Abstract
Organoids are three-dimensional culture systems generated from embryonic stem cells, induced pluripotent stem cells, and adult stem cells. They are capable of cell proliferation, differentiation, and self-renewal. Upon stimulation by signal factors and/or growth factors, organoids self-assemble to replicate the morphological and structural characteristics of the corresponding organs. They provide an extraordinary platform for investigating organ development and mimicking pathological processes. Organoid biobanks derived from a wide range of carcinomas have been established to represent different lesions or stages of clinical tumors. Importantly, genomic and transcriptomic analyses have confirmed maintenance of intra- and interpatient heterogeneities in organoids. Therefore, this technology has the potential to revolutionize drug screening and personalized medicine. In this review, we summarized the characteristics and applications of organoids in cancer research by the establishment of organoid biobanks directly from tumor organoids or from genetically modified non-cancerous organoids. We also analyzed the current state of organoid applications in drug screening and personalized medicine.
Keywords: Organoids, cancer research, heterogeneity, personalized medicine, clinical cancer therapy
Introduction
Cancer is the leading cause of death among noncommunicable diseases1 and the survivals of several cancers remain extremely low2. One of the reasons is a shortage of suitable preclinical tumor models, which hinders the development of anti-cancer drugs and precision treatments3. Existing preclinical models rely mainly on cancer cell lines and patient-derived xenografts (PDXs). Cancer cell lines, the earliest and most widely used tool in tumor research, are characterized by high manipulability, short culture cycle, and high throughput4. However, they lack the phenotype and genetic heterogeneity of the original tumor5. In comparison, PDX models preserve the structures, cell compositions, molecular characteristics, and complexities of the original tumors6,7, but are costly and time-consuming. Moreover, given that they are usually derived from a small number of tumor cells, they may not capture the original tumor’s heterogeneity8. Therefore, in recent years, cancer research has focused on new three-dimensional (3D) culture models. In 2009, Sato et al.9 used Lgr5+ stem cells from the bottom of small intestinal crypts to generate villus-like epithelial domains by culturing the cells in Matrigel with niche factors R-spondin, epidermal growth factor (EGF), noggin, and Wnt. This strategy yielded an organ-like structure, termed “organoid”9 and defined the starting point for organoid culturing of multiple mouse and human epithelia, as well as tumor-derived organoids.
Organoids are defined as a collection of organ-specific cell types produced from organ progenitors or stem cells that self-organize into the corresponding organ via 3D culture10. Compared to other preclinical models, they are superior in copying the phenotypes of the organs and reproducing the genetic heterogeneities of the original tumors11. In the 5 years following their discoveries, organoid models of the colon and small intestine12,13, retina14, brain14, liver15, stomach16–18 and mammary glands19 were successfully cultured. Gao et al.20 introduced organoid models for cancer research in 2014 by establishing systems with metastatic tissues and circulating tumor cells harvested from patients with advanced prostate cancers. Next, organoid models of colon carcinomas21, prostate carcinomas22, pancreatic carcinomas23,24, renal carcinomas25, gastric carcinomas26, breast carcinomas27, ovarian carcinomas28, primary liver carcinomas29, and bladder carcinomas30 were established. “Growing organoids” was selected as the “Breakthrough of the Year” by Science in 201331 and “Method of the Year” by Nature Methods in 201732. In 2018, Vlachogiannis et al.33 demonstrated the potential of organoids to predict patient clinical outcomes, suggesting their implementation in personalized medicine. Taken together, organoids provide a reliable method for preclinical disease modeling and drug screening, providing the bases for gene and stem cell therapies for cancer.
The development of organoid culture methods
Over the past decades, stem cells and/or progenitor cells have been used to reconstruct organs because of their capacities for self-renewal and differentiation into tissue-specific lineages10,34. Moreover, regenerative medicine has highlighted the ability of stem cells to differentiate into ≥ 1 cell types to repair damaged organs10,35,36. Eiraku et al.37,38 demonstrated the ability of cortical tissue to form complex optical cup structures in vitro, owing to the self-organizing property of pluripotent stem cells. Sato et al.9 showed that adult intestinal stem cells formed the main structures of intestinal organs, and the organoids could be propagated in culture for extended periods of time.
In recent years, organoids have been generated from tissue samples containing embryonic stem cells, induced pluripotent stem cells, and adult stem cells39,40. Embryonic stem cell-derived organoids have the advantage of simulating the morphological traits of organ development and transplantation in vivo; whereas organoids derived from the other 2 stem cell types play a fundamental role in precision medicine for modeling and drug screening of refractory diseases39.
Organoids are currently cultivated using a solid extracellular matrix to support cell proliferation and adhesion41; as well as animal-derived hydrogels, including Matrigel42 and collagen43, to promote organoid structure44. For example, Lgr5+ stem cells or crypts were coated in Matrigel and then cultured in medium containing EGF, R-spondin, and noggin to generate intestinal organoids9,45.
Recently, it has been reported that Matrigel-based cultivation might limit organoid growth and maturation by restricting gas and metabolite exchanges between organoids and the surrounding microenvironment46. Therefore, Matrigel has been replaced with sponge-like47 or fibrous reticular scaffolds48, which contain larger cavities. Robertson et al.49 used the internal vascular network of the liver to facilitate better transport to and from liver organoids, and thus increased their in vitro survival beyond 4 weeks. This breakthrough represented an important development for organoid generation and culture.
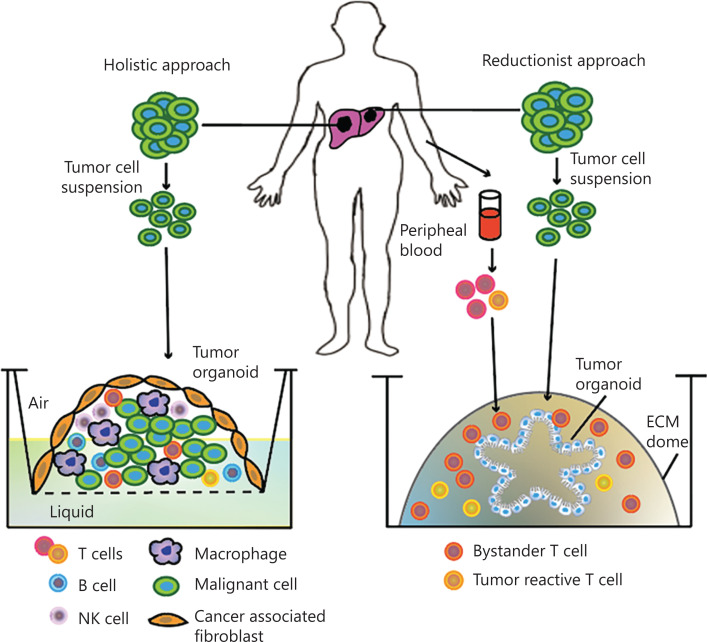
There is a growing interest in constructing a tumor-immune co-cultured organoid system that can preserve the tumor microenvironment50,51 (Figure 1). Several studies using a holistic52–54 or reductionist approach55 to build tumor-immune co-cultured ex vivo 3D models have been published. Neal et al.52 reported an air-liquid interface murine organoid model containing tightly integrated epithelial and stromal compartments, as well as specific tumor-infiltrating lymphocytes. Besides basal media, additional T cell activators such as inteleukin-2 were added to support the growth of immune cells. Consequently, variable subtypes, such as CD8+ T cells, CD4+ T cells, B cells, natural killer cells, and natural killer T cells, could be preserved over a period of days. In the reductionist approach, organoids are grown from tumor biopsies and are then co-cultured with autologous immune cells from the peripheral blood of the same patient to promote the serial expansion of tumor-reactive cells, allowing for the long-term culture and growth of the tumor epithelium (Figure 1)52. These co-cultured organoid systems enable investigations within the tumor microenvironment and facilitate personalized immunotherapy testing.
Figure 1.
Tumor-immune co-cultured organoid systems. In the holistic approach (left), a tumor cell suspension is cultured in an air-liquid interphase including all tumor cell types, endogenous immune cells, and fibroblasts, to generate tumor-immune organoids, facilitating culture of the entire tumor microenvironment, which closely resembles the in vivo situation. In the reductionist approach (right), tumor organoids are grown and expanded from a tumor cell suspension and then co-cultured with autologous immune cells from the peripheral blood of the same patient to promote the serial expansion of tumor-reactive T cells, which enables more extended investigations. ECM, extracellular matrix.
Organoid techniques facilitate research in developmental and cancer biology
Organoids are 3D cell cultures with unique biological characteristics, such as proliferation and differentiation into multifunctional cells under specific developmental conditions56 as well as self-assembly to mimic the structure of the parental organ57,58. In the case of primary liver cancer, Broutier et al.29 established organoids from hepatocellular carcinomas (HCCs), combined hepatocellular-cholangiocarcinomas (CHCs), and cholangiocarcinomas (CCs). Histological analysis demonstrated that HCC-derived organoids exhibited pseudoglandular rosettes as their parental tissues, whereas CC-derived organoids presented glandular lumens similar to the clinical morphology of the original patient, indicating genetic stability even after long-term expansion29. Moreover, organoids can recapitulate the physiological activity of the corresponding organs40,59. Huch et al.15 showed that, similar to primary hepatocytes, organoids from hepatocytes secreted albumin. In addition, organoids can mimic filtration, neural activity, and contraction processes10. Genomic and transcriptomic profiles have been investigated for a wide range of cancer-derived organoids. Although limited tumor evolution has been reported in organoid cultures, for the most part, profiles were similar between samples within individual organoid lines, indicating that tumor heterogeneity was well preserved30.
Based on these characteristics, basic research has been revolutionized by organoid technology, providing new possibilities for the study of development and organogenesis41. Organogenesis is a complex and interconnected process orchestrated via interactions across boundaries60,61. Its study has been limited by the availability of embryonic or fetal tissues and ethical concerns41. Organoids have helped demonstrate how individual neighboring tissues coordinate the onset of multi-organ structures62. Koike et al.62 reported that the anterior and posterior gut spheroids, differentiated from 3D-cultured human pluripotent stem cells, could interact with each other and form hepatic, biliary, and pancreatic organoids, recapitulating early morphogenetic events, such as invagination and branching of interconnected organ structures. Hence, organoids might offer an accessible model for the exploration of organogenesis and development.
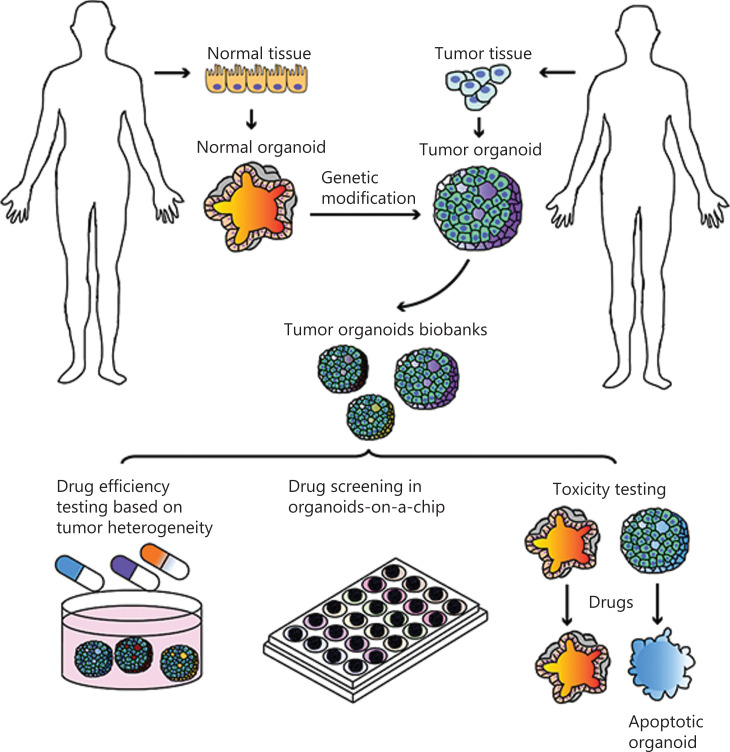
Organoids are of great importance in cancer research and personalized medicine. Theoretically, organoids allow the expansion of samples derived from tumor tissues of individual patients with a range of carcinomas63. Researchers have demonstrated that organoids can be established from surgical specimens63, fine-needle aspiration, and even from ascites liquid64, providing alternative sources of different organoid lines. In addition, CRISPR-based genetic modifications allow the engineering of non-cancerous organoids by introducing oncogenic alterations, providing ways to investigate tumor initiation and progression, which is vital in cancer research and personalized medicine. In this review, we analyzed the various applications of tumor organoids by using tumor biobanks and known genetic modifications of organoids, and summarized existing uses of organoids for personalized medicine (Figure 2).
Figure 2.
Organoids in cancer research and personalized medicine. Tumor organoid biobanks can be established from genetically modified noncancerous organoids or tumor organoids. These biobanks enable drug efficacy testing, drug screening, and toxicity testing, to facilitate personalized clinical medicine.
Development of tumor organoid biobanks
Tumor organoid biobanks are generated in 2 ways: via a direct tissue collection from patients by biopsy or surgical resection65–67 or modification of organoids derived from healthy tissues68,69. Tumor organoid biobanks allow passage, expansion, and cryopreservation of organoids derived from different carcinomas, as well as different lesions, grades, or stages of an indicated cancer. Established tumor organoid biobanks are listed in Table 1.
Table 1.
Overview of tumor organoid tissue banks
| Tumor organoid tissue banks | Source | No. of patients involved | No. of tumor organoid cell lines | Efficiency | Reference |
|---|---|---|---|---|---|
| Prostate cancer | Metastases and circulating tumor cells | 7 | 7 | ∼15%–20% | 20 |
| Colorectal cancer | Primary tumor | 20 | 22 | ∼90% | 70 |
| Metastases | 14 | 10 | 71% | 67 | |
| Primary tumor and metastases | 43 | 55 | 100% | 71 | |
| Breast cancer | Primary tumor | >100 | >150 | ∼80% | 27 |
| Gastric cancer | Primary tumor and metastases | 34 | 46 | >50% | 72 |
| Metastatic gastrointestinal cancer | 71 | 77 | 70% | 33 | |
| Bladder cancer | Primary tumor | 16 | 12 | ∼70% | 30 |
| Pancreatic cancer | Primary tumor and ascites specimens | 49 | 39 | ∼80% | 64 |
| Ovarian cancer | Primary tumor | 32 | 56 | 65% | 28 |
| Glioblastoma | Primary tumor | 53 | 70 | 91.4% | 73 |
| Liver cancer | Primary tumor | 8 | 7 | Not reported | 29 |
| Primary tumor | 8 | 10 | 26% | 65 | |
| Primary tumor | 5 | 27 | Not reported | 74 |
Tumor organoid biobanks can be established from tissue collection
The prostate cancer organoid biobank
The prostate cancer organoid biobank was established in 2014, introducing the use of organoids in cancer research20. Gao et al.20 cultured 6 organoid lines from prostate cancer metastatic lesions via tissue biopsy and 1 line from circulating tumor cells, resulting in 15%–20% efficiency. This small biobank included various subtypes of prostate cancers, such as high grade adenocarcinomas, mucinous adenocarcinomas, adenocarcinomas with extensive squamous differentiation, and adenocarcinomas with cribriform growth, which were consistent with the original patients. The histological and immunohistological patterns displayed in prostate cancer organoids were similar to those of the original patients. For example, organoids derived from bone marrow metastasis lesions recapitulated the intraductal pattern of primary cancer, with positive pan-cytokeratin and negative androgen receptor immunohistochemical staining. Furthermore, mRNA and protein analyses revealed that these organoid lines recapitulated the diversity observed in human prostate cancer20.
The colorectal cancer organoid biobank
Since their first establishment, organoids originating from the small intestine have been expanded to also generate colorectal carcinoma organoids9. Barker et al.75 reported the in vitro formation of germinal cystic organ-like structures derived from mice with intestinal adenomas. Fumagalli et al.76 analyzed the evolution of adenomas in vivo using organoids containing different combinations of colorectal cancer mutations. The results indicated gene changes in Wnt, EGF receptor, TP53, and transforming growth factor-beta signaling pathways, which led to tumor growth, migration, and metastasis.
Wetering et al.70 reported the establishment of 22 cancer organoids along with 19 organoids derived from adjacent non-cancerous tissues from 20 colorectal carcinoma patients70. The organoids were successfully established in ∼90% of cases, while >80% of them could be successfully frozen and thawed. Patient-specific morphologies such as cystic vs. solid organization of the epithelium were generally preserved, and expressions of colorectal cancer markers (Ki-67, OLFM4, and KRT20) in organoids reflected those in the patients.
To generate different subtypes of colorectal cancer organoids, Fujii et al.71 improved organoid culture methods based on niche factor requirements. They established 55 colorectal cancer organoids derived from a range of histological subtypes and clinical stages, as well as 41 counterpart organoids from noncancerous colorectal tissues, including hyperplastic polyps, sessile serrated adenoma/polyps, early cancers, advanced cancers, and metastatic cancer subtypes. By carefully adjusting ambivalent factors, Wnt activators, and other inhibitors, they were able to propagate colorectal carcinoma organoids with 100% efficiency. All these organoids preserved the histopathological structures and profiles of the parental tumors71.
The methods involved in establishing colorectal carcinoma biobanks facilitated the generation of different tumor subtypes and promoted the creation of individual models for personalized medicine. Since then, organoids derived from a wide range of tumor subtypes have been established in multiple cancers.
The breast cancer organoid biobank
Breast cancer comprises multiple pathologically and clinically distinct subtypes. Sachs et al.27 established 95 breast cancer organoids from 155 breast cancer specimens, with a success rate >80%. This organoid biobank included different histological subtypes, grades, and receptor statuses of the original tumor. Breast cancer-derived organoids showed a similar distribution of subtypes as clinical patients, i.e., 50%–80% invasive ductal carcinomas and 5%–15% invasive lobular carcinomas. These organoids presented significantly different morphologies, including sizes and cystic-like or grape-like phenotypes, in accordance with the original patients. Moreover, whole-genome sequencing (WGS) and RNA sequencing demonstrated that copy number variations, sequence changes, and expression profiling were similar to parental tumors even after long-term culture27.
The gastric cancer organoid biobank
Yan et al.72 reported the establishment of a primary gastric cancer organoid biobank, including noncancerous tissues, dysplastic tissues, primary tumors, and lymph node metastases from 63 specimens of 34 patients. The establishment percentage was >90% in noncancerous gastric organoids and >50% in cancer organoids72. The biobank encompassed most known molecular subtypes, including microsatellite instability, Epstein-Barr virus, intestinal (chromosome instability), and diffuse (genomically stable) subtypes. The subtypes displayed a variety of growth patterns, such as solid, glandular, or discohesive patterns, illustrating different degrees of gastric carcinomas. Protein expression analyses confirmed the overexpressions of known oncogene markers, such as HER2, ERBB2, and FGFR2, in these cancer organoids72.
In the same year, Vlachogiannis et al.33 generated an organoid biobank using 110 metastatic gastrointestinal cancer specimens from 71 patients enrolled in phase I or II clinical trials. In this case, organoids were successfully established in 70% of the specimens. After confirming the phenotypic and genotypic characteristics of organoids with patient tumors, they focused on the application of organoids in drug screening. This study highlighted how patient-derived organoids could be used to recapitulate patient responses in the clinic, which might have potential applications in personalized medicine33.
The bladder cancer organoid biobank
Lee et al.30 described an organoid biobank that recapitulated the histopathological and molecular diversities of human bladder cancers30. They cultured 22 bladder cancer organoids from 16 patients with an efficiency of ∼70%. The organoid morphology ranged from spheroidal to asymmetric, and 1 organoid line displayed features of squamous cell carcinoma, an uncommon histological subtype of bladder cancer similar to the parental tissue. These findings suggested the presence of multiple phenotypes in the bladder cancer organoid biobank. The biobank consisted of organoids collected directly from patients, serially passed organoids, organoids plated in orthotopic xenografts, and xenograft-derived organoids30. Using whole-genome and targeted exome sequencing, the organoids were found to retain parental tumor heterogeneity and exhibited a spectrum of genomic changes consistent with tumor evolution30.
The pancreatic cancer organoid biobank
Pancreatic cancer is characterized by elevated intra- and inter-tumor heterogeneities and highly malignant phenotypes, which makes it difficult to establish organoid biobanks. In this respect, a significant contribution was made by Boj et al.24 and Seino et al.64.
Boj et al.24 first established organoid models using non-cancerous human pancreatic tissues, as well as neoplastic murine and human pancreatic tissues. These organoids exhibited ductal- and disease-stage-specific characteristics24. Seino et al.64 established a human pancreatic cancer organoid biobank using surgical, fine needle aspiration, and ascites specimens. Their pancreatic cancer organoid library was comprised of 39 pancreatic ductal adenocarcinoma (PDAC) patient-derived organoids. They identified 3 functional subtypes according to stem cell niche factor dependency on Wnt or R-spondin, revealing functional heterogeneity in tumor progression. They also collected surrounding cancer-associated fibroblasts, which provided a Wnt niche for PDAC, and thus obtained a platform for investigating the tumor microenvironment using organoids64.
The ovarian cancer organoid biobank
Ovarian cancer is a heterogeneous disease with multiple subtypes. Kopper et al.28 established 56 organoid lines from 32 patients with a success of 65%, which represented all main subtypes of ovarian cancers, including serous borderline tumors (BT), mucinous BT, low grade serous BT, mucinous carcinomas, endometrioid, clear cell carcinomas, and the high grade serous subtypes. Consistent with the original patients, ovarian cancer organoids in this library displayed wide morphological variations between distinct histological subtypes. For example, most BT organoids were cystic; whereas mucinous carcinoma, low grade serous, endometrioid, and clear cell carcinoma organoids formed denser structures with multiple lumens. Moreover, the genomic features of each patient were well-preserved28, indicating that this biobank represented an excellent model for investigating ovarian cancer intra- and inter-patient heterogeneities.
The glioblastoma organoid biobank
Glioblastoma is the most common brain tumor in adults and is characterized by elevated invasiveness77. Heterogeneity between and within glioblastomas is the main reason for therapeutic resistance in clinical trials78–80. A recent report by Jacob et al.73 established a biobank consisting of 70 glioblastoma organoids from 53 patients. Transcriptome sequencing, whole-exome sequencing, and single cell transcriptome sequencing showed that intra- and inter-tumor heterogeneities are well maintained in glioblastoma organoids, reflecting the original tumors. A co-culture system including glioblastoma organoids and chimeric antigen receptor T (CAR-T) cells was used to determine the CAR-T cell treatment response, suggesting a possible application in personalized therapy73.
The liver cancer biobank
Broutier et al.29 first established primary liver cancer organoids in 2017. They included HCC, CC, and CHC subtypes from 8 surgically resected liver tumor tissues and were initially called “tumoroids”. Based on this culture method, Nuciforo et al.65 reported the generation of 13 patient-derived liver cancer organoids using 10 HCC needle biopsies from 8 patients and 3 intrahepatic cholangiocarcinoma needle biopsies from 3 patients. They demonstrated the replication of morphology and genetic heterogeneities of the original tumors65. Furthermore, in 2019, Li et al.74 generated 27 liver cancer organoid lines and tested them with 129 FDA-approved cancer drug libraries, illustrating the potential of cancer organoids in drug discovery. However, large-scale liver cancer organoid biobanks have not yet been reported.
Tumor organoids can be generated from genetically modified normal organoids
Along with organoids directly derived from patients diagnosed with carcinomas, organoids derived from non-cancerous tissue can also be used to establish biobanks for cancer research via genetic modification. Indeed, this method has been reported to mimic several carcinomas. Huang et al.23 differentiated human pluripotent stem cells into exocrine progenitor organoids via 3D cultures. KRAS or TP53 mutation-specific cancer organoids were successfully generated to model PDAC, and precision therapy strategies were identified. Furthermore, mutations in APC, SMAD4, and PIK3CA were edited by CRISPR/Cas9 technology in non-cancerous colon organoids to create colorectal carcinomas, illustrating the production of malignant phenotypes68,81.
CRISPR-based genetic modifications provide important clues for cancer initiation. Sun et al.82 used genetically reprogrammed human hepatocyte organoids to mimic the initial alterations in liver cancers. They found that c-Myc expression led to a malignant HCC morphology in these hepatocytes, whereas RASG12V, YAP5SA, IDH2R172K, and PTPN3A90P organoids developed morphological alterations similar to intrahepatic cholangiocarcinomas, indicating a tractable system in cancer modeling82.
CRISPR-based genetic modification enables the investigation of differentiated cancer subtypes and provides vital tools for tracing cancer progression. Seino et al.64 performed GATA6 short hairpin RNA-based knockdown and CRISPR-Cas9-based knockout experiments in Wnt-non-secreting PDAC organoids. They found that GATA6 expression levels were functionally relevant in PDAC subtypes64. Moreover, CRISPR-Cas9-based genomic editing of PDAC driver genes in organoids, such as KRAS, TP53, CDKN2A, and SMAD4, demonstrated non-genetic acquisition of Wnt niche independence during pancreatic cancer initiation and progression64.
Together, genetic modifications can accelerate the establishment of large-scale tumor organoid biobanks while allowing for the characterization of tumor initiation and progression.
Organoids preserve tumor heterogeneity of the original patient
Tumor heterogeneity is a common feature of multiple cancers. It can be divided into intra- and inter-tumor heterogeneities, and plays a vital role in tumor progression, tumor response to therapy, and the emergence of drug resistance83,84. Traditional cell culture and PDX models are not capable of replicating the heterogeneity of the original patient85. As a result, organoid models have been introduced to investigate cancer heterogeneity.
Organoids replicate the genomic profile of the original patient. For example, Wetering et al.70 performed WGS to investigate the heterogeneity of colorectal cancer in the biobank they established. They reported alterations in tumor suppressors, such as APC, FBXW7, TP53, and SMAD4, and activating mutations in KRAS and PIK3CA, which were consistent with previous reports on primary colorectal carcinomas70. Lee et al.30 demonstrated that bladder cancer organoids showed >80% concordance with the mutational profiles of parental tumors, such as mutations in TP53 and RB1 or the FGFR3-TACC3 fusion. Kopper et al.28 performed WGS in 40 ovarian cancer organoids from 22 patients and found that most copy number variants, somatic single nucleotide variants, and structural variants were preserved in cancer organoids, consistent with the original tumors, even after prolonged passaging. Broutier et al.29 demonstrated that ∼92% of the variants of each patient were retained in early organoid cultures (<2 months) and >80% of variants were maintained in late organoid cultures (>4 months), further suggesting that the genomic profiles of the original patients were preserved in cancer organoids.
Organoids replicate the transcriptome profiles of original patients. Yan et al.72 performed transcriptomic analyses of gastric cancer organoids derived from different histological subtypes and counterpart tumor tissues, and demonstrated a high concordance of gene expressions. In particular, several genes and pathways appeared upregulated in cancer organoids, such as the HOX family, the mitotic cell cycle, and Wnt pathway-associated genes72. Broutier et al.29 compared expression profiles using RNA sequencing and demonstrated that each organoid correlated with its corresponding tissue of origin. HCC markers, such as GPC3 and AFP, and hepatocyte markers, such as ALB, TTR, APOA1, and APOE, were highly expressed in HCC organoids and matched tissues; whereas CC and ductal markers, such as EPCAM, KRT19, and S100A1186, were highly expressed in CC organoids, which was consistent with the characteristics of clinical primary liver cancers29.
Cancer heterogeneity is largely maintained within organoids at different time points. Kopper et al.28 used a novel single cell DNA sequencing method to investigate recurrent ovarian cancer tumor samples derived from a single patient at different time points and the corresponding organoids. Distinct clusters revealed by copy number variant profiles overlapped with each other and did not generate new separate clusters, suggesting both their heterogeneity and resemblance to the corresponding tumor samples28. Notably, in some lines, cluster-represented diploid cells became fewer after passaging, indicating the gradual overgrowth of tumor cells compared with noncancerous cells, while maintaining tumor heterogeneity28.
Organoids mimic the evolution of heterogeneity both in vitro and in vivo. Heterogeneity evolution is a universal phenomenon in cancer87, which was challenging to manipulate in patients or xenografts before the application of effective organoid tools. Lee et al.30 performed deep sequencing to compare the mutational profiles of organoids in culture, during grafting or when re-establishing organoid lines from grafted models. They found that mutational profiles were largely maintained within individual lines, while a small subset of mutations underwent evolution. By analyzing the clonal composition of each line, they found that truncation mutations were retained, whereas subclonal mutations could be gained or lost, such as a gain of CTNNB1S45F in late passage organoids and losses of ERBB2D227N and JAK2H538Y during organoid culture, confirming clonal evolution during serial passaging30. Therefore, organoids can be used to investigate heterogeneous evolution in cancer.
Based on heterogeneity, organoids can contribute to the identification of potential prognostic biomarkers in carcinomas. Broutier et al.29 compared the similarities between transcriptomes of all primary liver cancer organoids to healthy liver-derived organoids and identified previously unknown genes. Specifically, overexpression of C19ORF48, DTYMK, or UBE2S in HCC and overexpression of C1QBP in CCs conferred poor survival prognoses29. This example highlighted the important role of organoid heterogeneity in identifying new targets with prognostic value, which could potentially be used in clinical cancer therapy.
Organoids in drug screening and personalized medicine
The lack of tumor models suitable for drug screening has shown the need for systems more representative to those in patients, which are more amenable to disease modeling and individual regimen design88. In recent years, organoid biobanks of multiple cancers have been used to mimic tumor heterogeneity of the original patients.
Based on tumor heterogeneity, organoids can be used to test drug efficiency in both clinical therapy and clinical trials. Wetering et al.70 found that organoids derived from the same patient, even those carrying the BRAF V600E mutation, differed in their drug response profiles, emphasizing the importance of drug screening based on tumor heterogeneity. Sachs et al.27 found that breast cancer organoids were sensitive to drugs that blocked the HER signaling pathway when HER2 was overexpressed or when other drug resistant genes existed in the absence of HER2. In addition, breast cancer organoid lines characterized by high BRCA1/2 signatures were sensitive to poly (ADP-ribose) polymerase inhibitors; whereas those with low BRCA1/2 signatures were not. These discordant drug efficacies were all based on the presence of different subpopulations27. Broutier et al.29 used liver cancer-derived organoids to screen 29 anti-cancer compounds, including drugs in clinical use or development. Notably, correlations between drug sensitivities and molecular profiling were observed. For example, HCC organoids harboring a mutated CTNNB1 were resistant to the porcupine inhibitor LGK974; whereas the Wnt-dependent organoid line of CC was sensitive. Wild-type KRAS organoid lines of HCC were sensitive to the EGF receptor inhibitor AZD8931, while organoids harboring mutated KRAS were resistant, suggesting guidance for personalized medicine based on tumor heterogeneity29.
Organoids can also be used to predict the drug response. Sachs et al.27 compared the response of breast cancer organoids to tamoxifen in patients administered standard clinical treatment and found that in vitro drug responses of breast cancer organoids matched those of the corresponding patients, indicating the potential use of organoids in predicting in vivo drug response27. Vlachogiannis et al.33 compared the response of organoids treated with anti-cancer agents with that of patients in clinical trials. Accordingly, organoids showed 100% sensitivity, 93% specificity, 88% positive predictive value, and 100% negative predictive value in forecasting the response to targeted agents or clinical chemotherapy, thus illustrating the ability of organoids to predict the drug response in personalized medicine.
Organoids can also be used for large-scale drug and toxicology screenings. Skardal et al.88 summarized the development of organoid-on-a-chip platforms. In this system, organoids generated from multiple cancer cells were encapsulated in Matrigel in separate chambers, while myeloblasts or fibroblasts were formed with alginate gels in additional chambers to mimic the tumor microenvironment89. These organoids-on-a-chip platforms supported organoid culture, fluid flow, and high-throughput testing, offering the potential for drug screening while taking into account a personalized tumor microenvironment88.
Importantly, organoids can be used for modeling personalized immunotherapy50. Immunotherapy in tumors has not been very effective and remains to be further developed52. With the improvement of tumor-immune co-cultured organoid systems, Fadi Jacob et al.73 co-cultured glioblastoma organoids with CAR-T cells designed to react specifically with cells expressing EGFRvIII. By testing the proliferation of T cells and death of tumor cells in the presence of EGFRvIII in this system, they demonstrated the utility of organoids for rapid testing of immunotherapy with the endogenous target in culture73. In addition, organoids can serve as a platform for investigating the functional response to anti-PD-1 or anti-PD-L152. Neal et al.52 established 20 organoids representing the immunotherapy-responsive neoplasms and treated organoids with the therapeutic PD-1 blocking antibody, nivolumab. As a result, nivolumab elicited significant induction of IFNG, PRF1 and/or granzyme B within organoid CD3+ tumor-infiltrating lymphocytes, showing functional in vitro recapitulation of checkpoint inhibition, which was consistent with personalized anti-PD-1 responses in clinical trials.
In addition, patient-derived organoid models can capture patient specific tumor evolution and acquired resistance to treatment33. Vlachogiannis et al.33 generated organoids using liver metastasis tissues from a colorectal cancer patient both before and after regorafenib treatments, which were used to establish xenografts. Consistent with clinical findings, CD31 immunostaining revealed a significant reduction in microvasculature in response to regorafenib in xenografts established with nontreated organoids, while no significant change was observed in xenografts established using regorafenib-treated organoids. More importantly, regorafenib treatment provided a selective survival benefit in mice with nontreated organoids33, suggesting the predictive role of organoids in reflecting cancer evolution upon treatment.
Another application of organoids in personalized medicine is the ability to target specific tumor cells while leaving healthy cells unharmed11. Drost and Clevers suggested the application of hepatocyte organoids in testing for hepatotoxicity, one of the leading causes of drug failure in clinical cancer therapy. They hypothesized that a suitable drug killed only tumor organoids without inducing hepatotoxicity11. Drug nephrotoxicity is another crucial cause of therapy failure in hospitalized patients90. By using organoids to demonstrate the toxicity of cisplatin in the proximal and distal tubules of the kidney, Morizane et al.91 established an important breakthrough in studies on nephrotoxicity. Furthermore, cardiac organoids derived from induced pluripotent stem cells have also been used to investigate cardiotoxicity92,93. These organoids provide patient-specific models for studying the toxicity of anti-cancer drugs in personalized medicine.
Challenge
Despite the advantages of organoids, there are still shortcomings compared with other models50. For example, organoids derived from tissue pieces indeed capture interpatient genetic heterogeneities and contain all cellular components of the tissue microenvironment; however, several organoid cell lines, especially tumor-immune co-cultured organoid systems, might show limited culture times94,95 when compared with cancer cell lines or PDX models. In addition, organoids generated from pluripotent stem cells showed the ability of serial passaging while not containing all cellular components of the tissue microenvironment, including fibroblasts, endothelial cells, and immune cells, when compared with the PDX models95,96. It is therefore important to determine whether organoid models are most appropriate in investigating tumor biology in specified conditions.
Although organoids derived from most carcinomas have been established, efforts still need to be made to improve the organoid model system. First, it is indispensable to reduce the cost of cultivating organoids to ensure their wider applicability. Gjorevski et al.97 replaced mouse-derived extracellular matrices with engineered synthetic matrices in mouse and human intestinal organoid culture systems. The new matrices still supported organoid growth and expansion, but at substantially reduced culture costs97, thus stimulating the search for alternatives to other ingredients employed in organoid cultures63. Organoids derived from several advanced cancers grow more slowly than those derived from non-cancerous epithelium, which may result in the outgrowth of healthy tissue and hinder cancer research. Hence, separating cancer organoids from noncancerous organoids remains a challenge58. In addition, vascular and neural systems supporting cancer growth and metastasis are not included in current organoid models58. However, a functional enteric nervous system was developed in intestinal organoids using induced pluripotent stem cell-derived tissue engineering, indicating the potential for developing more complex structures within organoids98. The introduction of vascular and neural systems and advanced structures in organoids will help replicate the tumor microenvironment to assess its role in cancer.
Outlook
In recent years, the rapid development of organoid platforms has enabled the successful establishment of cancer biobanks. Furthermore, CRISPR-based genetic modifications have allowed the exploration of cancer initiation and progression using organoid models. Based on genomic and transcriptomic characterizations, original intra- and inter-patient heterogeneities associated with a wide range of carcinomas have been successfully preserved in cancer organoids. Organoids are therefore predicted to play a decisive role in drug screening and personalized medicine, which will provide vital guidance for clinical cancer therapy.
Grant support
This work was supported by grants from the National Natural Science Foundation of China (Grant Nos. 31671421, 82030079, and 82003187).
Conflict of interest statement
No potential conflicts of interest are disclosed.
Author contributions
Conceived and designed the analysis: Hui Yang, Ning Zhang, and Pengyuan Wang.
Collected the data: Hui Yang, Yinuo Wang, and Peng Wang.
Contributed data or analysis tools: Hui Yang, Yinuo Wang, and Peng Wang.
Performed the analysis: Hui Yang, Yinuo Wang, and Peng Wang.
Wrote the paper: Hui Yang, Ning Zhang, and Pengyuan Wang.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450–62. doi: 10.1056/NEJMra1713263. [DOI] [PubMed] [Google Scholar]
- 3.Unger C, Kramer N, Walzl A, Scherzer M, Hengstschläger M, Dolznig H. Modeling human carcinomas: physiologically relevant 3D models to improve anti-cancer drug development. Adv Drug Deliv Rev. 2014;79-80:50–67. doi: 10.1016/j.addr.2014.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Gillet JP, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105:452–8. doi: 10.1093/jnci/djt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrne AT, Alférez DG, Amant F, Annibali D, Arribas J, Biankin AV, et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17:254–68. doi: 10.1038/nrc.2016.140. [DOI] [PubMed] [Google Scholar]
- 6.Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med. 2015;21:1318–5. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]
- 7.DeRose YS, Wang G, Lin YC, Bernard PS, Buys SS, Ebbert MT, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–20. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemper K, Krijgsman O, Cornelissen-Steijger P, Shahrabi A, Weeber F, Song JY, et al. Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol Med. 2015;7:1104–18. doi: 10.15252/emmm.201404914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 10.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science (New York, NY) 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 11.Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18:407–18. doi: 10.1038/s41568-018-0007-6. [DOI] [PubMed] [Google Scholar]
- 12.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–72. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 13.Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt-driven regeneration. Nature. 2013;494:247–50. doi: 10.1038/nature11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz JS, de Jonge HR, Forrest JN., Jr Value of organoids from comparative epithelia models. Yale J Biol Med. 2015;88:367–74. [PMC free article] [PubMed] [Google Scholar]
- 15.Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell. 2015;160:299–312. doi: 10.1016/j.cell.2014.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Bartfeld S, Bayram T, van de Wetering M, Huch M, Begthel H, Kujala P, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–36 e126. doi: 10.1053/j.gastro.2014.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katano T, Ootani A, Mizoshita T, Tanida S, Tsukamoto H, Ozeki K, et al. Establishment of a long-term three-dimensional primary culture of mouse glandular stomach epithelial cells within the stem cell niche. Biochem Biophys Res Commun. 2013;432:558–63. doi: 10.1016/j.bbrc.2013.02.051. [DOI] [PubMed] [Google Scholar]
- 19.Ewald AJ. Isolation of mouse mammary organoids for long-term time-lapse imaging. Cold Spring Harb Protoc. 2013;2013:130–3. doi: 10.1101/pdb.prot072892. [DOI] [PubMed] [Google Scholar]
- 20.Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–87. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimokawa M, Ohta Y, Nishikori S, Matano M, Takano A, Fujii M, et al. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature. 2017;545:187–92. doi: 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- 22.Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11:347–58. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, Holtzinger A, Jagan I, BeGora M, Lohse I, Ngai N, et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat Med. 2015;21:1364–71. doi: 10.1038/nm.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boj SF, Hwang CI, Baker LA, Chio II, Engle DD, Corbo V, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–38. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batchelder CA, Martinez ML, Duru N, Meyers FJ, Tarantal AF. Three dimensional culture of human renal cell carcinoma organoids. PLoS One. 2015;10:e0136758. doi: 10.1371/journal.pone.0136758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 27.Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–86 e310. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Kopper O, de Witte CJ, Lohmussaar K, Valle-Inclan JE, Hami N, Kester L, et al. An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat Med. 2019;25:838–49. doi: 10.1038/s41591-019-0422-6. [DOI] [PubMed] [Google Scholar]
- 29.Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, et al. Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nat Med. 2017;23:1424–35. doi: 10.1038/nm.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee SH, Hu W, Matulay JT, Silva MV, Owczarek TB, Kim K, et al. Tumor evolution and drug response in patient-derived organoid models of bladder cancer. Cell. 2018;173:515–28 e517. doi: 10.1016/j.cell.2018.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coontz R. Science’s Top 10 Breakthroughs of 2013. 2013 https://www.sciencemag.org/news/2013/12/sciences-top-10-breakthroughs-2013 .
- 32.Bonventre JV. Kidney organoids-a new tool for kidney therapeutic development. Kidney Int. 2018;94:1040–2. doi: 10.1016/j.kint.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 33.Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science. 2018;359:920–6. doi: 10.1126/science.aao2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–80. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Behfar A, Perez-Terzic C, Faustino RS, Arrell DK, Hodgson DM, Yamada S, et al. Cardiopoietic programming of embryonic stem cells for tumor-free heart repair. J Exp Med. 2007;204:405–20. doi: 10.1084/jem.20061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh SH, Witek RP, Bae SH, Zheng D, Jung Y, Piscaglia AC, et al. Bone marrow-derived hepatic oval cells differentiate into hepatocytes in 2-acetylaminofluorene/partial hepatectomy-induced liver regeneration. Gastroenterology. 2007;132:1077–87. doi: 10.1053/j.gastro.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–32. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–6. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- 39.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–97. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 40.Huch M, Koo BK. Modeling mouse and human development using organoid cultures. Development (Cambridge, England) 2015;142:3113–25. doi: 10.1242/dev.118570. [DOI] [PubMed] [Google Scholar]
- 41.Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet. 2018;19:671–87. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- 42.Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15:378–86. doi: 10.1016/j.semcancer.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Jabaji Z, Brinkley GJ, Khalil HA, Sears CM, Lei NY, Lewis M, et al. Type I collagen as an extracellular matrix for the in vitro growth of human small intestinal epithelium. PLoS One. 2014;9:e107814. doi: 10.1371/journal.pone.0107814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orkin RW, Gehron P, McGoodwin EB, Martin GR, Valentine T, Swarm R. A murine tumor producing a matrix of basement membrane. J Exp Med. 1977;145:204–20. doi: 10.1084/jem.145.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–4. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 46.Rambani K, Vukasinovic J, Glezer A, Potter SM. Culturing thick brain slices: an interstitial 3D microperfusion system for enhanced viability. J Neurosci Methods. 2009;180:243–54. doi: 10.1016/j.jneumeth.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fong ELS, Toh TB, Lin QXX, Liu Z, Hooi L, Mohd Abdul Rashid MB, et al. Generation of matched patient-derived xenograft in vitro-in vivo models using 3D macroporous hydrogels for the study of liver cancer. Biomaterials. 2018;159:229–40. doi: 10.1016/j.biomaterials.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 48.Tamai M, Adachi E, Tagawa Y. Characterization of a liver organoid tissue composed of hepatocytes and fibroblasts in dense collagen fibrils. Tissue Eng Part A. 2013;19:2527–35. doi: 10.1089/ten.tea.2012.0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson MJ, Soibam B, O’Leary JG, Sampaio LC, Taylor DA. Recellularization of rat liver: an in vitro model for assessing human drug metabolism and liver biology. PLoS One. 2018;13:e0191892. doi: 10.1371/journal.pone.0191892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bar-Ephraim YE, Kretzschmar K, Clevers H. Organoids in immunological research. Nat Rev Immunol. 2020;20:279–93. doi: 10.1038/s41577-019-0248-y. [DOI] [PubMed] [Google Scholar]
- 51.Yuki K, Cheng N, Nakano M, Kuo CJ. Organoid models of tumor immunology. Trends Immunol. 2020;41:652–64. doi: 10.1016/j.it.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neal JT, Li X, Zhu J, Giangarra V, Grzeskowiak CL, Ju J, et al. Organoid modeling of the tumor immune microenvironment. Cell. 2018;175:1972–88 e1916. doi: 10.1016/j.cell.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finnberg NK, Gokare P, Lev A, Grivennikov SI, MacFarlane AW, Campbell KS, et al. Application of 3D tumoroid systems to define immune and cytotoxic therapeutic responses based on tumoroid and tissue slice culture molecular signatures. Oncotarget. 2017;8:66747–57. doi: 10.18632/oncotarget.19965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng J, Wang ES, Jenkins RW, Li S, Dries R, Yates K, et al. CDK4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov. 2018;8:216–33. doi: 10.1158/2159-8290.CD-17-0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174:1586–98 e1512. doi: 10.1016/j.cell.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Y, Ding Q. Genome engineering of stem cell organoids for disease modeling. Protein Cell. 2017;8:315–27. doi: 10.1007/s13238-016-0368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–54. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science (New York, NY) 2013;340:1190–4. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 60.San Roman AK, Shivdasani RA. Boundaries, junctions and transitions in the gastrointestinal tract. Exp Cell Res. 2011;317:2711–8. doi: 10.1016/j.yexcr.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shih HP, Seymour PA, Patel NA, Xie R, Wang A, Liu PP, et al. A gene regulatory network cooperatively controlled by Pdx1 and Sox9 governs lineage allocation of foregut progenitor cells. Cell Rep. 2015;13:326–6. doi: 10.1016/j.celrep.2015.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koike H, Iwasawa K, Ouchi R, Maezawa M, Giesbrecht K, Saiki N, et al. Modelling human hepato-biliary-pancreatic organogenesis from the foregut-midgut boundary. Nature. 2019;574:112–6. doi: 10.1038/s41586-019-1598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364:952–5. doi: 10.1126/science.aaw6985. [DOI] [PubMed] [Google Scholar]
- 64.Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, et al. Human pancreatic tumor organoids reveal loss of stem cell Niche Factor dependence during disease progression. Cell Stem Cell. 2018;22:454–7 e456. doi: 10.1016/j.stem.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 65.Nuciforo S, Fofana I, Matter MS, Blumer T, Calabrese D, Boldanova T, et al. Organoid models of human liver cancers derived from tumor needle biopsies. Cell Rep. 2018;24:1363–6. doi: 10.1016/j.celrep.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tiriac H, Bucobo JC, Tzimas D, Grewel S, Lacomb JF, Rowehl LM, et al. Successful creation of pancreatic cancer organoids by means of EUS-guided fine-needle biopsy sampling for personalized cancer treatment. Gastrointest Endosc. 2018;87:1474–80. doi: 10.1016/j.gie.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weeber F, van de Wetering M, Hoogstraat M, Dijkstra KK, Krijgsman O, Kuilman T, et al. Preserved genetic diversity in organoids cultured from biopsies of human colorectal cancer metastases. Proc Natl Acad Sci U S A. 2015;112:13308–1. doi: 10.1073/pnas.1516689112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–62. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 69.Takeda H, Kataoka S, Nakayama M, Ali MAE, Oshima H, Yamamoto D, et al. CRISPR-Cas9-mediated gene knockout in intestinal tumor organoids provides functional validation for colorectal cancer driver genes. Proc Natl Acad Sci U S A. 2019;116:15635–44. doi: 10.1073/pnas.1904714116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–45. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–38. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 72.Yan HHN, Siu HC, Law S, Ho SL, Yue SSK, Tsui WY, et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell. 2018;23:882–97.e811. doi: 10.1016/j.stem.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 73.Jacob F, Salinas RD, Zhang DY, Nguyen PTT, Schnoll JG, Wong SZH, et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell. 2020;180:188–204 e122. doi: 10.1016/j.cell.2019.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li L, Knutsdottir H, Hui K, Weiss MJ, He J, Philosophe B, et al. Human primary liver cancer organoids reveal intratumor and interpatient drug response heterogeneity. JCI Insight. 2019;4:e121490. doi: 10.1172/jci.insight.121490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–11. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 76.Fumagalli A, Drost J, Suijkerbuijk SJ, van Boxtel R, de Ligt J, Offerhaus GJ, et al. Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc Natl Acad Sci U S A. 2017;114:E2357–64. doi: 10.1073/pnas.1701219114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet (London, England) 2018;392:432–46. doi: 10.1016/S0140-6736(18)30990-5. [DOI] [PubMed] [Google Scholar]
- 78.Tasdemir N, Bossart EA, Li Z, Zhu L, Sikora MJ, Levine KM, et al. Comprehensive phenotypic characterization of human invasive lobular carcinoma cell Lines in 2D and 3D Cultures. Cancer Res. 2018;78:6209–2. doi: 10.1158/0008-5472.CAN-18-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Qazi MA, Vora P, Venugopal C, Sidhu SS, Moffat J, Swanton C, et al. Intratumoral heterogeneity: pathways to treatment resistance and relapse in human glioblastoma. Ann Oncol. 2017;28:1448–6. doi: 10.1093/annonc/mdx169. [DOI] [PubMed] [Google Scholar]
- 80.Yang H, Zhang N, Liu YC. An organoids biobank for recapitulating tumor heterogeneity and personalized medicine. Chin J Cancer Res. 2020;32:408–13. doi: 10.21147/j.issn.1000-9604.2020.03.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–7. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 82.Sun L, Wang Y, Cen J, Ma X, Cui L, Qiu Z, et al. Modelling liver cancer initiation with organoids derived from directly reprogrammed human hepatocytes. Nat Cell Biol. 2019;21:1015–6. doi: 10.1038/s41556-019-0359-5. [DOI] [PubMed] [Google Scholar]
- 83.Rye IH, Trinh A, Saetersdal AB, Nebdal D, Lingjaerde OC, Almendro V, et al. Intratumor heterogeneity defines treatment-resistant HER2+ breast tumors. Mol Oncol. 2018;12:1838–55. doi: 10.1002/1878-0261.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dagogo-Jack I, Shaw AT. Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol. 2018;15:81–94. doi: 10.1038/nrclinonc.2017.166. [DOI] [PubMed] [Google Scholar]
- 85.Moreira L, Bakir B, Chatterji P, Dantes Z, Reichert M, Rustgi AK. Pancreas 3D organoids: current and future aspects as a research platform for personalized medicine in pancreatic cancer. Cell Mol Gastroenterol Hepatol. 2018;5:289–8. doi: 10.1016/j.jcmgh.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Andersen JB, Spee B, Blechacz BR, Avital I, Komuta M, Barbour A, et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology. 2012;142:1021–31 e1015. doi: 10.1053/j.gastro.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Turajlic S, Sottoriva A, Graham T, Swanton C. Resolving genetic heterogeneity in cancer. Nat Rev Genet. 2019;20:404–6. doi: 10.1038/s41576-019-0114-6. [DOI] [PubMed] [Google Scholar]
- 88.Skardal A, Shupe T, Atala A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov Today. 2016;21:1399–11. doi: 10.1016/j.drudis.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sung JH, Shuler ML. A micro cell culture analog (microCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anti-cancer drugs. Lab Chip. 2009;9:1385–94. doi: 10.1039/b901377f. [DOI] [PubMed] [Google Scholar]
- 90.Uchino S, Kellum JA, Bellomo R, Doig GS, Morimatsu H, Morgera S, et al. Acute renal failure in critically ill patients: a multinational, multicenter study. J Am Med Assoc. 2005;294:813–8. doi: 10.1001/jama.294.7.813. [DOI] [PubMed] [Google Scholar]
- 91.Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, Bonventre JV. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol. 2015;33:1193–200. doi: 10.1038/nbt.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eder A, Vollert I, Hansen A, Eschenhagen T. Human engineered heart tissue as a model system for drug testing. Adv Drug Deliv Rev. 2016;96:214–24. doi: 10.1016/j.addr.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 93.Voges HK, Mills RJ, Elliott DA, Parton RG, Porrello ER, Hudson JE. Development of a human cardiac organoid injury model reveals innate regenerative potential. Development. 2017;144:1118–27. doi: 10.1242/dev.143966. [DOI] [PubMed] [Google Scholar]
- 94.Ootani A, Li X, Sangiorgi E, Ho QT, Ueno H, Toda S, et al. Sustained in vitro intestinal epithelial culture within a Wnt-dependent stem cell niche. Nat Med. 2009;15:701–6. doi: 10.1038/nm.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Peng WC, Logan CY, Fish M, Anbarchian T, Aguisanda F, Alvarez-Varela A, et al. Inflammatory cytokine TNFalpha promotes the long-term expansion of primary hepatocytes in 3D Culture. Cell. 2018;175:1607–19 e1615. doi: 10.1016/j.cell.2018.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu H, Gehart H, Artegiani B, C LO-I, Dekkers F, Basak O, et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell. 2018;175:1591–606 e1519. doi: 10.1016/j.cell.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 97.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordonez-Moran P, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–4. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 98.Workman MJ, Mahe MM, Trisno S, Poling HM, Watson CL, Sundaram N, et al. Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat Med. 2017;23:49–59. doi: 10.1038/nm.4233. [DOI] [PMC free article] [PubMed] [Google Scholar]