Abstract
On the western border of Thailand, Plasmodium falciparum has become resistant to almost all antimalarial agents. The molecular basis of resistance in these parasite populations has not been well characterized. This study assessed genetic polymorphisms in the pfmdr1 gene in 54 parasites collected from the western border of Thailand to determine the relationship of pfmdr1 copy number and codon mutations with parasite sensitivities to mefloquine, chloroquine, halofantrine, quinine, and artesunate assessed in vitro. A point mutation at codon 86 (resulting in a change of Asn to Tyr) was associated with a significantly lower 50% inhibitory concentration (IC50) of mefloquine (median, 9 ng/ml versus 52.4 ng/ml; P = 0.003). Overall 35% of the isolates (19 of 54) had an increase in pfmdr1 copy number, and all 19 carried the wild-type allele at codon 86. Increased pfmdr1 copy number was associated with higher IC50s of mefloquine (P = 0.04) and artesunate (P = 0.005), independent of polymorphism at codon 86. The relationship between pfmdr1 and resistance to structurally distinct antimalarial agents confirms the presence of a true multidrug-resistant phenotype.
On the borders of Thailand, Plasmodium falciparum has become resistant to nearly all available antimalarial drugs (23, 33). Mefloquine has been useful for the treatment of uncomplicated falciparum malaria because it is relatively well tolerated and can be given in a single or split dose. In the past decade, however, resistance to mefloquine has developed rapidly (17). Increasing the dose from 15 mg/kg to 25 mg/kg gave some respite (28), but the decline in this parasite’s sensitivity to mefloquine has continued, and high-grade resistance is now sufficient to limit its use in monotherapy (21). Halofantrine, a related phenathrene-methanol, can cause major cardiovascular toxicity when given in the high doses necessary to treat mefloquine-resistant parasites (27). Quinine is a first-line treatment in the acute management of severe malaria, but its efficacy in the treatment of uncomplicated malaria when used alone has declined in the last few years to ∼50% (19a). Combination therapy, such as treatment with artesunate (for 3 days) and mefloquine, is one of the few approaches which has maintained acceptable levels of efficacy (>90% cure rate). This combination is now the treatment of choice for uncomplicated falciparum malaria on the northwestern border of Thailand (22).
The molecular basis for multidrug resistance in P. falciparum is still not fully understood. An association between pfmdr1, which encodes a transmembrane glycoprotein (Pgh1, for P-glycoprotein homologue 1), and the multidrug-resistant (MDR) phenotype was first reported in 1989 (15, 34). Point mutations in pfmdr1, most notably at codon 86, have been associated with decreased chloroquine sensitivity (1, 5, 11, 13, 14). However, this association is not a consistent finding (2, 3, 6, 7) and is therefore unlikely to account for all cases of chloroquine resistance (30). Furthermore, in a study involving a genetic cross, chloroquine resistance was found to segregate with cg2 (located on chromosome 7) rather than with pfmdr1 (located on chromosome 11) (26, 32). Amplification of the pfmdr1 gene copy number is associated with resistance to mefloquine and halofantrine, both in laboratory (10, 18) and field (35) isolates. However, since amplification of pfmdr1 is not a prerequisite for increased mefloquine resistance (16), the exact relationship of this gene to resistance to that drug and the clinical significance of this locus are still unknown.
This study was designed to test the hypothesis that an increased pfmdr1 copy number is associated with decreased susceptibility of P. falciparum to mefloquine by examining the relationship between pfmdr1 gene copy number and sensitivity to mefloquine in a large collection of field isolates. A secondary objective was to assess whether pfmdr1 copy number could be used as a molecular marker to monitor mefloquine drug resistance in areas where such resistance is emerging. To assess the role of the pfmdr1 gene with respect to a true MDR-like phenomenon, its relationship with sensitivity to other antimalarial drugs (i.e., chloroquine, quinine, halofantrine, and the structurally unrelated compound artesunate) was assessed and then correlated with other, more recently discovered genetic markers.
MATERIALS AND METHODS
Isolates of P. falciparum.
Fresh isolates of P. falciparum were obtained between April 1995 and December 1997 from patients with acute uncomplicated falciparum malaria (single-species infection) attending the clinics of the Shoklo Malaria Research Unit (SMRU) who agreed to be venesected. Patients were recruited from two camps (Shoklo and Maela) for displaced persons of the Karen ethnic minority situated in an area of forested hills on the Thai-Burmese border. Due to a decline in efficacy of mefloquine monotherapy, all patients treated at SMRU clinics after June 1994 received an artemisinin derivative alone or in combination with mefloquine or lumefantrine (21).
All parasite isolates collected were tested immediately for drug sensitivity or set up in continuous culture (within 4 to 8 h) upon their arrival at the laboratory, situated in the Thai town of Mae Sod, a 2-h drive from the study sites. Of the 264 isolates undergoing in vitro sensitivity testing without prior freezing, 125 were available for further molecular analysis. A random list of these isolates was generated, and specimens were then selected sequentially for assessment of pfmdr1 copy number and polymorphisms in codons 86, 184, 1034, 1042, and 1246. Molecular analysis was carried out without knowledge of results of in vitro drug assays. Mefloquine, artesunate, and quinine sensitivities were assessed for all specimens, whereas most halofantrine and chloroquine sensitivities were determined for isolates collected before 1996.
Drug sensitivity testing.
A microdilution radioisotope method was used to assess antimalarial drug susceptibility of P. falciparum isolates as described by Webster et al. (31). Briefly, whole blood (5 ml) was collected into a sterile tube containing 0.05 ml of 15% EDTA (Becton Dickinson) and transferred to the SMRU laboratory. After the plasma and the buffy coat were removed, the erythrocytes were washed three times in phosphate-buffered saline. If >90% of parasites were at the ring stage of infection and a parasitemia of >0.5% infected erythrocytes was evident, then the isolate was entered in the drug assay protocol. If these criteria were not met, infected erythrocytes were cultured in RPMI 1640 medium (Gibco BRL) supplemented with 10% heterologous serum of the patient’s blood group and incubated in an atmosphere of 5% CO2, 5% O2, and 90% N2 until a target parasitemia of 0.5% was achieved. In vitro drug testing was then carried out as described previously (31).
On the Western border of Thailand, the treatment failure rate following mefloquine monotherapy of uncomplicated falciparum malaria in 1995 was 50% (20). To define a clinically appropriate 50% inhibitory concentration (IC50) representing mefloquine resistance, we calculated the median IC50 (i.e., the concentration which bisects this parasite population). This value was 45 ng/ml, and parasites with IC50s higher than this were defined as being mefloquine resistant (8a).
The reproducibility of IC50 measurements was assessed over time by using the K1 isolate in drug sensitivity assays, performed six times for each drug, at the beginning and end of the study. To confirm the validity of these assays, interlaboratory variability was assessed using cryopreserved isolates, which were analyzed at the SMRU laboratory and, independently, at the Armed Forces Research Institute for Medical Sciences (AFRIMS), Bangkok, Thailand.
DNA extraction.
Venous blood was collected into heparinized tubes, transported to the laboratory, and washed twice in RPMI 1640 medium. Cells were pelleted and stored at −70°C for up to 14 months. After the pellet was thawed, hemolysate (200 μl) was purified by the use of a QIAamp Blood Kit (Qiagen) to yield 200 μl of genomic DNA. DNA was eluted and stored in elution buffer AE (Qiagen) and used for PCR immediately or stored at −20°C for up to 6 months or at −70°C for longer periods (maximum, 6 months).
Quantitative analysis by TC-PCR.
PCRs for pfmdr1 and the β-tubulin gene were performed separately in a total volumes of 25 μl containing PCR buffer (Cloned Pfu DNA 10× Reaction Buffer; Promega), 1.5 to 3.0 mM MgCl2, 100 μM each deoxynucleoside triphosphate, 0.4 μM each primer, and 0.625 U of Pfu polymerase (Promega). Reactions were carried out in a GeneAmp PCR System 2400 Thermocycler (Perkin-Elmer), using primers and conditions described previously (20).
To quantify each target in a given DNA extract, competitive PCRs were performed with reaction mixtures which contained serial dilutions of the competitor molecule RPTC1 (GenBank accession no. U72062) and a constant amount of DNA extract. The reaction products were digested with SacI or EcoRI, both of which recognize the altered unique restriction site in RPTC1 and discriminate the amplification product of the competitor molecule from that of the native target. The products were resolved on an agarose gel (1.8%) and visualized with ethidium bromide, using a transilluminator (White/Ultraviolet Transilluminator; UVP). The resulting image was captured and analyzed with Scion Image 1.60C and GelBase/GelBlot Pro Mac-based software. The C/T ratio was calculated for each reaction, and copy numbers were calculated as described previously (20).
Estimates of gene copy number were considered acceptable if there were at least three informative data points in the dilutional series for each target and the point of equivalence fell within a 15 to 85% range for the competitor-to-target ratio as determined from previous studies (20).
Detection of pfmdr1 polymorphism.
To determine the presence of sequence polymorphisms in pfmdr1, a PCR-restriction fragment length polymorphism method was used. Regions flanking codons 86, 184, 1034, 1042, and 1246 of the pfmdr1 gene were amplified by PCR and digested with specific restriction enzymes. This technique produces patterns corresponding to alternative polymorphisms following resolution on agarose gels. The region flanking codons 86 and 184 was amplified with the primer pair A4 (5′ AAA GAT GGT AAC CTC AGT ATC AAA GAA GAG 3′) and A2 (5′ GTC AAA CGT GCA TTT TTT ATT AAT GAC CAT TTA 3′), that flanking codons 1034 and 1042 was amplified with primers 1034f (5′ AGA ATT ATT GTA AAT GCA GCT TTA TGG GGA CTC 3′) and 1042r (5′ AAT GGA TAA TAT TTC TCA AAT GAT AAC TTA GCA 3′), and that flanking 1246 was amplified with primers 1246f (5′ ATG ATC ACA TTA TAT TAA AAA ATG ATA TGA CAA AT 3′) and O2 (5′ ATG ATT CGA TAA ATT CAT CTA TAG CAG CAA 3′). If natural restriction sites corresponding to specific polymorphisms were not already present, artificial ones were created by introducing nucleotide substitutions (bold typeface) in the 3′ ends of the primers as described before (12). Reaction conditions were exactly as described before (12).
Codon 86 (Asn or Tyr) is discriminated by digestion of the A4-A2 fragment with ApoI, which cuts if Asn is present and at an invariant site as a control for restriction digestion. Codon 184 (Tyr or Phe) is discriminated by digestion with DraI, which cuts when Phe is present and at other, invariant sites. Codons 1034 (Ser or Cys) and 1042 (Asn or Asp) are discriminated by digestion of the 1034f-1042r fragment with DdeI (which digests if Cys is present and at an invariant site) and VspI (which digests if Asn is present and at other invariant sites), respectively. Codon 1246 (Asp or Tyr) is identified by digestion with DpnII, which cuts if Asp is present and at another, invariant site. All restriction enzymes were from New England Biolabs. Resolution on gels composed of 1 to 3% agarose allows discrimination of each sequence polymorphism.
MSP-2 dimorphism.
MSP-2 dimorphisms were identified for each isolate by the method of Viriyakosol et al. (29). For the purpose of this assay, the samples were scored only on the basis of whether they belonged to the IC or the FC family of MSP-2 dimorphisms and not on the basis of size. Therefore, for each sample, three combinations were possible: either IC, FC, or a mixture of IC and FC alleles.
Assessment of clonality of infection of field isolates.
Three P. falciparum loci (MSP-1, MSP-2, and GLURP) which exhibit polymorphism in numbers of tandem repeats were amplified from each isolate by nested PCR (9). After separation of PCR products on an agarose gel (1.7%), the appearance of a double band at any one of the three loci implies the presence of at least two clones.
Statistical analysis.
Data were analyzed by using SPSS for Windows (SPSS Inc., Chicago, Ill.). Non-normally distributed data were described by median, range, and interquartile range (IQR), and comparisons were made by using the Mann-Whitney U test or Kruskal-Wallis analysis of variance. Box-Cox transformations were used to normalize distributions of IC50s, after which comparisons were made by using Student’s t test or one-way analysis of variance. Univariate analysis of IC50s was performed with each of the following variables: age, sex, prior in vitro culturing, parasitemia, clonality of infection, MSP-2 genotype, and type of infection (primary versus recrudescent). Data for these IC50 comparisons were described by using median, range, and IQR rather than the means of Box-Cox-transformed data since the latter are not immediately intuitive.
For categorical variables, percentages and corresponding 95% confidence intervals (CIs) were calculated. Tests for statistically significant associations between categorical variables were performed by using the χ2 test with Yates’ correction or by Fisher’s exact test (when an expected count in a 2 × 2 table was less than 5). Multiple linear regression modelling with variables which were correlated with IC50s in univariate analysis was performed on Box-Cox-transformed IC50 as the dependent variable.
Statistical significance was assumed if the P value was <0.05. For multiple comparisons, the level of significance was adjusted by using the Bonferroni correction.
Although a monoclonal infection will have integral values for pfmdr1 copy number, this is not so for polyclonal infections. In this study, the ratiometric technique used to assess copy number generated a figure representing a weighted composite of the copy numbers of genes in individual parasite clones. For this reason, the relationship between copy number and drug sensitivity was assessed by using a continuous variable for copy number as well as an ordinal variable in which copy number was rounded to the nearest integer. The reproducibility of the pfmdr1 copy number estimates (quoted as a repeatability coefficient) was calculated as described previously, using a continuous variable for copy number (8).
RESULTS
Between April 1995 and November 1998, 264 parasite isolates were collected and assayed for drug sensitivities without prior freezing (Table 1). At SMRU, repeated estimates for IC50 measurements with the reference strain K1 resulted in coefficients of variation of 7% for quinine, 12% for mefloquine, 14% for artesunate, and 27% for chloroquine. There were no significant differences in IC50 estimates over time or between the two different laboratories (SMRU and AFRIMS). Molecular analysis of pfmdr1 was attempted for 63 of 125 specimens available for further processing, although copy number could be assessed reliably in only 54 (86%) cases. In 20 cases, pfmdr1 copy number was estimated twice, with a mean difference between measurements of 0.04 and a repeatability coefficient of 0.56.
TABLE 1.
In vitro sensitivities to drugs for all specimens tested and the genotyped subgroup
| Drug | Specimens tested | n | IC50 (ng/ml)
|
||
|---|---|---|---|---|---|
| Median | Range | IQR | |||
| Mefloquine | All | 246 | 39.1 | 2.66–247 | 19.8–65.8 |
| Genotyped subgroup | 54 | 41.3 | 4.7–183 | 13.2–74.3 | |
| Chloroquine | All | 185 | 95.0 | 16.2–508 | 70.6–158 |
| Genotyped subgroup | 36 | 92.1 | 42.0–317 | 76.4–145 | |
| Halofantrine | All | 172 | 9.56 | 1.09–59.1 | 3.93–18.3 |
| Genotyped subgroup | 36 | 11.0 | 1.1–55.5 | 2.71–20.7 | |
| Quinine | All | 256 | 386 | 48.8–1259 | 253–622 |
| Genotyped subgroup | 53 | 347 | 48.8–1259 | 200–653 | |
| Artesunate | All | 256 | 2.10 | 0.18–10.4 | 1.29–3.55 |
| Genotyped subgroup | 51 | 1.98 | 0.25–8.85 | 1.36–3.61 | |
Of the 54 isolates for which pfmdr1 copy number was enumerated, 33 (61%) came from adults (>14 years), 20 (37%) were from children aged 5 to 14 years, and one (2%) was from a child under 5 years of age. Thirty-five (65%) came from patients with primary infections, and 19 (35%) were from persons with recrudescent infections. All patients received an artemisinin derivative, either alone or in combination with mefloquine or lumefantrine, and exhibited an excellent therapeutic response; only four patients were found to have recrudescent infections during subsequent followup. The geometric mean parasitemia at the time of venesection was 27,900 μl−1 (95% CI, 17,400 to 44,700 μl−1). A total of 27 isolates (54%) required in vitro culturing for a median of 3 days (range, 1 to 11 days) to achieve the required minimum parasitemia (0.5%) prior to drug sensitivity testing. There was no significant difference in the IC50s for any of the drugs between those isolates requiring prior in vitro culturing and those assayed immediately.
The IC50s for each drug tested are presented in Table 1. There was no significant difference in IC50s between the cases which were and those which were not genotyped, confirming that the subgroup in which genetic studies were carried out represented the general population of isolates. Correlations between the IC50s of antimalarial drugs are given in Table 2. IC50s for quinine, mefloquine, and halofantrine were highly correlated with each other (R > 0.65). IC50s of artesunate were moderately (R > 0.4) correlated with those of mefloquine and quinine (fewer data were available for halofantrine).
TABLE 2.
Pearson’s correlation matrix for IC50s
| Drug pair | No. of isolates | Correlation coefficient (R) | Pa |
|---|---|---|---|
| Mefloquine-artesunate | 51 | 0.43 | 0.018 |
| Mefloquine-halofantrine | 36 | 0.77 | <0.001 |
| Mefloquine-quinine | 53 | 0.79 | <0.001 |
| Mefloquine-chloroquine | 36 | 0.41 | NSb |
| Artesunate-halofantrine | 34 | 0.31 | NS |
| Artesunate-quinine | 50 | 0.41 | 0.03 |
| Artesunate-chloroquine | 33 | 0.18 | NS |
| Halofantrine-quinine | 35 | 0.66 | <0.001 |
| Halofantrine-chloroquine | 33 | 0.27 | NS |
| Quinine-chloroquine | 36 | 0.61 | 0.001 |
Bonferroni correction applied for multiple comparisons.
NS, not significant.
Molecular analysis.
Overall, 54% of infections (29 of 54) were found to be polyclonal by assessment of the MSP-1, MSP-2, and GLURP loci (25 with two clones, 3 with three clones, and 1 with four clones). In monoclonal infections there was no association between the MSP-2 dimorphism and in vitro sensitivity to any of the antimalarial drugs tested.
Amplification of pfmdr1 and mefloquine resistance.
pfmdr1 copy number (median, 1; range, 0.37 to 5.68) was correlated significantly with the IC50 of mefloquine (R = 0.37; P = 0.006). When rounded to the nearest integer, 35 isolates (65%) had a copy number of one. Of the remaining 19 with increased pfmdr1 copy number, 12 had two copies, 3 had three copies, 3 had four copies, and 1 had six copies. Copy number was not related to the clonality of infection, the sex or age of the patient, or the occurrence of recrudescent infections.
Parasite isolates with an increased pfmdr1 copy number had a significantly higher median mefloquine IC50 (65.6 ng/ml; IQR, 39.9 to 112) than isolates with a single copy of pfmdr1 (median IC50, 28.3 ng/ml; IQR, 9.6 to 68) (P = 0.01). In subsequent analyses, IC50s for isolates with a copy number of two or more were pooled because they were not significantly different. When assessing mefloquine resistance by using our predefined cutoff for sensitivity, 45 ng/ml (see Materials and Methods), 63% of isolates (12 of 19) with increased copy number were resistant to mefloquine, compared to 37% (13 of 35) of those with a single copy number (relative risk = 1.7 [95% CI, 1.0 to 3.0]; P = 0.06 [one-sided test for a directional hypothesis]). When using the standard cutoff for mefloquine resistance, 12 ng/ml, as previously published, the relative risk of resistance in parasites with multiple pfmdr1 copies compared to those with single copies fell to 1.3 (95% CI, 1.05 to 1.68; P = 0.04).
Amplification of pfmdr1 and other antimalarial agents.
Amplification of the pfmdr1 gene copy number was also associated with higher in vitro IC50s of artesunate and halofantrine. For these drugs, the IC50 was moderately correlated with pfmdr1 copy number (R = 0.34/P = 0.1 and R = 0.39/P = 0.18, respectively).
pfmdr1 codon mutations.
The Asn-to-Tyr mutation at codon 86 was detected in 11% (6 of 54) of isolates. Those with the Tyr mutation had significantly lower mefloquine IC50s (median, 9.0 ng/ml [IQR, 7.2 to 18.0] versus 52.4 [IQR, 15.8 to 84.3], respectively; P = 0.003), but this was not related to sensitivity to the other drugs tested. Nineteen isolates (35%) had the Tyr-to-Phe mutation at codon 184. Its presence was unrelated to pfmdr1 copy number or to in vitro sensitivity of the parasite to any drug tested. In two isolates, a Ser-to-Cys mutation at codon 1034 was detected. One of these had a pfmdr1 copy number of one, and the other had a copy number of two. Both were sensitive to mefloquine (IC50s, 9.1 and 15.3 ng/ml) and highly resistant to chloroquine (IC50s, >143 ng/ml). The Asn-to-Asp mutation at codon 1042 was detected in four isolates, only one of which was resistant to mefloquine (IC50, 68.0 ng/ml). No mutations were detected at codon 1246.
Combination of pfmdr1 copy number and codon 86 mutations.
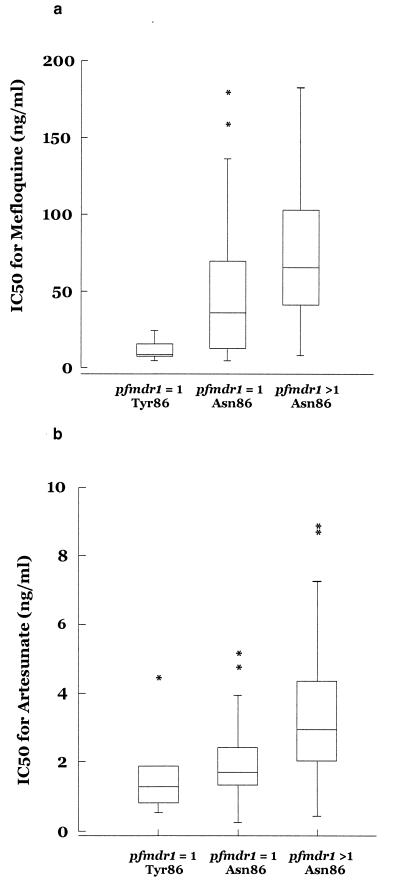
Six of 35 isolates with a single copy of pfmdr1 contained the Tyr86 mutation, compared with none of the 19 isolates with increased pfmdr1 copy number (P = 0.08). Isolates were therefore categorized into three groups according to pfmdr1 copy number and codon 86 allele: single pfmdr1 copy with the tyrosine mutation at codon 86, single pfmdr1 gene copy with wild-type (Asn) codon 86, and increased pfmdr1 copy number with wild-type codon 86. Results are tabulated in Tables 3 and 4 and presented in Fig. 1. There were significant differences in the IC50s of both mefloquine (P = 0.03) and artesunate (P = 0.003) between the three groups. Although there was a similar trend for halofantrine, this did not reach statistical significance (P = 0.07) because parasite numbers were smaller in this group.
TABLE 3.
Gene copy number and sequence analysis of pfmdr1 alleles from isolates of P. falciparum from the western border of Thailand
| Isolate | Mefloquine concn (ng/ml) | pfmdr1 copy no. | Amino acid at codona:
|
||||
|---|---|---|---|---|---|---|---|
| 86 | 184 | 1034 | 1042 | 1246 | |||
| 1 | 58.8 | 2.0 | Asn | Tyr | Ser | Asn | Asp |
| 2 | 88.6 | 1.0 | Asn | Tyr | Ser | Asn | Asp |
| 3 | 43.3 | 1.5 | Asn | Phe | Ser | Asn | Asp |
| 4 | 72.7 | 3.0 | Asn | Phe | Ser | Asn | Asp |
| 5 | 69.3 | 1.0 | Asn | Tyr | Ser | Asn | Asp |
| 6 | 24.7 | 0.5 | Tyr | Tyr | Ser | Asn | Asp |
| 7 | 11.4 | 0.9 | Asn | Phe | Ser | Asn | Asp |
| 8 | 34.7 | 0.9 | Asn | Tyr | Ser | Asn | Asp |
| 9 | 63.7 | 2.4 | Asn | Tyr | Ser | Asn | Asp |
| 10 | 157.9 | 1.1 | Asn | Phe | Ser | Asn | Asp |
| 11 | 68.0 | 0.5 | Asn | Phe | Ser | Asp | Asp |
| 12 | 36.1 | 1.0 | Asn | Tyr | Ser | Asn | Asp |
| 13 | 98.1 | 1.2 | Asn | Tyr | Ser | Asn | Asp |
| 14 | 8.6 | 2.8 | Asn | Tyr | Ser | Asn | Asp |
| 15 | 28.3 | 1.0 | Asn | Tyr | Ser | Asn | Asp |
| 16 | 65.7 | 2.3 | Asn | Tyr | Ser | Asn | Asp |
| 17 | 79.3 | 1.0 | Asn | Tyr | Ser | Asn | Asp |
| 18 | 32.3 | 0.6 | Asn | Tyr | Ser | Asn | Asp |
| 19 | 8.1 | 0.4 | Asn | Phe | Ser | Asp | Asp |
| 20 | 8.0 | 1.3 | Tyr | Tyr | Ser | Asn | Asp |
| 21 | 4.7 | 1.0 | Tyr | Tyr | Ser | Asn | Asp |
| 22 | 8.4 | 0.6 | Tyr | Tyr | Ser | Asn | Asp |
| 23 | 42.7 | 1.6 | Asn | Tyr | Ser | Asn | Asp |
| 24 | 13.0 | 0.7 | Asn | Tyr | Ser | Asn | Asp |
| 25 | 183.0 | 1.6 | Asn | Tyr | Ser | Asn | Asp |
| 26 | 58.3 | 1.0 | Asn | Phe | Ser | Asn | Asp |
| 27 | 7.2 | 1.0 | Asn | Phe | Ser | Asn | Asp |
| 28 | 38.3 | 0.6 | Asn | Phe | Ser | Asn | Asp |
| 29 | 15.8 | 1.0 | Tyr | Tyr | Ser | Asp | Asp |
| 30 | 137.3 | 1.6 | Asn | Tyr | Ser | Asn | Asp |
| 31 | 39.9 | 1.5 | Asn | Tyr | Ser | Asn | Asp |
| 32 | 71.1 | 1.3 | Asn | Phe | Ser | Asn | Asp |
| 33 | 111.7 | 4.1 | Asn | Tyr | Ser | Asn | Asp |
| 34 | 85.9 | 2.0 | Asn | Tyr | Ser | Asn | Asp |
| 35 | 17.4 | 2.6 | Asn | Phe | Ser | Asn | Asp |
| 36 | 46.5 | 0.5 | Asn | Tyr | Ser | Asn | Asp |
| 37 | 63.9 | 1.3 | Asn | Phe | Ser | Asn | Asp |
| 38 | 60.9 | 1.0 | Asn | Tyr | Ser | Asn | Asp |
| 39 | 115.4 | 3.8 | Asn | Tyr | Ser | Asn | Asp |
| 40 | 30.9 | 5.7 | Asn | Tyr | Ser | Asn | Asp |
| 41 | 9.1 | 0.7 | Asn | Tyr | Cys | Asn | Asp |
| 42 | 5.0 | 0.6 | Asn | Tyr | Ser | Asn | Asp |
| 43 | 179.1 | 0.5 | Asn | Tyr | Ser | Asn | Asp |
| 44 | 13.6 | 0.4 | Asn | Phe | Ser | Asn | Asp |
| 45 | 136.4 | 0.6 | Asn | Tyr | Ser | Asn | Asp |
| 46 | 13.3 | 0.5 | Asn | Phe | Ser | Asn | Asp |
| 47 | 8.8 | 0.4 | Asn | Phe | Ser | Asn | Asp |
| 48 | 12.3 | 0.6 | Asn | Tyr | Ser | Asn | Asp |
| 49 | 141.3 | 1.8 | Asn | Tyr | Ser | Asn | Asp |
| 50 | 9.6 | 1.0 | Tyr | Tyr | Ser | Asn | Asp |
| 51 | 15.3 | 1.7 | Asn | Phe | Cys | Asp | Asp |
| 52 | 17.5 | 0.9 | Asn | Phe | Ser | Asn | Asp |
| 53 | 65.6 | 2.2 | Asn | Phe | Ser | Asn | Asp |
| 54 | 94.2 | 3.5 | Asn | Phe | Ser | Asn | Asp |
Codon mutations are highlighted in boldface type.
TABLE 4.
IC50 profiles for each drug tested, categorized according to pfmdr1 genotype (single copy versus increased copy number) and the presence of a wild-type (Asn) or mutant (Tyr) allele at codon 86
| Drug | IC50 (ng/ml) for category:
|
Pd | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Single + mutateda
|
Single + wildb
|
Increased + wildc
|
||||||||
| Median | Range | IQR | Median | Range | IQR | Median | Range | IQR | ||
| Mefloquine | 9.0 | 4.7–24.7 | 8.0–15.8 | 36.1 | 5.0–179 | 13.0–69.3 | 65.6 | 8.6–183 | 41.3–103 | 0.001 |
| Artesunate | 1.3 | 0.55–4.43 | 0.83–1.88 | 1.71 | 0.25–5.15 | 1.35–2.42 | 2.94 | 0.45–8.85 | 2.04–4.37 | 0.007 |
| Halofantrine | 3.5 | 2.3–4.1 | 2.9–3.78 | 10.3 | 1.1–55.5 | 2.48–21.9 | 17.8 | 3.5–34.3 | 9.33–21.0 | 0.07 |
| Quinine | 156 | 99.9–766 | 133–350 | 302 | 49–1,259 | 203–637 | 404 | 149–945 | 224–662 | 0.38 |
| Chloroquine | 214 | 111–317 | 111–317 | 83 | 42–282 | 76–148 | 94 | 59.2–201 | 80.2–102 | 0.29 |
Single copy of pfmdr1, mutated codon 86 (Tyr).
Single copy of pfmdr1, wild-type codon 86 (Asn).
Increased pfmdr1 copy number, wild-type codon 86 (Asn).
One-way analysis of variance of normalized data.
FIG. 1.
Box plots of mean IC50s (in nanograms per milliliter) ± standard errors of the means for mefloquine (a) and artesunate (b), categorized according to pfmdr1 genotype (single copy versus increased copy number) and the presence of a wild-type (Asn) or mutated (Tyr) allele at codon 86. ∗, outlier.
A multiple linear regression model was constructed, using the IC50 as the dependent variable and codon 86 mutations and increased pfmdrf1 copy number as independent variables. No other assessed variables (e.g., age, prior culturing, clonality, baseline parasitemia, etc.) were correlated significantly with the IC50 in univariate analyses, and they were therefore not included in subsequent models. In this model, both increased pfmdr1 copy number and Asn86 were independently associated with higher mefloquine IC50s (P = 0.01 and P = 0.04, respectively). However, for a similar model created for artesunate, only pfmdr1 copy number, and not codon 86 mutation, was associated with artesunate IC50s (P = 0.005 and P = 0.5, respectively).
None of the 6 isolates with the Tyr mutation at codon 86 (Tyr86) were resistant to mefloquine, compared to 45% (13 of 29) of isolates with a single copy of wild-type pfmdr1 and 63% (12 of 19) of isolates with an increased number of copies of the wild-type pfmdr1 gene (P = 0.025). When a cutoff of 12 ng/ml was applied, the corresponding figures were 33% (2 of 6), 79% (23 of 29), and 95% (18 of 19) (P = 0.008).
DISCUSSION
The P. falciparum isolates in this study derive from an area on the western border of Thailand where high-grade multidrug-resistant falciparum malaria has been confirmed by both clinical and in vitro studies (33). In 1994 the cure rate for malaria patients given mefloquine monotherapy at the study site had fallen to 50%, with 15% of patients exhibiting treatment failure within the first week, and the use of this antimalarial agent as monotherapy could no longer be advocated (21). At this time, treatment protocols were changed to recommend a combination regimen consisting of mefloquine and a 3-day course of artesunate (MAS3). Although it has not been possible to assess the efficacy of mefloquine monotherapy since 1994, the efficacy of the MAS3 regimen, intrinsically dependent on the mefloquine component, has remained in excess of 95%.
The IC50s for our parasites were generally much higher than those observed in previous field studies of molecular markers of resistance in Southeast Asia (3, 35), sub-Saharan Africa, (5), and South America (36). The IC50 range defining resistant isolates has previously been derived, using African isolates, as being more than 2 standard deviations from the population’s mean IC50 (19). This assumes a symmetrical normal distribution of sensitivities and means that there will always be “resistant” isolates in any series. Using these criteria, 91% of our isolates would be classified as being resistant to mefloquine (>20 nM, 8.30 ng/ml), 75% would be said to be resistant to halofantrine (>5 nM, 2.68 ng/ml), 64% would be called resistant to quinine (>500 nM, 258.3 ng/ml), and 94% would be classed as resistant to chloroquine (>100 nM, 51.6 ng/ml).
Four previous studies have used field isolates to examine the relationship between an increase in pfmdr1 copy number and multidrug resistance (3, 5, 35, 36). In one of these studies, 10 of 11 parasite isolates were resistant to mefloquine, and for all 10, amplification of the pfmdr1 gene was associated with its overexpression (35). The three other studies failed to demonstrate an association between increased copy number and a drug-resistant phenotype; however, the numbers of isolates in which pfmdr1 copy number was assessed were small (13 or less), and nearly all of these isolates were sensitive to mefloquine in vitro (3, 5, 36). A further methodological difficulty in assessing gene copy number in field isolates is the relatively lower level of reproducibility of estimates obtained by Southern blot techniques. These methods also require microgram quantities of DNA, and therefore prior culturing of parasites is necessary.
In our study, pfmdr1 copy number was assessed in blood samples obtained directly from patients, therefore avoiding potential artifacts arising from culturing of parasites. Furthermore, the lack of an association between MSP-2 dimorphism and in vitro sensitivity to any of the antimalarial drugs tested indicates that the genetic population structure did not bias our interpretation of the contribution of pfmdr1 to drug resistance. pfmdr1 amplification was found in 35% of isolates and was associated significantly with increased IC50s of mefloquine and artesunate (Table 4 and Fig. 1). Although a similar trend was noted for halofantrine, this did not reach statistical significance (P = 0.07). This might be due to either the lack of an association or a lack of statistical power as a consequence of the examination of fewer parasite strains in the halofantrine group. The cross-correlations between mefloquine, halofantrine, and artesunate IC50s (Table 2) have been noted in previous in vitro (4, 24) and molecular (10, 18, 35) studies. In the present study, the relationship between pfmdr1 and the sensitivity to structurally distinct antimalarial drugs (e.g., mefloquine and artesunate) suggests the presence of a true MDR-like phenomenon.
The Asn-to-Tyr mutation at codon 86 was found in six isolates, each of which had a single copy of pfmdr1 and was sensitive to mefloquine. In contrast, isolates with increased pfmdr1 copy number all had the wild-type allele at codon 86, and 63% of them (12 of 19) were resistant to mefloquine (Fig. 1a). In multivariate analyses, both increased pfmdr1 copy number and the presence of the Asn86 allele were independently associated with higher mefloquine IC50s. These findings concur with in vitro studies of yeast cells, in which expression of wild-type pfmdr1 caused resistance to mefloquine and halofantrine but introduction of amino acid polymorphisms abolished this protection (25). Chloroquine resistance is most consistently associated with a tyrosine 86 mutation in pfmdr1 (30). Of the 36 isolates in our study for which chloroquine IC50s were available, only 2 had this mutation, and both of them were resistant to chloroquine (IC50s, 111 and 317 ng/ml). Taken together, these observations support the hypothesis that pfmdr1 acts to modulate the transport of drugs and that mutation or overexpression of pfmdr1 may alter its function.
pfmdr1 polymorphisms were used to predict in vitro mefloquine resistance. An increase in pfmdr1 copy number correctly identified resistant isolates (IC50, >45 ng/ml) with a sensitivity of 48% (12 of 25). The addition of mutations at codons 86, 1034, and 1042 to identify sensitive isolates allowed a discriminative test which correctly categorized 68% of isolates (19 of 28), identifying resistant isolates with a sensitivity of 92% (95% CI, 64 to 100) and a specificity of 60% (95% CI, 32 to 84). If the standard cutoff for mefloquine resistance (>12 ng/ml) was adopted, then sensitivity fell to 81% (95% CI, 58 to 95) and specificity rose to 85% (95% CI, 42 to 100). However, this limited genetic profile was unable to assign 48% of the isolates (26 of 54) to a category (i.e., all of the isolates with single copies of the wild-type gene), and 46% of these (12 of 26) were found to be resistant to mefloquine. Hence, while the findings strongly implicate a role for pfmdr1 in multidrug resistance, additional explanations are also required for a comprehensive understanding of drug resistance in this region. A number of studies have shown that an increase in pfmdr1 copy number is associated with increased expression of the protein (Pgh1) (10, 20, 35). However, the possibility of increased expression of Pgh1 in the presence of single copies of the pfmdr1 gene has not been described. Mefloquine resistance with single copies of wild-type pfmdr1 may arise either from increased gene expression in the absence of amplification of gene copy number or through other, as-yet-undefined, pfmdr1-unrelated molecular mechanisms. It is possible that the observed association of resistance and pfmdr1 is due to another gene closely linked to pfmdr1.
Although molecular techniques are increasingly being used to monitor pyrimethamine and sulfadoxine resistance, assessing in vitro mefloquine resistance will continue to depend on a combination of in vivo and in vitro techniques and may be improved by a more complete understanding of the molecular mechanisms of mefloquine resistance.
ACKNOWLEDGMENTS
Alan Brockman, Claire Cassar, and Manoj Duraisingh contributed equally to this project.
We thank the staff of the Shoklo Malaria Research Unit for the provision of the field samples. We also thank Alan Cowman for supplying the K1 isolate, David Warhurst for comments on the manuscript, and Paul Hellyer and Janine Laurie for logistical support.
Sanjeev Krishna is a Wellcome Trust Senior Research Fellow in Clinical Science. This work was funded by a project grant from the Medical Research Council (ID 41178).
REFERENCES
- 1.Adagu I S, Dias F, Pinheiro L, Rombo L, do Rosario V, Warhurst D C. Guinea Bissau: association of chloroquine resistance of Plasmodium falciparum with the Tyr86 allele of the multiple drug resistance gene Pfmdr1. Trans R Soc Trop Med Hyg. 1996;90:90–91. doi: 10.1016/s0035-9203(96)90491-5. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 2.Awad el Kariem F M, Miles M A, Warhurst D C. Chloroquine-resistant Plasmodium falciparum isolates from The Sudan lack two mutations in the pfmdr1 gene thought to be associated with chloroquine resistance. Trans R Soc Trop Med Hyg. 1992;86:587–589. doi: 10.1016/0035-9203(92)90140-8. [DOI] [PubMed] [Google Scholar]
- 3.Basco L K, Eldin de Pecoulas P, Le Bras J, Wilson C M. Plasmodium falciparum: molecular characterization of multidrug-resistant Cambodian isolates. Exp Parasitol. 1996;82:97–103. doi: 10.1006/expr.1996.0013. [DOI] [PubMed] [Google Scholar]
- 4.Basco L K, Le Bras J. In vitro activity of halofantrine and its relationship to other standard antimalarial drugs against African isolates and clones of Plasmodium falciparum. Am J Trop Med Hyg. 1992;47:521–527. doi: 10.4269/ajtmh.1992.47.521. [DOI] [PubMed] [Google Scholar]
- 5.Basco L K, Le Bras J, Rhoades Z, Wilson C M. Analysis of pfmdr1 and drug susceptibility in fresh isolates of Plasmodium falciparum from subsaharan Africa. Mol Biochem Parasitol. 1995;74:157–166. doi: 10.1016/0166-6851(95)02492-1. [DOI] [PubMed] [Google Scholar]
- 6.Basco L K, Ringwald P. Molecular epidemiology of malaria in Yaounde, Cameroon. Analysis of chloroquine resistance and point mutations in the multidrug resistance (pfmdr1) gene of Plasmodium falciparum. Am J Trop Med Hyg. 1998;59:577–581. doi: 10.4269/ajtmh.1998.59.577. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharya P R, Biswas S, Kabilan L. Alleles of the Plasmodium falciparum Pfmdr1 gene appear not to be associated with chloroquine resistance in India. Trans R Soc Trop Med Hyg. 1997;91:454–455. doi: 10.1016/s0035-9203(97)90283-2. [DOI] [PubMed] [Google Scholar]
- 8.Bland M, Altman D. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;i:307–310. [PubMed] [Google Scholar]
- 8a.Brockman, A. Unpublished data.
- 9.Brockman A, Paul R E L, Anderson T J C, Hackford I, Phaipun L, Looareesuwan S, Nosten F, Day K P. Application of genetic markers to the identification of recrudescent Plasmodium falciparum infections on the northwestern border of Thailand. Am J Trop Med Hyg. 1999;60:14–21. doi: 10.4269/ajtmh.1999.60.14. [DOI] [PubMed] [Google Scholar]
- 10.Cowman A F, Galatis D, Thompson J K. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc Natl Acad Sci USA. 1994;91:1143–1147. doi: 10.1073/pnas.91.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox Singh J, Singh B, Alias A, Abdullah M S. Assessment of the association between three pfmdr1 point mutations and chloroquine resistance in vitro of Malaysian Plasmodium falciparum isolates. Trans R Soc Trop Med Hyg. 1995;89:436–437. doi: 10.1016/0035-9203(95)90045-4. [DOI] [PubMed] [Google Scholar]
- 12.Duraisingh M T, Curtis J, Warhurst D C. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp Parasitol. 1998;89:1–8. doi: 10.1006/expr.1998.4274. [DOI] [PubMed] [Google Scholar]
- 13.Duraisingh M T, Drakeley C J, Muller O, Bailey R, Snounou G, Targett G A T, Greenwood B M, Warhurst D C. Selection for the tyrosine-86 mutation of the pfmdr1 gene of Plasmodium falciparum by chloroquine and amodiaquine. Parasitology. 1997;114:205–211. doi: 10.1017/s0031182096008487. [DOI] [PubMed] [Google Scholar]
- 14.Foote S J, Kyle D E, Martin R K, Oduola A M, Forsyth K P, Kemp D J, Cowman A F. Several alleles of the multidrug resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990;345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- 15.Foote S J, Thompson J K, Cowman A F, Kemp D J. Amplification of the multidrug resistance gene in some chloroquine-resistant isolates of P. falciparum. Cell. 1989;57:921–930. doi: 10.1016/0092-8674(89)90330-9. [DOI] [PubMed] [Google Scholar]
- 16.Lim A S, Galatis D, Cowman A F. Plasmodium falciparum: amplification and overexpression of pfmdr1 is not necessary for increased mefloquine resistance. Exp Parasitol. 1996;83:295–303. doi: 10.1006/expr.1996.0077. [DOI] [PubMed] [Google Scholar]
- 17.Nosten F, ter Kuile F O, Chongsuphajaisiddhi T, Luxemburger C, Webster H K, Edstein M, Phaipun L, Thew K L, White N J. Mefloquine-resistant falciparum malaria on the Thai-Burmese border. Lancet. 1991;337:1140–1143. doi: 10.1016/0140-6736(91)92798-7. [DOI] [PubMed] [Google Scholar]
- 18.Peel S A, Bright P, Yount B, Handy J, Baric R S. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am J Trop Med Hyg. 1994;51:648–658. doi: 10.4269/ajtmh.1994.51.648. [DOI] [PubMed] [Google Scholar]
- 19.Pradines B, Rogier C, Fusai T, Tall A, Trape J F, Doury J C. In vitro activity of artemether against African isolates (Senegal) of Plasmodium falciparum in comparison with standard antimalarial drugs. Am J Trop Med Hyg. 1998;58:354–357. doi: 10.4269/ajtmh.1998.58.354. [DOI] [PubMed] [Google Scholar]
- 19a.Price, R., and F. Nosten. Unpublished data.
- 20.Price R, Robinson G, Brockman A, Cowman A, Krishna S. Assessment of pfmdr1 gene copy number by tendem competitive polymerase chain reaction. Mol Biochem Parasitol. 1997;85:161–169. doi: 10.1016/s0166-6851(96)02822-8. [DOI] [PubMed] [Google Scholar]
- 21.Price R N, Nosten F, Luxemburger C, Am Kham, Brockman A, Chongsuphajaisiddhi T, White N J. Artesunate versus artemether in combination with mefloquine for the treatment of multidrug resistant falciparum malaria. Trans R Soc Trop Med Hyg. 1995;89:523–527. doi: 10.1016/0035-9203(95)90094-2. [DOI] [PubMed] [Google Scholar]
- 22.Price R N, Nosten F, Luxemburger C, van Vugt M, Phaipun L, Chongsuphajaisiddhi T, White N J. Artesunate/mefloquine treatment of multi-drug resistant falciparum malaria. Trans R Soc Trop Med Hyg. 1997;91:574–577. doi: 10.1016/s0035-9203(97)90032-8. [DOI] [PubMed] [Google Scholar]
- 23.Reacher M. Drug therapy for Plasmodium falciparum malaria resistant to pyrimethamine and sulfadoxine. Lancet. 1981;ii:1066–1068. doi: 10.1016/s0140-6736(81)91274-5. [DOI] [PubMed] [Google Scholar]
- 24.Ringwald P, Bickii J, Basco L K. In vitro activity of antimalarials against clinical isolates of Plasmodium falciparum in Yaounde, Cameroon. Am J Trop Med Hyg. 1996;55:254–258. doi: 10.4269/ajtmh.1996.55.254. [DOI] [PubMed] [Google Scholar]
- 25.Ruetz S, Delling U, Brault M, Schurr E, Gros P. The pfmdr1 gene of Plasmodium falciparum confers cellular resistance to antimalarial drugs in yeast cells. Proc Natl Acad Sci USA. 1996;93:9942–9947. doi: 10.1073/pnas.93.18.9942. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Su X Z, Kirkman L A, Fujioka H, Wellems T E. Complex polymorphisms in an ∼330 kDa protein are linked to chloroquine-resistant P. falciparum in Southeast Asia and Africa. Cell. 1997;91:593–603. doi: 10.1016/s0092-8674(00)80447-x. [DOI] [PubMed] [Google Scholar]
- 27.ter Kuile F O, Dolan G, Nosten F, Edstein M D, Luxemburger C, Phaipun L, Chongsuphajaisiddhi T, Webster H K, White N J. Halofantrine versus mefloquine in the treatment of multi-drug resistant falciparum malaria. Lancet. 1993;341:1044–1049. doi: 10.1016/0140-6736(93)92409-m. [DOI] [PubMed] [Google Scholar]
- 28.ter Kuile F O, Nosten F, Thieren M, Luxemburger C, Edstein M D, Chongsuphajaisiddhi T, Phaipun L, Webster H K, White N J. High dose mefloquine in the treatment of multidrug resistant falciparum malaria. J Infect Dis. 1992;166:1393–1400. doi: 10.1093/infdis/166.6.1393. [DOI] [PubMed] [Google Scholar]
- 29.Viriyakosol S, Siripoon N, Petcharapirat C, Petcharapirat P, Jarra W, Thaithong S, Brown K N, Snounou G. Genotyping of Plasmodium falciparum isolates by the polymerase chain reaction and potential uses in epidemiological studies. Bull W H O. 1995;73:85–95. [PMC free article] [PubMed] [Google Scholar]
- 30.von Seidlein L, Duraisingh M T, Drakeley C J, Bailey R, Greenwood B M, Pinder M. Polymorphism of the Pfmdr1 gene and chloroquine resistance in Plasmodium falciparum in The Gambia. Trans R Soc Trop Med Hyg. 1997;91:450–453. doi: 10.1016/s0035-9203(97)90281-9. [DOI] [PubMed] [Google Scholar]
- 31.Webster H K, Boudreau E F, Pavanand K, Yongvanitchit K, Pang L W. Antimalarial drug susceptibility testing of Plasmodium falciparum in Thailand using a microdilution radioisotope method. Am J Trop Med Hyg. 1985;34:228–235. doi: 10.4269/ajtmh.1985.34.228. [DOI] [PubMed] [Google Scholar]
- 32.Wellems T E, Panton L J, Gluzman I Y, do Rosario V E, Gwadz R W, Walker-Jonah A. Chloroquine resistance not linked to mdr-like genes in Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- 33.White N J. Antimalarial drug resistance: the pace quickens. J Antimicrob Chemother. 1992;30:571–585. doi: 10.1093/jac/30.5.571. . (Review.) [DOI] [PubMed] [Google Scholar]
- 34.Wilson C M, Serrano A E, Wasley A, Bogenschutz M P, Shankar A H, Wirth D F. Amplification of a gene related to mammalian mdr genes in drug-resistant Plasmodium falciparum. Science. 1989;244:1184–1186. doi: 10.1126/science.2658061. [DOI] [PubMed] [Google Scholar]
- 35.Wilson C M, Volkman S K, Thaithong S, Martin R K, Kyle D E, Milhous W K, Wirth D F. Amplification of pfmdr1 associated with mefloquine and halofantrine resistance in Plasmodium falciparum from Thailand. Mol Biol Parasitol. 1993;57:151–160. doi: 10.1016/0166-6851(93)90252-s. [DOI] [PubMed] [Google Scholar]
- 36.Zalis M G, Pang L, Silveira M S, Milhous W K, Wirth D F. Characterization of Plasmodium falciparum isolated from the Amazon region of Brazil: evidence for quinine resistance. Am J Trop Med Hyg. 1998;58:630–637. doi: 10.4269/ajtmh.1998.58.630. [DOI] [PubMed] [Google Scholar]