq libraries with the TruSeq Stranded Total RNA kit (Illumina) per the manufacturer’s protocol. The size selected libraries were analyzed for quality control with a high Sensitivity DNA chip on a Bioanalyzer 2100 (Agilent). The pooled libraries were sequenced with the HiSeq 3000 (Illumina) using 75-bp paired-end for an average of 40 million paired-end reads per sample at the MD Anderson Cancer Center Science Park NGS Core.
RNA-seq analysis and oscillation detection
Fastq files containing raw RNA-Seq reads were aligned to the mouse genome (mm10/NCBI38) using HISAT2 (2.1.0). Paired-end reads below MAPQ quality score 15 were removed. RNA-Seq quantification was performed with featureCounts (1.6.4, default parameters). Protein-coding genes were quantified by iGenome annotation (archive-2015-07-17-14-33-26). Quantification values were normalized with DESeq2 (2.11.40.2), followed by filtering out of zero expression genes in each replicate or time point. The values of normalized RNA-Seq data at each time point were used to determine the 12- and 24-hour oscillations via an independent rhythmicity analysis incorporating non-parametric method (RAIN) (Thaben and Westermark, 2014). The established rhythmic algorithms such as JTK_CYCLE and ARSER have a period pre-assignment bias, and thus are not capable of characterizing all superimposed oscillations (Hughes et al., 2009; Zhu et al., 2017). Therefore, we chose to use the RAIN method to provide unbiased detection of both 12-hour and 24-hour oscillations in transcript abundance. Briefly, RAIN as a non-parametric method for the detection of rhythms of prespecified periods and of arbitrary wave forms has been established for robustly identifying both circadian and ultradian rhythms (Thaben and Westermark, 2014, 2016). We used the same period criterion established by Hughes et al. (Hughes et al., 2009) for circadian genes and 12-hour genes for consistency and for better comparison and understanding of the prevalence of 12-hour genes: circadian genes (period between 20-hour to 28-hour), 12-hour genes (period between 10-hour to 14-hour). Because data from only one pooled RNA-Seq at each time point were used to determine the 12- and 24-hour oscillations, we performed a Venn analysis to compare the 12-hour transcriptome from this study with the superimposed 12-hour gene oscillations that were determined using the eigenvalue/pencil method (Matlab_R2019A) in our previous study to ensure the robustness (Antoulas et al., 2018; Meng et al., 2020; Zhu et al., 2017).
ChIP-seq data
Livers from the indicated mice were harvested at the designated time points. Formaldehyde treated chromatins from two biological replicates at each time point were combined for subsequent ChIP-Seq analysis. Mouse liver chromatin was prepared with the ChIP-IT Kit (Active Motif) per the manufacturer’s instruction with the following modifications. Briefly, mouse liver samples were dissected into small pieces in PBS containing 1% formaldehyde. The liver samples were then incubated for 15 min at room temperature, followed by the addition of 0.125 M glycine to stop the reaction. The formaldehyde-fixed samples were washed in PBS, followed by homogenization in lysis buffer. The released nuclei were pelleted and resuspended in digestion buffer for 5 minutes at 37°C. An appropriate amount of Micrococcal nuclease (MNase) (NEB) was added to enzymatically digest the chromatin. The optimal amount of incubation time was pre-determined to shear the chromatin into a 150-1000bp banded pattern. The reaction was stopped by 0.5 M EDTA, followed by chilling on ice for 10 minutes. Equal amount of the chromatins from two biological replicates at each time point were pooled prior to the following immunoprecipitation steps. Following chromatin quantification, aliquots of 100μg of mouse liver chromatin and 10μg of anti-SRC-3 antibody (Cell Signaling 5E11) or anti-XBP1s antibody (Biolegend Poly6195) were used for the two ZT0 and ZT8 time points, with appropriate input genomic DNA saved for the comprehensive analysis. For the set of SRC-3 and XBP1s ChIP-Seq from ZT0 every 4 hours for two complete 12-hour cycles, an aliquot of 50μg chromatin and 5μg anti-SRC-3 or 5μg anti-XBP1s antibody was used. The co-immunoprecipitated complexes were then washed and eluted with elution buffer. The genomic DNA input and ChIP DNA fragments were reverse crosslinked at 65°C overnight. The samples were treated with RNaseA and proteinase K, and then purified by phenol-chloroform extraction and ethanol precipitation. The comprehensive ZT0 and ZT8 DNA libraries were prepared at the MD Anderson Cancer Center, which generated ~40 million 75 bp paired-end reads per sample. The ZT0 to ZT24 time point DNA libraries were prepared at the MD Anderson Cancer Center Science Park NGS Core and generated ~30 million 75 bp single-end reads per sample.
ChIP-seq analysis
ChIP-Seq sequencing reads were mapped to the mouse genome (mm10/NCBI38) with BWA (0.7.17.4). The mapping allowed a maximum of one mismatch and removed duplicated reads to ensure only “unique” reads were allowed for output to the bam file. Unique reads with mapping scores higher than 15 were kept for subsequent ChIP-Seq analysis. MACS2 version 2.1.1.20160309 was used to perform the peak calling at ZT0 and ZT8 against the SRC-3 whole-body knockout ChIP controls. For the ZT0 to ZT24 4-hour interval ChIP-Seqs, heatmap and enrichment plotting were carried out against the ChIP-Seq of the SRC-3 whole-body knockout ChIP control. In all the sequencing analysis, the differential sequencing reads were “down-sampled” to the lowest number of uniquely mapped reads for the normalization of sequencing depth. The ChIP-Seq peak annotations and motif analysis were determined with Discriminative DNA Motif Discovery (DREAM) tool (version 4.9.1) or the SeqPos motif tool (version 0.590) using mouse reference genome mm10 as background.
Co-immunoprecipitation (Co-IP)
Nuclear extracts (NE) were made from mouse liver according to our published protocol (Lanz et al., 2010). Protein concentrations were determined by Bradford assays (Bio-Rad). Aliquots were snap-frozen in liquid nitrogen and stored at −80°C until usage. 200 μg of NE was used for per IP followed by Immunoblotting. 5 μg of anti-SRC-3 (Rabbit mAb #2126, 5E11, Cell signaling) or control IgG were coupled to 1.5 mg of magnetic Dynabeads (Life Technologies) for every IP using Dynabeads Antibody Coupling Kit (Life Technologies) per manufactures’ protocol. For purification and identification of targeted XBP1s protein complex, IP was carried out from 20 mg of total liver protein extracts and 20 μg of anti-XBP1s antibody (Biolegend Poly6195). Co-IP was essentially carried out as previously described (Stashi et al., 2014), except that coupled antibody Dynabeads was added to the NEs for incubation.
Protein identification by mass spectrometry
In-gel digested proteins are quantitatively analyzed by LC-MS/MS system using a nanoLC-1200 coupled to a Orbitrap Fusion Lumos ETD Tribrid (Thermo Fisher Scientific, San Jose, CA) mass spectrometer. The Coomassie blue stained proteins in SDS-PAGE gels in the indicated gel lanes were sliced and in-gel digested overnight at 37°C with trypsin. After digestion, peptides were extracted twice in 200 μL of acetonitrile with re-suspension in 20 μL of 2% formic acid prior to second extraction, dried in a Savant SpeedVac, and dissolved in a 5% methanol/0.1 % formic acid solution. The dissolved samples were loaded on a 2 cm C18 trap followed by 1 hour 0-30% acetonitrile gradients on a 10 cm C18 column (packed in-house with Reprosil-Pur Basic C18 3 μm beads; Dr. Maisch GmbH, Germany) and measured online with the Thermo Orbitrap Velos or QExactive instruments (Thermo Scientific, Germany). The MS data acquisition was operated in the top 30 peaks data-dependent mode under the direct control of Xcalibur software ver. 4.0 (Thermo Scientific). The raw files from the mass spectrometer were searched against the Mouse NCBI protein RefSeq database using Proteome Discoverer 2.1 interface (Thermo Fisher) with Mascot algorithm 2.4 (Matrix Science). Dynamic modification of oxidation, protein N-terminal acetylation, deamidation on asparagine and glutamine were allowed. The precursor mass tolerance was confined within 20 ppm with fragment mass tolerance of 0.5dalton and a maximum of two missed cleavage was allowed. The data was further grouped into gene products that were assigned homology and identification quality groups using an in-house developed algorithm. iBAQ quantification was performed after grouping peptides into gene protein products level using an in-house pipeline, gpGrouper (Saltzman et al., 2018). All protein gene products that were chosen for follow-up in this study were required to have at least one identification where a spectral match passing <1% FDR and >20 ion score or <5% FDR and >30 ion score thresholds was present. The process of identifying co-regulator proteins is through knowledge- and experience-based data analysis algorithms that excludes non-specific binders and generates a list of candidate proteins interactions.
Gene ontology and disease perturbation analysis
DAVID (https://david.ncifcrf.gov) was used to perform Gene Ontology analysis. Biological process relationships and disease perturbations, KEGG_PATHWAY analysis was performed as the primary pathway category using Mus musculus background.
Metabolic profiling using CLAMS
Real-time measurement of respiratory exchange ratio (RER) and food intake were monitored using Comprehensive Lab Animal Monitoring System Calorimetry (CLAMS, Columbus Instruments). Mice fed ad libitum were acclimated to the chambers for one week prior to data collection. Food intake was monitored for five consecutive days under a 12h light:12h dark cycle. CalR (version 1.2, https://calrapp.org) was used for statistical analysis of CLAMS experiments, with indirect calorimetry measured for physiological energy balance (Mina et al., 2018). Data for VO2 consumed, VCO2 released, and RER (calculated as VCO2/VO2) were monitored.
Body composition analysis (via MRI)
For measurement of whole-body fat mass, lean tissue mass, free fluid, and total body water, the nuclear magnetic resonance system EchoMRI-100™ (Echo Medical System, Houston, TX) was used without the need for anesthesia per the manufacturer’s instruction.
Locomotor activity monitoring
Home Cage Activity System (Omnitech Electronics, Inc) in a home-cage environment was used for real-time measurement of spontaneous locomotor activity. Mice fed ad libitum were acclimated to the home cage for at least one week prior to data collection. Spontaneous locomotor activity was measured under a 12h light:12h dark condition.
Immunoblotting
Immunoblotting analyses were performed using the total liver extracts and SDS/PAGE. Briefly, proteins separated by 4-20% gradient SDS/PAGE were used. The gels were transferred to nitrocellulose membranes, then blocked in tris-buffered saline with Tween-20 supplemented with 5% (wt/vol) bovine serum albumin (BSA). The membranes were incubated with anti-XBP1s antibody (Biolegend Poly6195, 0.5 μg per mL) rotating at 4°C overnight. Blots were incubated with an appropriate secondary antibody coupled to horseradish peroxidase (Digital anti-Rabbit-HRP, R1006, 1:1000, Kindle Biosciences), reacted with ECL reagents per the manufacturer’s instructions (Thermo), detected by autoradiography or with a KwikQuant Imager (Kindle Biosciences).
Plasma profiling
Mouse plasma glucose levels were measured with a handheld glucometer. Mouse plasma insulin TG, TC parameters were measured by the Metabolism Core Services at Baylor College of Medicine.
Lipidomics profiling
Mouse liver lipids were extracted using a modified Bligh-Dyer method (Bligh and Dyer, 1959; Vantaku et al., 2019). Briefly, 50 mg of crushed issue sample from mouse whole liver was used. A 2:2:2 volume ratio of water/methanol/dichloromethane was used for lipid extract at room temperature after spiking internal standards 17:0 LPC, 17:0PC, 17:0 PE, 17:0 PG, 17:0 ceramide, 17:0 SM, 17:0PS, 17:0PA, 17:0 TAG, 17:0MAG, DAG 16:0/18:1, CE 17:0. The organic layer was collected followed by a complete drying procedure under nitrogen. Before MS analysis, the dried extract was resuspended in 100 μL of Buffer B (10:5:85 Acetonitrile/water/Isopropyl alcohol) containing 10mM NH4OAc and subjected to LC/MS. The lipidome was separated using reverse-phase chromatography. High-performance Liquid Chromatography (LC) grade water, methanol, acetonitrile, dichloromethane, isopropanol from Fisher scientific were used per the manufacturer’s instructions. Mass spectrometry (MS) grade lipid standards from Avanti Polar Lipids (Alabaster, AL) and MS grade ammonium acetate from sigma Aldrich (St. Louis, MO) were used. For internal standards and quality controls, the lipid stock solution was prepared by weighing an exact amount of the lipid internal standards in Chloroform/Methanol/H2O, resulting in sample aliquots at concentration of 1mg/mL as stock solutions. The stock solutions were diluted to 100 pmol/μL by mixing an appropriate volume of the internal standards LPC 17:0/0:0, PG 17:0/17:0, PE 17:0/17:0, PC 17:0/17:0, TAG 17:0/17:0/17:0, SM 18:1/17:0, MAG 17:0, DAG 16:0/18:1, CE 17:0, ceramide d 18:1/17:0, PA 17:0, PI 17:0/20:4, and PS 17:0/17:0. To monitor instrument performance, 10 μL of a dried matrix-free mixture of the internal standards was used, reconstituted in 100 μL of buffer B (5% water, 85%Isopropanolol: 10%Acetonitrile in 10mM NH4OAc). To monitor the lipid extraction process, a standard pool representing the tissue sample aliquots was used.
Data acquisition through LC/MS analysis
Shimadzu CTO-20A Nexera X2 UHPLC systems was used for data acquisition, equipped with a degasser, binary pump, thermostat-regulated auto sampler, and a column oven for chromatographic separation. For lipid separation, 5 uL of the lipid extract was injected to a 1.8 μm particle 50 × 2.1 mm Acquity HSS UPLC T3 column (Waters, Milford, MA). The column heater temperature was set at 55°C. For chromatographic elution, a linear gradient was used over a 20 min total run time, with 60% Solvent A (acetonitrile/water (40:60, v/v) with 10 mM ammonium acetate) and 40% Solvent B (acetonitrile/water/isopropanol (10:5:85 v/v) with 10 mM ammonium acetate) gradient in the first 10 minutes. The gradient was ramped in a linear fashion to 100% Solvent B for 7 minutes. Then the system was switched back to 60% Solvent B and 40% Solvent A for 3 minutes. A flow rate of 0.4 mL/min was used at an injection volume of 5mL. The column was equilibrated for 3 min and run at a flow rate of 0.4 mL/min for a total run time of 20 min. The data acquisition of each sample was performed in both positive and negative ionization modes using a TripleTOF 5600 equipped with a Turbo VTM ion source (AB Sciex, Concord, Canada). The voltage of the source was set to 5500 V for positive ionization and 4500 V for negative ionization mode. The curtain gas flow, nebulizer, and heater gas were set to 30, 40, and 45 units respectively. TOF MS survey scans (150 ms) and 15 MS/MS scans with a total duty cycle time of 2.4 s were performed. The mass range in both modes was 50-1200 m/z. The acquisition of MS/MS spectra by data-dependent acquisition (DDA) function of the Analyst TF software (AB Sciex, Concord, Canada) was controlled with the following parameters: dynamic background subtraction, charge monitoring to exclude multiply charged ions and isotopes, and dynamic exclusion of former target ions for 9 s. Rolling collision energy spread was set whereby the software calculated the collision energy value to be applied as a function of m/z.
Data processing and lipidomics analysis
The raw data in .mgf format was converted using proteoWizard software (Chambers et al., 2012). The NIST MS PepSearch Program was used to search the converted files against LipidBlast libraries. The m/z width was determined via the mass accuracy of internal standards at 0.001 for positive mode and 0.005 for a negative mode at an overall mass error of less than 2 parts per million. The minimum match factor at 400 was set for the downstream data processing. The MS/MS identification results from all the files were combined using an in-house software tool to create a library for quantification (Vantaku et al., 2019). The raw data files were searched against this in-house library of known lipids with mass and retention time using Multiquant 1.1.0.26 (ABsciex, Concord, Canada). The lipid species identified in the positive or negative ion modes were analyzed separately using relative abundance of peak spectra for the downstream analyses. The identified lipids were quantified by normalizing against their respective internal standard. Quality controls were performed with internal control samples stated above to monitor the overall quality of the lipid extraction and mass spectrometry analyses.
The data were log2 transformed followed by normalization using the median normalization. The compound-by-compound t-test was applied to identify the top differentially regulated lipids that passed the nominal threshold P value of <0.05. The normalized concentration lipidomics data were processed with R version 4.0.2. To allow general-purpose adjustment for differences among samples, quantile normalization was performed before data analysis. The above-stated log data transformation and median-centered scaling divided by standard deviation of each variable made lipid features more comparable. Fold change (FC) analysis was performed with FC threshold 2, to tell the important features with p value threshold 0.05. Volcano plotting was performed for a combination demonstration of fold change (FC) analysis and t-tests. Correlation analysis (Spearman) was performed to visualize the overall correlations between different features. The R.prcomp package was used for principal component analysis (PCA) as an unsupervised method. R.hclust in this stat function package was used for hierarchical cluster analysis. Heatmaps were generated using Euclidean distance measurements (clustering algorithm: ward.D). To assess the significance of class discrimination, a discriminant analysis (PLS-DA) model-based permutation test was also conducted with the plsr function provided by the R pls package. The Benjamini-Hochberg procedure was used for false discovery rate (FDR) correction for multiple comparisons.
Membrane fluidity assay
Membrane fluidity assays were conducted using a membrane fluidity kit (Abcam). Briefly, the indicated cells (primary hepatocytes or MEFs) were perfused and incubated in the labeling solution (10 μM pyrenedecanoic acid and a final concentration of 0.08% Pluronic F-127 with perfusion buffer) at room temperature in the dark for 30 minutes. The cells were washed and then incubated in culture medium without phenol red for fluorescence reading. The values for membrane fluidity ratios were calculated by fluorescence (excitation at 350 nm and emission at 405 nm and 460 nm, respectively).
Mito stress assays
Mito stress assays were performed with Seahorse kits and the Seahorse XFe96 analyzer per the manufacturer’s protocol (Agilent). Briefly, cells were plated in a Seahorse XF96 cell culture plate and then incubated overnight at 37°C in a humidified 5% CO2 chamber. Seahorse XF cartridges were hydrated overnight with Seahorse XF calibrant in a 0% CO2 incubator. Compound injections for different stress tests (Seahorse XF Cell Mito Stress Test Kit) were added to the respective ports on the cartridge per the manufacturer’s instructions. Seahorse XF Report Generator (Agilent) was used to calculate the parameters of the Seahorse XF Cell Mito Stress results according to the manufacturer’s instruction.
Synchronization of MEFs
For tunicamycin (Tu) treatment, MEFs were plated in DMEM (4.5g/L glucose) supplemented with 10% FBS. The cultured cells were treated with 25ng/mL Tu for 2 hours, followed by washes with 1X PBS. The washed cells were then cultured in the culture medium.
siRNA transient transfections
MEFs were transfected with Lipofectamine RNAiMAX reagents (Life technologies) and 10μM of siRNAs (siGENOME Non-Targeting siRNA pool: Dharmacon, D-001206-13-05; siGENOME SMARTpool XBP1: Dharmacon, L-040825-00-0005) for 48 hours according to the manufacturer’s instructions.
Transient luciferase assay
MEFs were co-transfected with vector pRL-TK (internal control) and Eif2ak3-dluc or Eif2ak3-dluc with mutated XBP1s binding site. Lipofectamine 2000 was used with an increasing dose of SRC-3 over-expression vector per the manufacturer’s protocol (Life technologies). The transfected cells were lysed with 50μL of 1x passive lysis buffer. Dual-reporter luciferase assays were performed per the manufacturer’s protocol (Promega).
Real-time luminescence assay
Real-time luminescence assays were performed using a Lumicycle (Actimetrics) as previously described (Zhu et al., 2017). Stable Eif2ak3-dluc cells were cultured in DMEM (4.5g/L glucose) supplemented with 10% FBS. The cells were treated with 25ng/mL of Tu in high glucose DMEM with 10% FBS for 2 hour before the real-time luminescence assays. After synchronization of MEFs with Tu treatment, cells were washed with 1x PBS and cultured with DMEM (4.5g/L glucose) supplemented with 0.1 mM Luciferin and 10mM HEPES buffer in the absence of serum. The treated cells (in 35 mm tissue culture dishes) were transferred to the Lumicycle for immediate real-time luminescence analysis. The embedded Periodogram function in Lumicycle was used to determine the periods of oscillation. For siRNA treated MEFs, MEFs were transfected with non-targeting, SRC-3 or Xbp1 siRNA for 48 hours before synchronizing with Tu treatment, followed by the above-described real-time luminescence assay.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using GraphPad Prism 9. All data were tested for normal distribution of variables. All normally distributed data were analyzed as the means ± standard error of the mean (SEM), unless otherwise noted. Measurements between two groups were performed with an unpaired Student’s t-test (two-tailed p value) unless otherwise noted. Non-significant p values are shown as ns. Each in vivo and in vitro experiment was performed with the number of replicates specified in the figure legends. All data obtained are shown; all data are shown in the quantification.
Supplementary Material
Highlights.
SRC-3 coactivates XBP1 transcription and regulates hepatic 12-h gene oscillations
Transcriptional coactivation integrates the liver 12-h clock with lipid metabolism
Coactivation of the 12-h clock coordinates hepatic lipid metabolic processes
A fasting regimen reprograms cistromic SRC-3 coactivation of the 12-h clock
ACKNOWLEDGMENTS
The human pan-cancer prognostic results here are based upon data obtained from the TCGA Research Network: https://www.cancer.gov/tcga. We thank Vasanta Putluri in the Metabolomics Core at Baylor College of Medicine (BCM) supported by the CPRIT Core Facility Support Award RP170005 and RP210227, NCI Cancer Center Support Grant P30CA125123, NIH/NCI R01CA220297, and R01CA216426, and the Helis Foundation. We thank P.K. Saha and the MMPC core at BCM supported by NIH funds R01DK114356 and UM1HG006348. We thank the ADA award 1-18-JDF-025, NIGMS DP2GM140924, and NIDDK P30DK120531 to B.Z. We also thank the Science Park NGS Core at the MD Anderson Cancer Center, supported by CPRIT Core Facility Support Grant RP170002. This research work was supported by grants from the National Institutes of Health 1P01DK113954 and NICHD 5R01HD007857 and 5R01HD08188 to B.W.O. and Pilot Project Program under Award Number P30ES030285 to B.W.O. and H.M.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110491.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Ackerman D, and Simon MC (2014). Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 24, 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoulas AC, Zhu B, Zhang Q, York B, O’Malley BW, and Dacso CC (2018). A novel mathematical method for disclosing oscillations in gene transcription: a comparative study. PLoS One 13, e0198503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, and Dyer WJ (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917. [DOI] [PubMed] [Google Scholar]
- Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, et al. (2012). A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol 30, 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, and Koehler CM (2012). The complexity of cardiolipin in health and disease. Trends Biochem. Sci 37, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretenet G, Le Clech M, and Gachon F (2010). Circadian clock-coordinated 12 Hr period rhythmic activation of the IRE1alpha pathway controls lipid metabolism in mouse liver. Cell Metab. 11, 47–57. [DOI] [PubMed] [Google Scholar]
- Edidin M (2003). Lipids on the frontier: a century of cell-membrane bilayers. Nat. Rev. Mol. Cell Biol 4, 414–418. [DOI] [PubMed] [Google Scholar]
- Holthuis JC, and Menon AK (2014). Lipid landscapes and pipelines in membrane homeostasis. Nature 510, 48–57. [DOI] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, and Hogenesch JB (2009). Harmonics of circadian gene transcription in mammals. PLoS Genet. 5, e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolfsson HI, Melo MN, van Eerden FJ, Arnarez C, Lopez CA, Wassenaar TA, Periole X, de Vries AH, Tieleman DP, and Marrink SJ (2014). Lipid organization of the plasma membrane. J. Am. Chem. Soc 136, 14554–14559. [DOI] [PubMed] [Google Scholar]
- Lanz RB, Bulynko Y, Malovannaya A, Labhart P, Wang L, Li W, Qin J, Harper M, and O’Malley BW (2010). Global characterization of transcriptional impact of the SRC-3 coregulator. Mol. Endocrinol 24, 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Scapa EF, Cohen DE, and Glimcher LH (2008). Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320, 1492–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle S, Feldman A, Boyce MS, and Wilson WF (2008). Prey behavior, age-dependent vulnerability, and predation rates. Am. Nat 172, 712–725. [DOI] [PubMed] [Google Scholar]
- Lopez MS, Tan IS, Yan D, Kang J, McCreary M, Modrusan Z, Austin CD, Xu M, and Brown EJ (2017). Host-derived fatty acids activate type VII secretion in Staphylococcus aureus. Proc. Natl. Acad. Sci. U S A 114, 11223–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Gonzales NM, Lonard DM, Putluri N, Zhu B, Dacso CC, York B, and O’Malley BW (2020). XBP1 links the 12-hour clock to NAFLD and regulation of membrane fluidity and lipid homeostasis. Nat. Commun 11, 6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina AI, LeClair RA, LeClair KB, Cohen DE, Lantier L, and Banks AS (2018). CalR: a web-based analysis tool for indirect calorimetry experiments. Cell Metab. 28, 656–666 e651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel JV (1962). Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am. J. Hum. Genet 14, 353–362. [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Ballance H, Meng H, Gonzalez N, Kim SM, Abdurehman L, York B, Chen X, Schnytzer Y, Levy O, et al. (2020). 12-h clock regulation of genetic information flow by XBP1s. PLoS Biol. 18, e3000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patke A, Young MW, and Axelrod S (2020). Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol 21, 67–84. [DOI] [PubMed] [Google Scholar]
- Prentice AM, Rayco-Solon P, and Moore SE (2005). Insights from the developing world: thrifty genotypes and thrifty phenotypes. Proc. Nutr. Soc 64, 153–161. [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, and Santachiara-Benerecetti AS (1999). Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat. Genet 23, 437–441. [DOI] [PubMed] [Google Scholar]
- Reinke H, and Asher G (2019). Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol 20, 227–241. [DOI] [PubMed] [Google Scholar]
- Saltzman AB, Leng M, Bhatt B, Singh P, Chan DW, Dobrolecki L, Chandrasekaran H, Choi JM, Jain A, Jung SY, et al. (2018). gpGrouper: a peptide grouping algorithm for gene-centric inference and quantitation of bottom-up proteomics data. Mol. Cell Proteomics 17, 2270–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders CR, and Hutchison JM (2018). Membrane properties that shape the evolution of membrane enzymes. Curr. Opin. Struct. Biol 51, 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellayah D, Cagampang FR, and Cox RD (2014). On the evolutionary origins of obesity: a new hypothesis. Endocrinology 155, 1573–1588. [DOI] [PubMed] [Google Scholar]
- Speakman JR (2007). A nonadaptive scenario explaining the genetic predisposition to obesity: the "predation release" hypothesis. Cell Metab. 6, 5–12. [DOI] [PubMed] [Google Scholar]
- Speakman JR (2008). Thrifty genes for obesity, an attractive but flawed idea, and an alternative perspective: the ‘drifty gene’ hypothesis. Int. J. Obes. (Lond) 32, 1611–1617. [DOI] [PubMed] [Google Scholar]
- Stashi E, Lanz RB, Mao J, Michailidis G, Zhu B, Kettner NM, Putluri N, Reineke EL, Reineke LC, Dasgupta S, et al. (2014). SRC-2 is an essential coactivator for orchestrating metabolism and circadian rhythm. Cell Rep. 6, 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaben PF, and Westermark PO (2014). Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaben PF, and Westermark PO (2016). Differential rhythmicity: detecting altered rhythmicity in biological data. Bioinformatics 32, 2800–2808. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, and Feigenson GW (2008). Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol 9, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderkooi JM, and Callis JB (1974). Pyrene. A probe of lateral diffusion in the hydrophobic region of membranes. Biochemistry 13, 4000–4006. [DOI] [PubMed] [Google Scholar]
- Vantaku V, Dong J, Ambati CR, Perera D, Donepudi SR, Amara CS, Putluri V, Ravi SS, Robertson MJ, Piyarathna DWB, et al. (2019). Multi-omics integration analysis robustly predicts high-grade patient survival and identifies CPT1B effect on fatty acid metabolism in bladder cancer. Clin. Cancer Res 25, 3689–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmi de Toledo F, Buchinger A, Burggrabe H, Holz G, Kuhn C, Lischka E, Lischka N, Lutzner H, May W, Ritzmann-Widderich M, et al. (2013). Fasting therapy - an expert panel update of the 2002 consensus guidelines. Forsch Komplementmed 20, 434–443. [DOI] [PubMed] [Google Scholar]
- Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, and O’Malley BW (2000). The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/ TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl. Acad. Sci. U S A 97, 6379–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Hastings MH, Green EW, Tauber E, Sladek M, Webster SG, Kyriacou CP, and Wilcockson DC (2013). Dissociation of circadian and circatidal timekeeping in the marine crustacean Eurydice pulchra. Curr. Biol 23, 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, Dean A, Dacso CC, York B, and O’Malley BW (2015). Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Mol. Cell 60, 769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zhang Q, Pan Y, Mace EM, York B, Antoulas AC, Dacso CC, and O’Malley BW (2017). A cell-autonomous mammalian 12 hr clock coordinates metabolic and stress rhythms. Cell Metab. 25, 1305–1319 e1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-Seq and ChIP-Seq data have been deposited to the Gene Expression Omnibus (GEO), the lipid metabolomics data have been deposited to the NIH Common Fund’s National Metabolomics Data Repository (NMDR), the proteomics data have been deposited to the ProteomeXchange consortium member PeptideAtlas, and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
This paper does not report original code.
Any additional information required to re-analyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-SRC-3 | Cell Signaling Technology | Cat# 2126, RRID: AB_823642 |
| Rabbit polyclonal anti-XBP1s | Biolegend | Cat# 619501, RRID: AB_315907 |
| Rabbit monoclonal anti-ATF4 | Cell Signaling Technology | Cat# 11815S, RRID: AB_2616025 |
| Rabbit polyclonal anti-ATF6-α | Santa Cruz Biotechnology | Cat# sc-22799, RRID: AB_2242950 |
| Rabbit polyclonal anti-XBP1 | Santa Cruz Biotechnology | Cat# sc-7908, RRID: AB_2107620 |
| Rabbit polyclonal anti-BMAL1 | Abcam | Cat# ab3350, RRID: AB_303729 |
| Rabbit anti-beta-Actin Monoclonal | Cell Signaling Technology | Cat# 5125, RRID: AB_1903890 |
| Antibody, HRP Conjugated, Clone 13E5 | ||
| Bacterial and virus strains | ||
| Eif2ak3 promoter-pGreenfire-mCMV-dscGFP-EF1-Puro lentivirus | (Zhu et al., 2017) | N/A |
| XBP1s-pHAGE | A gift from Dr. Xi Chen at BCM | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Tunicamycin | EMD Millipore | Cat# 654380 |
| Eif2ak3 promoter-pGreenfire-mCMV-dscGFP-EF1-Puro lentivirus | (Zhu et al., 2017) | N/A |
| Hygromycin B | Thermo Fisher Scientific | Cat# 10687010 |
| Lipofectamine® 2000 Transfection Reagent | Thermo Fisher Scientific | Cat# 11668027 |
| Lipofectamine® RNAiMAX Transfection Reagent | Thermo Fisher Scientific | Cat# 13778030 |
| HEPES | Thermo Fisher Scientific | Cat# 15630080 |
| SuperScript® VILO™ Master Mix | Thermo Fisher Scientific | Cat# 11755050 |
| Micrococcal Nuclease | New England Biolabs | Cat# M0247S |
| Sodium Pyruvate | Thermo Fisher Scientific | Cat# 11360070 |
| PEG-it Virus Precipitation Solution | System Biosciences | Cat# LV810A-1 |
| RNase A, DNase and protease-free | Thermo Fisher Scientific | Cat# EN0531 |
| TaqMan® Universal Master Mix II, no UNG | Thermo Fisher Scientific | Cat# 4440047 |
| Power SYBR Green Master Mix | Thermo Fisher Scientific | Cat# 4367660 |
| Dialyzed Fetal Bovine Serum | Thermo Fisher Scientific | Cat# 26400036 |
| Fetal Bovine Serum | Thermo Fisher Scientific | Cat# 10437028 |
| D-glucose | Sigma | Cat# G8270 |
| Sodium Acetate | Sigma | Cat# S2889 |
| 2-Mercaptoethanol | Sigma | Cat# M3148 |
| DMSO | Sigma | Cat# D8418 |
| NP-40 | Sigma | Cat# N3516 |
| Sodium chloride | Sigma | Cat# S7653 |
| Tris/glycine/SDS buffer | Biorad | Cat# 161-0772 |
| TBST | Cell Signaling Technology | Cat# 9997 |
| 10% SDS | Gibco | Cat# 15553-035 |
| Triton X-100 | Promega | Cat# H5142 |
| Tween 20 | Fisher Scientific | Cat# BP337-100 |
| BSA | Sigma | Cat# A9647 |
| EDTA | Cell Signaling Technology | Cat# 7011S |
| Nonfat dry milk | Biorad | Cat# 170-6404 |
| BCA Protein Assay Reagent A | Thermo Fisher Scientific | Cat# 23223 |
| BCA Protein Assay Reagent B | Thermo Fisher Scientific | Cat# 23224 |
| iBlot Gel transfer stacks Nitrocellulose | Life Technologies | Cat# IB301001 |
| Sodium deoxycholate | Sigma | Cat# D6750 |
| Mouse Genomic DNA | Promega | Cat# G3091 |
| Ponceau S solution | Sigma | Cat# P7170 |
| VivoGlo™ Luciferin, In Vivo Grade | Promega | Cat# P1041 |
| Restore western blot stripping buffer | Thermo Fisher Scientific | Cat# 21063 |
| Penicillin Streptomycin | Thermo Fisher Scientific | Cat# 15070063 |
| Pellet Paint® Co-Precipitant | EMD Millipore | Cat# 69049 |
| Q5® High-Fidelity DNA Polymerase | New England Biolabs | Cat# M0491S |
| Criterion pre-cast gradient SDS-PAGE gel (4-20%) | Bio-Rad | Cat# 5671094 |
| TaKaRa Ex Taq® DNA Polymerase | Clontech | Cat# RR001A |
| T4 DNA Ligase | New England Biolabs | Cat# M0202 |
| Antarctic Phosphatase | New England Biolabs | Cat# M0289S |
| Ethanol 200 proof | Decon Labs | Cat# 2701 |
| Isopropanol | Sigma | Cat# I9516 |
| cOmplete™ Protease Inhibitor Cocktail | Roche | Cat# 11697498001 |
| PhosSTOP™ | Roche | Cat# 4906845001 |
| DMEM, high glucose | Thermo Fisher Scientific | Cat# 11965092 |
| DMEM, no glucose | Thermo Fisher Scientific | Cat# 11966025 |
| Proteinase K | Thermo Fisher Scientific | Cat# AM2544 |
| 0.45μM syringe filter | Thermo Fisher Scientific | Cat# 09-740-106 |
| Critical commercial assays | ||
| SimpleChIP® Plus Enzymatic Chromatin IP Kit | Cell Signaling Technology | Cat# 9005S |
| PureLink RNA mini kit | Life Technologies | Cat# 12183025 |
| Dual-luciferase Reporter assay system | Promega | Cat# E1960 |
| Insulin ELISA kit | EMD Millipore | Cat# EZRMI-13K |
| One touch ultra glucose meter and strips | LifeScan, Inc | N/A |
| Deposited data | ||
| GSE175598 | GEO public repository | GSE175598 |
| Study ST002079/PR001319 | NMDR public repository | datatrack_id:3065 study_id:ST002079; http://dev.metabolomicsworkbench.org:22222/data/DRCCMetadata.php?Mode=Study&StudyID=ST002079&Access=DoqF5000 |
| PASS01737 | PeptideAtlas (ProteomeXchange consortium member) | PASS01737; http://www.peptideatlas.org/PASS/PASS01737 |
| Experimental Models: Cell Lines | ||
| Immortalized mouse embryonic fibroblasts | (Zhu et al., 2017) | N/A |
| Eif2ak3-dluc or Eif2ak3-dluc with mutated XBP1s binding site stably expressing MEFs | (Zhu et al., 2017) | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J male mice | The Jackson Laboratory | JAX:000664 |
| Mouse: SRC-3 knockout male mice | (Xu et al., 2000) | N/A |
| Mouse: Xbp1 flx/flx male mice | A gift from Dr. Xi Chen at BCM (Lee et al., 2008) | N/A |
| Mouse: Xbp1 flx/flx;AlbCre male mice | (Meng et al., 2020) | N/A |
| Oligonucleotides | ||
| siRNA oligonucleotides | Dharmacon | Dharmacon, D-001206-13-05; Dharmacon, L-040825-00-0005 |
| See Table S5 for qRT-PCR and ChIP-qPCR Primer Sequences | N/A | N/A |
| Recombinant DNA | ||
| Eif2ak3-dluc | (Zhu et al., 2017) | N/A |
| Eif2ak3-dluc mutant | (Zhu et al., 2017) | N/A |
| Software and algorithms | ||
| RAIN | (Thaben and Westermark, 2014) | http://www.bioconductor.org/packages/release/bioc/html/rain.html |
| Eigenvalue/pencil | (Antoulas et al., 2018; Zhu et al., 2017) | N/A |
| Bioconductor 3.4 | R | https://www.bioconductor.org/news/bioc_3_4_release/ |
| Matlab R2019a | MathWorks | https://www.mathworks.com/products/new_products/latest_features.html |
| GraphPad Prism 9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| LumiCycle Analysis 2.54 | ActiMetrics | http://actimetrics.com/downloads/lumicycle/ |
| UCSC genome browser | UCSC | https://genome.ucsc.edu/cgi-bin/hgGateway |
| DAVID Bioinformatics Resources 6.8 | NIH | https://david.ncifcrf.gov/ |
| CLAMS data examination tool | Columbus Instruments | http://www.colinst.com/products/clams-hc-comprehensive-lab-animal-monitoring-system-for-home-cages#CLAX |
| VirtualDubMod 1.5.10 | Intel Software | https://sourceforge.net/projects/virtualdubmod/files/ |
| Venn Diagram Plotter v1.5.5228.29250 | Department of Energy and PNNL | https://omics.pnl.gov/software/venn-diagram-plotter |
| Gene Cluster 3.0 | Originally writtern by Michael Eisen at Stanford University | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm#ctv |
| Java TreeView 3.0 alpha03 | Open Source software | https://bitbucket.org/TreeView3Dev/treeview3/ |
| Other | ||
| NCBI Aceview (AceView offers a comprehensive annotation of human, mouse and nematode genes) | NIH | https://www.ncbi.nlm.nih.gov/IEB/Research/Acembly/index.html |