Abstract
Non-coding centromeres, which dictate kinetochore formation for proper chromosome segregation, are extremely divergent in DNA sequences across species but are under active transcription carried out by RNA polymerase (RNAP) II. The RNAP II-mediated centromeric transcription has been shown to facilitate the deposition of the centromere protein A (CENP-A) to centromeres, establishing a conserved and critical role of centromeric transcription in centromere maintenance. Our recent work revealed another role of centromeric transcription in chromosome segregation: maintaining centromeric cohesion during mitosis. Interestingly, this role appears to be fulfilled through ongoing centromeric transcription rather than centromeric transcripts. In addition, we found that centromeric transcription may not require some of the traditional transcription initiation factors, suggestive of “uniqueness” in its regulation. In this review, we discuss the novel role and regulation of centromeric transcription as well as the potential underlying mechanisms.
Keywords: centromere, centromeric cohesion, centromeric transcription, chromosome segregation, mitosis
INTRODUCTION
The non-coding centromere, a specific region of a chromosome, varies in size among eukaryotes. It can be as short as ≈120 base pairs (bp) in budding yeast and as long as several mega bp in human.[1] Centromeric DNA sequences are also extremely divergent and are not conserved across species. In higher eukaryotes, the centromere is usually composed of highly repetitive DNA sequences. For example, the human centromere contains repetitive DNA sequences termed α-satellite DNA of 171 bp in length. These α-satellite repeats are assembled into higher order repeats, which can be further organized into satellite arrays.[1] In spite of the diversity and complexity, all the centromeres in eukaryotes play a critical and conserved role in chromosome segregation during cell cycle: dictate kinetochore formation. It seems that the size and DNA sequence of the centromere are not important for its functions; instead, the centromere-specific histone H3 variant CENP-A is required. CENP-A at centromeres recruits other centromere proteins to initiate kinetochore formation, which is essential for faithful chromosome segregation during mitosis.[2] Unlike canonical histones, which are mainly incorporated into chromatin in a DNA duplication-dependent manner during S phase, CENP-A is deposited into centromeres independently of DNA replication during anaphase or telophase/early G1.[3,4] CENP-A deposition into centromeres have been shown to be regulated by polymerase (RNAP)II-catalyzed transcription or Plk1.[5,6] Thus, centromeric transcription is crucial for centromere functions and chromosome segregation.
During mitosis, RNAPII is released from chromosomes and transcription was thought to be silenced.[7,8] However, increasing evidence suggests that transcription is still undergoing albeit in a reduced manner.[9,10] Surprisingly, RNAPII is remained on centromeres and it is actively transcribing centromeres.[11–14] We have demonstrated that centromeric transcription promotes centromeric cohesion during mitosis in human cells.[11,13] This novel function seems to be dependent on ongoing centromeric transcription rather than centromeric transcripts. Moreover, we also found that some of general transcriptional initiation factors may be dispensable for centromeric transcription, suggesting that the regulation of centromeric transcription is distinct from that of gene transcription.[11] In this review, we discuss the novel role and regulation of centromeric transcription and the potential underlying mechanisms.
ONGOING CENTROMERIC TRANSCRIPTION MAINTAINS CENTROMERIC COHESION
A major function of centromeric transcription during interphase is to facilitate CENP-A deposition into centromeric chromatin.[15] What is the function of centromeric transcription during mitosis? When cells enter mitosis, the majority of RNA polymerase (RNAP)II dissociates from chromatin,[7] thus largely decreasing global transcription levels albeit low-level transcription is still detected.[9,10] Interestingly, actively elongating RNAPII (RNAPII phospho-Ser2) is retained on mitotic kinetochores,[12–14] suggesting that RNAPII is still actively transcribing centromeres. Addition of α-amanitin into mitotic cells, a specific RNAPII inhibitor, induced an increase in anaphase lagging chromosomes, and weakened centromeric cohesion.[12,13] Thus, centromeric transcription seems to be important for proper chromosome segregation. However, the efficacy of α-amanitin on centromeric transcription in those studies was not rigorously assessed,[12,13] challenging the notion that the observed phenotypes are indeed the consequences of α-amanitin inhibited centromeric transcription. In addition, using triptolide that inhibits the activity of the transcription initiation factor TFIIH, another study concluded that mitotic transcription is not important for mitotic progression.[16] These disagreeing studies provoked us to re-look into the approaches that were utilized to assess centromeric transcription. As abundant centromeric transcripts are stored in human nucleoli,[17] only measuring the amount of total α-satellite RNAs seems impractical to monitor a rapid change in centromeric transcription within a short period of time. Therefore, we developed an approach to monitor ongoing centromeric transcription by measuring the amount of 5′-ethynyl uridine (EU)-labeled nascent α-satellite RNAs.[11] By doing so, the efficacy of a given transcriptional inhibitor on the suppression of centromeric transcription can be determined. Consistent with the previous studies, we found that α-amanitin treatment for a few hours barely affected the amount of total α-satellite RNAs, but largely decreased the amount of nascent total α-satellite RNAs, indicating that α-amanitin is an efficient inhibitor to centromeric transcription.[11] In contrast, triptolide treatment for a few hours did not decrease the amounts of both total and nascent α-satellite RNAs although it largely inhibited gene transcription, suggesting that triptolide is not an effective inhibitor to centromeric transcription. These interesting results well explain why the treatment of α-amanitin, not triptolide, induced mitotic defects including lagging chromosomes and impaired centromeric cohesion in human cells in the previous studies.[12,13,16] We therefore propose that centromeric transcription maintains centromeric cohesion during mitosis. This notion is further strengthened by our evidence that ectopically increased centromeric transcription enhanced centromeric cohesion.[11]
Of note, our results indicated that the robustness of centromeric cohesion is positively corelated with the level of ongoing centromeric transcription, much less with the amount of total centromeric transcripts. Thus, ongoing centromeric transcription maintains centromeric cohesion during mitosis. As ongoing centromeric transcription also plays an important role in CENP-A deposition to centromeres in interphase,[6] constantly maintaining ongoing transcription at centromeres possibly throughout the cell cycle could be critical for centromere functions. In spite of the importance of ongoing centromeric transcription, centromeric transcripts may also be involved in the regulation of centromere functions as various centromere proteins have been shown to physically interact with centromeric transcripts.[15] Potential functions and regulation of centromeric transcripts will be discussed in the next section.
FUNCTIONS OF CENTROMERIC RNA TRANSCRIPTS
Functions of centromeric RNA transcripts are not well understood mainly due to the lack of a comprehensive characterization of these transcripts. Centromeric RNAs have been demonstrated to localize at distinct places within cells: centromeres/kinetochores, nucleoli, and independent foci. These different localization patterns may suggest diverse functions of centromeric RNAs played within cells. Centromeric RNAs were previously detected on centromeres/kinetochores.[17–20] Interestingly, they can exist either in trans in Xenopus egg extracts or in cis in human RPE-1 cells.[18,21] Functionally, the centromere-localized centromeric RNAs may serve as scaffolds to facilitate the recruitment or maintenance of some of centromere proteins, thus regulating centromere functions. For example, Centromere-encoded RNAs were suggested to be integral components in Mazie.[22] Moreover, Aurora B and Sgo1 were shown to bind centromeric RNAs either in vitro or in cells.[13,21,23–25] These physical interactions might also regulate the recruitment of Aurora B and Sgo1 to centromeres or modulate Aurora B kinase activity although further evidence is needed. More recently, a study found that human centromeric RNAs appear as multiple foci, which do not associate with centromeres.[26] The levels of these RNA foci vary over the cell cycle and across cell lines. However, the biochemical nature of these α-satellite RNA foci and their functions remain to be addressed. Centromeric RNAs were also previously shown to localize within the nucleolus.[17,27] Still, these nucleolar centromeric RNAs are mysterious in their functionality and regulation. Considering the conserved and essential roles of nucleoli in ribosome biogenesis and stress response, the centromeric RNAs might function to regulate these processes.
Why had only one localization pattern of centromeric RNAs been detected by each of these research groups? One possible reason is that different RNA-FISH probes used in these studies might differentially recognize distinct pools of centromeric RNAs. This is just like “Blind men and an elephant.” Therefore, a comprehensive characterization on centromeric transcripts, such as types and length, would be needed to address these discrepancies as well as understand the functionality of these centromeric RNAs. Recent accomplishment of human centromere assembly and long-read RNA-Seq may help identify a variety of types of centromeric transcripts.[28]
UNDERLYING MECHANISMS OF CENTROMERIC TRANSCRIPTION-MAINTAINED CENTROMERIC COHESION
Our previous results suggested that centromeric transcription may promote centromeric cohesion through Sgo1 in human cells.[11,13] Sgo1 directly binds cohesin at inner centromeres to protect centromeric cohesion during mitosis.[29–32] In order to do so, Sgo1 firstly binds phospho-T120 H2A-containing nucleosomes and is thus recruited to the kinetochore-proximal region at early mitosis.[30] Then RNA polymerase (RNAP)II-mediated transcription facilitates Sgo1 relocation to inner centromeres, where Sgo1 binds to cohesin[13] (Figure 1). This idea is supported by two pieces of evidence.[13] Firstly, treatment of transcriptional inhibitors on mitotic cells stalled Sgo1 on the kinetochore-proximal region; and as a result, centromeric cohesion was significantly weakened. Secondly, Sgo1 physically interacted with RNAPII both in vitro and in vivo. Thus, RNAPII could carry Sgo1 as a cargo to gradually approach inner centromeres, where it binds with cohesin to protect centromeric cohesion[33] (Figure 1). This notion may explain why ongoing transcription, not transcripts themselves, plays an important role in maintaining centromeric cohesion in our study.[11] However, Sgo1 physically binding centromeric transcripts is also indicative of a potential role of centromeric transcripts in regulating Sgo1.[13] Functionally, the Sgo1-RNA interaction might facilitate the Sgo1-nuclosome binding, thus regulating Sgo1 recruitment to the kinetochore-proximal region. We had previously demonstrated that RNA directly interacts with the same region of Sgo1 that also binds to phospho-T120 H2A-containing nucleosomes.[13] Therefore, in order to understand how centromeric RNAs regulate Sgo1, rigorous mutagenesis on Sgo1 to separate these two binding activities or knockdown of centromeric RNAs using siRNAs or antisense oligos are needed.
FIGURE 1.
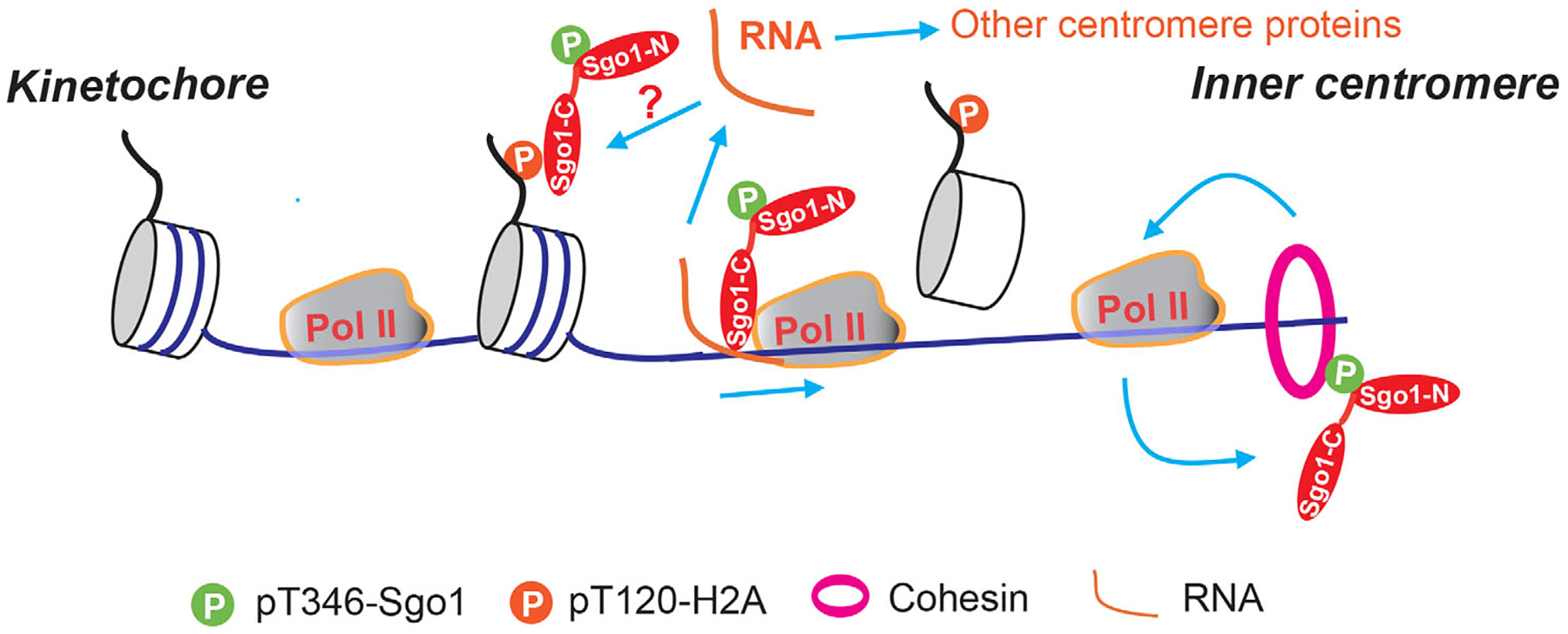
Working model of centromeric transcription installing Sgo1 onto inner centromeres. At early mitosis, Sgo1 is recruited to the kinetochore-proximal region through binding to phospho-T120 H2A-containing nucleosomes and then relocated by RNAPII-dependent transcription to inner centromeres, where it binds to cohesin to protect centromeric cohesion. Cohesin conversely promotes centromeric transcription, which may further facilitate Sgo1 relocation.
The findings from a more recent study suggested that the relationships among Sgo1, cohesion and transcription seem more complicated.[14] In that study, increased cohesion by Wapl deletion retained more elongating RNAPII on whole chromosomes including centromeres in miotic human cells, indicative of a positive role of cohesin in regulating transcription during mitosis. Considering the established roles of cohesin in the regulation of gene expression during interphase,[34] this is not surprising. Thus, taking all the findings together, a positive feedback loop may be formed among cohesin, transcription, and Sgo1 to regulate centromeric cohesion during mitosis (Figure 1). At early mitosis, RNAPII-mediated transcription relocates Sgo1 to inner centromeres to preserve cohesin at centromeres and the centromeric cohesin may in turn promote transcription to further facilitate Sgo1 relocation. However, it is equally possible that transcription might also directly manipulate cohesin to regulate centromeric cohesion. In future, exploring how Sgo1 and cohesin are distributed along centromeres using ChIP-Seq in response to transcriptional inhibition would offer some insights into the relationships.
THE “UNIQUENESS” OF CENTROMERIC TRANSCRIPTION
The regulation of centromeric transcription is poorly understood, especially in human cells that contain tens of thousands of repetitive DNA sequences in their centromeres. This is partially due to the lack of the assembly of complete centromere sequences. Recent advances on centromere assembly in human cells will likely provide a solid foundation to comprehensively understand centromeric transcription.[28] Our recent results from transcriptional inhibitors suggested a unique regulation of centromeric transcription, which is distinct from that of canonical gene transcription.[11] In our studies, treatment of α-amanitin and flavopiridol, both of which inhibit transcription elongation, largely suppressed ongoing centromeric transcription in human cells; whereas treatment of triptolide and THZ1, which both inhibit transcription initiation, did not decrease ongoing centromeric transcription. These results strongly suggest that centromeric transcription may not require some, if not all, of transcriptional initial factors. This notion is further supported by the findings from us and others that elongating RNAPII (phospho-Ser2) rather than initiating RNAPII (phospho-Ser5) is present at centromeres during mitosis.[11–14] Of note, several previous studies using transcription inhibitors appeared to yield distinct impacts on centromeric transcription.[14,26] However, transcriptional activity (ongoing transcription) at centromeres were not rigorously examined in those studies as either only total centromeric RNAs or centromere localized RNAs (might not be generated from centromeres) were interrogated. Therefore, it is not clear whether ongoing centromeric transcription was indeed repressed. In addition, differential application durations and doses of transcription inhibitors could also contribute to the observed difference. Then, why are some of transcriptional factors not required for centromeric transcription? The answers might lie in the specific structures of centromeric DNA. Although they are very divergent across species, centromeric DNA sequences contain a number of dyad symmetries.[35] Dyad symmetries tend to make centromeres adopt non-B form DNA conformations such as stem loop and cruciform, which might allow centromeric transcription to be less dependent on some transcriptional initiation factors (Figure 2). In addition, R-loops of an RNA-DNA duplex have been found to be present at centromeres,[36] which might also promote transcriptional activation at centromeres.[37,38] In future, it will be interesting to screen all the known transcriptional initiation factors for their requirement for centromeric transcription. Such a study would provide a more comprehensive view of the “unique” centromeric transcription. As transcription inhibitors had similar effects on transcription at both mitotic and interphase centromeres in our studies, it is very likely that centromeric transcription is subject to similar regulation in both mitosis and interphase (Figure 2). However, it is also possible that centromeric transcription initiates in interphase and then maintains in mitosis. Notably, unlike human cells, triptolide that inhibits TFIIH largely suppressed ongoing centromeric transcription of X chromosomes in drosophila S2 cells,[6,11] suggesting that some of drosophila centromeres are subject to transcriptional initiation regulation. Thus, centromeric DNA structures might vary a lot across species. In future, it would also be interesting to survey for the dependency of centromeric transcription on transcriptional initiation among distinct organisms, which may help us further understand the evolution of centromeric DNA structures.
FIGURE 2.
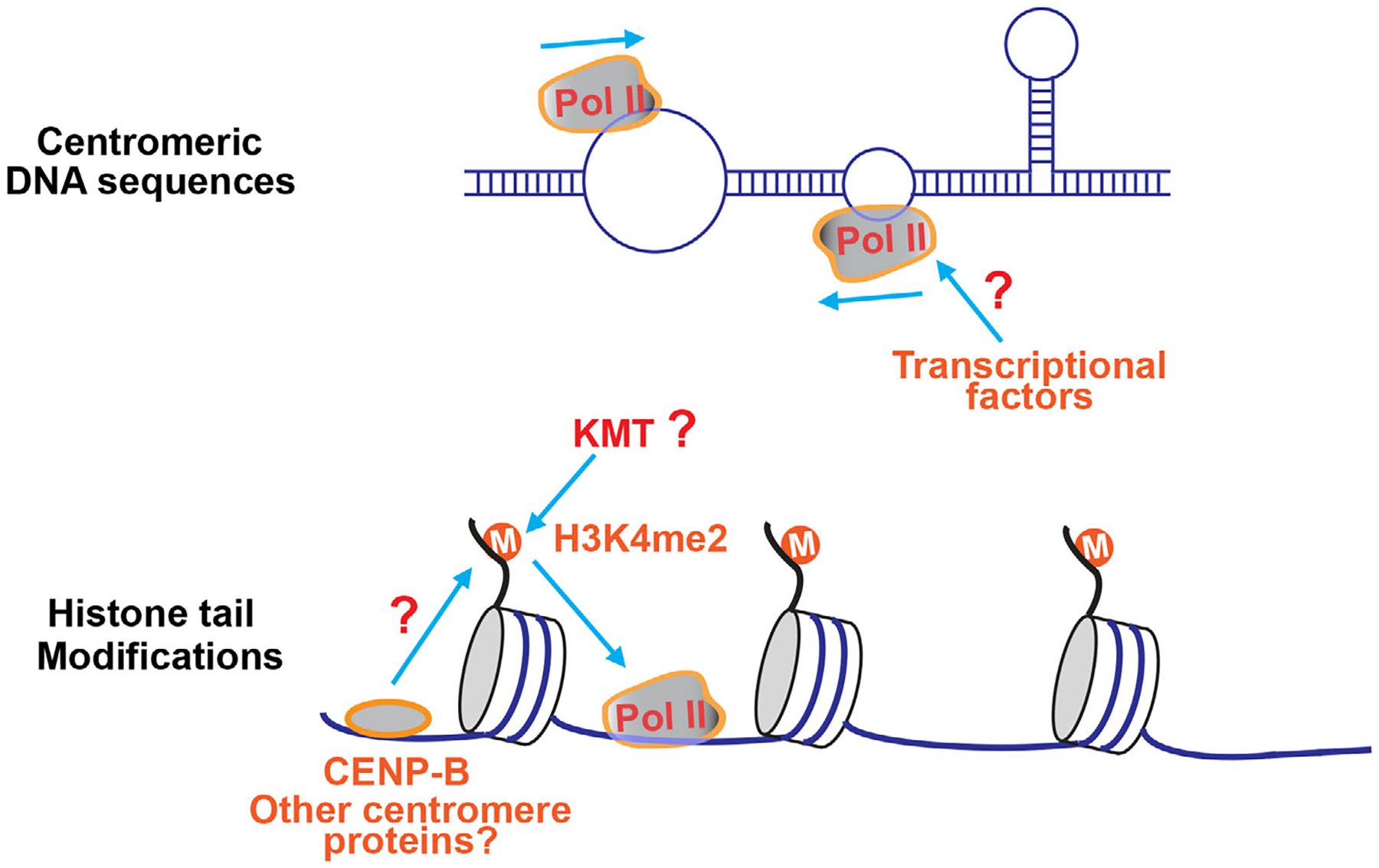
Centromeric transcription may be regulated at the levels of DNA sequences and histone tail modifications. Top panel: Centromeres tend to adopt non-B form DNA conformations such as stem loop, which might allow centromeric transcription to be less dependent on some transcriptional initiation factors. Bottom panel: The histone modification H3K4me2 renders the centromeric chromatin “transcription-permissive,” which may be required for centromeric transcription. Centromere proteins including CENP-B might be involved in the regulation of H3K4me2. The lysine methyltransferase(s) (KMT) responsible for H3K4me2 at centromeres is yet to be identified.
In addition to the DNA sequence-based regulation, centromeric transcription is also subject to epigenetic regulation by histone tail modifications (Figure 2). Centromeric chromatin is marked with histone H3 lysine 4 and lysine 36 methylations (H3K4me1, H3K4me2, H3K36me2, and H3K36me3).[39–41] These transcriptionally active histone modifications render the centromeric chromatin “transcription-permissive,” but how they regulate centromeric transcription is still vague although they seem to be important for transcription at human artificial centromeres.[42] We recently demonstrated that expression of the CENP-B DNA-binding domain significantly increased H3K4me2 levels at centromeres, not at gene regions[11]; accordingly, increased centromeric transcription was also observed. These findings provide strong evidence to support the positive role of H3K4me2 in regulating centromeric transcription and also suggest that CENP-B might be involved in the regulation of H3K4me2 at centromeres (Figure 2). In addition to H3K4 methylation and CENP-B, other histone modifications and regulatory factors were recently found to be involved in the regulation of centromeric transcription.[43,44] In future, it would be intriguing to identify more of such histone modifications or regulatory factors that are important for centromeric transcription and then determine how they work to regulate centromeric transcription.
CONCLUSION AND OUTLOOK
It has been established that centromeric transcription promotes CENP-A deposition into centromeric chromatin, thus maintaining centromere identity. We have recently discovered a novel role of centromeric transcription in centromere functions in human cells: maintaining centromeric cohesion. Interestingly, these two processes appear to require ongoing centromeric transcription. Based on the current findings, we propose the following working model to explain how centromeric transcription maintains centromeric cohesion: at early mitosis, Sgo1 is carried by RNAPII to approach inner centromeres, where it binds cohesin to preserve centromeric cohesion (Figure 1). The retained cohesin may conversely promote centromeric transcription, which further facilitates Sgo1 relocation. This potential positive feedback loop might thus provide a critical mechanism to enable timely and rapid protection of centromeric cohesion at early mitosis. In further, it would be important to understand in great details how this positive loop functions to protect centromeric cohesion. Comprehensively characterizing the centromeric RNA transcripts is necessary for further understanding of their functions. In this regard, long-read RNA-Seq and the recent accomplishment of human centromere assembly could offer a great help. In addition, mass spectrometry can be applied to identify the binding proteins of centromeric RNAs, thus helping understand the functions of centromeric RNAs. Finally, as suggested by our results, the centromere may not require some of transcriptional initiation factors for its transcription in human cells. This “uniqueness” might render the regulation of centromeric transcription not as restrictive as that of gene transcription, thus ensuring ongoing transcription to constantly occur at centromeres during the cell cycle to fulfill its duties: deposition of CENP-A into centromeric chromatin and maintenance of centromeric cohesion. As stated above, the specific non-B type DNA structures that centromeres can form probably determine the “unique” regulation of centromeric transcription. In future, identifying the transcriptional initiation factors that are not required for centromeric transcription and reconstructing centromeric transcription in vitro would provide a comprehensive understanding of the “unique” centromeric transcription. As aberrant overexpression of centromeric RNAs were found in human cancer tissues,[45] the understanding of centromeric transcription regulation could offer new therapeutic strategies to combat human malignancies.
ACKNOWLEDGMENT
Research in the Liu laboratory is supported by grants from Tulane Startup and NIH (R01GM124018 and R01GM141123).
Footnotes
CONFLICT OF INTEREST
The authors declare no competing financial interests.
REFERENCES
- 1.Thakur J, Packiaraj J, Henikoff S, (2021). Sequence, chromatin and evolution of satellite DNA. International Journal of Molecular Sciences, 22(9), 4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mckinley KL, & Cheeseman IM (2016). The molecular basis for centromere identity and function. Nature Reviews Molecular Cell Biology, 17(1), 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen LET, Black BE, Foltz DR, & Cleveland DW (2007). Propagation of centromeric chromatin requires exit from mitosis. Journal of Cell Biology, 176(6), 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuh M, Lehner CF, & Heidmann S (2007). Incorporation of Drosophila CID/CENP-A and CENP-C into centromeres during early embryonic anaphase. Current Biology, 17(3), 237–243. [DOI] [PubMed] [Google Scholar]
- 5.Swartz SZ, Mckay LS, Su K-C, Bury L, Padeganeh A, Maddox PS, Knouse KA, & Cheeseman IM (2019). Quiescent cells actively replenish CENP-A nucleosomes to maintain centromere identity and proliferative potential. Developmental Cell, 51(1), 35–48.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bobkov GOM, Gilbert N, & Heun P (2018). Centromere transcription allows CENP-A to transit from chromatin association to stable incorporation. Journal of Cell Biology, 217(6), 1957–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teves SS, An L, Bhargava-Shah A, Xie L, Darzacq X, & Tjian R (2018). A stable mode of bookmarking by TBP recruits RNA polymerase II to mitotic chromosomes. Elife, 7, e35621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsons GG, & Spencer CA (1997). Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Molecular and Cellular Biology, 17(10), 5791–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palozola KC, Donahue G, Liu H, Grant GR, Becker JS, Cote A, Yu H, Raj A, & Zaret KS (2017). Mitotic transcription and waves of gene reactivation during mitotic exit. Science, 358(6359), 119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palozola KC, Liu H, Nicetto D, & Zaret KS (2018). Low-level, global transcription during mitosis and dynamic gene reactivation during mitotic exit. Cold Spring Harbor Symposia on Quantitative Biology, 82, 197–205. [DOI] [PubMed] [Google Scholar]
- 11.Chen Y, Zhang Q, Teng Z, & Liu H (2021). Centromeric transcription maintains centromeric cohesion in human cells. Journal of Cell Biology, 220(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan FL, Marshall OJ, Saffery R, Won Kim B, Earle E, Choo KHA, & Wong LH (2012). Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proceedings of the National Academy of Sciences of the United States of America, 109(6), 1979–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Qu Q, Warrington R, Rice A, Cheng N, & Yu H (2015). Mitotic transcription installs Sgo1 at centromeres to coordinate chromosome segregation. Molecular Cell, 59(3), 426–436. [DOI] [PubMed] [Google Scholar]
- 14.Perea-Resa C, Bury L, Cheeseman IM, Blower MD (2020). Cohesin removal reprograms gene expression upon mitotic entry. Molecular Cell, 78(1), 127–140.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Talbert PB, & Henikoff S (2018). Transcribing centromeres: Non-coding RNAs and kinetochore assembly. Trends in Genetics, 34(8), 587–599. [DOI] [PubMed] [Google Scholar]
- 16.Novais-Cruz M, Alba Abad M, van IJcken WF, Galjart N, Jeyaprakash AA, Maiato H, Ferras C (2018). Mitotic progression, arrest, exit or death relies on centromere structural integrity, rather than de novo transcription. Elife, 7, e36898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, & Choo KHA (2007). Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Research, 17(8), 1146–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mcnulty SM, Sullivan LL, & Sullivan BA (2017). Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Developmental Cell, 42(3), 226–240.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li F, Sonbuchner L, Kyes SA, Epp C, & Deitsch KW (2008). Nuclear non-coding RNAs are transcribed from the centromeres of Plasmodium falciparum and are associated with centromeric chromatin. Journal of Biological Chemistry, 283(9), 5692–5698. [DOI] [PubMed] [Google Scholar]
- 20.Carone DM, Longo MS, Ferreri GC, Hall L, Harris M, Shook N, Bulazel KV, Carone BR, Obergfell C, O’neill MJ, & O’neill RJ (2009). A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma, 118(1), 113–125. [DOI] [PubMed] [Google Scholar]
- 21.Blower MD (2016). Centromeric transcription regulates Aurora-B localization and activation. Cell reports, 15(8), 1624–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Topp CN, Zhong CX, & Dawe RK (2004). Centromere-encoded RNAs are integral components of the maize kinetochore. Proceedings of the National Academy of Sciences of the United States of America, 101(45), 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jambhekar A, Emerman AB, Schweidenback CTH, & Blower MD (2014). RNA stimulates Aurora B kinase activity during mitosis. PLoS ONE, 9(6), e100748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferri F, Bouzinba-Segard H, Velasco G, Hubé F, & Francastel C (2009). Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Research, 37(15), 5071–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ideue T, Cho Y, Nishimura K, & Tani T (2014). Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes to Cells, 19(6), 528–538. [DOI] [PubMed] [Google Scholar]
- 26.Bury L, Moodie B, Ly J, Mckay LS, Miga KH, Cheeseman IM (2020). Alpha-satellite RNA transcripts are repressed by centromere-nucleolus associations. Elife, 9, e59770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koo D-H, Zhao H, & Jiang J (2016). Chromatin-associated transcripts of tandemly repetitive DNA sequences revealed by RNA-FISH. Chromosome Research, 24(4), 467–480. [DOI] [PubMed] [Google Scholar]
- 28.Logsdon GA, Vollger MR, Hsieh P, Mao Y, Liskovykh MA, Koren S, Nurk S, Mercuri L, Dishuck PC, Rhie A, De Lima LG, Dvorkina T, Porubsky D, Harvey WT, Mikheenko A, Bzikadze AV, Kremitzki M, Graves-Lindsay TA, Jain C, … Eichler EE (2021). The structure, function and evolution of a complete human chromosome 8. Nature, 593(7857), 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Rankin S, & Yu H (2013). Phosphorylation-enabled binding of SGO1-PP2A to cohesin protects sororin and centromeric cohesion during mitosis. Nature Cell Biology, 15(1), 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Jia L, & Yu H (2013). Phospho-H2A and cohesin specify distinct tension-regulated Sgo1 pools at kinetochores and inner centromeres. Current Biology, 23(19), 1927–1933. [DOI] [PubMed] [Google Scholar]
- 31.Hara K, Zheng Ge, Qu Q, Liu H, Ouyang Z, Chen Z, Tomchick DR, & Yu H (2014). Structure of cohesin subcomplex pinpoints direct shugoshin-Wapl antagonism in centromeric cohesion. Nature Structural & Molecular Biology, 21(10), 864–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q, & Liu H (2020). Functioning mechanisms of Shugoshin-1 in centromeric cohesion during mitosis. Essays in Biochemistry, 64(2), 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu H (2016). Insights into centromeric transcription in mitosis. Transcription, 7(1), 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perea-Resa C, Wattendorf L, Marzouk S, & Blower MD (2021). Cohesin: behind dynamic genome topology and gene expression reprogramming. Trends in Cell Biology, 31(9), 760–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasinathan S, & Henikoff S (2018). Non-B-form DNA is enriched at centromeres. Molecular Biology and Evolution, 35(4), 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabeche L, Nguyen HD, Buisson R, & Zou L (2018). A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science, 359(6371), 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boque-Sastre R, Soler M, Oliveira-Mateos C, Portela A, Moutinho C, Sayols S, Villanueva A, Esteller M, & Guil S (2015). Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proceedings of the National Academy of Sciences of the United States of America, 112(18), 5785–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee C-Y, Mcnerney C, Ma K, Zhao W, Wang A, & Myong S (2020). R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation. Nature Communications, 11(1), 3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergmann JH, Rodríguez MG, Martins NMC, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LET, & Earnshaw WC (2011). Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO Journal, 30(2), 328–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sullivan BA, & Karpen GH (2004). Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nature Structural & Molecular Biology, 11(11), 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hall LE, Mitchell SE, & O’neill RJ (2012). Pericentric and centromeric transcription: A perfect balance required. Chromosome Research, 20(5), 535–546. [DOI] [PubMed] [Google Scholar]
- 42.Molina O, Vargiu G, Abad MA, Zhiteneva A, Jeyaprakash AA, Masumoto H, Kouprina N, Larionov V, & Earnshaw WC (2016). Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nature Communications, 7:13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ishikura S, Nakabayashi K, Nagai M, Tsunoda T, & Shirasawa S (2020). ZFAT binds to centromeres to control noncoding RNA transcription through the KAT2B-H4K8ac-BRD4 axis. Nucleic Acids Research, 48(19), 10848–10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herlihy CP, Hahn S, Hermance NM, Crowley EA, & Manning AL (2021). Suv420 enrichment at the centromere limits Aurora B localization and function. Journal of Cell Science, 134(15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, Contino G, Deshpande V, Iafrate AJ, Letovsky S, Rivera MN, Bardeesy N, Maheswaran S, & Haber DA (2011). Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science, 331(6017), 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]


