SUMMARY
Targeted delivery of therapeutic proteins toward specific cells and across cell membranes remains major challenges. Here, we develop protein-based delivery systems utilizing detoxified single-chain bacterial toxins such as diphtheria toxin (DT) and botulinum neurotoxin (BoNT)-like toxin, BoNT/X, as carriers. The system can deliver large protein cargoes including Cas13a, CasRx, Cas9, and Cre recombinase into cells in a receptor-dependent manner, although delivery of ribonucleoproteins containing guide RNAs is not successful. Delivery of Cas13a and CasRx, together with guide RNA expression, reduces mRNAs encoding GFP, SARS-CoV-2 fragments, and endogenous proteins PPIB, KRAS, and CXCR4 in multiple cell lines. Delivery of Cre recombinase modifies the reporter loci in cells. Delivery of Cas9, together with guide RNA expression, generates mutations at the targeted genomic sites in cell lines and induced pluripotent stem cell (iPSC)-derived human neurons. These findings establish modular delivery systems based on single-chain bacterial toxins for delivery of membrane-impermeable therapeutics into targeted cells.
Graphical Abstract
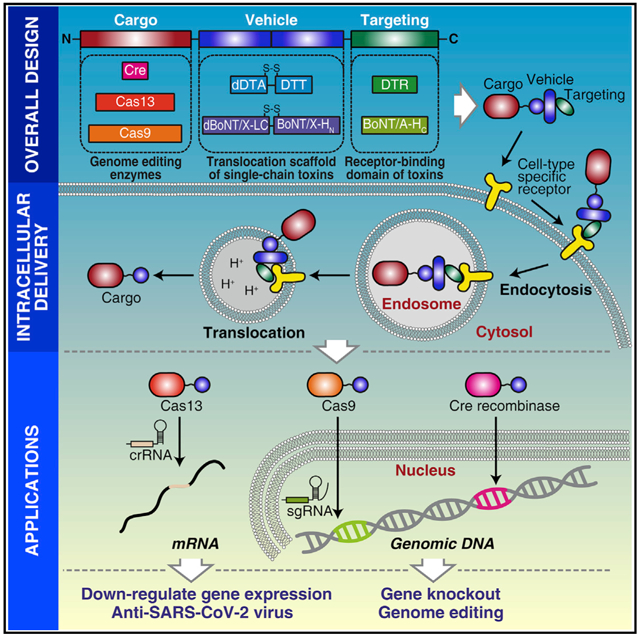
In brief
Targeted intracellular delivery of therapeutic proteins toward specific cells is challenging. Tian et al. present a protein delivery system utilizing detoxified single-chain bacterial toxins as carriers. This system can deliver large protein cargoes including Cas13a, CasRx, Cas9, and Cre into cell lines and iPSC-derived human neurons for editing mRNAs and genomes.
INTRODUCTION
Development of protein-based biological therapeutics has revolutionized treatment for many human diseases, yet since the cell membrane is a formidable barrier, these therapeutics are largely limited to cell surface targets. Developing effective delivery methods that can mediate entry of membrane-impermeable macromolecules into cells is essential to unlock the therapeutic potential of targeting intracellular substrates.
CRISPR-Cas (clustered regularly interspaced short palindromic repeats-CRISPR-associated) systems are a major class of potential therapeutic tools. CRISPR-Cas systems, an adaptive immune system widespread in bacteria and archaea (Barrangou et al., 2007), are comprised of Cas proteins and CRISPR RNA (crRNA), which together target and cleave invading genetic materials through RNA-guided endonuclease activity. Among the diverse CRISPR-Cas systems, CRISPR-Cas9 from Streptococcus pyogenes is the most popular (Koonin et al., 2017). Cas9 can bind to single-guide RNA (sgRNA) containing an ~20-nucleotide sequence designed to match the targeted genes (Cong et al., 2013; Jiang and Doudna, 2017; Jinek et al., 2012; Mali et al., 2013). The resulting ribonucleoprotein (RNP) targets the DNA substrate containing the complementary sequence, and Cas9 then cleaves the DNA, generating site-specific double-strand breaks (DSBs) that can be repaired through non-homologous end-joining (NHEJ) or homology-directed repair (HDR) in cells. NHEJ often leads to nucleotide insertions or deletions (indels) around the DSB site (Jinek et al., 2012), disrupting the open reading frame of the target gene. HDR requires exogenous DNA templates that can direct the replacement of the native DNA sequence with the designed one. CRISPR-Cas9 and its derivatives have been extensively developed for therapeutic applications (Doudna, 2020; Fellmann et al., 2017; Jinek et al., 2012; Knott and Doudna, 2018; Ran et al., 2015; Shalem et al., 2014; Zhou et al., 2014).
Besides targeting DNAs, there are also CRISPR-Cas systems targeting RNA, such as the Cas13 family, including Cas13a (previously known as C2c2), Cas13b, Cas13c, and Cas13d (Abudayyeh et al., 2017; Cox et al., 2017; East-Seletsky et al., 2017; Knott et al., 2017). Cas13a from Leptotrichia wadei is a well-characterized member, which uses a single crRNA containing an ~36-nucleotide direct repeat (DR) and a 28- to 30-nucleotide spacer to target the spacer complementary region within single-stranded RNA (ssRNA) substrates (Abudayyeh et al., 2016, 2017; East-Seletsky et al., 2016). The CRISPR-Cas13a system has been utilized for various applications including messenger RNA (mRNA) knockdown, mRNA live imaging, and RNA base editing (Abudayyeh et al., 2017, 2019; Cox et al., 2017). Several studies have shown that CRISPR-Cas13 systems can be utilized to develop ultra-sensitive and easily programmable assays to detect viral RNAs as well as to target and cleave viral RNAs within mammalian cells (Abbott et al., 2020; Abudayyeh et al., 2017; Ackerman et al., 2020; Cox et al., 2017; Freije et al., 2019; Gootenberg et al., 2017). Furthermore, there have been recent reports of proof-of-principle experiments utilizing the CRISPR-Cas13-based approaches to target and reduce influenza viruses and the pandemic severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in vivo in mouse and hamster models (Blanchard et al., 2021).
To realize the therapeutic potential of Cas9 and Cas13, it is necessary to deliver them into human cells. Viral-based delivery platforms have often been utilized and can provide sustained expression of Cas proteins (Ran et al., 2015; Wang et al., 2020). However, these virus-based methods are associated with several limitations and safety concerns, including risks of integration into the host genome, existing immunity against viral vectors, and limitations in the size of the DNA fragment that can be packaged efficiently (Hanlon et al., 2019; Lino et al., 2018; Wang et al., 2020; Yip, 2020). Furthermore, sustained expression of the CRISPR-Cas components in cells increases the chance of off-target cleavage (Hanlon et al., 2019; Vakulskas and Behlke, 2019). Non-viral methods based on liposomes, nanoparticles, cell-penetration peptides, and physical methods such as electroporation have been utilized to deliver Cas proteins, RNP, or mRNA-encoding CRISPR-Cas systems (Blanchard et al., 2021; Lino et al., 2018; Ramakrishna et al., 2014; Rouet et al., 2018; Shalaby et al., 2020; Yip, 2020; Zuris et al., 2015). These methods provide the transient presence of Cas nucleases in cells, thus reducing off-target effects and immune responses. However, they generally showed low efficacy in targeting specific cells such as neurons and stem cells due to a lack of intrinsic machinery for recognizing cells and escaping from endosomes.
Many of the most potent bacterial exotoxins are elegant nanomachines for targeting cells and delivering protein cargoes into the cell cytosol (Alouf, 2005). Once released from bacteria, these toxins act autonomously as “guided missiles”: targeting cells by binding to receptors, crossing the endosomal membrane, and delivering an enzymatic domain into the cytosol. These multiple steps of toxin action are achieved with specialized domains arranged in an “AB” paradigm, with the A domain the enzymatic cargo and the B domain a delivery tool composed of the receptor-binding domain and membrane translocation domain (Barth et al., 2004; Odumosu et al., 2010). There are many variations within the AB toxin concept, including single-chain toxins such as diphtheria toxin (DT) and botulinum neurotoxins (BoNTs) that contain A and B domains within a single polypeptide (Dong et al., 2019; Pirazzini et al., 2017), toxins composed of separate A and B domains such as anthrax toxin (Young and Collier, 2007), and toxins that do not contain their own translocation domain such as Shiga toxin and cholera toxin, which enter the ER via retrograde sorting and escape from the ER into the cytosol using the host machinery (Johannes and Romer, 2010).
Many of these toxins have previously been explored for delivery of protein cargoes into cells (Auger et al., 2015; Bade et al., 2004; Barth et al., 2002; Beilhartz et al., 2017; Chen et al., 2015; Fahrer et al., 2013; Ho et al., 2011; McNutt et al., 2021; Miyashita et al., 2021; Ng Ang et al., 2019; Roderer et al., 2019; Ryou et al., 2016; Schmit et al., 2019; Vidimar et al., 2020). In general, the cargo proteins are fused with either the whole or a fragment of the A domain. For instance, a well-characterized and widely utilized delivery system is based on anthrax toxin. A wide range of cargoes fused with the N-terminal fragment of the anthrax toxin A domain can be delivered into cultured cells through the transmembrane pore formed by its B domain (Dyer et al., 2015; Liao et al., 2014; McCluskey and Collier, 2013; Milne et al., 1995; Rabideau and Pentelute, 2016; Verdurmen et al., 2015). Furthermore, cell-type specificity can be changed by altering the receptor-binding domain (McCluskey and Collier, 2013; Miyashita et al., 2021). However, anthrax toxin B domain forms a rigid heptameric transmembrane pore, and the cargo proteins must unfold within endosomes to pass through this pore, limiting the choice of cargo proteins (Beilhartz et al., 2017; Verdurmen et al., 2015). In contrast, delivery systems based on DT and Pseudomonas exotoxin A may deliver a wider range of cargoes (Auger et al., 2015; Verdurmen et al., 2015), although their potential for delivery of large proteins such as Cas9 (~163 kDa) and Cas13a (~143 kDa) has yet to be explored.
Here we developed versatile single-chain AB toxin-based platforms for delivering Cas9 and Cas13 proteins into cells, utilizing DT and a BoNT-like toxin (BoNT/X) as examples (Zhang et al., 2017). We validated that these engineered delivery platforms can effectively target cells in a receptor-dependent manner and deliver large proteins across the cell membrane into cultured cell lines and human induced pluripotent stem cell (iPSC)-derived neurons. Both cargoes and receptor-binding domains of these single-chain toxin-based platforms can be re-programmed with distinct specificity, offering protein-based tools that can achieve cell-type-specific intracellular delivery of large Cas proteins for scientific and therapeutic applications.
RESULTS
DT and BoNT/X-based delivery platforms
DT (58 kDa) is the archetype for single-chain AB toxins. Its B domain is composed of a membrane translocation domain (DTT, ~20 kDa) and a receptor-binding domain (DTR, ~17 kDa). DT recognizes widely expressed heparin-binding epidermal growth factor precursor (HBEGF) as a receptor (Naglich et al., 1992). The A domain (DTA, ~21 kDa) blocks protein synthesis in cells by ADP-ribosylation of elongation factor 2 (Collier, 1967). The A and B domains are produced as a single polypeptide in bacteria, and the linker region between the two domains is then cleaved by proteases such as cell surface furin in a process that activates the toxicity of DT (Figure 1A) (Greenfield et al., 1983). The two domains remain connected via a single disulfide bond. DT recognizes and enters cells via receptor-mediated endocytosis. Endosomal acidification then induces conformational changes in DT, resulting in translocation of DTA across the endosomal membrane into the cytosol, where the disulfide bond is reduced and DTA is released into cells (Cheung et al., 1985; Donovan et al., 1981, 1982, 1985).
Figure 1. Delivery of Cas13a into cells using DT-based and BoNT/X-based systems.
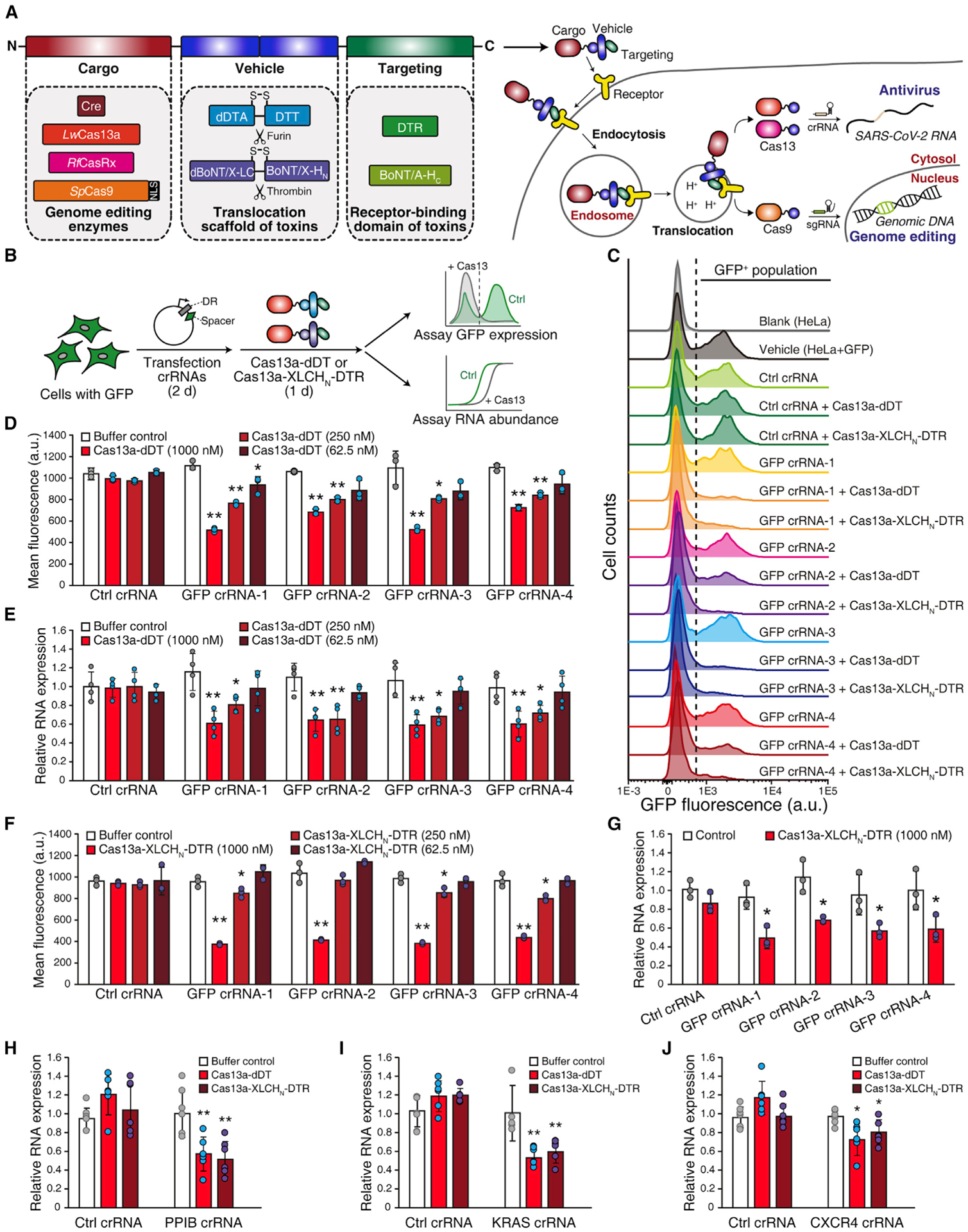
(A) Schematic illustrations of single-chain AB toxin-based delivery systems. The chimera proteins contain three domains: the N-terminal cargo, the middle translocation unit, and the C-terminal cell-targeting domain. Four enzymes were selected as cargoes: Cre recombinase, Cas13a, CasRx, and Cas9. The two translocation units are the deactivated diphtheria toxin enzymatic domain (dDTA) plus translocation domain (DTT), and the deactivated botulinum neurotoxin-like toxin enzymatic domain (dBoNT/X-LC) plus translocation domain (BoNT/X-HN). There is an endogenous furin cleavage site between dDTA and DTT and an engineered thrombin cleavage site between dBoNT/X-LC and BoNT/X-HN. The receptor-binding domain of diphtheria toxin (DTR) and BoNT type A (BoNT/A-HC) were used as cell-targeting domains. NLS, nuclear localization signal; crRNA, CRISPR RNA; sgRNA, single-guide RNA.
(B) Schematic illustration of the GFP reporter assay. Cells over-expressing GFP were generated via lentiviral transduction. The plasmids expressing GFP-targeting crRNAs were transfected into cells for 48 h. Cas13a-dDT or activated Cas13a-XLCHN-DTR was then added to cells for 24 h. GFP protein expression was analyzed by flow cytometry, and GFP mRNA levels were assessed by qRT-PCR. DR, direct repeat.
(C) Representative flow cytometry histograms. GFP over-expressing HeLa cells (HeLa + GFP) have ~50% GFP-positive (GFP+) population. Cells treated with 1 μM Cas13a-dDT or activated Cas13a-XLCHN-DTR together with GFP-targeting crRNAs but not non-targeting crRNA (Ctrl crRNA) showed lower GFP+ population.
(D–G) Quantification of the reduction of GFP by flow cytometry histogram and qRT-PCR. HeLa + GFP cells treated with crRNAs and Cas13a-dDT (D and E) or activated Cas13a-XLCHN-DTR (F and G) at the indicated concentrations. The reduction of mean fluorescence intensity (MFI) in GFP crRNA groups shown as concentration-dependent (D and F). Total RNA was extracted, and the relative expression level of GFP mRNA was quantified by qRT-PCR (E and G). GAPDH was used as housekeeping control. Error bars indicate mean ± SD; N = 3 (biological replicates); *p < 0.05; **p < 0.01 (Student’s t test).
(H–J) Knockdown endogenous transcripts by delivered Cas13a. HEK293 cells were transfected with crRNAs targeting endogenous PPIB (H), KRAS (I), or CXCR4 (J) for 48 h, followed by treatment with 1 μM Cas13a-dDT or activated Cas13a-XLCHN-DTR for 24 h. The total RNA was extracted, and the relative mRNA levels of PPIB, KRAS, or CXCR4 were quantified by qRT-PCR. GAPDH was used as housekeeping control. Error bars indicate mean ± SD; N = 6 (biological replicates); *p < 0.05; **p < 0.01 (Student’s t test).
To utilize DT as a delivery tool, three point-mutations (K51E, G52E, and E138K) were designed and introduced into DTA to abolish its ADP-ribosylation activity and toxicity (Fu et al., 1997). We then constructed a fusion protein of Cas13a fused to the N terminus of this detoxified DT (dDT), with a short GSGSGSGSGS linker separating Cas13a and dDT. The fusion protein (Cas13a-dDT, ~205 kDa; Figure 1A) was expressed and purified with a C-terminal His6-tag using Escherichia coli (Figure S1A).
To further generalize our approach, we also utilized a BoNT-like toxin known as BoNT/X to build a chimeric toxin-based platform. BoNT/X is a homolog of BoNT family members and shares the conserved three-domain modules: a receptor-binding domain (HC), a translocation domain (HN), and an enzymatic domain (also known as light chain, LC; Figure 1A) (Dong and Stenmark, 2021; Zhang et al., 2017, 2018). In contrast to BoNTs, BoNT/X does not target motor neurons in animals, and its natural target remains unknown (Zhang et al., 2017, 2018). We recently showed that a delivery system can be constructed by replacing the BoNT/X-HC with a HC of a BoNT, and the BoNT/X-LC can be detoxified by introducing three point-mutations (Miyashita et al., 2021). Such a delivery system can efficiently target motor neurons and deliver fused nanobodies into the cytosol of neurons in vitro and in vivo (Miyashita et al., 2021). The reason we selected BoNT/X instead of a BoNT is that previous studies have suggested that BoNTs have residual toxicity in vivo for unknown reasons even after their LCs are deactivated (Miyashita et al., 2021; Vazquez-Cintron et al., 2017). In contrast, chimeric proteins composed of detoxified BoNT/X-LCHN fused with a BoNT-HC showed no toxicity in vivo (Miyashita et al., 2021).
Here, we constructed a modular fusion protein by replacing BoNT/X-HC with DTR and fusing Cas13a to the N terminus of detoxified BoNT/X-LC, generating the ~267-kDa chimera protein Cas13a-XLCHN-DTR (Figures 1A and S1B). A thrombin cleavage site was introduced to the linker region between the LC and HN; thus, Cas13a-XLCHN-DTR can be specifically activated by thrombin treatment in vitro, with the two fragments (Cas13a-XLC and XHN-DTR) remaining connected via a disulfide bond (Figure S1N). Cas13a-XLC would be the cargo protein delivered into the cytosol of cells.
Cas13a is delivered into target cells
To assess cytosolic delivery of Cas13a, we first examined its ability to target and suppress green fluorescent protein (GFP) expression in cells (Figure 1B). GFP was expressed in HeLa cells (HeLa-GFP) via lentiviral transduction. HeLa-GFP cells were seeded into 24-well plates. Four crRNAs targeting distinct regions in the GFP mRNA (crRNA-1–4) or a control non-targeting crRNA (Ctrl crRNA) were expressed in HeLa-GFP cells via transient transfection. Cas13a-dDT protein (62.5 nM to 1 μM) was then added to fresh cell culture medium and incubated for 24 h. GFP protein in cells was analyzed by flow cytometry, and its mRNA was quantified using real-time PCR (qRT-PCR; Figure 1B). Incubation with Cas13a-dDT reduced both GFP protein and mRNA in cells that express crRNAs targeting GFP but not in cells that express control crRNA (Figures 1C–1E). Quantification of the mean fluorescence intensity (MFI) of cells showed that exposure to 250 nM Cas13a-dDT reduced MFI by ~26%, 22%, 23%, and 19%, respectively, in cells that express each of the four GFP-targeting crRNAs, and 1 μM Cas13a-dDT reduced MFI 30%–50% (Figure 1D). Similar results were replicated on multiple cell lines, including human bladder cancer cell line 5637 (Figures S2A and S2B), human lung cancer cell line A549 (Figures S2C and S2D), and green monkey Vero cells (Figures S2E and S2F).
Similar to Cas13a-dDT, exposure to Cas13a-XLCHN-DTR suppressed GFP in HeLa cells that express GFP-targeting crRNAs (Figure 1C). For instance, incubation with 1 μM Cas13a-XLCHN-DTR reduced GFP MFI by ~60% and mRNA by ~40% in cells that express GFP-targeting crRNA-1–4 (Figures 1F and 1G). Cell viability was not affected by either Cas13a-dDT or Cas13a-XLCHN-DTR at the highest concentration tested (5 μM; Figure S2G).
Besides targeting GFP, we further analyzed endogenous protein targets including peptidylprolyl isomerase B (PPIB), KRAS, and C-X-C motif chemokine receptor 4 (CXCR4). Previously validated crRNA sequences targeting these proteins were expressed in cells via transient transfection (Abudayyeh et al., 2017). As shown in Figures 1H–1J, 1 μM Cas13a-dDT treatment reduced PPIB, KRAS, and CXCR4 mRNAs by 42%, 45%, and 28%, respectively, whereas these mRNAs were not reduced in cells expressing non-targeting control crRNAs. Similarly, 1 μM Cas13a-XLCHN-DTR reduced endogenous mRNAs of PPIB, KRAS, and CXCR by 50%, 38%, and 19%, respectively (Figures 1H–1J). These findings demonstrate that Cas13a can be delivered into the cytosol of cells within a fusion protein with dDT or XLCHN-DTR.
We further assessed delivery of another member of the Cas13 family, Cas13d from Ruminococcus flavefaciens (CasRx in short) (Konermann et al., 2018; Yan et al., 2018). We constructed and purified CasRx-dDT (173 kDa; Figure S1E) and CasRx-XLCHN-DTR (234 kDa, Figures S1F and S1N). Delivery of CasRx is less efficient than Cas13a, despite CasRx being a smaller protein (~112 kDa) than Cas13a (Figures S3A–S3E). For instance, only HeLa cells incubated with 1 μM of activated CasRx-XLCHN-DTR showed reduction in GFP levels (Figures S3D and S3E).
Delivery depends on receptor recognition and membrane translocation
To determine whether delivery of Cas13a utilizes the translocation domain of the toxin, we performed four sets of experiments. First, Cas13a protein alone (Figure S1C) did not affect GFP in HeLa cells expressing crRNAs (Figures 2A and 2B). Second, Cas13a-XLCHN-DTR without thrombin activation did not reduce GFP in cells, as Cas13a-XLC would remain fused with the translocation domain (Figures 2A and 2B). Third, we introduced a mutation (E349K) to DT that disrupts the function of its translocation domain (O’Keefe et al., 1992), and this mutant Cas13a-dDT(E349K) showed lower ability to affect GFP in cells compared with Cas13a-dDT (Figures S1D, 2C, and 2D). Fourth, pre-treatment with the endosomal acidification inhibitor Bafilomycin A1 (BafA) protected GFP levels in cells, likely by blocking translocation of Cas13a-dDT (Figures 2C and 2D). These data indicate that delivery of Cas13a into cells involves translocation across endosomal membranes.
Figure 2. Delivery of Cas13a depends on receptor recognition and membrane translocation.
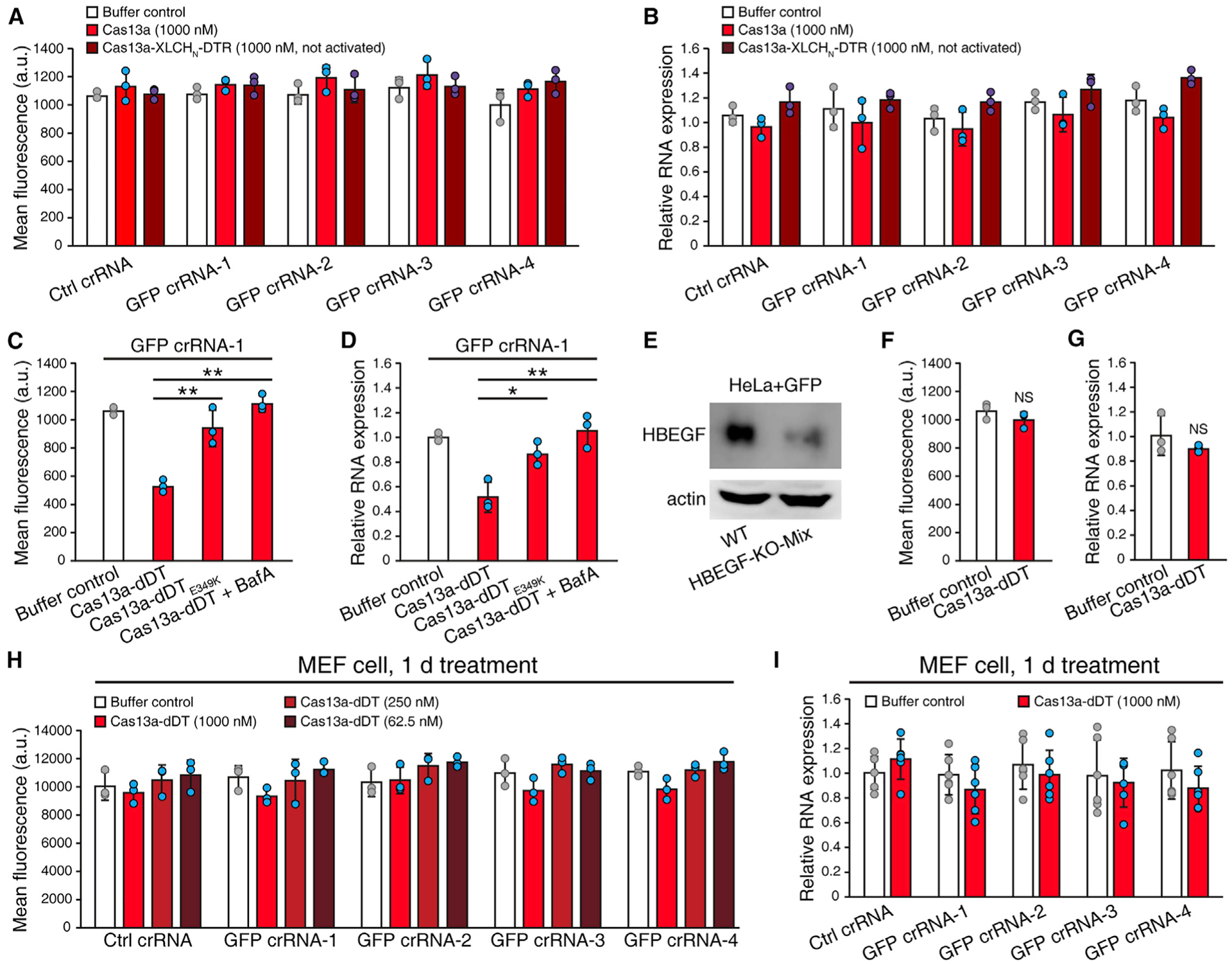
(A and B) Cas13a and Cas13a-XLCHN-DTR without thrombin activation could not reduce GFP in cells. HeLa + GFP cells were transfected with Ctrl or GFP-targeting crRNAs and treated with 1 μM Cas13a or Cas13a-XLCHN-DTR without thrombin activation. GFP protein was measured by flow cytometry and quantified by MFI (A). GFP mRNA was analyzed by qRT-PCR (B). Error bars indicate mean ± SD; N = 3 (biological replicates).
(C and D) Delivery of Cas13a is dependent on DT translocation. HeLa + GFP cells were transfected with GFP crRNA-1 and treated with 1 μM Cas13a-dDT, 1 μM Cas13a-dDT(E349K), or 1 μM Cas13a-dDT combined with Bafilomycin A1 (BafA) pretreatment. GFP protein was measured by flow cytometry and quantified by MFI (C) and GFP mRNA was analyzed by qRT-PCR (D). Error bars indicate mean ± SD; N = 3 (biological replicates); *p < 0.05; **p < 0.01 (Student’s t test).
(E–G) Delivery of Cas13a is dependent on HBEGF receptor. HBEGF was knocked out via CRISPR in HeLa + GFP cells (E). HBEGF-KO-Mix cells were transfected with GFP crRNA-1 and treated with 1 μM Cas13a-dDT or buffer control. GFP protein was measured by flow cytometry and quantified by MFI (F), and GFP mRNA was analyzed by qRT-PCR (G). Error bars indicate mean ± SD; N = 3 (biological replicates); NS, not significant (Student’s t test).
(H and I) GFP-expressing mouse MEF cells were transfected with Ctrl or GFP-targeting crRNAs and treated with different concentrations of Cas13a-dDT for 24 h. GFP protein was measured by flow cytometry and quantified by MFI (H), and GFP mRNA was analyzed by qRT-PCR (I). Error bars indicate mean ± SD; N = 6 (biological replicates).
To assess whether entry of Cas13a-dDT is receptor dependent, we generated HBEGF knockout (KO) HeLa cells (HBEGF-KO-Mix) by CRISPR-Cas9 approach. The reduction of HBEGF in cells was confirmed by immunoblot analysis of cell lysates (Figure 2E). Cas13a-dDT (1 μM) failed to reduce the GFP level in these HBEGF KO cells (Figures 2F and 2G). We also took advantage of the well-established finding that rodent cells lack functional receptors for DT due to residue changes in rodent HBEGF (Mitamura et al., 1995). Consistently, Cas13a-dDT did not affect GFP in mouse embryonic fibroblast cells (MEFs, Figures 2H and 2I). These findings demonstrate that binding and entry of Cas13a-dDT into cells depend on HBEGF as its receptor.
Delivery of RNPs was not successful
We next examined whether RNP composed of both Cas13a and guide RNA can be delivered by fusion with dDT or XLCHN-DTR (Figure 3A). The control (Ctrl) and GFP-targeting crRNAs were incubated with Cas13a-dDT or activated Cas13a-XLCHN-DTR proteins in vitro to form RNPs. We found that exposure to these RNPs did not reduce GFP expression at the protein or mRNA levels (Figures 3B–3E). As controls, delivery of these Cas13a-dDT RNP and Cas13a-XLCHN-DTR RNP using lipid nanoparticle-based transfection reduced GFP in cells (Figures 3F and 3G). These results suggest that Cas13-crRNA RNPs cannot be delivered efficiently into cells within Cas13a-dDT and Cas13a-XLCHN-DTR proteins. The crRNA component would have to be delivered separately.
Figure 3. DT-based and BoNT/X-based systems cannot deliver Cas13a RNP.
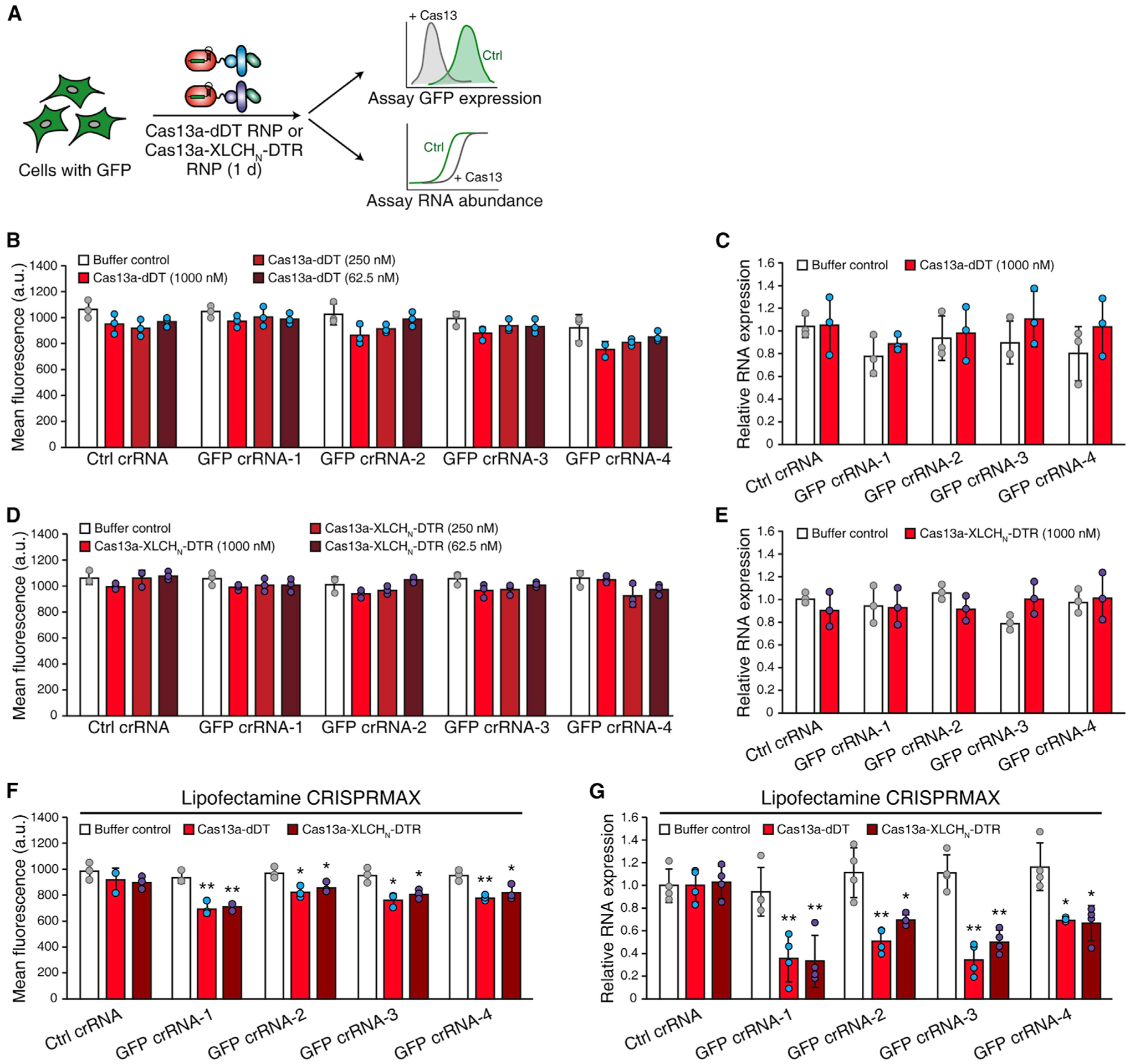
(A) Schematic of the RNP delivery experiment. HeLa + GFP cells were treated with ribonucleoproteins (RNPs) formed by GFP-targeting crRNAs and Cas13a-dDT or activated Cas13a-XLCHN-DTR for 24 h. GFP expression on the protein and mRNA levels were analyzed by flow cytometry and qRT-PCR, respectively.
(B–E) RNP was generated by mixing protein and RNA (5:1 ratio, w/w) at room temperature for 5 min. HeLa + GFP cells were treated with different concentrations of RNP of Cas13a-dDT (B and C) or activated Cas13a-XLCHN-DTR (D and E) for 24 h. GFP protein was measured by flow cytometry and quantified by MFI (B and D), and GFP mRNA was analyzed by qRT-PCR (C and E). Error bars indicate mean ± SD; N = 3 (biological replicates).
(F and G) RNP can be delivered by lipid nanoparticle-based transfection. Lipid nanoparticle-based transfection was performed using a commercial kit (Lipofectamine CRISPRMAX, Invitrogen). GFP protein was measured by flow cytometry and quantified by MFI (F), and GFP mRNA was analyzed by qRT-PCR (G). Error bars indicate mean ± SD; N = 4 (biological replicates); *p < 0.05; **p < 0.01 (Student’s t test).
Delivering Cas13a for degrading SARS-CoV-2 RNA in cells
To assess whether delivery of Cas13a can target and degrade viral RNAs in cells, we utilized a GFP reporter system (Abbott et al., 2020). Two reporter genes were constructed, with one (Reporter-I) expressing mRNA encoding GFP and a fragment of SARS-CoV-2 RNA-dependent RNA polymerase, and the other (Reporter-II) expressing mRNA encoding GFP and a fragment of nucleocapsid (Figure S4A). Two separate pools of crRNAs, with each pool containing four distinct crRNAs, were utilized to target Reporter-I or Reporter-II expressed in HEK293 cells by transient transfection (Figures 4A and S4A). Treatment with Cas13a-dDT or activated Cas13a-XLCHN-DTR reduced MFI of GFP and the mRNA encoding Reporter-I or II in cells that express targeting crRNA, but not in cells expressing the non-targeting control crRNA (Figures 4B–4F and S4B). We further introduced reporters into cells through lentiviral transduction to mimic natural viral infection (Figure 4A) (Abbott et al., 2020). Cas13a-dDT reduced 40% MFI and 37% mRNA levels of Reporter-I and 22% MFI and 22% mRNA for Reporter-II, while activated Cas13a-XLCHN-DTR repressed 62% MFI and 34% mRNA of Reporter-I and 19% of MFI and 25% mRNA for Reporter II in cells that express pooled crRNAs. As controls, Reporter-I and -II in cells expressing non-targeting crRNA were not affected (Figures 4G–4K and S4C).
Figure 4. Delivered Cas13a targets and inhibits SARS-CoV-2 reporters in cells.
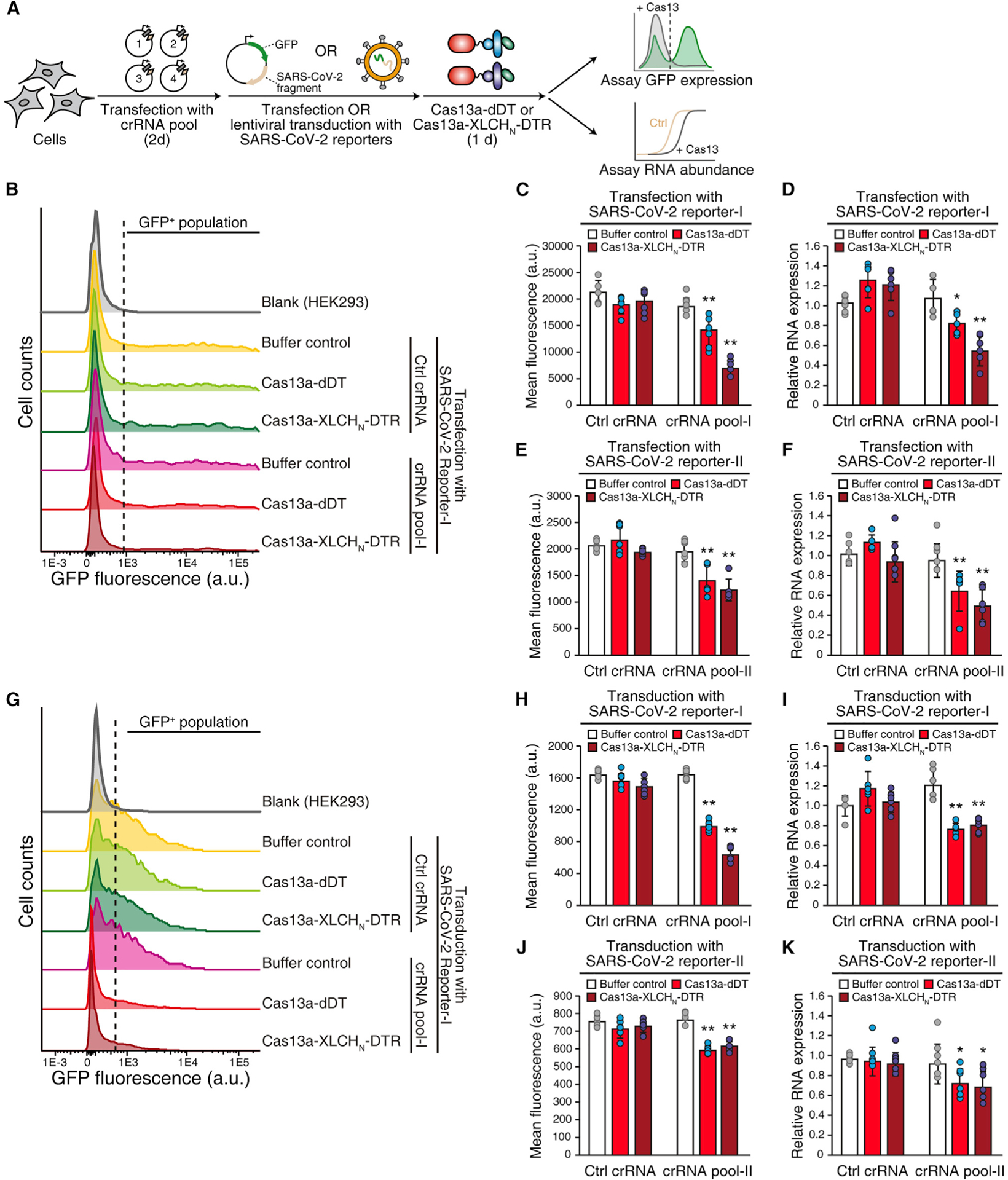
(A) Schematic of the experiment. HEK293 cells were transfected with two pools of crRNAs targeting SARS-CoV-2 fragments (four crRNAs per pool) for 48 h. The SARS-CoV-2 reporters were delivered into cells via either transient transfection or lentiviral transduction. Cas13a-dDT or activated Cas13a-XLCHN-DTR (1 μM) was then added to cells for 24 h. The protein level of GFP and the mRNA level of SARS-CoV-2 fragments were analyzed by flow cytometry and qRT-PCR, respectively.
(B–F) SARS-CoV-2 reporter-I (B–D) or reporter-II (E and F) were delivered into HEK293 cells by transient transfection. Upon treatment with crRNAs and Cas13a-dDT or activated Cas13a-XLCHN-DTR, GFP expression was measured by flow cytometry (B) and quantified by MFI (C and E), and the mRNA level of SARS-CoV-2 fragments was analyzed by qRT-PCR (D and F). GAPDH was used as housekeeping control. Error bars indicate mean ± SD; N = 6 (biological replicates); *p < 0.05; **p < 0.01 (Student’s t test).
(G–K) Lentiviral transduction of reporters. SARS-CoV-2 reporter-I (G–I) or reporter-II (J and K) were delivered into HEK293 cells by lentiviral transduction. Upon treatment with crRNAs and Cas13a-dDT or activated Cas13a-XLCHN-DTR, GFP expression was measured by flow cytometry (G) and quantified by MFI (H and J), the mRNA level of SARS-CoV-2 fragments was analyzed by qRT-PCR (I and K). GAPDH was used as housekeeping control. Error bars indicate mean ± SD; N = 6 (biological replicates); *p < 0.05; **p < 0.01 (Student’s t test).
Delivery of Cre recombinase using dDT
We next explored whether single-chain AB toxin-based systems can deliver proteins that must enter the nucleus for genome editing. We started with Cre recombinase (38 kDa), taking advantage of an available reporter cell line (HEK293 cells containing a loxP-Stop-LoxP-mCherry cassette). Cre-mediated recombination results in expression of red fluorescent protein mCherry. Cre recombinase was directly fused to the N terminus of dDT, and the fusion protein was purified from E. coli (Figure S1G). Incubation with 50 nM Cre-dDT resulted in ~10% mCherry-positive cells, whereas incubation with 1 μM Cre-dDT generated ~30% mCherry-positive cells (Figures S5A and S5B).
We also evaluated the translocation defect form Cre-dDT(E349K) (Figure S1H) and Cre alone (Figure S1I). Exposure to Cre-dDT(E349K) generated Cre-mediated recombination with lower efficiency than Cre-dDT (Figure S5B). Exposure to Cre alone at low concentrations (50 nM) showed no mCherry expression (Figure S5B). Incubation with a high dose of Cre (1 μM) resulted in ~10% mCherry-positive cells, which is consistent with previous observations that Cre itself can enter cells at high concentrations (Will et al., 2002).
To further confirm that binding and entry of Cre-dDT requires HBEGF, we knocked down HBEGF expression in reporter cells by using RNA interference (RNAi). The HBEGF-targeting or non-targeting control siRNAs were transfected into cells (Figure S5C). Cre-dDT (1 μM) treatment resulted in ~10% mCherry-positive cells in HBEGF knockdown (KD) cells, which is similar to incubation with Cre alone and far less than the efficacy on wild-type reporter cells (~30%; Figure S5D). These results demonstrate that HBEGF mediates binding and entry of Cre-dDT into cells.
Delivery of Cas9 for genome editing
To explore whether single-chain toxin-based systems can deliver Cas9, we constructed and purified two chimera proteins: ~221-kDa Cas9-dDT (Figure S1J) and 283-kDa Cas9-XLCHN-DTR (Figures S1K and S1N). We first validated the activity of fused Cas9 protein using an in vitro DNA cleavage assay. RNPs were generated by pre-mixing Cas9-dDT or Cas9-XLCHN-DTR with sgRNAs targeting GFP gene or non-targeting control sgRNA and then incubation with GFP DNA fragments (Figure S6A). Both Cas9-dDT and Cas9-XLCHN-DTR cleaved substrate DNA fragments when complexed with the GFP-targeting sgRNAs but not with the control non-targeting sgRNA (Figures S6B and S6C).
We then examined whether Cas9 can be delivered to modify genomes in HeLa and HEK293 cells. The human EMX1 locus is a well-established target for validating genomic editing with Cas9. A plasmid encoding a sgRNA targeting EMX1 was expressed in cells via transient transfection, and cells were then incubated with 1 μM Cas9-dDT or activated Cas9-XLCHN-DTR for 48 h. The genomic DNA in cells was extracted, and the sequence in the sgRNA targeted region was analyzed by next-generation sequencing (NGS; Figure 5A). The frequencies of indels (insertions colored red, deletions colored blue) identified at individual position were plotted as bar graphs. Both Cas9-dDT and activated Cas9-XLCHN-DTR generated indels with the highest frequencies at ~0.06% and 0.04%, respectively, in HeLa cells (Figure 5B). The same on-site indels could also be identified in HEK293 cells treated with Cas9-dDT or activated Cas9-XLCHN-DTR, with the highest mutation frequencies at 0.31% and 0.15%, respectively (Figure 5C). Within the identified mutations, the one-nucleotide insertion at the DSB is the most frequent genotype (Figure 5D). As negative controls, neither sgRNA alone, Cas9-dDT alone, Cas9-XLCHN-DTR alone, nor a combination of sgRNA with Cas9 (Figure S1L) generated on-site indels (Figures 5B and 5C). Furthermore, cell viability was not affected by either Cas13a-dDT or Cas13a-XLCHN-DTR (5 μM; Figure S2G). As positive controls, Cas9 were expressed directly via lentiviral transduction or transient transfection, which generated indels with ~100-fold higher frequency (4%–5% in HeLa cells, 30%–40% in HEK293 cells; Figures 5B and 5C) than Cas9-dDT and Cas9-XLCHN-DTR. Indels were more frequent in HEK293 than HeLa cells, which may be due to the higher plasmid transfection efficiency in HEK293 cells.
Figure 5. Cas9 can be delivered into cells and edit genomic DNA.
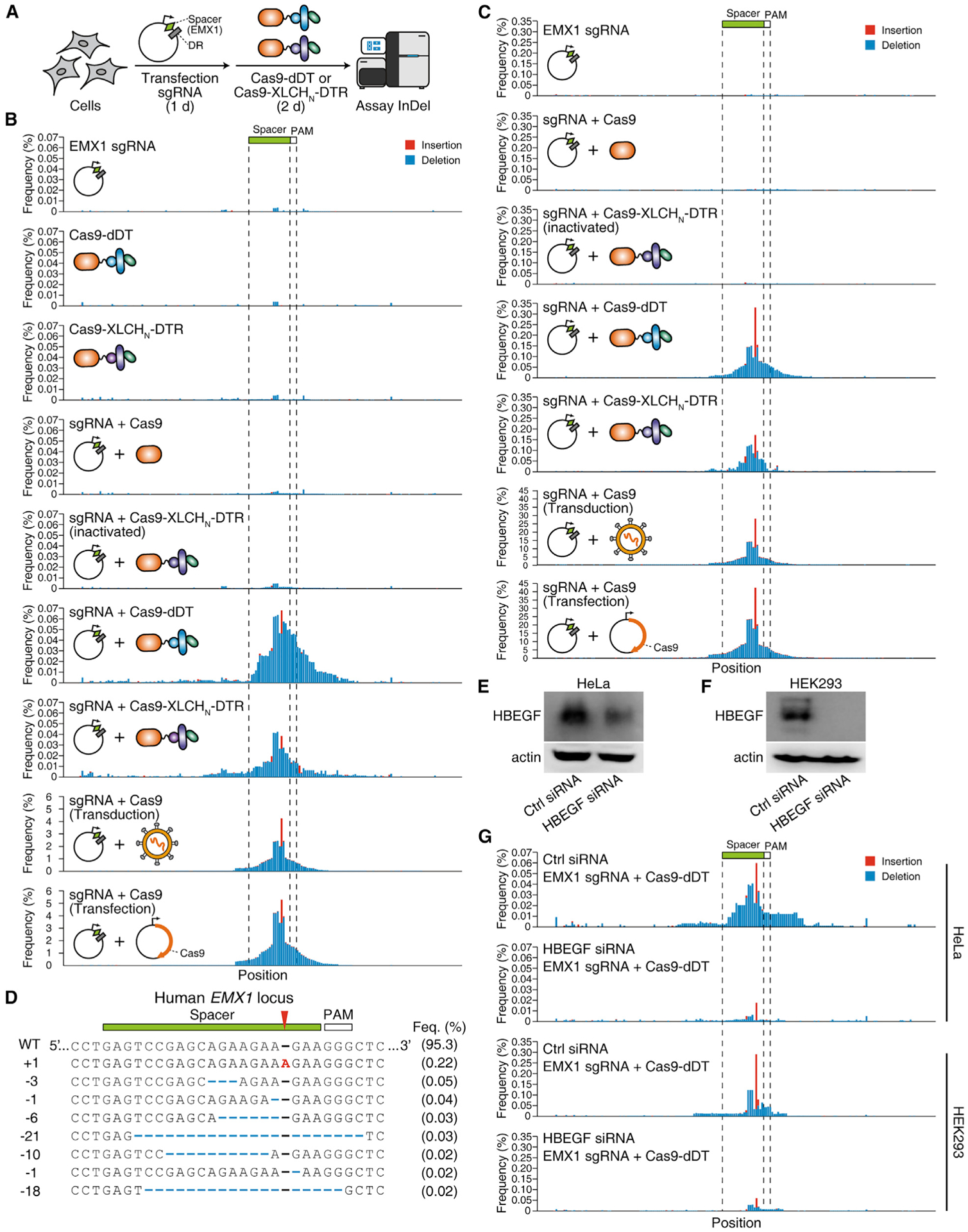
(A) Schematic illustration of the experiment. Cells were transfected with a plasmid that expresses sgRNA targeting the human EMX1 locus for 24 h, Cas9-dDT (1 μM) or activated Cas9-XLCHN-DTR (1 μM) was then added to medium and incubated for 48 h. The genomic DNA was extracted. The sgRNA-targeting region on EMX1 locus was PCR amplified and assayed by next-generation sequencing (NGS) for detection of indels (insertion or deletion). DR, direct repeat.
(B and C) Genome editing for EMX1 locus. The amplicon sequence was plotted as x axis, and the frequency of insertion (red) and deletion (blue) at individual position were plotted as y axis. The positions of spacer and PAM (protospacer adjacent motif) of the sgRNA were marked. HeLa (B) or HEK293 (C) cells co-treated with sgRNA and 1 μM Cas9-dDT or activated Cas9-XLCHN-DTR show indel peaks around the spacer region. Treatment with sgRNA alone and Cas9-dDT/Cas9-XLCHN-DTR alone, or co-treatment with sgRNA and Cas9 or inactivated Cas9-XLCHN-DTR failed to generate an indel peak. Representative histograms from one of three independent experiments are shown.
(D) Representative genotypes of Cas9-dDT-edited human EMX1 locus in HEK293 cells. The +1 bp mutation at the DBS is the most frequent genotype.
(E and F) The HBEGF-targeting or non-targeting control siRNAs were transfected into HeLa cells (E) and HEK293 cells (F). Cell lysates were analyzed by immunoblot detecting HBEGF. Actin served as a loading control.
(G) Knocking down HBEGF via siRNA reduced editing frequency at the EMX1 site in HeLa and HEK293 cells after treatment with Cas9-dDT (1 μM). The amplicon sequence of the sgRNA-targeting region was plotted on the x axis, and the frequency of insertion (red) and deletion (blue) at individual positions were plotted on the y axis. Representative histograms from one of three independent experiments are shown.
To confirm that Cas9 is delivered via receptor-mediated binding and entry, we knocked down HBEGF expression by RNAi in HeLa and HEK293 cells. Reduction in HBEGF expression was validated by immunoblot of cell lysates (Figures 5E and 5F). We then tested Cas9-dDT on these KD cells and found drastically lower genome editing frequency compared with wild-type cells (Figure 5G; reduced from 0.30% to 0.05% in in HEK293 and from 0.06% to 0.01% for HeLa). These results demonstrate that targeting and entry of Cas9-dDT is dependent on HBEGF.
Besides targeting EMX1, Cas9-dDT and activated Cas9-XLCHN-DTR were also shown to target and generate indels at the CCR5 gene locus (Figures S6D and S6E). Finally, we treated HeLa cells with the RNPs formed by sgRNA and Cas9-dDT or activated Cas9-XLCHN-DTR. However, no on-site indels were detected (Figure S6F), indicating that Cas9-sgRNA-RNP cannot be delivered effectively into cells within Cas9-dDT or Cas9-XLCHN-DTR fusion proteins.
Targeted delivery of Cas9 into iPSC-derived human neurons
Targeting and delivering Cas9 into human neurons remains a major challenge. As the receptor-binding domain of BoNTs naturally targets neurons, we next examined neuronal-specific delivery of Cas9 into human iPSC-derived neurons (Dolmetsch and Geschwind, 2011) utilizing the receptor-binding domain of BoNT/A (AHC) to target neurons. An ~316-kDa chimeric protein, Cas9-XLCHN-AHC, was constructed, purified, and activated (Figures S1M and S1O). The sgRNA targeting human EMX1 locus was expressed in human iPSC-derived neurons by lentiviral transduction. Then, 500 nM activated Cas9-XLCHN-AHC was added to culture medium for 48 h, and the indels were analyzed by NGS (Figure 6A). The combination of sgRNA and activated Cas9-XLCHN-AHC generated an on-site indel peak around the spacer region, with the highest indel frequency at ~0.04% (Figure 6B). As a positive control, Cas9 expressed via lentiviral transduction generated indels at the same position with ~5% frequency (Figure 6B). As negative controls, neither sgRNA alone nor Cas9-XLCHN-AHC alone generated onsite indels. These findings illustrate an application of our targeted delivery system for sending Cas9 protein into hard-to-transfect/infect human neurons.
Figure 6. Targeted delivery of Cas9 into iPSC-derived human neurons.
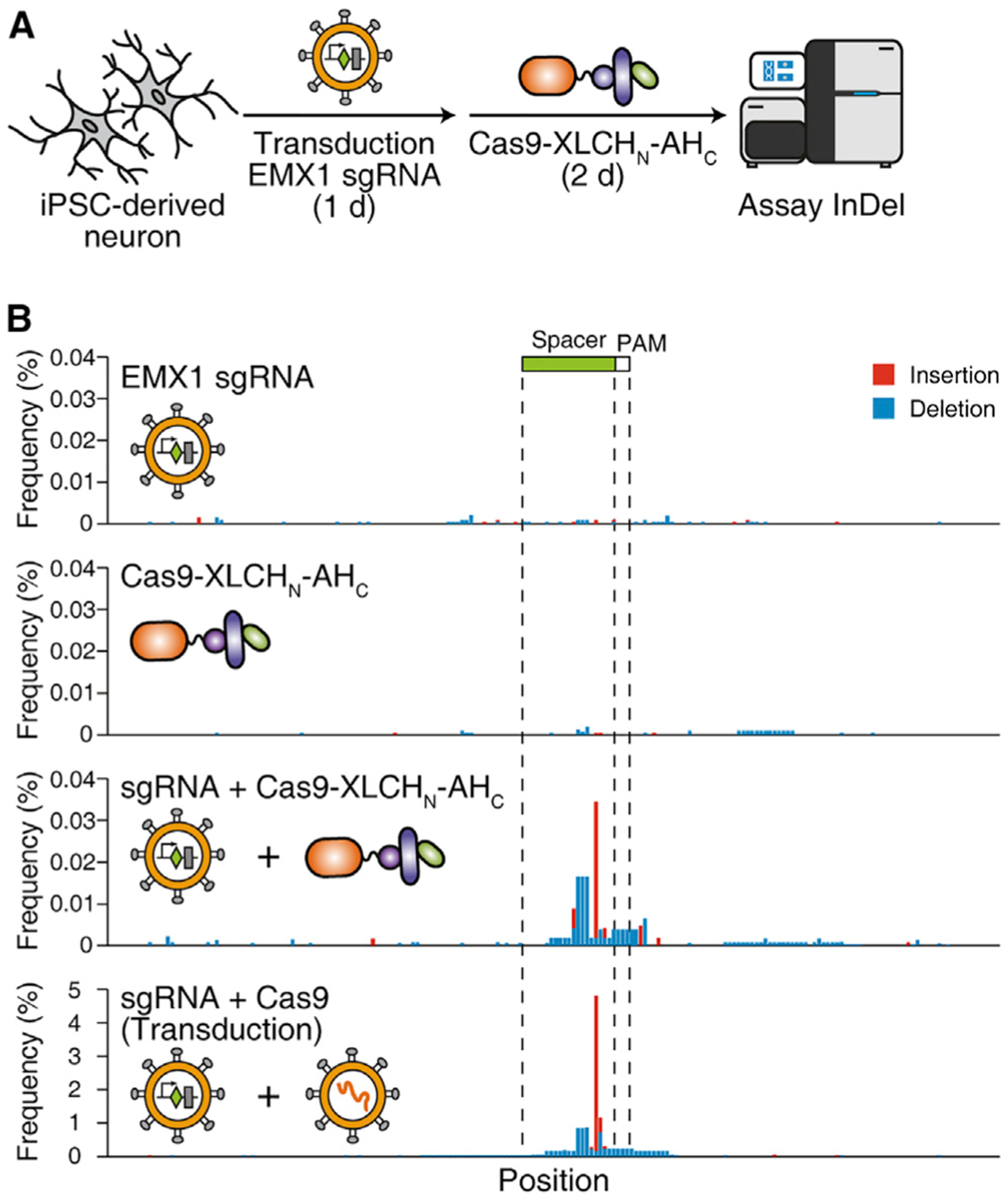
(A) Schematic illustration of the experiment. Human iPSC-derived neurons were lentiviral transduced with the sgRNA-targeting human EMX1 locus for 24 h. Activated Cas9-XLCHN-AHC (500 nM) were then added to the medium and incubated for 48 h. The genomic DNA was extracted, and the indel on the sgRNA-targeting region was assayed as described above.
(B) Genome editing for EMX1 locus. The amplicon sequence was plotted on the x axis, and the frequency of insertion (red) and deletion (blue) at individual position were plotted on the y axis. The position of spacer and PAM (protospacer adjacent motif) of the sgRNA were marked. The iPSC-derived neurons co-treated with sgRNA and activated Cas9-XLCHN-AHC show significant indel peaks around the spacer region. Cas9 delivered by lentiviral transduction generated the on-site indel peak more frequently than delivery of Cas9 using Cas9-XLCHN-AHC. Representative histograms from one of three independent experiments are shown.
DISCUSSION
Here, we established modular protein-based delivery systems based on “detoxified” single-chain AB-type bacterial toxins and validated the feasibility of this approach with DT-based and BoNT/X-based systems that can target specific cell types and deliver Cas13a, CasRx, Cas9, or Cre recombinase into cells. This protein-based delivery system is composed of three moieties: a cargo, a toxin translocation unit (including both the detoxified enzymatic domain and translocation domain), and a cell-targeting domain. It offers several advantages over existing drug delivery tools: (1) it is produced as a single fusion protein; (2) it delivers Cas proteins into cells transiently and thus minimizes the chance of off-target effects; and (3) it contains functional domains to target cells and escape from endosomes.
In this study, DTR and AHC were shown to be interchangeable as the cell-targeting domain. We expect that the receptor-binding domain of other toxins, as well as specific antibody-derived and antibody-like binders such as nanobodies, could be utilized to target specific cell surface receptors (Figure 1A) (McCluskey and Collier, 2013). Two sets of translocation units, one derived from DT and the other from BoNT/X, have been validated in this study. In our current sets of constructs, the cargo proteins are directly fused with the toxin enzymatic domain and remain fused in the cytosol. It would be possible to use linkers that can separate the cargo proteins and the toxin enzymatic domain, for instance by a peptide fragment containing a disulfide bond with a protease activation site between the two cysteines, so the cargo protein and the enzymatic domain would be connected by a disulfide bond after protease treatment and separate from each other after reaching the cytosol.
Utilizing bacterial toxin-based systems to deliver cargoes into the cytosol of cells has been explored with various toxins such as DT (Arnold et al., 2020; Auger et al., 2015; Vidimar et al., 2020), BoNTs (Bade et al., 2004; McNutt et al., 2021; Miyashita et al., 2021), anthrax toxin (Milne et al., 1995; Young and Collier, 2007), Pseudomonas exotoxin A (Ryou et al., 2016; Verdurmen et al., 2015), heat-labile enterotoxin (Chen et al., 2015), Shiga toxins (Ryou et al., 2016; Schmit et al., 2019), Clostridium botulinum C2 toxin (Barth et al., 2002; Fahrer et al., 2013), and Photorhabdus luminescens Tc toxins (Ng Ang et al., 2019; Roderer et al., 2019). The mechanisms of membrane translocation differ among these toxins. The anthrax toxin-based system fuses the cargo with the N-terminal fragment of its enzymatic subunit (A subunit), which docks and translocates across a rigid β-barrel pore formed by its B subunit (Milne et al., 1995; Young and Collier, 2007). The major limitation of AT-based systems is that, since the AT pore is a rigid structure, the cargoes have to be fully unfolded in order to pass through (Young and Collier, 2007). Heat-labile enterotoxin and Shiga toxins mediate trafficking of the cargo proteins retrogradely into the ER and utilize the host ER-associated protein degradation machinery to send the cargo protein into the cytosol. DT and BoNTs are typical single-chain AB toxins that translocate into the cytosol from endosomes. The molecular mechanism of translocation for these single-chain toxins remains to be fully elucidated. Here, we showed that cargo proteins as large as Cas9 can be effectively delivered using DT-and BoNT/X-based systems, suggesting that translocation of these toxins may not rely on rigid proteinaceous pores. Thus, single-chain AB toxins might be particularly suitable for delivering large and stably folded cargoes.
Limitations of the study
The inability to deliver RNPs is a major limitation of our delivery platforms. This is possibly because the RNA components dissociate from Cas proteins during membrane translocation; there-fore, the crRNA components must be co-delivered by other methods. A recent study showed that a DT-based system can be used to deliver siRNA into cells (Arnold et al., 2020), suggesting that crRNA may be delivered using AB toxin-based systems.
Our studies were not able to directly detect and quantify the amount of Cas13 and Cas9 proteins delivered into the cytosol. The overall efficacy for delivering Cas13 and Cas9 could be rather low, as micromolar concentrations of fusion proteins are often required under our experimental conditions. Since native toxins operate at much lower concentrations, fusion with Cas13 and Cas9 likely reduces the efficacy of membrane translocation.
Another key limitation for in vivo application is the development of neutralizing antibodies against delivery tools and/or cargo proteins. Most people are already immunized against DT, limiting the potential use of DT-based delivery. The recently reported DT-like toxins (Mansfield et al., 2018) may be explored as an alternative. BoNT/X may offer a better choice for in vivo applications, as there is no pre-existing immunity against it in human populations. Pre-existing immunity against Cas9 in human populations has been reported (Charlesworth et al., 2019), which could reduce the efficacy of Cas9 delivery in vivo. Recent advances in protein engineering to “de-immunize” the surface epitopes (King et al., 2014; Nagata and Pastan, 2009) may reduce the immunogenicity of therapeutic proteins. Finally, suitable in vivo applications for any protein-based delivery of Cas9 and Cas13 are likely those that do not require repeated dosing. Examples of such indications include delivery of Cas9 to fix genetic defects within neurons and stem cells, and delivery of Cas13 against acute viral infections such as SARS-CoV-2. Neurons and stem cells represent major therapeutic targets for somatic genome editing and have been difficult to selectively target using available delivery tools. Our modular toxin-based delivery system combines the advantages of selective targeting and efficient endosomal escape, providing an effective means to deliver Cas proteins into sparsely distributed and hard-to-reach neurons.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Min Dong (min.dong@childrens.harvard.edu).
Materials availability
Plasmids and purified proteins sequences can be generated upon execution of a material transfer agreement (MTA) with inquiries directed to Dr. Min Dong.
Data and code availability
All data supporting the findings of this study are available upon request. The sequencing data has been deposited to Dryad datasets (https://doi.org/10.5061/dryad.dncjsxm1p).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
Cell lines were all originally obtained from ATCC: HeLa (CCL-2), A549 (CRM-CCL-185), 5637 (HTB-9), HEK293 (CRL-1573), HEK293T (CRL-3216), Vero (CCL-81), and MEF (SCRC-1040). Cre reporter cells were generously provided by Dr. Carlos Lois (California Institute of Technology). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, 11965–118) with 10% fetal bovine serum (FBS, Gibco, 10437–028), 100 U penicillin, and 0.1 mg/mL streptomycin in a humidified atmosphere of 95% air and 5% CO2 at 37°C.
Induced pluripotent stem cells (iPSCs)
Neurons were cultured from human induced pluripotent stem cells (hiPSCs) using an adapted protocol as previously described (Zhang et al., 2013). An established hiPSC line (Coriell GM23338) was integrated with inducible gene expression cassettes containing the human transcription factor NGN3 using Piggybac transposon (SystemsBio). NGN3-hiPSCs were cultured in mTeSR1 medium (Stemcell Technologies).
METHOD DETAILS
cDNA constructs
The selected crRNA sequences (Ctrl: AGTGGAGGAGTGTCTTTTCAATTACTTG; GFP-1: AGGATGGGCACCACCCCGGTGAA-CAGCT; GFP-2: CACCCCGGTGAACAGCTCCTCGCCCTTG; GFP-3: GACACGCTGAACTTGTGGCCGTTTACGT; GFP-4: ATGA ACTTCAGGGTCAGCTTGCCGTAGG; PPIB: TCCTTGATTACACGATGGAATTTGCTGT; KRAS: AATTTCTCGAACTAATGTATA GAAGGCA; CXCR4: ATGATAATGCAATAGCAGGACAGGATGA; pool-I-1: ATATATGTGGTACCATGTCACC; pool-I-2: ATTACCTT CATCAAAATGCCTT; pool-I-3: CTTGATTATCTAATGTCAGTAC; pool-I-4: AAGAATCTACAACAGGAACTCC; pool-II-1: TTGAATCT GAGGGTCCACCAAA; pool-II-2: CCCCACTGCGTTCTCCATTCTG; pool-II-3: TGAACCAAGACGCAGTATTATT; and pool-II-4: AACGCCTTGTCCTCGAGGGAAT) were cloned into LwCas13a-guide-U6 vector (Addgene, #91906) or CasRx-guide-U6 vector (Addgene, #109049), respectively. The selected sgRNA sequences (Ctrl: GTATTACTGATATTGGTGGG; GFP-1: CAACTACAA GACCCGCGCCG; GFP-2: GGGCGAGGAGCTGTTCACCG; GFP-3: GGCCACAAGTTCAGCGTGTC; GFP-4: GAGCTGGACGGC GACGTAAA, HBEGF: CTATGACCACACAACCATCC, EMX1: GAGTCCGAGCAGAAGAAGAA, and CCR5: TCATCCTCCTGACAATC GAT) were cloned into LentiGuide-Puro vector (Addgene, #52963). The cDNAs of DT (#11081), Cas13a (#91865), CasRx (#109049), and Cas9 (#104997) were obtained from Addgene. Mutagenesis of DT (K51E, G52E, E148K) was generated by QuikChange (Agilent, 210519). pEGFP-N1 (Clontech) was used as template for GFP. The cDNA of Cre was synthesized by a commercial vendor (Gen-script). The cDNA of dBoNT/X-LCHN was produced as previously described (Miyashita et al., 2021). Cre-dDT was cloned into pET28a vector (Novagen) with N-terminal His-tag by Gibson Assembly (NEB, E2621). Cas13a-dDT, CasRx-dDT, Cas9-dDT, Cas13a, Cas9, Cre, and Cas13a-XLCHN-DTR were cloned into pET28a vector (Novagen) with C-terminal His-tag by Gibson Assembly (NEB, E2621). CasRx-XLCHN-DTR, Cas9-XLCHN-DTR, and Cas9-XLCHN-AHC were cloned into pHis1522 vector (MoBiTec) with C-terminal His-tag by Gibson Assembly. Cas13a-dDT(E349K) and Cre-dDT(E349K) were generated using the QuickChange mutagenesis kit (Agilent, 200517). The SARS-CoV-2 reporter-I was purchased from Addgene (#155303) and subcloned into pLenti-Hygro vector (Addgene, #17484). The SARS-CoV-2 reporter-II was cloned by fusion of GFP with a fragment of nucleocapsid (used Addgene #153201 as template) and inserted into pLenti-Hygro vector via Gibson Assembly. The sequences of all constructs were confirmed by Sanger sequencing (Genewiz). All plasmid sequences are available upon request.
Recombinant proteins
The expression constructs of Cre-dDT, Cas13a-dDT, CasRx-dDT, Cas9-dDT, Cas13a, Cas9, Cre, Cas13a-dDT(E349K), Cre-dDT(E349K), and Cas13a-XLCHN-DTR were transformed into E. coli (BL21 strain) and grown in LB media at 37°C, induced at mid-log phase (OD600 ~0.6) with 0.4 mM IPTG (Sigma, I6758), and then transferred to 18°C for overnight expression. The expression constructs of CasRx-XLCHN-DTR, Cas9-XLCHN-DTR, and Cas9-XLCHN-AHC were transformed in Bacillus megaterium following the manufacturer’s protocol (MoBiTec) and grown in LB media at 37°C, induced at OD600 ~0.4 with 0.5% D-(+)-Xylose (w/w, Sigma, X1500), and then transferred to 18°C for overnight expression. The recombinant proteins were purified as His6-tagged proteins. Briefly, cells were resuspended in lysis buffer (20 mM PBS, pH 7.4, 500 mM NaCl, 10 mM Imidazole, and 1 mM PMSF), lysed by sonication, and clarified by centrifugation at 25,000 × g. Cell lysate was incubated with Ni-NTA agarose (MCLAB, NINTA-300) at 4°C for 2 h, washed with wash buffer (20 mM PBS, pH 7.4, 500 mM NaCl, 20 mM Imidazole), and eluted by elution buffer (20 mM PBS, pH 7.4, 500 mM NaCl, 500 mM Imidazole). The eluted proteins were buffer-exchanged into Dulbecco’s phosphate buffered saline (DPBS, Corning, 55–031-pc), sterilized by passing through a membrane filter, and quantified by BCA assay (Thermo Fisher, 23225) or SDS-PAGE (for Cas13a-XLCHN-DTR, whose purity is low). Thrombin cleavage of XLCHN was carried out by mixing 6 μg purified proteins with 0.2 U thrombin (EMD Millipore, 605157) in 20 μL DPBS. The mixture was incubated at 4°C overnight and analyzed by SDS-PAGE under reduction conditions (with 20 mM DTT).
Lentiviral transduction
The lentiviruses were produced as previously described (Peng et al., 2013). Briefly, 3×106 HEK293T cells were co-transfected with pMD2.G (Addgene, #12259), psPAX2 (Addgene, #12260), and the transfer plasmids at 1:10:10 ratio (w/w/w). The lentivirus-containing medium was collected 48 h post-transfection, filtered through 0.45 μm filters, and stored at ‒80°C. For the transduction, 3×105 cells were seeded into 6-well plates and cultured overnight. Polybrene (Santa Cruz, sc-134220, 8 μg/mL) and a serial dilution of the stock of lentivirus were added to the cells. The GFP-overexpressing cells were selected with 200 μg/mL Hygromycin B (EMD Millipore, 400051) at 24 h post-transduction. When the selection of Hygromycin B was complete (all cells were dead in the virus-free well), the approximate MOI (multiplicity of infection) was calculated; the well transduced at MOI <0.5 was selected for the following study. HeLa + GFP cells were utilized for generating HBEGF-KO cells via lentiviral transduction of LentiCas9-Blast (Addgene, #52962) and LentiPuro-HBEGF. Mixed populations of infected cells were selected with Blasticidin S (10 mg/mL) and Puromycin (5 μg/mL).
Flow cytometry
GFP-expressing cells were washed three times with ice-cold DPBS and collected in FACS buffer (DPBS, 10% FBS, 5 mM EDTA). Cells were stained by 1 μg/mL DAPI (Sigma, D9542) and subjected to single-cell flow cytometry analysis using a Canto II FACS system (BD Biosciences). Wild-type cells without GFP expression were used as controls. Data was analyzed using FlowJo software (version 10). The raw MFI data were summarized in Data S1.
Cell viability assay
HeLa cells were seeded in 96-well plates (70% confluence) and incubated with serial dilutions (2-fold) of the indicated proteins (Figure S2G) for 2 days. MTT (0.5 mg/mL, RPI, M92050–1.0) was added and incubated at 37°C for 4 h. Solubilization solution (10% SDS in 0.01 M HCl) was then added to each well and incubated overnight. The absorbance of formazan formed was measured at 580 nm by a microplate reader (BMG Labtech, FLUOstar Omega). A vehicle control (without exposure to the indicated proteins) and a blank of medium were analyzed in parallel. The cytotoxicity curves were analyzed and fitted using Origin software (version 8.5).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, 15596026) according to the manufacturer’s instructions. RNA was reverse transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystem, 4375575) according to the manufacturer’s instructions. One-step qRT-PCR was carried out using FastStart Universal SYBR Green Master (Roche, 4913850001) and a QuantStudio 3 Real-Time PCR system (Applied Biosystems) with primers as follows: GAPDH_F (CTGGGCTA CACTGAGCACC); GAPDH_R (AAGTGGTCGTTGAGGGCAATG); GFP_F (ATGAGCAAGGGCGAGGAGCT); GFP_R (CAGGGT CAGCTTGCCGTAGGT); PPIB_F (AAGTCACCGTCAAGGTGTATTTT); PPIB_R (TGCTGTTTTTGTAGCCAAATCCT); KRAS_F (GCC TGTTTTGTGTCTACTGTTCT); KRAS_R (GGACTGGGGAGGGCTTTCT); CXCR4_F (AAGTCACCGTCAAGGTGTATTTT); CXCR4_R (TGCTGTTTTTGTAGCCAAATCCT); Reporter-I_F (AACGGGTTTGCGGTGTAAGT); Reporter-I_R (AATTTAGCAAAACCAGCTACTT TATCATTGTAG); Reporter-II_F (ATGTCTGATAATGGACCCCAAAATCAGCG); Reporter-II_R (GACGCAGTATTATTGGGTAAA CCTTGGGG). For each sample, the CT (cycle threshold) of the house-keeping gene (GAPDH) and target genes were measured. Relative changes in gene expression were calculated based on the ΔΔCT method (Livak and Schmittgen, 2001) and normalized to control sample (cells without treatment). The raw CT data were summarized in Data S1.
Immunoblot analysis
Cells were scraped, washed, and lysed with RIPA buffer (50 mM Tris, pH 7.5, 1% NP-40, 150 mM NaCl, 0.5% sodium deoxycholate, 1% SDS, protease inhibitor cocktail) on ice for 30 min. The protein amounts in cell lysate were determined by BCA assay (Thermo Scientific, 23225). The cell lysates were mixed with loading dye (50 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 0.01% bromophenol blue, 20 mM DTT) and heated for 5 min, analyzed by SDS-PAGE, and transferred onto a nitrocellulose membrane (GE Healthcare, 10600002). The membrane was blocked with TBST buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 0.1 % Tween 20) containing 5% skim milk at room temperature for 40 min. The membrane was then incubated with the primary antibodies for 1 h, then washed and incubated with secondary antibodies for 1 h. Signals were detected using the enhanced chemiluminescence method (Thermo Fisher Scientific, 34080) with a Fuji LAS3000 imaging system.
Knockdown HBEGF by RNAi
The HBEGF targeting siRNA was purchased from Santa Cruz (sc-39420). The non-targeting scramble siRNA was purchased from Life Technologies. Cells were seeded in 24-well plates for 24 h. When confluency reached 70%, cells were incubated in serum-free medium for 8 h. The siRNAs were transfected into cells using Lipofectamine RNAiMAX (ThermoFisher). Experiments were carried out 48 h later. The knockdown efficiency was validated by immunoblot analysis.
Microcopy
Cells were seeded onto glass coverslips (Hampton, HR3–239) in 24-well plates and incubated to 70% confluence. Cells were incubated with Cre-dDT, Cre-dDT(E349K), or Cre at indicated conditions. Cells were washed by PBS and fixed with 4% paraformaldehyde (PFA) at room temperature for 20 min. Slides were sealed within DAPI-containing mounting medium (SouthernBiotech, 0100–20). Fluorescent images were captured using an Olympus DSU-IX81 Spinning Disk Confocal System. Images were pseudo-colored and analyzed using ImageJ software (version 1.52o). The raw quantification data were summarized in Data S1.
In vitro RNA transcription and Cas9 cleavage assay
The transcription templates of crRNA or sgRNA were generated by adding a T7 promoter to the 5′ end of the RNA-expressing cassette on the RNA-expression constructs via PCR. The PCR product was purified via agarose gel electrophoresis. In vitro transcription was performed using HiScribe T7 Quick High Yield RNA Synthesis Kit (NEB, E2050) following the manufacturer’s protocol. The DNA template was removed by treatment with DNase I (NEB, M0303). The RNA products were subsequently purified using Monarch RNA Cleanup Kit (NEB, T2040) and quantified by Nanodrop. The RNP of Cas13a, CasRx, or Cas9 was produced by mixing protein and RNA at 5:1 ratio (w/w) at room temperature for 5 min. The DNA substrate (containing GFP ORF) for Cas9 in vitro cleavage assay was amplified by PCR and purified by agarose gel electrophoresis. 400 ng DNA substrate was mixed with 300 ng RNP (250 ng protein plus 50 ng RNA) and incubated at 37°C for 1 h. Agarose gel electrophoresis was used to analyze the cleaved products.
Culture iPSC-derived neurons
Neurons were cultured from human induced pluripotent stem cells (hiPSCs) using an adapted protocol as previously described (Zhang et al., 2013). An established hiPSC line (Coriell GM23338) was integrated with inducible gene expression cassettes containing the human transcription factor NGN3 using Piggybac transposon (SystemsBio). Day 0: seed 6×105 NGN3-hiPSCs into a 6-well plate previously coated with Matrigel (Corning) in mTeSR1 medium (Stemcell Technologies) containing Y-27632 (10 μM, Stemcell Technologies) and doxycycline (DOX, 1 μg/μL, Clontech). Day 1: change medium to mTeSR1 plus DOX. Day 2–3: change medium which is composed of DMEM/F12 (LifeTech), B27 (LifeTech), Nonessential amino acids (LifeTech), growth factors (BDNF, PeproTech), NT-3 (PeproTech), Laminin (LifeTech), and DOX. Day 4–7: change medium to Neurobasal-A media (LifeTech), B27 (LifeTech), Glutamax (LifeTech), growth factors (BDNF, PeproTech), AraC (Sigma), and DOX. Day 8: dissociate neurons with DNaseI (Worthington Biochemical) and Papain (Worthington Biochemical), plate >1×106 cells into plates pre-coated with Poly-D-Lysine (Sigma) and laminin in Conditioned Sudhof Medium, which is composed of Neurobasal-A media, FBS, Glutamax, B27, 45% glucose, 8% NaHCO3, and 50 mg/mL Transferrin (Sigma). Day 9: half media change with Conditioned Sudhof Medium. Days 10–28: Half media change with Conditioned Sudhof Media twice per week.
Assay InDel by next-generation sequencing
The genomic DNA of cells was extracted using a commercial kit (Qiagen, 13323). The DNA fragments of the human EMX1 locus and CCR5 locus were amplified by PCR using primers EMX1_GT_F (CTGGGGGCCTCCTGAGTTTC), EMX1_GT_R (CGTGGG TTTGTGGTTGCCCA), CCR5_GT_F (CTCTACTCACTGGTGTTCATCTTTGGTTTTGTG), and CCR5_GT_R (CTGTATGGAAAATGA GAGCTGCAGGTG). Next-generation sequencing and data analysis were performed by a commercial vendor (Genewiz, Amplicon-EZ and genoTYPER-NEXT Analysis). DNA Library Preparation, clustering, and sequencing reagents were used throughout the process using NEBNext Ultra DNA Library Prep kit following the manufacturer’s recommendations (Illumina, San Diego, CA, USA). End repaired adapters were ligated after adenylation of the 3′ends followed by enrichment by limited cycle PCR. DNA libraries were validated on the Agilent TapeStation (Agilent Technologies, Palo Alto, CA, USA) and quantified using the Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA) and multiplexed in equal molar mass. The pooled DNA libraries were loaded on the Illumina instrument according to manufacturer’s instructions. The samples were sequenced using a 2 × 250 paired-end (PE) configuration. Image analysis and base calling were performed using the Illumina Control Software on the Illumina instrument.
QUANTIFICATION AND STATISTICAL ANALYSIS
At least three independent biological replicates were performed for all experiments. The quantification data were analyzed using the OriginPro (OriginLab, v8.5) and Excel (Microsoft, 2007) software. Bar-charts were constructed with mean ± SD (standard deviation). p values were calculated by unpaired two-tailed Student’s t-test using Excel software. Data were considered significant when p < 0.05. These statistical details can be found in the figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Actin | Aves Labs | ACT-1010; RRID:AB_2313504 |
| Mouse monoclonal anti-HBEGF | Santa Cruz | sc-365182; RRID:AB_10708568 |
| Chemicals, peptides, and recombinant Proteins | ||
| All the other chemicals | Sigma | N/A |
| PolyJet | SignaGen | SL100688 |
| Polybrene | Santa Cruz | sc-134220 |
| Lipofectamin CRISPRMAX | Invitrogen | CMAX00003 |
| Lipofectamine RNAiMAX | Invitrogen | 13778100 |
| Ni-NTA agarose | MCLAB | NINTA-300 |
| DMEM | Life technologies | Cat#11995-065 |
| Fetal bovine serum | Life technologies | Cat#26140-079 |
| Penicillin/streptomycin | Life technologies | Cat#15140-122 |
| Puromycin | ThermoFisher | A1113830 |
| Blasticidin S | RPI | B12150.01 |
| Hygromycin B | EMD Millipore | 400051 |
| Protease Inhibitor Cocktail | Roche | 4693159001 |
| Nitrocellulose membrane | GE Healthcare | 10600002 |
| DAPI | Sigma | D9542 |
| DAPI-containing mounting medium | SouthernBiotech | 0100-20 |
| Thrombin | EMD Millipore | 605157 |
| TRIzol reagent | Invitrogen | 15596026 |
| Piggybac transposon system | SystemsBio | PB220PA-1 |
| Matrigel | Corning | DLW356231 |
| mTeSR1 medium | Stemcell Technologies | 100-0276 |
| Y-27632 | Stemcell Technologies | 72302 |
| Doxycycline | Clontech | 3P 631311 |
| DMEM/F12 | Life technologies | 11320033 |
| B27 | Life technologies | 17504044 |
| Nonessential amino acids | Life technologies | 11140050 |
| Growth factors BDNF | PeproTech | AF-450-02 |
| NT-3 | PeproTech | 450-03 |
| Laminin | Life technologies | A29249 |
| Neurobasal-A media | Life technologies | 10888022 |
| Glutamax | Life technologies | 35050061 |
| Papain | Worthington Biochemical | LS003126 |
| Poly-D-Lysine | Sigma | P6407 |
| Transferrin | Sigma | 10652202001 |
| Bafilomycin A1 | Sigma | 19-148 |
| MTT | RPI | M92050-1.0 |
| Cas13a-dDT | This paper | N/A |
| Cas13a-XLCHN-DTR | This paper | N/A |
| Cas13a | This paper | N/A |
| Cas13a-dDTE349K | This paper | N/A |
| CasRx-dDT | This paper | N/A |
| CasRx-XLCHN-DTR | This paper | N/A |
| Cre-dDT | This paper | N/A |
| Cre-dDTE349K | This paper | N/A |
| Cre | This paper | N/A |
| Cas9-dDT | This paper | N/A |
| Cas9-XLCHN-DTR | This paper | N/A |
| Cas9 | This paper | N/A |
| Cas9-XLCHN-AHC | This paper | N/A |
| Critical commercial assays | ||
| Gibson Assembly | NEB | E2621 |
| QuikChange | Agilent | 210519 |
| Bacillus Expression Systems | MoBiTec GmbH | BMEG04 |
| BCA assay kit | ThermoFisher | 23225 |
| Genomic DNA extraction kit | Qiagen | 13323 |
| Enhanced chemiluminescence kit | ThermoFisher | 34080 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystem | 4375575 |
| FastStart Universal SYBR Green Master | Roche | 4913850001 |
| HiScribe T7 Quick High Yield RNA Synthesis Kit | NEB | E2050 |
| DNase I | NEB | M0303 |
| Monarch RNA Cleanup Kit | NEB | T2040 |
| Deposited data | ||
| InDel sequencing data | Dryad | https://doi.org/10.5061/dryad.dncjsxm1p |
| Experimental models: Cell lines | ||
| HeLa | ATCC | CCL-2 |
| A549 | ATCC | CRM-CCL-185 |
| 5637 | ATCC | HTB-9 |
| HEK293 | ATCC | CRL-1573 |
| HEK293T | ATCC | CRL-3216 |
| Vero | ATCC | CCL-81 |
| MEF | ATCC | SCRC-1040 |
| HeLa+GFP | This paper | N/A |
| A549+GFP | This paper | N/A |
| 5637+GFP | This paper | N/A |
| Vero+GFP | This paper | N/A |
| MEF+GFP | This paper | N/A |
| HeLa+GFP-HBEGF-KO-Mix | This paper | N/A |
| human induced pluripotent stem cells | Coriell | GM23338 |
| Oligonucleotides | ||
| See Table S1 for the primers used in this study | This paper | N/A |
| Control siRNA | Life Technologies | Synthesized |
| HBEGF siRNA | Santa Cruz | sc-39420 |
| Recombinant DNA | ||
| pET28a | Novagen | 69864 |
| pHIS1522 | MoBiTec GmbH | BMEG12 |
| pMD2.G | Addgene | #12259 |
| pSPAX2 | Addgene | #12260 |
| LentiGuide-puro | Addgene | #52963 |
| Lenti-SpCas9 blast | Addgene | #104997 |
| pLenti-Hygro | Addgene | #17484 |
| pEGFP-N1 | Clontech | 6085-1 |
| pC016-LwCas13a-guide-U6 | Addgene | #91906 |
| pUC19-CasRx-guide-U6 | Addgene | #109049 |
| pET22b-DT-51E/148K | Addgene | #11081 |
| p2CT-His-MBP-LwaCas13a | Addgene | #91865 |
| EF1a-CasRx-2A-EGFP | Addgene | #109049 |
| SARS-CoV-2 reporter-I | Addgene | #155303 |
| SARS-CoV-2 nucleocapsid | Addgene | #153201 |
| Lentiguide-GFP-1 | This paper | N/A |
| Lentiguide-GFP-2 | This paper | N/A |
| Lentiguide-GFP-3 | This paper | N/A |
| Lentiguide-GFP-4 | This paper | N/A |
| Lentiguide-Ctrl | This paper | N/A |
| Lentiguide-HBEGF | This paper | N/A |
| Lentiguide-EMX1 | This paper | N/A |
| Lentiguide-CCR5 | This paper | N/A |
| pC016-LwCas13a-GFP-1 | This paper | N/A |
| pC016-LwCas13a-GFP-2 | This paper | N/A |
| pC016-LwCas13a-GFP-3 | This paper | N/A |
| pC016-LwCas13a-GFP-4 | This paper | N/A |
| pC016-LwCas13a-Ctrl | This paper | N/A |
| pC016-LwCas13a-RdRP-1 | This paper | N/A |
| pC016-LwCas13a-RdRP-2 | This paper | N/A |
| pC016-LwCas13a-RdRP-3 | This paper | N/A |
| pC016-LwCas13a-RdRP-4 | This paper | N/A |
| pC016-LwCas13a-PPIB | This paper | N/A |
| pC016-LwCas13a-CXCR4 | This paper | N/A |
| pC016-LwCas13a-KRAS | This paper | N/A |
| pC016-LwCas13a-N-1 | This paper | N/A |
| pC016-LwCas13a-N-2 | This paper | N/A |
| pC016-LwCas13a-N-3 | This paper | N/A |
| pC016-LwCas13a-N-4 | This paper | N/A |
| pUC19-CasRx-GFP-1 | This paper | N/A |
| pUC19-CasRx-GFP-2 | This paper | N/A |
| pUC19-CasRx-GFP-3 | This paper | N/A |
| pUC19-CasRx-GFP-4 | This paper | N/A |
| pUC19-CasRx-Ctrl | This paper | N/A |
| LentiHygro-GFP | This paper | N/A |
| pET28a-Cas13a-dDT-His | This paper | N/A |
| pET28a-CasRx-dDT-His | This paper | N/A |
| pET28a-Cas9-dDT-His | This paper | N/A |
| pET28a-Cas13a-XLCHN-DTR-His | This paper | N/A |
| pHis1522-CasRx-XLCHN-DTR-His | This paper | N/A |
| pHis1522-Cas9-XLCHN-DTR-His | This paper | N/A |
| pHis1522-Cas9-XLCHN-AHC-His | This paper | N/A |
| pET28a-Cas9-His | This paper | N/A |
| pET28a-Cas13a-His | This paper | N/A |
| pET28a-Cas13a-dDTE349K-His | This paper | N/A |
| pET28a-His-Cre-dDtx | This paper | N/A |
| pET28a-His-Cre-dDtxE349K | This paper | N/A |
| pET28a-Cre-His | This paper | N/A |
| LentiHygro-Reporter-I | This paper | N/A |
| pcDNA3.1-Reporter-II | This paper | N/A |
| LentiHygro-Reporter-II | This paper | N/A |
| Software and algorithms | ||
| SnapGene Viewer | SnapGene | V4.0.8 |
| OriginPro | OriginLab | v8.5 |
| FlowJo | https://www.flowjo.com/ | V10 |
| Excel | Microsoft | 2007 |
| ImageJ | imagej.nih.gov/ij | Version 1.52o |
| Other | ||
| Spinning Disk Confocal System | Olympus | DSU-IX81 |
| Canto II FACS system | BD Biosciences | Canto II |
| QuantStudio 3 Real-Time PCR system | Applied Biosystems | QuantStudio 3 |
| Fuji LAS3000 imaging system | Fuji | LAS3000 |
| Microplate reader | BMG Labtech | FLUOstar Omega |
| Next-generation sequencing | Genewiz | Amplicon-EZ |
| NGS data analysis | Genewiz | genoTYPER-NEXT Analysis |
Highlights.
Development of modular delivery platforms based on diphtheria toxin and BoNT/X
Targeted intracellular delivery of Cas13a, CasRx, Cas9, and Cre proteins is validated
Delivery of Cas13 reduces mRNAs of targeted genes and SARS-CoV-2 reporters in cells
Delivery of Cas9 generates on-site mutations in cell lines and iPSC-derived neurons
ACKNOWLEDGMENTS
We thank Dr. Kuanwei Sheng for assisting in NGS analysis and members of the Dong lab for discussion. E.A. and G.M.C. were supported by the FunGCAT program from the Office of the Director of National Intelligence Intelligence Advanced Research Projects Activity (IARPA), via the Army Research Office (grant number W911NF-17-2-0089) and the EGL Charitable Foundation. This study was partially supported in part by grants from National Institutes of Health (NIH) (R01NS080833, R01NS117626, and R21NS106159 to M.D.). M.D. acknowledges support by the NIH-funded Harvard Digestive Disease Center (P30DK034854) and Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center (P30HD18655). M.D. holds the Investigator in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.110476.
DECLARATION OF INTERESTS
The authors declare no conflicts of interest.
REFERENCES
- Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, Chemparathy A, Chmura S, Heaton NS, Debs R, et al. (2020). Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell 181, 865–876 e812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. (2017). RNA targeting with CRISPR-Cas13. Nature 550, 280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Franklin B, Koob J, Kellner MJ, Ladha A, Joung J, Kirchgatterer P, Cox DBT, and Zhang F (2019). A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. (2016). C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman CM, Myhrvold C, Thakku SG, Freije CA, Metsky HC, Yang DK, Ye SH, Boehm CK, Kosoko-Thoroddsen TF, Kehe J, et al. (2020). Massively multiplexed nucleic acid detection with Cas13. Nature 582, 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alouf JE (2005). A 116-year story of bacterial protein toxins (1888–2004): from “diphtheritic poison” to molecular toxinology. In The Comprehensive Sourcebook of Bacterial Protein Toxins, 3rd edition, Alouf M.P. Joseph, ed., pp. 3–21. [Google Scholar]
- Arnold AE, Smith LJ, Beilhartz GL, Bahlmann LC, Jameson E, Melnyk RA, and Shoichet MS (2020). Attenuated diphtheria toxin mediates siRNA delivery. Sci. Adv 6, eaaz4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger A, Park M, Nitschke F, Minassian LM, Beilhartz GL, Minassian BA, and Melnyk RA (2015). Efficient delivery of structurally diverse protein cargo into mammalian cells by a bacterial toxin. Mol. Pharm 12, 2962–2971. [DOI] [PubMed] [Google Scholar]
- Bade S, Rummel A, Reisinger C, Karnath T, Ahnert-Hilger G, Bigalke H, and Binz T (2004). Botulinum neurotoxin type D enables cytosolic delivery of enzymatically active cargo proteins to neurones via unfolded translocation intermediates. J. Neurochem 91, 1461–1472. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, and Horvath P (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. [DOI] [PubMed] [Google Scholar]
- Barth H, Aktories K, Popoff MR, and Stiles BG (2004). Binary bacterial toxins: biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol. Mol. Biol. Rev 68, 373–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth H, Roebling R, Fritz M, and Aktories K (2002). The binary Clostridium botulinum C2 toxin as a protein delivery system: identification of the minimal protein region necessary for interaction of toxin components. J. Biol. Chem 277, 5074–5081. [DOI] [PubMed] [Google Scholar]
- Beilhartz GL, Sugiman-Marangos SN, and Melnyk RA (2017). Repurposing bacterial toxins for intracellular delivery of therapeutic proteins. Biochem. Pharmacol 142, 13–20. [DOI] [PubMed] [Google Scholar]
- Blanchard EL, Vanover D, Bawage SS, Tiwari PM, Rotolo L, Beyersdorf J, Peck HE, Bruno NC, Hincapie R, Michel F, et al. (2021). Treatment of influenza and SARS-CoV-2 infections via mRNA-encoded Cas13a in rodents. Nat. Biotechnol 39, 717–726. [DOI] [PubMed] [Google Scholar]
- Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, Vakulskas CA, Collingwood MA, Zhang L, Bode NM, et al. (2019). Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med 25, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Przedpelski A, Tepp WH, Pellett S, Johnson EA, and Barbieri JT (2015). Heat-labile enterotoxin IIa, a platform to deliver heterologous proteins into neurons. mBio 6, e00734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung NK, Saarinen UM, Neely JE, Landmeier B, Donovan D, and Coccia PF (1985). Monoclonal antibodies to a glycolipid antigen on human neuroblastoma cells. Cancer Res. 45, 2642–2649. [PubMed] [Google Scholar]
- Collier RJ (1967). Effect of diphtheria toxin on protein synthesis: inactivation of one of the transfer factors. J. Mol. Biol 25, 83–98. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, and Zhang F (2017). RNA editing with CRISPR-Cas13. Science 358, 1019–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolmetsch R, and Geschwind DH (2011). The human brain in a dish: the promise of iPSC-derived neurons. Cell 145, 831–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Masuyer G, and Stenmark P (2019). Botulinum and tetanus neurotoxins. Annu. Rev. Biochem 88, 811–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, and Stenmark P (2021). The structure and classification of botulinum toxins. Handb. Exp. Pharmacol 263, 11–33. [DOI] [PubMed] [Google Scholar]
- Donovan JJ, Simon MI, Draper RK, and Montal M (1981). Diphtheria toxin forms transmembrane channels in planar lipid bilayers. Proc. Natl. Acad. Sci. U S A 78, 172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan JJ, Simon MI, and Montal M (1982). Insertion of diphtheria toxin into and across membranes: role of phosphoinositide asymmetry. Nature 298, 669–672. [DOI] [PubMed] [Google Scholar]
- Donovan JJ, Simon MI, and Montal M (1985). Requirements for the translocation of diphtheria toxin fragment A across lipid membranes. J. Biol. Chem 260, 8817–8823. [PubMed] [Google Scholar]
- Doudna JA (2020). The promise and challenge of therapeutic genome editing. Nature 578, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer PDR, Shepherd TR, Gollings AS, Shorter SA, Gorringe-Pattrick MAM, Tang CK, Cattoz BN, Baillie L, Griffiths PC, and Richardson SCW (2015). Disarmed anthrax toxin delivers antisense oligonucleotides and siRNA with high efficiency and low toxicity. J. Control Release 220, 316–328. [DOI] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Burstein D, Knott GJ, and Doudna JA (2017). RNA targeting by functionally orthogonal type VI-A CRISPR-cas enzymes. Mol. Cell 66, 373–383 e373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East-Seletsky A, O’Connell MR, Knight SC, Burstein D, Cate JH, Tjian R, and Doudna JA (2016). Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrer J, Rausch J, and Barth H (2013). A cell-permeable fusion protein based on Clostridium botulinum C2 toxin for delivery of p53 tumorsuppressor into cancer cells. PLoS One 8, e72455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellmann C, Gowen BG, Lin PC, Doudna JA, and Corn JE (2017). Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat. Rev. Drug Discov 16, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freije CA, Myhrvold C, Boehm CK, Lin AE, Welch NL, Carter A, Metsky HC, Luo CY, Abudayyeh OO, Gootenberg JS, et al. (2019). Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell 76, 826–837 e811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Blanke SR, Mattheakis LC, and Collier RJ (1997). Selection of diphtheria toxin active-site mutants in yeast. Rediscovery of glutamic acid-148 as a key residue. Adv. Exp. Med. Biol 419, 45–52. [DOI] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, et al. (2017). Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield L, Bjorn MJ, Horn G, Fong D, Buck GA, Collier RJ, and Kaplan DA (1983). Nucleotide sequence of the structural gene for diphtheria toxin carried by corynebacteriophage beta. Proc. Natl. Acad. Sci. U S A 80, 6853–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon KS, Kleinstiver BP, Garcia SP, Zaborowski MP, Volak A, Spirig SE, Muller A, Sousa AA, Tsai SQ, Bengtsson NE, et al. (2019). High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun 10, 4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M, Chang LH, Pires-Alves M, Thyagarajan B, Bloom JE, Gu Z, Aberle KK, Teymorian SA, Bannai Y, Johnson SC, et al. (2011). Recombinant botulinum neurotoxin A heavy chain-based delivery vehicles for neuronal cell targeting. Protein Eng. Des. Sel 24, 247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, and Doudna JA (2017). CRISPR-Cas9 structures and mechanisms. Annu. Rev. Biophys 46, 505–529. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes L, and Romer W (2010). Shiga toxins–from cell biology to biomedical applications. Nat. Rev. Microbiol 8, 105–116. [DOI] [PubMed] [Google Scholar]
- King C, Garza EN, Mazor R, Linehan JL, Pastan I, Pepper M, and Baker D (2014). Removing T-cell epitopes with computational protein design. Proc. Natl. Acad. Sci. U S A 111, 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott GJ, and Doudna JA (2018). CRISPR-Cas guides the future of genetic engineering. Science 361, 866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott GJ, East-Seletsky A, Cofsky JC, Holton JM, Charles E, O’Connell MR, and Doudna JA (2017). Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme. Nat. Struct. Mol. Biol 24, 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, and Hsu PD (2018). Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676 e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, and Zhang F (2017). Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol 37, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Rabideau AE, and Pentelute BL (2014). Delivery of antibody mimics into mammalian cells via anthrax toxin protective antigen. Chembiochem 15, 2458–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino CA, Harper JC, Carney JP, and Timlin JA (2018). Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 25, 1234–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, and Church GM (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield MJ, Sugiman-Marangos SN, Melnyk RA, and Doxey AC (2018). Identification of a diphtheria toxin-like gene family beyond the Corynebacterium genus. FEBS Lett. 592, 2693–2705. [DOI] [PubMed] [Google Scholar]
- McCluskey AJ, and Collier RJ (2013). Receptor-directed chimeric toxins created by sortase-mediated protein fusion. Mol. Cancer Ther 12, 2273–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNutt PM, Vazquez-Cintron EJ, Tenezaca L, Ondeck CA, Kelly KE, Mangkhalakhili M, Machamer JB, Angeles CA, Glotfelty EJ, Cika J, et al. (2021). Neuronal delivery of antibodies has therapeutic effects in animal models of botulism. Sci. Transl. Med 13, eabd7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JC, Blanke SR, Hanna PC, and Collier RJ (1995). Protective antigen-binding domain of anthrax lethal factor mediates translocation of a heterologous protein fused to its amino- or carboxy-terminus. Mol. Microbiol 15, 661–666. [DOI] [PubMed] [Google Scholar]
- Mitamura T, Higashiyama S, Taniguchi N, Klagsbrun M, and Mekada E (1995). Diphtheria toxin binds to the epidermal growth factor (EGF)-like domain of human heparin-binding EGF-like growth factor/diphtheria toxin receptor and inhibits specifically its mitogenic activity. J. Biol. Chem 270, 1015–1019. [DOI] [PubMed] [Google Scholar]
- Miyashita SI, Zhang J, Zhang S, Shoemaker CB, and Dong M (2021). Delivery of single-domain antibodies into neurons using a chimeric toxin-based platform is therapeutic in mouse models of botulism. Sci. Transl. Med 13, eaaz4197. [DOI] [PubMed] [Google Scholar]
- Nagata S, and Pastan I (2009). Removal of B cell epitopes as a practical approach for reducing the immunogenicity of foreign protein-based therapeutics. Adv. Drug Deliv. Rev 61, 977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglich JG, Metherall JE, Russell DW, and Eidels L (1992). Expression cloning of a diphtheria toxin receptor: identity with a heparin-binding EGF-like growth factor precursor. Cell 69, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Ng Ang AP, Ebner JK, Plessner M, Aktories K, and Schmidt G (2019). Engineering Photorhabdus luminescens toxin complex (PTC) into a recombinant injection nanomachine. Life Sci. Alliance 2, e201900485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe DO, Cabiaux V, Choe S, Eisenberg D, and Collier RJ (1992). pH-dependent insertion of proteins into membranes: B-chain mutation of diphtheria toxin that inhibits membrane translocation, Glu-349––Lys. Proc. Natl. Acad. Sci. U S A 89, 6202–6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odumosu O, Nicholas D, Yano H, and Langridge W (2010). AB toxins: a paradigm switch from deadly to desirable. Toxins (Basel) 2, 1612–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Liu H, Ruan H, Tepp WH, Stoothoff WH, Brown RH, Johnson EA, Yao WD, Zhang SC, and Dong M (2013). Cytotoxicity of botulinum neurotoxins reveals a direct role of syntaxin 1 and SNAP-25 in neuron survival. Nat. Commun 4, 1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirazzini M, Rossetto O, Eleopra R, and Montecucco C (2017). Botulinum neurotoxins: biology, pharmacology, and toxicology. Pharmacol. Rev 69, 200–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabideau AE, and Pentelute BL (2016). Delivery of non-native cargo into mammalian cells using anthrax lethal toxin. ACS Chem. Biol 11, 1490–1501. [DOI] [PubMed] [Google Scholar]
- Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, and Kim H (2014). Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 24, 1020–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. (2015). In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roderer D, Schubert E, Sitsel O, and Raunser S (2019). Towards the application of Tc toxins as a universal protein translocation system. Nat. Commun 10, 5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet R, Thuma BA, Roy MD, Lintner NG, Rubitski DM, Finley JE, Wisniewska HM, Mendonsa R, Hirsh A, de Onate L, et al. (2018). Receptor-mediated delivery of CRISPR-cas9 endonuclease for cell-type-specific gene editing. J. Am. Chem. Soc 140, 6596–6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryou JH, Sohn YK, Hwang DE, Park WY, Kim N, Heo WD, Kim MY, and Kim HS (2016). Engineering of bacterial exotoxins for highly efficient and receptor-specific intracellular delivery of diverse cargos. Biotechnol. Bioeng 113, 1639–1646. [DOI] [PubMed] [Google Scholar]
- Schmit NE, Neopane K, and Hantschel O (2019). Targeted protein degradation through cytosolic delivery of monobody binders using bacterial toxins. ACS Chem. Biol 14, 916–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalaby K, Aouida M, and El-Agnaf O (2020). Tissue-specific delivery of CRISPR therapeutics: strategies and mechanisms of non-viral vectors. Int. J. Mol. Sci 21, 7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakulskas CA, and Behlke MA (2019). Evaluation and reduction of CRISPR off-target cleavage events. Nucleic Acid Ther. 29, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Cintron EJ, Beske PH, Tenezaca L, Tran BQ, Oyler JM, Glotfelty EJ, Angeles CA, Syngkon A, Mukherjee J, Kalb SR, et al. (2017). Engineering botulinum neurotoxin C1 as a molecular vehicle for intraneuronal drug delivery. Sci. Rep 7, 42923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdurmen WP, Luginbuhl M, Honegger A, and Pluckthun A (2015). Efficient cell-specific uptake of binding proteins into the cytoplasm through engineered modular transport systems. J. Control. Release 200, 13–22. [DOI] [PubMed] [Google Scholar]
- Vidimar V, Beilhartz GL, Park M, Biancucci M, Kieffer MB, Gius DR, Melnyk RA, and Satchell KJF (2020). An engineered chimeric toxin that cleaves activated mutant and wild-type RAS inhibits tumor growth. Proc. Natl. Acad. Sci. U S A 117, 16938–16948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Zhang F, and Gao G (2020). CRISPR-based therapeutic genome editing: strategies and in vivo delivery by AAV vectors. Cell 181, 136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will E, Klump H, Heffner N, Schwieger M, Schiedlmeier B, Ostertag W, Baum C, and Stocking C (2002). Unmodified Cre recombinase crosses the membrane. Nucleic Acids Res. 30, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, and Scott DA (2018). Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 70, 327–339 e325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip BH (2020). Recent advances in CRISPR/Cas9 delivery strategies. Biomolecules 10, 839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, and Collier RJ (2007). Anthrax toxin: receptor binding, internalization, pore formation, and translocation. Annu. Rev. Biochem 76, 243–265. [DOI] [PubMed] [Google Scholar]
- Zhang S, Lebreton F, Mansfield MJ, Miyashita SI, Zhang J, Schwartzman JA, Tao L, Masuyer G, Martinez-Carranza M, Stenmark P, et al. (2018). Identification of a botulinum neurotoxin-like toxin in a commensal strain of Enterococcus faecium. Cell Host Microbe 23, 169–176 e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Masuyer G, Zhang J, Shen Y, Lundin D, Henriksson L, Miyashita SI, Martinez-Carranza M, Dong M, and Stenmark P (2017). Identification and characterization of a novel botulinum neurotoxin. Nat. Commun 8, 14130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. (2013). Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, and Wei W (2014). High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509, 487–491. [DOI] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, and Liu DR (2015). Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol 33, 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available upon request. The sequencing data has been deposited to Dryad datasets (https://doi.org/10.5061/dryad.dncjsxm1p).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


