Abstract
Schizophrenia is a clinically and genetically heterogeneous neuropsychiatric disorder, with a polygenic basis but identification of the specific determinants is a continuing challenge. In this study we analyzed a multigenerational family, with all healthy individuals in the first two generations, and four progeny affected with schizophrenia in the subsequent two generations, using whole exome sequencing. We identified five rare protein sequence altering heterozygous variants, in five different genes namely SMARCA5, PDE1B, TNIK, SMARCA2 and FLRT shared among all affected members and predicted to be damaging.
Variants in SMARCA5 and PDE1B were inherited from the unaffected father whereas variants in TNIK, SMARCA2 and FLRT1 were inherited from the unaffected mother in all the three affected individuals in the third generation; and notably all these five variants were transmitted by an affected mother to her affected son. Microsatellite based analysis lent a modest linkage support (LOD score of 1.2; θ=0 at each variant). Of note, analysis of exome data of an ancestry matched unrelated schizophrenia cohort (n=350), revealed a total of 16 rare variants (MAF<0.01) in these five genes. Interestingly, these five genes involved in neurodevelopmental and/or neurotransmitter signaling processes are implicated in the etiology of schizophrenia previously. This study provides good evidence for a likely cumulative contribution of multiple rare variants from disease relevant genes with a threshold effect in disease development and seems to explain the unusual disease transmission pattern generally witnessed in such conditions, but warrants extensive replication efforts in families with similar complex disease inheritance profiles.
Introduction
Schizophrenia (SZ) is a severe neuropsychiatric disorder, commonly believed to arise from a complex interplay between genetic and environmental risk factors that influences early brain development and causes significant disability and early mortality. With onset of illness in early adolescence (Crome, 2017; Hjorthøj et al., 2017; Kahn et al., 2015; Pulver et al., 1990; Walker et al., 2015) it is expressed as a combination of positive symptoms such as hallucinations, delusions and negative symptoms including social withdrawal, affective flattening, anhedonia, diminished initiative and energy (McGlashan and Fenton, 1992). More than 98% of SZ patients also suffer from neurocognitive impairment (Heinrichs and Zakzanis, 1998; Zai et al., 2017). Deficits in synaptic formation and function due to early neurodevelopmental abnormalities are also reported (Corroon, 2005; Stachowiak et al., 2013).
Knowledge underlying disease etiology is still limited, but altered dopamine, glutamate, serotonin and Gamma Amino Butyric Acid (GABA) activity have been reported in SZ (Frohlich and Van Horn, 2014; Harrison and Weinberger, 2005; Schiffer, 2002; Stone, 2011; Stone et al., 2008; Thornberg and Saklad, 1996; Wang et al., 2011). On the other hand, a large number of family, twin and adoption studies in the pre-molecular genetics era, using a range of ascertainment and assessment rules, have consistently shown the importance of genetic factors in SZ (Gejman et al., 2011). However, in spite of high heritability (Cannon et al., 1998; Lichtenstein et al., 2009), and notable advances in technology from early linkage analysis, through candidate gene and genome-wide association studies (GWASs) to whole exome/genome sequencing, identification of genetic risk remains a challenge. GWAS performed by the Schizophrenia Working Group of the Psychiatric Genomics Consortium, has identified 108 risk loci with genome-wide significance (Ripke et al., 2014), but this barely explains the total heritability (Loh et al., 2015). A directed shift to exome sequencing that allows for identification of DNA variants within the 1–2% protein-coding regions in the genome, has substantially increased the likelihood of identification of putative causal/risk conferring rare variants at single-base resolution (Henriksen et al., 2017). This tool has since contributed notably to the spectrum of genetic risk variants for SZ. This has also garnered support by a previous study using the same approach wherein a polygenic burden of very rare (<1/10,000) disruptive variants distributed across many genes in a set of 2546 genes previously implicated in SZ has been put forth (Purcell et al., 2014). Furthermore, enrichment of genes involved in chromatin remodeling, glutamatergic postsynaptic proteins, N-methyl-D-aspartate receptor (NMDAR) etc. have been highlighted in studies reporting rare de novo variants in SZ (Fromer et al., 2014; McCarthy et al., 2014). Importantly, recent family based studies wherein we (John et al., 2019, 2018a, 2018b, 2017) and others (Egawa et al., 2016; Homann et al., 2016; Hornig et al., 2017; Hoya et al., 2017; Shirzad et al., 2016; Steinberg et al., 2017; Timms et al., 2013; Zhou et al., 2016) have identified inherited known/novel rare variants across several disease relevant genes have been informative. In the present study, rare variant analysis in an informative four generation family with multiple members affected with SZ was undertaken. Analysis uncovered five heterozygous inherited rare protein sequence altering variants in yet another group of functionally relevant genes namely SMARCA5, PDE1B, TNIK, SMARCA2 and FLRT shared among the affected individuals, and most notably, contributed in parts from the two unaffected parents. This observation supports the cumulative contribution of rare protein sequence altering variants in a few disease relevant genes and also a threshold effect for disease manifestation. Furthermore, it seems to provide likely explanation for the complex mode of inheritance commonly seen in complex traits, but whether this can be generalized remains to be validated in additional families with such diseases.
Methods
Sample ascertainment and recruitment
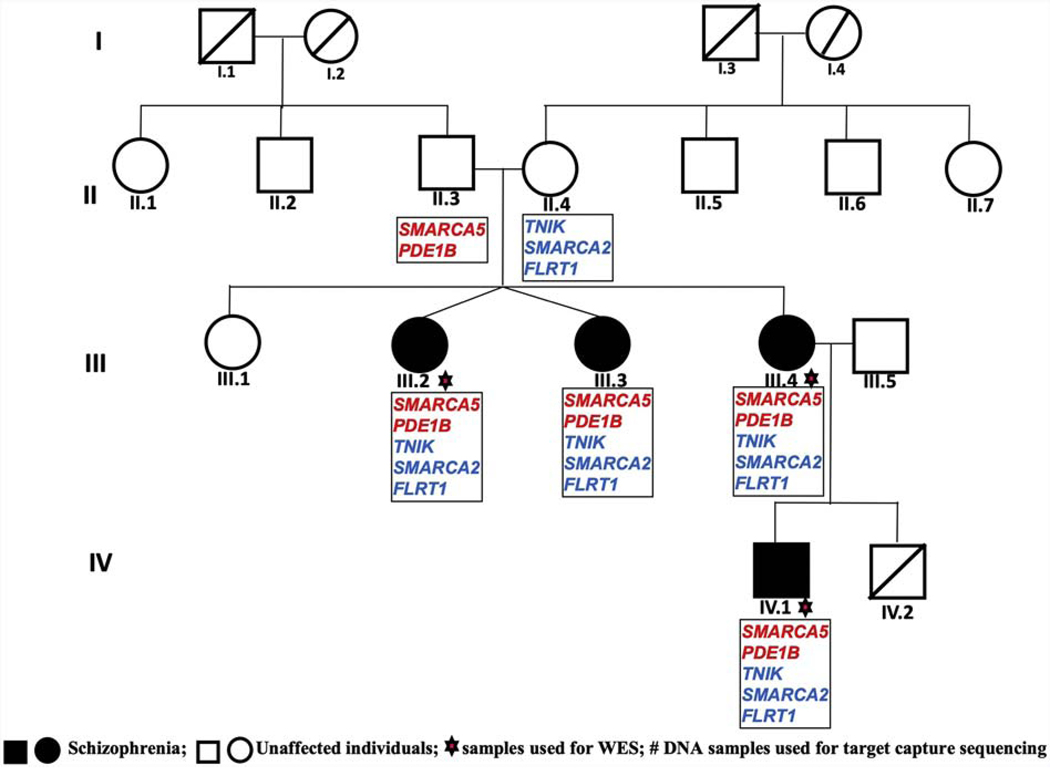
The institutional review committee of both participating institutes approved the study namely Dr. RML Hospital New Delhi India and University of Delhi South Campus, New Delhi, India. It was also approved by the University of Pittsburgh IRB. A four generation family of north Indian origin, with multiple members affected with SZ, was recruited from Dr. RML Hospital New Delhi, India. Diagnosis of SZ in this family was made according to DSM IV and relevant information for genetic studies collected by administering the Hindi version of DIGS and FIGS to the family members, as described previously (John et al., 2016; Kukshal et al., 2013a, 2013b). The four-generation family comprised of three siblings affected with SZ in the third generation (III.2 and III.3 are dizygotic twins), and one in the fourth generation along with 14 unaffected individuals across all the four generations. All four affected individuals and three unaffected parents from second to fourth generations were available for the study (Fig. 1). Venous blood samples were collected from these individuals and DNA isolated using conventional phenol chloroform method.
Figure 1:
Pedigree of a family used for whole exome sequencing, showing prioritized genes encompassing rare variants and their transmission pattern.
Whole exome sequencing
Assuming that gene sharing among distantly related individuals will be less, sequencing such members is expected to yield a much smaller number of shared variants for further segregation analysis in the family. With this rationale, two affected members from the third generation and one affected from the fourth generation were used for WES. Agilent SureSelectXT Human All Exon V5 + UTRs kit was used for enrichment of exonic regions and library preparation from blood derived DNA samples. Paired end (2×100) sequencing of the prepared libraries was performed in a commercial facility (Medgenome Labs Pvt. Ltd, Bangalore, India) using Illumina HiSeq 2000.
WES data analysis
The raw fastQ data obtained from the service providers were processed as per the recommendation provided by GATK (McKenna et al., 2010) best practices for germline variant discovery and which has been detailed previously (John et al., 2018b, 2017). Good quality reads were aligned to the reference genome (hg19), followed by removal of PCR duplicates and realignment of the mapped reads was performed. Subsequently combined variant calling of the three samples was performed using GATK. Called single nucleotide variants (SNVs), insertions and deletions (indels) were annotated using KGGSeq (Li et al., 2012) against RefSeq hg19 gene definitions.
Variant prioritisation
Prioritisation of the variants was based on the published recommendations and guidelines (Dashti and Gamieldien, 2017; Kircher et al., 2014; Richards et al., 2015) that we had also employed previously (John et al., 2019, 2018b, 2018a, 2017). Briefly, all the variants from the protein coding regions were extracted from the annotated data. Variants reported in public databases (1000G, ExAC, Exome Variant Server, dbSNP, gnomAD browser) with minor allele frequency (MAF) >0.01, synonymous variants and the variants that are not shared among the three affected and exome sequenced individuals were removed to start with. The remaining variants were screened among the four other family members available for the study, by target custom capture sequencing. Variants from segmentally duplicated regions and from genes that are known to produce false positive variants (Fuentes Fajardo et al., 2012) were then removed. Further, variants with MAF >0.01 in the non SZ exome data of north Indian population available in-house and variants which were not shared with the fourth affected individual in the family were eliminated, followed by removal of missense variants that are not predicted to be damaging by any two in silico software and with a CADD score<10. Finally, shared deleterious variants thus narrowed down were further prioritised based on their i) expression in brain; ii) circumstantial biochemical evidence; iii) previous genetic linkage/association findings; and iv) supporting evidence from published animal studies. Each of these steps and the variants obtained thereafter were tabulated (Supplementary Table 1) and variants confirmed by Sanger sequencing.
Screening for additional rare (MAF ≤0.01) variants in the prioritized genes in an independent SZ cohort
In order to identify index or additional rare variants in genes prioritized above, we used WES data from an unrelated cohort of 350 SZ cases and 220 non-psychotic controls of matched ethnicity available in the lab and which have been used in our previous studies (John et al., 2018b, 2017).
Results
QC analysis of WES data from three exome sequenced individuals in the family (Fig. 1) was performed using Qualimap (García-Alcalde et al., 2012). >99.8% target region was covered with >1X; >97.03% with 10X; and >88.9% with 20X; a 57.4X mean on target depth and mean mapping quality of 46.96 across these samples were observed.
A total of 45 rare heterozygous protein sequence altering variants were shared among all four affected individuals in the family (Supplementary Table 1), of which 32 were predicted to be damaging (Supplementary Table 2). Of note, though none of the family members were affected in the first and second generations, suggestive of an autosomal recessive mode of inheritance, we did not identify any rare homozygous/compound-heterozygous protein sequence altering variants shared among the affected individuals, in the third and also fourth generations. This necessitated us to consider the other well accepted oligogenic/polygenic hypothesis in SZ in the study family. To assess this approach, we focused on rare variants from genes previously implicated in SZ and other psychiatric disorders based on genetic and functional studies, as described elsewhere (Purcell et al., 2014).
Following this strategy, we identified five rare heterozygous missense variants in five different genes, namely TNIK (NM_001161562:c.1660C>A:p.P554T:exon15); SMARCA5 (NM_003601:c.1628A>G:p.N543S:exon13); SMARCA2(NM_001289396:c.210G>A:p.M70I:exon2); FLRT1 (NM_013280:c.1184C>T:p.T395M:exon2); and PDE1B (NM_001315534:c.47C>G:p.A16G:exon4) in all the three affected children in the third generation. Of note, variants in SMARCA5 and PDE1B were seen to be inherited from the unaffected father, while those in TNIK, SMARCA2 and FLRT1 were inherited from the unaffected mother. The same set of five variants was observed in the affected child in the fourth generation, having been inherited through the affected mother (Fig 1). All these five variants were confirmed by Sanger sequencing. p.P554T in TNIK is a novel variant and the other four are rare with MAF <0.004(maximum observed) in different public databases. All these five variants were predicted to be damaging using several in silico tools (Supplementary table 2).
Based on merely the absence of comparable contextual support for the role of other 27 variants (prioritized) in disease development, they cannot be dismissed as non-contributors to SZ. Therefore, the complete list of variants with all the annotations and genotypes of the members are documented in Supplementary Table 2, but not considered further in this study. It may be mentioned here that since homozygous or compound-heterozygous variants could not be identified as risk conferring in this family, we explored an oligogenic/polygenic model. The motivation behind checking this approach was not to provide a complete explanation for the genetic underpinning of the disease in the family but to check whether a few heterozygous variants from functionally relevant genes coming together but in parts from two unaffected parents can provide some insight into the unclear mode of disease inheritance in the family and whether it will support an oligogenic contribution or not.
Linkage analysis
To confirm that the genes/loci encompassing the five inherited rare variants shared among the affected individuals in the study family are indeed linked to the disease/phenotype, linkage analysis was performed using microsatellite (MS) markers (ABI Prism Linkage Mapping Set MD 10-ver. 2.5 kit) following the manufacturer’s instructions. Based on chromosomal location of the five variants identified to be segregating in the family, informative MS markers flanking each of them were selected for genotyping. These included D3S1614 & D3S1262 for TNIK; D4S424 & D4S413 for SMARCA5; D9S286 for SMARCA2; D11S905 & D11S987 for FLRT1; and D12S85, D12S368 & D12S351 for PDE1B. Phase established based on the haplotypes generated from the respective markers complemented the results obtained from WES data, which had shown the inheritance of the variants in FLRT1, SMARCA2 & TNIK from the unaffected mother and variants in PDE1B & SMARCA5 from the unaffected father (Fig. 1). Two-point LOD scores of 1.2 at θ 0.0 (the maximum expected for the study family size) supports a suggestive linkage of each of the markers except D3S1262, wherein a recombinant (III-2) was identified.
Additional rare variants
On screening for index and or additional rare variants in these five genes in exome data of an independent SZ cohort (n=350), 16 rare (MAF<0.01) protein sequence altering variants were identified. These included three missense variants (in five individuals) in TNIK; one missense variant (in one individual) in SMARCA5; two missense variants (in three individuals) in SMARCA2; six missense variants (in 11 individuals) in FLRT1; and three missense and a start-loss variant (in four individuals) in PDE1B. Of these, only two variants were present in the in-house control data with MAF <0.005. All these variants with their complete annotations are provided in the Supplementary Table 3.
Screening for variants in the PGC GWAS summary statistics
On screening PGC2 SZ GWAS (Ripke et al., 2014) summary statistics for each gene of interest, we identified a large number of SNPs (MAF>0.05) with p-value <0.05. SNPs with lowest p-value from each gene as follows, rs34134627, SMARCA5 (p=0.0003); rs10757249, SMARCA2 (p=0.0004); rs76321458, TNIK (p=0.004); rs3824854, FLRT1 (p=0.01); and rs1249951, PDE1B (p=0.03).
Discussion
Considering the enigmatic genetic etiology of SZ, the recent WES based rare variant identification along with or without linkage analysis support, from family based studies seems to be insightful. Findings of inherited rare variants in five disease relevant genes in a four generation family (Fig. 1; Supplementary Table 2), supported by suggestive linkage (LOD score 1.2 at θ 0.0), hint at the likely involvement of these five rare variants in SZ etiology in the family, and thus an oligogenic inheritance. Furthermore, a cumulative genetic contribution of rare variants to disease is evident by the five rare variants being shared among all affected and those with lesser number of variants (Fig. 1) being unaffected. All these rare variants are predicted to be damaging by a large number of in silico tools. All the five genes encompassing these variants are promising candidates from pathways of relevance in SZ etiology based on biochemical, in vitro and animal model studies and receive unambiguous support as likely risk conferring, as they can all be directly connected with well documented disease pathology and hypothesis (Walker et al., 2004). In addition, a number of SNPs from all the five genes are reported to have p-value <0.05 in PGC2 SZ GWAS (Ripke et al., 2014). Functions of each of these genes, which are of relevance to SZ and also for other brain related activities, are detailed below.
TNIK encodes for a serine-threonine kinase and is an activator of the Wnt signaling pathway. It has been found to be involved in responses to environmental stress (Potkin et al., 2009; Ryu et al., 2008). This kinase is essential for dendritic arborization, morphological integrity of dendrites and synaptic transmission and AMPA surface expression receptors (Hussain et al., 2010). It interacts with NMDA receptor via AKAP9 and both NMDA and metabotropic receptors bidirectionally regulate TNIK phosphorylation (Coba et al., 2012). It is a key synaptic partner of DISC1and regulates synaptic composition and activity by stabilizing the levels of key postsynaptic density proteins (Wang et al., 2011). Knockdown of TNIK in vitro is known to disrupt neuronal network physiology (MacLaren et al., 2011). TNIK has been shown to be required in synaptic plasticity, neuronal development and specific aspects of higher order cognition through both synaptic and nuclear signaling pathways in mice. Knockout mice have shown impaired dentate gyrus neurogenesis along with marked elevation in GSK3β on postsynaptic substrates and impaired cognitive functions. These animals also displayed hyperlocomotor behavior, that could be rapidly reversed by GSK3β inhibitors (Coba et al., 2012). De novo mutation (NM_015028.2: c.1208G>A: p.R403Q) in this gene in Autism has also been reported (Iossifov et al., 2014). Convergent functional genomics based approach identified TNIK as one of the most promising genes involved in pathophysiology and as a target for therapeutic intervention (Ayalew et al., 2012). SNP rs6444970 in TNIK (p=4·85 × 10−8) is associated with antipsychotic treatment response (Yu et al., 2018).
SMARCA5 encodes for ATP dependent chromatin remodeling protein SNF2H, which is also a member of SWI/SNF family of proteins. It is involved in chromatin remodeling and thus regulates the expression of certain genes. The protein plays a significant role during development particularly during neural development while transiting from a committed progenitor state to a differential cell state(Lazzaro and Picketts, 2001).Brains isolated from conditional knockout mice were found to be reduced in size, with striking cerebellar hypoplasia; along with this the forebrain also showed a modest size difference. Postnatal external granule cell layer within cerebellum of the mice showed reduced proportion of cycling, mitotic, and S-phase cells indicating its important role in granule neuron progenitor (GNP) expansion. However, Purkinje neurons were found abundantly but were highly disorganized and showed poor dendritic arborisation. Collectively all these evidences suggest an important role of SMARCA5 in neural cell differentiations. The conditional knockout mice showed motor and cognitive dysfunction(Alvarez-Saavedra et al., 2014; Goodwin and Picketts, 2018). De novo mutation (NM_003601.3:c.329C>A: p.A110D) in this gene has also been reported in autism (De Rubeis et al., 2014).
SMARCA2 is also a member of the SWI/SNF family of proteins. Through the interaction with various transcription factors and other DNA-binding proteins it is involved in chromatin structural modification and thus epigenetic regulation of gene and their expression. Expression of the protein was low in neural precursor cells, but it was increased during differentiation to neurons and astrocytes, suggesting its role in neuronal differentiation(Machida et al., 2001). It is involved in dendrite spine morphology and dendrite growth. The gene is known to interact with SZ-associated (GWAS) genes namely CSF2RA, NOTCH4, NRGN, HIST1H2BJ, ZNF804A, SHOX and TCF4. The gene and its interacting partners are enriched among the genes showing positive selection in primates and in human lineage (Loe-Mie et al., 2010). Knockout mice showed impaired prepulse inhibition (SZ like behavior) and social interaction. Two SNPs (rs2296212; P = 5.8×10−5; rs3793490; P = 2.0 ×10−6) in this gene are reported to be associated with SZ in Japanese population (Koga et al., 2009). De novo in-frame deletion and missense mutations in patients with intellectual disability, epilepsy etc (Wolff et al., 2012) and a de novo mutation (NM_003070.3: c.4046G>A: p.R1349Q) in a patient with autism(Yuen et al., 2017)have also been reported.
FLRT1 is a member of the fibronectin leucine rich transmembrane protein (FLRT) family. It is involved in cell adhesion and receptor signaling. Through the FGFR1 mediated signaling cascade this protein activates MAP kinases and promotes the neurite outgrowth (Haines et al., 2006; O’Sullivan et al., 2012; Wheldon et al., 2010). Role of FGFR1 is previously implicated in psychiatric disorders (Narla et al., 2017; Stachowiak et al., 2007) and we have also recently reported rare variants segregating with SZ and other psychiatric disorders in a family (John et al., 2019). Adhesion proteins that contain such Leucine-rich repeats (LRR) have been found to play as key organizers of excitatory and inhibitory synapses and regulate the assembly of specific connectivity patterns across neural circuits thus contributing towards the diverse structural and functional properties of synapses(Schroeder and de Wit, 2018). This gene is also involved in various neurodevelopmental processes (del Toro et al., 2017). A de novo mutation (NM_013280.4; c.598C>G; p.L200V) has been reported in an autism patient (Krumm et al., 2015).
PDE1B is a calcium-dependent cyclic nucleotide phosphodiesterase and activated by a calcium-calmodulin complex. The protein is highly expressed in striatum, hippocampus, and prefrontal cortex and colocalized with dopamine D1 receptors (Lakics et al., 2010). Pde1b knockout mice showed increased baseline exploratory activity and exaggerated locomotor response when challenged with amphetamine (AMPH) and methamphetamine (METH), increased dopamine turnover and serotonin (5-HT) levels in striatum and impaired spatial learning (Reed et al., 2002; Siuciak et al., 2007). After dopamine D1 receptor agonist or forskolin stimulation in striatal slices from the knockout mice showed higher levels of phospho-Ser845 GluR1 and phosphor-Thr34 DARPP-32 and augmentation of cyclic nucleotide signaling (Reed et al., 2002). Increased striatal Pde10a mRNA expression and resistance to acute stress-induceddepression-like behavior has been reported (J. R. Hufgard et al., 2017; Jillian R. Hufgard et al., 2017). A de novo mutation (NM_000924.3; c.640G>A: p.G214S) in an autism patient has been reported(De Rubeis et al., 2014).
How these five protein act together and will develop the disease is not able to explain completely, however based on the already known function of these genes it can be hypothesized that since except PDE1B all other genes are playing important role in various neurodevelopmental process, malfunction of these genes may disrupt the normal brain development during early life, along with the abnormal neurotransmitter signaling in the earlier and later stages of the life by the disruption of PDEB1 and TNIK normal function may leads to the development of the disease
Like all other reports of this kind, this study also has a few limitations: i) even though we performed linkage analysis with several markers, the LOD score of 1.2 at θ 0.0, is low and only suggestive of linkage and thus lacking strong statistical support; ii) despite the use of hypothesis free WES approach to identify putative risk conferring variants, genes finally included are based on a priori literature support and those with variants predicted to be deleterious (similar to our previous studies) and thus a few genes may have been missed; and iii) all the five variants were either novel or very rare in various public data bases, predicted to be damaging and located at evolutionarily conserved positions suggesting their normal protein function altering effect but lack functional validation. iv) the unaffected individual in the third generation (iii.1) were not ready to participate in the study and lack of genotype availability of that individual, is one of the limitation which may helped to check the contribution of these five variants in the etiology of the disease in the family. v) Replication cohort used in the study (350 SZ cases and 220 controls) is small for case-control based rare variant analysis, so these findings need to validated in a larger cohort. Conversely, the possible involvement of all the five variants in disease development derives ample support at variant and gene levels. Besides their rare/novel but highly conserved nature, and predicted to be damaging, all the five genes are known to be involved in neurodevelopment/neurotransmitter signaling processes; and based on previous expression/exome sequencing studies are also known to be involved in SZ and/or other psychiatric disorders.
In summary, with contextual support at the variant and gene levels, the multiple heterozygous protein sequence altering variants identified in disease relevant genes may contribute in a cumulative manner to the development of disease in the family, Besides supporting an oligogenic mode of inheritance, the observations seem to explain the complex mode inheritance observed in the families with psychiatric disorders and other complex diseases, generalization of which remains to be validated. Though such studies augment our current knowledge of SZ genetics, newer analytical paradigms and novel functional screens are highly desired for effective use of the fast accumulating vast genome data.
Supplementary Material
Acknowledgements
Gratefully acknowledge Junior and Senior Research Fellowship (09/045(1166)/2012-EMR-I) to JJ; Junior Research Fellowship (# 325541-2015, to NY) from Council for Scientific and Industrial Research (CSIR), New Delhi; Junior Research Fellowship (to UB) from the Department of Genetics, under UGC-Special Assistance Program Meritorious Award scheme; and DSK-PDF (BL/13-14/0404) (to PK) from University Grants commission (UGC), New Delhi. We are thankful for the study sample collection by trained and dedicated staff at Dr RML Hospital, DNA isolation by Mrs. Anjali Dabral at the University of Delhi South Campus (UDSC)and computational facility provided by Central Instrumentation Facility, UDSC. We gratefully acknowledge infrastructure support provided by the UGC, New Delhi, through Special Assistance Programme and Department of Science and Technology, New Delhi, through FIST and DU-DST PURSE programmes to the Department of Genetics, UDSC.
Role of funding sources
This work was supported in part by grants #BT/MB/Project-Schizophrenia/2012–2013 and #BT/PR2425/Med13/089/2001 to Prof. B.K. Thelma and Prof. S. N. Deshpande from the Department of Biotechnology, Government of India, New Delhi; Grant #MH093246, #MH063480 and #TW009114 to Prof. V. L. Nimgaonkar from NIMH, the Fogarty International Center, USA. All funding sources had no further role in in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Footnotes
Conflicts of Interest
The authors declare that there is no conflict of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Alvarez-Saavedra M, De Repentigny Y, Lagali PS, Raghu Ram EVS, Yan K, Hashem E, Ivanochko D, Huh MS, Yang D, Mears AJ, Todd MAM, Corcoran CP, Bassett EA, Tokarew NJA, Kokavec J, Majumder R, Ioshikhes I, Wallace VA, Kothary R, Meshorer E, Stopka T, Skoultchi AI, Picketts DJ, 2014. Snf2h-mediated chromatin organization and histone H1 dynamics govern cerebellar morphogenesis and neural maturation. Nat. Commun 5, 4181. 10.1038/ncomms5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, Winiger E, Breier A, Shekhar A, Amdur R, Koller D, Nurnberger JI, Corvin A, Geyer M, Tsuang MT, Salomon D, Schork NJ, Fanous AH, O’Donovan MC, Niculescu AB, 2012. Convergent functional genomics of schizophrenia: From comprehensive understanding to genetic risk prediction. Mol. Psychiatry 17, 887–905. 10.1038/mp.2012.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Kaprio J, Lönnqvist J, Huttunen M, Koskenvuo M, 1998. The genetic epidemiology of schizophrenia in a Finnish twin cohort: A population-based modeling study. Arch. Gen. Psychiatry 55, 67–74. 10.1001/archpsyc.55.1.67 [DOI] [PubMed] [Google Scholar]
- Coba MP, Komiyama NH, Nithianantharajah J, Kopanitsa MV, Indersmitten T, Skene NG, Tuck EJ, Fricker DG, Elsegood KA, Stanford LE, Afinowi NO, Saksida LM, Bussey TJ, O’Dell TJ, Grant SGN, 2012. TNiK Is Required for Postsynaptic and Nuclear Signaling Pathways and Cognitive Function. J. Neurosci 32, 13987–13999. 10.1523/JNEUROSCI.2433-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corroon EB, 2005. A Review of the Neurodevelopmental Hypothesis of Schizophrenia. Trinity Student Med. J 6, 39–43. [Google Scholar]
- Crome I, 2017. Substance misuse, in: Psychiatry by Ten Teachers, Second Edition. Nature Publishing Group, pp. 114–128. 10.1201/9781315380612 [DOI] [Google Scholar]
- Dashti MJS, Gamieldien J, 2017. A practical guide to filtering and prioritizing genetic variants. Biotechniques 62, 18–30. 10.2144/000114492 [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, Singh T, Klei L, Kosmicki J, Fu SC, Aleksic B, Biscaldi M, Bolton PF, Brownfeld JM, Cai J, Campbell NG, Carracedo A, Chahrour MH, Chiocchetti AG, Coon H, Crawford EL, Crooks L, Curran SR, Dawson G, Duketis E, Fernandez BA, Gallagher L, Geller E, Guter SJ, Hill RS, Ionita-Laza I, Gonzalez PJ, Kilpinen H, Klauck SM, Kolevzon A, Lee I, Lei J, Lehtimäki T, Lin CF, Ma’ayan A, Marshall CR, McInnes AL, Neale B, Owen MJ, Ozaki N, Parellada M, Parr JR, Purcell S, Puura K, Rajagopalan D, Rehnström K., Reichenberg A, Sabo A, Sachse M, Sanders SJ, Schafer C, Schulte-Rüther M., Skuse D, Stevens C, Szatmari P, Tammimies K, Valladares O, Voran A, Wang LS, Weiss LA, Willsey AJ, Yu TW, Yuen RKC, Cook EH, Freitag CM, Gill M, Hultman CM, Lehner T, Palotie A, Schellenberg GD, Sklar P, State MW, Sutcliffe JS, Walsh CA, Scherer SW, Zwick ME, Barrett JC, Cutler DJ, Roeder K, Devlin B, Daly MJ, Buxbaum JD, 2014. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215. 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Toro D, Ruff T, Cederfjäll E, Villalba A, Seyit-Bremer G, Borrell V, Klein R, 2017. Regulation of Cerebral Cortex Folding by Controlling Neuronal Migration via FLRT Adhesion Molecules. Cell. 10.1016/j.cell.2017.04.012 [DOI] [PubMed] [Google Scholar]
- Egawa J, Hoya S, Watanabe Y, Nunokawa A, Shibuya M, Ikeda M, Inoue E, Okuda S, Kondo K, Saito T, Kaneko N, Muratake T, Igeta H, Iwata N, Someya T, 2016. Rare UNC13B variations and risk of schizophrenia: Whole-exome sequencing in a multiplex family and follow-up resequencing and a case–control study. Am. J. Med. Genet. Part B Neuropsychiatr. Genet 171, 797–805. [DOI] [PubMed] [Google Scholar]
- Frohlich J, Van Horn JD, 2014. Reviewing the ketamine model for schizophrenia. J. Psychopharmacol 10.1177/0269881113512909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, Dwyer S, Gormley P, Georgieva L, Rees E, Palta P, Ruderfer DM, Carrera N, Humphreys I, Johnson JS, Roussos P, Barker DD, Banks E, Milanova V, Grant SG, Hannon E, Rose SA, Chambert K, Mahajan M, Scolnick EM, Moran JL, Kirov G, Palotie A, McCarroll SA, Holmans P, Sklar P, Owen MJ, Purcell SM, O’Donovan MC, O’Donovan MC, O’Donovan MC, 2014. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–84. 10.1038/nature12929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes Fajardo KV, Adams D, Mason CE, Sincan M, Tifft C, Toro C, Boerkoel CF, Gahl W, Markello T, 2012. Detecting false-positive signals in exome sequencing. Hum. Mutat 33, 609–613. 10.1002/humu.22033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Götz S, Tarazona S, Dopazo J, Meyer TF, Conesa A, 2012. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 28, 2678–2679. 10.1093/bioinformatics/bts503 [DOI] [PubMed] [Google Scholar]
- Gejman PV, Sanders AR, Kendler KS, 2011. Genetics of schizophrenia: new findings and challenges. Annu. Rev. Genomics Hum. Genet 12, 121–44. 10.1146/annurev-genom-082410-101459 [DOI] [PubMed] [Google Scholar]
- Goodwin LR, Picketts DJ, 2018. The role of ISWI chromatin remodeling complexes in brain development and neurodevelopmental disorders. Mol. Cell. Neurosci 10.1016/j.mcn.2017.10.008 [DOI] [PubMed] [Google Scholar]
- Haines BP, Wheldon LM, Summerbell D, Heath JK, Rigby PWJ, 2006. Regulated expression of FLRT genes implies a functional role in the regulation of FGF signalling during mouse development. Dev. Biol 297, 14–25. 10.1016/j.ydbio.2006.04.004 [DOI] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR, 2005. Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol. Psychiatry 10.1038/sj.mp.4001558 [DOI] [PubMed] [Google Scholar]
- Heinrichs RW, Zakzanis KK, 1998. Neurocognitive deficit in schizophrenia: A quantitative review of the evidence. Neuropsychology 12, 426–445. 10.1037/0894-4105.12.3.426 [DOI] [PubMed] [Google Scholar]
- Henriksen MG, Nordgaard J, Jansson LB, 2017. Genetics of Schizophrenia: Overview of Methods, Findings and Limitations. Front. Hum. Neurosci 11, 322. 10.3389/fnhum.2017.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorthøj C, Stürup AE, McGrath JJ, Nordentoft M, 2017. Years of potential life lost and life expectancy in schizophrenia: a systematic review and meta-analysis. The Lancet Psychiatry 4, 295–301. 10.1016/S2215-0366(17)30078-0 [DOI] [PubMed] [Google Scholar]
- Homann OR, Misura K, Lamas E, Sandrock RW, Nelson P, Mcdonough SI, De Lisi LE, 2016. Whole-genome sequencing in multiplex families with psychoses reveals mutations in the SHANK2 and SMARCA1 genes segregating with illness. Mol. Psychiatry 21, 1690–1695. 10.1038/mp.2016.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornig T, Grüning B, Kundu K, Houwaart T, Backofen R, Biber K, Normann C, 2017. GRIN3B missense mutation as an inherited risk factor for schizophrenia: Whole-exome sequencing in a family with a familiar history of psychotic disorders. Genet. Res. (Camb) 99, e1. 10.1017/S0016672316000148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoya S, Watanabe Y, Hishimoto A, Nunokawa A, Kaneko N, Muratake T, Shinmyo N, Otsuka I, Okuda S, Inoue E, Igeta H, Shibuya M, Egawa J, Orime N, Sora I, Someya T, 2017. Rare PDCD11 variations are not associated with risk of schizophrenia in Japan. Psychiatry Clin. Neurosci 71, 780–788. 10.1111/pcn.12549 [DOI] [PubMed] [Google Scholar]
- Hufgard Jillian R., Williams MT, Skelton MR, Grubisha O, Ferreira FM, Sanger H, Wright ME, Reed-Kessler TM, Rasmussen K, Duman RS, Vorhees CV, 2017. Phosphodiesterase-1b (Pde1b) knockout mice are resistant to forced swim and tail suspension induced immobility and show upregulation of Pde10a. Psychopharmacology (Berl). 234, 1803–1813. 10.1007/s00213-017-4587-8 [DOI] [PubMed] [Google Scholar]
- Hufgard JR, Williams MT, Vorhees CV, 2017. Phosphodiesterase-1b deletion confers depression-like behavioral resistance separate from stress-related effects in mice. Genes, Brain Behav. 16, 756–767. 10.1111/gbb.12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain NK, Hsin H, Huganir RL, Sheng M, 2010. MINK and TNIK Differentially Act on Rap2-Mediated Signal Transduction to Regulate Neuronal Structure and AMPA Receptor Function. J. Neurosci 30, 14786–14794. 10.1523/JNEUROSCI.4124-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, Smith JD, Paeper B, Nickerson DA, Dea J, Dong S, Gonzalez LE, Mandell JD, Mane SM, Murtha MT, Sullivan CA, Walker MF, Waqar Z, Wei L, Willsey AJ, Yamrom B, Lee YH, Grabowska E, Dalkic E, Wang Z, Marks S, Andrews P, Leotta A, Kendall J, Hakker I, Rosenbaum J, Ma B, Rodgers L, Troge J, Narzisi G, Yoon S, Schatz MC, Ye K, McCombie WR, Shendure J, Eichler EE, State MW, Wigler M, 2014. The contribution of de novo coding mutations to autism spectrum disorder. Nature 515, 216–221. 10.1038/nature13908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J, Bhatia T, Kukshal P, Chandna P, Nimgaonkar VL, Deshpande SN, Thelma BK, 2016. Association study of MiRSNPs with schizophrenia, tardive dyskinesia and cognition. Schizophr. Res 174, 29–34. 10.1016/j.schres.2016.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J, Kukshal P, Bhatia T, Chowdari KV, Nimgaonkar VL, Deshpande SN, Thelma BK, 2017. Possible role of rare variants in Trace amine associated receptor 1 in schizophrenia. Schizophr. Res 189, 190–195. 10.1016/j.schres.2017.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J, Kukshal P, Bhatia T, Nimgaonkar VL, Deshpande SN, Thelma BK, 2019. Rare variant based evidence for oligogenic contribution of neurodevelopmental pathway genes to schizophrenia. Schizophr. Res 10.1016/J.SCHRES.2018.12.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J, Kukshal P, Sharma A, Bhatia T, Nimgaonkar VL, Deshpande SN, Thelma BK, 2018a. Rare variants in Protein tyrosine phosphatase, receptor type A (PTPRA) in schizophrenia: Evidence from a family based study. Schizophr. Res 10.1016/J.SCHRES.2018.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John J, Sharma A, Kukshal P, Bhatia T, Nimgaonkar VL, Deshpande SN, Thelma BK, 2018b. Rare Variants in Tissue Inhibitor of Metalloproteinase 2 as a Risk Factor for Schizophrenia: Evidence From Familial and Cohort Analysis. Schizophr. Bull 10.1093/schbul/sbx196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RS, Sommer IE, Murray RM, Meyer-Lindenberg A, Weinberger DR, Cannon TD, O’Donovan M, Correll CU, Kane JM, van Os J, Insel TR, 2015. Schizophrenia. Nat. Rev. Dis. Prim 1, 15067. 10.1038/nrdp.2015.67 [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J, Roak BJO, Cooper GM, Shendure J, 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet 46, 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga M, Ishiguro H, Yazaki S, Horiuchi Y, Arai M, Niizato K, Iritani S, Itokawa M, Inada T, Iwata N, Ozaki N, Ujike H, Kunugi H, Sasaki T, Takahashi M, Watanabe Y, Someya T, Kakita A, Takahashi H, Nawa H, Muchardt C, Yaniv M, Arinami T, 2009. Involvement of SMARCA2/BRM in the SWI/SNF chromatin-remodeling complex in schizophrenia. Hum. Mol. Genet 18, 2483–2494. 10.1093/hmg/ddp166 [DOI] [PubMed] [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, Leal SM, Bernier R, Eichler EE, 2015. Excess of rare, inherited truncating mutations in autism. Nat. Genet 47, 582–588. 10.1038/ng.3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukshal P, Bhatia T, Bhagwat AM, Gur RE, Gur RC, Deshpande SN, Nimgaonkar VL, Thelma BK, 2013a. Association study of Neuregulin-1 gene polymorphisms in a north Indian schizophrenia sample. Schizophr. Res 144, 24–30. 10.1016/j.schres.2012.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukshal P, Kodavali VC, Srivastava V, Wood J, McClain L, Bhatia T, Bhagwat AM, Deshpande SN, Nimgaonkar VL, Thelma BK, 2013b. Dopaminergic gene polymorphisms and cognitive function in a north Indian schizophrenia cohort. J. Psychiatr. Res 47, 1615–22. 10.1016/j.jpsychires.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakics V, Karran EH, Boess FG, 2010. Quantitative comparison of phosphodiesterase mRNA distribution in human brain and peripheral tissues. Neuropharmacology 59, 367–374. 10.1016/j.neuropharm.2010.05.004 [DOI] [PubMed] [Google Scholar]
- Lazzaro MA, Picketts DJ, 2001. Cloning and characterization of the murine Imitation S witch (ISWI) genes: Differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J. Neurochem 77, 1145–1156. 10.1046/j.1471-4159.2001.00324.x [DOI] [PubMed] [Google Scholar]
- Li MX, Gui HS, Kwan JSH, Bao SY, Sham PC, 2012. A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res 40, e53–e53. 10.1093/nar/gkr1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM, 2009. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 373, 234–239. 10.1016/S0140-6736(09)60072-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loe-Mie Y, Lepagnol-Bestel AM, Maussion G, Doron-Faigenboim A, Imbeaud S, Delacroix H, Aggerbeck L, Pupko T, Gorwood P, Simonneau M, Moalic JM, 2010. SMARCA2 and other genome-wide supported schizophrenia-associated genes: Regulation by REST/NRSF, network organization and primate-specific evolution. Hum. Mol. Genet 19, 2841–2857. 10.1093/hmg/ddq184 [DOI] [PubMed] [Google Scholar]
- Loh P-R, Bhatia G, Gusev A, Finucane HK, Bulik-Sullivan BK, Pollack SJ, Schizophrenia Working Group of Psychiatric Genomics Consortium, de Candia TR, Lee SH, Wray NR, Kendler KS, O’Donovan MC, Neale BM, Patterson N, Price AL, 2015. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat. Genet 47, 1385–92. 10.1038/ng.3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y, Murai K, Miyake K, Iijima S, 2001. Expression of chromatin remodeling factors during neural differentiation. J. Biochem 129, 43–49. 10.1093/oxfordjournals.jbchem.a002834 [DOI] [PubMed] [Google Scholar]
- MacLaren EJ, Charlesworth P, Coba MP, Grant SGN, 2011. Knockdown of mental disorder susceptibility genes disrupts neuronal network physiology in vitro. Mol. Cell. Neurosci 47, 93–99. 10.1016/j.mcn.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy SE, Gillis J, Kramer M, Lihm J, Yoon S, Berstein Y, Mistry M, Pavlidis P, Solomon R, Ghiban E, Antoniou E, Kelleher E, O’Brien C, Donohoe G, Gill M, Morris DW, McCombie WR, Corvin A, 2014. De novo mutations in schizophrenia implicate chromatin remodeling and support a genetic overlap with autism and intellectual disability. Mol. Psychiatry 19, 652–8. 10.1038/mp.2014.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlashan TH, Fenton WS, 1992. The Positive-Negative Distinction in Schizophrenia. Arch. Gen. Psychiatry 49, 63. 10.1001/archpsyc.1992.01820010063008 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA, 2010. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narla ST, Lee YW, Benson CA, Sarder P, Brennand KJ, Stachowiak EK, Stachowiak MK, 2017. Common developmental genome deprogramming in schizophrenia — Role of Integrative Nuclear FGFR1 Signaling (INFS). Schizophr. Res 185, 17–32. 10.1016/j.schres.2016.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan ML, de Wit J, Savas JN, Comoletti D, Otto-Hitt S, Yates JR, Ghosh A, 2012. FLRT Proteins Are Endogenous Latrophilin Ligands and Regulate Excitatory Synapse Development. Neuron 73, 903–910. 10.1016/j.neuron.2012.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD, Mathalon D, Ford J, Lauriello J, Macciardi F, 2009. A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophr. Bull 35, 96–108. 10.1093/schbul/sbn155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver AE, Brown CH, Wolyniec P, McGrath J, Tam D, Adler L, Carpenter WT, Childs B, 1990. Schizophrenia: age at onset, gender and familial risk. Acta Psychiatr. Scand 82, 344–351. 10.1111/j.1600-0447.1990.tb01399.x [DOI] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kähler A, Duncan L, Stahl E, Genovese G, Fernández E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PKE, Banks E, Shakir K, Garimella K, Fennell T, Depristo M, Grant SGN, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA, Sklar P, 2014. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190. 10.1038/nature12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed TM, Repaske DR, Snyder GL, Greengard P, Vorhees CV, 2002. Phosphodiesterase 1B knock-out mice exhibit exaggerated locomotor hyperactivity and DARPP-32 phosphorylation in response to dopamine agonists and display impaired spatial learning. J. Neurosci 22, 5188–5197. https://doi.org/22/12/5188 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, 2015. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med 17, 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JTR, Farh KH, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H, Pers TH, Agartz I, Agerbo E, Albus M, Alexander M, Amin F, Bacanu SA, Begemann M, Belliveau RA, Bene J, Bergen SE, Bevilacqua E, Bigdeli TB, Black DW, Bruggeman R, Buccola NG, Buckner RL, Byerley W, Cahn W, Cai G, Campion D, Cantor RM, Carr VJ, Carrera N, Catts SV, Chambert KD, Chan RCK, Chen RYL, Chen EYH, Cheng W, Cheung EFC, Chong SA, Cloninger CR, Cohen D, Cohen N, Cormican P, Craddock N, Crowley JJ, Curtis D, Davidson M, Davis KL, Degenhardt F, Del Favero J, Demontis D, Dikeos D, Dinan T, Djurovic S, Donohoe G, Drapeau E, Duan J, Dudbridge F, Durmishi N, Eichhammer P, Eriksson J, Escott-Price V, Essioux L, Fanous AH, Farrell MS, Frank J, Franke L, Freedman R, Freimer NB, Friedl M, Friedman JI, Fromer M, Genovese G, Georgieva L, Giegling I, Giusti-Rodríguez P, Godard S, Goldstein JI, Golimbet V, Gopal S, Gratten J, De Haan L, Hammer C, Hamshere ML, Hansen M, Hansen T, Haroutunian V, Hartmann AM, Henskens FA, Herms S, Hirschhorn JN, Hoffmann P, Hofman A, Hollegaard MV, Hougaard DM, Ikeda M, Joa I, Julià A, Kahn RS, Kalaydjieva L, Karachanak-Yankova S, Karjalainen J, Kavanagh D, Keller MC, Kennedy JL, Khrunin A, Kim Y, Klovins J, Knowles JA, Konte B, Kucinskas V, Kucinskiene ZA, Kuzelova-Ptackova H, Kähler AK, Laurent C, Keong JLC, Lee SH, Legge SE, Lerer B, Li M, Li T, Liang KY, Lieberman J, Limborska S, Loughland CM, Lubinski J, Lönnqvist J, Macek M, Magnusson PKE, Maher BS, Maier W, Mallet J, Marsal S, Mattheisen M, Mattingsdal M, McCarley RW, McDonald C, McIntosh AM, Meier S, Meijer CJ, Melegh B, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mokrab Y, Morris DW, Mors O, Murphy KC, Murray RM, Myin-Germeys I, Müller-Myhsok B, Nelis M, Nenadic I, Nertney DA, Nestadt G, Nicodemus KK, Nikitina-Zake L, Nisenbaum L, Nordin A, O’Callaghan E, O’Dushlaine C, O’Neill FA, Oh SY, Olincy A, Olsen L, Van Os J, Pantelis C, Papadimitriou GN, Papiol S, Parkhomenko E, Pato MT, Paunio T, Pejovic-Milovancevic M, Perkins DO, Pietiläinen O, Pimm J, Pocklington AJ, Powell J, Price A, Pulver AE, Purcell SM, Quested D, Rasmussen HB, Reichenberg A, Reimers MA, Richards AL, Roffman JL, Roussos P, Ruderfer DM, Salomaa V, Sanders AR, Schall U, Schubert CR, Schulze TG, Schwab SG, Scolnick EM, Scott RJ, Seidman LJ, Shi J, Sigurdsson E, Silagadze T, Silverman JM, Sim K, Slominsky P, Smoller JW, So HC, Spencer CCA, Stahl EA, Stefansson H, Steinberg S, Stogmann E, Straub RE, Strengman E, Strohmaier J, Stroup TS, Subramaniam M, Suvisaari J, Svrakic DM, Szatkiewicz JP, Söderman E, Thirumalai S, Toncheva D, Tosato S, Veijola J, Waddington J, Walsh D, Wang D, Wang Q, Webb BT, Weiser M, Wildenauer DB, Williams NM, Williams S, Witt SH, Wolen AR, Wong EHM, Wormley BK, Xi HS, Zai CC, Zheng X, Zimprich F, Wray NR, Stefansson K, Visscher PM, Adolfsson R, Andreassen OA, Blackwood DHR, Bramon E, Buxbaum JD, Børglum AD, Cichon S, Darvasi A, Domenici E, Ehrenreich H, Esko T, Gejman PV, Gill M, Gurling H, Hultman CM, Iwata N, Jablensky AV, Jönsson EG, Kendler KS, Kirov G, Knight J, Lencz T, Levinson DF, Li QS, Liu J, Malhotra AK, McCarroll SA, McQuillin A, Moran JL, Mortensen PB, Mowry BJ, Nöthen MM, Ophoff RA, Owen MJ, Palotie A, Pato CN, Petryshen TL, Posthuma D, Rietschel M, Riley BP, Rujescu D, Sham PC, Sklar P, St Clair D, Weinberger DR, Wendland JR, Werge T, Daly MJ, Sullivan PF, O’Donovan MC, 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu J, Futai K, Feliu M, Weinberg R, Sheng M, 2008. Constitutively Active Rap2 Transgenic Mice Display Fewer Dendritic Spines, Reduced Extracellular Signal-Regulated Kinase Signaling, Enhanced Long-Term Depression, and Impaired Spatial Learning and Fear Extinction. J. Neurosci 28, 8178–8188. 10.1523/JNEUROSCI.1944-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer HH, 2002. Glutamate receptor genes: Susceptibility factors in schizophrenia and depressive disorders? Mol. Neurobiol 25, 191–212. 10.1385/MN:25:2:191 [DOI] [PubMed] [Google Scholar]
- Schroeder A, de Wit J, 2018. Leucine-rich repeat-containing synaptic adhesion molecules as organizers of synaptic specificity and diversity. Exp. Mol. Med 50, 10. 10.1038/s12276-017-0023-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirzad H, Beiraghi N, Ataei Kachoui M, Akbari MT, 2016. Family-Based Whole-Exome Sequencing for Identifying Novel Variants in Consanguineous Families with Schizophrenia. Iran. Red Crescent Med. J. In press, 1–8. 10.5812/ircmj.35788 [DOI] [Google Scholar]
- Siuciak JA, McCarthy SA, Chapin DS, Reed TM, Vorhees CV, Repaske DR, 2007. Behavioral and neurochemical characterization of mice deficient in the phosphodiesterase-1B (PDE1B) enzyme. Neuropharmacology 53, 113–124. 10.1016/j.neuropharm.2007.04.009 [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Kucinski A, Curl R, Syposs C, Yang Y, Narla S, Terranova C, Prokop D, Klejbor I, Bencherif M, Birkaya B, Corso T, Parikh A, Tzanakakis ES, Wersinger S, Stachowiak EK, 2013. Schizophrenia: A neurodevelopmental disorder - Integrative genomic hypothesis and therapeutic implications from a transgenic mouse model. Schizophr. Res 10.1016/j.schres.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Stachowiak MK, Maher PA, Stachowiak EK, 2007. Integrative Nuclear Signaling in Cell Development—A Role for FGF Receptor-1. DNA Cell Biol. 26, 811–826. 10.1089/dna.2007.0664 [DOI] [PubMed] [Google Scholar]
- Steinberg S, Gudmundsdottir S, Sveinbjornsson G, Suvisaari J, Paunio T, Torniainen-Holm M, Frigge ML, Jonsdottir GA, Huttenlocher J, Arnarsdottir S, Ingimarsson O, Haraldsson M, Tyrfingsson T, Thorgeirsson TE, Kong A, Norddahl GL, Gudbjartsson DF, Sigurdsson E, Stefansson H, Stefansson K, 2017. Truncating mutations in RBM12 are associated with psychosis. Nat. Genet 49, 1251–1254. 10.1038/ng.3894 [DOI] [PubMed] [Google Scholar]
- Stone JM, 2011. Glutamatergic antipsychotic drugs: a new dawn in the treatment of schizophrenia? Ther. Adv. Psychopharmacol 1, 5–18. 10.1177/2045125311400779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone JM, Erlandsson K, Arstad E, Squassante L, Teneggi V, Bressan RA, Krystal JH, Ell PJ, Pilowsky LS, 2008. Relationship between ketamine-induced psychotic symptoms and NMDA receptor occupancy - A [123I]CNS-1261 SPET study. Psychopharmacology (Berl). 197, 401–408. 10.1007/s00213-007-1047-x [DOI] [PubMed] [Google Scholar]
- Thornberg SA, Saklad SR, 1996. A review of NMDA receptors and the phencyclidine model of schizophrenia. Pharmacotherapy 16, 82–93. [PubMed] [Google Scholar]
- Timms AE, Dorschner MO, Wechsler J, Choi KY, Kirkwood R, Girirajan S, Baker C, Eichler EE, Korvatska O, Roche KW, Horwitz MS, Tsuang DW, 2013. Support for the N-methyl-d-aspartate receptor hypofunction hypothesis of schizophrenia from exome sequencing in multiplex families. JAMA Psychiatry 70, 582–590. 10.1001/jamapsychiatry.2013.1195 [DOI] [PubMed] [Google Scholar]
- Walker E, Kestler L, Bollini A, Hochman KM, 2004. Schizophrenia: Etiology and Course. Annu. Rev. Psychol 55, 401–430. 10.1146/annurev.psych.55.090902.141950 [DOI] [PubMed] [Google Scholar]
- Walker ER, McGee RE, Druss BG, 2015. Mortality in Mental Disorders and Global Disease Burden Implications. JAMA Psychiatry 72, 334. 10.1001/jamapsychiatry.2014.2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Charych EI, Pulito VL, Lee JB, Graziane NM, Crozier RA, Revilla-Sanchez R, Kelly MP, Dunlop AJ, Murdoch H, Taylor N, Xie Y, Pausch M, Hayashi-Takagi A, Ishizuka K, Seshadri S, Bates B, Kariya K, Sawa A, Weinberg RJ, Moss SJ, Houslay MD, Yan Z, Brandon NJ, 2011. The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol. Psychiatry 16, 1006–1023. 10.1038/mp.2010.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheldon LM, Haines BP, Rajappa R, Mason I, Rigby PW, Heath JK, 2010. Critical Role of FLRT1 Phosphorylation in the Interdependent Regulation of FLRT1 Function and FGF Receptor Signalling. PLoS One 5, e10264. 10.1371/journal.pone.0010264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff D, Endele S, Azzarello-Burri S, Hoyer J, Zweier M, Schanze I, Schmitt B, Rauch A, Reis A, Zweier C, 2012. In-frame deletion and missense mutations of the c-terminal helicase domain of SMARCA2 in three patients with nicolaides-baraitser syndrome. Mol. Syndromol 2, 237–244. 10.1159/000337323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Yan H, Wang L, Li J, Tan L, Deng W, Chen Q, Yang G, Zhang F, Lu T, Yang J, Li K, Lv L, Tan Q, Zhang H, Xiao X, Li M, Ma X, Yang F, Li L, Wang C, Li T, Zhang D, Yue W, Chinese Antipsychotics Pharmacogenomics Consortium, 2018. Five novel loci associated with antipsychotic treatment response in patients with schizophrenia: a genome-wide association study. The Lancet Psychiatry 5, 327–338. 10.1016/S2215-0366(18)30049-X [DOI] [PubMed] [Google Scholar]
- Yuen RKC, Merico D, Bookman M, Howe JL, Thiruvahindrapuram B, Patel RV, Whitney J, Deflaux N, Bingham J, Wang Z, Pellecchia G, Buchanan JA, Walker S, Marshall CR, Uddin M, Zarrei M, Deneault E, D’Abate L, Chan AJS, Koyanagi S, Paton T, Pereira SL, Hoang N, Engchuan W, Higginbotham EJ, Ho K, Lamoureux S, Li W, MacDonald JR, Nalpathamkalam T, Sung WWL, Tsoi FJ, Wei J, Xu L, Tasse AM, Kirby E, Van Etten W, Twigger S, Roberts W, Drmic I, Jilderda S, Modi BM, Kellam B, Szego M, Cytrynbaum C, Weksberg R, Zwaigenbaum L, Woodbury-Smith M, Brian J, Senman L, Iaboni A, Doyle-Thomas K, Thompson A, Chrysler C, Leef J, Savion-Lemieux T, Smith IM, Liu X, Nicolson R, Seifer V, Fedele A, Cook EH, Dager S, Estes A, Gallagher L, Malow BA, Parr JR, Spence SJ, Vorstman J, Frey BJ, Robinson JT, Strug LJ, Fernandez BA, Elsabbagh M, Carter MT, Hallmayer J, Knoppers BM, Anagnostou E, Szatmari P, Ring RH, Glazer D, Pletcher MT, Scherer SW, 2017. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci 20, 602–611. 10.1038/nn.4524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zai G, Robbins TW, Sahakian BJ, Kennedy JL, 2017. A review of molecular genetic studies of neurocognitive deficits in schizophrenia. Neurosci. Biobehav. Rev 72, 50–67. 10.1016/J.NEUBIOREV.2016.10.024 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hu ZZ, Zhang L, Hu ZZ, Liu H, Liu Z, Du J, Zhao J, Zhou L, Xia K, Tang B, Shen L, 2016. Identification of RELN variation p.Thr3192Ser in a Chinese family with schizophrenia. Sci. Rep 6, 24327. 10.1038/srep24327 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.