ABSTRACT
Objective.
To evaluate molecular tools to detect low-level parasitemia and the five species of Plasmodium that infect humans for use in control and elimination programs, and in reference laboratories.
Methods.
We evaluated 145 blood samples from patients who tested positive by nested polymerase chain reaction (nPCR), from asymptomatic individuals and from the WHO Global Malaria Programme/United Kingdom National External Quality Assessment Service. Samples were assayed using the genus-specific RealStar® Malaria PCR Kit 1.0 (alt-Gen; altona Diagnostics) and the RealStar® Malaria Screen & Type PCR Kit (alt-S&T; altona Diagnostics). The results from the molecular tests were compared with those from quantitative PCR (qPCR), nPCR and thick blood smear.
Results.
The levels of parasitemia ranged from 1 to 518 000 parasites/µL, depending on the species. Compared with nPCR, alt-S&T had a sensitivity of 100%, except for identifying P. falciparum, for which the sensitivity was 93.94%. All samples positive by alt-Gen were also positive by nPCR. When comparing alt-Gen to qPCR, the sensitivity was 100% for P. vivax, P. malariae and P. falciparum. For all Plasmodium species, the correlation between cycle threshold values of alt-S&T and alt-Gen compared with qPCR was significant (P < 0.0001, Spearman’s test), with r = 0.8621 for alt-S&T and r = 0.9371 for alt-Gen. When all Plasmodium species were considered, there was a negative correlation between the level of parasitemia and real-time PCR cycle threshold values (P < 0.0001). In this study, only 2 of 28 samples from asymptomatic individuals were positive by thick blood smear; however, all 28 of these samples were positive by alt-S&T.
Conclusions.
The alt-Gen and alt-S&T assays are suitable for detecting submicroscopic infections for distinct epidemiological purposes, such as for use in surveys and reference laboratories, and screening in blood banks, which will contribute to global efforts to eliminate malaria.
Keywords: Malaria, Plasmodium, real-time polymerase chain reaction
RESUMEN
Objetivo.
Evaluar herramientas moleculares para detectar bajos niveles de parasitemia y las cinco especies de Plasmodium que infectan a los seres humanos, a fin de emplearlas en los programas de control y eliminación y en los laboratorios de referencia.
Métodos.
Se evaluaron 145 muestras de sangre de pacientes positivos por reacción en cadena de la polimerasa anidada (nPCR), de individuos asintomáticos y de muestras del Programa Mundial de Malaria de la Organización Mundial de la Salud/Servicio Nacional de Evaluación Externa de Calidad del Reino Unido. Las muestras se analizaron con el kit de PCR RealStar® Malaria 1.0 (alt-Gen; altona Diagnostics), específico para cada género, y con el kit de PCR RealStar® Malaria Screen & Type (alt-S&T; altona Diagnostics). Se compararon los resultados de las pruebas moleculares con los de la PCR cuantitativa (qPCR), la nPCR y el frotis de gota gruesa.
Resultados.
Los niveles de parasitemia oscilaron entre 1 y 518 000 parásitos/µl, según la especie. En comparación con la nPCR, la prueba alt-S&T tuvo una sensibilidad del 100%, excepto para la identificación de P. falciparum, para el cual la sensibilidad fue del 93,94%. Todas las muestras positivas por alt-Gen lo fueron también por nPCR. Al comparar alt-Gen con la qPCR, la sensibilidad fue del 100% para P. vivax, P. malariae y P. falciparum. Para todas las especies de Plasmodium, la correlación entre los valores del umbral de ciclo de alt-S&T y alt-Gen en comparación con la qPCR fue significativa (P < 0,0001, prueba de Spearman), con r = 0,8621 para alt-S&T y r = 0,9371 para alt-Gen. Cuando se consideraron todas las especies de Plasmodium hubo una correlación negativa entre el nivel de parasitemia y los valores de umbral de ciclo de PCR en tiempo real (P < 0,0001). En este estudio, solo 2 de las 28 muestras de individuos asintomáticos fueron positivas por frotis de gota gruesa; sin embargo, las 28 muestras fueron positivas por alt-S&T.
Conclusiones.
Los ensayos alt-Gen y alt-S&T son adecuados para detectar infecciones submicroscópicas con distintos fines epidemiológicos, como su uso en investigaciones y laboratorios de referencia y el cribado en bancos de sangre, lo que contribuirá a los esfuerzos mundiales para eliminar la malaria.
Palabras clave: Malaria, Plasmodium, reacción en cadena en tiempo real de la polimerasa
RESUMO
Objectivo.
Avaliar ferramentas moleculares para detectar parasitemia de baixo nível e as cinco espécies de Plasmodium que infectam humanos, para utilização em programas de controlo e eliminação e em laboratórios de referência.
Métodos.
Avaliámos 145 amostras de sangue de doentes que testaram positivo por reacção em cadeia da polimerase aninhada (nPCR), de indivíduos assintomáticos, e do Programa Global de Paludismo da Organização Mundial de Saúde/Serviço Nacional de Avaliação da Qualidade Externa do Reino Unido. As amostras foram ensaiadas utilizando o RealStar® Malaria PCR Kit 1.0 (alt-Gen; altona Diagnostics) e o RealStar® Malaria Screen & Type PCR Kit (alt-S&T; altona Diagnostics). Os resultados dos testes moleculares foram comparados com os resultados da PCR quantitativa (qPCR), nPCR e exame da gota espessa.
Resultados.
Os níveis de parasitemia variaram de 1 a 518 000 parasitas/µL, dependendo da espécie. Em comparação com a nPCR, alt-S&T tinha uma sensibilidade de 100%, excepto na identificação de P. falciparum, para a qual a sensibilidade era de 93,94%. Todas as amostras positivas por alt-Gen foram também positivas por nPCR. Ao comparar alt-Gen com qPCR, a sensibilidade foi de 100% para P. vivax, P. malariae e P. falciparum. Para todas as espécies Plasmodium, a correlação entre os valores limiares de ciclo de alt-S&T e alt-Gen comparados com qPCR foi significativa (P < 0,0001, teste de Spearman), com r = 0,8621 para alt-S&T e r = 0,9371 para alt-Gen. Quando todas as espécies de Plasmodium foram consideradas, houve uma correlação negativa entre o nível de parasitemia e os valores limiares do ciclo de PCR em tempo real (P < 0,0001). Neste estudo, apenas 2 de 28 amostras de indivíduos assintomáticos foram positivas por exame da gota espessa; no entanto, todas estas 28 amostras foram positivas por alt-S&T.
Conclusões.
Os ensaios alt-Gen e alt-S&T são adequados para a detecção de infecções submicroscópicas para fins epidemiológicos distintos, tais como para utilização em inquéritos e laboratórios de referência e o rastreio em bancos de sangue, o que contribuirá para os esforços globais de eliminação da malária.
Palavras-chave: Malária, Plasmodium, reação em cadeia da polimerase em tempo real
Malaria is an infectious disease transmitted by Anopheles infected by the Plasmodium parasite. There are five Plasmodium species known to infect humans: P. falciparum, P. vivax, P. malariae, P. ovale and P. knowlesi. According to the World Health Organization (WHO), there were an estimated 229 million cases of malaria in 2019 in 87 malaria-endemic countries, with 409 000 deaths. Although the WHO Region of the Americas had a 40% reduction in malaria cases from 2000 to 2019, the Region was impacted by an increase in the burden of malaria in the Bolivarian Republic of Venezuela, from 35 500 cases in 2000 to more than 467 000 by 2019. Brazil, Colombia and Venezuela (Bolivarian Republic of) notify more than 86% of all cases in the Region (1).
In Brazil, 99% of malaria cases occur in the Amazon region, which notified 157 454 cases in 2019 (2). In the same year, 542 cases were reported from outside the Amazon region, with 55 cases classified as autochthonous transmission in areas of the Atlantic Forest (3).
Infection with some species, such as P. falciparum and P. knowlesi, can lead to fatal malaria if not diagnosed and treated promptly. The gold standard for detecting the parasite is still the thick blood smear (TBS) with Giemsa stain because it is low cost and easy to perform (4). Furthermore, the technique allows for the quantification of parasites and the differentiation of species, which is important since treatment differs by species (5). However, sensitivity and specificity depend on the microscopist, and even experienced examiners cannot detect <50 parasites/µL (6). Malaria can also be diagnosed using rapid diagnostic tests (RDTs), which use immunochromatographic techniques, are easy to perform and do not require equipment, such as a microscope or thermocycler (7). However, the minimum that most RDTs can detect is 100 parasites/µL (6). Although WHO recommends using TBS and RDTs as primary diagnostic tools (5), both techniques are not sensitive in detecting low-level parasitemia, and more accurate protocols are essential for malaria control, as false-negative results contribute to maintaining sources for mosquito infections (8). Moreover, even in reference laboratories, species determination is not always easy due to morphological similarities (9).
A wide variety of molecular approaches is available for diagnosing Plasmodium infection, such as polymerase chain reaction (PCR), and these molecular approaches are significantly more sensitive than microscopy and RDTs (8). These techniques identify Plasmodium infections using different gene targets. Snounou and colleagues first used PCR to detect four species of Plasmodium (10) targeting 18S ribosomal RNA (18S rRNA), with a limit of detection (LoD) of 10 parasites/µL (11). Since then, new technologies have been developed, including multiplex PCR with an LoD of 0.2–5 parasites/µL (12). However, conventional PCR is time-consuming and specimens may be easily contaminated due to the amplicons produced. To overcome these disadvantages, several real-time PCR protocols have been developed. Lima et al. described a sensitive real-time protocol with an LoD of 1 parasite/µL (13). More recently, a loop-mediated isothermal amplification assay for detecting Plasmodium has been described, with an LoD of 1–8 parasites/µL, depending on the species (14). However, these assays (13, 14) are genus-specific, and additional assays are needed for species differentiation. Therefore, there is a need for more sensitive and easy-to-use molecular techniques for diagnosis, especially for asymptomatic infections that pose a challenge to efforts to control and eliminate transmission.
This study aimed to evaluate molecular tools to detect low-level parasitemia and the five species of Plasmodium known to infect humans for use in control and elimination programs, and in reference laboratories. We evaluated the RealStar® Malaria PCR Kit 1.0 (alt-Gen; altona Diagnostics, Germany) and RealStar® Malaria Screen & Type PCR Kit (alt-S&T; altona Diagnostics, Germany), which are two real-time PCR assays for screening for, respectively, the genus Plasmodium and the five species of Plasmodium that infect humans. The results from these assays were compared with in-house genus-specific quantitative PCR (qPCR) testing (13), nested PCR (nPCR) (11) and TBS.
METHODS
All procedures were performed in accordance with good laboratory practices in a reference laboratory for malaria diagnosis.
Characterization of samples
The minimum sample size was calculated according to Banoo et al. (15), considering an expected sensitivity of 97% (p), with 90% minimum acceptable sensitivity (po), obtaining a minimum sample of 62.3, where Z1 = 1.96 for α = 0.05 and Z2 = 1.28, giving a 90% power for the test:
We evaluated 111 blood samples collected in ethylenediaminetetraacetic acid (EDTA) tubes or dried blood spots (DBS). Samples were obtained from: (a) patients admitted to the Núcleo de Estudos em Malária/Hospital das Clínicas/Secretaria de Saúde de São Paulo who tested positive for Plasmodium infection and (b) asymptomatic individuals from areas of low endemicity in the state of São Paulo during surveillance activities carried out from June 2017 to June 2019, all of which were tested by TBS, according to Brazilian guidelines (16). The use of the samples for this study was approved by the Ethics Committee of the Hospital das Clínicas and the Medical School of the University of São Paulo (approval nos. 0493/10, 0791/08 and 446/11). All individuals provided informed consent and their anonymity was guaranteed. In addition, 34 samples from the WHO Global Malaria Programme in partnership with the United Kingdom National External Quality Assessment Service (10) were used as references: 12 samples of P. falciparum, 6 of P. vivax, 6 of P. ovale, 3 of P. knowlesi, 2 of P. malariae and 5 negative samples (Figure 1). Of the 145 blood samples previously tested by at least one of the techniques (qPCR, nPCR, TBS), 140 were positive for Plasmodium and 5 were negative; 105 were collected in EDTA and 40 in DBS. The levels of parasitemia for P. falciparum ranged from 1 to 100 000 parasites/μL (median: 500), for P. vivax from 1 to 41 760 parasites/μL (median: 15 120) and for P. malariae from 1 to 518 000 parasites/μL (median: 5130). Twenty-six samples from asymptomatic individuals from areas of low endemicity in the state of São Paulo were negative by TBS.
FIGURE 1. Flowchart of positive, negative and reference samples used in the study, by test, with nested polymerase chain reaction as the reference test and 5 negative samples from the WHO Global Malaria Programme external quality assessment scheme (WHO/UK EQA).
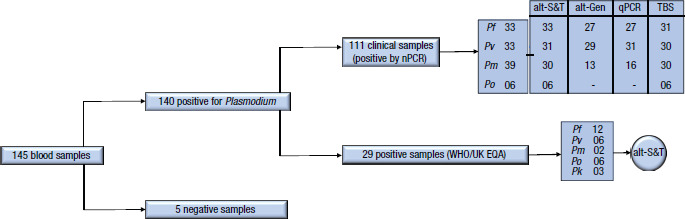
alt-Gen: RealStar® Malaria PCR Kit 1.0
alt-S&T: RealStar® Malaria Screen & Type PCR Kit
nPCR: nested polymerase chain reaction
qPCR: quantitative PCR
Pf: Plasmodium falciparum; Pv: Plasmodium vivax; Pm: Plasmodium malariae; Po: Plasmodium ovale; Pk: Plasmodium knowlesi
Source: Figure 1 was created by the authors based on the results of the study.
DNA extraction
For samples collected in EDTA, DNA was extracted using the QIAamp® DNA Blood Mini Kit (QIAGEN, Germany), according to the manufacturer’s instructions: to a microtube containing 20 µL of protease, 200 µL of blood was added along with 200 µL of AL lysis buffer, with incubation at 56 °C for 10 minutes. The next step was to add 200 µL of ethanol, after which the sample was vortexed and applied to the silica spin column. After centrifugation (6000 × g for 1 minute), the column was inserted into a clean collection tube and the filtrate discarded; 500 µL of wash buffer AW1 was added, and the column was centrifuged (6000 × g for 1 minute). The column was then inserted into a clean collection tube and the filtrate discarded; 500 µL wash buffer AW2 was added; the column was centrifuged (20 000 × g for 3 minutes) and the filtrate discarded. A 200 µL volume of AE elution buffer was added to the column, which was then incubated at room temperature for 5 minutes, centrifuged (6000 × g for 1 minute) and the eluted DNA stored at −20 °C. For DBS samples, DNA was extracted using the Chelex 100® protocol (Bio-Rad, United States of America) (17). Of the 34 reference samples from the WHO external quality assurance scheme, 24 were extracted from EDTA and 10 from DBS.
Molecular assays
Quantitative PCR. The protocol described by Lima and colleagues (13) was used for genus-specific amplification targeting the 18S rRNA genes of Plasmodium. The primers M60 and M61 and the M62 probe were used, with 2.5 μL of genomic DNA, 12.5 μL of 2X TaqMan® Universal PCR Master Mix (Applied Biosystems, USA), 500 nM of each primer and 300 nM of FAM™- and TAMRA™-labeled probes (Applied Biosystems, USA). Amplification reactions were performed under the following conditions: 50 ºC for 2 minutes and 95 ºC for 10 minutes; these were followed by 40 cycles at 94 ºC for 30 seconds and a final cycle at 60 ºC for 1 minute. Duplicate samples were assayed in the 7500 Real Time PCR System™ (Applied Biosystems, USA). All reactions were assayed with both positive controls (DNA from P. falciparum culture diluted to 1, 10 and 100 parasites/ μL) and negative controls (DNA from uninfected individuals).
Nested PCR. The protocol described by Snounou and colleagues (11) was used, targeting the 18S rRNA genes. The first reaction used the genus-specific primers rPLU5 and rPLU6, and the second reaction used the species-specific primers rFAL1 and rFAL2 for P. falciparum, rVIV1 and rVIV2 for P. vivax, rMAL1 and rMAL2 for P. malariae, and rOVA1 and rOVA2 for P. ovale. Reactions were prepared with 2 mM MgCl2, 50 mM KCl, 10 mM Tris at pH 8.3, 250 nM of each primer, 125 μM of deoxynucleotide triphosphates, 0.4 U Taq polymerase and 2 μL of DNA. The amplicons were resolved by electrophoresis in 1.5% agarose gel in Tris-borate-EDTA buffer, stained with Blue Green Loading Dye I (LGC Biotecnologia, Brazil) and visualized through ultraviolet light in the UVI-doc HD2 System (UVITEC, United Kingdom). All reactions included both positive controls (DNA from P. falciparum, P. vivax, P. malariae and P. ovale isolates) and negative controls (DNA from uninfected individuals).
RealStar® Malaria PCR Kit. The alt-Gen protocol for detecting Plasmodium-specific DNA involved preparing the specimen with 5 μL of Master A and 15 μL of Master B, both containing PCR buffer, DNA polymerase, magnesium salt, specific primers and probes, 1 μL internal control and 10 μL DNA or positive and negative controls. The fluorescence detectors (dyes) used were FAM™ for Plasmodium-specific DNA and JOE™ for the internal control as reporter. ROX™ was used as passive reference. Using the 7500 Real Time PCR System™, reactions were performed with a denaturation step at 95 ºC for 10 minutes followed by 45 cycles at 95 ºC for 15 seconds, 58 ºC for 45 seconds and 72 ºC for 15 seconds.
RealStar® Malaria Screen & Type PCR Kit. The alt-S&T protocol targeting specific genes from the five species of Plasmodium that infect humans was conducted in two separate steps. One reaction was prepared with 5 μL of Master Mix A and 15 μL of Master Mix B to detect P. ovale, P. malariae and P. knowlesi, with both Master Mixes containing PCR buffer, DNA polymerase, magnesium salt, specific primers and probes, 1 μL internal control and 10 μL DNA or positive and negative controls. A separate reaction used 5 μL of Master Mix A and 15 μL of Master Mix B to detect P. vivax and P. falciparum; it also contained PCR buffer, DNA polymerase, magnesium salt, specific primers and probes, 1 μL internal control and 10 μL DNA or positive and negative controls. The fluorescence detectors (dyes) used were Cy5® for P. ovale and P. vivax, FAM™ for P. malariae and P. falciparum, ROX™ for P. knowlesi, and JOE™ for the internal control as reporter. No passive reference was used. Using the 7500 Real Time PCR System, reactions occurred with a denaturation step at 95 ºC for 2 minutes followed by 45 cycles at 95 ºC for 15 sec, 58 ºC for 45 seconds and 72 ºC for 15 seconds.
Statistical analyses
The results were analyzed using Microsoft Excel 2016, GraphPad Prism 9.0 and GraphPad QuickCalcs (GraphPad Software Inc., USA). The sensitivity of the alt-Gen and alt-S&T tests with 95% confidence intervals (CI) and the correlation between the positive results of the different tests were compared using McNemar’s test. The correlation among the cycle threshold (Ct) values for all real-time PCR tests and the correlation between the level of parasitemia and Ct were calculated using Spearman’s r test. According to Bonett and Wright (18) the minimum sample for α = 0.05, a desired confidence interval width of 0.3 and a Spearman’s correlation value of 0.9, is 16.
The agreement among the techniques was assessed by using Cohen’s ƙ index with the 95% confidence interval. Proportions were compared using the χ2 test. Differences were considered statistically significant when P < 0.05 (α < 0.05).
RESULTS
Sensitivity of tests, using nested PCR as the reference
Compared with nPCR, alt-S&T had a sensitivity of 100% (95% CI: 86.91% to 100%) for P. vivax; 100% for P. malariae (95% CI: 86.53% to 100%) and 93.94% for P. falciparum (95% CI: 79.40% to 99.32%). The six samples of P. ovale were positive by both nPCR and alt-S&T. The sensitivity of qPCR for P. falciparum was 77.78% (95% CI: 58.90% to 89.74%) and 100% for P. vivax (95% CI: 86.91% to 100%) and P. malariae (95% CI: 77.31% to 100%). Regardless of the species, all samples positive by alt-Gen were also positive by the reference test.
The sensitivity of TBS was higher for P. falciparum (100%; 95% CI: 86.91% to 100%) and P. vivax (93.33%; 95% CI: 77.63% to 99.20%) than for P. malariae (53.33%; 95% CI: 36.14% to 69.77%) (P < 0.0001, χ2 test). When P. ovale specimens were tested, all were positive by TBS and nPCR; however, four P. ovale isolates were misidentified by TBS as P. malariae (3 samples) and P. vivax (1 sample). One sample identified as P. vivax by TBS was identified as P. malariae by nPCR. (Table 1). When all species of Plasmodium were considered, TBS had significantly lower sensitivity than the molecular techniques (P < 0.0001, χ2 test), due mainly to the low sensitivity obtained in P. malariae infections.
TABLE 1. Sensitivity of thick blood smear, two molecular tests and quantitative polymerase chain reaction for malaria infection, using nested polymerase chain reaction as the reference test.
|
Species of Plasmodium |
Test |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Thick blood smear |
alt-S&T |
qPCR |
alt-Gen |
|||||
|
No. positive/total |
Sensitivityd |
No. positive/total |
Sensitivity |
No. positive/total |
Sensitivity |
No. positive/total |
Sensitivity |
|
|
P. falciparum |
31/31 |
100% (86.91% to 100%) |
31/33 |
93.94% (79.40% to 99.32%) |
21/27 |
77.78% (58.90% to 89.74%) |
27/27 |
100% (85.24% to 100%) |
|
P. vivax |
28/30 |
93.33% (77.63% to 99.20%) |
31/31 |
100% (86.91% to 100%) |
31/31 |
100% (86.91% to 100%) |
29/29 |
100% (86.13% to 100%) |
|
P. malariae |
16/30a |
53.33% (36.14% to 69.77%) |
30/30 |
100% (86.53% to 100%) |
16/16 |
100% (77.31% to 100%) |
13/13 |
100% (73.41% to 100%) |
|
P. ovale |
6/6b |
100% (55.72% to 100%)e |
6/6 |
100% (55.72% to 100%)e |
ND |
ND |
5/5 |
100% (51.09% to 100%)e |
|
Total |
81/97c |
83.51% (74.77% to 89.69%) |
98/100 |
98% (92.56% to 99.89%) |
68/74 |
91.89% (83.11% to 96.54%) |
74/74 |
100% (94.09% to 100%) |
alt-Gen: RealStar® Malaria PCR Kit 1.0; alt-S&T: RealStar® Malaria Screen & Type PCR Kit; ND: not done; qPCR: quantitative polymerase chain reaction.
One sample was misidentified as P. vivax.
Four samples were misidentified: 3 as P. malariae and 1 as P. vivax.
P < 0.0001 (χ2 test) compared with molecular techniques.
Values are sensitivity (95% confidence interval).
The large width of the confidence interval is due to the small number of samples.
Source: Table 1 was created by the authors based on the results of the study.
Sensitivity of the genus-specific assays
When the results from alt-Gen were compared with those of qPCR, for P. vivax samples the sensitivity was 100% (95% CI: 88.3% to 100%; ƙ = 1.0). For P. falciparum, although the sensitivity was 100% (95% CI: 82.41% to 100%), the agreement was low (ƙ = 0.5; 95% CI: 0.201 to 814; P = 0.04, McNemar’s test) due to six samples that tested negative by qPCR but were positive by alt-Gen. For P. malariae, the six samples were positive by both tests (Table 2).
TABLE 2. Comparison of the sensitivity of the genus-specific assays and the species-specific assays for detecting Plasmodium, by species.
|
Test and sensitivity |
Comparison of genus-specific assays |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Quantitative PCR | ||||||||
|
P. falciparum |
P. vivax |
P. malariae |
P. ovale |
|||||
|
Positive |
Negative |
Positive |
Negative |
Positive |
Negative |
Positive |
Negative |
|
|
alt-Gen |
18 |
6 |
29 |
0 |
6 |
0 |
ND |
|
|
|
0 |
0 |
0 |
0 |
0 |
0 |
|
|
|
Sensitivity (95% CI) |
100% (82.41% to 100%) |
100% (88.3% to 100%) |
100% (60.97% to 100%)b |
ND |
||||
|
ƙ |
0.5 |
1.0 |
1.0 |
|
||||
|
McNemar’s test |
P = 0.04a |
NA |
NA |
|
||||
|
Test and sensitivity |
Comparison of species-specific assays |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Nested PCR | ||||||||
|
P. falciparum |
P. vivax |
P. malariae |
P. ovale |
|||||
|
Positive |
Negative |
Positive |
Negative |
Positive |
Negative |
Positive |
Negative |
|
|
alt-S&T |
31 |
0 |
31 |
0 |
30 |
0 |
6 |
0 |
|
|
2 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Sensitivity (95% CI) |
93.94% (80.39% to 98.32%) |
100% (88.97% to 100%) |
100% (86.65% to 100%) |
100% (60.97% to 100)b |
||||
|
ƙ |
0.8 |
1.0 |
1.0 |
1.0 |
||||
|
McNemar’s test |
P = 0.4795 |
NA |
P = 1.00 |
NA |
||||
alt-Gen: RealStar® Malaria PCR Kit 1.0; alt-S&T: RealStar® Malaria Screen & Type PCR Kit; CI: confidence interval; NA: not applicable; ND: not done; PCR: polymerase chain reaction.
The result is statistically significant.
The large width of the confidence interval is due to the small number of samples.
Source: Table 2 was created by the authors based on the results of the study.
Sensitivity of the species-specific assays
When using nPCR as the reference, alt-S&T had a sensitivity of 100% for P. vivax (95% CI: 88.97% to 100%) and P. malariae (95% CI: 86.65% to 100%). For P. falciparum, two samples were negative by alt-S&T, giving a sensitivity of 93.94% (95% CI: 80.39% to 98.32%; with ƙ = 0.8; 95% CI: 0.543 to 1.000 and P = 0.4795, McNemar’s test). For P. ovale, the six samples were positive by both tests (Table 2).
Correlation of cycle threshold values in real-time PCR
For all Plasmodium species, the correlation of Ct values from alt-S&T and alt-Gen with qPCR was significant (P < 0.0001, Spearman’s test), with r = 0.8621 (95% CI: 0.7851 to 0.9128) for alt-S&T and r = 0.9374 (95% CI: 0.8918 to 0.9641) for alt-Gen (Figure 2). Considering the Cts of Plasmodium species independently, for P. falciparum testing there was a significant correlation between qPCR and alt-S&T (r = 0.7204; 95% CI: 0.4277 to 0.8763; P = 0.0001) and between qPCR and alt-Gen (r = 0.7688; 95% CI: 0.4592 to 0.9119; P = 0.0002). Similar results were obtained for P. vivax, with the correlation between qPCR and alt-S&T of r = 0.741 (95% CI: 0.5163 to 0.8703; P<0.0001) and between qPCR and alt-Gen of r = 0.888 (95% CI: 0.7685 to 0.9477; P < 0.0001). For P. malariae, the correlation was calculated only between qPCR and alt-S&T, with r = 0.9338 (95% CI: 0.8171 to 0.9770; P < 0.0001), due to the small number of samples (n = 6) tested by both qPCR and alt-Gen.
FIGURE 2. Spearman’s correlation coefficient for the relationship of cycle threshold values in real-time polymerase chain reaction for all species of Plasmodium tested.
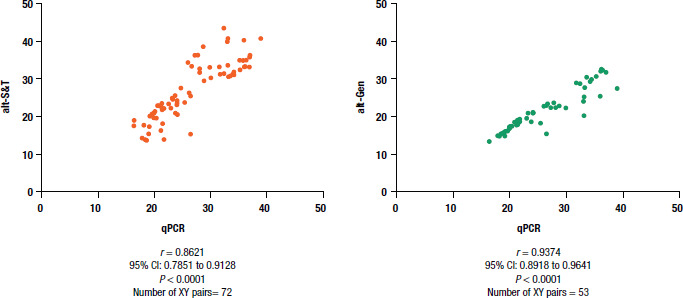
alt-Gen: RealStar® Malaria PCR Kit 1.0
alt-S&T: RealStar® Malaria Screen & Type PCR Kit
CI: confidence interval
qPCR: quantitative polymerase chain reaction
Source: Figure 2 was created by the authors based on the results of the study.
Correlation between parasitemia and cycle threshold values in real-time PCR
When all Plasmodium species were considered, a negative correlation was observed between parasitemia and real-time PCR Ct values: for alt-S&T, r = −0.7988 (95% CI: −0.8666 to −0.7019; P < 0.0001), for alt-Gen, r = −0.8766 (95% CI: −0.9245 to −0.8013; P < 0.0001) and for qPCR, r = −0.8756 (95% CI: −0.9266 to −0.7930; P < 0.0001). When analysed individually, negative correlations were also observed for P. falciparum, P. vivax and P. malariae (Table 3).
TABLE 3. Spearman’s correlation coefficient for level of parasitemia and cycle threshold values in real-time polymerase chain reaction for Plasmodium species.
|
Test |
Plasmodium species |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
P. falciparum |
P. vivax |
P. malariae |
||||||||||
|
r |
95% CI |
P |
No. of pairs |
r |
95% CI |
P |
No. of pairs |
r |
95% CI |
P |
No. of pairs |
|
|
alt-Gen |
‒0.548 |
‒0.7727 to ‒0.2003 |
0.003 |
27 |
‒0.7319 |
‒0.8728 to ‒0.4783 |
<0.0001 |
27 |
‒0.9324 |
‒0.9807 to ‒0.7772 |
<0.0001 |
13 |
|
alt-S&T |
‒0.381 |
‒0.6431 to ‒0.03839 |
0.026 |
34 |
‒0.7461 |
‒0.8730 to ‒0.5246 |
<0.0001 |
31 |
‒0.7990 |
‒0.8931 to ‒0.6380 |
<0.0001 |
38 |
|
qPCR |
‒0.621 |
‒0.8343 to ‒0.2456 |
0.003 |
21 |
‒0.7393 |
‒0.8728 to ‒0.5028 |
<0.0001 |
29 |
‒0.5518 |
‒0.8277 to ‒0.06111 |
0.0267 |
16 |
alt-Gen: RealStar® Malaria PCR Kit 1.0; alt-S&T: RealStar® Malaria Screen & Type PCR Kit; CI: confidence interval; qPCR: quantitative polymerase chain reaction.
Source: Table 3 was created by the authors based on the results of the study.
Performance on WHO reference samples
Thirty four reference samples – 12 P. falciparum, 6 P. vivax, 6 P. ovale, 3 P. knowlesi, 2 P. malariae and 5 negative – from the WHO external quality assurance scheme were analyzed by alt-S&T. The alt-S&T assay was able to accurately detect each of the Plasmodium species. All known negative isolates were also negative by alt-S&T.
DISCUSSION
The WHO framework for malaria elimination aims to reduce by 2030 mortality from and the incidence of malaria by 90% from the number of cases detected in 2015; this strategy is based on three pillars: ensuring global access to prevention, diagnosis, and treatment; accelerating elimination efforts; and establishing malaria surveillance as the primary intervention (19).
Diagnosis is one of the most critical measures in malaria control and elimination programs. Although microscopy is the primary diagnostic tool supporting these goals, malaria elimination requires more sensitive techniques to detect asymptomatic infections, mainly to prevent transmission (8).
In this study, when using nPCR as a reference, TBS showed good sensitivity for P. falciparum and P. ovale; however, for P. vivax and P. malariae, the sensitivity was lower (93.33% and 53.33%, respectively). Furthermore, five TBS specimens were misidentified when compared with results from nPCR. The accuracy and sensitivity of microscopy depend on the experience of the microscopist and the quality of the slides (4).
When compared with nPCR, alt-Gen had 100% sensitivity for all species, and alt-S&T had 100% sensitivity for most Plasmodium species except for P. falciparum (93.94%). These results agree with those of Frickmann and colleagues (20) in a study that evaluated alt-S&T and found slightly reduced sensitivity for P. falciparum infections. In our study, all P. falciparum samples were collected in DBS and, despite the manufacturer’s recommendation to use well-established protocols for DNA extraction from whole blood, we obtained excellent results using Chelex 100® (sensitivity: 93.94%) in contrast to the results obtained by Ataei and colleagues (21), who detected DNA in 42.7% of Plasmodium samples from DBS and 46.7% from whole blood. Likewise, lower rates of positivity by PCR have been reported when using DNA extracted from DBS (11.2% positivity) than from whole blood (24.5% positivity) (22).
When comparing the sensitivity of alt-Gen with that of qPCR, the results for P. vivax and P. malariae showed perfect agreement. However, for P. falciparum, qPCR failed to detect the species in six samples in which it was detected by alt-Gen. When comparing the results obtained by alt-S&T with those from nPCR, the sensitivities for P. vivax, P. malariae and P. ovale were 100%. The alt-S&T assay failed to detect P. falciparum in two samples with low levels of parasitemia (25 and 100 parasites/µL), which could possibly be explained by an irregular distribution of blood in the DBS, which was collected in field conditions. The alt-S&T assay showed high specificity when testing the WHO external quality assurance reference samples, correctly identifying all species evaluated, as well the negative samples. The inclusion of the panel containing samples from a quality control program contributed to the validation of the alt-S&T assay, showing that it is able to detect the five plasmodia that infect humans. Furthermore, no amplification was observed when alt-S&T was tested on the small number of negative reference samples.
The molecular protocols used in this study are based on the amplification of different targets. The qPCR, alt-Gen and nPCR techniques amplify 18S rRNA gene sequences, of which there are 5 to 10 copies in the Plasmodium genome (23). The alt-S&T assay uses three different targets in other regions to differentiate species. Although protocols based on different genes have greater sensitivity due to their higher copy number, assays based on 18S rRNA genes are most commonly used for molecular diagnosis (11, 24).
There was good correlation among Ct values for qPCR and the alt-Gen and alt-S&T assays, except for the comparison between qPCR and alt-Gen for P. malariae samples, probably due to the small number of sample pairs available for statistical analysis. In addition, there was a correlation between the level of parasitemia and Ct for real-time PCR, with parasitemia densities inversely proportional to Ct values. Similar results were obtained by Mischlinger and colleagues (25), who evaluated samples from malaria patients and found a good correlation (Pearson’s r > 0.9) between level of parasitemia and alt-S&T Ct values. In contrast, Frickmann and colleagues (20) obtained a weak correlation (Pearson’s r > 0.23) when they tested blood samples from German patients with suspected malaria who had traveled to an endemic area.
In this study, of the 28 samples from asymptomatic individuals from areas of low endemicity, only 2 were positive by TBS, one P. vivax and one P. malariae. All 28 samples were positive by alt S&T (5 P. vivax and 23 P. malariae). Detection of Plasmodium reservoirs is an important issue in malaria control, as these asymptomatic individuals are not treated unless submicroscopic parasitemia is revealed by more sensitive assays. Through molecular protocols, it is possible to detect these submicroscopic infections, which pose a risk to malaria elimination efforts, as this population represents a source of gametocytes, and thus maintains malaria transmission. According to Okell and colleagues (26), in low-endemicity areas, individuals with submicroscopic infections are responsible for 20% to 50% of parasite transmission to the mosquito.
Accurate diagnosis is one of the pillars of malaria control programs. Although microscopy and RDTs are not suitable for detecting low levels of parasitemia, they are the main tools used for routine diagnosis due to their cost and ease of use. Although sensitive and specific molecular tests are expensive and not available in remote areas, they should be available for surveillance activities and in reference laboratories, thus allowing the detection of asymptomatic cases, as well as determination of the five species of Plasmodium that infect humans. Despite the availability of conventional PCR tests, the need for several rounds of amplification is time-consuming, and there is an additional risk of contamination. These disadvantages may be overcome by qPCR tests that allow for accurate and fast results in a closed system that reduces the risk of contamination. When comparing the time taken to perform each type of test, reading slides takes 4.5 times longer than an alt-S&T assay. For qPCR the time necessary is similar to that for alt-S&T; however, to our knowledge, no qPCR test for detecting the five Plasmodium species is available. Finally, the time necessary for the nPCR assay is twice as long as that for alt-S&T.
The results of this study have to be seen in the light of some limitations, such as the lack of sufficient DNA volume to perform all tests with some samples and the low number of P. ovale and P. knowlesi samples, which made statistical comparisons unfeasible for these two species. Cost–effectiveness studies would be appropriate. In conclusion, the alt-Gen and alt-S&T protocols evaluated in this study are suitable for detecting all Plasmodium species and submicroscopic infections for distinct epidemiological purposes, such as surveys, diagnosis in reference laboratories and screening in blood banks; thus they can contribute to global efforts to eliminate malaria.
Disclaimer.
Authors hold sole responsibility for the views expressed in the manuscript, which may not necessarily reflect the opinion or policy of the Revista Panamericana de Salud Pública/Pan American Journal of Public Health or the Pan American Health Organization (PAHO).
Acknowledgments
We thank the staff of Núcleo de Estudos em Malária/SUCEN/IMT–FMUSP for their support. We also acknowledge the support of Laboratório de Investigação Médica/HCFMUSP (LIM 49). We are especially grateful to altona Diagnostics (Germany) and to the WHO Global Malaria Programme partnership with the United Kingdom National External Quality Assessment Service.
Funding Statement
Funding for this study was received from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES; grant no. 88882.376831/2019-01 to MA); from the São Paulo Research Foundation (FAPESP; grant nos. 2011/07380-8, 2012/18014-5, 2018/07890-5); from Núcleo de Estudos em Malaria/SUCEN/Instituto de Medicina Tropical de São Paulo–FMUSP; and from LIM 49/HCFMUSP. The sponsors had no influence on the design of the study, the data collection, the analysis, the writing and the decision to publish these results.
Footnotes
Author’s contributions.
MA performed the statistical analyses of the data and wrote the manuscript; MCAS performed the statistical analysis and revised the manuscript; MJCN and MLRNF performed the tests; ADH, GFMCL and JI performed the nested PCR testing; JEL designed the study and revised the manuscript; SMDS designed the study, analyzed the data and wrote and reviewed the manuscript. All authors reviewed and approved the final version of the manuscript.
Conflicts of interest.
The authors declare that they have no financial or other relationships that might lead to any conflict of interest. This study was sponsored by altona Diagnostics (Germany); however, the sponsor did not have any role in interpreting the data or in writing the manuscript. No payment was made to any of the authors.
REFERENCES
- 1.World Health Organization . Geneva: WHO; 2020. World malaria report 2020: 20 years of global progress and challenges.https://apps.who.int/iris/handle/10665/337660 [Google Scholar]; 1. World Health Organization. World malaria report 2020: 20 years of global progress and challenges. Geneva: WHO; 2020. https://apps.who.int/iris/handle/10665/337660
- 2.Ministério da Saúde, Brasil Secretaria de Vigilância em Saúde. [[cited 2021 Nov 25]];Boletim Epidemiológico: malária 2020 [Epidemiological Report – malaria 2020] Brasília: Ministério da Saúde. 2020 Available from: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/boletins-epidemiologicos-especiais/2020/boletim_especial_malaria_1dez20_final.pdf. [Google Scholar]; 2. Ministério da Saúde, Brasil; Secretaria de Vigilância em Saúde. Boletim Epidemiológico: malária 2020 [Epidemiological Report – malaria 2020] Brasília: Ministério da Saúde; 2020 [cited 2021 Nov 25]. Available from: https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/boletins-epidemiologicos-especiais/2020/boletim_especial_malaria_1dez20_final.pdf
- 3.Ministério da Saúde, Brasil . [Public data from Sivep-Malaria and Sinan data sources, for notifications from Brazil from 2007 to 2021] Brasília: Ministério da Saúde; 2021. [[cited 2021 Nov 29]]. Secretaria de Vigilância em Saúde “Dados para cidadão a partir da fonte de dados do Sivep-Malária e do Sinan, para notificações do Brasil de 2007 a 2021”. Available from: https://public.tableau.com/app/profile/mal.ria.brasil/viz/Dadosparacidado_201925_03_2020/Incio. [Google Scholar]; 3. Ministério da Saúde, Brasil; Secretaria de Vigilância em Saúde. “Dados para cidadão a partir da fonte de dados do Sivep-Malária e do Sinan, para notificações do Brasil de 2007 a 2021” [Public data from Sivep-Malaria and Sinan data sources, for notifications from Brazil from 2007 to 2021]. Brasília: Ministério da Saúde; 2021 [cited 2021 Nov 29]. Available from: https://public.tableau.com/app/profile/mal.ria.brasil/viz/Dadosparacidado_201925_03_2020/Incio
- 4.World Health Organization . Geneva: World Health Organization; 2016. Malaria microscopy quality assurance manual, version 2.https://apps.who.int/iris/handle/10665/204266 [Google Scholar]; 4. World Health Organization. Malaria microscopy quality assurance manual, version 2. Geneva: World Health Organization; 2016. https://apps.who.int/iris/handle/10665/204266
- 5.World Health Organization . 3rd ed. Geneva: World Health Organization; 2015. Guidelines for the treatment of malaria.https://apps.who.int/iris/handle/10665/162441 [Google Scholar]; 5. World Health Organization. Guidelines for the treatment of malaria. 3rd ed. Geneva: World Health Organization; 2015. https://apps.who.int/iris/handle/10665/162441
- 6.Moody A. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev. 2002;15:66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]; 6. Moody A. Rapid diagnostic tests for malaria parasites. Clin Microbiol Rev. 2002;15:66-78. [DOI] [PMC free article] [PubMed]
- 7.Murray CK, Bell D, Gasser RA, Wongsrichanalai C. Rapid diagnostic testing for malaria. Trop Med Int Health. 2003;8:876–883. doi: 10.1046/j.1365-3156.2003.01115.x. [DOI] [PubMed] [Google Scholar]; 7. Murray CK, Bell D, Gasser RA, Wongsrichanalai C. Rapid diagnostic testing for malaria. Trop Med Int Health. 2003;8:876-83. [DOI] [PubMed]
- 8.Berzosa P, de Lucio A, Romay-Barja M, Herrador Z, González V, García L, et al. Comparison of three diagnostic methods (microscopy, RDT, and PCR) for the detection of malaria parasites in representative samples from Equatorial Guinea. Malar J. 2018;17(1):333. doi: 10.1186/s12936-018-2481-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; 8. Berzosa P, de Lucio A, Romay-Barja M, Herrador Z, González V, García L, et al. Comparison of three diagnostic methods (microscopy, RDT, and PCR) for the detection of malaria parasites in representative samples from Equatorial Guinea. Malar J. 2018;17(1):333. [DOI] [PMC free article] [PubMed]
- 9.Silva RSU. Ocorrência de malária causada por Plasmodium malariae no Município de Cruzeiro do Sul, Estado do Acre, Brasil [Occurence of Plasmodium malariae malaria in the Municipality of Cruzeiro do Sul, Acre State, Brazil] Rev Pan Amaz Saude. 2010;1:105–106. [Google Scholar]; 9. Silva RSU. Ocorrência de malária causada por Plasmodium malariae no Município de Cruzeiro do Sul, Estado do Acre, Brasil [Occurence of Plasmodium malariae malaria in the Municipality of Cruzeiro do Sul, Acre State, Brazil]. Rev Pan Amaz Saude. 2010;1:105-6.
- 10.Cunningham JA, Thomson RM, Murphy SC, de la Paz Ade M, Ding XC, Incardona S, et al. WHO malaria nucleic acid amplification test external quality assessment scheme: results of distribution programmes one to three. Malar J. 2020;19(1):129. doi: 10.1186/s12936-020-03200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; 10. Cunningham JA, Thomson RM, Murphy SC, de la Paz Ade M, Ding XC, Incardona S, et al. WHO malaria nucleic acid amplification test external quality assessment scheme: results of distribution programmes one to three. Malar J. 2020;19(1):129. [DOI] [PMC free article] [PubMed]
- 11.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pieiro L, do Rosario VE, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-b. [DOI] [PubMed] [Google Scholar]; 11. Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pieiro L, do Rosario VE, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315-20. [DOI] [PubMed]
- 12.World Health Organization . Geneva: World Health Organization; 2014. Evidence Review Group on malaria diagnosis in low transmission settings.https://www.who.int/malaria/mpac/mpac_mar2014_diagnosis_low_transmission_settings_report.pdf?ua=1 [Google Scholar]; 12. World Health Organization. Evidence Review Group on malaria diagnosis in low transmission settings. Geneva: World Health Organization; 2014. https://www.who.int/malaria/mpac/mpac_mar2014_diagnosis_low_transmission_settings_report.pdf?ua=1
- 13.Lima GF, Levi JE, Geraldi MP, Sanchez MC, Segurado AA, Hristov AD, et al. Malaria diagnosis from pooled blood samples: comparative analysis of real-time PCR, nested PCR and immunoassay as a platform for the molecular and serological diagnosis of malaria on a large-scale. Mem Inst Oswaldo Cruz. 2011;106(6):691–700. doi: 10.1590/s0074-02762011000600008. [DOI] [PubMed] [Google Scholar]; 13. Lima GF, Levi JE, Geraldi MP, Sanchez MC, Segurado AA, Hristov AD, et al. Malaria diagnosis from pooled blood samples: comparative analysis of real-time PCR, nested PCR and immunoassay as a platform for the molecular and serological diagnosis of malaria on a large-scale. Mem Inst Oswaldo Cruz. 2011;106(6):691-700. [DOI] [PubMed]
- 14.Lucchi NW, Ljolje D, Silva-Flannery L, Udhayakumar V. Use of malachite green-loop mediated isothermal amplification for detection of Plasmodium spp parasites. PLOS ONE. 2016;11(3):e0151437. doi: 10.1371/journal.pone.0151437. [DOI] [PMC free article] [PubMed] [Google Scholar]; 14. Lucchi NW, Ljolje D, Silva-Flannery L, Udhayakumar V. Use of malachite green-loop mediated isothermal amplification for detection of Plasmodium spp. parasites. PLOS ONE. 2016;11(3):e0151437. [DOI] [PMC free article] [PubMed]
- 15.Banoo S, Bell D, Bossuyt P, Herring A, Mabey D, Poole F, et al. Evaluation of diagnostic tests for infectious diseases: general principles. Nat Rev Microbiol. 2010;8(12 Suppl):S17–S29. [PubMed] [Google Scholar]; 15. Banoo S, Bell D, Bossuyt P, Herring A, Mabey D, Poole F, et al. Evaluation of diagnostic tests for infectious diseases: general principles. Nat Rev Microbiol. 2010;8(12 Suppl):S17-29. [PubMed]
- 16.Ministério da Saúde, Brasil . Manual de diagnóstico laboratorial da malária. 2nd ed. Brasília: Ministério da Saúde; 2009. Secretaria de Vigilância em Saúde. [Google Scholar]; 16. Ministério da Saúde, Brasil; Secretaria de Vigilância em Saúde. Manual de diagnóstico laboratorial da malária. 2nd ed. Brasília: Ministério da Saúde; 2009.
- 17.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]; 17. Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52:565-8. [DOI] [PubMed]
- 18.Bonett DG, Wright TA. Sample size requirements for estimating Pearson, Kendall and Spearman correlations. Psychometrika. 2000;65:23–28. [Google Scholar]; 18. Bonett DG, Wright TA. Sample size requirements for estimating Pearson, Kendall and Spearman correlations. Psychometrika. 2000;65:23-8.
- 19.World Health Organization . Geneva: World Health Organization; 2017. A framework for malaria elimination.https://apps.who.int/iris/handle/10665/254761 [Google Scholar]; 19. World Health Organization. A framework for malaria elimination. Geneva: World Health Organization; 2017. https://apps.who.int/iris/handle/10665/254761
- 20.Frickmann H, Wegner C, Ruben S, Behrens C, Kollenda H, Hinz R, et al. Evaluation of the multiplex real-time PCR assays RealStar malaria S&T PCR kit 1.0 and FTD malaria differentiation for the differentiation of Plasmodium species in clinical samples. Travel Med Infect Dis. 2019;31:101442. doi: 10.1016/j.tmaid.2019.06.013. [DOI] [PubMed] [Google Scholar]; 20. Frickmann H, Wegner C, Ruben S, Behrens C, Kollenda H, Hinz R, et al. Evaluation of the multiplex real-time PCR assays RealStar malaria S&T PCR kit 1.0 and FTD malaria differentiation for the differentiation of Plasmodium species in clinical samples. Travel Med Infect Dis. 2019;31:101442. [DOI] [PubMed]
- 21.Ataei S, Nateghpour M, Hajjaran H, Edrissian GH, Foroushani AR. High specificity of semi-nested multiplex PCR using dried blood spots on DNA Banking Card in comparison with frozen liquid blood for detection of Plasmodium falciparum and Plasmodium vivax. J Clin Lab Anal. 2011;25(3):185–190. doi: 10.1002/jcla.20454. [DOI] [PMC free article] [PubMed] [Google Scholar]; 21. Ataei S, Nateghpour M, Hajjaran H, Edrissian GH, Foroushani AR. High specificity of semi-nested multiplex PCR using dried blood spots on DNA Banking Card in comparison with frozen liquid blood for detection of Plasmodium falciparum and Plasmodium vivax. J Clin Lab Anal. 2011;25(3):185-90. [DOI] [PMC free article] [PubMed]
- 22.Strøm GE, Moyo S, Fataki M, Langeland N, Blomberg B. PCR targeting Plasmodium mitochondrial genome of DNA extracted from dried blood on filter paper compared to whole blood. Malar J. 2014;13:137. doi: 10.1186/1475-2875-13-137. [DOI] [PMC free article] [PubMed] [Google Scholar]; 22. Strøm GE, Moyo S, Fataki M, Langeland N, Blomberg B. PCR targeting Plasmodium mitochondrial genome of DNA extracted from dried blood on filter paper compared to whole blood. Malar J. 2014;13:137. [DOI] [PMC free article] [PubMed]
- 23.Zimmerman PA. Howes RE. Malaria diagnosis for malaria elimination. Curr Opin Infect Dis. 2015;28:446–454. doi: 10.1097/QCO.0000000000000191. [DOI] [PubMed] [Google Scholar]; 23. Zimmerman PA, Howes RE. Malaria diagnosis for malaria elimination. Curr Opin Infect Dis. 2015;28:446-54. [DOI] [PubMed]
- 24.Haanshuus CG, Mohn SC, Mørch K, Langeland N, Blomberg B, Hanevik K. A novel, single-amplification PCR targeting mitochondrial genome highly sensitive and specific in diagnosing malaria among returned travellers in Bergen, Norway. Malar J. 2013;12:26. doi: 10.1186/1475-2875-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]; 24. Haanshuus CG, Mohn SC, Mørch K, Langeland N, Blomberg B, Hanevik K. A novel, single-amplification PCR targeting mitochondrial genome highly sensitive and specific in diagnosing malaria among returned travellers in Bergen, Norway. Malar J. 2013;12:26. [DOI] [PMC free article] [PubMed]
- 25.Mischlinger J, Pitzinger P, Veletzky L, Groger M, Zoleko-Manego R, Adegnika AA, et al. Use of capillary blood samples leads to higher parasitemia estimates and higher diagnostic sensitivity of microscopic and molecular diagnostics of malaria than venous blood samples. J Infect Dis. 2018;218(8):1296–1305. doi: 10.1093/infdis/jiy319. [DOI] [PubMed] [Google Scholar]; 25. Mischlinger J, Pitzinger P, Veletzky L, Groger M, Zoleko-Manego R, Adegnika AA, et al. Use of capillary blood samples leads to higher parasitemia estimates and higher diagnostic sensitivity of microscopic and molecular diagnostics of malaria than venous blood samples. J Infect Dis. 2018;218(8):1296-1305. [DOI] [PubMed]
- 26.Okell LC, Bousema T, Griffin JT, Ouédraogo AL, Ghani AC, Drakeley CJ. Factors determining the occurrence of submicroscopic malaria infections and their relevance for control. Nat Commun. 2012;3:1237. doi: 10.1038/ncomms2241. [DOI] [PMC free article] [PubMed] [Google Scholar]; 26. Okell LC, Bousema T, Griffin JT, Ouédraogo AL, Ghani AC, Drakeley CJ. Factors determining the occurrence of submicroscopic malaria infections and their relevance for control. Nat Commun. 2012;3:1237. [DOI] [PMC free article] [PubMed]


