Abstract
The outbreak of novel corona virus 2019 (COVID-19) has been treated as a public health crisis of global concern by the World Health Organization (WHO). COVID-19 pandemic hugely affected countries worldwide raising the need to exploit novel, alternative and emerging technologies to respond to the emergency created by the weak health-care systems. In this context, Artificial Intelligence (AI) techniques can give a valid support to public health authorities, complementing traditional approaches with advanced tools. This study provides a comprehensive review of methods, algorithms, applications, and emerging AI technologies that can be utilized for forecasting and diagnosing COVID-19. The main objectives of this review are summarized as follows. (i) Understanding the importance of AI approaches such as machine learning and deep learning for COVID-19 pandemic; (ii) discussing the efficiency and impact of these methods for COVID-19 forecasting and diagnosing; (iii) providing an extensive background description of AI techniques to help non-expert to better catch the underlying concepts; (iv) for each work surveyed, give a detailed analysis of the rationale behind the approach, highlighting the method used, the type and size of data analyzed, the validation method, the target application and the results achieved; (v) focusing on some future challenges in COVID-19 forecasting and diagnosing.
Keywords: COVID-19, Artificial intelligence, Machine learning, Deep learning, Forecasting, Diagnosing
1. Introduction
After recognizing the COVID-19 outbreak in Wuhan, China, as a Public Health Emergency of International Concern in the month of December 2019, the World Health Organization (WHO) declared it an epidemic on January 30th, 2020, and a pandemic on March 12th, 2020. Since then, COVID-19 spread exponentially all over the world, causing worldwide travel restrictions, as well as mandatory lockdown in many cities.
COVID-19 severely affected countries worldwide, causing enormous problems in the health systems due to the exceptional magnitude of the pandemic. Overwhelmed hospitals, exhausted physicians and nurses, shortage of medical supplies and of detection tools, among which COVID-19 testing kits and screening tools, made the battle against the pandemic difficult and often ineffective, not able to reduce the disease spreading.
COVID-19 pandemic made clear that advanced and emerging technologies are required to respond to the emergency and to tackle the challenges due to the weak health-care systems and financial burden.
In this context, Artificial Intelligence (AI) techniques can give a valid support to public health authorities, complementing traditional approaches with advanced tools, in the difficult tasks of COVID-19 detection, spreading, monitoring, diagnosing, screening, surveillance, and contact tracing.
Artificial intelligence is defined as a technology that allows computers to imitate human intelligence to perform tasks commonly associated with intelligent beings, such as learning and problem solving.
In the recent years, the use of AI-based tools is having a key role in improving the management and the solution of several issues and problems of the health sector like medical image inspection, precision medicine, epidemics prevention and spreading, as well as for disease detection and prevention. COVID-19 created a global health emergency where the importance and key role of AI-driven intelligent systems has been drastically increased in the last year. As such, the challenges arisen by the COVID-19 emergency in terms of tracing of the infection or the prediction of its diffusion and the way it would spread, together with the evaluation of the effects of restrictive measures and lockdown, have produced several and promising research activities using artificial intelligence techniques.
In this context, disease prediction and spreading is still a tough concern in the healthcare field. Providing prediction systems that can accurately anticipate and diagnose virus spreading remains a challenging task. Nevertheless, the integration of medical expertises within AI-driven algorithms could represent an effective solution to address the challenges and issues arisen by COVID-19. The huge availability of data produced by the pervasiveness of IT tools and devices, together with the ever-increasing computing power, made possible the implementation of AI-based solutions that exhibited an exceptional performance in addressing many of the above mentioned issues.
1.1. Scope of the survey and contributions
The purpose of this review is to provide a comprehensive description of the application and effectiveness of AI technologies for forecasting, detecting, and diagnosing COVID-19. The study investigates and discusses an extensive collection of papers published in the last year with the aim of giving an overview of how AI can help fighting COVID-19 pandemic.
In particular, the review has been conducted by trying to answer to the following questions: (i) which are the emerging AI technologies used to forecast COVID-19? (ii) What is the effectiveness of such methodologies? We explore these questions, with the following contributions:
-
•
This is the first exhaustive and focused survey concerning only a very specific topic that is COVID-19 forecasting through AI techniques. In fact, even if there are some other interesting surveys focusing on the role of AI techniques in the battle against COVID-19, they cover a more broader spectrum of applications and topics. For example, the survey of Dagliati et al. [1] focuses on collaborative data infrastructures to support COVID-19 research. The survey of Combi et al. [2] presents a taxonomy based on methodologies and techniques for classifying intelligent information systems and AI techniques state-of-the-art for COVID-19 data-intensive applications.
-
•
In comparison with other similar surveys focusing only on AI based approaches for forecasting COVID-19, we provide an extensive background description of such techniques, which can help non-expert to better understand and grasp the underlying concepts.
-
•
For each work surveyed, we provide a detailed analysis of the rationale behind the approach, highlighting the method used, the type and size of data analyzed, the validation method, the target application and the results achieved.
-
•
We summarize the main research contributions related to the role of AI in the COVID-19 forecasting by reporting in Table 3 the main features of the approaches, in order to guide the reader through the principal literature results about the targeted topics.
-
•
Finally, based on the selected literature review we conclude that, even if several applications addressing COVID-19 issues have been proposed, only few of them are currently mature enough to be used in practice. We report the main limitations of current approaches, including interpretability and learning from limited labeled data. We also draw some suggestion for future works.
Table 3.
Summary of methods.
| Publication | Method | Data Types | Data size | Output | Validation method | Result |
|---|---|---|---|---|---|---|
| Abdulaal et al. [154] | ANN | Demographics, comorbidities, smoking history | 398 patients - hospital admissions for SARS-CoV-2, February 2–April 22, 2020, West London teaching hospital | Patient-specific mortality risk | Acc, R, SPC, P, NPV, ROC AUC | 86.25%Acc, 87.50% R, 85.94% SPC, 60.87% P, 96.49% NPV, 90.12% AUC |
| Abdulkareem et al. [68] | SVM,RF,NB | 18 laboratory findings from the Hospital Israelita Albert Einstein at Sao Paulo, Brazil | 600 patients | COVID-19 cases | Acc, P, R, AUC, F-measure | SVM outperforms other methods: 95% Acc, 95% AUC, 94% F-measure |
| Ahamad et al. [61] | XGB, DT, RF, SVM, GBM | COVID-19 patients clinical data | 6512 patients from 7 provinces in China | Prediction of positive patients | P, R, F-measure, AUC | XGB outperforms other methods |
| Alakus et al. [51] | ANN, CNN, LSTM, RNN, CNNLSTM, CNNRNN | 18 laboratory findings from the Hospital Israelita Albert Einstein, Sao Paulo, Brazil | 600 patients | COVID-19 cases | Acc, P, R, AUC, and F-measure | CNNLSTM outperforms other methods: 92.30% Acc, 90% AUC, 93% F-measure |
| Aljame et al. [77] | RF, LoR, XGB | Routine blood tests | 5644 data samples from Albert Einstein Hospital, Brazil | COVID-19 diagnosis | Acc, AUC, R, SPC | XGBoost outperforms other methods: 99.88% Acc, 99.38% AUC |
| Ardabili et al. [87] | LoR, LR, logarithmic, quadratic, cubic, compound, power and exponential regressors, MLP, ANFIS | COVID-19 daily cases in Italy, Germany, Iran, USA, and China | 30 days | COVID-19 daily cases | RMSE, Correlation coefficient | MLP outperforms other methods |
| Arpaci et al. [63] | NB, LoR, IBk, CR, PART, DT | 14 clinical features | 114 patients | COVID-19 daily cases | CCI, P, R, F-measure, AUC | CR outperforms other methods |
| Assaf et al. [64] | ANN, RF, CR, DT | Patients demographics, clinical data | 6995 patients | COVID-19 Death and critical cases | R, SPC, P, NPV, Acc, AUC | RF outperforms other methods: R, SPC, P, NPV and Acc of 75.0%, 95.8%, 75.0%, 95.8% and 92.9%, respectively, 93%AUC |
| Brinati et al. [65] | DT, ExtraTrees, KNN, LoR, NB, RF | Routine blood exams | 279 patients | Positive or negative cases | Acc, P, R, SPC, AUC | RF outperforms the other classifers: 82% Acc, 92% R, 83%P, 65% SPC, 84% AUC |
| Burdick et al. [76] | XGBC | Clinical data (textual) | 197 patients of 5 US hospitals, March 24 to May 4, 2020 | Prediction of need for invasive mechanical ventilation of COVID-19 patients within 24 h of an initial encounter | AUC, P, R, SPC | Outperforms MEWS approach |
| Casiraghi et al. [80] | RF | Symptoms, clinical history, comorbidities, laboratory measurements, saturation/oxygen values, patients data | 301 patients, March 7–April 10, 2020 | COVID risk prediction | AUC, R, SPC, Acc, F-measure | Outperforms linear models |
| Chakraborty et al. [59] | LR, PR | COVID-19 cases and deaths per day | January 30–April 19, 2020 | COVID cases and deaths | R2, MAE, RMSE, MSE | PR outperforms LR |
| Chaurasia et al. [66] | NB, MA, ES, Holt's linear, Holt-Winters, ARIMA | Number of COVID-19 cases and deaths per day | January 22 to May 28, 2020 | COVID-19 deaths | RMSE | NB outperforms other methods |
| Devaraj et al. [48] | ARIMA, LSTM, SLSTM, Prophet | COVID-19 cases per day from John Hopkins University, World Weather Page, Wikipedia | Global-wide, country and city specific analysis data from 22nd Jan 2020 to 8th May 2020. Simulated dataset for seven cities for the months of May, June, July, August 2020. All countries data from January 2020 to September 2020. | COVID-19 cases, deaths, recovery for India and Chennai | RMSE, MAE, MAPE, R2 | SLSTM outperforms other methods, ARIMA outperforms LSTM. |
| dos Santos Gomes et al. [71] | Kalman filter, fuzzy clustering | Daily deaths in Brazil | February 29–May 18, 2020 | Death cases | RMSE, MAE, RMSPE, R2, MAPE | Outperforms contestants methods |
| Farooq et al. [52] | ANN | COVID-19 cases, deaths and recoveries, rate of vaccination | 30th January - 13 June 2020, total cases in India is 308,993 with 154,330,154,330 recoveries and 8884 deaths | Rate of infection, rate of recovery, rate of death | Simulation | Reduce the number of deaths to 1.3 million from 55 million, if mobility and contact is made 5 times to that of the lockdown period |
| Gao et al. [62] | Mortality Risk Prediction ensemble Model (MRPMC) including: EL, LoR, SVM, GBDT, NN | EHRs in 4 China hospitals | 2160 patients | Prediction of physiological deterioration and death up to 20 days in advance | Acc, AUC | AUC ranges from 0.9186 to 0.9762 in the three validation cohorts |
| Gupta et al. [53] | LSTM | Time series of number of COVID-19 cases and deaths | January 22–October 9, 2020 | COVID-19 cases and deaths | RMSE | RMSE: 0.0766–0.0533, outperforms SVM and DT |
| Hasan et al. [79] | ANN, EEMD | COVID-19 cases, deaths, recovery | January 22–May 18, 2020, Center for Systems Science and Engineering (CSSE) at the Johns Hopkins University | Confirmed, death, recovery cases | MSE, R2, Acc | Outperforms traditional statistical analysis |
| Hazarika and Gupta [88] | MLP | COVID-19 daily cases in Brazil, India, Peru, Russia, USA | April 11–July 10, 2020 | COVID-19 cases | RMSE, MAE, R2, PSNR, SC, MD, LMSE, NAE | Comparable or better results than SVR and RVFL |
| Hernandez et al. [46] | ARIMA | COVID-19 daily cases | 145 countries, 1 M people. From the day each country presented the first case of COVID-19 to May 28, 2020 | COVID-19 daily cases | RMSE | RMSE: 144.81 |
| Khanda et al. [67] | SVM, NB, LoR, DT, RF, EL | Clinical textual reports | 212 patients | Classification of texts into four different categories: COVID, SARS, ARDS and both COVID, and ARDS. | Acc, P, R, F-measure | LoR and NB showed better results than other methods with 96.2% Acc, 94% P, 96% R, 95% F-measure. |
| Kumar et al. [42] | ARIMA, Prophet | COVID-19 cases in SP, IT, FR, DE, RS, Iran, UK, Turkey,India | March 1st-May 20th, 2020 | COVID-19 confirmed, active, recovered, death cases | MAE, RMSE, RRSE, MAPE | ARIMA outperforms Prophet |
| Meng et al. [54] | CNN, LoR | Clinical data and CT images | 366 severe or critical COVID-19 patients | Survival probability | AUC, Acc, R, SPC, Kaplan-Meier analysis | AUC: 0.952 (95% confidence interval, 0.928–0.977) on the training set and 0.943 (0.904–0.981) |
| Pinter et al. [60] | ANFIS, MLP-ICA | COVID-19 cases and death rate in Hungary | 4 March - 19 April 2020 | COVID-19 cases and deaths | R2, MAPE, RMSE | MLP-ICA outperforms ANFIS |
| Pourhomayoun et al. [78] | SVM, ANN, RF, DT, LR, KNN | Clinical, demographic and physiological data | 2,670,000 laboratory-confirmed COVID-19 patients from 146 countries around the world including 307,382 labeled samples | Predict the mortality of patients with COVID-19 | Acc, R, SPC | 89.98% Acc |
| Ramchandani et al. [83] | NN | Census data, intra-county mobility, inter-county mobility, social distancing data, past growth of infection in US | April 5th-June 28th | 4 COVID cases increase classes: negligible, moderately low, moderately high, significantly high | Acc | 63.7% Acc on the test set: 12–28 June |
| Ren et al. [81] | Singular spectral analysis | COVID-19 daily cases | January 22–April 11, 2020, | COVID-19 Daily cases | Singular spectral analysis | Efficay of the proposed model |
| Ribeiro et al. [69] | ARIMA, CUBIST,RF, RIDGE, SVR, EL | COVID-19 daily cases in Brazil | February 24th - April 19th, 2020 | COVID-19 cases ahead in one, three, and six-days | MAE, sMAPE | SVR and EL outperform other methods |
| Rostami et al. [82] | MLR | COVID-19 daily cases and number of daily phone calls | East Midlands region of England between 18 March −19 October 2020 | COVID-19 daily cases | ME, MAE, RMSE | Outperforms ARIMA, ETS, Seasonal Naive, Prophet and a regression model without call data |
| Rustam et al. [58] | LR,LASSO SVM,ES | Worldwide COVID-19 patients | January 22nd-March 27th | COVID-19 cases, deaths, recovery in the next 10 days | MAE,RMSE R2score RAdjusted2 | EA Outperforms other methods |
| Shahid et al. [47] | ARIMA SVR,GRU LSTM,Bi-LSTM | COVID-19 cases | January 22nd - June 27th | COVID-19 cases in Brazil, DE, IT, SP UK, China, India, Israel, Russia,USA | MAE MAPE R2 | BiLSTM outperforms other methods |
| Shastri et al. [49] | ConvLSTM Stacked LSTM Bi-directional LSTM | COVID-19 cases in USA and India | February 7th-July 7th | COVID-19 cases and deaths | MAE, P, R, F-measure | Conv-LSTM outperforms the other two methods |
| Shastri et al. [86] | Deep-LSTM ensemble model | COVID-19 cases and deaths in India | 29th January - 1st September 2020 | COVID-19 cases and death | MAPE, Acc, P, R, F-measure | 97.59% Acc for confirmed cases and 98.88% Acc for deaths. MAPE is for confirmed and death cases of 2.40 and 1.11. |
| Singh et al. [44] | ARIMA LS-SVM | COVID-19 cases IT,SP,FR,UK,USA | January 21st - May 9th, 2020 | COVID-19 cases in | MAE,MAPE RMSE,R2 | LS-SVM outperforms ARIMA |
| Wang et al. [45] | LoR, Prophet | COVID-19 cases in Brazil, Russia, India Peru,Indonesia | February to June 16, 2020 | COVID-19 cases | Acc | Comparing the predicted cases with the real cases, the approach sensibly underestimates the true value. |
| Zeroual et al. [50] | RNN, LSTM, BiLSTM GRU,VAE | COVID-19 cases | January 22 to June 17 | COVID-19 cases | RMSE, MAE MAPE, RMSLE | VAE outperforms other methods |
| Zheng et al. [85] | LSTM NLP | COVID-19 cases | January 23rd-February 18th | COVID-19 cases | MAE MAPE | LSTM+NLP outperforms each model alone |
The paper is organized as follows. In the next section, an overview of the already published reviews on the use of AI techniques for COVID-19 is reported and the main differences with this survey are highlighted. In Section 3 we describe the main AI learning techniques used by researchers to deal with the coronavirus pandemic and recall the evaluation measures used to assess the results of the experimentations. Section 4 provides summary statistics and information regarding the algorithms described in the survey, including publication venues. In Section 5 we give a detailed review of the works in the literature discussing models, methods and results obtained for COVID-19 forecasting and tracking. Section 6 provides a conclusive discussion about the main limitations of the reviewed approaches also outlining some advices for future researches. Section 7 concludes the survey.
2. Related reviews
To the date of writing this paper, a number of researches conducted reviews of the approaches proposed for tackling the pandemic by exploiting artificial intelligence methods. Most of the related reviews cover different medical research aspects to help fighting against COVID-19, such as screening, image analysis, vaccine and drug development. In the following, a description of the most significant and recent reviews is reported.
Chen et al. [3] performed a review where different areas in which AI has been used are discussed. In this survey, authors investigated the main scope and contributions of AI in combating COVID-19 from the aspects of disease detection and diagnosis to virology and pathogenesis, drug and vaccine development, epidemic and transmission prediction. In addition, they also summarize the available data and resources that can be used for AI-based COVID-19 research. Finally, the main challenges and potential directions of AI in fighting against COVID-19 are discussed.
In Naude et al. [4] the limitations, constraints and pitfalls for application of AI in battling the disease are discussed. State-of-the-arts of a wide range of applications of AI and big data for the pandemic is presented in [5]. In Alamo et al. [6], data-driven methods for monitoring, modeling and forecasting the pandemic are described. A discussion on how big data can help to manage the pandemic is presented in [7]. In [8], a review on the data science approaches to combat the disease is presented.
The survey of Dagliati et al. [1] focuses on collaborative data infrastructures to support COVID-19 research. The authors highlighted the current state-of-the art and the open issues of data sharing, data privacy regulations and governance, pointing out the problem of data interoperability due to the heterogeneity in terms of data formats and standards, healthcare processes modeling and representation, shared procedures.
In Combi et al. [2] is presented a survey about the state-of- the art of AI and clinical information systems to support the management of COVID-19 patients. The authors proposed a taxonomy based on methodologies and techniques for classifying intelligent information systems and AI techniques for COVID-19 data-intensive applications. According to such a taxonomy, in the paper are described the main features of the applications like data collection, machine learning, natural language processing, process mining and pathway identification, decision support systems. With respect to other surveys, Combi et al. provided a slightly more technically-oriented survey mainly focusing on computer science oriented bibliography source.
A systematic review on the diagnosis and prognosis of COVID-19 can be found in Wynantsm et al. [9]. The review aimed at appraising the validity and usefulness of published and preprint reports of prediction models for diagnosing COVID-19 in patients with suspected infection, for prognosis of patients with COVID-19, and for detecting people in the general population at increased risk of COVID-19 infection or being admitted to hospital with the disease. Another review of machine learning and AI algorithms for managing the pandemic with respect to different application scenarios is performed in [10].
In the work of Kumar et al. [11] AI approaches in tackling COVID-19 under different perspectives and addressing several research topics, spanning from epidemiology to tracking and prediction are described. In Bullock et al. [12] an overview of recent studies using ML and, more broadly, AI, to deal with the many aspects of the COVID- 19 crisis is presented. Authors identified applications that address challenges posed by COVID-19 at different scales, including: molecular, by identifying new or existing drugs for treatment; clinical, by supporting diagnosis and evaluating prognosis based on medical imaging and non-invasive measures; and societal, by tracking both the epidemic and the accompanying infodemic using multiple data sources. Authors also review datasets, tools, and resources needed to facilitate Artificial Intelligence research, and discuss strategic considerations related to the operational implementation of multidisciplinary partnerships and open science.
The review in Alrazaq et al. [13] focused on AI methods for diagnosis, treatment and vaccine discovery, epidemiological modeling, patient outcome related tasks, and infodemiology. The goal of the work in Kamalov et al. [14] is to present the advances in machine learning research applied to COVID-19 by covering four major areas of research: forecasting, medical diagnostics, drug development, and contact tracing. In Nayak et al. [15] an in-depth analysis has been performed on the significance of deep learning for COVID-19.
In Tayarani [16] a detailed overview on the applications of AI in a variety of fields, including diagnosis of the disease via different types of tests and symptoms, monitoring patients, identifying severity of a patient, processing COVID-19 related imaging tests, epidemiology, pharmaceutical studies, etc. is presented. The aim of this study is to perform a comprehensive survey on the applications of AI in battling against COVID-19, covering every way that AI approaches have been employed.
In the survey in [17] Hussain et al. summarize the current state of AI applications against COVID-19. The study overviews several techniques and methods that can be applied to various types of medical information-based pandemic. Specifically, the study classifies the existing AI techniques in clinical data analysis, including neural systems, classical SVM, and edge significant learning. Also, an emphasis has been made on regions that utilize AI-oriented cloud computing in combating various viruses similar to COVID-19.
Differently from other existing surveys on the subject, this paper proposes a perspective from a point of view of applications for outbreak forecasting and spread tracking, framing the problem as a prediction problem by looking at the history of infections, deaths, recovery and other information to predict the future diffusion of the pandemics by means of AI techniques. Since we focus on a very specific area, the review presents a high level overview of the current research, which is sufficiently detailed to provide an informed insight. Conversely, since the above cited reviews address a broader scope, they do not discuss in detail the different approaches in the literature, but provide only an overview of the leading ones.
Nevertheless, as highlighted from all the above reviews, the development of AI-based models to forecast, diagnose and predict COVID-19 infections is still an open research problem. This motivated us to conduct a very specific review study of the current approaches proposed to forecast and tracking the spread and evolution of COVID-19.
Before giving a detailed picture of the papers reviewed in this survey, we provide a comprehensive description of the AI techniques adopted by researches to deal with COVID-19 pandemic and the evaluation indexes used to assess the quality of the results they obtained.
3. AI techniques to predict COVID-19 outbreak
Artificial intelligence (AI) is a computer science research field, coined by the American scientist John MacCarthy in 1956, whose aim is to build computers that imitate human intelligence when performing tasks. One of the primary goals of AI is learning. Machine Learning (ML) and Deep Learning (DL) are two of the main learning areas in AI.
ML is the use and development of algorithms that are able to learn and adapt automatically through experience and by the use of data, algorithms and statistical models to analyze and draw inferences from patterns in data. Supervised learning algorithms, in particular, build a model based on sample data, known as training data, in order to make predictions or decisions without being explicitly programmed to do so.
Deep learning is part of a broader family of learning methods based on artificial neural networks in which multiple layers of processing are used to extract progressively higher level features from data. Deep-learning architectures include deep neural networks, deep belief networks, deep reinforcement learning, recurrent neural networks and convolutional neural networks.
In recent years AI technology has been receiving a lot of interest in many application fields, including medicine to assist physicians and authorities in image inspection, surgery, medical data integration, hospital management, disease-assisted diagnosis, to name a few.
In the following, we recall and describe the main AI learning techniques used by researchers to forecast the propagation of the coronavirus infection and its effects on new cases, recoveries, deaths, and diagnosis. A summary of the described approaches, along with the acronym used for denoting them, is reported in Table 1 .
Table 1.
Summary of the described AI techniques and their abbreviation.
| Method | Abbreviation | Description | |
|---|---|---|---|
| Machine Learning | Statistics | LR | Linear Regression |
| MLR | Multiple Linear Regression | ||
| PR | Polinomial Regression | ||
| LoR | Logistic Regression | ||
| LASSO | Least Absolute Shrinkage and Selection Operator | ||
| Time Series | WMA | Weighted Moving Average | |
| ES | Exponential smoothing | ||
| AR | Autoregressive process | ||
| MA | Moving Average | ||
| ARMA | AutoRegressive Moving Average | ||
| ARIMA | AutoRegressive Integrated Moving Average | ||
| Prophet | Modular regression model developed a Facebook | ||
| Classification | SVM | Support Vector Machine | |
| LS-SVM | Least Square Support Vector Machine | ||
| SVR | Support vector regression | ||
| NB | Naive Bayes | ||
| EL | Ensemble Learning | ||
| XGB | Extreme Gradient Boosting | ||
| HMM | Hidden Markov | ||
| IBL | Instance-Based Learning | ||
| KNN | K-Nearest Neighbor | ||
| DT | Decision trees | ||
| CR | Classification via Regression | ||
| RF | Random Forest | ||
| Extra Trees | Extremely Randomized Trees | ||
| Deep Learning | Artificial Neural Netwoks | RVFL | Random Vector Functional Link Network |
| RNNs | Recurrent Neural Networks | ||
| DNNs | Deep Neural Networks | ||
| LSTMs | Long Short-Term Memory Networks | ||
| BiLSTMs | Bidirectional Long short-term Memory Networks | ||
| SLSTMs | Stacked Long Short-Term Memory Networks | ||
| ConvLSTM | Convolutional LSTM | ||
| GRU | Gate Recurrent Unit | ||
| CNN | Convolutional Neural Network | ||
| GAN | Generative Adversarial Network | ||
| VAE | Variational Autoencoder |
3.1. Regression
Regression analysis [18] is a supervised learning technique based on statistical concepts which allows to estimate the relationships between a dependent variable and one or more independent variables and to model the future relationship between them.
3.1.1. Linear regression
The idea at the base of regression analysis for forecasting a time series Y is that there is a linear relationship with other time series X. Y is called regressand, forecast or dependent variable, while X the regressors, predictors or independent variables. In the simplest case, the forecast variable has a linear relationship with a single variable:
| (1) |
The Eq. (1) represents a straight line where β 0 is the Y-intercept, β 1 is the slope, called the regression coefficient, and ε is the error term. The goal of the prediction is thus to find the values of the coefficients β i to obtain the best-fit regression line. The extension to the multivariable regression model is obtained as
| (2) |
with the regression coefficients β i computed for each independent variable X i. These coefficients measure the effect of each predictor by considering the effects of all the other predictors. In order to build a model, the regression coefficients must be estimated. The least square principle allows to choose the values of the coefficients by minimizing the sum of squared errors:
| (3) |
Fitting (or training or learning) the models then means finding the best estimates of the regression coefficients which minimize the sum of squared errors. The prediction of Y can thus be obtained by substituting the estimated coefficients through Eq. (3) in the Eq. (2) by setting ε = 0, i.e.
| (4) |
3.1.2. LASSO and RIDGE regression
LASSO (Least Absolute Shrinkage and Selection Operator) and RIDGE regression are regression methods that use regularization techniques for obtaining more accurate predictions. Regularization is a technique that reduces overfitting when data has high variation. To achieve less variance on the test data, a penalty term is added to the best fit obtained from the training set and compresses the coefficients of the predictor variables to reduce their influence on the output variable. Thus, the number of variables is the same but the magnitude of their coefficients is reduced.
The LASSO regression applies a shrinkage technique that shrinks the extreme values of the features towards the central values. LASSO considers one feature at a time and uses it only if it improves the fit. The process penalizes such features by setting their coefficients to a very small value, potentially equal to zero. Thus the important features are automatically selected. The method optimizes the following equation
| (5) |
where λ is the amount of shrinkage. When λ = 0 all the features are considered and the LASSO reduces to the linear regression that uses the residual sum of squares to build the predictive model.
The RIDGE regression differs from the LASSO one in the penalty function which considers the square of the absolute values of the coefficients:
| (6) |
3.1.3. Logistic regression
Logistic regression is a predictive analysis technique used when the dependent variable is binary, like presence/absent, yes/no. Consider the simplest case with two predictors, X 1 and X 2, and a binary variable Y. Let p denote the probability that Y = 1 (p = P(Y = 1)). It is assumed a linear relationship between the predictor variables and the log-odds (also called logit) of the event that Y = 1. In statistics, the logit function is the logarithm of the odds (a measure of the likelihood of a particular outcome) of the result . This relationship can be written as:
| (7) |
by exponentiating the log-odds we obtain
| (8) |
and applying algebraic manipulations the probability that Y = 1 is given as
| (9) |
Thus, if the coefficients are fixed, it is possible to compute the outcome of the Y variable.
3.2. Time series prediction
A time series [19], [20] is defined as a collection of data observed sequentially over time. A time series is modeled as a sequence of random variables Y = {Y t : t ∈ T}, with T an index set. Y is called stochastic process and it is assumed to satisfy the assumption of stationarity, i.e. the probability laws of the process do not change over time.
Time series analysis aims to model the stochastic mechanism that generates the observed series and to forecast the future values of the series on the base of the known history of that series. Often, a time series is decomposed into three components: the trend, which considers the variable movements without taking into account seasonality or irregularities; the seasonality, i.e. the periodic fluctuation of the variables; the residual, which is the unexplainable part of the time series. Moreover, time series can be univariate and multivariate. The former contains a single observation stored sequentially over time, the latter are used when more variables and their interactions are considered.
3.2.1. Weighted Moving Average (WMA)
A general linear process, {Y t}, is a weighted linear combination
where e t are independent random variables with zero-mean. ψ j are weights assumed to form an exponentially decaying sequence ψ j = ϕ j, where −1 ≤ ϕ ≤ 1. Then
| (10) |
If the number of weights is finite, the process is called a moving average process, and it is denoted as
| (11) |
This series is called a moving average of order q and it is abbreviated to MA(q).
3.2.2. Autoregressive process (AR)
An autoregressive process obtains the current value of the series Y t by using its past values. More in detail, a p-order auto regressive process Y t is obtained as a linear combination of the most recent past p values plus a new term, thus is satisfies the following equation:
| (12) |
where e t is assumed to be independent of the past Y t, for every t.
3.2.3. AutoRegressive Moving Average (ARMA)
If the series is partly autoregressive and partly moving average, we obtain a mixed Autoregressive Moving Average Model satisfying the equation:
| (13) |
{Y t} is called a mixed autoregressive moving average process of orders p and q and denoted as ARMA(p, q).
3.2.4. AutoRegressive Integrated Moving Average (ARIMA)
The above models assume stationarity, i.e. the process has a deterministic trend that will persist in the future. However, in many applications such an assumption is not realistic, and time series are non-stationary, thus do not have a constant mean over time.
A time series {Y t} is said to follow an integrated autoregressive moving average model if the d-th difference {W t} of {Y t} is a stationary ARMA process. If {W t} follows an ARMA(p, q) model, {Y t} is said an ARIMA(p, d, q) process [21]. For practical uses, d = 1 or at most d = 2 are adopted. When d = 1, {W t = Y t − Y t−1} and ARIMA(p, 1, q) can be expressed as:
| (14) |
The ARIMA(p, d, q) model is thus an extension of the ARMA (p, q) model which combines the Auto-Regressive (AR(p)) and the Moving Average (MA(q)) time series models with a differencing parameter d used to convert a non-stationary time series into a stationary series.
3.2.5. Exponential smoothing
Exponential smoothing is a time series forecasting method which, differently from the moving average family, assigns exponentially decreasing weights over time to the past observations. The simplest form of exponential smoothing forecasts the current value of Y t as
where α is called the smoothing factor, with 0 ≤ α ≤ 1, e t is the actual value, and Y t−1 is the previous forecast value.
3.2.6. Prophet
Prophet is a method developed at Facebook by Taylor and Letham [22], available as open source software in Python and R. It is based on a modular time series model having three components, trend, seasonality, and holidays. These components are combined as the equation
where G t is the trend function modeling non-periodic changes in the time series values, S t represents periodic changes, such as weekly and yearly seasonality, and H t represents the effects of holidays. ε t is the error term due to changes that the model cannot contemplate. As the authors point out, Prophet shows some advantages with respect to other methods, such as flexibility, which allows to make different assumptions regarding trends and multiple periods of seasonality. Moreover, differently from ARIMA, measurements do not need to be regularly spaced, thus it is not necessary to interpolate missing values.
3.3. Classification Classification
Machine learning (ML) is a branch of artificial intelligence that finds the underlying relationships among data and information [23]. Arthur Samuel [24] in 1959 defined ML as the field of study that gives computers the ability to learn without being explicitly programmed. Supervised machine learning algorithms use training examples to obtain a hypothesis, named also model, that estimates a class membership, able to generalize the hypothesis on unseen data by predicting their unknown class. Models can be either deterministic or probabilistic. More formally, let and be the input and the output domains, respectively. A deterministic model is a function
| (15) |
with , and θ = {θ 1, …, θ D} a set of real parameters.
A probabilistic model assumes that data input and output are random variables drawn from a probabilistic distribution p(x, y), which is the ground truth. A model distribution, which approximates the ground truth, is built from the data. It is then possible to compute the probability of a class label given an input p(y| x). This procedure is called marginalization. A probabilistic model refers to either discriminative model distribution
| (16) |
or generative model distribution
| (17) |
over the data. A generative model obtains the distribution from the dataset.
3.3.1. Naive Bayes (NB)
Bayesian Learning[25] is as very popular approach to learning based on the famous Bayes rule:
| (18) |
where a and b are random variables and p(a| b) is conditional probability of a given b, defined as
| (19) |
p(a, b) is the probability that both a and b occur. The term p(b| a) is called the likelihood, p(a) the prior, and p(a| b) the posterior. In the machine learning context, given a training set D with m examples, the input x and the output y random variables, the aim is to find a probabilistic model p(x, y| D) which produces the data. It is possible to apply the Bayes rule by replacing y by the unknown parameters θ. Thus we get:
| (20) |
where p(D| θ) is the likelihood of parameters θ, p(θ) is the prior probability of θ, and p(θ| D) is the posterior of θ given data D.
3.3.2. Support Vector Machine (SVM)
Support Vector Machine (SVM) is a classification technique introduced by Boser et al. [26] that maximizes the margin between the training data and the decision boundary. SVM solves a binary classification problem by using the concept of separation hyperplane and finding the maximum separation margin that correctly classifies the training data as much as possible. The optimal hyperplane is represented with the support vectors.
One of the main characteristics of SVM is the use of the so-called kernel trick [27]. Since often data is not linearly separable in the original input space, data is mapped into a higher-dimensional space by using a kernel function ϕ. In this new space a linear separator is able to better discriminate between the different classes.
Given a training set D = {(x m, y m)}m=1 M, with x m ∈ R n, y m ∈ {−1, 1}, an SVM classifier satisfies the inequalities:
| (21) |
where ϕ is a kernel function. Several kernel functions can be used for the mapping, such as linear, polynomial, Gaussian, exponential, and Sigmoid. Changing the kernel, allows to build new models. SVM has been shown to be one of the most powerful classifiers in machine learning.
3.3.3. Least Square Support Vector Machine (LS-SVM)
Least Square Support Vector Machine is a variation of SVM introduced by Suykens and Vandewalle [28] which solves a set of linear equations instead of the inequalities (21). The main advantage of this formulation of SVM is the higher efficiency since it transforms the task of solving a complex quadratic program to that of finding a solution of a set of linear equations.
3.3.4. Support Vector Regression (SVR)
SVM can be used also to deal with regression problems. As described in Section 3.1, in a regression problem the model returns a continuous-valued output instead of a set of discrete values, thus regression is a generalization of the classification problem.
Support Vector Regression is an extension of SVM which introduces a region, named tube, around the function to optimize with the aim of finding the tube that best approximates the continuous-valued function, while minimizing the prediction error, that is, the difference between the predicted and the true class label. SVR uses an ε-insensitive loss function which penalizes predictions farther than ε from the desired output. Different loss functions can be used, such as linear or quadratic. The value of ε determines the width of the tube.
3.3.5. Instance-based learning
Instance-based learning (IBL) is a group of algorithms that build an hypothesis directly from the training instances, and perform generalization by comparing a new instance with instances seen in training, already stored in memory. These algorithms are referred to as lazy, since computation is postponed until a new instance is observed. An example of IBL classifier is the K-Nearest Neighbor (KNN), which, in order to assign an instance to a class, computes the similarity between the current instance and the k nearest training instances.
3.3.6. Decision Trees (DTs)
Decision Trees (DTs)[29] is one of the most known classification method which predicts the class label of unknown instances after generating a tree from a set of training examples. The nodes of the tree are the attributes of the training set and a branch from a node corresponds to one of the possible values of that attribute. A new instance is classified by starting from the root of the tree, testing the value of its attributes, and following the branch down along the tree having the same attribute value of that example.
Classification via Regression (CR) is a variant of a decision tree classifier proposed by Frank et al. [30], which has linear regression functions at the leaves of the tree.
3.4. Ensemble Learning
Ensemble Learning is a machine learning methodology which uses multiple learning methods, named weak learners or base models, to improve the predictive capability of each constituent learning algorithm. There are two main ensemble strategies: bagging [31] and boosting [32]. In bagging (bootstrap aggregating) the models have equal weights and are trained on bootstrap samples of the same training set size, i.e. examples are randomly chosen by allowing replacement, meaning that an example can occur multiple times.
Random Forests (RFs) is a bagging ensemble learning method that generates several decision trees during the training phase and returns as result the mean prediction of the individual trees. Extremely Randomized Trees, also referred as extra trees, is another ensemble method that changes the tree generation by introducing more variation, such as tree depth.
In boosting weak learners are sequentially combined in an adaptive way, i. e. each model gives more importance to the misclassified examples by assigning lower weights to correctly classified examples and higher weights to examples difficult to classify. AdaBoost [33] is the most known boosting method.
Gradient Boosting (GB) is an ensemble method that builds weak learners by optimizing a suitable cost function. XGBoost is an efficient implementation of Gradient Boosting which obtains more accurate predictions.
Stacking ensemble is a variation of ensemble learning whose main characteristic is the combination of different types of weak learners.
3.5. Artificial Neural Networks (ANNs)
Artificial Neural Networks have been proposed since 1940s as a simplified model of the human brain. However, it was only in 2006, after the paper of Hinton et al. [34] proposing the deep neural networks (DNNs), that the research in the field propagated very fast. Let w be a vector containing the parameters and x the input, ANNs can be mathematically considered as a nonlinear regression model f(x) = φ(w, x), where φ is a nonlinear model function.
Perceptrons are the basic units of ANNs. Their model function is computed as
| (22) |
The nonlinear function φ is called activation function. Training the perception model is done by updating the weights as follows:
| (23) |
where η is the learning rate. The perception model, however, is not able to deal with nonlinearly separable functions, thus since its definition in early 1960s, several extensions have been proposed.
Feed-Forward Neural Networks where one of the first ones, with a model function of the form
| (24) |
where φ 1, φ 2, … are nonlinear activation functions, W i are weight matrices containing the parameters, and the points indicate that there can be an arbitrary number of nested functions. Popular activation functions are the Rectified Linear Unit ReLU φ(x) = max (0, x), the hyperbolic tangent, the sigmoid .
For a labeled training D = {(x m, y m)}m=1 M, the square loss for a single hidden layer is computed as
| (25) |
Random Vector Functional Link Netwok (RVFL) is a special single hidden layer feed-forward neural network proposed by Pao et al. [35],where the output weights are chosen as an adaptable parameter.
3.5.1. Recurrent neural networks (RNNs)
Feed-forward neural networks are organized in layers where information is fed forward through layers. A recurrent neural network is represented as a graph of units, all connected to each other, which are updated in discrete time steps at the same time. Thus the input is interpreted as the set of units at time t = 0 while the output is given by some units at time T. The hidden units have the role of computational function. The state of the G hidden units can be described as h t = (h 1 t, …, h G t)T, and after each update, it is given by:
| (26) |
where W is the G × G matrix containing the weights and φ is a nonlinear activation function.
3.5.2. Long Short-term Memory Networks (LSTMs) and variants
RNN are particularly apt to analyze sequential data and thus for temporal forecasting applications. However training could be challenging because of the problem of exploding and vanishing gradients and they are unable to model long term dependencies [36]. Long Short-term Memory networks [37] tries to address these problems by introducing the concepts of memory cells c t and new gate units, i.e. the input gate i t, the output gate o t, and the forget gate f t. The forget gate decides what can be propagated from the previous memory units, the input gate which information must be accepted, the output gate generates the new long-term memory. Given the input sequence x t and the number h of hidden units, the gates are defined as follows:
-
•
input gate: i t = σ(x t W xi + H t−1 W hi + b i)
-
•
forget gate: f t = σ(x t W xf + H t−1 W hf + b f)
-
•
output gate: o t = σ(x t W xo + H t−1 W ho + b o)
-
•
intermediate cell state:
-
•
cell state
-
•
new state: h t = o t ∘ tanh (c t)
where ∘ is the element wise multiplication, W xi, W xf, W xo, W hi, W hf, W ho are the weight parameters, and b i, b f, b o the bias parameters. The sigmoid σ and tangent functions tanh are the activation functions.
The bi-directional LSTM (BiLSTM) is an extension of LSTM which takes into account not only the backward context, but also the forward one.
The Gate Recurrent Unit (GRU) [38] model improves the LSTM performance by reducing the number of LSTM parameters and by merging the input and forget gates from the LSTM model.
Stacked LSTM[39] is another extension of LSTM, also known as multilayer fully connected structure, which combines multiple LSTM layers, where each intermediate layer output is used as an input for next LSTM layer. Stacked LSTM gives the output for each time stamp and not a unique output for all time stamps.
3.5.3. Convolutional Neural Networks (CNNs)
Convolutional Neural Network[40] is a feed-forward network using three main layers: the convolutional and the pooling, which are used to reduce the complexity, and the fully connected layer, which is a flattened layer connected to the output. The term convolutional comes from the mathematical convolution operation, which, given two functions, produces a new function providing how the shape of one is modified by the other. In can be considered as a specialized type of linear operator. Convolutional operation is used in place of matrix multiplication.
Generative Adversarial Network GAN is a learning model composed of two neural networks, the generative network which generates candidate solutions, and the discriminative network which evaluates them.
An autoencoder is an artificial neural network which learns efficient codings, generally using dimensionality reduction techniques, of unlabeled data. The encoding is evaluated and improved by trying to regenerate the input from the encoding. The model is trained with the objective of minimizing the error between the encoded-decoded data and the original data.
A Variational Autoencoder (VAE) is an autoencoder which regularizes the training to avoid overfitting and improves the generative process.
3.6. Evaluation metrics
In this section we summarize the evaluation indexes adopted in the described papers for assessing the quality of the results obtained by the approaches. The main evaluation metrics are reported in Table 2 , along with their mathematical equation defining them.
Table 2.
Evaluation metrics for assessing the results of the reviewed algorithms.
| Evaluation Metric | Equation |
|---|---|
| Mean Absolute Error (MAE) | MAE= |
| Normalized Absolute Error (NAE) | NAE= |
| Maximum Difference (MD) | MD= |
| Mean Square Error (MSE) | MSE= |
| Laplacian Mean Square Error (LMSE) | LMSE= |
| Mean Absolute Percentage Error (MAPE) | MAPE= |
| Structural Content (SC) | SC= |
| Peak Signal to Noise Ratio | PSNR = 10 log 10 |
| Relative Mean Bias Error (rMBE) | rMBE= |
| Root Mean Square Error (RMSE) | RMSE= |
| Root Mean Square Log Error (RMSLE) | RMSLE= |
| relative Root Mean Square Error (rRMSE) | rRMSE= |
| Coefficient of determination R squared | |
| Adjusted Coefficient of determination R squared | |
| Accuracy (Acc) | |
| Precision (P) or Positive Predictive Value (PPV) | |
| Recall (R) or Sensitivity | |
| F-measure | |
| Specificity (SPC) | |
| Negative Predictive Value (NPV) | |
| Matthews Correlation Coefficient (MCC) |
Let y t, t = 1, …, T, be the actual values of a measurement, their mean value, the predicted values, k the number of regressors, and T the number of measurements. Moreover, let TP denote the true positive cases, i.e. the number of persons that truly have COVID-19 infection, FP the false positive cases, i.e. the number of persons that don't have the infection, but a classifier mistakenly identified them as infected, TN the true negative cases, i.e. the number of persons that don't have COVID-19, and the classifier correctly identified them, FN the false negative cases, i.e. the number of persons that have COVID-19, but the classifier did not identify them.
The Receiving Operating Characteristics (ROC) is a graphical representation of the ability of a binary classifier when a discrimination threshold is varied. The ROC curve plots the true positive rates against increasing values of the false positive rate. The Area Under the Curve (AUC) measures the performance of the binary classifier. Its value ranges between 0.5 and 1, where 0.5 means that the classifier behaves like a random classifier, while 1 that it is perfect, i.e. its error rate is zero.
4. Material and method
This study aimed at providing a comprehensive review of methods, algorithms, applications, and emerging technologies that can be utilized for forecasting, monitoring, diagnosing, and tracking COVID-19.
Given the fast-moving nature of the epidemics, we attempted to be comprehensive in the literature coverage. For this reason, many of the articles cited are still preprints at the time of writing. The review was guided by the procedures stated by [41] namely; search strategy, study selection (inclusion/exclusion criteria), study eligibility, and quality assessment. The review of literature is carried out on databases of ScienceDirect (SD), IEEE Xplore, Web of Science (WoS), Google Scholar, Scopus, PubMed, ACM Digital Library, arXiv and medRxiv. The search has been conducted using keywords related to the detection and prediction of COVID-19 under the concept of AI like: Coronavirus, artificial intelligence, machine learning, deep learning, COVID-19, forecasting, prediction, tracking, spreading, time-series prediction.
A comprehensive literature search was conducted in the above mentioned databases for English language papers published from February 2020 to March 2021. We selected peer-reviewed articles, both journal and conference papers, and pre-prints. These articles were further screened based on title and abstract to check their compatibility with the targeted topics. The main focus of the search was on systems, algorithms, methods and techniques for the forecasting of COVID-19 spread. The applications mainly target on the detection, diagnosis, classification and prediction of COVID-19 cases, in terms of daily new cases, number of deaths, number of recovered. We excluded incomplete articles, and application papers with limited achieved results. The relevant articles were subjected to a full reading process for collecting and extracting relevant research publications for the review. As result of the search, we collected the latest research about forecasting of COVID-19 exploiting artificial intelligence methods.
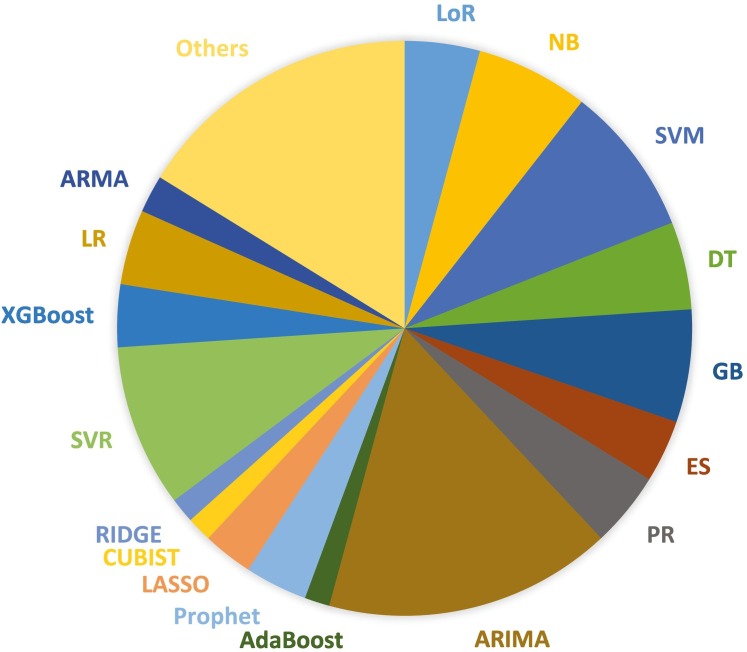
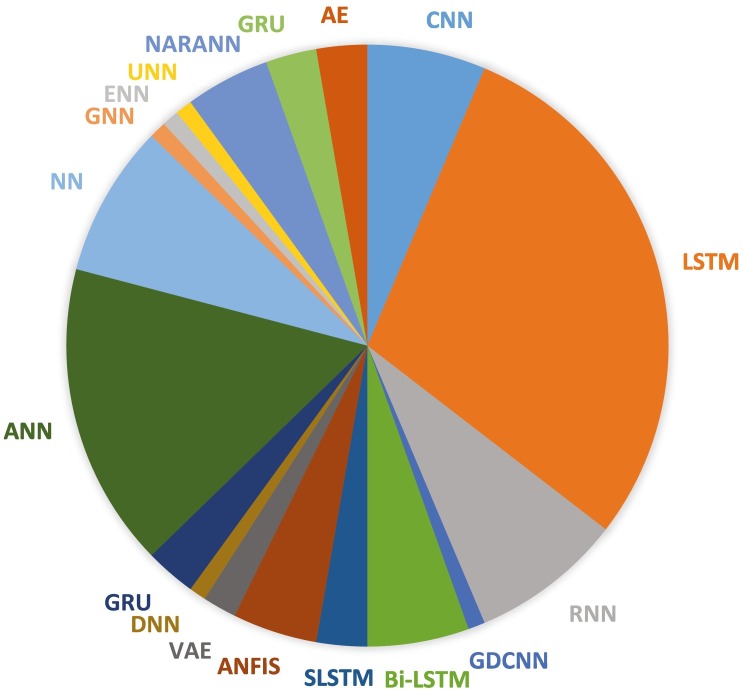
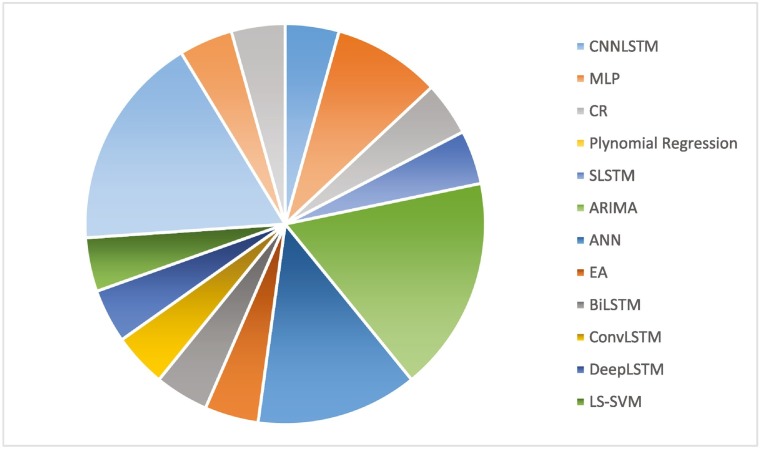
After applying the inclusion as well as exclusion procedures, a total number of 146 papers have been considered for the final study. Different techniques of ML and DL have been used in the papers: 61% of the techniques are ML related, and 39% are DL related. The graphs in Fig. 1, Fig. 2 show the specific ML or DL methods used, respectively. Fig. 1 highlights that ARIMA is the most used technique with a percentage of 13%, followed by SVR (8%), SVM (7%), Naive Bayes and MLP (5%). Fig. 2 shows that LSTM is the most used deep learning method with a percentage of 29%, followed by ANN with 16%, RNN and NN with 8%, CNN 6%, and Bi-LSTM 5%.
Fig. 1.
ML techniques used for COVID-19 forecasting.
Fig. 2.
DL techniques used for COVID-19 forecasting.
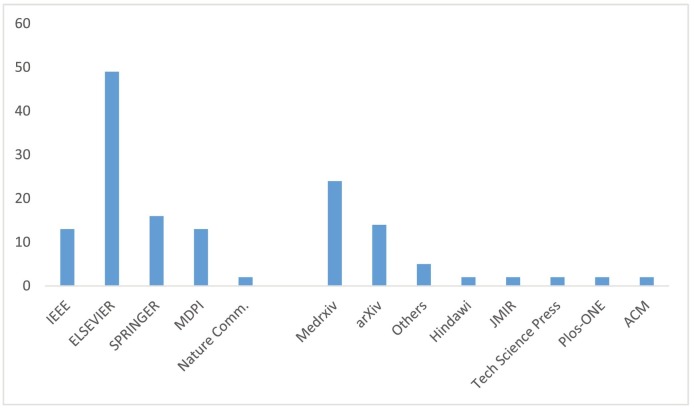
The selected papers are published by different editors like IEEE, Elsevier, Springer, MDPI, and some others, while for preprints Medrxiv and arXiv are considered; 26% of the papers are preprints and 74% are peer reviewed articles and conference proceedings. Fig. 3 shows that Elsevier largely surpasses all others with 33% of publications, followed by Springer with 10% and IEEE with 8%.
Fig. 3.
Editor distribution of the publications.
Of the 146 papers selected, we removed preprints and chose the most significant and representative for each of the main ML and DL techniques. In total 38 papers have been selected to undergo a more in-depth study. An analytical review of those 38 papers is reported in Section 5 and summarized in Table 3 . Papers have been categorized on the basis of the AI method they used and described in the appropriate section according to the classification. However, some of the papers implemented more AI methods, thus, the classification includes the different methods used. In Section 5.5 the remaining 146 papers, either preprint at the time of writing, or reporting experimentations on few available data at the publication time, are overviewed.
5. Literature review
In this section we review the works in the literature discussing models, methods and applications of machine learning or deep learning techniques for COVID-19 forecasting and tracking, selected following the procedures discussed in Section 4. Differently from previous reviews regarding COVID-19, the one proposed in this paper focuses on a very specific topic, that is COVID-19 forecasting exploiting DL and ML methods. Accordingly, data extraction and classification of the selected studies, were conducted to evaluate the efficacy of the approaches in terms of COVID-19 detection, diagnosis, forecasting and spreading throughout AI enhancements, such as learning, regression and prediction. In particular, a detailed analysis of the 38 selected papers is provided throughout the section, grouping the different works according to the AI category employed. For each study in the literature, we extracted the most important features like the method implemented, the data type and size used, the evaluation methods adopted, the accuracy for each method, the results achieved. For each study, the features are summarized in Table 3, while in Section 5.5 the rest of the 146 papers, including preprints, are overviewed.
5.1. Time series methods
Kumar et al. [42] present an evaluation study for predicting COVID-19 cases in the 10 counties which had the higher number of infected people in the early 2020, namely US, Spain, Italy, France, Germany, Russia, Iran, United Kingdom, Turkey, and India. The authors collected the reported daily COVID-19 confirmed, recovered, death, and active cases for these 10 countries from March 1st until May 20th, downloading them from the COVID-19 data repository managed by the Johns Hopkins University Center for Systems Science and Engineering (JHU CSSE) [43]. The authors considered two times series forecasting models, ARIMA [21] and Prophet [22], to obtain predictions and evaluated the quality of the results by using statistical measures. The results showed that ARIMA obtains better performance than Prophet, for most of the countries. For instance, the MAPE value of the active cases in Iran is 2% while that of Prophet is 82%. However, the main problem of ARIMA is that the authors used a different value for the order of the autoregression, i.e. the number of previous days necessary for finding the parameters.
Singh et al. [44] performed an experimental study to predict the daily cases of infections for five countries, namely Italy, Spain, France, United Kingdom, United States of America, by applying as prediction models the autoregressive integrated moving average (ARIMA) and the least square support vector machine (LS-SVM). The prediction results were validated by comparing them with the true confirmed data and computing the mean absolute error (MAE), the mean square error (MSE), the root mean square error (RMSE), and the coefficient of determination (R 2). The results showed that the values obtained by using the LS-SVM model are much better than those obtained by applying ARIMA.
Wang et at. [45] propose to combine the Logistic model of population growth and the Prophet model with the aim of improving the long-term prediction capability of the time series model in order to obtain a reliable epidemic curve and trend of the epidemic. The authors consider epidemiological data from January until June 16, 2020, and present results for global countries, Brazil, Russia, India, Peru and Indonesia.
The logistic growth forecasting model is based on the following equation:
| (27) |
where Q is the population size, r the intrinsic growth rate, and K the maximum population size that the environment could carry. dQ/dt represents the growth of the population. r and K are constants, while the value of Q follows a typical S-shaped curve with a rapid increase at the beginning reaching a maximum value, denoted as cap. This cap marks a critical point after which the disease transmission begins to decline. The cap value computed by the logistic model is given as input to Prophet to obtain the epidemic curve and predict the epidemic trend. Prediction results are reported until December 2020. However, it worth pointing out that by comparing the prediction of cumulated cases with the actual cumulated cases of infections, the approach sensibly underestimates the true value.
In Hernandez et al. [46] an algorithm to perform and evaluate the ARIMA model for 145 countries, distributed into 6 continents, is proposed. The authors construct a model for these continents using the ARIMA parameters, the population per 1 M people, the number of cases, and polynomial functions. The time series start on the day when each country presented the first case of COVID-19 and finish on May 28. The proposal is able to predict the COVID-19 cases with a RMSE average of 144.81. The main outcome of this paper is showing a relation between COVID-19 behavior and population in a continent, pointing out the opportunity to create more models to predict the COVID-19 behavior using variables as humidity, climate, culture, among others.
5.2. Deep learning methods
Shahid et al. [47] evaluated the capability of times series and deep learning methods in predicting the number of confirmed, deaths, and recovered cases in ten countries, namely Brazil, Germany, Italy, Spain, UK, China, India, Israel, Russia, and USA. The authors used autoregressive integrated moving average (ARIMA), support vector regression (SVR), long short term memory (LSTM), bidirectional long short term memory (Bi-LSTM), gated recurrent unit (GRU). The performance of the models has been measured by computing the mean absolute error MAE, root mean square error RMSE, and R 2 score. The dataset used for the experiments consists of the number of confirmed, deaths and recovered cases of 158 samples in the period January 22nd until June 27th, 2020. The 110 cases from 1/22/2020 to 5/10/2020 have been used for training the models, and to predict the 48 cases from 5/11/2020 to 6/27/2020. The experimentation results showed that the Bi-LSTM model outperforms, in the majority of cases, all the other methods. This method showed to be more robust and to obtain higher prediction accuracy. The ranking of the models, in decreasing order of performance, is Bi-LSTM, LSTM, GRU, SVR and ARIMA.
In [48] Devaraj et al. aim to predict the cumulative confirmed, death and recovered global cases by using different models with Auto-Regressive Integrated Moving Average (ARIMA), Long Short-Term Memory (LSTM), Stacked Long Short-Term Memory (SLSTM) and Prophet approaches. For long-term forecasting of COVID-19 cases, multivariate LSTM models is employed. The performance metrics are computed for all the models and the prediction results are subjected to comparative analysis to identify the most reliable model. From the results, it is evident that the Stacked LSTM algorithm yields higher accuracy with an error of less than 2% as compared to the other considered algorithms for the studied performance metrics. Country-specific analysis and city-specific analysis of COVID-19 cases for India and Chennai, respectively, are predicted and analyzed in detail.
Shastri et al. [49] present a comparative analysis of deep learning methods to predict COVID-19 cases for one month ahead in USA and India. The DL methods used for the experimentation are Stacked LSTM, Bi-directional LSTM and Convolutional LSTM. The datasets of confirmed and death cases of COVID-19 taken into consideration ranges from February 7th till July 7th 2020. The experiments showed that Convolution LSTM outperforms the other two models in predicting the COVID-19 cases. In fact, the values obtained by Convolution LSTM of precision, recall, and f-measure are higher than those returned by Stacked LSTM, Bi-directional LSTM, while the MAPE error is lower.
Zeroual et al. [50] present a comparative evaluation of deep learning methods for predicting the number of new and recovered cases. The methods used for the experimentation are Recurrent Neural Network (RNN), Long short-term memory (LSTM), Bidirectional LSTM (BiLSTM), Gated recurrent units (GRUs) and Variational AutoEncoder (VAE). The study considered the number of daily confirmed and recovered cases coming from Italy, Spain, France, China, USA, and Australia in the period January 22nd till June 17th, 2020. The methods have been trained with data until May 31st, and then testing has been performed on the next 17 days. The accuracy of the models has been measured by computing RMSE, MAE, MAPE, and RMSLE. Results showed the better performance of the VAE compared to the other algorithms.
In [51] Alakus et al. developed an application to predict COVID-19 exploiting laboratory findings and using six different deep learning models, like: Artificial Neural Network (ANN), Convolutional Neural Networks (CNN), Long-Short Term Memory (LSTM), Recurrent Neural Networks (RNN), CNN-LSTM, and CNN-RNN. For the experimental evaluation a dataset of 600 patients and 18 laboratory findings from the Hospital Israelita Albert Einstein at Sao Paulo Brazil has been used. Performance of the models is measured with accuracy, precision, recall, AUC, and F1-scores. To validate the models, 10 fold cross-validation has been applied. Best results are observed from the LSTM deep learning model with accuracy of 86.66%, recall of 99.42%, and AUC score of 62.50%. All the deep learning models experimented in the study showed an accuracy of over 84%. Similar inferences can be made for precision and recall values.
Farooq et al. [52] proposed an Artificial Neural Network (ANN) based real-time online incremental learning technique to estimate the parameters of a data stream guided analytical model of COVID-19 based on traditional epidemiological model. The COVID-19 data from India has been taken as the case study, during the period 30th January 2020–13 June 2020, with a total number of cases reported in India equals to 308,993. Using this model, authors simulated preventive measures like lockdown, vaccination and herd immunity to study their impact on the evolution of COVID-19 disease. Finally, they proposed a method to reduce the number of deaths caused by the pandemic in case a vaccine is not available at the mass level. The impact of this strategy has been simulated and it has been shown that the number of deaths can be reduced from 55 million to 1.3 million if the population compartmentalization starts tomorrow and ends on day 300 of the pandemic in India. During this period, the mobility and contact in low risk group have to be made five times as compared to the lockdown period and upon remixing of the two groups the mobility and contact should be reduced to 2 times from 5.
Gupta et al. [53] proposed a model based on a deep learning algorithm with two long short-term memory (LSTM) layers for predicting the number of confirmed and death cases of COVID-19. The paper considers the available infection cases of COVID-19 in India from January 22, 2020, till October 9, 2020. The model predicts coronavirus cases and deaths for the next 30 days, taking the data of the previous 260 days of duration of the pandemic. It has been compared with other popular prediction methods (Support Vector Machine, Decision Tree and Random Forest) showing a lower normalized RMSE. This work also compares COVID-19 with other previous diseases (SARS, MERS, H1N1, Ebola, and 2019-nCoV). Based on the mortality rate and virus spread, this study concludes that the novel coronavirus (COVID-19) is more dangerous than other diseases.
Meng et a. [54] developed a prognostic tool to identify high-risk patients and assist in the formulation of treatment plans. They retrospectively collected 366 severe or critical COVID-19 patients from four centers, including 70 patients who died within 14 days (labeled as high-risk patients) since their initial CT scan and 296 who survived more than 14 days or were cured (labeled as low-risk patients). They developed a 3D densely connected convolutional neural network (termed De-COVID19-Net) to predict the probability of COVID-19 patients belonging to the high-risk or low-risk group, combining CT and clinical information. The area under the curve (AUC) is 0.952 (95% confidence interval, 0.928–0.977) on the training set and 0.943 (0.904–0.981) on the test set. The Kaplan-Meier analysis revealed that the model could significantly categorize patients into high-risk and low-risk groups.
Hu [55] proposed a modified stacked auto-encoder for modeling the transmission dynamics of the epidemics in China. In [56], Rizk et al. proposed an improved Multi-layer Feed-forward Neural Network (ISACL-MFNN) model, which uses an improved Interior Search Algorithm (ISA) to optimize model parameters and a Chaotic Learning (CL) strategy to enhance ISA performance. From the official COVID-19 data set reported by the WHO, data from January 22, 2020, to April 3, 2020, in the United States, Italy, and Spain were collected to train the ISACL-MFNN model and to predict the confirmed cases within the next 10 days.
In Cabras et al. [57], a deep learning algorithm (LSTM) and a Bayesian Poisson-Gamma model are used to estimate the evolution of the pandemic in Spain.
5.3. Machine Learning methods
Rustam et al. [58] present a study on the capability of four machine learning methods to predict the number of newly infected cases, of deaths, and of recoveries in the upcoming 10 days. The authors for the experimentation use linear regression, LASSO regression, support vector machine, and exponential smoothing. The performance of each models has been evaluated by computing the R 2 score, R Adjusted 2 score, mean square error (MSE), mean absolute error (MAE), and root mean square error (RMSE). The data used for the training is relative the worldwide COVID-19 patients provided by Johns Hopkins University. The dataset has been preprocessed and divided into training set (85%) and testing set (15%) form January 22nd until March 27th. The results pointed out that the ES forecasting model outperforms all the others.
In [59] Chakraborty et al. proposed to use Linear Regression, Polynomial Regression and a granular computing based regression model, the Granular Box Regression (GBR), to predict the daily increase of new COVID-19 cases in India. GBR finds the relationship between independent variables and a dependent variable by using multidimensional boxes. Its objective is of surrounding the data objects with boxes as closely as possible and then use the diagonal of these boxes as linear regression lines. A comparative analysis is conducted to evaluate the performance of these three regression models on three COVID-19 Indian datasets, collected from api.covid19india.org in the period January 30 to April 19. The performance of the different models has been evaluated using R 2, Mean Absolute Error, Root Mean Square Error, and Mean Square Error values. The experimental results showed that the Polynomial regression model outperforms the other two regression models.
Pinter et al. [60] propose a hybrid machine learning approach to predict COVID-19 using data from Hungary. The hybrid machine learning method consists of a network-based fuzzy inference system (ANFIS) and a multi-layered perceptron-imperialist competitive algorithm (MLP-ICA). The methods are used to predict time series of infected individuals and mortality rate. Evaluations were conducted by computing the determination coefficient, root mean square error and mean absolute percentage error values. The dataset used for the evaluations consists of COVID-19 cases and mortality rate of Hungary from 4-March to 19-April 2020. Two scenarios were proposed. Scenario 1 considered sampling the odd days and Scenario 2 used even days for training the data. Both scenarios were used for training the two machine learning models ANFIS and MLP-ICA and to find the best set of parameters to use for predicting outbreaks on the validation samples. The validation is performed for nine days with promising results which confirms the model accuracy. MLP-ICA outperformed ANFIS.
In Ahamad et al. [61] a model that employs supervised machine learning algorithms to identify the features predicting the COVID-19 disease with high accuracy is presented. Features examined included age, gender, observation of fever, history of travel, and clinical details such as the severity of cough and incidence of lung infection. Authors applied different machine learning algorithms and found that the XGBoost algorithm performed with the highest accuracy (≥85%) in predicting and selecting features detecting the COVID-19 status, independently from the age. A statistical analysis pointed out that the most frequent and significant predictive symptoms are fever (41.1%), cough (30.3%), lung infection (13.1%) and runny nose (8.43%). The authors observe that, since a high percentage of people do not develop any symptoms, their approach could be used for diagnosing COVID-19 presence, also at the early stages of infection.
Gao et al. [62] presents a Mortality Risk Prediction Model of COVID-19, named MRPMC, exploiting patient's clinical data on admission able to predict death up to 20 days in advance. MRPMC is an ensemble model including Logistic Regression, Support Vector Machine, Gradient Boosted Decision Tree, and Neural Network. To train and validate MRPMC, the authors considered 2520 COVID-19 patients with known outcomes (discharge or death) from two affiliated hospitals of Tongji Medical College, Huazhong University of Science and Technology, including Sino-French New City Campus of Tongji Hospital (SF) and Optical Valley Campus of Tongji Hospital (OV), and The Central Hospital of Wuhan (CHWH) between January 27, 2020 and March 21, 2020. 360 patients out of the total were excluded, while the remaining 2160 COVID-19 patients were considered for the analysis. Participants from SF were randomly partitioned 50% for training and 50% for internal validation. Participants from OV and CHWH were used as two external validation sets. MRPMC outperformed the baseline models in predicting the mortality risk of COVID-19 on the SFV and CHWH groups. It achieved an area under the receiver operating characteristics (ROC) curve (AUC) of 0.9621 in identifying the of non survivors with an accuracy of 92.4% in the SFV cohort. Regarding the prediction of prognosis, MRPMC obtained an AUC of 0.9760 and an accuracy of 95.5% on the OV data, while an AUC of 0.9246 and an accuracy of 87.9% on the CHWH cohort.
Arpaci et al. [63] presented a study for COVID-19 diagnosis that implements six predictive models using six different classifiers based on 14 clinical features. This research employs machine learning classification algorithms, including Bayes classifier, logistic-regression, lazy-classifier, meta-classifier, Classification via Regression (CR), rule-learner, and decision-tree. Domain experts selected 14 attributes, out of 170, to be included in the predictive model. These features were then used to build a predictive model. In particular, the IBM SPSS statistical software platform is used to generate the descriptive statistics of the patients, while the data mining tool Weka to analyze the data and test the predictive model. This study considered 114 cases from the Taizhou hospital of Zhejiang Province in China from January 17, 2020 to February 1, 2020. The models were validated using Accuracy, TP rate, FP rate, precision, recall, F-measure, and ROC area. The results showed that the CR meta-classifier is the most accurate classifier for predicting the positive and negative COVID-19 cases with an accuracy of 84.21%.
In Assaf et al. [64] three different machine-learning models were used to predict COVID-19 patient deterioration. The authors considered Neural Networks, Random Forest, and Classification and Regression Decision Tree and compared the results obtained by the models to known predictor parameters (e.g., patients demographics, clinical data) and to the Acute Physiology and Chronic Health Disease Classification System II (APACHE II) risk prediction score, which is a measure of the disease severity for adult patients admitted to intensive care units. Among 6995 patients evaluated, 162 were hospitalized with non-severe COVID-19, of them, 25 (15.4%) patients deteriorated to critical COVID-19. Machine-learning models outperformed all other parameters, including the APACHE II score (ROC AUC of 0.92 vs. 0.79, respectively), reaching 88.0% sensitivity, 92.7% specificity and 92.0% accuracy in predicting critical COVID-19. Machine-learning models demonstrated high efficacy in predicting critical COVID-19 compared to the most efficacious available tools.
Brinati et al. [65] developed machine learning classification models using hematochemical values from blood exams drawn from 279 patients who, after being admitted to the San Raffaele Hospital (Milan, Italy) emergency room with COVID-19 symptoms. 177 patients resulted positive, whereas 102 were negative. The authors considered different machine learning classifiers: decision tree (DT), extremely randomized Trees (ET), K-nearest neighbors (KNN), Logistic Regression (LR), Naive Bayes (NB), Random Forest (RF), Support Vector Machines (SVM), three-way Random Forest classifier (TWRF), a modification of the Random Forest algorithm. Machine learning models are able to discriminate between patients who are either positive or negative to the SARS-CoV-2: their accuracy ranges between 82% and 86%, and sensitivity between 92% e 95%, so comparably well with respect to the gold standard. Authors also developed an interpretable Decision Tree model as a simple decision aid for clinician interpreting blood tests (even off-line) for COVID-19 suspect cases.
The approach in Chaurasia et al. [66] aims to predict the future spread of COVID-19 using the dataset taken from Data WHO Coronavirus COVID-19 cases and deaths-WHO-COVID- 19-global-data. Data includes confirmed cases, deaths, and recovered cases from all countries (https://data.humdata.org/dataset/coronavirus-covid-19-cases-and-deaths 2020). The data period is from January 22 to May 28, 2020. Several forecasting techniques have been implemented: naive method, which assumes that the next expected value to predict is equal to the last observed, simple average, moving average, single exponential smoothing, two variations of exponential smoothing, and ARIMA, for comparison, and compared by computing the Root mean square error score. The authors found that no method outperforms the others.
In [67], Khanday et al. classified textual clinical reports into four classes by using classical and ensemble machine learning algorithms. The data consists of clinical reports in the form of text, texts are classified into four different categories of diseases such that it can help in detecting coronavirus from earlier clinical symptoms. Data concern about 212 patients which have shown symptoms of coronavirus and other viruses. The information regarding each patient are stored in 24 attributes, i.e. patient id, offset, sex, age, finding, survival, intubated, went-icu, needed-supplemental-O2, extubated, temperature, pO2-saturation, leukocyte count, neutrophil count, lymphocyte count, view, modality, date, location, folder, filename, DOI, URL, License Clinical notes and other notes. Different supervised machine learning techniques are used for classifying the text into four different categories: COVID, SARS (severe acute respiratory syndrome), ARDS (acute respiratory distress syndrome) and both COVID, and ARDS. The classifiers are: support vector machine (SVM), multinomial Naive Bayes (MNB), logistic regression, decision tree, random forest, bagging, Adaboost and stochastic gradient boosting. Preprocessing on clinical reports by using techniques like Term frequency/inverse document frequency (TF/IDF), Bag of words (BOW) and report length, allowed to select 40 relevant features which were used by the methods to obtain the classification. Logistic regression and Multinomial Naive Bayes showed better results than other ML algorithms by having 96.2% testing accuracy, precision 94%, recall 96%, and F1 score 95%.
Abdulkareem et al. [68] propose a model based on Machine Learning (ML) and Internet of Things (IoT) for Smart Hospital Environments which could help physicians to diagnose patients with COVID-19. The dataset used is that of Alakus et al. [51], consisting of 600 patients and 18 laboratory findings from the Hospital Israelita Albert Einstein at Sao Paulo Brazil. The clinical data measured for patients are red blood cells, hemoglobin, platelets, hematocrit, aspartate transaminase, lymphocytes, monocytes, sodium, urea, basophils, creatinine, serum glucose, alanine transaminase, leukocytes, potassium, eosinophils, C reactive protein, and neutrophils. These laboratory findings are considered as the features on which the ML models are trained to classify patients as either normal or COVID-19 cases. A feature extraction process is also performed to detect the most discriminative features and improve classifier prediction. The ML models adopted for the experimentation are Naive Bayes (NB), Random Forest (RF), and Support Vector Machine (SVM). Experimentation on the benchmark dataset showed that the SVM model outperforms all the others by producing a correct diagnosis performance of up to 95%.
Ribeiro et al. [69] present an experimentation of several methods for short-term forecasting of COVID-16 cases in Brazil. The authors employ autoregressive integrated moving average (ARIMA), cubist regression (CUBIST) [70], random forest (RF), ridge regression (RIDGE), support vector regression (SVR), and stacking-ensemble learning. All these models are evaluated for time series forecasting in ten Brazilian states having high incidence of daily confirmed cases in the period February 25 till March 19, 2020, with one, three, and six-days ahead. Regarding the stacking-ensemble learning approach, Gaussian process (GP) is used as meta-learner, while the CUBIST regression, RF, RIDGE, and SVR models as base-learners. The prediction capabilities of the models has been evaluated by computing the mean absolute error, and the symmetric mean absolute percentage error. The results show that SVR and stacking-ensemble learning obtain better performance when compared to the other models. The models achieve errors in a range 0.87%–3.51%, 1.02%–5.63%, and 0.95%–6.90% in one, three, and six-days-ahead, respectively. The ranking of models, from the best to the worst regarding accuracy, in all scenarios is The ranking of the models, with respect to the obtained accuracy in decreasing order, is SVR, stacking-ensemble learning, ARIMA, CUBIST, RIDGE, and RF.
In [71] dos Santos Gomes et al. propose a new machine learning computational tool for adaptive tracking and real time forecasting of COVID-19 death cases. The approach combines Kalman filters and an interval type-2 fuzzy clustering algorithm which adopts an adaptive similarity distance mechanism. The dataset used for the experimentation, extracted from the official report by Ministry of Health of Brazil, consists of the daily deaths reports in the period ranging from 29 of February 2020 to 18 of May 2020. The method has been compared with LASSO, ARIMA and LSTM recurrent neural network, Wavelet-Coupled Random Vector Functional Link (WCRVFL) of [72], and the approach in [73], based on ARIMA. The evaluation indexes computed for the comparison are RMSE (Root Mean Square Error), MAE (Mean Absolute Error), RMSPE (Root Mean Square Percentage Error), R 2 (coefficient of determination), MAD (Median Absolute Deviation) and MAPE (Mean Absolute Percentage Error). Experimental results showed the efficiency and better performance of proposed methodology when compared to the other approaches.
Cheng et al. [74] introduced an ML-based technique that forecasts the Intensive Care Unit transfer for the COVID patients. The authors have utilized an RF model along with nursing assessments, laboratory information, electrocardiograms, and time series as input types. The proposed model proved the significance of inflammation, shock, renal, and respiratory failure in COVID-19 advancement. It obtained 72.8% sensitivity, 76.3% specificity, 79.9% AUC, and 76.2% accuracy respectively. Based on the experiments, the authors conclude that the ML-based forecasting technique can be utilized as a testing tool to recognize the threat of COVID-19 patients and to improve hospital resource management thus providing more effective care to these patients.
In Nemati et al. [75] several statistic and machine learning techniques are implemented to analyze the survival characteristics of 1182 hospitalized patients. The survival analysis can be applied to predict patient length of stay in the hospital. The authors, for the discharge-time prediction of COVID-19 patients, used the statistical estimators Kaplan-Meier estimator KM, Cox Proportional Hazard CoxPH, Coxnet, Accelerated Failure Time model (IPCRidge), and three machine learning methods Stagewise Gradient Boosting, Componentwise Gradient Boosting, and Support Vector Machine (SVM). Model performances in discharge-time prediction are compared by using the Concordance index (C-index) metric. C-index is a metric to evaluate the predictions of algorithms in survival analysis which computes the percentage of concordant pairs among all feasible evaluation pairs. The computational results agree with the outcome reported in early clinical reports released for a group of patients from China that confirmed a higher mortality rate in men compared with women and in older age groups. The results indicate that the Gradient Boosting survival model outperforms other models for patient survival prediction in this study. The model is not only more accurate when compared with the other boosting methods, but also it outperforms other algorithms in discharge-time prediction in terms of accuracy.
Burdick et al. [76] proposed a machine learning-based method to predict the need for ventilation in COVID-19 patients to help triage patients, allocate resources, and prevent emergency intubations and their associated risks. In a multicenter clinical trial, the authors evaluated the performance of a Gradient Boosting algorithm (XGBoost Classifier) for the prediction of invasive mechanical ventilation of 197 patients with a COVID-19 diagnosis within 24 h of their hospitalization into five United States health systems, between March 24 and May 4, 2020. The algorithm demonstrated a good accuracy in predicting the need for mechanical ventilation within 24 h. Further, the algorithm identified 16% more patients than a standard scoring system minimizing also the false positive rate.
In Aljame et al. [77] an ensemble learning model for COVID-19 diagnosis, named ERLX, from blood tests is proposed. The model uses three well-known diverse classifiers, extra trees, random forest and logistic regression, which have different architectures and learning characteristics at the first level, and then combines their predictions by using a second level extreme gradient boosting (XGBoost) classifier to achieve a better performance. For data preparation, the proposed methodology employs a KNNImputer algorithm to handle null values in the dataset, isolation forest (iForest) to remove outlier data, and a synthetic minority oversampling technique (SMOTE) to balance data distribution. The model was trained and evaluated by using a publicly available data set from Albert Einstein Hospital in Brazil, which consisted of 5644 data samples with 559 confirmed COVID-19 cases. The ensemble model achieved a very good performance with an overall accuracy of 99.88%, AUC of 99.38%, a sensitivity of 98.72% and a specificity of 99.99%.
Pourhomayoun et al. [78] proposed an AI model to help hospitals and medical facilities to decide which patients needs immediate attention, who must be hospitalized first, triage patients when the system is overwhelmed by overcrowding, and reduce delays in giving the necessary care. Several machine learning algorithms, including Support Vector Machine, Artificial Neural Networks, Random Forest, Decision Tree, Logistic Regression, and K-Nearest Neighbor, have been used to predict the mortality rate in patients with COVID-19. In the study authors used a dataset of more than 2,670,000 laboratory-confirmed COVID-19 patients from 146 countries around the world including 307,382 labeled samples. The results showed an overall accuracy of 89.98% in predicting the mortality rate. In this study, the most alarming symptoms and features were also identified. A separate dataset of COVID-19 patients was used to evaluate the model accuracy.
In Hasan [79] a hybrid model that incorporates ensemble empirical mode decomposition (EEMD), a method that decomposes non-linear and non-stationary time series, and artificial neural network (ANN) for predicting the COVID-19 epidemic is proposed. A real-time COVID-19 time series data has been used in the period January 22 to May 18, 2020. The cumulative global data of daily level information by country were retrieved from the Center for Systems Science and Engineering (CSSE) at the Johns Hopkins University GitHub repository (https://github.com/CSSEGISandData/COVID-19) accessed on 19/05/2020. The time-series data has first been decomposed using EEMD to produce sub-signals and make original data denoised, then an ANN has been built to train the denoised data. The results have been compared with some traditional statistical analysis methods and showed that the proposed model outperforms statistical approaches. The validation metrics used were MSE, R 2 and accuracy.
5.4. Methodological breakthrough
The approaches described in the previous sections mainly apply the existing AI techniques on available data and compare the different methods to experimentally evaluate them, without introducing significant novel ideas. In this section, we report some of the approaches in both DL and ML domains that did not simply experimented existing techniques, rather they introduced original new ideas aiming at advancing methodologies in the field of AI for COVID-19 forecasting.
The work of Casiraghi et al. [80] proposed an interesting methodological approach to identify abnormalities in chest radiographs (CXR) and, thus, improving patient risk prediction. To this purpose they designed an explainable machine learning system which may provide simple decision criteria to be used by clinicians as a support for early assessment of COVID-19 risk prediction estimated by both expert radiologists and by specialized state- of-the-art deep neural networks. A novel feature selection algorithm is proposed that combines the Boruta algorithm with permutation based feature selection methods to select variables that are most relevant for COVID-19 risk prediction. The most important variables are then selected to train a RF classifier, whose rules may be extracted, simplified, and pruned to finally build an associative tree. Results show that the radiological score automatically computed through a neural network is highly correlated with the score computed by radiologists, and that laboratory variables, together with the number of comorbidities, aid risk prediction. This study was performed on clinical, comorbidity, laboratory, and anterior-posterior (A-P) or posterior-anterior (P-A) CXR data from patients referred to the ED of an urban multicenter health system, from March, 7, 2020, to April, 10, 2020. All patients in the cohort were RT-PCR positive for COVID-19. With this setting, the patient set included, 207 and 94 adult men and women with a mean age of 61 years, and with a number of days with symptoms from COVID-19 that were on average 7. Among them 214 patients were at low risk, while 87 patients were at high risk. Results show that the prediction performance of the approach was compared to that of generalized linear models and shown to be effective. The proposed machine learning-based computational system can be easily deployed and used in emergency departments for rapid and accurate risk prediction in COVID-19 patients.
Ren et al. [81] proposed a computational model to predict epidemiological trends of COVID-19, with the model parameters enabling an evaluation of the impact of non-pharmacological interventions (NPIs) such as lockdowns for effective management of the disease and control of its spread. By representing the number of daily confirmed cases (NDCC) as a time-series, authors assumed that, with or without NPIs, the pattern of the pandemic satisfies a series of Gaussian distributions according to the central limit theorem. The underlying pandemic trend is first extracted using a singular spectral analysis (SSA) technique, which decomposes the NDCC time series into the sum of a small number of independent and interpretable components such as a slow varying trend, oscillatory components and structureless noise. After that a mixture of Gaussian fitting (GF) has been used to derive a novel predictive model for the SSA extracted NDCC incidence trend, with the overall model termed SSA-GF. The model is shown to accurately predict the NDCC trend, peak daily cases, the length of the pandemic period, the total confirmed cases and the associated dates of the turning points on the cumulated NDCC curve. The predictive model is validated using available data from China and South Korea, and new predictions are made, partially requiring future validation, for the cases of Italy, Spain, the UK and the USA. Comparative results demonstrate that the introduction of consistent control measures across countries can lead to development of similar parametric models.
Gao et al. [62] developed a mortality risk prediction model for COVID-19 that utilizes clinical data in EHRs to stratify patients by mortality risk on admission. Six ML models including LR, support vector machine (SVM), gradient boosted decision tree (GBDT), neural network (NN), K-nearest neighbor (KNN), and random forest (RF) displayed varying but promising performances to predict mortality risk in the three validation cohorts in terms of discrimination and calibration. To build a predictive model with augmented prognostic implications, the authors integrated the top four best predictive models (LR, SVM, GBDT, and NN) to create an ensemble model called MRPMC. The validated capability of enabling expeditious and accurate mortality risk stratification of COVID-19 may facilitate more responsive health systems that are conducive to high-risk COVID-19 patients via early identification.
In [82] Rostami et al. exploited phone call data to forecast daily confirmed cases. They proposed a multiple linear regression model that exploits the relationship between the confirmed cases and the phone call data. In particular, the regression model expresses the number of confirmed cases as a function of past lags of confirmed cases, current and past lags of the number of phone calls, the effect of weekends and the trend. Data used in this paper comprised the number of daily COVID-19 confirmed cases and the number of daily phone calls received at the National Health Service 111 in the East Midlands region of England between 18 March 2020 and 19 October 2020. The simplicity, interpretability and reliability of the model, obtained in a careful forecasting exercise, is a meaningful contribution to decision makers at local level who acutely need to organise resources in already strained health services. The authors show that their approach outperforms ARIMA, ES, Seasonal Naive, a simple method which forecasts the number of confirmed cases as equal to the last observed confirmed cases from the same day of the previous week, Prophet and a regression model without call data.
In Ramchandani et al. [83] is presented DeepCOVIDNet, a deep learning approach to predict COVID cases in the next seven days by using several features, such as census data, intra-county mobility, inter-county mobility, social distancing data, past growth of infection. These features are grouped on the base of their similarity and classified as constant, time-dependent, and cross-county time-dependent. DeepCOVIDNet is composed of two modules: the embedding module and the DeepFM module, a factorization based neural network proposed by Guo et al. [84]. The task of the embedding module is to provide an embedding for each group of features of the same dimension. These embeddings are given as input to the DeepFM module which computes higher order interactions between the embeddings and outputs a probability distribution of the current rise in cases, i.e. those with the greatest probability. The period considered for the experimentation is April 5th through June 28th. The authors trained the model by using data until June 11, considering four classes to categorize the growth in the number of new cases: negligible increase, moderately low increase, moderately high increase, and significantly high increase. Then tested the model on the 17 days from June 12 through June 28. The average accuracy on these 17 days is 63.7% on the four output classes. The code is available at https://github.com/urban-resilience-lab/deepcovidnet.
Zheng et al. [85] propose a hybrid artificial intelligence model for the prediction of COVID-19 cases in China by embedding the long short-term memory (LSTM) network with the Natural Language Processing (NLP) module into a traditional model of virus spreading. The model assumes a retrospective approach which uses the ratio of the number of new confirmed cases at time t to the cumulative number of new confirmed cases before time t to compute the infection rate. Moreover, in order to take into account the effects of government control measures, the reports of media on the subject, and the awareness of people on epidemic prevention, features from relevant news of various provinces and cities are extracted by using pretrained NLP models. These features are then combined with the LSTM network to correct the deviation of the infection rate estimated by the ISI model. Experimental results on the epidemic data of Beijing, Shanghai, Zhejiang, and Hunan show that the proposed hybrid model outperforms traditional epidemic models.
Shastri et al. [86] proposed a nested ensemble model using deep learning methods based on long short term memory (LSTM). The proposed Deep-LSTM ensemble model is evaluated on intensive care COVID-19 confirmed and death cases of India, with different classification metrics, namely accuracy, precision, recall, f-measure and mean absolute percentage error. COVID-19 confirmed and death cases of India are taken from World Health Organization. Confirmed cases are taken from 29th January to 1st September 2020 and death cases are taken from 12th March to 1st September 2020. The deep-LSTM ensemble model using convolutional and bi-directional LSTM obtains high accuracy to forecast COVID-19. The error in the model is calculated in terms of mean absolute percentage error (MAPE). The COVID-19 confirmed and death cases are predicted for one month ahead. For COVID-19 confirmed cases the accuracy is of 97.59% and for death cases is 98.88%, while the MAPE value for the confirmed and death cases are 2.40 and 1.11, respectively.
In [87] Ardabili et al. presented a comparative analysis of machine learning and regression models to predict the COVID-19 outbreak. Data is collected from the website https : //www. worldometers. info/coronavirus/ # countries for five countries, namely Italy, Germany, Iran, USA, and China, for a period of 30 days from January 22, 2020. The authors use a regression model with different types of relationships among the independent variables, i.e. logistic, linear, logarithmic, quadratic, cubic, compound, power, and exponential. Estimation of the parameters was performed by using evolutionary algorithms such as the genetic algorithm (GA), particle swarm optimizer (PSO), and grey wolf optimizer (GWO). Through parameter tuning authors determined the optimal performance of the models. Experiments showed that logistic model outperforms the other regression methods. In particular, the logistic model using GWO for parameter tuning outperformed that based on PSO and GA. As machine learning models two types of artificial neural networks were used: Multi-Layered Perceptron (MLP) and an adaptive neuro fuzzy inference system (ANFIS), a particular kind of ANN based on a fuzzy system. For these methods two scenarios were proposed. Scenario 1 considers three weeks of previous data, while Scenario 2 the previous 5 days. The performance of both ML models for the selected countries varied for the two different scenarios. By considering the average values of the RMSE and correlation coefficient values, Scenario 1 is more suitable than Scenario 2, and MLP is more suitable than ANFIS for outbreak prediction.
Hazarika and Gupta [88] present the WCRVFL model which hybridizes random vector functional link (RVFL) network with 1-D discrete wavelet transform and a wavelet-coupled RVFL network. RVLF is a multilayer perceptron (MLP) approach. The RVFL model receives as input the wavelet decomposed time-series data. The prediction performance of WCRVFL is compared with the state-of-the-art support vector regression (SVR) model and the RVFL model on five countries, Brazil, India, Peru, Russia and the USA in the period April 11 until June 10, 2020. Moreover, a 60 days ahead of daily COVID-19 spread forecasting is also reported. The performance evaluation has been performed by computing the coefficient of determination R 2, root mean square error (RMSE), mean absolute error (MAE), the ratio between the sum of squared error and the total sum of squares (SSE/SST), peak signal to noise ratio (PSNR), structural content (SC), the maximum difference (MD), Laplacian mean Square error (LMSE) and normalized absolute error (NAE). Experimental results show that proposed WCRVFL model obtains good prediction values of COVID-19 spread.
In Kim et al. [89] is proposed Hi-COVIDNet, which takes advantage of the geographic hierarchy to predict the number of COVID-19 cases. Hi-COVIDNet is based on a neural network with two-level components, namely, country-level and continent-level encoders, which understand the complex relationships among foreign countries and derive their respective contagion risk to the destination country. An in-depth case study in South Korea with real-world COVID-19 datasets confirmed the effectiveness and practicality of Hi-COVIDNet.
5.5. Miscellaneous
The number of papers published in the last year is really huge, thus an exhaustive review and description of each of them is not possible. In this section we briefly describe approaches published as preprint at the time of writing, and thus not yet accepted after peer revision, or published in the early 2020s experimenting their methods on the few available data in the consider period. We report them since we used for computing statistics reported in Section 4.
5.5.1. Preprints
In [90] Huang et al. used 4 DL models (CNN, LSTM, GRU, and MLP) to train and predict COVID-19 cases from 7 severely epidemic cities in China. The input of these DL models are the features of COVID-19 cases, including the number of confirmed cases, cured cases, and deaths. Based on the input of the previous 5 days, each model can predict the number of COVID-19 cases in the following few days.
Punn et al. [91] used two machine learning models SVR and PR and three deep learning regression models, DNN, LSTM, and RNN to predict real-time COVID-19 cases.
Sarkar et al. [92] used the RF model to analyze the records of 433 patients with COVID-19 from Kaggle, and identified the important features and their impact on mortality. Experimental results show that patients over 62 years of age have a higher risk of death.
In [93], [94], Yan et al. analyzed a blood sample data set of 404 patients with COVID-19 in Wuhan, China, and used the XGBoost classification method to select three important biomarkers to predict the survival rate of individual patients. Experimental results with an accuracy of 90% indicate that higher LDH levels seem to play an important role in distinguishing the most critical COVID-19 cases.
In Kolozsv et al. [95], a recurrent neural network is proposed to predict the epidemic curve. Two prediction models are created in this work, first the data is fed to a dense neural network and then a consequent regression output layer is used to predict the value.
In Li et al. [96], a recurrent NN is proposed to build a model of the pandemic in Italy. Kapoor [97] proposed a novel forecasting approach for COVID-19 case prediction that uses Graph Neural Networks and mobility data. In contrast to existing time series forecasting models, the proposed approach learns from a single large-scale spatio-temporal graph, where nodes represent the region-level human mobility, spatial edges represent the human mobility based inter-region connectivity, and temporal edges represent node features through time.
A combination of XGBoost, K-means and LSTM algorithms is used in Vadyala et al. [98] to build a model to predict the pandemic in Louisiana, USA. In Javod et al. [99], polynomial regression and neural network algorithms are used with the data made available by John Hopkins University to build a model of the pandemic. In [100], exponential smoothing and ARIMA are used to predict the pandemic in India.
In Zandavi et al. [101], LSTM with dynamic behavioral model is adopted which considers the effect of multiple factors to enhance the accuracy of the prediction across top 10 most affected countries. In order to build a predictive model for the pandemic, a new architecture for DNN is proposed in Direkoglu et al. [102], which consists of a LSTM layer, dropout layer and fully connected layers to predict regional and worldwide forecasts.
In Karimuzzaman et al. [103], ARIMA is used along with Multi-Layer-Perceptron (MLP), Extreme Learning Machine (ELM) and Generalized Linear count time series Model (GLM) to model the behavior of the pandemic. To predict the epidemic growth rare, an LSTM method is proposed in Yudistira [104]. In Rani et al. [105] ARIMA is combined with LSTM to predict the pandemic.
In Melin et al. [106] a multiple ensemble neural network model with fuzzy response aggregation for the COVID-19 time series is presented. Ensemble neural networks are composed of a set of modules, which are used to produce several predictions under different conditions. Fuzzy logic is then used to aggregate the responses of several predictor modules, in this way, improving the final prediction by combining the outputs of the modules.
In Tian et al. [107] an approach integrating LSTM and Gated Recurrent Unit to predict the trajectory of the pandemic is proposed.
LSTM algorithm, combined with a recurrent neural network, is used in Kolozsvari et al. [108] to build two prediction models of the pandemic in India. In Amo-Boateng [109], a 1D CNN is applied to the time-series data of confirmed COVID-19 cases to track and classify progress of the pandemic in different countries. In Zhao et al. [110], various Recurrent Neural Networks, including the LSTM and 10 types of slim LSTM are presented to predict the pandemic in the US.
In Huang et al. [90] a Convolutional Neural Network is proposed to analyze and predict the number of confirmed cases in China. A machine learning algorithm is proposed in Kumar et al. [111], to predict the number of daily cases. The algorithm combines three machine learning algorithms, namely decision tree, support vector machine and Gaussian process regression. In Mathur et al. [112], 24 variables linked to COVID-19 are used to build a model with CatBoost regression and random forest algorithms to predict mortality in the US.
In order to build a predictive model of COVID-19, three machine learning models, namely hidden Markov chain model (HMM), hierarchical Bayes model, and LSTM is proposed in Tian et al. [113]. In Liu et al. [114] a clustering algorithm is used to process data from Internet searches and news alerts to perform a real-time forecasting of the outbreak.
In order to study the epidemic behavior in different zones in New York city, a clustering algorithm is proposed in [115] Khmaissia et al., that models the outbreak in the city. In Suzuki et al. [116], XGBoost is used to predict the number of infections in South Korea.
In Pereira et al. [117], a clustering algorithm is applied to the world regions for which epidemic data are available and the pandemic is at an advanced stage. Then a set of features representing the countries response to the early spread of the pandemic are used to train an Autoencoder Network to predict the future of the pandemic in Brazil.
Two machine learning algorithms, neural network and Prophet, are used in Balde et al. [118] to study the impact of nation-wide measures on the pandemic.
An empirical top-down modeling algorithm is proposed in Uhlig et al. [119], which uses a combination of epidemiological, statistical and neural network applications. In this approach, a neural network is used to develop leading indicators for different regions. These indicators are used to assess the risk of an outbreak, determine the effectiveness of the measures, predict the outbreak with the associated uncertainty.
In Dandekar et al. [120], an epidemiological model augmented with a neural network approach is proposed to study the effect of quarantine and isolation measures implemented in Wuhan on the reproduction number, R0.
5.5.2. Other publications
In this section are outlined articles that are still preliminary works.
In da Silva et al. [121] Bayesian regression neural network, cubist regression, k-nearest neighbors, quantile random forest, and support vector regression, are used stand-alone, and coupled with the variational mode decomposition (VMD) employed to decompose the time series into several intrinsic mode functions. All AI techniques are evaluated in the task of time-series forecasting with one, three, and six-days-ahead the cumulative COVID-19 cases in five Brazilian and American states, with a high number of cases up to April 28th, 2020. Previous cumulative COVID-19 cases and exogenous variables as daily temperature and precipitation were employed as inputs for all forecasting models.
In Barnerjee et al. [122] early diagnoses of COVID-19 is obtained through blood counts exploiting several ML approaches (LR, ANN, Lasso).
Giuliani et al. [123] collected the number of infected people in various provinces of Italy, and used the SGLMM Spatial Generalized Linear Mixed Models to simulate and predict the spatial and temporal distribution of COVID-19 infection in Italy. They collected daily epidemic data and saved them in a time series data format, and then used LR and LSTM models to make predictions, thereby obtaining the outbreak and spread trend of COVID-19.
In Braga et al. [124] an approach based on artificial neural networks (ANN) for the daily and cumulative forecasts of cases and deaths caused by COVID-19 in the Brazilian Amazon, and the forecast of demand for hospital beds, is proposed. Six scenarios with different periods were used to identify the quality of the generated forecasting and the period in which they start to deteriorate. Results indicated that the computational model adapted capably to the training period and was able to make consistent short-term forecasts, especially for the cumulative variables and for demand hospital beds.
An ANN is used in Pirouz et al. [125] to predict the number of cases in Hubei, China. Khakharia et al. [126] propose an approach for outbreak prediction of COVID-19 for dense and populated countries by using machine learning algorithms. Ghany et al. [127] exploits LSTM for COVID-19 prediction. In Khan et al. [128] ARIMA and a nonlinear autoregressive neural network are deployed to build a model of the epidemic to predict the behavior of the epidemic. In Kumar et al. [129], ARIMA and ANN are used to predict the pandemic in Italy, Spain and France.
In Car et al. [130] an ANN is used on a publicly available dataset that contain information on infected, recovered and deceased patients. In this work, the data is transformed into a regression dataset and used in a multilayer perceptron to build a model of the number of patients across all locations.
In Fong et al. [131] a case study of using Composite Monte-Carlo (CMC) simulation forecasting, enhanced with deep learning network and fuzzy rule induction for gaining better stochastic insights about the epidemic development is experimented. Instead of applying simplistic and uniform assumptions for a MC which is a common practice, a deep learning-based CMC is used in conjunction with fuzzy rule induction techniques. As a result, decision makers are benefited from a better fitted MC outputs complemented by min-max rules that foretell about the extreme ranges of future possibilities with respect to the epidemic. In another work [132] Fong et al. used traditional time series data analysis methods (such as ARIMA, Exponential, and Holt-Winters), ML methods (such as KR, SVM, and DT), and AI methods (such as PNN) to analyze and predict future outbreaks.
In Hartono [133], Neural Networks and LSTM are used to build a model to forecast the pandemic all over the world. In [134], a multi-layer perceptron and vector aggression method are used to design a forecasting model for the epidemic in India.
An unsupervised neural network algorithm called self-organizing map is proposed in Melin et al. [135], which spatially groups together the countries that are similar to one another with respect to the pandemic, so can benefit from using similar strategies.
A multilayer perceptron neural network is used in Mollato et al. [136] to predict the incidence rate of the pandemic in United States. In Tamang et al. [137], an ANN-based curve fitting algorithm is presented for forecasting the number of cases in India, US, France and the UK, considering the progressive trends of China and South Korea. In Torrealba et al. [138], neural networks are used to predict the number of COVID-19 cases in Mexico.
In Distante et al. [139] a modified autoencoder is proposed to predict the epidemic curve of different regions in Italy. Statistical and AI-based approaches are combined in Saba et al. [140] to model and forecast the prevalence of the pandemic in Egypt. The work integrates ARIMA and Non linear Auto Regressive Artificial Neural Networks (NARANN).
In Chatterjee et al. [141], LSTM, vanilla, stacked and bidirectional LSTM were used to predict the pandemic in the world. The LSTM networks are used in Chimmula et al. [142], to build a predicting model for the trend and possible finishing time of the outbreak in Canada.
In another work Aldhyani et al. [143], LSTM algorithm and Holt-trend are applied to predict confirmed number of death cases. In Tomar et al. [144], LSTM and curve fitting methods are used for the prediction of the number of cases in India.
In Mohammad et al. [145], LSTM is used to model the data obtained from Google Trends website and estimate the number of positive COVID- 19 cases. The authors report that the most effective predictive factors are the search frequency of hand-washing, hand sanitizer and antiseptic topics.
A shallow long short-term memory based neural network is proposed in Pal et al. [146] to predict the epidemic in different countries. The authors use a Bayesian optimization framework to optimize the network. In Malki et al. [147] an approach using various regressor machine learning models and exploiting the relationship between the spread of the disease and factors like weather variables, temperature and humidity is proposed.
In order to investigate the role of environmental parameters, the climate and urban parameters of four cities in Italy are studied in Haghshenas2 et al. [148]. The authors use ANN, PSO and DE optimization algorithms for prioritizing climate and urban parameters.
Different learning algorithms, including ARIMA, Non linear Autoregression Neural Network (NARNN) and LSTM approaches are used in Kirbas et al. [149], to predict the number of new cases in Denmark, Belgium, Germany, France, United Kingdom, Finland, Switzerland and Turkey.
In order to predict the spread of the virus, analyze the growth rate, predict how the epidemic will end and correlate the pandemic with weather conditions, a novel Support Vector Regression method is proposed in Yadav et al. [150].
In Peng et al. [151] support vector regression is applied to predict the number of COVID-19 cases in 12 most affected countries.
An improved version of Adaptive Neuro Fuzzy Inference System (ANFIS) is used in Al-qaness et al. [152] to predict the spread of the virus in Italy, Iran, Korea and USA. The proposed algorithm uses marine predator algorithm to optimize the parameters of ANFIS. In order to model the behavior of the pandemic, in another work Al-qaness et al. [153] extended the proposed ANFIS model with a flower pollination algorithm and a swarm algorithm to optimize the model parameters.
5.6. Summary of the main features of the selected papers
Table 3 summarizes the main features of the 38 reviewed papers. The features mainly concern AI-related characteristics, data related information, the topic addressed by the method, the experimental methodology adopted and the results obtained. Specifically, we have a column about the kind of ML or DL model used, or what could be considered more in general belonging to AI; other two columns reporting information about the dataset, and about the type of data and the time interval in which the COVID-19 related data was collected. The remaining columns specify the output produced, the validation method and the results achieved.
Different ML and DL methods have been employed for COVID-19 forecasting and tracking including ARIMA, LSTM, LR, RNN. For example, Multilayer Perceptron (MLP) and Adaptive Network-based Fuzzy Inference System (ANFIS), among DL methods resulted to be very efficient achieving high correlation levels. Novel approaches combining temporal and spatial data resulted to be very powerful like, for example, using graph neural networks and Google mobility data to uncover the rich interactions between time and space that is often present in the spread of pandemic. Several ML models are compared to forecast confirmed cases. The models under consideration include Bayesian neural network, cubist regression, kNN, random forest, and support vector regression (SVR). Numerical experiments produce mixed results with no clear favorite. Other contributions compare statistical and ML approaches to time series forecasting. In particular, they study autoregressive integrated moving average (ARIMA), SVR, and LSTM models to forecast the number of infections, deaths, and recoveries.
The experimental results reported in the revised papers show that LSTM models generally outperform ARIMA and SVR. However, ML approaches do not always outperform traditional methods, as shown in other studies that compared classic statistical methods to SVR and other traditional ML methods, to predict the number of positive cases, death rate, and recovery rate. Such studies show that statistical models outperform SVR, RF.
Long short-term memory (LSTM) networks have been shown to outperform the traditional time series models, such ARIMA. As a result, LSTMs have been used in various applications involving time series projections. Another promising approach, even if not well explored, resulted to be the use of ensemble neural network to predict the number of confirmed cases and deaths.
The models were evaluated on the basis of their accuracy and efficacy for different prediction lead times and employed different types of data from different countries in their study. Experiments have been validated following well known metrics in literature used for the evaluation of prediction performance, like prediction accuracy measured in terms of AUC, ROC curves, specificity, sensitivity, precision, correlation and prediction error measures in term of MAPE, MAE, RMSE.
The analyzed papers address COVID-19 forecasting by looking at different factors and covering different scopes and topics. Fig. 5 shows that the majority of the papers focuses on COVID-19 daily cases forecasting, with a relevant prevalence compared to the other tackled topics (42%). Mortality risk prediction was another topic widely studied in the selected literature papers, it has been found in the 30% of the works, followed by the prediction of recovery cases that reaches about 8%. COVID-19 risk prediction and diagnosis both reach 5%. Critical cases prediction and positive and negative cases prediction are around 3%. Since COVID-19 daily cases forecasting is the most widely studied topic, we show in Fig. 6 the most performing ML and DL methods used by researchers. ARIMA and LSTM with both a percentage of 17% resulted to be the most successful AI methods for COVID-19 daily cases forecasting, followed by ANN with 13% and MLP with 9%. As can be argued from the figure, the rest of the approaches used other variants of LSTM like BiLSTM, DeepLSTM, which in total represents 37% of the approaches. This confirms the fact that LSTM-based approaches turned out to be the most successful for COVID-19 cases prediction.
Fig. 5.
COVID-19 Forecasting- Topics.
Fig. 6.
Most performing ML and DL methods used for COVID-19 cases forecasting.
Fig. 4 shows the data types used for COVID-19 forecasting. Different data types have been exploited including demographics, comorbidities, clinical data, blood tests, number of daily cases, number of daily deaths, number of recovery cases, vaccination rate, physiological data and number of daily phone calls. Of those types the most widely used is the number of daily cases with a percentage of 41%, followed by clinical data with the 19%, the number of daily deaths with 14%, demographics with 7% and then all other types with smaller percentage.
Fig. 4.
Data Types used for COVID-19 cases forecasting.
While these studies show how a range of different methodological choices can be made when building forecasting models, they demonstrate the complexities involved in choosing between such models and the non-trivial interplay between methods, hyperparameters, and datasets. Moreover, since much of the data collected for COVID-19 modeling tasks is limited, the choice of models and datasets can have significant effects on overall performance.
6. Discussion
Artificial Intelligence algorithms play a key role in rapid forecasting, detection, classification, screening, and diagnosis of COVID-19 infection cases.
Currently, AI mainly focuses on medical image inspection, genomics, drug development, and transmission prediction, and thus AI still has great unexplored potential mainly in terms of number of new cases and deaths prediction. In fact, even if many applications addressing COVID-19 forecasting and diagnoses have been proposed, only few of them are currently mature enough to be effective in real-world scenarios.
Till end of 2020 AI was not fully explored on tracking and prediction of COVID-19 cases due to the lack of a vast amount of historical data to train the AI models. Accordingly, earlier papers that were published after few months of the worldwide COVID-19 outbreak, reported results of limited relevance due both to the lack of sufficient data to train the AI techniques in an appropriate way, but also because of the quality of the data themselves. In fact, due to the rapid diffusion of COVID-19, there was insufficient data at disposal as well as extensive labeled datasets not yet available. Training models on unrepresentative datasets lead to poor and even misleading outcomes as the fast-moving nature of the problem can make it difficult to perform informed model selection and parameters. This severely affected the performance and accuracy of the forecasting models.
Today the availability of COVID-19 surveillance data in terms of number of daily and cumulative cases, number of deaths and number of recovery is not an issue anymore. In fact, after two years since COVID-19 outbreak, several collections of detailed data are available from different sources, like for example the one gathered by the Coronavirus Research Center of the Johns Hopkins University. Therefore, it would be very interesting if the authors of those early works could re-execute the proposed approaches using the high volumes of data now available and validate their approach on the new data.
Another limitation is that many of the analyzed works do not exploit any exogenous variable in the forecasting process. Accounting restrictive measures like lockdown, quarantine, traveling limitations could enhance the prediction accuracy. Furthermore, the availability of vaccination data could be integrated in the forecasting models, greatly improving the performance of the prediction. Accordingly, a future research line could be to extend the proposed forecasting models with exogenous variables like the ones just discussed.
Still concerns remain for the use of clinical data for COVID-19 early diagnosis and early symptoms prediction. There are several limitations to the feasible applications of AI methods for COVID-19 prediction on such kind of data. We outlined some of them as follows:
-
1.
Lack of available large-scale training data. Most AI methods rely on large-scale annotated training data. In addition, annotating training samples is very time-consuming and requires professional medical personnel.
-
2.
The distributed and heterogeneous nature of many data sources contributes to data scarcity. In fact, the different data formats together with the lack of data standardization and interoperability and missing values, make the application of AI methods on such data often inaccurate and unreliable. As highlighted in Dagliati et al. [1], interoperability is a key concept: COVID-19 pandemic made clear that unified frameworks for sharing and exchanging digital epidemiological data together with data protection are necessary. Data federation, data integration and data fusion could be applied to overcome data heterogeneity, as well as the use of common standards at international level. Another suggestion could be the design of analytical models tailored to work with the specific issues of the current available clinical data related to COVID-19.
-
3.
Data imbalance between positive and negative samples. Indeed, the possibility to collect only few positive COVID-19 samples can impact the accuracy of COVID-19 diagnosis.
-
4.
As pointed out in Combi et al. [2], most of the research carried out so far is on Machine Learning rather than on Natural Language Processing (NLP) and Decision Support Systems (DSS), this because while both DSS and NLP require a major effort in developing, ML is based on the application of well-known techniques on COVID-19 data. Accordingly, there are still space for the design and development of both NLP methods and DSS applications specifically tailored to deal with COVID-19 peculiarities.
-
5.
Lack of interdisciplinary cooperation. The use of AI techniques for COVID-19 diagnosis and forecasting requires integration of different expertises from multiple disciplines like computer science, medical imaging, virology, medical doctors in general. Therefore, a key point is the cooperation of researchers belonging to the different disciplines to combine and supplement the various knowledge in order to be more incisive in the fight against COVID-19.
-
6.
Privacy, anonymity and ethic issues are key concerns, which need to be addressed so as to enable effective contact tracing between citizens as well as effectively preserving their privacy. Privacy matters are also relevant when dealing with specific type of data like, for example, social media data that is often exposed to privacy violation as reported by Combi et al. [2].
As final remark we want to underline that still there is space to exploit advanced ML algorithms, like ensemble methods such as bagging, boosting, stacking, etc. for COVID-19 forecasting of new infections, deaths and recovery. Furthermore, the applicability of AI for early symptoms detection and disease diagnosing is not fully exploited yet. For example, supervised classification methods could be better adapted and explored for detection and classification of the different symptoms associated with COVID-19.
7. Conclusion
The paper presented a systematic and comprehensive survey of the application of AI technologies for forecasting, detecting, and diagnosing COVID-19. The study examined and reviewed an extensive collection of state-of-the-art COVID-19 prediction and diagnosis algorithms, providing a detailed background description of the AI techniques used for COVID-19. For each work surveyed, is provided a detailed analysis of the rationale behind the approach, highlighting the method used, the type and size of data analyzed, the validation method, the target application and the results achieved.
Despite all the significant progress in the application of AI in addressing COVID-19 issues, there is still a need for further implementation of these technologies for detecting, monitoring, and diagnosing. Future work should focus on strengthening the current technologies mostly for early differential diagnosis of COVID-19 on clinical data. Also, future work should consider the issues related to privacy preserving and security of sensible health and personal data of citizens.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Dagliati A., Malovini A., Tibollo V., Bellazzi R. Health informatics and EHR to support clinical research in the COVID-19 pandemic: an overview. Briefings Bioinform. 2021;22(2):812–822. doi: 10.1093/bib/bbaa418. URL doi:10.1093/bib/bbaa418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Combi C., Pozzi G. 9th IEEE International Conference on Healthcare Informatics, ICHI 2021, Victoria, BC, Canada, August 9-12, 2021. IEEE; 2021. Health informatics: clinical information systems and artificial intelligence to support medicine in the COVID-19 pandemic; pp. 480–488. [DOI] [Google Scholar]
- 3.Chen J., Li K., Zhang Z., Li K., Yu P.S. A survey on applications of artificial intelligence in fighting against COVID-19. ACM Comput Surv. 2022;54(8):158:1–158:32. [Google Scholar]
- 4.W N. Artificial intelligence vs COVID-19: limitations, constraints and pitfalls. AI Soc. 2020:1–5. doi: 10.1007/s00146-020-00978-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pham Q.-V., Nguyen D.C., Huynh-The T., Hwang W.-J., Pathirana P.N. Artificial intelligence (ai) and big data for coronavirus (COVID-19) pandemic: a survey on the state-of-the-arts. IEEE Access. 2020;8:130820–130839. doi: 10.1109/ACCESS.2020.3009328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.T. Alamo D. G. Reina P. Millán, Data-Driven Methods to Monitor, Model, Forecast and Control COVID-19 Pandemic: Leveraging Data Science, Epidemiology and Control Theory, arXiv:2006.01731.
- 7.N. L. Bragazzi H. Dai G. Damiani M. Behzadifar M. Martini J. Wu, How big data and artificial intelligence can help better manage the COVID-19 pandemic, International Journal of Environmental Research and Public Health 17 (9). [DOI] [PMC free article] [PubMed]
- 8.Latif S., Usman M., Manzoor S., Iqbal W., Qadir J., Tyson G., Castro I., Razi A., Boulos M.N.K., Weller A., Crowcroft J. Leveraging data science to combat COVID-19: a comprehensive review. IEEE TransArtifIntell. 2020;1(1):85–103. doi: 10.1109/TAI.2020.3020521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L. Wynants B. Van Calster G. S. Collins R. D. Riley G. Heinze E. Schuit M. M. J. Bonten D. L. Dahly J. A. Damen T. P. A. Debray V. M. T. de Jong M. De Vos P. Dhiman M. C. Haller M. O. Harhay L. Henckaerts P. Heus M. Kammer N. Kreuzberger A. Lohmann K. Luijken J. Ma G. P. Martin D. J. McLernon C. L. Andaur Navarro J. B. Reitsma J. C. Sergeant C. Shi N. Skoetz L. J. M. Smits K. I. E. Snell M. Sperrin R. Spijker E. W. Steyerberg T. Takada I. Tzoulaki S. M. J. van Kuijk B. C. T. van Bussel I. C. C. van der Horst F. S. van Royen J. Y. Verbakel C. Wallisch J. Wilkinson R. Wolff L. Hooft K. G. M. Moons M. van Smeden, Prediction models for diagnosis and prognosis of COVID-19: systematic review and critical appraisal, BMJ 369:m1328. [DOI] [PMC free article] [PubMed]
- 10.Lalmuanawma S., Hussain J., Chhakchhuak L. Applications of machine learning and artificial intelligence for COVID-19 (sars-cov-2) pandemic: a review. Chaos, SolitonsFractals. 2020;139 doi: 10.1016/j.chaos.2020.110059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar A., Gupta P.K., Srivastava A. A review of modern technologies for tackling COVID-19 pandemic. Diabetes Metab Syndr. 2020;14(4):569–573. doi: 10.1016/j.dsx.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bullock J., Luccioni A., Pham K., Lam C., Luengo-Oroz M. Mapping the landscape of artificial intelligence applications against COVID-19. JArtifIntellRes. 2020;69:807–845. [Google Scholar]
- 13.Abd-Alrazaq A., Alajlani M., Alhuwail D., Schneider J., Al-Kuwari S., Shah Z., Hamdi M., Househ M. Artificial intelligence in the fight against COVID-19: scoping review. J Med Internet Res. 2020;22(12) doi: 10.2196/20756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamalov F., Cherukuri A., Sulieman H., Thabtah F., Hossain A. 2021. Machine learning applications for COVID-19: a state-of-the-art review. arXiv:2101.07824. [Google Scholar]
- 15.J. Nayak B. Naik P. Dinesh K. Vakula P. B. Dash D. Pelusi, Significance of deep learning for COVID-19: state-of-the-art review, Research Biomedical Engineering, doi:10.1007/s42600-021-00135-6.
- 16.Tayarani N. M.-H. Applications of artificial intelligence in battling against COVID-19: a literature review. Chaos, Solitons Fractals. 2021;142 doi: 10.1016/j.chaos.2020.110338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain A.A., Bouachir O., Al-Turjman F., Aloqaily M. AI techniques for COVID-19. IEEE Access. 2020;8:128776–128795. doi: 10.1109/ACCESS.2020.3007939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freedman D.A. Cambridge University Press; 2005. Theory and practice. [Google Scholar]
- 19.Cryer J.D., Chan K.-S. Springer; 2008. Time series analysis with applications in R. [Google Scholar]
- 20.Hyndman R.J., Athanasopoulos G. OTexts; Melbourne, Australia: 2018. Forecasting: principles and practice. [Google Scholar]
- 21.Box G.E.P., Jenkins G.M., Reinsel G.C., Ljung G.M. 5th edition. Wiley; 2015. Time series analysis, forecasting and control. [Google Scholar]
- 22.Taylor S.J., Letham B. Forecasting at scale. PeerJ. 2017;5 Prepr. [Google Scholar]
- 23.Awad M., Khanna R. Springer; 2015. Efficient learning machines. [Google Scholar]
- 24.Samuel A.R. Some studies in machine learning using the game of checkers. IBM JResDev. 1959;44(1.2):1210–1229. [Google Scholar]
- 25.Murphy K.P. MIT Press; Cambridge: 2012. Machine learning: a probabilistic perspective. [Google Scholar]
- 26.Boser B.E., Guyon I.M., Vapnik V.N. Proceedings of the Fifth Annual Workshop on Computational Learning Theory, COLT '92. Association for Computing Machinery; New York, NY, USA: 1992. A training algorithm for optimal margin classifiers. p. 144?152. [Google Scholar]
- 27.Schölkopf B., Herbrich R., Smola A.J. Computational Learning Theory, 14th Annual Conference on Computational Learning Theory, COLT 2001 and 5th European Conference on Computational Learning Theory, EuroCOLT 2001, Amsterdam, The Netherlands, July 16-19, 2001, Proceedings. 2001. A generalized representer theorem; pp. 416–426. [Google Scholar]
- 28.Suykens J.A.K., Vandewalle J. Least squares support vector machine classifiers. Neural Process Lett. 1999;9(3):293–300. [Google Scholar]
- 29.Mitchell T.M. McGraw-Hill; 1997. Machine learning. [Google Scholar]
- 30.Frank E., Wang Y., Inglis S., Holmes G., Witten I. Using model trees for classification. MachLearn. 1998;32(1):63–76. [Google Scholar]
- 31.Breiman L. Bagging predictors. MachLearn. 1996;24(2):123–140. [Google Scholar]
- 32.Schapire R.E. Boosting a weak learning by majority. InformComput. 1996;121(2):256–285. [Google Scholar]
- 33.Freund Y., Scapire R. Proceedings of the 13th Int. Conference on Machine Learning. 1996. Experiments with a new boosting algorithm; pp. 148–156. [Google Scholar]
- 34.Hinton G.E., Osindero S., Teh Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18(7):1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- 35.Pao Y.-H., Park G.-H., Sobajicn D.J. Learning and generalization characteristics of the random vector functional-link net. Neurocomputing. 1994;6(2):163–180. [Google Scholar]
- 36.<collab>G. I. B.Y., C. A.</collab> . MIT Press; 2016. Deep learning. [Google Scholar]
- 37.Hochreiter S., Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 38.Cho K., van Merrienboer B., Gülçehre Ç., Bahdanau D., Bougares F., Schwenk H., Bengio Y. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, EMNLP 2014, October 25-29, 2014, Doha, Qatar, A meeting of SIGDAT, a Special Interest Group of the ACL. 2014. Learning phrase representations using RNN encoder-decoder for statistical machine translation; pp. 1724–1734. [Google Scholar]
- 39.Jahromi A.N., Hashemi S., Dehghantanha A., Parizi R.M., Choo K.R. An enhanced stacked LSTM method with no random initialization for malware threat hunting in safety and time-critical systems. IEEE TransEmergTopComputIntell. 2020;4(5):630–640. [Google Scholar]
- 40.LeCun Y., Bengio Y. MIT Press; Cambridge, MA, USA: 1998. Convolutional networks for images, speech, and time series. p. 255?258. [Google Scholar]
- 41.Kitchenham B., Kitchenham . 2004. 2004 procedures for performing systematic reviews. arXiv:https://www.medrxiv.org/content/early/2020/04/22/2020.04.17.20069666.full.pdf. [Google Scholar]
- 42.Kumar N., Susan S. 11th International Conference on Computing, Communication and Networking Technologies, ICCCNT 2020, Kharagpur, India, July 1-3, 2020. 2020. COVID-19 pandemic prediction using time series forecasting models; pp. 1–7. [Google Scholar]
- 43.GitHub Inc. COVID-19 cases. https://github.com/cssegisanddata/covid-19
- 44.Singh S., Makkhan S.J.S., Kaur J., Peshoria S., Kumar J., Parmar K.S. Study of ARIMA and least square support vector machine (LS-SVM) models for the prediction of SARS-CoV-2 confirmed cases in the most affected countries. Chaos, SolitonsFractals. 2020;139 doi: 10.1016/j.chaos.2020.110086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang P., Zheng X., Li J., Zhu B. Prediction of epidemic trends in COVID-19 with logistic model and machine learning technics. Chaos, SolitonsFractals. 2020;139 doi: 10.1016/j.chaos.2020.110058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hernandez-Matamoros A., Fujita H., Hayashi T., Perez-Meana H. Forecasting of COVID19 per regions using arima models and polynomial functions. Appl Soft Comput. 2020;96 doi: 10.1016/j.asoc.2020.106610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shahid F., Zameer A., Muneeb M. Predictions for COVID-19 with deep learning models of lstm, gru and bi-lstm. Chaos, SolitonsFractals. 2020;140 doi: 10.1016/j.chaos.2020.110212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Devaraj J., Madurai Elavarasan R., Pugazhendhi R., Shafiullah G., Ganesan S., Jeysree A.K., Khan I.A., Hossain E. Forecasting of COVID-19 cases using deep learning models: is it reliable and practically significant? ResultsPhys. 2021;21 doi: 10.1016/j.rinp.2021.103817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shastri S., Singh K., Kumar S., Kour P., Mansotra V. Time series forecasting of covid-19 using deep learning models: India-USAcomparative case study. Chaos, SolitonsFractals. 2020;140 doi: 10.1016/j.chaos.2020.110227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeroual A., Harrou F., Dairi A., Sun Y. Deep learning methods for forecasting COVID-19 time-series data: a comparative study. Chaos, SolitonsFractals. 2020;140 doi: 10.1016/j.chaos.2020.110121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.T. Alakus I. Turkoglu, Comparison of deep learning approaches to predict covid-19 infection, Chaos, Solitons and Fractals 140. [DOI] [PMC free article] [PubMed]
- 52.J. Farooq M. Bazaz, A novel adaptive deep learning model of covid-19 with focus on mortality reduction strategies, Chaos, Solitons and Fractals 138. [DOI] [PMC free article] [PubMed]
- 53.Gupta M., Jain R., Taneja S., Chaudhary G., Khari M., Verdú E. Real-time measurement of the uncertain epidemiological appearances of COVID-19 infections. Appl Soft Comput. 2021;101 doi: 10.1016/j.asoc.2020.107039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meng L., Dong D., Li L., Niu M., Bai Y., Wang M., Qiu X., Zha Y., Tian J. A deep learning prognosis model help alert for COVID-19 patients at high-risk of death: a multi-center study. IEEE J Biomed Health Inform. 2020;24(12):3576–3584. doi: 10.1109/JBHI.2020.3034296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Z., Ge Q., Li S., Jin L., Xiong M. Artificial intelligence forecasting of COVID-19 in China. IntJEducExcell. 2020;6(1):71–94. doi: 10.3389/frai.2020.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rizk-Allah R.M., Hassanien A.E. Studies in computational intelligence. Vol. 1005. 2022. COVID-19 forecasting based on an improved interior search algorithm and multi-layer feed forward neural network, medical informatics and bioimaging using artificial intelligence; pp. 129–152. [Google Scholar]
- 57.S. Cabras, A Bayesian - deep learning model for estimating COVID-19 evolution in Spain, Mathematics 9 (22).
- 58.Rustam F., Reshi A.A., Mehmood A., Ullah S., On B., Aslam W., Choi G.S. COVID-19 future forecasting using supervised machine learning models. IEEE Access. 2020;8:101489–101499. [Google Scholar]
- 59.Chakraborty M., Mukhopadhyay A., Maulik U. 2020 IEEE 5th International Conference on Computing Communication and Automation (ICCCA) 2020. A comparative analysis of different regression models on predicting the spread of covid-19 in India; pp. 519–524. [DOI] [Google Scholar]
- 60.G. Pinter I. Felde A. Mosavi P. Ghamisi R. Gloaguen, Covid-19 pandemic prediction for Hungary; a hybrid machine learning approach, Mathematics (6).
- 61.Ahamad M.M., Aktar S., Rashed-Al-Mahfuz M., Uddin S., Liò P., Xu H., Summers M.A., Quinn J.M., Moni M.A. A machine learning model to identify early stage symptoms of sars-cov-2 infected patients. Expert SystApplic. 2020;160 doi: 10.1016/j.eswa.2020.113661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.G. Y. al, Machine learning based early warning system enables accurate mortality risk prediction for covid-19, Nature Communications. [DOI] [PMC free article] [PubMed]
- 63.I. Arpaci S. Huang M. Al-Emran M. Al-Kabi M. Peng, Predicting the covid-19 infection with fourteen clinical features using machine learning classification algorithms, Multimedia Tools and Applications. [DOI] [PMC free article] [PubMed]
- 64.D. Assaf Y. Gutman Y. Neuman G. Segal S. Amit S. Gefen-Halevi N. Shilo A. Epstein R. Mor-Cohen A. Biber G. Rahav I. Levy A. Tirosh, Utilization of machine-learning models to accurately predict the risk for critical covid-19, Internal and Emergency Medicine 15 (8). [DOI] [PMC free article] [PubMed]
- 65.D. Brinati A. Campagner D. Ferrari M. Locatelli G. Banfi F. Cabitza Detection of covid-19 infection from routine blood exams with machine learning: a feasibility study, Journal of Medical Systems 135 (44). [DOI] [PMC free article] [PubMed]
- 66.Chaurasia V., Pal S. Application of machine learning time series analysis for prediction covid-19 pandemic. Res Biomed Eng. 2020:1–13. Special Issue: Emerging Technologies for Fighting COVID-19. [Google Scholar]
- 67.A. Khanday S. Rabani Q. Khan N. Rouf M. Mohi Ud Din, Machine learning based approaches for detecting covid-19 using clinical text data, International Journal of Information Technology (Singapore) 12 (3). [DOI] [PMC free article] [PubMed]
- 68.K. H. Abdulkareem M. A. Mohammed A. Salim M. Arif O. Geman D. Gupta A. Khanna, Realizing an effective covid-19 diagnosis system based on machine learning and iot in smart hospital environment, IEEE Internet of Things Journal DOI 10.1109/JIOT.2021.3050775. [DOI] [PMC free article] [PubMed]
- 69.M. H. D. M. Ribeiro R. G. da Silva V. C. Mariani L. dos Santos Coelho, Short-term forecasting covid-19 cumulative confirmed cases: Perspectives for Brazil, Chaos, Solitons and Fractals 135 (109853). [DOI] [PMC free article] [PubMed]
- 70.Quinlan J.R. Machine Learning, Proceedings of the Tenth International Conference, University of Massachusetts, Amherst, MA, USA, June 27-29, 1993. 1993. Combining instance-based and model-based learning; pp. 236–243. [Google Scholar]
- 71.dos Santos Gomes D.C., de Oliveira Serra G.L. Machine learning model for computational tracking and forecasting the COVID-19 dynamic propagation. IEEE J Biomed Health Inform. 2021;25(3):615–622. doi: 10.1109/JBHI.2021.3052134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.B. B. Hazarika D. Gupta, Modelling and forecasting of covid-19 spread using wavelet-coupled random vector functional link networks, Applied Soft Computing 96 (106626). [DOI] [PMC free article] [PubMed]
- 73.Sahai A.K., Rath N., Sood V., Singh M.P. Arima modelling and forecasting of covid-19 in top five affected countries. <sb:contribution><sb:title>Diabetes Metab Syndr Clin</sb:title></sb:contribution><sb:host><sb:issue><sb:series><sb:title>Res Rev</sb:title></sb:series></sb:issue></sb:host>. 2020;14(5):1419–1427. doi: 10.1016/j.dsx.2020.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheng F.-Y., Joshi H., Tandon P., Freeman R., Reich D.L., Mazumdar M., Kohli-Seth R., Levin M.A., Timsina P., Kia A. Using machine learning to predict icu transfer in hospitalized COVID-19 patients. J Clin Med. 2020;9(6):1668. doi: 10.3390/jcm9061668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nemati M., Ansary J., Nemati N. Machine-learning approaches in COVID-19 survival analysis and discharge-time likelihood prediction using clinical data. Patterns. 2020;1(5) doi: 10.1016/j.patter.2020.100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.H. Burdick C. Lam S. Mataraso A. Siefkas G. Braden R. Dellinger A. McCoy J. Vincent A. Green-Saxena G. Barnes J. Hoffman J. Calvert E. Pellegrini R. Das, Prediction of respiratory decompensation in COVID-19 patients using machine learning: The ready trial, Computers in Biology and Medicine 124. [DOI] [PMC free article] [PubMed]
- 77.AlJame M., Ahmad I., Imtiaz A., Mohammed A. Ensemble learning model for diagnosing COVID-19 from routine blood tests. InformMedUnlocked. 2020;21 doi: 10.1016/j.imu.2020.100449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pourhomayoun M., Shakibi M. Predicting mortality risk in patients with COVID-19 using machine learning to help medical decision-making. Smart Health. 2021;20 doi: 10.1016/j.smhl.2020.100178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hasan N. A methodological approach for predicting COVID-19 epidemic using eemd-ann hybrid model. InternetThings. 2020;11 doi: 10.1016/j.iot.2020.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Casiraghi E., Malchiodi D., Trucco G., Frasca M., Cappelletti L., Fontana T., Esposito A.A., Avola E., Jachetti A., Reese J., Rizzi A., Robinson P.N., Valentini G. Explainable machine learning for early assessment of COVID-19 risk prediction in emergency departments. IEEE Access. 2020;8:196299–196325. doi: 10.1109/ACCESS.2020.3034032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ren J., Yan Y., Zhao H., Ma P., Zabalza J., Hussain Z., Luo S., Dai Q., Zhao S., Sheikh A., Hussain A., Li H. A novel intelligent computational approach to model epidemiological trends and assess the impact of non-pharmacological interventions for COVID-19. IEEE J Biomed Health Inform. 2020;24(12):3551–3563. doi: 10.1109/JBHI.2020.3027987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rostami-Tabar B., Rendon-Sanchez J.F. Forecasting COVID-19 daily cases using phone call data. Appl Soft Comput. 2021;100 doi: 10.1016/j.asoc.2020.106932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ramchandani A., Fan C., Mostafavi A. Deepcovidnet: an interpretable deep learning model for predictive surveillance of COVID-19 using heterogeneous features and their interactions. IEEE Access. 2020;8:159915–159930. doi: 10.1109/ACCESS.2020.3019989. URL doi:10.1109/ACCESS.2020.3019989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guo H., Tang R., Ye Y., Li Z., He X. Proceedings of the Twenty-Sixth International Joint Conference on Artificial Intelligence, IJCAI 2017, Melbourne, Australia, August 19-25, 2017. 2017. Deepfm: a factorization-machine based neural network for CTR prediction; pp. 1725–1731. [Google Scholar]
- 85.Zheng N., Du S., Wang J., Zhang H., Cui W., Kang Z., Yang T., Lou B., Chi Y., Long H., Ma M., Yuan Q., Zhang S., Zhang D., Ye F., Xin J. Predicting COVID-19 in China using hybrid AI model. IEEE TransCybern. 2020;50(7):2891–2904. doi: 10.1109/TCYB.2020.2990162. [DOI] [PubMed] [Google Scholar]
- 86.S. Shastri K. Singh S. Kumar P. Kour V. Mansotra, Deep-lstm ensemble framework to forecast COVID-19: an insight to the global pandemic, International Journal of Information Technology (Singapore). [DOI] [PMC free article] [PubMed]
- 87.S. F. Ardabili A. Mosavi P. Ghamisi F. Ferdinand A. R. Varkonyi-Koczy U. Reuter T. Rabczuk P. M. Atkinson, Covid-19 outbreak prediction with machine learning, Algorithms 13 (10).
- 88.Hazarika B.B., Gupta D. Modelling and forecasting of COVID-19 spread using wavelet-coupled random vector functional link networks. Appl Soft Comput. 2020;96 doi: 10.1016/j.asoc.2020.106626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim M., Kang J., Kim D., Song H., Min H., Nam Y., Park D., Lee J.-G. Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD '20. Association for Computing Machinery; New York, NY, USA: 2020. Hi-covidnet: deep learning approach to predict inbound COVID-19 patients and case study in South Korea; pp. 3466–3473. [Google Scholar]
- 90.C.-J. Huang Y.-H. Chen Y. Ma P.-H. Kuo, Multiple-input deep convolutional neural network model for COVID-19 forecasting in China, medRxiv 2020.03.23.20041608.
- 91.N. S. Punn S. K. Sonbhadra S. Agarwal COVID-19 epidemic analysis using machine learning and deep learning algorithms, medRxiv 2020.04.08.20057679.
- 92.J. Sarkar P. Chakrabarti, A machine learning model reveals older age and delayed hospitalization as predictors of mortality in patients with COVID-19, medRxiv 2020.03.25.20043331.
- 93.L. Yan H.-T. Zhang Y. Xiao M. Wang Y. Guo C. Sun X. Tang L. Jing S. Li M. Zhang Y. Xiao H. Cao Y. Chen T. Ren J. Jin F. Wang Y. Xiao S. Huang X. Tan N. Huang B. Jiao Y. Zhang A. Luo Z. Cao H. Xu Y. Yuan, Prediction of criticality in patients with severe COVID-19 infection using three clinical features: a machine learning-based prognostic model with clinical data in wuhan, medRxiv 2020.02.27.20028027.
- 94.L. Yan H.-T. Zhang J. Goncalves Y. Xiao M. Wang Y. Guo C. Sun X. Tang L. Jin M. Zhang X. Huang Y. Xiao H. Cao Y. Chen T. Ren F. Wang Y. Xiao S. Huang X. Tan N. Huang B. Jiao Y. Zhang A. Luo L. Mombaerts J. Jin Z. Cao S. Li H. Xu Y. Yuan, A machine learning-based model for survival prediction in patients with severe COVID-19 infection, medRxiv 2020.02.27.20028027.
- 95.Kolozsvári L.R., Bérczes T., Hajdu A., Gesztelyi R., Tiba A., Varga I., Szöllösi G.J., Harsányi S., Garbóczy S., Zsuga J. 2020. Predicting the epidemic curve of the coronavirus (sars-cov-2) disease (COVID-19) using artificial intelligence. arXiv:https://www.medrxiv.org/content/early/2020/04/22/2020.04.17.20069666.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li Z., Zheng Y., Xin J., Zhou G. 2020. A recurrent neural network and differential equation based spatiotemporal infectious disease model with application to COVID-19. arXiv:https://www.medrxiv.org/content/early/2020/07/22/2020.07.20.20158568.full.pdf. [Google Scholar]
- 97.Kapoor A., Ben X., Liu L., Perozzi B., Barnes M., Blais M., O'Banion S. 2020. Examining COVID-19 forecasting using spatio-temporal graph neural networks. arXiv:2007.03113. [Google Scholar]
- 98.Vadyala S.R., Betgeri S.N., Sherer E.A., Amritphale A. Prediction of the number of COVID-19 confirmed cases based on k-means-lstm. Array. 2021;11 doi: 10.1016/j.array.2021.100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Javid A.M., Liang X., Venkitaraman A., Chatterjee S. 2020. Predictive analysis of COVID-19 time-series data from Johns Hopkins University. arXiv:2005.05060. [Google Scholar]
- 100.Poonia N., Azad S. 2020. Short-term forecasts of COVID-19 spread across Indian states until 1 May 2020. [Google Scholar]
- 101.Zandavi S.M., Rashidi T.H., Vafaee F. 2020. Forecasting the spread of COVID-19 under control scenarios using lstm and dynamic behavioral models. [Google Scholar]
- 102.Direkoglu C., Sah M. 2020. Worldwide and regional forecasting of coronavirus (COVID-19) spread using a deep learning model. arXiv:https://www.medrxiv.org/content/early/2020/05/26/2020.05.23.20111039.full.pdf. [DOI] [Google Scholar]
- 103.Karimuzzaman M., Afroz S., Hossain M.M., Rahman A. 2020. Forecasting the COVID-19 pandemic with climate variables for top five burdening and three South Asian countries. [Google Scholar]
- 104.Yudistira N. COVID-19 growth prediction using multivariate long short term memory. IAENG IntJComputSci. 2020;47(4):829–837. [Google Scholar]
- 105.J V.R., Jakka A. 2020 International Conference on Smart Technologies in Computing, Electrical and Electronics (ICSTCEE) 2020. Forecasting COVID-19 cases in india using machine learning models; pp. 466–471. [Google Scholar]
- 106.P. Melin J. C. Monica D. Sanchez O. Castillo, Multiple ensemble neural network models with fuzzy response aggregation for predicting COVID-19 time series: The case of Mexico, Healthcare 8 (2). [DOI] [PMC free article] [PubMed]
- 107.Tian T., Jiang Y., Zhang Y., Li Z., Wang X., Zhang H. 2020. COVID-net: a deep learning based and interpretable predication model for the county-wise trajectories of COVID-19 in the United States. arXiv:https://www.medrxiv.org/content/early/2020/05/27/2020.05.26.20113787.full.pdf, doi:10.1101/2020.05.26.20113787. URL https://www.medrxiv.org/content/early/2020/05/27/2020.05.26.20113787. [Google Scholar]
- 108.L. R. Kolozsvári T. Bérczes A. Hajdu R. Gesztelyi A. Tiba I. Varga G. J. Szöllösi S. Harsányi S. Garbóczy J. Zsuga, Predicting the epidemic curve of the coronavirus (sars-cov-2) disease (COVID-19) using artificial intelligence, medRxiv:2020.04.17.20069666. [DOI] [PMC free article] [PubMed]
- 109.M. Amo-Boateng, Tracking and classifying global COVID-19 cases by using 1d deep convolution neural networks, medRxiv 2020.06.09.20126565.
- 110.Z. Zhao K. Nehil-Puleo Y. Zhao, How well can we forecast the COVID-19 pandemic with curve fitting and recurrent neural networks?, medRxiv 2020.06.09.20126565.
- 111.Kumar A., Khan F.M., Gupta R., Puppala H. 2020. Preparedness and mitigation by projecting the risk against COVID-19 transmission using machine learning techniques. [Google Scholar]
- 112.P. Mathur T. Sethi A. Mathur K. Maheshwari J. B. Cywinski A. K. Khanna S. Dua F. Papay, Explainable machine learning models to understand determinants of COVID-19 mortality in the united states, medRxiv:2020.05.23.20110189.
- 113.Tian Y., Luthra I., Zhang X. 2020. Forecasting COVID-19 cases using machine learning models. [Google Scholar]
- 114.Liu D., Clemente L., Poirier C., Ding X., Chinazzi M., Davis J.T., Vespignani A., Santillana M. 2020. A machine learning methodology for real-time forecasting of the 2019-2020 COVID-19 outbreak using internet searches, news alerts, and estimates from mechanistic models. arXiv:2004.04019. [Google Scholar]
- 115.Khmaissia F., Haghighi P.S., Jayaprakash A., Wu Z., Papadopoulos S., Lai Y., Nguyen F.T. 2020. An unsupervised machine learning approach to assess the zip code level impact of COVID-19 in nyc. [Google Scholar]
- 116.Suzuki Y., Suzuki A., Nakamura S., Ishikawa T., Kinoshita A. 2020. Machine learning model estimating number of COVID-19 infection cases over coming 24 days in every province of South Korea (xgboost and multioutputregressor) [Google Scholar]
- 117.Pereira I.G., Guerin J.M., Júnior A.G.Silva, Distante C., Garcia G.S., Gonçalves L.M.G. 2020. Forecasting COVID-19 dynamics in Brazil: a data driven approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Balde M.A.M.T., Balde C., Ndiaye B.M. 2020. Impact studies of nationwide measures COVID-19 anti-pandemic: compartmental model and machine learning. [Google Scholar]
- 119.Uhlig S., Nichani K., Uhlig C., Simon K. 2020. Modeling projections for COVID-19 pandemic by combining epidemiological, statistical, and neural network approaches. [Google Scholar]
- 120.Dandekar R., Barbastathis G. 2020. Neural network aided quarantine control model estimation of COVID spread in Wuhan, China. [Google Scholar]
- 121.da Silva R.G., Ribeiro M.H.D.M., Mariani V.C., Coelho L.d.S. Forecasting Brazilian and American COVID-19 cases based on artificial intelligence coupled with climatic exogenous variables. Chaos, Solitons Fractals. 2020;139 doi: 10.1016/j.chaos.2020.110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Banerjee A., Ray S., Vorselaars B., Kitson J., Mamalakis M., Weeks S., Baker M., Mackenzie L.S. Use of machine learning and artificial intelligence to predict sars-cov-2 infection from full blood counts in a population. Int Immunopharmacol. 2020;86 doi: 10.1016/j.intimp.2020.106705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.D. Giuliani M. M. Dickson G. Espa F. Santi, Modelling and predicting the spatio-temporal spread of coronavirus disease 2019 (COVID-19) in Italy, BMC Infect Dis 20 (700). [DOI] [PMC free article] [PubMed]
- 124.Braga M.d.B., Fernandes R.d.S., Souza G.N.d., Rocha J.E.C.d., Jr., Dolácio C.J.F., Tavares I.d.S., et al. Artificial neural networks for short-term forecasting of cases, deaths, and hospital beds occupancy in the COVID-19 pandemic at the Brazilian Amazon. PLOS ONE. 2021;16(3):1–27. doi: 10.1371/journal.pone.0248161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.B. Pirouz S. Shaffiee Haghshenas S. Shaffiee Haghshenas P. Piro, Investigating a serious challenge in the sustainable development process: Analysis of confirmed cases of COVID-19 (new type of coronavirus) through a binary classification using artificial intelligence and regression analysis, Sustainability 12 (6).
- 126.Khakharia A., Shah V., Jain S., Shah J., Tiwari A., Daphal P., Warang M., Mehendale N. Outbreak prediction of COVID-19 for dense and populated countries using machine learning. <sb:contribution><sb:title>Ann Data</sb:title></sb:contribution><sb:host><sb:issue><sb:series><sb:title>Sci</sb:title></sb:series></sb:issue></sb:host>. 2021;8:1–19. doi: 10.1007/s40745-020-00314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ghany K.K.A., Zawbaa H.M., Sabri H.M. COVID-19 prediction using lstm algorithm: Gcc case study. InformMedUnlocked. 2021;23 doi: 10.1016/j.imu.2021.100566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Khan F.M., Gupta R. Arima and nar based prediction model for time series analysis of COVID-19 cases in India. JSafSciResilience. 2020;1(1):12–18. [Google Scholar]
- 129.R. K. Singh M. Rani A. S. Bhagavathula R. Sah A. J. Rodriguez-Morales H. Kalita C. Nanda S. Sharma Y. D. Sharma A. A. Rabaan J. Rahmani P. Kumar, Prediction of the COVID-19 pandemic for the top 15 affected countries: Advanced autoregressive integrated moving average (arima) model, JMIR Public Health Surveill 6 (2). [DOI] [PMC free article] [PubMed]
- 130.Car Z., Šegota S.B., Andelić N., Lorencin V.M.Ivan. Modeling the spread of COVID-19 infection using a multilayer perceptron. Comput Math Methods Med. 2020;2020:1–10. doi: 10.1155/2020/5714714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fong S.J., Li G., Dey N., Crespo R.G., Herrera-Viedma E. Composite Monte Carlo decision making under high uncertainty of novel coronavirus epidemic using hybridized deep learning and fuzzy rule induction. Appl Soft Comput. 2020;93 doi: 10.1016/j.asoc.2020.106282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Fong S.J., Li G., Dey N., Gonzalez-Crespo R., Herrera-Viedma E. Finding an accurate early forecasting model from small dataset: a case of 2019-ncov novel coronavirus outbreak. IntJInteractMultimediaArtifIntell. 2020;6(1):132. [Google Scholar]
- 133.Hartono P. Similarity maps and pairwise predictions for transmission dynamics of COVID-19 with neural networks. InformMedUnlocked. 2020;20 doi: 10.1016/j.imu.2020.100386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sujath R., Chatterjee J., Hassanien A. Stoch environ res risk assess. IEEE Access. 2020;34:959–972. doi: 10.1007/s00477-020-01827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Melin P., Monica J.C., Sanchez D., Castillo O. Analysis of spatial spread relationships of coronavirus (COVID-19) pandemic in the world using self organizing maps. Chaos, SolitonsFractals. 2020;138 doi: 10.1016/j.chaos.2020.109917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.A. Mollalo K. M. Rivera B. Vahedi, Artificial neural network modeling of novel coronavirus (COVID-19) incidence rates across the continental united states, International Journal of Environmental Research and Public Health 17 (12). [DOI] [PMC free article] [PubMed]
- 137.Tamang S., Singh P., Datta B. Forecasting of COVID-19 cases based on prediction using artificial neural network curve fitting technique. Glob J Environ Sci Manag. 2020;6(Special Issue (COVID-19)):53–64. [Google Scholar]
- 138.Torrealba-Rodriguez O., Conde-Gutiérrez R., Hernández-Javier A. Modeling and prediction of COVID-19 in Mexico applying mathematical and computational models. Chaos, SolitonsFractals. 2020;138 doi: 10.1016/j.chaos.2020.109946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Distante C., Pereira I.G., Gonçalves L.M.G., Piscitelli P., Miani A. 2020. Forecasting COVID-19 outbreak progression in Italian regions: a model based on neural network training from Chinese data. [DOI] [Google Scholar]
- 140.Saba A.I., Elsheikh A.H. Forecasting the prevalence of COVID-19 outbreak in Egypt using nonlinear autoregressive artificial neural networks. Process SafEnvironProt. 2020;141:1–8. doi: 10.1016/j.psep.2020.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.A. Chatterjee M. W. Gerdes S. G. Martinez, Statistical explorations and univariate timeseries analysis on COVID-19 datasets to understand the trend of disease spreading and death, Sensors 20 (11). [DOI] [PMC free article] [PubMed]
- 142.Chimmula V.K.R., LeiZhang Time series forecasting of COVID-19 transmission in Canada using lstm networks. Chaos, Solitons Fractals. 2020;135 doi: 10.1016/j.chaos.2020.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aldhyani T.H.H., Alrasheed M., Al-Adaileh M.H., Alqarni A.A., Alzahrani M.Y., Alahmadi A.H. Deep learning and holt-trend algorithms for predicting COVID-19 pandemic. ComputMaterContinua. 2021;67(2):2141–2160. [Google Scholar]
- 144.Tomar A., Gupta N. Prediction for the spread of COVID-19 in India and effectiveness of preventive measures. Sci Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.S. M. Ayyoubzadeh S. M. Ayyoubzadeh H. Zahedi M. Ahmadi S. R Niakan Kalhori, Predicting COVID-19 incidence through analysis of google trends data in Iran: Data mining and deep learning pilot study, JMIR Public Health Surveill 6 (2). [DOI] [PMC free article] [PubMed]
- 146.Pal R., Sekh A.A., Kar S., Prasad D.K. Neural network based country wise risk prediction of COVID-19. ApplSci. 2020;10(18):6448. [Google Scholar]
- 147.Malki Z., Atlam E.-S., Hassanien A.E., Dagnew G., Elhosseini M.A., Gad I. Association between weather data and COVID-19 pandemic predicting mortality rate: machine learning approaches. Chaos, SolitonsFractals. 2020;138 doi: 10.1016/j.chaos.2020.110137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.S. Shaffiee Haghshenas B. Pirouz S. Shaffiee Haghshenas B. Pirouz P. Piro K.-S. Na S.-E. Cho Z. W. Geem, Prioritizing and analyzing the role of climate and urban parameters in the confirmed cases of COVID-19 based on artificial intelligence applications, International Journal of Environmental Research and Public Health 17 (10). [DOI] [PMC free article] [PubMed]
- 149.İsmail Kırbaş A., Sözen A.D., Tuncer F.Şinasi, Kazancıoğlu Comparative analysis and forecasting of COVID-19 cases in various European countries with arima, narnn and lstm approaches. Chaos, SolitonsFractals. 2020;138 doi: 10.1016/j.chaos.2020.110015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yadav M., Perumal M., Srinivas M. Analysis on novel coronavirus (COVID-19) using machine learning methods. Chaos, SolitonsFractals. 2020;139 doi: 10.1016/j.chaos.2020.110050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Peng Y., Nagata M.H. An empirical overview of nonlinearity and overfitting in machine learning using COVID-19 data. Chaos, SolitonsFractals. 2020;139 doi: 10.1016/j.chaos.2020.110055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.M. A. A. Al-qaness A. A. Ewees H. Fan L. Abualigah M. Abd Elaziz, Marine predators algorithm for forecasting confirmed cases of COVID-19 in Italy, USA, Iran and Korea, International Journal of Environmental Research and Public Health 17 (10). [DOI] [PMC free article] [PubMed]
- 153.M. A. A. Al-qaness A. A. Ewees H. Fan M. Abd El Aziz, Optimization method for forecasting confirmed cases of COVID-19 in China, Journal of Clinical Medicine 9 (3). [DOI] [PMC free article] [PubMed]
- 154.A. A., P. A., C. E., D. S., M. N., M. L., Prognostic modeling of covid-19 using artificial intelligence in the united kingdom: Model development and validation., J Med Internet Res. 22 (8). [DOI] [PMC free article] [PubMed]