Abstract
Gummosis, one of the most detrimental diseases to the peach industry worldwide, can be induced by Lasiodiplodia theobromae. Ethylene (ET) is known to trigger the production of gum exudates, but the mechanism underlying fungus-induced gummosis remains unclear. In this study, L. theobromae infection triggered the accumulation of ET and jasmonic acid (JA) but not salicylic acid (SA) in a susceptible peach variety. Gaseous ET and its biosynthetic precursor increased gum formation, whereas ET inhibitors repressed it. SA and methyl-jasmonate treatments did not influence gum formation. RNA-seq analysis indicated that L. theobromae infection and ET treatment induced a shared subset of 1808 differentially expressed genes, which were enriched in the category “starch and sucrose, UDP-sugars metabolism”. Metabolic and transcriptional profiling identified a pronounced role of ET in promoting the transformation of primary sugars (sucrose, fructose, and glucose) into UDP-sugars, which are substrates of gum polysaccharide biosynthesis. Furthermore, ethylene insensitive3-like1 (EIL1), a key transcription factor in the ET pathway, could directly target the promoters of the UDP-sugar biosynthetic genes UXS1a, UXE, RGP and MPI and activate their transcription, as revealed by firefly luciferase and yeast one-hybrid assays. On the other hand, the supply of SA and inhibitors of ET and JA decreased the lesion size. ET treatment reduced JA levels and the transcription of the JA biosynthetic gene OPR but increased the SA content and the expression of its biosynthetic gene PAL. Overall, we suggest that endogenous and exogenous ET aggravate gummosis disease by transactivating UDP-sugar metabolic genes through EIL1 and modulating JA and SA biosynthesis in L. theobromae-infected peach shoots. Our findings shed light on the molecular mechanism by which ET regulates plant defense responses in peach during L. theobromae infection.
Introduction
Gummosis, a broad defense response to abiotic (environmental adversity and physical wounds) and biotic (insect and pathogen infestation) stresses, is characterized by the formation and exudation of gum in the shoots of several plant species, such as the stone fruit trees of Prunus spp. Over the past decades, gummosis caused by microbes has been reported in stone fruits (including peach, nectarine, plum and cherry), citrus, and grape [1–3]. Peach (Prunus persica) fungal gummosis is one of the most prevalent and detrimental diseases in this tree crop and limits the growth and yield of peach orchards throughout southern China, the United States and Japan [2, 4]. The causal agents colonize permanent woody structures of peach trees (trunks, limbs, branches, and twigs) via wounds and lenticels, causing large amounts of gum exudation, wood necrosis, and stem-bark cracking, eventually leading to poor tree vigor and reduced lifespan [1, 4, 5]. Four species of Botryosphaeriaceae, Lasiodiplodia theobromae, Botryosphaeria dothidea, Diplodia seriata, and Neofusicoccum parvum, have been reported to cause peach gummosis [2, 6]. Among these species, L. theobromae has a broad host range, similar to that of biotic agents of disease identified in other economically valuable fruit trees, such as apple ring rot and grapevine bot canker [2, 7]. Very recently, a dominant resistance allele to peach fungal gummosis was identified in Prunus using interspecific crosses between Kansu peach (Prunus kansuensis), almond (Prunus dulcis), and peach (P. persica) [8].
Ethylene (ET) plays sophisticated roles in plant defense responses against various pathogens; it could steer defense signaling to activate or suppress large and complex defense networks that function during plant-pathogen interactions [9]. On the one hand, ET activates defense pathways to enhance plant resistance against pathogen attacks [10, 11]. For example, Arabidopsis ET-deficient acs [lesioned in 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, ACS] mutants exhibit compromised resistance mediated by pathogen-associated molecular patterns (PAMPs) and effector-triggered immunity, rendering the plants more susceptible to Pseudomonas syringae (Pst) infection [10]. On the other hand, ET is also exploited by microbes to disturb the defense signaling network and increase plant susceptibility [12, 13]. Ethylene insensitive3 (EIN3) and EIN3-like1 (EIL1), two ET signaling components, negatively regulate PAMP-triggered immunity and thus enhance susceptibility to Pst invasion through the downregulation of salicylic acid (SA) biosynthesis [12]. In addition, increasing evidence suggests that ET modulates gum formation in response to stress-related stimuli in plants. The application of ethephon (2-chloroethylphosphonic acid, an ET-releasing compound) induces gum formation in sour cherry [14], grape hyacinth [15] and peach [16]; it also initiates gum duct formation to facilitate gum exudation in Prunus mume [17]. Moreover, the simultaneous application of ethephon and methyl-jasmonate (MeJA) substantially enhances gummosis in peach [16], cherry [14], and hyacinth [15].
Previous studies have demonstrated that L. theobromae infection can cause tissue necrosis and significantly induce gum exudation in peach shoots [2, 18]. Peach gum is a heteropolysaccharide that mainly consists of arabinose, xylose, mannose, galactose, rhamnose, and glucuronic acid at different molar ratios [19]. These sugars can be transformed into UDP sugars, which are necessary for the biosynthesis of gum polysaccharides, by different types of interconverting enzymes [20]. During L. theobromae-induced gummosis, the concentrations of soluble sugars (such as mannose, arabinose, and xylose) are increased [18], and the expression of genes involved in sugar metabolism changes significantly in peach shoots, as revealed by transcriptome profiling [21]. In this pathosystem, as in the ones mentioned above, pre- and posttreatments with ethephon indicate a dual role for ethephon in regulating gummosis by affecting both the peach sugar contents (sucrose, glucose, and fructose) and the mycelial growth of pathogenic fungi [22]. However, the use of ethephon (which can be phytotoxic) can lead to nonspecific responses, leaving a certain degree of uncertainty in the interpretation of the results [23]. Thus, in addition to these limited insights, the molecular mechanisms by which ET affects peach fungal gummosis remain poorly understood, especially in the shoot disease of perennial woody trees.
In this study, the role of ET in peach fungal gummosis due to L. theobromae infection was investigated through comparative transcriptome profiling and metabolite measurements to understand the defense responses of peach shoots at the physiological, molecular, and metabolic levels. We first quantified ET, JA and SA and investigated the effect of treatments with exogenous ET, MeJA, SA or their inhibitors on peach gummosis progression. The transcriptional and metabolic responses of hormones and sugar pathways to ET supply were then investigated in L. theobromae-infected shoots. We further identified an ET signaling component, EIL1, that could bind to the promoters of UDP-sugar metabolic genes and activate them, as assessed by firefly luciferase-based dynamic bioluminescence imaging and yeast one-hybrid (Y1H) assays. These findings elucidate a regulatory role played by ET during the defense response of peach against L. theobromae infection and provide insights into the molecular mechanisms by which ET promotes gum formation in peach gummosis.
Results
L. theobromae inoculation induces ET production
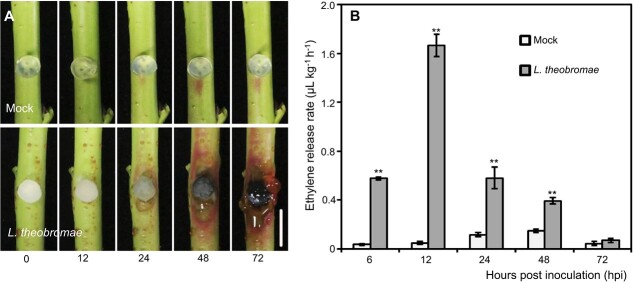
The inoculation of L. theobromae onto the shoots of the peach variety “Spring snow” led to brown necrotic lesions at 24 hours postinoculation (hpi); the lesions enlarged with time (Fig. 1A). In particular, L. theobromae-inoculated shoots displayed typical gum release symptoms at 72 hpi (Fig. 1A). Another obvious morphological change was that the tissues around the lesion turned red at 24 hpi (Fig. 1A). In contrast, shoots mock-inoculated with sterile PDA plugs did not develop necrotic lesions or exhibit gum release and color change at any time point. Given the previously reported effects of ethephon treatment on L. theobromae infection [22] and the transcriptional response of ET pathway genes during gummosis [21], we measured ET accumulation in peach shoots over time after L. theobromae inoculation. The amount of ET released in L. theobromae-inoculated shoots was consistently and significantly higher than that in the mock control throughout the infection period and until 48 hpi. Notably, ET was sharply and significantly induced at 6 hpi, peaked at 12 hpi, and then gradually dropped in L. theobromae-inoculated shoots (Fig. 1B).
Figure 1.
Symptoms of peach gummosis and quantification of the ethylene content in peach shoots inoculated with Lasiodiplodia theobromae strain JMB-122. (A) Morphological progression of gummosis in current-year peach shoots wounded and inoculated with sterile potato dextrose agar (PDA) plugs (mock) or L. theobromae. Bar represents 1 cm. (B) Time-course assay of the ethylene release rate in mock- and L. theobromae-inoculated shoots. Values represent the means ± SD of three independent replicates, with 40 individual shoots per replicate. Asterisks on top of paired bars indicate statistical significance with ** P < 0.01.
ET treatment aggravates L. theobromae-induced gummosis
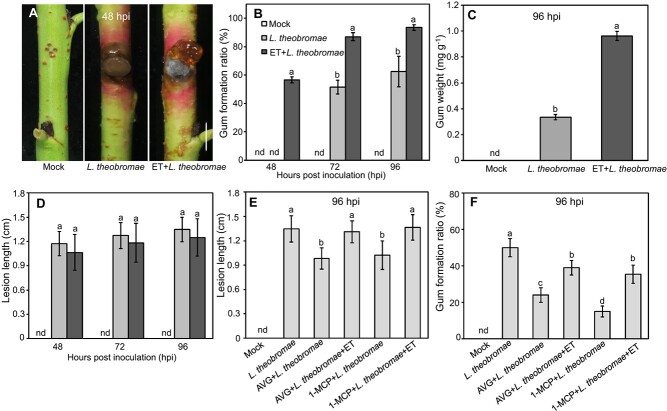
When peach shoots were treated with ET for 24 hours before L. theobromae inoculation, released gum was apparent at 48 hpi but was still barely detectable in untreated L. theobromae- and mock-inoculated shoots (Fig. 2A). Quantitatively, gum formation was consistently and significantly higher in L. theobromae-inoculated shoots treated with ET than in nontreated shoots. At 48 hpi, the shoot gummosis ratio was measurable (at approximately 60%) in ET-treated and inoculated shoots; similar values were reached in L. theobromae-inoculated shoots at 72 and 96 hpi, while ET-pretreated samples still showed values that were approximately 30% higher (Fig. 2B). The total dry weight of the gum of ET-treated and L. theobromae-inoculated shoots was 2.9-fold higher than that of L. theobromae-inoculated shoots at 96 hpi (Fig. 2C). However, no significant difference in lesion length was observed at any time point among inoculated shoots, irrespective of ET pretreatment (Fig. 2D).
Figure 2.
Ethylene (ET) and ET inhibitor treatment of L. theobromae-induced gummosis in peach shoots. (A) Gummosis progression at 48 hpi. Bar represents 1 cm. (B) Ratio of gum formation in peach shoots at 48, 72 and 96 hpi. (C) Gum weight analysis at 96 hpi. The gum weight was calculated by dividing the total dry weight of the gum by the fresh weight of the experimental shoots. (D) Lesion length analysis in ET-treated and inoculated peach shoots at 96 hpi. (E-F) Effects of ET and/or ET inhibitor treatments on lesion size and the gum formation ratio, respectively. The detached current-year peach shoots were wounded and inoculated with a sterile PDA plug (mock), L. theobromae treated with 10 μL L-1 gaseous ET for 24 h before inoculation (ET + L. theobromae), L. theobromae treated with AVG (aminoethoxyvinylglycine, an ET biosynthetic inhibitor) or 1-MCP (1-methylcyclopropene, an ET signaling inhibitor) for 24 h before inoculation (AVG or 1-MCP + L. theobromae), or L. theobromae treated with AVG or 1-MCP and gaseous ET (AVG or 1-MCP + L. theobromae+ET). nd stands for not detected. Values are the means ± SD of three independent replicates, with 40 shoots per replicate. Different letters on top of bars indicate statistical significance at P < 0.05.
Next, either an inhibitor of ET synthesis (aminoethoxyvinylglycine, AVG) or signaling (1-methylcyclopropene, 1-MCP) was sprayed on peach shoots 24 h before L. theobromae inoculation. The lesion length was significantly decreased by 27% in AVG-treated shoots compared to nontreated shoots (Fig. 2E). In infected tissues treated with both AVG and ET, the lesion length was comparable to infected tissues not pretreated with ET (Fig. 2E). A similar trend of lesion development was observed in 1-MCP-treated shoots (Fig. 2E). The gum formation ratio in AVG- or 1-MCP-treated infected shoots was decreased by 26 and 35% compared to that in nontreated shoots, respectively (Fig. 2F). This drop was approximately 15% in similar samples to which exogenous ET was also added (Fig. 2F). Taken together, these results indicate that ET positively affects peach gummosis progression both in terms of lesion size and gum release.
Transcriptome profiling of peach shoots treated with L. theobromae and ET+ L. theobromae
To elucidate the molecular network underlying the effects of ET in promoting peach gummosis, RNA-seq was performed on samples from mock- and L. theobromae-inoculated shoots, both ET-pretreated and not, at 0, 6, 12, 24 and 48 hpi. The quality of the transcriptome data is shown in Table S1. An average of 7.53 Gb of clean reads (Q20 > 97%; Q30 > 92%) were obtained for each sample, of which 91% were uniquely mapped to the peach genome (Table S1). In the principal component analysis, samples at different time points from L. theobromae-inoculated shoots were highly dispersed, whereas L. theobromae-inoculated samples collected at 24 and 48 hpi clustered with ET-pretreated, L. theobromae-inoculated samples harvested at 12 and 24 hpi (Fig. S1). To validate the RNA-seq data, 16 randomly selected differentially expressed genes (DEGs) were subjected to quantitative reverse-transcriptase PCR (qRT–PCR) analysis, and the results were highly consistent with the corresponding FPKM values from RNA-seq (Fig. S2).
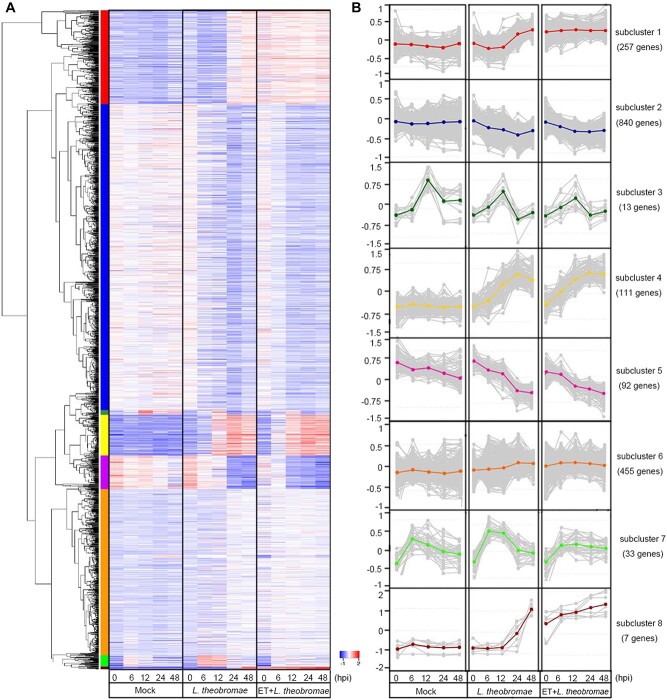
A total of 7045 DEGs were identified between the L. theobromae- and mock-inoculated treatments (Fig. S3A). In addition, 2733 DEGs were obtained between the L. theobromae-inoculated samples that were pretreated or not with ET (Fig. S3B). Venn analysis identified 1808 common DEGs shared between the two DEG datasets (Fig. S3 and Fig. S4A). We further carried out GO and KEGG enrichment analyses of the 1808 and 7045 DEGs, respectively. In total, 972 and 3707 DEGs in the L. theobromae-inoculated samples pretreated or not with ET, respectively, were enriched in the GO categories “catalytic activity”, “oxidoreductase activity”, and “carbohydrate metabolic process” (Fig. S4B and C). In the KEGG analysis, 702 and 2627 DEGs in the L. theobromae-inoculated samples pretreated or not with ET, respectively, were highly enriched in “phenylpropanoid biosynthesis” and “starch, sucrose metabolism” pathways, followed by “glycolysis/gluconeogenesis”, “amino or nucleotide sugar metabolism”, “pentose and glucuronate interconversions” and “plant-pathogen interaction” (Fig. S4D and E).
To better understand the transcriptomic data, hierarchical clustering analysis was performed, and a heat map was generated. The 7045 DEGs in the mock- vs. L. theobromae-inoculated samples were classified into five subclusters (Fig. S5). The DEGs related to “environmental adaptation”, “carbohydrate metabolism” and “secondary metabolite biosynthesis” were dramatically upregulated in subclusters 1, 3, 4, and 5, whereas DEGs related to “photosynthesis” were downregulated in subcluster 2 (Table S2). For the 1808 common DEGs in the untreated vs. ET-pretreated, L. theobromae-inoculated samples, eight transcriptional subclusters were defined (Fig. 3A and B). The upregulated DEGs were markedly enriched in “starch and sucrose metabolism”, “pentose and glucuronate interconversions”, and “amino or nucleotide sugar metabolism”, whereas the downregulated DEGs were enriched in “cutin, suberin and wax biosynthesis” and “fatty acid elongation” (Table S3).
Figure 3.
Overview of the transcriptome of peach shoots treated with ET and/or L. theobromae, as well as mock-inoculated shoots. In total, 1808 common differentially expressed genes (DEGs) were identified between the mock vs. L. theobromae-inoculated and L. theobromae-inoculated vs. ET + L. theobromae-treated shoot samples. (A) Temporal expression profiles of the common DEGs across different samples (false discovery rate < 0.05, absolute log2 fold change ≥1). In the heatmap, each row represents a gene, and each column represents the sample indicated. The colors of the heatmap cells indicate scaled gene expression levels across different samples. The color gradient ranging from blue to white to red represents low, medium, and high levels of gene expression. (B) Subclusters of 1808 DEGs across different samples. Each box corresponds to the expression of the module eigengene. Different colors of the centroid lines in B represent different gene expression patterns and correspond to subclusters identified in A by the colored bar on the left of the heatmap.
ET interacts with JA and SA during L. theobromae infection
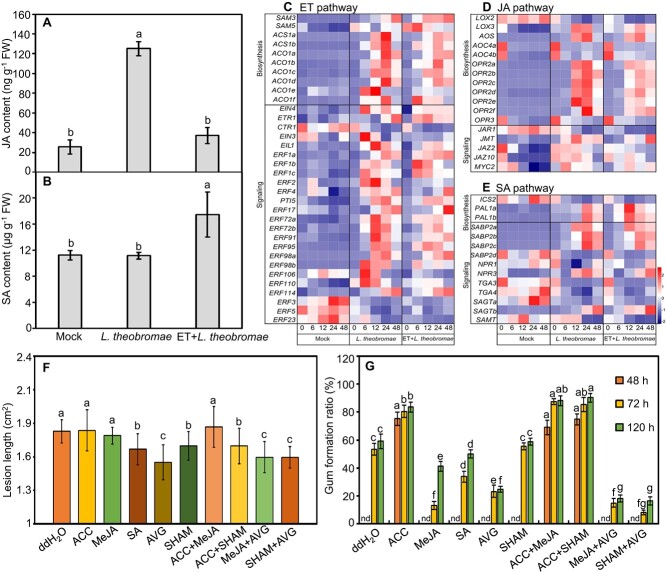
ET is known to interact with JA and SA to modulate the plant defense response against pathogen invasion [9]. Here, we quantified the metabolite levels and gene transcripts for SA and JA in L. theobromae-inoculated peach shoots that were either pretreated with ET or not (Fig. 4A-E). The JA content of inoculated shoots was 5.5-fold higher than that of the mock control; ET pretreatment significantly decreased JA to levels similar to those of the mock-inoculated control (Fig. 4A). In contrast, the SA content was not changed in response to L. theobromae inoculation alone relative to the mock control but was significantly enhanced if ET pretreatment was performed before inoculation (Fig. 4B).
Figure 4.
Metabolic and transcriptional changes in the hormones ethylene (ET), jasmonic acid (JA) and salicylic acid (SA) and their impact on L. theobromae-induced gummosis. (A and B) Contents of JA and SA in peach shoots at 12 hpi; (C-E) Expression profiles of genes related to ET, JA and SA biosynthesis and perception in different treatments. Different colors of heatmap cells indicate scaled expression levels of genes across different samples. Gene expression is color coded from blue (low) to white (medium) to red (high). (F and G) Lesion length and gum formation ratio in peach shoots treated with different chemicals for 24 hours before L. theobromae inoculation. ddH2O: the control; ACC: 1-aminocyclopropanecarboxylic acid, the ET precursor; MeJA: methyl-jasmonate; SA: salicylic acid; and SHAM: salicylhydroxamic acid, a JA biosynthesis inhibitor. nd indicates not detected. Values are the means ± SD of three biological replicates. Different letters on top of the bars indicate statistical significance between treatments at the same time points at P < 0.05.
As the heatmap analysis shows, all putative paralogs of genes encoding ACS and ACO (ACC oxidase), the ET core biosynthetic genes, were significantly upregulated at 12 and 24 hpi in inoculated shoots, irrespective of ET pretreatment (Fig. 4C). In the ET signal transduction cascade, the transcripts of ETR and EIN4 (ethylene receptor and ethylene insensitive4, respectively; both encoding endoplasmic reticulum-associated receptors) and of EIN3 and EIL1 were significantly upregulated in L. theobromae-inoculated shoots (Fig. 4C). The APETALA2 (AP2)/ethylene response factor (ERF) superfamily plays an important role in the biotic stress response [24]. The transcripts of most ERFs were upregulated, whereas the transcripts of ERF3, ERF5 and ERF23 were consistently downregulated in the inoculated shoots compared with the mock-inoculated shoots (Fig. 4C).
In ET-pretreated and inoculated tissues, the transcripts of the JA pathway-related genes OPR, JAR1, JAZ and MYC2 were decreased, whereas the transcripts of the SA biosynthetic gene PAL were sharply and significantly induced at 6 hpi and peaked earlier (at 12 hpi) than they did in L. theobromae-inoculated tissues (Fig. 4D and E). The L. theobromae-induced transcriptional activation of NPR3 (coding for nonexpresser of pathogenesis-related 3, a key SA signaling component) [25] was slightly delayed by ET pretreatment and was undetectable at 12 hpi (Fig. 4E). The relative transcript abundance of LOX2, OPR2e, JAR1, and PAL1a in ET inhibitor-treated and inoculated shoots showed an opposite profile to the ET-treated and inoculated samples (Fig. S6).
To further assess the effect of JA and SA on gummosis and thus the relevance of the effects of ET on their levels in terms of L. theobromae infection, the detached peach shoots were treated with different chemicals (ACC, MeJA, SA and/or their biosynthetic inhibitors) before L. theobromae inoculation. The lesion length of ACC (an ET biosynthetic precursor)- and/or MeJA-treated peach shoots was comparable to that of the control, whereas it was significantly decreased by SA treatment (Fig. 4F). Treatment with AVG and/or SHAM (salicylhydroxamic acid, a JA inhibitor) significantly repressed lesion expansion compared to untreated, L. theobromae-infected shoots (Fig. 4F). When peach shoots were treated with ACC for 24 hours before L. theobromae inoculation, gum was observed at 48 hpi but was still barely detectable in MeJA- or SA-treated shoots (Fig. 4G). In addition, the gum formation ratio was not affected in SHAM-treated peach shoots but was suppressed by AVG treatment, compared with L. theobromae-infected shoots (Fig. 4G).
ET participates in sugar metabolism in L. theobromae-infected shoots
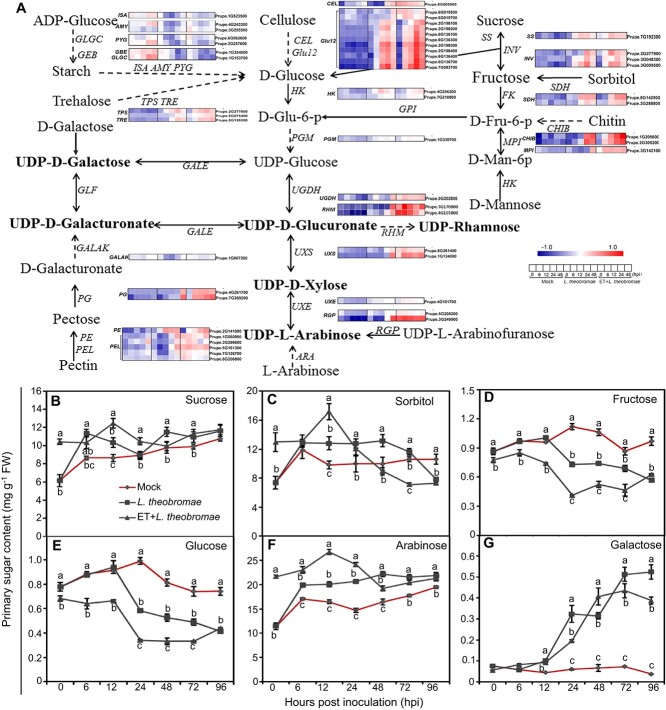
To understand the effect of ET on gum formation, transcripts of the DEGs involved in primary sugar metabolism were compared between L. theobromae-infected samples, either ET-pretreated or not, and represented via schematic illustration and heatmap insets (Fig. 5A). To this end, we identified 106 total DEGs related to UDP-sugar metabolism (32 DEGs), pentose and glucuronate interconversions (13 DEGs), and starch and sucrose metabolism (61 DEGs). Inoculated peach shoots consistently had higher transcripts of SS (encoding sucrose synthase) than the mock shoots from 6 hpi to the last time point, concomitant with the increase in sucrose content (Fig. 5A and B). Expression of INV genes (coding for invertases, which catabolize sucrose into glucose and fructose) was induced and peaked at 12 and 24 hpi in infected tissues, irrespective of ET pretreatment (Fig. 5A). Sucrose content was higher in ET-pretreated tissues at 0, 12 and 24 hpi, followed by a rapid decline, compared with nonpretreated, infected tissues (Fig. 5B). The sorbitol content in ET-pretreated, infected tissues was higher than that in L. theobromae-treated tissues at 0 and 12 hpi, but the transcripts of SDH genes (encoding sorbitol dehydrogenases) were comparable between L. theobromae and ET + L. theobromae treatments (Fig. 5A and C). In addition, both fructose and glucose levels rapidly decreased from 12 hpi onwards in infected samples, but their content in ET-pretreated tissues was significantly lower than that in nonpretreated tissues at all time points except 96 hpi (Fig. 5D and E). The contents of arabinose and galactose rapidly increased in peach shoots after inoculation, and particularly within 24 hpi, the arabinose content was much higher in ET-pretreated, inoculated tissues than in nonpretreated, inoculated tissues (Fig. 5F and G).
Figure 5.
Expression profiles of differentially expressed genes associated with the metabolism of primary sugars and sugar content changes in mock-treated, ET-treated and/or L. theobromae-infected shoots. (A) Schematic illustration of UDP-sugar, starch, and sucrose metabolism, as well as pentose and glucuronate interconversions. Gene transcript patterns are presented as heatmaps on the basis of log2(FPKM) values. (B-G) Primary sugar concentrations at the indicated time points in mock, L. theobromae-infected and L. theobromae-infected, ET-pretreated tissues. Data are the means ± SD of three independent replicates. Different letters represent significant differences in different treatments at the same time point at P < 0.05.
UDP sugars are the most important precursors for polysaccharide biosynthesis and are essential for cell wall and gum formation [26, 27]. The transcript levels of UXS, UXE, UGDH, RGP, MPI, and RHM (coding for UDP-glucuronate decarboxylase, UDP-arabinose 4-epimerase, UDP-glucose 6-dehydrogenase, reversibly glycosylated polypeptide, mannose-6-phosphate isomerase, and UDP-rhamnose synthase, respectively) were upregulated at 48 hpi in L. theobromae-inoculated samples and peaked earlier with ET pretreatment (Fig. 5A).
ET triggers sugar transport and cell wall degradation
Since sugars synthesized in healthy tissues are presumably translocated to sites of pathological changes for the biosynthesis of UDP-sugars, which are then transported to the Golgi lumen for polysaccharide formation [18, 28], we analyzed the transcript levels of sugar transporter-encoding genes (Fig. S7). As expected, one SUC (sucrose carrier-encoding gene), four STPs (sugar transport protein-encoding genes), three GONSTs (GDP-mannose transporter-encoding genes), one ERD6-like (sugar transporter ERD6-like gene), two URGTs (UDP-galactose transporter-encoding genes) and SWEET, a bidirectional sugar transporter, showed higher transcripts at 48 hpi in L. theobromae-inoculated samples compared with the mock control (Fig. S7). The transcript levels of GONSTs and URGTs (except for GONST1) were increased by ET pretreatment at 6 or 12 hpi (Fig. S7).
The plant cell wall is mainly composed of polysaccharides, which are crucial for physical defense [29], so we examined the expression of genes involved in cell wall degradation (Fig. 5 and S8). Transcripts of Glu12s and CEL, encoding the important cellulose degradation enzymes 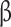 -glucosidase12s and cellulase, respectively, were significantly upregulated at 6, 12 and 24 hpi in ET-pretreated samples compared to merely infected samples (Fig. 5A). Transcripts of genes encoding the pectin degradation-related enzymes pectate lyase (PEL), polygalacturonase (PG), and pectinesterase (PE) peaked earlier in ET-pretreated than in nonpretreated inoculated samples (Fig. 5A). Moreover, the genes coding for expansins, endo-1,4-
-glucosidase12s and cellulase, respectively, were significantly upregulated at 6, 12 and 24 hpi in ET-pretreated samples compared to merely infected samples (Fig. 5A). Transcripts of genes encoding the pectin degradation-related enzymes pectate lyase (PEL), polygalacturonase (PG), and pectinesterase (PE) peaked earlier in ET-pretreated than in nonpretreated inoculated samples (Fig. 5A). Moreover, the genes coding for expansins, endo-1,4- -glucanase, galactosidase and xylosidase (Exps, EGases, Gal, and Xyl) had consistently and significantly higher transcripts at 0, 6 and 12 hpi in ET-pretreated, L. theobromae-infected samples compared with nonpretreated samples (Fig. S9).
-glucanase, galactosidase and xylosidase (Exps, EGases, Gal, and Xyl) had consistently and significantly higher transcripts at 0, 6 and 12 hpi in ET-pretreated, L. theobromae-infected samples compared with nonpretreated samples (Fig. S9).
Suppression of the ET pathway decreases the transcript levels of UDP sugar- and cell wall degradation-related genes
The transcripts of the UXS1a, UXE, MPI, RGP, UXS1b and UGDH genes involved in UDP-sugar biosynthesis were constantly and significantly upregulated by L. theobromae inoculation compared to the mock treatment, as expected (Fig. 6A-F). Furthermore, their transcripts were constantly and significantly repressed in AVG-treated tissues in relation to the corresponding nontreated tissues but were still significantly higher than those in the mock samples (except for MPI) (Fig. 6A-F). A similar transcriptional change was observed in 1-MCP-treated shoots. In AVG- or 1-MCP-treated samples, the expression of the cell wall-degrading genes PEL, PG and Glu12 was significantly downregulated relative to the corresponding L. theobromae treatment (Fig. 6G-I). These results indicate that the expression levels of UDP-sugar biosynthetic- and, to a greater degree, cell wall degradation-related genes positively correlate with the presence of ET.
Figure 6.
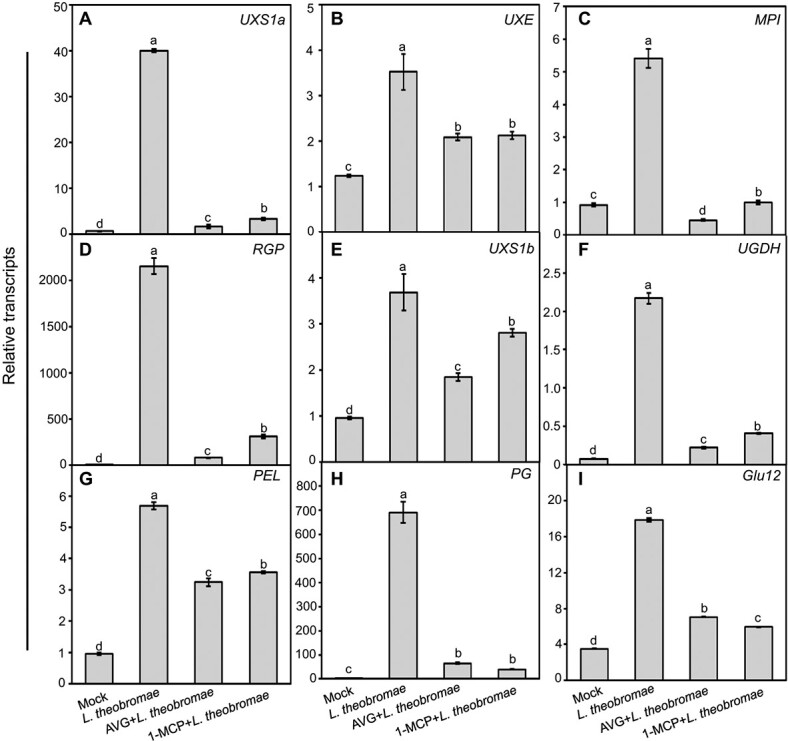
Impact of the ET inhibitor AVG or 1-MCP treatment on transcripts of genes related to UDP-sugar metabolism and cell wall degradation in L. theobromae-inoculated peach shoots. (A-F) Transcript levels of genes involved in UDP-sugar metabolism (UXS1a, UXE, MPI, RGP, UXS1b and UGDH). (G-I) Transcript abundance of genes related to cell wall degradation (PEL, PG and Glu12) in peach shoots treated with AVG or 1-MCP before L. theobromae inoculation. Relative transcripts of the tested genes were normalized to the transcript of the reference gene TEF2 and are displayed in relation to the transcript levels in mock samples at time 0 (which was therefore set to 1). Different letters on top of the bars indicate a statistically significant difference among treatments at P < 0.05.
EIL1 directly binds to the promoters of UDP-sugar metabolic genes and activates their transcription
We analyzed the promoters of UDP metabolism genes and found that these promoters contained putative EIN3/EIL1 binding sites (EBS, TACAT) [30] (Fig. 7A). As shown in Fig. 4C, the transcription of EIL1 was significantly upregulated in L. theobromae-infected peach shoots. Accordingly, we further examined whether EIL1 could activate UDP-sugar-related gene transcription. Reporter constructs for LUC expression (driven by the UXS1a, UXE, RGP or MPI promoter) and constructs for an effector fused with EIL1 were created for the firefly luciferase assay (Fig. 7A). Transient expression in fully expanded leaves of 4-week-old tobacco plants showed that the coexpression of the EIL1-containing effector and any of the reporter constructs significantly increased LUC activity compared with the control (Fig. 7B), indicating that EIL1 acts as a transcriptional activator of UXS1a, UXE, RGP and MPI.
Figure 7.
EIL1 targets the promoters of the UDP-sugar metabolic genes UXS1a, RGP, UXEand MPI. (A) Schematic diagram of effectors and reporters used in the luciferase (LUC) activity assay. Black rectangles represent the EIN3/EIL1 binding sites in the promoters of the test genes. The numbers below the fragments (UXS1a, RGP, UXE and MPI) indicate the positions of the nucleotides at the 5′-3′ end of each fragment relative to the translation start site in the reporter. (B) Transient expression assays showing that EIL1 activates transcription driven by the promoters of UXS1a, RGP, UXE and MPI. Luminescence imaging of Nicotiana tabacum leaves is shown 48 h after coinfiltration with reporter and effector construct pairs. (C) Schematic diagram of the prey and bait used in the Y1H assay. The numbers inside the fragments (UXS1a, RGP, UXE and MPI) indicate the positions of the nucleotides at the 5′-3′ end of each fragment relative to the translation start site in the bait. (D) Interaction of EIL1 with fragments of the UXS1a, RGP, UXE or MPI promoters. Transformed yeast cells containing both prey and bait were plated on selective medium (SD/-Ura-Leu/AbA). AbA, aureobasidin A. Cotransformation of pGADT7–53 and pAbAi–p53 was employed as a positive control, and cotransformation of pGADT7 and pAbAi-bait was employed as a negative control.
Furthermore, a Y1H assay was performed to test the physical interaction between EIL1 and the UXS1a, UXE, RGP and MPI promoters. The generation of the prey combining an activation domain (AD) fused with EIL1 (EIL1-AD) and of the baits containing the UXS1a, UXE, RGP and MPI promoters is schematically described in Fig. 7C. The prey EIL1-AD and baits of the tested sequences were cotransformed into Y1H gold yeast cells. The results showed that EIL1-AD dramatically activated AbA (aureobasidin A) resistance and that yeast cells grew well on the selective medium (SD/-Ura-Leu + 200 ng/ml AbA) (Fig. 7D).
Discussion
ET and JA enhance peach shoot susceptibility to L. theobromae infection
Phytohormones regulate plant development and play central roles in the sensing and signaling of diverse biotic and abiotic stresses and in the priming of the plant defense response [13, 31]. ET plays complicated roles in plant interactions with numerous pathogens, conferring either resistance or susceptibility to host plants depending on the pathogen lifestyle and plant genotypes [9]. High levels of ET increase the susceptibility of mature peach fruits to Monilinia laxa [32]. In wheat, suppression of the ET pathway attenuates resistance to hemibiotrophic Fusarium graminearum in resistant genotypes, whereas artificial applications of ET lower the susceptibility to infection in susceptible genotypes [11]. Based on our results, a schematic model (Fig. 8) is proposed for the role of ET in the peach defense response during L. theobromae infection, which significantly triggered the accumulation of ET and JA but not SA in the susceptible peach variety “Spring snow” (Figs. 1 and 4).
Figure 8.
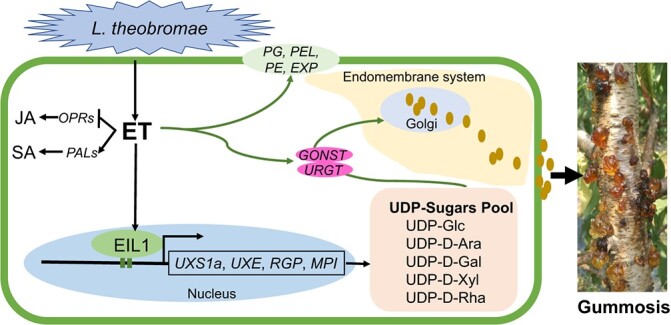
Proposed model for the role of ET in defense responses during L. theobromae-induced peach gummosis. The inoculation of L. theobromae onto peach shoots dramatically induces ET production. Exogenous ET represses the JA pathway by downregulating OPR expression but increases salicylic acid (SA) levels by upregulating PAL expression. On the other hand, ET treatment transcriptionally increases UDP-sugar metabolism, transport (GONST, URGT) and cell wall degradation (PG, PEL, PE, EXP), leading to excessive polysaccharide accumulation and cell damage due to gum outflow. EIL1, a key transcription factor of ethylene signaling, directly binds to promoters of the UDP-sugar biosynthetic genes UXS1a, UXE, RGP and MPI and transactivates their expression. Solid brown dots represent the gum biosynthesized in the Golgi being transported through the endomembrane system and out of the cells. The arrows and blunt-head lines indicate activation and suppression, respectively.
ET is a well-known modulator of plant immunity, either directly or via crosstalk with SA and JA networks, and activates specific and systemic defense responses [9]. Generally, SA mediates resistance against biotrophic and hemibiotrophic pathogens, whereas JA and ET act synergistically to enhance resistance to necrotrophs in dicots [33]. As L. theobromae is a hemibiotrophic pathogen [34], the plant defense response to it is poorly understood. Although ET, ACC and MeJA supply had no direct effect on lesion length, blocking the ET or JA pathways with their inhibitors (AVG, 1-MCP or SHAM) significantly decreased the lesion size upon L. theobromae infection (Figs. 2 and 4). It is possible that the use of a highly virulent isolate of L. theobromae and the susceptible peach variety “Spring snow” in our pathosystem resulted in a rapid expansion of necrotic lesions, which could not be further enhanced by ET, ACC and MeJA treatments. In addition, transcripts of PR-1a, PR-3, and PR-4 were higher, while the expression of cell wall degradation genes was repressed in shoots treated with ET inhibitors compared with those merely infected with L. theobromae (Figs. 6 and S9). For gum formation, several previous studies have shown that MeJA could induce gum formation in peach shoots [16, 35]. However, in our pathosystem, the MeJA inhibitor SHAM had no significant effect on gum formation, whereas both ET and ACC increased the gum formation ratio, irrespective of MeJA pretreatment (Figs. 2 and 4). These results demonstrate that ET plays a core role in gum formation, while both ET and JA positively regulate lesion expansion in peach fungal gummosis.
Pretreatment with exogenous ET also rapidly and markedly upregulated PAL transcripts and thus increased SA accumulation upon L. theobromae infection (Fig. 4B and E). It also downregulated the JA biosynthetic gene OPR and decreased JA levels (Figs. 4A, 4D, and S6). SA treatment decreased lesion length but not gum formation in infected peach shoots (Fig. 4F and G). Although ET supply facilitated SA accumulation, there were no significant differences between ET-treated and untreated inoculated peach shoots, but there was a slight decrease in lesion size in the treated set (Figs. 2 and 4). We speculate that ET itself could also positively promote lesion spread (Fig. 2), which could mask, to some degree, SA-induced resistance during the defense response of susceptible peach varieties against L. theobromae. These results demonstrated that both the ET and JA pathways could increase peach susceptibility to the hemibiotrophic fungal pathogen L. theobromae, while SA could promote resistance, consistent with previous studies [33]. Our results indicate that ET could participate in complex antagonistic and synergistic interactions with JA and SA biosynthesis, but the interplay and relative contribution of these hormones in peach defense responses to L. theobromae require further in-depth exploration.
ET boosts primary sugars flowing into UDP-sugar metabolism for gum polysaccharide synthesis during peach gummosis
The phytohormones ET and JA are essential factors known to induce gum formation in plants [15]. Both ethephon and MeJA could stimulate gum formation in tulip [15], sour cherry [36], culinary rhubarb [37], and peach [16, 22]. Additionally, ethephon, rather than MeJA, induced gum duct formation in the xylem adjacent to the cambium in Ailanthus excelsa, Cerasus tomentosa, P. mume and Liquidambar styraciflua [17]. Ethephon is widely used in agricultural practices as fruit ripening inducers and herbicides, but high concentrations of ethephon are phytotoxic to plants, and its decomposition products may cause nonspecific responses [23]. To advance our understanding of ET action while avoiding such potential pitfalls linked to ethephon, we used gaseous ET in our experimental system. Consistent with previous studies, ET treatment before L. theobromae inoculation accelerated and aggravated gummosis symptoms in the current work; this was nicely complemented by the effects of ET inhibitor treatment (Fig. 2). Metabolic analysis showed that sucrose and sorbitol accumulated around the lesion area at 12 hpi, and high sucrose levels were still detected at 72 hpi after the release of large gum amounts in L. theobromae-inoculated peach shoots (Fig. 5B and C). In the same samples, transcripts of SUC and STP transporter-encoding genes were upregulated (Fig. S7). This result suggests that high local sucrose could be due to existing sucrose being transported from healthy tissues to the lesion area and to new sucrose being synthesized via the upregulation of SS expression.
ET treatment increased the content of arabinose, which is a main component of heteropolysaccharides [19]; moreover, it significantly and sharply (at 24 hpi) reduced the contents of fructose and glucose, which could be broken down during respiration or converted into other sugars, such as UDP sugars and sucrose (Fig. 5). The formation of UDP sugars by different types of interconverting enzymes (such as epimerases, decarboxylases and dehydrogenases) is an important step for polysaccharide biosynthesis [20]. Herein, ET pretreatment induced transcripts of UDP-sugar metabolic genes such as RHM, UXS, UXE, RGP, PGM and UGDH [38, 39], and they peaked significantly earlier than in nonpretreated, L. theobromae-infected tissues (Fig. 5A), which was in accordance with the results from ET inhibitor treatment (Fig. 6A-G). Our results also demonstrated that EIL1 could directly bind to the promoter of the UDP-sugar biosynthetic genes UXS1a, UXE, RGP and MPI and thus activate their transcription (Fig. 7). As a result, ET could promote UDP-sugar metabolism through the action of EIL1, leading to high accumulation of gum polysaccharide precursors.
ET may facilitate the transformation of UDP sugars into gum polysaccharides
The plant cell wall is a complex structure composed of polysaccharides and plays an important role in protecting plants against pathogens [29]. A previous study showed that peach gum might be generated from the degradation of the cell wall in the periderm and vascular cambium within the lesion area [1]. Interestingly, however, gum was exuded from incisions 5 cm away from the inoculation site (as well as out of the lesion surrounding it) on the 4th day post L. theobromae inoculation in peach shoots [40], suggesting that polysaccharide synthesis underlying gum formation in L. theobromae-infected peach shoots may not be fueled by local cell wall degradation.
UDP-sugar metabolism and transport are responsible for the assembly of polysaccharides and for pectin biosynthesis during the remodeling of the cell wall structure, which is directly affected by changes in the UDP-sugar pools [28]. However, the cell wall structure has been shown to be seriously degraded, and related enzymes accumulate during peach gummosis progression [40]. In our study, ET application accelerated UDP-sugar metabolism around the lesion (Fig. 5) and shifted the transcript peak of genes encoding cell wall-degrading enzymes earlier relative to infection alone, an effect suppressed by ET inhibitor treatment (Figs. 6G-I and S8). The essential transporters URGT and GONST are responsible for importing UDP-sugar substrates into the Golgi lumen to synthesize polysaccharides [41], and ET pretreatment significantly increased the transcripts of the corresponding genes induced by L. theobromae infection (Fig. S7). Hence, based on our previously published data [18, 22] and the results of this work, we suggest that ET stimulates the metabolism and transport of UDP sugars that are then transformed into gum polysaccharides, in addition to accelerating cell wall degradation in the lesion area.
Conclusions
L. theobromae inoculation sharply and clearly triggered ET production in peach shoots. ET supply dramatically accelerated gum polysaccharide accumulation; consistently, ET inhibitor treatments slowed it down. A key transcription factor in the ET pathway, EIL1, could directly bind to the promoters of UDP-sugar metabolic genes and activate them; similarly, we propose that EIL1 may activate the expression of these same genes during L. theobromae infection. Blocking the ET pathway decreased the lesion size; thus, we suggest that this regulation may happen by the transcriptional downregulation of cell wall-degradation genes in inoculated peach shoots and reflects better infection containment. Furthermore, ET treatment altered JA and SA synthesis and possibly perception; this finding offers further mechanistic insight into the hormonal interplay, ultimately defining the severity of symptoms (tissue lesions, gum formation) upon L. theobromae infection. Our study provides evidence that ET acts as a negative modulator of symptom development during gummosis progression and provides information on the larger hormonal network regulating disease progression.
Materials and methods
Biological materials and growth conditions
The peach (P. persica L. Batsch) variety “Spring snow”, a genotype that is highly susceptible to L. theobromae [42], was grafted onto wild peach rootstocks and used in this study. The phenotype of orchard-grown plants during the growth or dormant period is shown in Fig. S10. Lignified, current-year shoots with a length of 50–60 cm and a diameter of 0.5 cm were selected from 4-year-old peach trees in the experimental orchard of Huazhong Agricultural University (Wuhan, China) between July and August 2017. The pathogen L. theobromae strain JMB-122 was previously isolated from a peach tree laden with gummosis in Hubei Province, China [2]. Prior to inoculation, L. theobromae was cultured on potato dextrose agar (PDA) medium at 28°C for 3 days under a 12 h dark/12 h light cycle (300 μmol s−1 m−2).
Experimental design and sample preparation
Inoculation was performed as described previously [40, 43], with minor modifications. In brief, current-year peach shoots were cut into 15-cm segments, and leaves were stripped off. After surface sterilization with 70% ethanol, shoot segments were wounded at the midpoint using sterilized needles, and then a single 5 mm-diameter agar plug covered in L. theobromae mycelium was placed onto the wound. In parallel, a sterile PDA plug was placed onto the wounded site as a mock control. The inoculated/mock shoot segments were separately placed in an upright position in 1800 mL plastic bottles that contained 200 mL of sterilized water, and the bottles were covered with transparent plastic film to maintain humidity. The water in the bottles was refreshed daily. Unless otherwise specified, bottles containing shoot segments were placed in a growth chamber at 28°C with 90% relative humidity and 12 h of light (300 μmol s−1 m−2).
To assess the hormonal effects on gummosis, the previously prepared peach shoots were pretreated with either 500 μM ACC (Sigma, MO, USA), 10 μL L−1 gaseous ET (Newradar gas, Wuhan, China), 15 ng mL−1 AVG (Sigma, MO, USA), 0.1 mg mL−1 1-MCP (Yuanye Biotechnology, Shanghai, China), 500 μM SA (Sigma, MO, USA), 100 μM MeJA (Sigma, MO, USA) or 500 μM SHAM (Sigma, MO, USA) before inoculation with L. theobromae. For each chemical treatment, 40 detached current-year shoots per replicate were placed in separate plastic bottles and thoroughly sprayed twice, 12 hours apart, with 5 mL of each aforementioned chemical solution (except for ET) and water (control). For ET treatment, 10 μL L−1 gaseous ET was injected into sealed plastic bottles containing 40 peach shoot segments per replicate for four times at 6-h intervals before L. theobromae inoculation. In parallel, AVG- or 1-MCP-pretreated peach shoots were inoculated with L. theobromae concomitantly with the injection of gaseous ET.
L. theobromae inoculation was set as the starting time point in all the treatments. Each treatment was run with three independent biological replicates. In each treatment, 15 shoot segments at every time point per replicate were pooled as one analytical sample. At harvest, the shoot tissues within 0.5–1.0 cm of the lesion were sampled from the different treatments at 0, 6, 12, 24, 48, 72 and 96 hpi. The shoot samples were immediately frozen in liquid nitrogen and stored at −80°C for physiological and molecular analysis.
For gummosis phenotyping, 40 individual peach shoots per replicate were used for gum formation ratio and lesion length measurement in each treatment as mentioned above. The peach shoots with visible gum were divided by total experimental shoots, which was defined as the gum formation ratio. At 96 hpi, the gum was collected and dried at 65°C until weight stabilization. Gum weight was calculated by dividing the total dry weight of gum by the fresh weight of the experimental shoots. The data are shown as the means of three independent biological replicates ± standard deviation (SD).
ET, JA and SA analysis
ET produced by inoculated peach shoots was measured as described previously [44] with minor changes. Forty inoculated shoot segments were enclosed in an airtight bottle as aforementioned at the indicated time points. A headspace gas sample (1 mL) was withdrawn using a syringe and then submitted to gas chromatography (GC; 7890B, Agilent, DE, USA) using a machine equipped with a DB-624 column (J & W GDX-502) and a flame ionization detector (FID) for ET analysis. The temperatures of the front inlet, FID, and column were set at 230, 180 and 90°C, respectively. Nitrogen and hydrogen at rates of 350 and 45 mL min−1, respectively, were used as carrier gases. ET was quantified using an external standard (2% ET in N2) and expressed as μL kg−1 h−1.
The contents of JA and SA were determined as described previously [45], with some modifications. Briefly, 0.1 g of shoot tissues was manually ground to fine powder in liquid nitrogen and extracted with 750 μL of cold extraction buffer (methanol: water: acetic acid, 80:19:1, v:v:v) containing the internal standards H2-JA (10 ng mL−1; Sigma, MO, USA) and d4-SA (10 ng mL−1; Yuanye Biotechnology, Shanghai, China). The mixture was incubated on a shaker at 4°C for 16 hours. After centrifugation at 12000 g for 15 min at 4°C, the supernatant was transferred to a new 1.5 mL tube, and the pellet was re-extracted with another 450 μL of extraction buffer without internal standards. The two supernatants were combined and passed through a 0.22 μm filter (Nylon 66; Jinteng experiment equipment, Tianjin, China). The filtrate was dried under nitrogen flow, and residues were dissolved in 200 μL methanol for ultrafast liquid chromatography combined with electrospray ionization tandem mass spectrometry (UFLC-ESI-MS/MS; Shimadzu Corporation, Kyoto, Japan) analysis. Samples were further diluted 200 times with methanol for SA analysis. Phytohormones were measured at each sampling point of each treatment, with three biological replicates.
Quantification of primary sugars
An analysis of primary sugars and nontargeted metabolite profiling were performed as previously reported [46], with minor modifications. Briefly, a 0.2 g powdered sample of fresh shoot tissues was extracted in a 5 mL glass tube with 2.7 mL of a mixture containing 2.4 mL of methanol and 0.3 mL of adonitol solution (0.2 mg mL−1 as an internal standard). The samples were vortexed and incubated at 70°C for 15 min and then cooled down to −20°C for 30 min. After centrifugation at 5000 g at 4°C for 15 min, the pellets were dried under speed vacuum and then incubated with 80 μL of 20 mg mL−1 methoxamine hydrochloride (Sigma, MO, USA) at 30°C for 1.5 h, and 80 μL of 1% N-methyl-N-(trimethylsilyl)-trifluoroacetamide (Sigma, MO, USA) was added to the mixture for another 30 min incubation. Samples were subjected to gas chromatography mass spectrometry (GC–MS; Thermo Fisher Scientific, MA, USA) using a machine coupled to a DSQ II mass spectrometer (Thermo Fisher Scientific, MA, USA) for sugar analysis. The primary sugars were identified by comparing their MS spectra with authentic reference standards or with those available in the National Institute of Standards and Technology library. All of the sugar quantifications were performed with three biological replicates.
RNA-seq and data processing
Total RNA of current-year peach shoots was extracted using the EASY Spin Plus RNA Kit (Aidlab, Beijing, China), along with DNase digestion columns, according to the manufacturer’s instructions. The quantity, quality and integrity of RNA samples were assessed using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, MA, USA) and an Agilent 2100 Bioanalyzer (Agilent, DE, USA). Total RNA samples from the mock, L. theobromae and ET + L. theobromae treatments at 5 time points (0, 6, 12, 24 and 48 hpi) in triplicate were used for RNA-seq with an Illumina HiSeq 4000 at Majorbio Biopharm Technology (Shanghai, China). The samples were prepared using the Illumina Truseq™ RNA Sample Preparation Kit. After sequencing, the raw reads containing adapters, poly N > 10%, or low-quality reads (> 50% of the bases with a quality value Q ≤ 5) were filtered to obtain clean reads. The GC content, Q20 and Q30 of the clean reads were calculated. The resulting clean reads were then aligned to the reference genome of P. persica [47] using the software TopHat2 and HISAT2, allowing two bp mismatches per read. Gene expression levels were calculated using the FPKM (fragments per kilobase per million reads) method as described previously [48]. Differential expression of genes in these samples was analyzed using DESeq2, and only those genes with log2 expression level changes ≥1-fold and false discovery ratios <0.05 for at least two time points between mock vs. L. theobromae and L. theobromae vs. ET + L. theobromae treatments were defined as differentially expressed genes (DEGs). The transcriptome data were deposited in the Sequence Read Archive of NCBI (Accession number: PRJNA687321).
cDNA synthesis and quantitative reverse-transcriptase PCR (qRT–PCR)
For qRT–PCR analysis, 0.8 μg of isolated total RNA was reverse transcribed into single-stranded DNA using the primer Script® RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer’s instructions. qRT-PCRs were set up in 10 μL volumes using Hieff™ qPCR SYBR Green Master Mix (Low Rox Plus, Yeasen, Shanghai, China) on a QuantStudio 6 Flex system (Applied Biosystems, MA, USA). The putative gene sequences used in this study were obtained from NCBI (http://www.ncbi.nlm.nih.gov/). Gene-specific primers were designed with the Primer3 Plus program (http://www.primer3plus.com) and synthesized by Tsingke Biological Technology (Wuhan, China). The primer sequences used for qRT–PCR are listed in Table S4. Thermocycling was conducted with the following settings: 95°C for 10 min, followed by 40 cycles at 95°C for 15 sec, 60°C for 20 sec and 72°C for 20 sec. The gene transcript levels were normalized with the corresponding values for translation elongation factor 2 (TEF2) transcripts as the internal standard in peach [21, 44]. The fold-change of transcript abundance was determined by the 2-ΔΔCT method [49]. The qRT–PCR results are shown as the means of three independent biological replicates and four technical replicates ± SD.
Firefly luciferase assay
The coding sequence (CDS) of EIL1 was inserted into pGreenII 62-SK to construct an effector, and promoter fragments of the target genes were fused into the pGreenII0800-LUC vector. The reporter and effector constructs, together with the pSoup19 vector, were transfected into Agrobacterium tumefaciens (GV3101). After culturing for 12–16 hours, the agrobacterium cultures harboring pGreenII-62-SK-EIL1 and pGreenII0800-promoter-LUC were resuspended and mixed at a 10:1 (v:v) ratio. The mixed resuspension was infiltrated into 4-week-old Nicotiana benthamiana leaves. In parallel, the agrobacterium culture mix harboring the pGreenII-62-SK vector and pGreenII0800-LUC promoter was used as a control. The experimental combinations and the corresponding controls were injected into the same leaf on the opposite sides of the main rib. Two days after coinfiltration, luciferase activity was measured using the VivoGloTM Luciferin, In Vivo Grade Kit (Promega, WI, USA) and imaged using a NightSHADE LB985 system (Berthold Technologies, Stuttgart, Germany).
Yeast one-hybrid (Y1H) assay
For the Y1H assay, the full-length CDS of EIL1 was cloned into the pGADT7 vector containing a GAL4 transcriptional activation domain to generate a prey. The promoter fragments of UXS1a, UXE, MPI and RGP containing the bait sequence were individually cloned into the pAbAi vector. After verification of the integration of bait vectors into the Y1H gold yeast strain (Shanghai Weidi Biotech, China), the prey vector was transformed into the strain. The cotransformed yeast cells were cultivated in SD/-Ura-Leu medium with or without AbA at 30°C for 3–4 d. The Y1H assay was performed according to the manufacturer’s protocol (Matchmaker One-Hybrid System; Clontech). Plates were incubated at 30°C for 3–5 days, after which yeast growth was assessed.
Statistical analysis
Statistical significance analysis (one-way ANOVA and Duncan’s post hoc test) was performed with the program SPSS13 (IBM, USA) for the gummosis phenotype, phytohormone, primary sugar and transcript abundance by qRT–PCR data. The transcriptome data were analyzed on the Majorbio I-Sanger Cloud Platform (www.i-sanger.com).
Supplementary Material
Acknowledgments
This work was financially supported by the National Key Research and Development Program [grant 2019YFD1000100] to JL, the Natural Science Foundation of China [grant No. 31471840] and the China Agriculture Research System of MOF and MARA [grant number CARS-30] to GL. The authors thank Prof Yongzhong Liu and Dr Jie Luo (Huazhong Agricultural University, China) and Dr Anqi Xing (Clemson University, USA) for critical reading of the manuscript.
Contributions
DZ, JY, JWL and GL designed the experiments; DZ performed experiments with occasional help from XS; DZ, XH, JY and JWL analyzed the data; and DZ, HH, JY, FC, JHL and JWL wrote the manuscript. All authors read and approved the final manuscript.
Data availability
The raw data of the sequenced samples were deposited to the NCBI under the accession number PRJNA687321. All data supporting this research result can be obtained within this paper and its Supplementary Materials online.
Conflict of interest
The authors declare no competing interests.
Supplementary data
Supplementary data is available at Horticulture Research Journal online.
References
- 1. Biggs AR. Presymptom histopathology of peach trees inoculated with Botryosphaeria obtusa and B. dothidea. Phytopathology. 1988;78:1109–18. [Google Scholar]
- 2. Wang F, Zhao L, Li G.et al. Identification and characterization of Botryosphaeria spp. causing gummosis of peach trees in Hubei Province, Central China. Plant Dis. 2011;95:1378–84. [DOI] [PubMed] [Google Scholar]
- 3. Adesemoye A, Mayorquin JS, Wang DH.et al. Identification of species of Botryosphaeriaceae causing bot gummosis in citrus in California. Plant Dis. 2014;98:55–61. [DOI] [PubMed] [Google Scholar]
- 4. Beckman TG, Pusey PL, Bertrand PF. Impact of fungal gummosis on peach trees. HortScience. 2003;38:1141–43. [Google Scholar]
- 5. Ezra D, Hershcovich M, Shtienberg D. Insights into the etiology of gummosis syndrome of deciduous fruit trees in Israel and its impact on tree productivity. Plant Dis. 2017;101:1354–61. [DOI] [PubMed] [Google Scholar]
- 6. Gao L, Zhang H, Cheng Y.et al. First report of Neofusicoccum parvum causing gummosis of peach trees in Hubei province, Central China. Plant Dis. 2019;103:2673–4. [Google Scholar]
- 7. Urbez-Torres JR, Gubler WD. Susceptibility of grapevine pruning wounds to infection by Lasiodiplodia theobromae and Neofusicoccum parvum. Plant Pathol. 2011;60:261–70. [Google Scholar]
- 8. Mancero-Castillo D, Beckman TG, Harmon PF.et al. A major locus for resistance to Botryosphaeria dothidea in Prunus. Tree Genet Genomes. 2018;14:26. [Google Scholar]
- 9. Broekgaarden C, Caarls L, Vos I.et al. Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiol. 2015;169:2371–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guan R, Su J, Meng X.et al. Multilayered regulation of ethylene induction plays a positive role in Arabidopsis resistance against pseudomonas syringae. Plant Physiol. 2015;169:299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Foroud NA, Pordel R, Goyal RK.et al. Chemical activation of the ethylene signaling pathway promotes resistance in detached wheat heads. Phytopathology. 2019;109:796–803. [DOI] [PubMed] [Google Scholar]
- 12. Chen HM, Xue L, Chintamanani S.et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress salicylic acid induction deficient2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell. 2009;21:2527–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casteel CL, De Alwis M, Bak A.et al. Disruption of ethylene responses by turnip mosaic virus mediates suppression of plant defense against the green peach aphid vector. Plant Physiol. 2015;169:209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miyamoto K, Saniewski M, Ueda J. Gummosis and leaf abscission in Yoshino cherry (Prunus yedoensis): relevance to hormonal regulation and chemical composition of gums. Acta Hortic. 2019;1235:467–74. [Google Scholar]
- 15. Miyamoto K, Kotake T, Boncela AJ.et al. Hormonal regulation of gummosis and composition of gums from bulbs of hyacinth (Hyacinthus orientalis). J Plant Physiol. 2015;174:1–4 [DOI] [PubMed] [Google Scholar]
- 16. Li M, Liu M, Peng F.et al. Influence factors and gene expression patterns during MeJA-induced gummosis in peach. J Plant Physiol. 2015;182:49–61. [DOI] [PubMed] [Google Scholar]
- 17. Carolina A, Kusumoto D. Gum duct formation mediated by various concentrations of ethephon and methyl jasmonate treatments in Cerasus x yedoensis, Prunus mume and Liquidambar styraciflua. IAWA J. 2020;41:98–108. [Google Scholar]
- 18. Li Z, Gao L, Wang YT.et al. Carbohydrate metabolism changes in Prunus persica gummosis infected with Lasiodiplodia theobromae. Phytopathology. 2014b;104:445. [DOI] [PubMed] [Google Scholar]
- 19. Simas FF, Gorin PAJ, Wagner R.et al. Comparison of structure of gum exudate polysaccharides from the trunk and fruit of the peach tree (Prunus persica). Carbohydr Polym. 2008;71:218–28. [Google Scholar]
- 20. Lazarowski ER, Harden TK. UDP-sugars as extracellular signaling molecules: cellular and physiologic consequences of P2Y(14) receptor activation. Mol Pharmacol. 2015;88:151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao L, Wang Y, Li Z.et al. Gene expression changes during the gummosis development of peach shoots in response to Lasiodiplodia theobromae infection using RNA-Seq. Front Physiol. 2016;7:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Z, Zhu W, Fan YC.et al. Effects of pre- and post-treatment with ethephon on gum formation of peach gummosis caused by Lasiodiplodia theobromae. Plant Pathol. 2014;63:1306–15. [Google Scholar]
- 23. Zhang W, Wen CK. Preparation of ethylene gas and comparison of ethylene responses induced by ethylene, ACC, and ethephon. Plant Physiol Biochem. 2010;48:45–53. [DOI] [PubMed] [Google Scholar]
- 24. Huang PY, Catinot J, Zimmerli L. Ethylene response factors in Arabidopsis immunity. J Exp Bot. 2016;67:1231–41. [DOI] [PubMed] [Google Scholar]
- 25. Liu Y, Sun T, Sun Y.et al. Diverse roles of the salicylic acid receptors NPR1 and NPR3/NPR4 in plant immunity. Plant Cell. 2020;32:4002–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kohlberger M, Thalhamer T, Weiss R.et al. Arabidopsis MAP-kinase 3 phosphorylates UDP-glucose dehydrogenase: a key enzyme providing UDP-sugar for cell wall biosynthesis. Plant Mol Biol Rep. 2018;36:870–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zablackis E, Huang J, Muller B.et al. Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 1995;107:1129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Caffall KH, Mohnen D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res. 2009;344:1879–900. [DOI] [PubMed] [Google Scholar]
- 29. Malinovsky FG, Fangel JU, Willats WGT. The role of the cell wall in plant immunity. Front Plant Sci. 2014;5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kosugi S, Ohashi Y. Cloning and DNA-binding properties of a tobacco ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res. 2000;28:960–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Han X, Kahmann R. Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front Plant Sci. 2019;10:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balsells-Llaurado M, Silva CJ, Usall J.et al. Depicting the battle between nectarine and Monilinia laxa: the fruit developmental stage dictates the effectiveness of the host defenses and the pathogen's infection strategies. Hortic Res. 2020;7:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol. 2005;43:205–27. [DOI] [PubMed] [Google Scholar]
- 34. Félix C, Meneses R, Goncalves MFM.et al. A multi-omics analysis of the grapevine pathogen Lasiodiplodia theobromae reveals that temperature affects the expression of virulence- and pathogenicity-related genes. Sci Rep. 2019;9:13144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saniewski M, Miyamoto K, Ueda J. Methyl jasmonate induces gums and stimulates anthocyanin accumulation in peach shoots. J Plant Growth Regul. 1998;17:121–4. [Google Scholar]
- 36. Olien WC, Bukovac MJ. Ethephon-induced gummosis in sour cherry (Prunus cerasus L.). I. Effect on xylem function and shoot water status. Plant Physiol. 1982;70:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goraj-Koniarska J, Saniewski M, Ueda J.et al. Effect of methyl jasmonate on gummosis in petioles of culinary rhubarb (Rheum rhabarbarum L.) and the determination of sugar composition of the gum. Acta Physiol Plant. 2018;40:8. [Google Scholar]
- 38. Seifert GJ. Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr Opin Plant Biol. 2004;7:277–84. [DOI] [PubMed] [Google Scholar]
- 39. Schaper S, Wendt H, Bamberger J.et al. A bifunctional UDP-sugar 4-epimerase supports biosynthesis of multiple cell surface polysaccharides in Sinorhizobium meliloti. J Bacteriol. 2019;201:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Z, Wang YT, Gao L.et al. Biochemical changes and defence responses during the development of peach gummosis caused by Lasiodiplodia theobromae. Eur J Plant Pathol. 2014a;138:195–207. [Google Scholar]
- 41. Parker JL, Newstead S. Structural basis of nucleotide sugar transport across the Golgi membrane. Nature. 2017;551:521–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yang W, Zhao L, Hao L.et al. Evaluating the susceptibility of six peach cultivars to gummosis. Journal of Huazhong Agricultural University. 2015;34:13–8. [Google Scholar]
- 43. Zhang H, Zhang D, Wang F.et al. Lasiodiplodia theobromae-induced alteration in ROS metabolism and its relation to gummosis development in Prunus persica. Plant Physiol Biochem. 2020;154:43–53. [DOI] [PubMed] [Google Scholar]
- 44. Zhang L, Kou X, Huang X.et al. Peach-gum: a promising alternative for retarding the ripening and senescence in postharvest peach fruit. Postharvest Biol Technol. 2020;161:111088. [Google Scholar]
- 45. Liu HB, Li XH, Xiao JH.et al. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: application in study of rice-bacterium interaction. Plant Methods. 2012;8:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yun Z, Gao H, Liu P.et al. Comparative proteomic and metabolomic profiling of citrus fruit with enhancement of disease resistance by postharvest heat treatment. BMC Plant Biol. 2013;13:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Verde I, Abbott AG, Scalabrin S.et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet. 2013;45:487–94. [DOI] [PubMed] [Google Scholar]
- 48. Mortazavi A, Williams BA, McCue K.et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. [DOI] [PubMed] [Google Scholar]
- 49. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods. 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of the sequenced samples were deposited to the NCBI under the accession number PRJNA687321. All data supporting this research result can be obtained within this paper and its Supplementary Materials online.