Abstract
RNAi is a gene-silencing mechanism conserved in the vast majority of eukaryotes. It is widely used to study gene function in animals due to the ease of eliciting gene knockdown. Beyond research applications, RNAi technology based on exogenous dsRNA is a promising candidate for next generation insect pest control. An advantage of using RNAi is that design of dsRNA essentially requires only the sequence of the target gene. The greatest challenge, however, is dsRNA delivery for large scale insect control. Delivery methods that have widely been used are oral, injection, or via soaking. Unfortunately, each insect presents its own challenges owing to the differences in the presence of dsRNA degrading enzymes, cellular uptake efficiency, expression of core RNAi machinery, the nature of the target gene, the concentration and persistence of the dsRNA, as well as the particular way of feeding of each insect, which together cause variations in the efficiency of RNAi. In this chapter, a protocol for the synthetic production of dsRNA is described along with three methods for delivery that have been successful in one of the more problematic insects, Diaphorina citri.
Keywords: RNAi, dsRNA design, dsRNA synthesis, feeding, topical-feeding, soaking, insect
1. Introduction
Interference RNA (RNAi) is a highly conserved cellular mechanism present in a vast majority of eukaryotes (1). This mechanism is triggered through the processing of long precursor double-strand RNA (dsRNA) to yield small RNA fragments which load into Argonaute (Ago) family proteins. Ago proteins loaded with small RNAs form the RNA induced silencing complex (RISC). In this complex, the small RNA serves as a specificity factor to direct the complex to target nucleic acid molecules through complementary base-pairing. This leads to either mRNA degradation or repression of translation (1, 2) (Fig. 1).
Figure 1. siRNA pathway.
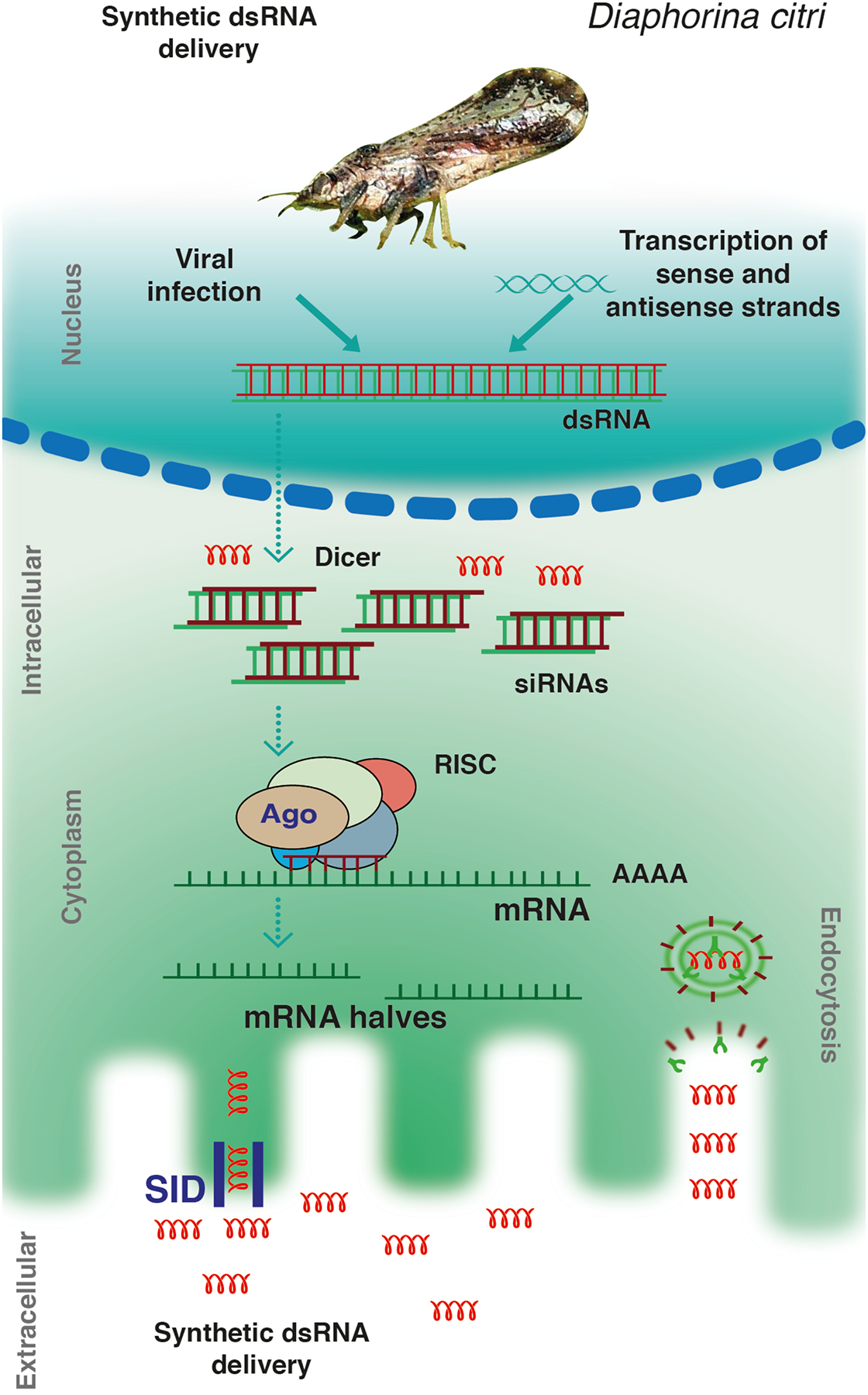
After the synthetic dsRNA taken up by the cell, long dsRNA is cleaved by a RNaseIII-enzyme called DICER into small interfering RNA (siRNA) molecules of 20–25 nucleotides long. Subsequently, these siRNA associate with an Argonaute protein to form the RNA-induced silencing complex (RISC), which then targets and destroys mRNA complementary to the siRNA in the RISC.
Since its discovery, RNAi has been used for studying gene function at the cellular and organismal levels (3–5). It has facilitated large scale functional genomics in human cells and in other model organism systems (6, 7). It is also being used in the clinic as a gene modulatory therapy through negative regulation of the expression of specific genes in cancerous cells (8). Furthermore, in order to try to expand its use, RNAi has been studied in infectious disease, signaling, and ageing (9–12). However, the only use of RNAi approved by FDA, so far, is Patisiran in amyloidosis (13). In non-model organisms where genetic techniques are not available, RNAi has been essential to enabling functional genomics (14).
In insects, RNAi has been increasingly emphasized as an approach for insect pest management due to its high specificity to target genes and therefore species, as well as the lack of environmental persistence. RNAi shows significant promise in combating coleopteran insects. Indeed, in 2017 the transgenic corn product SmartStax Pro, which in addition to 6 transgenes producing Bacillus thuringiensis (Bt) toxin incorporates Diabrotica virgifera Snf7 (DvSnf7) dsRNA. DvSnf7 protein is a class E vacuolar sorting protein, shown to be essential for transmembrane protein sorting. Transgenic maize produce DvSnf7 dsRNA which when consumed by D. virgifera and other corn rootworm species enters the RNAi pathway leading to down-regulation of the targeted DvSnf7 gene and death (15–19). This shows the promise of RNAi for pest management, which will only be realized through effective delivery methods.
Successful RNAi is dependent on design, synthesis and delivery of dsRNA (20). Typically, dsRNAs are between 300–600 bp in size with homology to coding regions of the target gene. Ideally, the sequence should contain no more than 19 nucleotides of contiguous homology with non-target genes within its own species and others, lest the dsRNA could be non-specific (15, 21–23). There are many options to produce dsRNA such as chemical synthesis for siRNAs which mimic products of DICER cleavage. Other strategies include expression in microbes or in crop cells via transgenes. In this chapter, in vitro synthesis based on T7 RNA polymerase kits is described (24, 25). Regardless of method of production, insects will likely encounter the molecules through diet or environmental exposure. In this chapter we discussed feeding strategies along with methods for topical delivery and dsRNA soaking.
Delivery of dsRNA requires maintenance of integrity during application to uptake by cells, so that it can ultimately be processed by the RNAi machinery (26). In cell lines assays, the dsRNA is typically introduced via endocytosis (27). In insects, the soaking delivery method has been widely used in S2 cell lines derived from Drosophila melanogaster embryos (28–31), or Sf21 cells derived from ovaries Spodoptera frugiperda (32). Better results of gene silencing were observed upon combining soaking dsRNA delivery with different transfection reagents in Sf21 cells as well as CiE1, a cell line derived from Chrysodeixis includens embryos (33, 34). Although the S12 cell derived from Spodoptera littoralis was also used for dsRNA uptake, no efficient silencing was observed in this lepidopteran cell nor in Bm5 cell line derived from Bombyx mori (35). Thus generally, insect cells readily uptake dsRNA and mount an RNAi response, however, this suggests in some cases additives may be required to fully elicit gene silencing.
In whole insects, dsRNA delivery methods such as injection, microinjection, oral delivery and soaking have been tested or applied (3, 36–39) (see Table 1). Injection of various insect species, such as Tribolium castaneum (40, 41), Locusta migratoria (42–45), and Blattella germanica (35, 46) showed success in down regulations of target genes. The advantages of injection include the immediate, direct delivery of a known amount and concentration of dsRNA into the insect body at various developmental stages or even to specific body parts (43) as well as avoidance of the structural barriers such as the integument, that prevent penetration of dsRNA. However, injection is more time-consuming and is sometimes challenging due to the small body size of some insects and is not applicable to control insect pests in the field.
Table 1.
List of genes targeted by RNAi and dsRNA delivery methods in different insect orders
| dsRNA Delivery Methods | ||||||
|---|---|---|---|---|---|---|
| Orden | Specie | Target | Oral | Injection/Soaking | Stage | Ref |
| Coleoptera | Tribolium castaneum | V-ATPase E | Artificial diet | Neonates | (49) | |
| Chewing-feed | Diabrotica virgifera | Multiple targets | Artificial diet | Neonates | (68) | |
| Virgifera | Snf 7 | Artificial diet | Neonates | (15) | ||
| Bol and vgr | Transgenic Plant | Larva/Adults | (69) | |||
| Leptinotarsa | Multiple targets | Leaf tissue | Neonates | (68) | ||
| decemlineata | V-ATPase A and E | TransgenicEcoli | 2nd Instar | (70) | ||
| Diabrotica | Snf 7 | Artificial diet | Neonates | (15) | ||
| undecimpuctata | V- ATPase A and E | Artificial diet | Neonates | (68) | ||
| howardi | α-Tubulin | Artificial diet | Neonates | (68) | ||
| Phyllotreta striolata | Arginine kinase | Leaf tissue | Adults | (71) | ||
| Diptera | Aedes aegypti | V-ATPase A Multiple targets | Artificial diet | Adults | (72) | |
| Water | 1st Instar | (50) | ||||
| Blood-nectar plant-insect juice Sucking-Feed |
ATP-dependent efflux | Water | 2nd Instar | (73) | ||
| Pump | ||||||
| Anhopeles gambiae | Chitin synthase 1, 2 | Artificial diet | 3rd Instar | (74) | ||
| Anopheles stephensi | 3-HKT | Transgenic algae | 1st Instar | (75) | ||
| Bactrocera dorsalis | Multiple targets | Transgenic E.coli | Adults | (76) | ||
| Glossina morsitans | Tsetse EP | Blood meal | Male Adults | (77) | ||
| Morsitans | Transferrin | Blood meal | Male Adults | (77) | ||
| Hemiptera | Acyrthosiphon pisum | Aquaporin | Artificial diet | 6-day-old nymphs | (78) | |
| V-ATPase E | Artificial diet | 1st Instar | (49) | |||
| Phloem/Xylem Sucking-feed | V-ATPase E | Artificial diet | Neonates | (79) | ||
| Hunchback (hb) | Artificial diet | Neonates | (80) | |||
| Salivary protein COO2 | Microinjection | Adults | (81) | |||
| Gut digestive enzyme cathepsin-L | Artificial diet | Microinjection | 1st, 3rd Instar | (82) | ||
| Structural sheath protein SHP | Microinjection | Adults | (83) | |||
| Angiotensin-converting enzymes ACE1 and ACE2 | Microinjection | Adults | (84) | |||
| Peroxiredoxin 1 gene ApPrxl | Microinjection | Adults | (85) | |||
| Macrophage migration inhibitory factor ApMIF1 | Microinjection | Adults | (86) | |||
| Calreticulin, Cathepsin-L | Microinjection | Adults | (87) | |||
| Myzus persicae | Rack1 and COO2 | Transgenic Plants | Adults | (88) | ||
| Aphis gossypii | Carboxylesterase geneCar | Artificial diet | Adults | (89) | ||
| Cytochrome P450 monooxygenase gene CYP6A2 | Artificial diet | 3rd instar, Adults | (90) | |||
| Odorant-binding protein 2 AgOBP2 | Artificial diet | Adults | (91) | |||
| Bactericera cockerelli | Multiple targets | Artificial diet | Adults | (92) | ||
| Bemisia tabaci | V-ATPase subunit A, rpL19 V-ATPase subunit A AChE and EcR |
Artificial diet | Adults | (93) | ||
| Transgenic Plants | Adults | (94) | ||||
| Transgenic Plants | Adults | (95) | ||||
| Nilaparvata lugens | Trehalose PO4 synthase | Artificial diet | 3rd instar | (74) | ||
| V-ATPase E | Artificial diet | 2nd Instar | (96) | |||
| Grain aphid | Catalase gene CAT | Artificial diet | 3rd instar | (97) | ||
| Acetylcholinesterase gene SaAce1 | Microinjection | Adults | (98) | |||
| Multiple targets | Artificial diet | 3rd instar | (99) | |||
| Multiple targets | Artificial diet | Adults | (100) | |||
| Olfactory coreceptor gene SaveOrco | Artificial diet | Adults | (90, 101) | |||
| Rhopalosiphum padi | Acetylcholinesterase gene RpAcel | Microinjection | Adult s | (98) | ||
| Schizaphis graminum | Salivary protein C002 | Artificial diet | Adults | (102) | ||
| Peregrinus maidis | V-ATPase B and D | Artificial diet | 3rd instar | (103) | ||
| Rhodnius prolixus | Nitrophorin 2 | Artificial diet | 2nd Instar | (104) | ||
| Lygus lineolaris | Inhibitor of apoptosis | Artificial diet | Neonates | (105) | ||
| Diaphorina citri | awd gene muscle protein 20 carboxyesterases CYP4 genes sucrose hydrolase boule homologue gene glutathione S-transferase transformer 2 homologue | Topical | 5th instar | (106) | ||
| Water | 4rd, 5th instar | (63) | ||||
| Topical | 4rd, 5th instar | (62) | ||||
| Topical | Adults | (107) | ||||
| Topical | 4–5th instar, Adults | (44) | ||||
| Artificial diet | Adults | (64) | ||||
| Artificial diet | Adults | (108) | ||||
| Artificial diet | Female Adults | (66) | ||||
| Hymenoptera | Apis mellifera | Vitellogenin | Natural diet | 2nd Instar | (109) | |
| Chewing and Sucking-Feed | Solenopsis invicta | PBAN/pyrokinin | Artificial diet | 4rd instar | (110) | |
| Guanine nucleotide binding GNBP | Artificial diet | Workers | (111) | |||
| Isoptera | Reticulitermes flavipes | Cellulase | Paper disc | Workers | (112) | |
| Chewing-Feed | Hexamerin | Paper disc | Workers | (112) | ||
| Lepidoptera | Chilo infuscatellus | CiHR3 molting factor | Corn kernels | 3rd instar | (113) | |
| Chewing and Sucking-Feed | Epiphyas postvittana | Carboxylesterase | Droplet | 3rd instar | (36) | |
| Pheromone bp | Droplet | 3rd instar | (36) | |||
| Helicoverpa armigera | AchE receptor | Artificial diet | Neonates | (114) | ||
| AchE receptor | Leaf tissue | Neonates | (114) | |||
| Ecdysone receptor EcR | Artificial diet | 3rd instar | (115) | |||
| HaHR3 molting factor | Artificial diet | 3rd instar | (116) | |||
| CYP6B6 | Artificial diet | 3rd instar | (26) | |||
| CYP6AE14 | Transgenic Plant | Adults | (117) | |||
| Ultraspiracle protein, EcR | Artificial diet | 3rd instar | (118) | |||
| Manduca sexta | V-ATPase E | Artificial diet | Neonates | (49) | ||
| Ostrinia nubilalis | Chitinase | Artificial diet | Neonates | (119) | ||
| Plutella xylostella | CYP6BG1 | Artificial diet | 4rd instar | (120) | ||
| Rieske protein | Artificial diet | 2nd Instar | (121) | |||
| AchE receptor | Artificial diet | 2nd Instar | (122) | |||
| Sesamia nonagrioides | JH esterase JHER | Droplet | 1st – 6th instars | (123) | ||
| Spodoptera exigua | Chitin synthase A β1 integrin subunit | Leaf tissue | Neonates | (124) | ||
| Leaf tissue | 4rd instar | (125) | ||||
| Spodoptera litura | Aminopeptidase N | Artificial diet (Ec) | Neonates | (126) | ||
| Bombix mori | atg gene bursicon gene | Injection | Larva | (127) | ||
| Injection | Pupae | (128) | ||||
| Spodoptera frugiperda | Allatostatin C | Droplet | Injection | 1st instar, larva | (129) | |
| Allototropin 2 | Droplet | 1st instar | (129) | |||
| SfT6 serine protease | Droplet | 4rd instar | (130) | |||
| Orthoptera | Gryllus bimaculatus | Sulfakinins | Droplet | Adults | (131) | |
| Chewing-Feed | Locusta migratoria | Multiple targets | Artificial diet | 4rd instar | (132) | |
| Schistocerca gregaria | Tubulin, GAPDH | Artificial diet | Adults | (133) | ||
Modified 744 of Kumar et al., 2018
In contrast, synthetic dsRNA can be introduced with food or artificial diet for oral delivery to insects, which has been applied with success in some lepidopterans (36, 47)), coleopterans (48), hemipterans (49) and dipterans (50). In contrast, neither artificial diet that was coated with dsRNA nor food that was mixed with dsRNA could induce RNAi in four Drosophila species (D. melanogaster, D. pseudoobscura, D. sechellia, D. yakuba) (49).
Two more ways of oral dsRNA delivery are dsRNA droplet-feeding and topical feeding (36, 38). Although the techniques are similar, the difference is that during droplet-feeding, the insects are placed in a petri dish with a drop so they may acquire the dsRNA when ingesting the drop. In topical-feeding, the insects are placed face up and the drop containing dsRNA is applied over their mouthparts. In both methods, the gene silencing effect has been observed in lepidopterans and hemipterans (36, 38). However, one disadvantage to the methods of oral delivery is that the dsRNA can be degraded by nucleases in the insect saliva, gut lumen or hemolymph preventing take up by cells. Several solutions have emerged to counteract these effects. One is that following identification of nucleases genes (dsRNases) in the insect of interest co-administer the dsRNA target to the dsRNases alongside dsRNA against the main target as seen in work with Bemisia tabaci (51). Feeding insects with a diet containing microorganism such as Escherichia coli and Saccharomyces cerevisiae or with endosymbionts that expressed the dsRNA of interest are strategies that protect dsRNA from endonucleases and improve oral delivery (20, 26). In transgenic plant feeding delivery a concern is dsRNA processing in plants cells, which hampers the effectiveness. One possible solution could be expression from chloroplasts (REF).
Soaking dsRNA delivery method may be the most convenient, especially for functional genomics in the laboratory. It was first reported in Caenorhabditis elegans that RNAi can be induced by soaking nematodes in dsRNA solution. Owing to the ease of this technique, soaking has been used in large-scale analysis of gene function by high-throughput RNAi (52, 53). On the other hand, delivery of dsRNA in whole insect bodies is possible, despite the extra barriers such as the insect cuticle, provided the development state of the insect is considered. For example, considerable mortality ranging between 40–70% correlated with downregulation of target gene expression was found after spray of dsRNA on newly hatched Ostrinia furnacalis larvae (54) when the cuticle was soft. On the other hand, spray chitosan or other nanocarrier-based dsRNA nanoparticles increase the stability of the dsRNA as well as cellular uptake in insects (20). More recently, guanidine-containing polymers, nanocarrier/dsRNA/detergent formulation, and branched amphiphilic peptide bilayer conjugated gold nanoparticles have been reported to protect dsRNA against nucleolytic degradation (55, 56), facilitate dsRNA penetration through the insect body wall (57), and likely improve the cellular uptake and endosomal escape of dsRNA (58).
The efficiency of all dsRNA delivery methods in insects, is subject to the presence of dsRNA degrading enzymes, cellular uptake efficiency, the expression of the core components of the RNAi machinery, the nature of the target gene, the concentration and persistent of the dsRNA, as well as the feeding behavior of each insect. For these reasons, it is common to observe variations in the effectiveness of RNAi and even to find no effect whatsoever. In this sense in depth study of RNAi pathways biogenesis in insects may help to identify the most effective mechanism to generate superior methodologies for the control of insect pests.
In this chapter, two enzymatic synthesis methods for obtaining dsRNA and three delivery methods are described that have been successful in one of the most challenging insects, Diaphorina citri, a major pest devastating the citrus industry (59–61). Its manner of feeding and size make successful delivery of dsRNA to D. citri difficult. The three methods for delivering dsRNA are: feeding, topical-feeding and soaking. Each is able to elicit silencing of target genes, leading to lifespan reduction and high mortality (62–66). A part of success in D. citri may be due to insufficient dsRNase activity in the digestive tract and hemolymph as well as efficient cell uptake of dsRNA (67).
2. Materials
-
1.
Acid-Phenol-Chloroform, 5:1 solution, pH 4.5+/−0.2 (99%) Ambion.
-
2.
Ethanol 70 and 100 % (v/v).
-
3.
2-Propanol, Isopropanol, Fisher Scientific.
-
4.
D-Sucrose, biological molecular grade, Fisher Scientific.
-
5.
Q5 high-fidelity DNA Polymerase (New England, BioLabs).
-
6.
Phire Hot Start II DNA Polymerase (Thermo Fisher).
-
7.
gBlock Gene Fragment. Integrated DNA Technologies (IDT) company.
-
8.
Primers. Integrated DNA Technologies (IDT).
-
9.
QIAquick PCR Purification Kit (QIAGEN). Supplied with the Kit: pH indicator, buffer PE, buffer PB, Filter Cartridge and collection tubes.
-
10.
MEGAScript RNAi Kit (Ambion). Supplied with the Kit: T7 Enzyme, 10X T7 Reaction Buffer, ATP, GTP, CTP, UTP Solution, 10X Binding Buffer, Elution Solution, RNase, DNase I, 10X Digestion Buffer, 2X wash solution, Nuclease free water, Filter Cartridge and collection tubes.
-
11.
MEGAScript Kit (Thermo Fisher). Supplied with the Kit: T7 Enzyme, 10X T7 Reaction Buffer, ATP, GTP, CTP, UTP Solution, 10X Binding Buffer, Elution Solution, RNase, DNase I, 10X Digestion Buffer and 2X wash solution, Nuclease free water, ammonium acetate stop solution, TURBO DNase.
-
12.
Eppendorf Mastercylcer Pro 6321 PCR Thermal Cycler w/vapo protect lid.
-
13.
Sorvall Legend Micro 21R Centrifuge, Thermo Scientific.
-
14.
Growth room.
-
15.
Insect aspirator # 654135, CAROLINA.
-
16.
Carbon dioxide (CO2).
-
17.
Parafilm.
-
18.
Distillate water.
-
19.
Plants of Citrus machrophyla (small size).
-
20.
Rearing cages (12 × 12 × 12 inches).
-
22.
Petri dishes (100 × 15 mm).
-
23.
Stereoscope.
-
24.
Concave slides.
-
25.
2115 Economical Camel Hair Touch Up Brush size 2.
-
26.
Filter paper 320 mm (Whatman).
-
27.
Pipet of 200 μl.
-
28.
Small Berlese funnel trap.
-
29.
10 μl Hamilton syringe.
-
30.
PB600 Dispenser.
-
31.
DNase-RNase free-water.
3. Methods
D. citri belongs to the order Hemiptera, which feed exclusively on sucrose rich plant sap (phloem) through the introduction of its stylet. To avoid damaging the stylet before removing insects from Citrus macrophyla, the plants must first be shaken. This provokes the insects to pull their stylet back so as not to break it during the aspiration. In this way, the feeding structure remains intact. In the case of nymphs, they can be gently touched with a paint brush before being carefully removed from the plants. The identification of the 4–5 instar is done under stereoscope. For each methodology, the insects were kept without food for 3 hours before dsRNA delivery and were kept in temperature-controlled growth rooms set at 25 ± 3°C, 60 % ± 5 % relative humidity (RH), and with a 16:8 (light/dark) photoperiod.
3.1. dsRNA: enzymatic synthesis from gBlock Gene Fragment.
Use NCBI database to identify the Open Reading Frame (ORF) of the gene of interest.
Copy the ORF sequences in Primer Plus 3 free on-line software to pick up the primers from the ORF sequences (see Note 1).
Run a Blast of the DNA sequence of the target against the genome of the insect that you want to avoid as target.
Do a second pair of primers using the sequences of the primers already designed and add the T7 promotor sequence (see Note 2)
Order the gBlock Gene Fragment (see Note 3).
- Re-suspend the gBlock following the instructions from the IDT and do two PCR reaction using the primers with the T7 attached:
-
6.1Prepare the reaction:
Component uL Concentration Q5 high-fidelity 2X 25 1 X Forward Primer T7 X 2.5 0.5 μM Reverse Primer T7 X 2.5 0.5 μM Template DNA (gBlock) 2 <1.000 ng DNase-RNase free-water 18 TOTAL VOL 50 Gently mix the reaction and run the PCR. -
6.2Run an agarose gel in order to check the integrity of the DNA. Then mix the two PCR reactions
-
6.1
- Purify the PCR using QIAquick Kit
-
7.1Mix (1:250 volume) pH indicator I and Buffer PB. Add 5 vol of this mix at 1 vol of PCR.
-
7.2Transfer the mix to the column QIAquick (load in the center) to bind the DNA and centrifuge at 8,944 × g for 1 min. Discard flow-through and re-used the tube
-
7.3Add 750 μL of Buffer PE at the column to wash and centrifuge and repeat twice.
-
7.4Transfer the column to a clean 1.5 mL microcentrifuge tube, add 30 μL of DNase-RNase free-water to elute DNA and centrifuge. (see Note 4)
-
7.5Read on nanodrop and do an agarose gel in order to get the quantity and quality of the DNA.
-
7.1
- Synthesize the dsRNA using MEGAScript RNAi Kit.
-
8.1Prepare the in vitro transcription reaction. (X: uL necessary to reach 2 μg or 20 μL)
Component μL PCR + gBlock (2 μg) X 10x T7 Reaction Buffer 2 ATP Solution 2 CTP Solution 2 GTP Solution 2 UTP Solution 2 T7 Enzyme Mix 2 Nuclease-Free Water X TOTAL VOL 20 μL -
8.2Incubate at 37°C overnight (~16 hrs).
-
8.3Transfer the reaction into thermocycler and heat it at 75°C for 5 min. Then cool to room temperature for around 15 min (see Note 5).
-
8.4Prepare the following reaction on ice and incubate at 37°C for 1 hr (do not continue this incubation for longer than 2 hrs):
Component μL dsRNA reaction 20 Nuclease-free water 21 10X Digestion buffer 5 DNase I 2 RNase 2 TOTAL VOL 50 μL -
8.5Purify of dsRNA
Component μL dsRNA reaction from Step 4 50 10X Binding buffer 50 Nuclease-free water 150 100% Ethanol 250 Total Volume 500 μL Gently mix the reaction by pipetting up and down. -
8.6Transfer the dsRNA reaction into the Filter Cartridge and collection tubes. Centrifuge at 12,878 × g for 2 min
-
8.7Wash the Filter twice with 500 μL of 2X wash solution and centrifuge (12,878 × g for 2 min)
-
8.8Centrifuge one more time. Then transfer the filter into the new collection tube.
-
8.9Add 50 – 100 μL of hot Elution Solution into the Filter and incubate at Room Temperature for 10 min. Then centrifuge 12,878 × g for 3 min and read in nanodrop. Do an agarose gel in order to check the dsRNA integrity (see Note 6).
-
8.1
3.2. dsRNA: enzymatic synthesis from genomic DNA.
Use NCBI database to select a region exon or transcript in the Genome Browser Viewer.
Copy the sequence in the Plasmid Editor APE (https://jorgensen.biology.utah.edu/wayned/ape/), for design Forward and Reverse Primers to have fragment size between 300–600 bp and add the T7 promotor sequence to each primer.
- Amplify through PCR the target fragment using the primers previously designed and the genomic DNA extracted from D. virgifera.
-
3.1Prepare the following reaction. (X: uL necessary to reach 25 μL)
Component μL Final Concentration 5X Phire Reaction Buffer 4 1x 10mM dNTPs 0.4 200 μM each T7 Forward Primer 1 0.5 μM T7 Reverse Primer 1 0.5 μM gDNA 50ng DNase-RNase free water X Phire Hot Start II enzyme 0.4 DNA Polymerase TOTAL VOL 25 Check the PCR through agarose gel
-
3.1
- Synthesize the dsRNA using MEGAScript Kit. Use directly the PCR.
-
4.1Prepare the in vitro transcription reaction. (X: uL necessary to reach 2 μg or 20 μL)
Compound μL T7 10x Reaction buffer 2 T7 ATP solution 2 T7 CTP solution 2 T7 GTP solution 2 T7 UTP solution 2 DNA template (PCR: 2μg) X Nuclease free water. X T7 Enzyme Mix 2 TOTAL VOL 20 -
4.2Incubate the reaction for 4 hrs. at 37°C.
-
4.1
- Clean up the reaction:
-
5.1Add 1μL of TURBO DNase to the in vitro transcription reaction (20 μL) and incubate the reaction at 37°C for 10 min
-
5.2Add 20 μL of ammonium acetate stop solution and 100 μL of DNase-RNase free water
-
5.3Add 200 μL 99% of Acid-Phenol-Chloroform, take aqueous part and transfer in new tube
-
5.4Add 600 μL of isopropanol and chill for 20 min at 80°C
-
5.5Speed at 17,530 × g for 15 min and remove supernatant
-
5.6Wash three times with 70% of Ethanol
-
5.7Spin down at 17,530 × g for 10 min and discard the supernatant
-
5.8Let the sample dry and resuspend in 20 μL of DNase-RNase free-water
-
5.9Read concentration in nanodrop and do an agarose gel in order to check the integrity of the dsRNA.
-
5.1
3.3. Feeding dsRNA delivery in adults (Fig. 2A)
Figure 2. dsRNA delivery methods in Diaphorina citri.
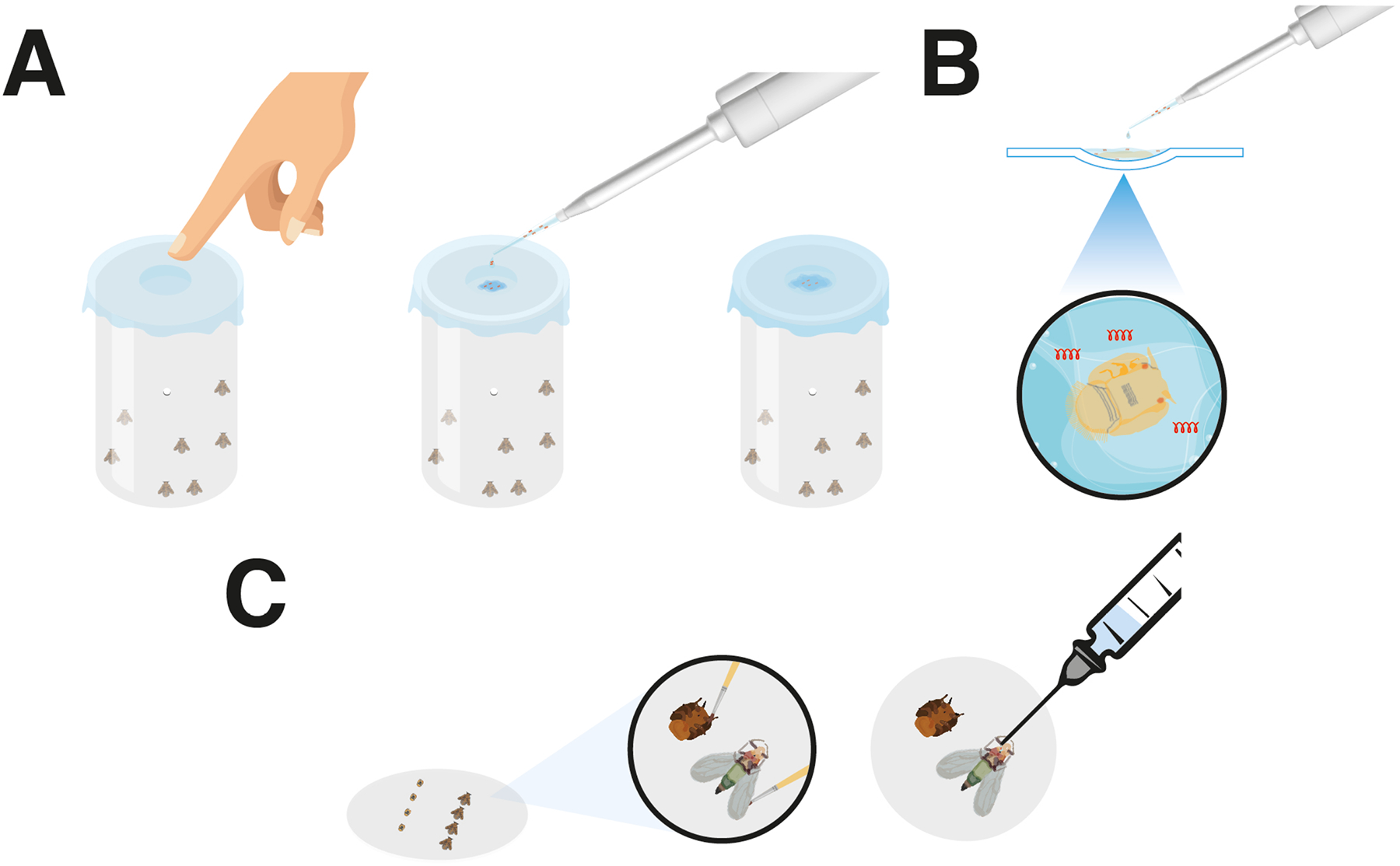
A) Artificial diet-feeding, B) Soaking and C) Topical-feeding.
Collect 30 insects from their colonies with the aspirator. The aspirator come with small cylindrical containers, each with a hole bored into each side so that oxygen may enter throughout the ingestion period for dsRNA.
Prepare the artificial diet which consists of 20 % (w:v) D-sucrose mixed with dsRNA. Use DNase-RNase free water to prepare the solution of sucrose.
Plug the holes in the container with hand and add a little CO2 in order to immobilize the insects for a couple of minutes and thus prevent them from flying out when the lid is opened.
Stretch a layer of parafilm over the mouth of the container and create a slight depression in this cover by pressing down with the thumb in the center.
Add 150 μl of dsRNA into the depression and place a second layer of well-stretched parafilm on top.
Return the insects with the container to the growth room, and they will feed from the dsRNA solution.
Change the dsRNA every 24 hrs. for three days. Remove only the second layer to add a new 150 μl of dsRNA (see Note 7).
Transfer the adults of D. citri to rearing cages in the growth room that contain plants with small sized C. macrophylia (see Note 8)
3.4. Soaking dsRNA delivery in Nymphs in 4–5th instar (Fig. 2B)
Remove nymphs in 4–5 instar from leaves of the plants and maintain them in petri dishes.
Prepare the dsRNA with DNase-RNase free-water at the desired concentration.
Place 4 nymphs in the concave slide under stereoscope and immediately add 50 μl of dsRNA with pipet. Wait 20 min (see Note 9).
Remove all the excess solution and put it in a new Eppendorf tube. Put the filter paper in the floor of the petri dish and transfer the nymphs with camel hair brush and cover it (see Note 10).
Place 4 more nymphs on the same slide and add the excess solution of dsRNA reserved in the Eppendorf tube. Add the quantity of solution necessary to again reach 50 μl (see Note 11).
Transfer the nymphs with the camel hair brush to a petri dish furnished with tender C. macrophylla leaves to feed nymphs. Make sure the leaves are placed face down (see Note 12).
Cover the petri dish with well-stretched parafilm. Use a pin to make small holes in the parafilm.
Repeat the application of dsRNA after 24 hrs.
Transfer the nymphs with camel hair brush to the leaves and stems of the C. macrophylla plant and place on a small Berlese funnel trap. Transfer the trap to the growth room (see Note 13).
3.5. Topical-feeding dsRNA delivery in Nymphs in 4–5 instar and adults (Fig 2C)
Prepare the solution of dsRNA in DNase-RNase free water.
Place the filter paper under the stereoscope and moisten it with distilled water using a camel hair brush (see Note 14).
Place the nymphs onto the filter paper in ventral position with the help of the camel hair brush. The adults must first be immobilized with CO2 and then placed face up with the brush.
Moisten the nymphs passing the camel hair brush several times over the bodies gently. Adults must be principally moistened on the wings (see Note 15).
Place lines of ten nymphs for more control. Adults should be placed in lines of five (see Note 16).
Have the syringe ready with the dispenser calibrated to apply 0.2 μl of solution of dsRNA.
Press the button of the dispenser to form a drop on the tip of the needle.
Gently bring the drop to the mouth of the insect and just touch it (see Note 17).
Keep the insect in that position feeding on the drop for 20 minutes (see Note 18).
Transfer the nymphs and the adults on the leaves with the help of the camel hair brush and close the small Berlese funnel trap. Transfer the trap to the growth room (see Note 19).
4. Notes
The final fragment should be around between 300–500 bp which is the gBlock. (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) this is the page of Primer Plus 3 free on-line software.
This pair of primers will use to amplify the gBlock with the T7 promotor through PCR.
https://www.idtdna.com/pages/products/genes-and-gene-fragments/double-stranded-dna-fragments/gblocks-gene-fragments this is the page to get all the information of the company IDT. Following the indications of the company (IDT) send the information of DNA sequence target for synthetized the gBlock Fragments and primers. The cost depends on the size of the fragments.
One recommendation is elute the DNA with DNase-RNase free-water.
Never let it cool down on ice.
In this step, it is necessary to heat at 95°C in the thermocycler the Elution Solution and apply it hot in the center of the column.
The insects will stick their stylet through the parafilm to ingest the artificial diet of dsRNA. The layer of parafilm mimics the leaf and, given that stylet is in the ventral part, they must be placed face up to feed. When changing the dsRNA, it is possible to see small bubbles between the layers of the parafilm, which indicates that the insects have stuck their stylets through there.
From this moment, it is possible to consider doing survival studies a/o evaluate the silencing of the gene of interest.
The nymphs will try to escape rapidly. To keep them in the solution, it’s important to push them below the surface with the camel hair brush.
Place a weight on the lid of the petri dish, because the nymphs are very active and will try to get out.
Repeat this procedure until the number of desired insects is reached in each repetition.
It is very important that the leaves be soft and in very good state so that they may hold up for 13 hours. If they are dry, the insects will die during this period.
Funnel trap is removed to place the small plants contained in standard round pots. Reinforce with fine screen the airway in the upper part. This will prevent the nymphs or some emerged adult from escaping during the treatment.
It’s important to maintain the paper filter moist during the application time.
It’s important to moisten the bodies of the nymphs and the wings of the adults during the application because this, together with the moistened paper filter, keeps them adhered to the paper filter.
With a little practice, it is possible to place up to 50 nymphs in rows of 25 and quickly apply the drop to each one. With adults, as many as 10 is manageable, while always keeping the wings moist. Otherwise, they dry, the insects flip over, and they fly away.
The mouth in both nymphs and adults is easily identifiable under stereoscope due to its appearance as a black beak. Moistening the insect just before applying the drop helps to avoid lifting the insect with the attraction effect
In some cases, the insects turn over or the drop gets dissipated by the moisture already in the paper filter. If this happens, place the insect in the ventral position and place the drop again.
It is advisable to not apply the dsRNA to nymphs and adults at the same time.
Acknowledgement:
We acknowledge Graphic designer Cuauhtémoc Moises Hernandez Castelán for his help in the graphics. YSO and ASF are supported by NSF MCB 1845978
References
- 1.Carthew RW and Sontheimer EJ (2009) Origins and Mechanisms of miRNAs and siRNAs. Cell 136:642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Großhans H and Filipowicz W (2008) Molecular biology: The expanding world of small RNAs. Nature 451:414–416 [DOI] [PubMed] [Google Scholar]
- 3.Nandety RS, Kuo YW, Nouri S, et al. (2014) Emerging strategies for RNA interference (RNAI) applications in insects. Bioeng Bugs 6:8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Amar A, Daldoul S, Reustle GM, et al. (2016) Reverse Genetics and High Throughput Sequencing Methodologies for Plant Functional Genomics. Curr Genomics 17:460–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki T, Nunes MA, España MU, et al. (2017) RNAi-based reverse genetics in the chelicerate model Tetranychus urticae: A comparative analysis of five methods for gene silencing. PLoS One 12:1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elbashir S, Harborth J, Lendeckel W, et al. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–498 [DOI] [PubMed] [Google Scholar]
- 7.Andrews OE, Cha DJ, Wei C, et al. (2014) RNAi-mediated gene silencing in zebrafish triggered by convergent transcription. Sci Rep 4:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conde J, Bao C, Tan Y, et al. (2015) Dual Targeted Immunotherapy via In Vivo Delivery of Biohybrid RNAi-Peptide Nanoparticles to Tumor-Associated Macrophages and Cancer Cells. Adv Funct Mater 25:4183–4194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seyhan AA and Ryan TE (2010) RNAi Screening for the Discovery of Novel Modulators of Human Disease. Curr Pharm Biotechnol 11:735–756 [DOI] [PubMed] [Google Scholar]
- 10.Wolters NM and MacKeigan JP (2008) From sequence to function: Using RNAi to elucidate mechanisms of human disease. Cell Death Differ 15:809–819 [DOI] [PubMed] [Google Scholar]
- 11.Ni Z and Lee SS (2010) RNAi screens to identify components of gene networks that modulate aging in Caenorhabditis elegans. Briefings Funct Genomics Proteomics 9:53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cherry S (2009), What have RNAi screens taught us about viral-host interactions?. Curr Opin Microbiol 12(4):446–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kristen AV, Ajroud-Driss S, Conceição I, et al. (2019) Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener Dis Manag 9:5–23 [DOI] [PubMed] [Google Scholar]
- 14.Boettcher M and McManus MT (2015) Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Mol Cell 58:575–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolognesi R, Ramaseshadri P, Anderson J, et al. (2012) Characterizing the Mechanism of Action of Double-Stranded RNA Activity against Western Corn Rootworm (Diabrotica virgifera virgifera LeConte). PLoS One 7(10):1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Head G, Campbell LA, Carroll M, et al. (2014) Movement and survival of corn rootworm in seed mixtures of SmartStax® insect-protected corn. Crop Prot 58:14–24 [Google Scholar]
- 17.Head GP, Carroll M, Evans S, et al. (2017) Evaluation of SmartStax and SmartStax PRO against WCR and NCR. Pest Manag Sci 73:1883–1899 [DOI] [PubMed] [Google Scholar]
- 18.Khajuria C, Ivashuta S, Wiggins E, et al. (2018) Development and characterization of the first dsRNA-resistant insect population from western corn rootworm, Diabrotica virgifera virgifera LeConte. 13(5):1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Epa US and Programs P (2017) US EPA, Pesticide Product Label, MON 89034 X TC1507 X MON 87411 X DAS-59122–7,06/08/2017.
- 20.Zhu KY and Palli SR (2020) Mechanisms, applications, and challenges of insect RNA interference. Annu Rev Entomol 65:293–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachman PM, Bolognesi R, Moar WJ, et al. (2013) Characterization of the spectrum of insecticidal activity of a double-stranded RNA with targeted activity against Western Corn Rootworm (Diabrotica virgifera virgifera LeConte). Transgenic Res 22:1207–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christiaens O, Swevers L, and Smagghe G (2014) DsRNA degradation in the pea aphid (Acyrthosiphon pisum) associated with lack of response in RNAi feeding and injection assay. Peptides 53:307–314 [DOI] [PubMed] [Google Scholar]
- 23.Christiaens O, Whyard S, Vélez AM, et al. (2020) Double-Stranded RNA Technology to Control Insect Pests: Current Status and Challenges. Front Plant Sci 11:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.La Fauce K and Owens L (2012) RNA interference with special reference to combating viruses of crustacea. Indian J Virol 23:226–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganbaatar O, Cao B, Zhang Y, et al. (2017) Knockdown of Mythimna separata chitinase genes via bacterial expression and oral delivery of RNAi effectors. BMC Biotechnol 17:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang YL, Zhang SZ, Kulye M, et al. (2012) Silencing of molt-regulating transcription factor gene, CiHR3, affects growth and development of sugarcane stem borer, Chilo infuscatellus. J Insect Sci 12:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huvenne H and Smagghe G (2010) Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J Insect Physiol 56:227–235 [DOI] [PubMed] [Google Scholar]
- 28.Clemens JC, Worby CA, Simonson-Leff N, et al. (2000) Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A 97:6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caplen NJ, Fleenor J, Fire A, et al. (2000) dsRNA-mediated gene silencing in cultured Drosophila cells: A tissue culture model for the analysis of RNA interference. Gene 252:95–105 [DOI] [PubMed] [Google Scholar]
- 30.Peng T, Pan Y, Yang C, et al. (2016) Over-expression of CYP6A2 is associated with spirotetramat resistance and cross-resistance in the resistant strain of Aphis gossypii Glover. Pestic Biochem Physiol 126:64–69 [DOI] [PubMed] [Google Scholar]
- 31.Shah C and Förstemann K (2008) Monitoring miRNA-mediated silencing in Drosophila melanogaster S2-cells. Biochim Biophys Acta - Gene Regul Mech 1779:766–772 [DOI] [PubMed] [Google Scholar]
- 32.Sivakumar S, Rajagopal R, Venkatesh GR, et al. (2007) Knockdown of aminopeptidase-N from Helicoverpa armigera larvae and in transfected Sf21 cells by RNA interference reveals its functional interaction with Bacillus thuringiensis insecticidal protein Cry1Ac. J Biol Chem 282:7312–7319 [DOI] [PubMed] [Google Scholar]
- 33.Valdes VJ, Sampieri A, Sepulveda J, et al. (2003) Using double-stranded RNA to prevent in vitro and in vivo viral infections by recombinant baculovirus. J Biol Chem 278:19317–19324 [DOI] [PubMed] [Google Scholar]
- 34.Johnson JA, Bitra K, Zhang S, et al. (2010) The UGA-CiE1 cell line from Chrysodeixis includens exhibits characteristics of granulocytes and is permissive to infection by two viruses. Insect Biochem Mol Biol 40:394–404 [DOI] [PubMed] [Google Scholar]
- 35.Terenius O, Papanicolaou A, Garbutt JS, et al. (2011) RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J Insect Physiol 57:231–245 [DOI] [PubMed] [Google Scholar]
- 36.Turner CT, Davy MW, MacDiarmid RM, et al. (2006) RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol Biol 15:383–391 [DOI] [PubMed] [Google Scholar]
- 37.Joga MR, Zotti MJ, Smagghe G, et al. (2016) RNAi efficiency, systemic properties, and novel delivery methods for pest insect control: What we know so far. Front Physiol 7:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Killiny N and Kishk A (2017) Delivery of dsRNA through topical feeding for RNA interference in the citrus sap piercing-sucking hemipteran, Diaphorina citri. Arch Insect Biochem Physiol 95:1–13 [DOI] [PubMed] [Google Scholar]
- 39.Palli SR (2014) RNA interference in Colorado potato beetle: Steps toward development of dsRNA as a commercial insecticide. Curr Opin Insect Sci 6:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu Y, Park Y, Gao X, et al. (2012) Cholinergic and non-cholinergic functions of two acetylcholinesterase genes revealed by gene-silencing in Tribolium castaneum. Sci Rep 2:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomoyasu Y, Miller SC, Tomita S, et al. (2008) Exploring systemic RNA interference in insects: A genome-wide survey for RNAi genes in Tribolium. Genome Biol 9:1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin C, Shen G, Guo D, et al. (2016) InsectBase : a resource for insect genomes and transcriptomes. 44:801–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang T, Liu W, Li D, et al. (2018) LmCht5-1 promotes pro-nymphal molting during locust embryonic development. Insect Biochem Mol Biol 101:124–130 [DOI] [PubMed] [Google Scholar]
- 44.Santos-Ortega Y and Killiny N (2018) Silencing of sucrose hydrolase causes nymph mortality and disturbs adult osmotic homeostasis in Diaphorina citri (Hemiptera: Liviidae). Insect Biochem Mol Biol 101:131–143 [DOI] [PubMed] [Google Scholar]
- 45.Li Z, Zeng B, Ling L, et al. (2015) Enhancement of larval RNAi efficiency by over-expressing argonaute2 in bombyx mori. Int J Biol Sci 11:176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martín D, Maestro O, Cruz J, et al. (2006) RNAi studies reveal a conserved role for RXR in molting in the cockroach Blattella germanica. J Insect Physiol 52:410–416 [DOI] [PubMed] [Google Scholar]
- 47.Asokan R, Sharath Chandra G, Manamohan M, et al. (2014) Response of various target genes to diet-delivered dsRNA mediated RNA interference in the cotton bollworm, Helicoverpa armigera. J Pest Sci (2004) 87:163–172 [Google Scholar]
- 48.Lü J, Liu Z, Guo W, et al. (2020) Oral delivery of ds Hvlwr is a feasible method for managing the pest Henosepilachna vigintioctopunctata (Coleoptera: Coccinellidae). Insect Science doi:101111/1744–7917.12784 [DOI] [PubMed] [Google Scholar]
- 49.Whyard S, Singh AD, and Wong S (2009) Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem Mol Biol 39:824–832 [DOI] [PubMed] [Google Scholar]
- 50.Singh AD, Wong S, Ryan CP, et al. (2013) Oral delivery of double-stranded RNA in larvae of the yellow fever mosquito, aedes aegypti: Implications for pest mosquito control. J Insect Sci 13:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo Y, Chen Q, Luan J, et al. (2017) Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem Mol Biol 88:21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tabara H, Grishok A, and Mello CC (1998) the Genome Sequence T “. Science 282:430–431 [DOI] [PubMed] [Google Scholar]
- 53.Maeda I, Kohara Y, Yamamoto M, et al. (2001) Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol 11:171–176 [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Zhang H, Li H, et al. (2011) Second-Generation Sequencing Supply an Effective Way to Screen RNAi Targets in Large Scale for Potential Application in Pest Insect Control. PLoS One 6(4):1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zotti M, dos Santos EA, Cagliari D, et al. (2018), RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag Sci 74(6):1239–1250 [DOI] [PubMed] [Google Scholar]
- 56.Parsons KH, Mondal MH, McCormick CL, et al. (2018) Guanidinium-Functionalized Interpolyelectrolyte Complexes Enabling RNAi in Resistant Insect Pests. Biomacromolecules 19:1111–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng Y, Hu Y, Yan S, et al. (2019) A polymer/detergent formulation improves dsRNA penetration through the body wall and RNAi-induced mortality in the soybean aphid Aphis glycines. Pest Manag Sci 75:1993–1999 [DOI] [PubMed] [Google Scholar]
- 58.Natarajan P, Sukthankar P, Changstrom J, et al. (2018) Synthesis and Characterization of Multifunctional Branched Amphiphilic Peptide Bilayer Conjugated Gold Nanoparticles. ACS Omega 3:11071–11083 [Google Scholar]
- 59.Grafton-Cardwell EE, Stelinski LL, and Stansly PA (2013) Biology and Management of Asian Citrus Psyllid, Vector of the Huanglongbing Pathogens. Annu Rev Entomol 58:413–432 [DOI] [PubMed] [Google Scholar]
- 60.Boina DR and Bloomquist JR (2015) Chemical control of the Asian citrus psyllid and of huanglongbing disease in citrus. Pest Manag Sci 71:808–823 [DOI] [PubMed] [Google Scholar]
- 61.Tabachnick WJ (2015) Diaphorina citri (Hemiptera: Liviidae) Vector Competence for the Citrus Greening Pathogen “Candidatus Liberibacter Asiaticus.” J Econ Entomol 108:839–848 [DOI] [PubMed] [Google Scholar]
- 62.Kishk A, Anber HAI, AbdEl-Raof TK, et al. (2017) RNA interference of carboxyesterases causes nymph mortality in the Asian citrus psyllid, Diaphorina citri. Arch Insect Biochem Physiol 94:1–13 [DOI] [PubMed] [Google Scholar]
- 63.Yu X, Gowda S, and Killiny N (2017) Double-stranded RNA delivery through soaking mediates silencing of the muscle protein 20 and increases mortality to the Asian citrus psyllid, Diaphorina citri. Pest Manag Sci 73:1846–1853 [DOI] [PubMed] [Google Scholar]
- 64.Yu X and Killiny N (2018) Effect of silencing a boule homologue on the survival and reproduction of Asian citrus psyllid Diaphorina citri. Physiol Entomol 43:268–275 [Google Scholar]
- 65.Santos-Ortega Y and Killiny N (2018) Silencing of sucrose hydrolase causes nymph mortality and disturbs adult osmotic homeostasis in Diaphorina citri (Hemiptera: Liviidae). Insect Biochem Mol Biol 101:131–143 [DOI] [PubMed] [Google Scholar]
- 66.Yu X and Killiny N (2018) RNA interference of two glutathione S-transferase genes, Diaphorina citri DcGSTe2 and DcGSTd1, increases the susceptibility of Asian citrus psyllid (Hemiptera: Liviidae) to the pesticides fenpropathrin and thiamethoxam. Pest Manag Sci 74:638–647 [DOI] [PubMed] [Google Scholar]
- 67.Taning CNT, Andrade EC, Hunter WB, et al. (2016) Asian Citrus Psyllid RNAi Pathway-RNAi evidence. Sci Rep 6:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baum JA, Bogaert T, Clinton W, et al. (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25:1322–1326 [DOI] [PubMed] [Google Scholar]
- 69.Niu X, Kassa A, Hu X, et al. (2017) Control of Western Corn Rootworm (Diabrotica virgifera virgifera) Reproduction through Plant-Mediated RNA Interference. Sci Rep 7:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu F, Xu J, Palli R, et al. (2011) Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag Sci 67:175–182 [DOI] [PubMed] [Google Scholar]
- 71.Zhao Y, Yang G, Wang-Pruski G, et al. (2008) Phyllotreta striolata (Coleoptera: Chrysomelidae): Arginine kinase cloning and RNAi-based pest control. Eur J Entomol 105:815–822 [Google Scholar]
- 72.Coy MR, Sanscrainte ND, Chalaire KC, et al. (2012) Gene silencing in adult Aedes aegypti mosquitoes through oral delivery of double-stranded RNA. J Appl Entomol 136:741–748 [Google Scholar]
- 73.Figueira-Mansur J, Ferreira-Pereira A, Mansur JF, et al. (2013) Silencing of P-glycoprotein increases mortality in temephos-treated Aedes aegypti larvae. Insect Mol Biol 22:648–658 [DOI] [PubMed] [Google Scholar]
- 74.Chen J, Zhang D, Yao Q, et al. (2010) Feeding-based RNA interference of a trehalose phosphate synthase gene in the brown planthopper, Nilaparvata lugens. Insect Mol Biol 19:777–786 [DOI] [PubMed] [Google Scholar]
- 75.Kumar Anil Ou Ruguang, Samrakandi Mustapha, Beerntsen Brenda T. and Richard T Sayre SW (2013) Development of an RNAi based microalgal larvicide to control mosquitoes. 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu K, Tsujimoto H, Cha SJ, et al. (2011) Aquaporin water channel AgAQP1 in the malaria vector mosquito Anopheles gambiae during blood feeding and humidity adaptation. Proc Natl Acad Sci U S A 108:6062–6066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Walshe DP, Lehane SM, Lehane MJ, et al. (2009) Prolonged gene knockdown in the tsetse fly Glossina by feeding double stranded RNA. Insect Mol Biol 18:11–19 [DOI] [PubMed] [Google Scholar]
- 78.Shakesby AJ, Wallace IS, Isaacs HV., et al. (2009) A water-specific aquaporin involved in aphid osmoregulation. Insect Biochem Mol Biol 39:1–10 [DOI] [PubMed] [Google Scholar]
- 79.Christiaens O and Smagghe G (2014) The challenge of RNAi-mediated control of hemipterans. Curr Opin Insect Sci 6:15–21 [DOI] [PubMed] [Google Scholar]
- 80.Mao J and Zeng F (2014) Plant-mediated RNAi of a gap gene-enhanced tobacco tolerance against the Myzus persicae. Transgenic Res 23:145–152 [DOI] [PubMed] [Google Scholar]
- 81.Mutti NS, Park Y, Reese JC, et al. (2006) RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. J Insect Sci 6:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sapountzis P, Duport G, Balmand S, et al. (2014) New insight into the RNA interference response against cathepsin-L gene in the pea aphid, Acyrthosiphon pisum: Molting or gut phenotypes specifically induced by injection or feeding treatments. Insect Biochem Mol Biol 51:20–32 [DOI] [PubMed] [Google Scholar]
- 83.Will T and Vilcinskas A (2015) The structural sheath protein of aphids is required for phloem feeding. Insect Biochem Mol Biol 57:34–40 [DOI] [PubMed] [Google Scholar]
- 84.Yin C, Shen G, Guo D, et al. (2015) InsectBase: a resource for insect genomes and transcriptomes. Nucleic Acids Res 44:801–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang Y and Lu Z (2015) Peroxiredoxin 1 protects the pea aphid Acyrthosiphon pisum from oxidative stress induced by Micrococcus luteus infection. J Invertebr Pathol 127:115–121 [DOI] [PubMed] [Google Scholar]
- 86.Naessens E, Dubreuil G, Giordanengo P, et al. (2015) A Secreted MIF Cytokine Enables Aphid Feeding and Represses Plant Immune Responses. Curr Biol 25:1898–1903 [DOI] [PubMed] [Google Scholar]
- 87.Jaubert-Possamai S, Le Trionnaire G, Bonhomme J, et al. (2007) Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC Biotechnol 7:7–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pitino M, Coleman AD, Maffei ME, et al. (2011) Silencing of aphid genes by dsRNA feeding from plants. PLoS One 6:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gong YH, Yu XR, Shang QL, et al. (2014) Oral Delivery Mediated RNA Interference of a Carboxylesterase Gene Results in Reduced Resistance to Organophosphorus Insecticides in the Cotton Aphid, Aphis gossypii Glover. PLoS One 9:23–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fan W, Wei Z, Zhang M, et al. (2015) Resistance to Ditylenchus destructor infection in sweet potato by the expression of small interfering RNAs targeting unc-15, a movement-related gene. Phytopathology 105:1458–1465 [DOI] [PubMed] [Google Scholar]
- 91.Rebijith KB, Asokan R, Hande HR, et al. (2016) RNA Interference of Odorant-Binding Protein 2 (OBP2) of the Cotton Aphid, Aphis gossypii (Glover), Resulted in Altered Electrophysiological Responses. Appl Biochem Biotechnol 178:251–266 [DOI] [PubMed] [Google Scholar]
- 92.Wuriyanghan H, Rosa C, and Falk BW (2011) Oral delivery of double-stranded RNAs and siRNAs induces RNAi effects in the potato/tomato psyllid, Bactericerca cockerelli. PLoS One 6(11):e27736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Upadhyay SK, Chandrashekar K, Thakur N, et al. (2011) RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J Biosci 36:153–161 [DOI] [PubMed] [Google Scholar]
- 94.Thakur N, Upadhyay SK, Verma PC, et al. (2014) Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase a gene. PLoS One 9:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Malik HJ, Raza A, Amin I, et al. (2016) RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci Rep 6:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li X, Zhang M, and Zhang H (2011) RNA interference of four genes in adult Bactrocera dorsalis by feeding their dsRNAs. PLoS One 6(3):e17788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deng F and Zhao Z (2014) Influence of catalase gene silencing on the survivability of sitobion avenae. Arch Insect Biochem Physiol 86:46–57 [DOI] [PubMed] [Google Scholar]
- 98.Xiao D, Lu YH, Shang QL, et al. (2015) Gene silencing of two acetylcholinesterases reveals their cholinergic and non-cholinergic functions in Rhopalosiphum padi and Sitobion avenae. Pest Manag Sci 71:523–530 [DOI] [PubMed] [Google Scholar]
- 99.Zhang X, Liu X, Ma J, et al. (2013) Silencing of cytochrome P450 CYP6B6 gene of cotton bollworm (Helicoverpa armigera) by RNAi. Bull Entomol Res 103:584–591 [DOI] [PubMed] [Google Scholar]
- 100.Wang W, Luo L, Lu H, et al. (2015) Angiotensin-converting enzymes modulate aphid-plant interactions. Sci Rep 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Fan J, Zhang Y, Francis F, et al. (2015) Orco mediates olfactory behaviors and winged morph differentiation induced by alarm pheromone in the grain aphid, Sitobion avenae. Insect Biochem Mol Biol 64:16–24 [DOI] [PubMed] [Google Scholar]
- 102.Zhang J, Kha, Hasse C, et al. (2015) Pest control. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science (80- ) 27:991–4 [DOI] [PubMed] [Google Scholar]
- 103.Yao J, Rotenberg D, Afsharifar A, et al. (2013) Development of RNAi Methods for Peregrinus maidis, the Corn Planthopper. PLoS One 8(8):e70243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Araujo RN, Santos A, Pinto FS, et al. (2006) RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem Mol Biol 36:683–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Allen ML and Walker WB (2012) Saliva of Lygus lineolaris digests double stranded ribonucleic acids. J Insect Physiol 58:391–396 [DOI] [PubMed] [Google Scholar]
- 106.El-Shesheny I, Hajeri S, El-Hawary I, et al. (2013) Silencing Abnormal Wing Disc Gene of the Asian Citrus Psyllid, Diaphorina citri Disrupts Adult Wing Development and Increases Nymph Mortality. PLoS One 8:2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hijaz F and Killiny N (2014) Collection and chemical composition of phloem sap from Citrus sinensis L. Osbeck (sweet orange). PLoS One 9:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yu X and Killiny N (2018) Effect of parental RNA interference of a transformer-2 homologue on female reproduction and offspring sex determination in Asian citrus psyllid. Physiol Entomol 43:42–50 [Google Scholar]
- 109.Nunes FMF and Simões ZLP (2009) A non-invasive method for silencing gene transcription in honeybees maintained under natural conditions. Insect Biochem Mol Biol 39:157–160 [DOI] [PubMed] [Google Scholar]
- 110.Meer V (2012) (12) Patent Application Publication (10) Pub. No.: US 2012/0148524 A1. 1
- 111.Vander Meer K. and Choi MY (2011) Formicidae (Ant) Control Using Double-Stranded RNA Constructs, US Patent No. 8,575,328
- 112.Zhou X, Wheeler MM, Oi FM, et al. (2008) RNA interference in the termite Reticulitermes flavipes through ingestion of double-stranded RNA. Insect Biochem Mol Biol 38:805–815 [DOI] [PubMed] [Google Scholar]
- 113.Zhang H, Li HC, and Miao XX (2013) Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci 20:15–30 [DOI] [PubMed] [Google Scholar]
- 114.Kumar M, Gupta GP, and Rajam MV (2009) Silencing of acetylcholinesterase gene of Helicoverpa armigera by siRNA affects larval growth and its life cycle. J Insect Physiol 55:273–278 [DOI] [PubMed] [Google Scholar]
- 115.Zhu JQ, Liu S, Ma Y, et al. (2012) Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect-associated gene EcR. PLoS One 7(6):e38572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xiong Y, Zeng H, Zhang Y, et al. (2013) Silencing the HaHR3 gene by transgenic plant-mediated RNAi to disrupt Helicoverpa armigera development. Int J Biol Sci 9:370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mao YB, Cai WJ, Wang JW, et al. (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25:1307–1313 [DOI] [PubMed] [Google Scholar]
- 118.Yang J and jun Han Z (2014) Efficiency of Different Methods for dsRNA Delivery in Cotton Bollworm (Helicoverpa armigera). J Integr Agric 13:115–123 [Google Scholar]
- 119.Khajuria C, Buschman L, Chen M, et al. (2010) A gut-specific chitinase gene essential for regulation of chitin content of peritrophic matrix and growth of Ostrinia nubilalis larvae. Insect Biochem Mol Biol 40:621–629 [DOI] [PubMed] [Google Scholar]
- 120.Bautista MAM, Miyata T, Miura K, et al. (2009) RNA interference-mediated knockdown of a cytochrome P450, CYP6BG1, from the diamondback moth, Plutella xylostella, reduces larval resistance to permethrin. Insect Biochem Mol Biol 39:38–46 [DOI] [PubMed] [Google Scholar]
- 121.Gong L, Yang X, Zhang B, et al. (2011) Silencing of Rieske iron-sulfur protein using chemically synthesised siRNA as a potential biopesticide against Plutella xylostella. Pest Manag Sci 67:514–520 [DOI] [PubMed] [Google Scholar]
- 122.Gong L, Chen Y, Hu Z, et al. (2013) Testing Insecticidal Activity of Novel Chemically Synthesized siRNA against Plutella xylostella under Laboratory and Field Conditions. PLoS One 8:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kontogiannatos D, Swevers L, Maenaka K, et al. (2013) Functional characterization of a juvenile hormone esterase related gene in the moth Sesamia nonagrioides through RNA interference. PLoS One 8(9):e73834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tian H, Peng H, Yao Q, et al. (2009) Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS One 4(7):e6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Surakasi VP, Mohamed AAM, and Kim Y (2011) RNA interference of β1 integrin subunit impairs development and immune responses of the beet armyworm, Spodoptera exigua. J Insect Physiol 57:1537–1544 [DOI] [PubMed] [Google Scholar]
- 126.Rajagopal R, Sivakumar S, Agrawal N, et al. (2002) Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J Biol Chem 277:46849–46851 [DOI] [PubMed] [Google Scholar]
- 127.Tian L, Ma L, Guo E, et al. (2013) 20-hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy 9:1172–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huang J, Zhang Y, Li M, et al. (2007) RNA interference-mediated silencing of the bursicon gene induces defects in wing expansion of silkworm. FEBS Lett 581:697–701 [DOI] [PubMed] [Google Scholar]
- 129.Griebler M, Westerlund SA, Hoffmann KH, et al. (2008) RNA interference with the allatoregulating neuropeptide genes from the fall armyworm Spodoptera frugiperda and its effects on the JH titer in the hemolymph. J Insect Physiol 54:997–1007 [DOI] [PubMed] [Google Scholar]
- 130.Rodríguez-Cabrera L, Trujillo-Bacallao D, Borrás-Hidalgo O, et al. (2010) RNAi-mediated knockdown of a Spodoptera frugiperda trypsin-like serine-protease gene reduces susceptibility to a Bacillus thuringiensis Cry1Ca1 protoxin. Environ Microbiol 12:2894–2903 [DOI] [PubMed] [Google Scholar]
- 131.Meyering-Vos M and Müller A (2007) RNA interference suggests sulfakinins as satiety effectors in the cricket Gryllus bimaculatus. J Insect Physiol 53:840–848 [DOI] [PubMed] [Google Scholar]
- 132.Luo Y, Wang X, Wang X, et al. (2013) Differential responses of migratory locusts to systemic RNA interference via double-stranded RNA injection and feeding. 574–583 [DOI] [PubMed] [Google Scholar]
- 133.Wynant N, Verlinden H, Breugelmans B, et al. (2012) Tissue-dependence and sensitivity of the systemic RNA interference response in the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol 42:911–917 [DOI] [PubMed] [Google Scholar]


