Abstract
Housefly (Musca domestica) is an excellent candidate for the distribution of susceptible and resistant bacterial strains that potentially threaten public health. To date, there is a paucity of information on the global distribution of pathogenic bacteria of medical and veterinary importance from diverse environmental settings. Therefore, this study was undertaken to conduct a systemic review and meta‐analysis to estimate occurrence of various bacterial species of medical and veterinary importance harboured by houseflies around the world. Published articles from 1980 to 2020 were retrieved from electronic databases and assessed for eligibility according to Preferred Reporting Items for Systemic Reviews and Meta‐Analysis guidelines. Seventy‐eight studies were included in the review with only 44 studies being eligible for meta‐analysis. Results indicated that eligible studies used in this review were from four continents, i.e., Asia (47.4%) America (23.1%), Africa (20.5%) and Europe (8.9%). The majority of the studies (56.4%) used the culture method for the identification of bacterial pathogens, while 30.7% used both culture and PCR techniques. For meta‐analysis, we focused on five pathogenic bacterial species including Escherichia coli, Enterococcus faecium, Klebsiella pneumoniae, Pseudomonas aeruginosa and Staphylococcus aureus. High heterogeneity was found among studies investigating different pathogens including E. coli (Q = 10,739.55; I 2 = 99.60; Q‐p 0.0001), E. faecium (Q = 317.61; I 2 = 86.46; Q‐p < 0.0001), K. pneumonia (Q = 1,576.61; I 2 = 97.27; Q‐p < 0.0001), S. aureus (Q = 2,439.12; I 2 = 98.24; Q‐p < 0.0001) and P. aeruginosa (Q = 1,283.0; I 2 = 96.65; Q‐p < 0.0001). Furthermore, it was observed that houseflies carried a considerable number of susceptible and antibiotic‐resistant bacterial strains that pose considerable threats to public health. Findings from this study have provided more insight on the vectoral potential of houseflies in the transmission of significant bacterial pathogens from different regions across the world. Further investigation is required to quantify the bacterial contamination and dissemination by houseflies.
Keywords: animals, bacteria, houseflies, humans
Houseflies are in abundance in different human and animal settings due to their behavioural characteristics. Houseflies carry a considerable number of susceptible and antibiotic‐resistant bacteria, including multiple‐antibiotic resistant strains that could pose severe threat to public health.
Impacts.
Houseflies are in abundance in different human and animal settings due to their behavioural characteristics.
Houseflies carry a considerable number of susceptible and antibiotic‐resistant bacteria, including multiple‐antibiotic resistant strains that could pose severe threat to public health.
Majority of studies have used culture methods for detection of bacteria from houseflies, but recently there is an increase in use of DNA‐based techniques.
1. INTRODUCTION
Houseflies exist as major pests of humans, poultry and livestock surroundings and facilities where they transmit vector‐borne infections globally (Ahmed et al., 2013; Akter et al., 2020; Iqbal et al., 2014). Based on World Health Organization (WHO) records, the major vector‐borne infections account for almost 17% of communicable diseases that occurs per annum worldwide, with high incidence in tropical and subtropical regions (WHO, 2017).
Houseflies are common and occur in high abundance in close association with human activities. These insects are attracted to human and animal waste for feeding and reproduction (Awache & Forouk, 2016). Human (i.e., hospitals, food markets and restaurants) and animal environments (i.e., farms, feedlots and slaughterhouses) are the major points of focus and sampled areas for houseflies. Owing to their diverse habitat preference, indiscriminate movement, the ability to fly long distance and attraction to both decaying organic materials, houseflies greatly amplify the risk of human exposure to variety of pathogens including bacteria (Barreiro et al., 2013; Gupta et al., 2012).
The reproduction habits and feeding mechanisms of houseflies involve microbe‐rich substrates (Park et al., 2019). Thus, houseflies harbour bacteria in both immature and mature life stages (Zurek et al., 2000) and are widely considered a significant source of bacterial diversity (Wei et al., 2013). The bacterial species found to colonize houseflies include Salmonella spp., Yersinia enterocolitica, Edwardsiella tarda, Shigella sonnei, Escherichia coli, Klebsiella spp., Staphylococcus aureus, Pseudomonas aeruginosa and Enterococcus faecalis (Rahuma et al., 2005), to name a few. The characteristics of bacterial genera carried by houseflies are dependent on the sampled area (Khamesipour et al., 2018). Thus, houseflies can transport a diverse and active microbial community from different reservoirs (Barreiro et al., 2013; Gupta et al., 2012). Therefore, these pathogens may cause serious infections and diseases to susceptible humans and animals (Ahmed et al., 2013; Malik et al., 2007).
The risk of houseflies to disseminate the antibiotic resistant bacterial strains from hospital settings, livestock and poultry farms to public residents is of great concern (Zhang et al., 2018). According to Zhang et al. (2018), houseflies sampled from these areas with extensive usage of antibiotics are commonly colonized by antibiotic‐resistant bacteria including bacterial species that are multidrug resistant. An increasing frequency of antibiotic resistance has been reported from all over the world (Davari et al., 2010). Thus, the emergence of bacteria that is resistant to critically important antimicrobials is of a major concern in human and veterinary medicine (Solà‐Ginés et al., 2015).
Houseflies colonized with these bacterial species may also be associated with the dissemination of antibiotic resistant genes or exposure to virulent bacterial strains within the same environment (Bouamama et al., 2010; Zhang et al., 2018). Transmission takes place mostly when the houseflies come in contact with animals, people and their food (WHO, 1997). Thus, houseflies contribute to the mechanical transmission of foodborne pathogens and other serious diseases that can affect humans and animals. These diseases include anthrax, ophthalmia, typhoid fever, tuberculosis, cholera and infantile diarrhoea (Khamesipour et al., 2018; WHO, 1997).
The current systematic review and meta‐analysis was carried out to determine the pathogenic bacteria isolated from houseflies by various studies conducted around the world, and how frequent these studies report on the antibiotic susceptibility profiles and resistant genes in common bacterial pathogens. This was performed to generate evidence and knowledge necessary to help formulate preventative measures against infectious bacteria harboured by houseflies.
2. MATERIALS AND METHODS
2.1. Systematic search strategy and selection criteria of the journal articles
This review was done following the recommended methodology as outlined by the preferred reporting items for systematic reviews (PRISMA) as shown in Figure 1 (Moher et al., 2015). The literature search was performed to identify journal articles published from 1980 to 2020 reporting on housefly or Musca domestica and bacteria. The phrase ‘Musca domestica’ was searched following the combination of free text and thesaurus terms in different variations: houseflies and bacterial pathogens. These keywords were used to retrieve studies from electronic databases including PubMed (https://pubmed.ncbi.nlm.nih.gov/, from 01/09/2019 to 08/06/2020); Science Direct (https://www.sciencedirect.com/ from 06/09/2019 to 08/06/2020); and Google scholar (https://scholar.google.com/ from 11/09/2019 to 08/06/2020) with the last search done on the 8th of June 2020. The systematic search was accomplished following the combination of free text and wordlist terms in diverse distinctions: ‘Houseflies’ AND/OR, ‘Musca domestica’, ‘Bacterial pathogens’ AND/OR ‘Bacterial isolation’. Thereafter, titles and abstract were scanned and potential journal articles were reviewed and downloaded. The reference list from the potential searched published journal articles was also screened for journal articles relating to this review.
FIGURE 1.
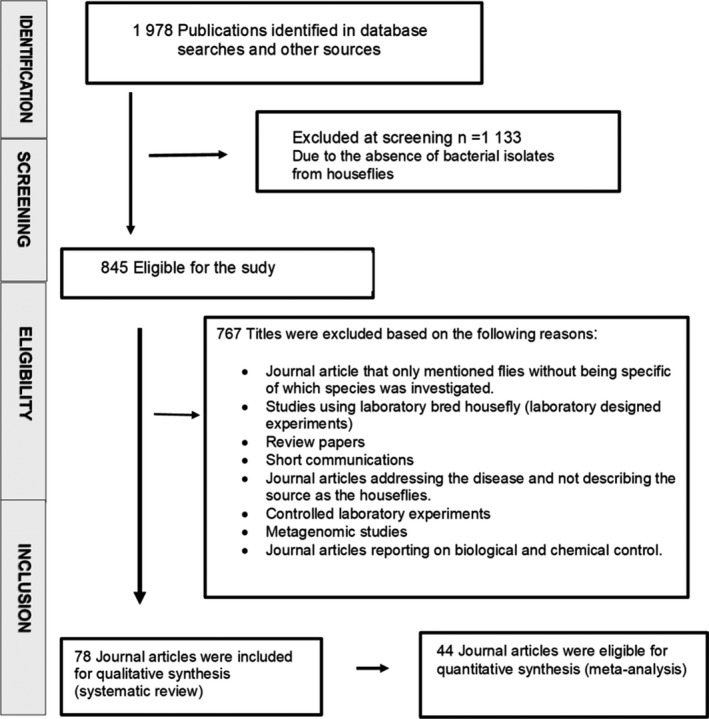
Selection process of eligible studies following PRISMA guidelines
2.2. Inclusion and exclusion criteria for systematic review
The search was limited to only to the English language and conducted from 1 September 2019 to 8 June 2020. The abstracts were assessed and the scientific papers that met a preceding inclusion criterion were selected. Preliminary screening of titles and abstracts was carried out for eligibility and relevance to this systematic review and meta‐analysis according to the inclusion and exclusion criteria. The published journal articles selected in this study were read for data extraction and screened further by a detailed review of the full text. The articles had to fulfil the following criteria to be included: (a) original research articles from around the world, (b) the availability of the full texts, and (c) journal articles published in English language (d) conducted between 1 September 2019 and 8 June 2020; (e) study design was cross‐sectional/prevalence study; (f) the laboratory examination methods used was clearly stated; (g) the geographical location of the study was clearly stated; (h) the species of M. domestica was clearly stated; (i) the number of M. domestica with bacterial positive cases and sample size were provided; (j) the study screened for all bacterial species. Articles were excluded if they were published only in abstract form, or if they were editorials, letters, comments or reviews. Furthermore, the excluded articles were those reporting pathogens isolated from flies in general without specifying the fly species, studies using laboratory‐reared M. domestica (laboratory designed experiments), journal articles addressing the disease and not describing the source as M. domestica, controlled laboratory experiments and metagenomic studies (Figure 1). No attempt was made to contact the authors of the original manuscripts for any additional information or retrieval of unpublished studies. The data was retrieved by one investigator and then confirmed by another investigator.
2.3. Data extraction and meta‐analysis
Key characteristics of each included article were summarized in a spreadsheet (Microsoft Excel®), with the following variables: author name, country; sites of sample collection, diagnostic technique and bacterial isolates. Several bacteria species were reported from all the included studies. For the sampling locations, closely related sampled places were grouped together. Only journal articles which were definite with the species of bacteria were included for meta‐analysis. Five selected pathogenic bacteria, namely, E. coli, E. faecium, Klebsiella pneumoniae, P. aeruginosa and S. aureus were focused on. The prevalence for the subgroups was calculated as follows: the number of bacterial species divided by the sample size with the results expressed in percentages (Table 1). However, some studies used duplicates or replicate from the same fly homogenates. Thus, the number of positive bacterial species was recorded at 100% prevalence if the number recorded exceeds the sample size.
TABLE 1.
Meta‐analysis of eligible studies reporting on selected pathogenic bacteria isolated from houseflies in different regions
| Study reference | Region | Method of diagnosis utilized | Sample size | E. coli no. of positives: (prevalence) | E. faecium no. of positives: (Prevalence) | S. aureus no. of positives: (Prevalence) | K. pneumonia no. of positives: (Prevalence) | P. aeruginosano. of positives: (Prevalence) |
|---|---|---|---|---|---|---|---|---|
| Fotedar, Banerjee, Shriniwas, and Verma (1992) | India | Culture | 113 | 22: (19.5%) | — | 24: (21.2%) | — | 5: (4.4%) |
| Bouamama et al. (2010) | Morocco | Culture | 600 | 26: (4.3%) | — | 8: (1.3%) | — | — |
| Fotedar, Banarjee, Samantray, and Shriniwas (1992) | India | Culture | 240 | — | — | — | 45: (18.8%) | — |
| Hemmatinezhad et al. (2015) | Iran | Culture PCR | 600 | — | — | — | — | 53: (8.8%) |
| Macovei and Zurek (2006) | USA | PCR | 268 | — | 14: (5.2%) | — | — | — |
| Macovei et al. (2008) | USA | Culture PCR | 75 | — | 3: (4%) | — | — | — |
| Solà‐Ginés et al. (2015) | Spain | Culture | 615 | 41: (6.7%) | — | — | — | — |
| Mohammed et al. (2016) | Egypt | Culture | 22 | 6: (27.3%) | — | 2: (9.1%) | — | — |
| Nmorsi et al. (2007) | Nigeria | Culture | 737 | 194: (26.3%) | — | — | — | — |
| Ranjbar et al. (2016) | Iran | Culture PCR | 600 | — | — | — | 68: (11.3%) | — |
| Sobur et al. (2019) | Bangladesh | Culture PCR | 300 | 40: (13.3%) | — | — | — | — |
| Songe et al. (2016) | Zambia | Culture PCR | 418 | 307: (73.4%) | — | — | — | — |
| Usui et al. (2013) | Japan | Culture PCR | 91 | 41: (45.1%) | — | — | — | — |
| Usui et al. (2015) | Japan | Culture PCR | 74 | 13: (17.6%) | — | — | — | — |
| Yosboonruang et al. (2019) | Thailand | Culture PCR | 60 | 60: (100%) | — | — | — | — |
| Al‐Yousef. (2014) | Saudi Arabia | Culture | 200 | 85: (42,5%) | — | 21: (10.5%) | — | 22: (11%) |
| Baker et al. (2018) | Iraq | Culture | 150 | 36: (24%) | — | 25: (1.3%) | 11: (7.3%) | — |
| Burrus et al. (2017) | USA | Culture PCR | 35 | 27: (77.1%) | — | — | — | — |
| Doud et al. (2014) | USA | PCR | 276 | — | 60: (21.7%) | — | — | — |
| Manandhar and Gokhale (2017) | Nepal | Culture PCR | 100 | 8: (8%) | — | 4: (4%) | — | — |
| Nazari et al. (2017) | Iran | Culture | 300 | 46: (15.3%) | — | 25; (8.3%) | — | — |
| Neupane et al. (2019) | USA | Culture PCR | 60 | — | — | — | — | — |
| Pohlenz et al. (2018) | USA | Culture | 224 | 205: (91.1%) | — | — | — | — |
| Puri‐Giri et al. (2017) | USA | Culture PCR | 463 | 159: (34.3%) | — | — | — | — |
| Rosef and Kapperud (1983) | Norway | Culture | 608 | — | — | — | — | — |
| Vaziriannzadeh et al. (2008) | Iran | Culture | 230 | 230: (100%) | — | 169: (73.5%) | — | — |
| Tsangaan et al. (2015) | Japan | PCR | 267 | 114: (42.7%) | — | — | — | — |
| Szalanski et al. (2004) | USA | Culture PCR | 2,486 | 160: (6.4%) | — | — | — | — |
| Rahuma et al. (2005) | Libya | Culture | 150 | 58: (38.7%) | — | — | — | — |
| Yalli et al. (2017) | Nigeria | Culture | 150 | 11: (7.3%) | — | 12: (8%) | — | — |
| Nwankwo et al. (2020) | Nigeria | Culture | 150 | 35: (23.3%) | — | 23: (15.3%) | — | 9: (6%) |
| Davari et al. (2010) | Iran | Culture | 908 | 45: (4.95%) | — | — | — | 77: (8.5%) |
| Awache and Forouk (2016) | Nigeria | Culture | 1,059 | 177: (16.7%) | — | — | 113: (10.7%) | — |
| Akter et al. (2020) | Bangladesh | Culture PCR | 140 | 72: (51.4%) | — | 110: (78.6%) | — | — |
| Mawak and Olukose (2006) | Nigeria | Culture | 200 | 30: (15%) | — | — | — | — |
| Chaiwong et al. (2014) | Thailand | culture | 1,349 | 94: (6.96%) | — | 13: (0.96%) | — | 97: (7.2%) |
| Ghalehnoo (2015) | Iran | Culture | 450 | — | — | 87: (19.3%) | — | — |
| Kababian et al. (2020) | Iran | Culture | 160 | 59: (36.9%) | — | 27: (16.9%) | — | 33: (20.6%) |
| Moriya et al. (1999) | Japan | Culture PCR | 89 | 10: (11.2%) | — | — | — | — |
| Odentoyin et al. (2020) | Nigeria | Culture | 25 | 25: (100%) | — | 11: (44%) | 17: (68%) | 3: (12%) |
| Akter et al. (2017) | Bangladesh | Culture | 15 | 15: (100%) | — | — | — | — |
| Brits et al. (2016) | South Africa | Culture PCR | 10 | — | — | — | — | — |
| Alam and Zurek (2004) | USA | Culture PCR | 3,440 | 75: (2.2%) | — | — | — | — |
| Choo et al. (2011) | Malaysia | Culture | 60 | — | — | — | — | — |
Abbreviations: PCR, polymerase chain reaction; USA, United States of America.
2.4. Statistical analysis
Meta‐analysis was carried out using Comprehensive Meta‐Analysis Version 3.0. Between the studies, Cochran's heterogeneity (Q) within studies as well as percentage variation in prevalence was assessed using Higgin's I 2 (inverse variance) and Cochran's Q method. I 2 values of 25%, 50% and 75% were considered low, moderate and high heterogeneity, respectively (Higgins & Thompson, 2002). Subgroup analysis was also conducted by the location of study, study sites and diagnosis method used. The effect size and their corresponding confidence interval (95% CI) for each subgroup were calculated using MedCalc® statistical software and expressed on the forest plots. A funnel plot was generated to evaluate the potential for asymmetry or publication bias present.
3. RESULTS
3.1. Study selection
Following a systematic approach and bibliographic search, 1,978 references were retrieved from the initial search strategy (Figure 1). Citation searches were based on published journal articles. Overall, 1,133 studies were excluded following the initial screening based on titles or abstracts and duplicate journal articles were also uninvolved. The remaining 845 journal articles were further screened for eligibility, and 767 studies were excluded for various reasons stipulated in Figure 1.
3.2. Study characteristics of eligible studies
Characteristics of the eligible studies includes; the number of houseflies collected, bacterial species isolated, identification method, country and sampled sites as presented in Table S1. All the journal articles were published between the years 1980 and 2020. A handful of studies have been conducted globally that reports on bacterial pathogens isolated from houseflies. Studies included in this review were from four different continents namely; Africa, Americas, Asia and Europe (Figure 2). Asia (47.4%) was observed to have the highest number of studies followed by Americas (23.1%) and Africa (20.5%), and Europe (8.9%) being the least. The sample size from eligible studies ranged from 10 houseflies to 3, 440 individual houseflies' specimens. Samples were collected from diverse sites including hospitals, water treatment facilities, slaughterhouses, garbage dumps, farms and residential areas. Thus, 38 (48.7%) studies were sampled from more than one site, while 39 (50%) of the studies sampled only from on one specific site and one (1.3%) study failed to report on their sampled site (Table S1).
FIGURE 2.
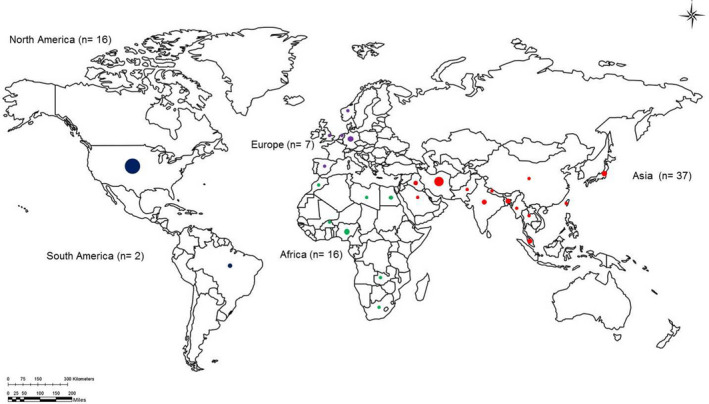
Number of countries, number of studies and sample size per continent. The size of the circle indicates the number of studies conducted in that country with different colours for different continents whereby green is for Africa, red for Asia, blue for North America and black is for South America
The techniques employed by the eligible studies to isolate and identify bacterial species included culture‐dependent (microbiological culture) and culture‐independent (polymerase chain reaction [PCR]) respectively. However, other studies that made use of both techniques simultaneously were also included in the analysis. At least 56.4% (44/78) of the studies used culture method for the identification of pathogens and 30.7% (24/78) used the combination of both culture and PCR diagnostic technique for the identification and only 7.7% (6/78) used PCR method for the identification. However, one study failed to mention their diagnostic technique. Of 39/78 (50%) studies, reported on antibiotic susceptibility profile for different bacterial genera and only 13/78 (16.7%) studies reported on antibiotic‐resistant genes. The bacterial species reported in this review are recognized pathogens including E. coli, E. faecium, K. pneumoniae, P. aeruginosa and S. aureus. Amplification of some common resistance genes for colistin, sulphonamide, tetracycline, beta‐lactam, penicillin, quinolones and integrons were observed in this study (Table S1).
3.3. Meta‐analysis
A total of 44 eligible studies were included in the quantitative synthesis (Table 1); this was owing to the study being definite with bacterial species. Substantial heterogeneity was observed between the eligible studies. The pooled estimates and heterogeneities of the eligible studies are presented in Tables 2, 3, 4, 5, 6 for the five selected pathogenic bacteria including E. coli, E. faecium, K. pneumoniae, P. aeruginosa and S. aureus. In all meta‐analyses conducted, the overall effect estimates and their corresponding 95% confidence intervals (CI) did not overlap point estimate (Figures 3, 4, 5, 6, 7). Similarly, the latter was observed in all the subgroups (Figures S1–S4).
TABLE 2.
Proportion of Escherichia coli isolated from houseflies sampled from regions, screening methods and sampling locations
| Number of studies | Pooled estimate | Measure of heterogeneity | ||||
|---|---|---|---|---|---|---|
| Sample size | Positive | Q | I 2 (95% CI) | Significance level Q‐p | ||
| Bacterial species | ||||||
| Escherichia coli | 44 | 20,455 | 2,711 | 10,739.5523 | 99.60 (99.57–99.63) | <0.0001 |
| Region | ||||||
| Africa | 11 | 13,581 | 875 | 3,569.6776 | 99.72 (99.68–99.76) | <0.0001 |
| America | 9 | 5,097 | 41 | 2,599.8385 | 99.69 (99.64–99.74) | <0.0001 |
| Asia | 22 | 196 | 1,003 | 1,487.0151 | 98.59 (98.32–98.81) | <0.0001 |
| Europe | 2 | 2 | 717 | 279.5474 | 99.64 (99.42–99.78) | <0.0001 |
| Sampling locations | ||||||
| Dumping sites | 3 | 9,906 | 17 | 86.2819 | 97.68 (95.57–98.79) | <0.0001 |
| Farms | 18 | 1,705 | 797 | 2,075.0757 | 99.18 (99.04–99.30) | <0.0001 |
| Hospitals | 13 | 634 | 213 | 738.0189 | 98.37 (97.93–98.72) | <0.0001 |
| Market | 4 | 393 | 351 | 40.9600 | 92.68 (84.47–96.55) | <0.0001 |
| No specific location | 3 | 509 | 155 | 13.6833 | 85.38 (57.08–95.02) | =0.0011 |
| Restaurant | 6 | 493 | 245 | 6,303,3,062 | 99.75 (99.72–99.77) | <0.0001 |
| Slaughterhouse (abattoir) | 7 | 253 | 94 | 206.0165 | 97.09 (95.62–98.06) | <0.0001 |
| University campus/residential houses | 7 | 426 | 186 | 134.0283 | 95.52 (92.86–97.19) | <0.0001 |
| Wastewater/broken sewage | 2 | 388 | 9 | 35.1989 | 97.16 (92.69–98.90) | <0.0001 |
| Diagnostic technique | ||||||
| Culture | 25 | 319 | 1,537 | 2,698.8736 | 99.11 (98.98–99.23) | <0.0001 |
| PCR | 2 | 549 | 114 | 173.0078 | 99.42 (98.97–99.67) | <0.0001 |
| Culture & PCR | 17 | 11,587 | 985 | 6,303.3062 | 99.75 (99.72–99.77) | <0.0001 |
Abbreviations: CI, confidence interval; I 2, inverse variance; PCR, polymerase chain reaction; Q‐p, Cochran's.
TABLE 3.
Proportion of Staphylococcus aureus isolated from houseflies sampled from regions, screening methods and sampling locations
| Number of studies | Pooled estimate | Measure of heterogeneity | ||||
|---|---|---|---|---|---|---|
| Sample size | Positives | Q | I 2 (95% CI) | Significance level Q‐p | ||
| Bacterial species | ||||||
| Staphylococcus aureus | 44 | 20,455 | 561 | 2,439.1220 | 98.24 (97.99–98.45) | <0.0001 |
| Region | ||||||
| Africa | 11 | 13,581 | 56 | 230.0741 | 95.65 (93.77–96.97) | <0.0001 |
| Asia | 22 | 196 | 505 | 1,414.2219 | 98.52 (98.23–98.76) | <0.0001 |
| Sampling locations | ||||||
| Dumping sites | 3 | 9,906 | 7 | 33.2093 | 93.98 (85.80–97.47) | <0.0001 |
| Farms | 18 | 1705 | 132 | 777.4544 | 97.81 (97.27–98.25) | <0.0001 |
| Hospitals | 13 | 634 | 31 | 79.6692 | 84.94 (75.80–90.62) | <0.0001 |
| Market | 4 | 393 | 4 | 17.0149 | 82.53 (54.61–93.15) | =0.0007 |
| No specific location | 3 | 509 | 12 | 38.1214 | 94.75 (88.02–97.70) | <0.0001 |
| Restaurant | 6 | 493 | 37 | 518.4722 | 96.91 (96.01–97.61) | <0.0001 |
| Slaughterhouse (abattoir) | 7 | 253 | 3 | 13.5396 | 55.69 (00.00–80.97) | =0.0352 |
| University campus/residential houses | 7 | 426 | 44 | 42.3304 | 85.83 (72.83–92.61) | <0.0001 |
| Wastewater/broken sewage | 2 | 388 | 6 | 22.6646 | 95.59 (87.18–98.48) | <0.0001 |
| Diagnostic technique | ||||||
| Culture | 25 | 319 | 447 | 1,603.7790 | 98.50 (98.23–98.73) | <0.0001 |
| Culture & PCR | 17 | 11,587 | 114 | 518.4722 | 96.91 (96.01–97.61) | <0.0001 |
Abbreviations: CI, confidence interval; I 2, inverse variance; PCR, polymerase chain reaction; Q‐p, Cochran's.
TABLE 4.
Proportion of Pseudomonas aeruginosa isolated from houseflies sampled from regions, screening methods and sampling locations
| Number of studies | Pooled estimate | Measure of heterogeneity | ||||
|---|---|---|---|---|---|---|
| Sample size | Positives | Q | I 2 (95% CI) | Significance level Q‐p | ||
| Bacterial species | ||||||
| Pseudomonas aeruginosa | 44 | 20,455 | 299 | 1,283.2227 | 96.65 (96.07–97.15) | <0.0001 |
| Region | ||||||
| Africa | 11 | 13,581 | 12 | 52.8707 | 81.09 (67.19–89.10) | <0.0001 |
| Asia | 22 | 196 | 287 | 772.7609 | 97.28 (96.63–97.81) | <0.0001 |
| Sampling locations | ||||||
| Dumping sites | 3 | 9,906 | 5 | 23.9932 | 91.66 (78.72–96.74) | <0.0001 |
| Farms | 18 | 1,705 | 53 | 235.3866 | 92.78 (90.03 94.77) | <0.0001 |
| Hospitals | 13 | 634 | 107 | 282.6736 | 95.75 (94.12–96.94) | <0.0001 |
| Slaughterhouse (abattoir) | 7 | 253 | 7 | 51.2794 | 88.30 (78.30–93.69) | <0.0001 |
| University campus/residential houses | 7 | 426 | 22 | 31.5330 | 80.97 (61.56–90.58) | <0.0001 |
| Wastewater/broken sewage | 2 | 388 | 4 | 14.9064 | 93.29 (78.01–97.95) | =0.0001 |
| Diagnostic technique | ||||||
| Culture | 25 | 319 | 246 | 595.1690 | 95.97 (94.94–96.79) | <0.0001 |
| Culture & PCR | 17 | 11,587 | 53 | 490.0658 | 96.74 (95.76–97.48) | <0.0001 |
Abbreviations: CI, confidence interval; I 2, inverse variance; PCR, polymerase chain reaction; Q‐p, Cochran's.
TABLE 5.
Proportion of Klebsiella pneumoniae isolated from houseflies sampled from regions, screening methods and sampling locations
| Number of studies | Pooled estimate | Measure of heterogeneity | ||||
|---|---|---|---|---|---|---|
| Sample size | Positives | Q | I 2 (95% CI) | Significance level Q‐p | ||
| Bacterial species | ||||||
| Klebsiella pneumoniae | 44 | 20,455 | 279 | 1,576.6073 | 97.27 (96.83–97.65) | <0.0001 |
| Region | ||||||
| Africa | 11 | 13,581 | 17 | 79.1374 | 87.36 (79.31–92.28) | <0.0001 |
| Asia | 22 | 196 | 262 | 1,314.0651 | 98.40 (98.09–98.67) | <0.0001 |
| Sampling locations | ||||||
| Farms | 18 | 1,705 | 51 | 262.1432 | 93.51 (91.14–95.26) | <0.0001 |
| Hospitals | 13 | 634 | 164 | 487.8350 | 97.54 (96.76–98.13) | <0.0001 |
| Restaurant | 6 | 493 | 2 | 628.8196 | 97.46 (96.76–98.00) | <0.0001 |
| Slaughterhouse (abattoir) | 7 | 253 | 37 | 91.3,336 | 93.43 (88.91–96.11) | <0.0001 |
| University campus/residential houses | 7 | 426 | 4 | 17.1543 | 65.02 (21.35–84.45) | =0.0087 |
| Diagnostic technique | ||||||
| Culture | 25 | 319 | 211 | 889.4596 | 97.30 (97.70–97.79) | <0.0001 |
| Culture & PCR | 17 | 11,587 | 68 | 628.8196 | 97.46 (96.76–98.00) | <0.0001 |
Abbreviations: CI, confidence interval; I 2, inverse variance; PCR, polymerase chain reaction; Q‐p, Cochran's.
TABLE 6.
Proportion of Enterococcus faecium isolated from houseflies sampled from regions, screening methods and sampling locations
| Number of studies | Pooled estimate | Measure of heterogeneity | ||||
|---|---|---|---|---|---|---|
| Sample size | Positive | Q | I 2 (95% CI) | Significance level Q‐p | ||
| Bacterial species | ||||||
| Enterococcus faecium | 44 | 20,455 | 77 | 317.6145 | 86.46 (82.70–89.41) | <0.0001 |
| Region | ||||||
| Americas | 9 | 1,581 | 77 | 178.3214 | 95.51 (93.27–97.01) | <0.0001 |
| Sampling locations | ||||||
| Dumping sites | 3 | 9,906 | 4 | 20.4559 | 90.22 (74.03–96.32) | <0.0001 |
| Farms | 18 | 1,705 | 3 | 15.6957 | 00.00 (00.00–45.86) | =0.5455 |
| Restaurant | 17 | 11,587 | 14 | 25.1695 | 36.43 (00.00–64.46) | =0.0669 |
| Wastewater/broken sewage | 2 | 388 | 60 | 13.7057 | 92.70 (75.50–97.83) | =0.0002 |
| Diagnostic technique | ||||||
| Culture | 25 | 7,970 | 60 | 228.5404 | 89.85 (86.19–92.54) | <0.0001 |
| PCR | 2 | 549 | 14 | 30.1038 | 96.68 (91.08–98.76) | <0.0001 |
| Culture & PCR | 17 | 11,587 | 3 | 25.1695 | 36.43 (00.00–64.46) | =0.0669 |
Abbreviations: CI, confidence interval; I 2, inverse variance; PCR, polymerase chain reaction; Q‐p, Cochran's.
FIGURE 3.
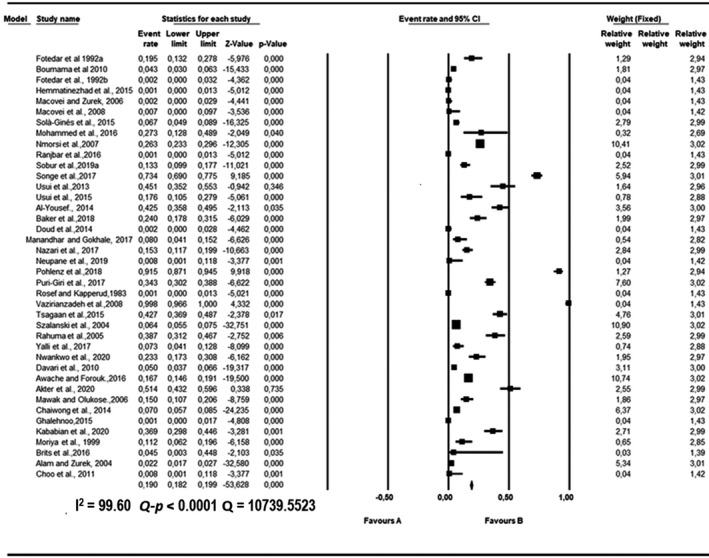
Forest plot showing the pooled estimates of Escherichia coli proportion isolated from houseflies. The squares demonstrate the individual point estimate. The diamond at the base indicates the pooled estimates from the overall studies
FIGURE 4.
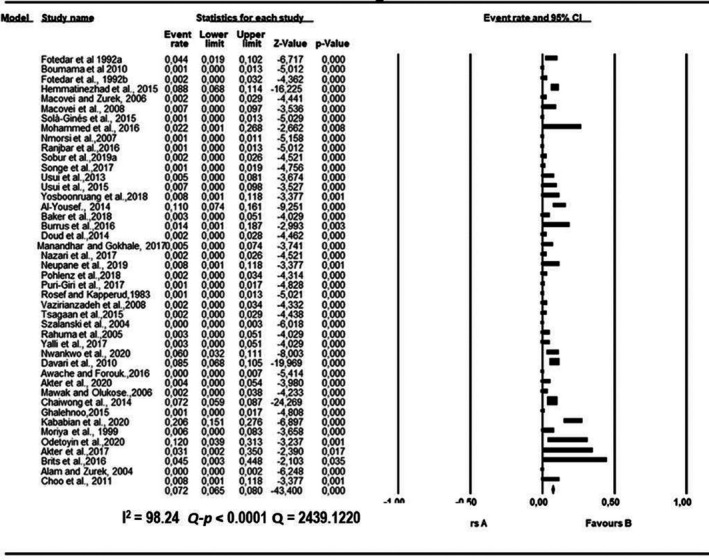
Forest plot showing the pooled estimates of Staphylococcus aureus proportion isolated from houseflies. The squares demonstrate the individual point estimate. The diamond at the base indicates the pooled estimates from the overall studies
FIGURE 5.
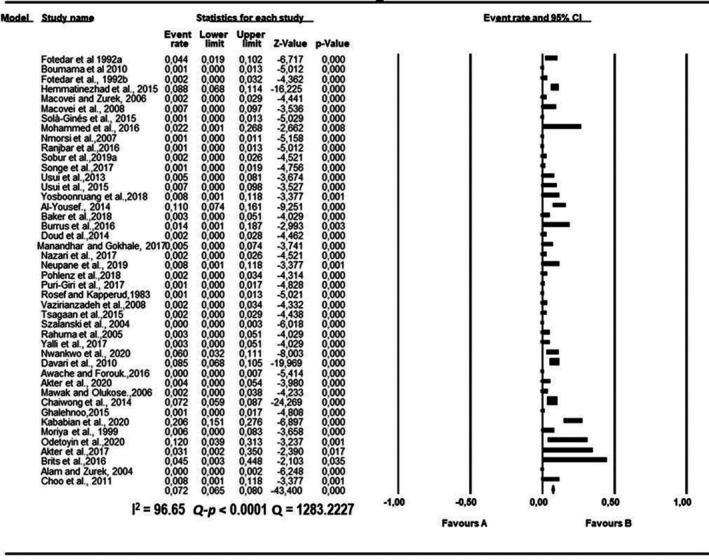
Forest plot showing the pooled estimates of Pseudomonas aeruginosa proportion isolated from houseflies. The squares demonstrate the individual point estimate. The diamond at the base indicates the pooled estimates from the overall studies
FIGURE 6.
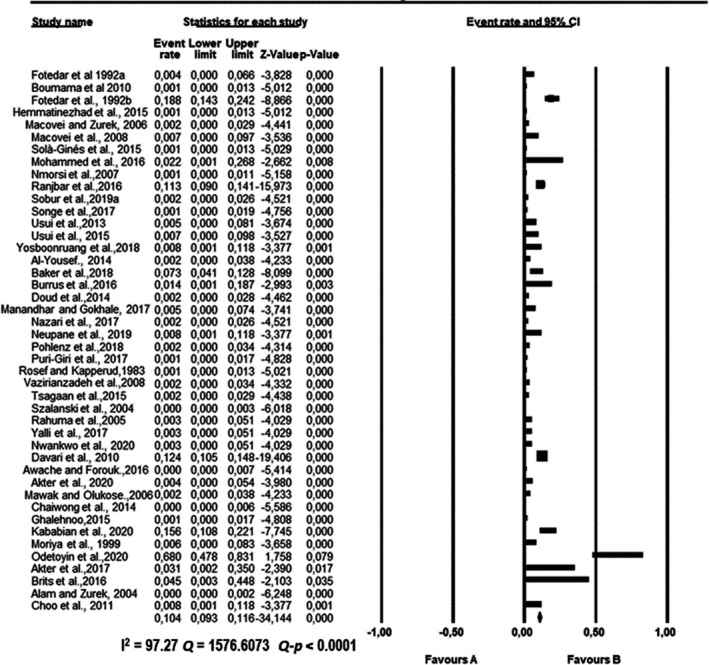
Forest plot showing the pooled estimates of Klebsiella pneumoniae proportion isolated from houseflies. The squares demonstrate the individual point estimate. The diamond at the base indicates the pooled estimates from the overall studies
FIGURE 7.
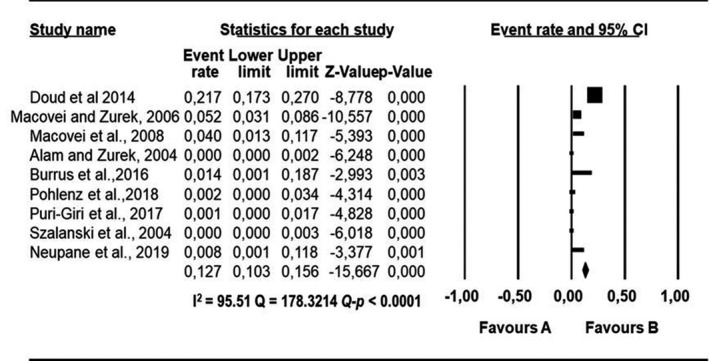
Forest plot showing the pooled estimates of Enterococcus faecium proportion isolated from houseflies. The squares demonstrate the individual point estimate. The diamond at the base indicates the pooled estimates from the overall studies
3.3.1. Pooling and heterogeneity estimates
Pooled proportions and their corresponding heterogeneities are presented in Tables 2, 3, 4, 5, 6 for E. coli, E. faecium, K. pneumoniae, P. aeruginosa and S. aureus respectively. Substantial heterogeneity was observed for the selected pathogens as well as in the subgroup analysis. For E. coli, high heterogeneity was observed (Q = 10,739.5523; I 2 = 99.60; 95% CI: 99.57–99.63; Q‐p < 0.0001; Table 2), followed by S. aureus (Q = 2,439.1220; I 2 = 98.24; 95% CI: 97.99–98.45; Q‐p < 0.0001; Table 3), P. aeruginosa (Q = 1,283.2227; I 2 = 96.65; 95% CI: 96.07–97.15; Q‐p < 0.0001; Table 4), K. pneumoniae (Q = 1,576.6073; I 2 = 97.27; 95% CI: 96.83–97.65; Q‐p < 0.0001; Table 5) and E. faecium (Q = 317.6145; I 2 = 86.46; 95% CI: 82.70–89.41; Q‐p < 0.0001; Table 6) being the least. The forest plot depicting the point estimate for individual studies reporting the occurrence of E. coli, E. faecium, K pneumoniae, P. aeruginosa and S. aureus are graphically presented (Figures 3, 4, 5, 6, 7).
3.3.2. Subgroup analysis
Subgroup analyses were done for study locations (Africa, America, Asia and Europe), study sites (Dumping sites, farms, hospitals, market, no specific location, restaurant, slaughterhouse (Abattoir), university campus/residential houses and wastewater/broken sewage), and diagnosis methods used (culture and molecular techniques [PCR]). Different subgroup analysis was undertaken for the five selected bacteria. The forest plot showing the point estimates results of the subgroup analysis are depicted in Figures S1–S4.
3.3.3. Prevalence based on study region
High degree of heterogeneity with respect to E. coli was observed in four continents, whereas other pathogens including P. aeruginosa and S. aureus were observed in both Africa and Asia while E. faecium was observed only in America (Tables 2, 3, 4, 5, 6). With regards to E. coli identified in Africa, a high degree of heterogeneity was observed (Q = 3,569.6776; I 2 = 99.72; 95% CI: 99.68–99.76; Q‐p < 0.0001) followed by America with (Q = 2,599.8385; I 2 = 99.69; 95% CI: 99.64–99.74; Q‐p < 0.0001), while the least observed was in Asia (Q = 1,487.0151; I 2 = 98.59; 95% CI: 98.32–98.81; Q‐p < 0.0001) (Table 2). The results were at variance for S. aureus and P. aeruginosa with high heterogeneity (Q = 1,414.2219; I 2 = 98.52; 95% CI: 98.23–98.76; Q‐p < 0.0001) and (Q = 772.7609; I 2 = 97.28; 95% CI: 96.63–97.81; Q‐p < 0.0001) in Asia.
3.3.4. Prevalence based on sampling locations
High heterogeneity was found among studies investigating different pathogens including E. coli, E. faecium, K. pneumoniae, S. aureus and P. aeruginosa from different sampling locations (Tables 2, 3, 4, 5, 6). The sampling locations include the farms, hospital, dumping site, market, restaurant, slaughterhouse/abattoir, university campus/residential houses and wastewater/ broken sewage. High heterogeneity was observed (I 2 > 75%) among the studies, with studies investigating E. coli and P. aeruginosa from the farms having the highest heterogeneity (Q = 2,075.0757; I 2 = 99.18; 95% CI: 99.04–99.30; p < 0.0001) and (Q = 2,075.0757; I 2 = 99.18; 95% CI: 99.04–99.30; p < 0.0001), respectively. Also, high heterogeneity was observed from the studies investigating E. faecium from wastewater/broken sewage (Q = 13.7057; I 2 = 92.70; 95% CCI: 75.50–97.83; Q‐p < 0.0001), respectively. However, studies investigating S. aureus in slaughterhouse/abattoir had moderate heterogeneity (Q = 13.5396; I 2 = 55.69; 95% CI: 0.00–80.97; Q‐p < 0.0352) and studies investigating K. pneumoniae in university campus/residential houses had moderate heterogeneity (Q = 17.1543; I 2 = 65.02; 95% CI: 21.35–84.45; Q‐p < 0.0087).
3.3.5. Prevalence based on diagnostic methods
Two different diagnostic methods were used by the eligible studies to identify pathogens from houseflies, the combination of two diagnostic methods; culture and PCR had the highest heterogeneity in studies investigating E. coli and P. aeruginosa (Q = 6,303.3062; I 2 = 99.75; 95% CI: 99.72 – 99.77; Q‐p < 0.0001) and (Q = 490.0658; I 2 = 96.74; 95% CI: 95.76–97.48; Q‐p < 0.0001). Lowest heterogeneity (Q = 25.1695; I 2 = 36.43; 95% CI: 0.00–64.46 Q‐p = 0.0669 was observed with the studies investigating E. faecium using both culture and PCR techniques; Tables 2, 3, 4, 5, 6).
3.3.6. Publication bias
According to the data observed from the funnel plot by virtual inspection, for all bacterial species, namely, E. coli, E. faecium, K. pneumoniae, P. aeruginosa and S. aureus, no publication bias was observed as the funnel was symmetrical (Figure 8). Thus, there was significant asymmetry for the analysis of all the selected bacterial pathogens.
FIGURE 8.
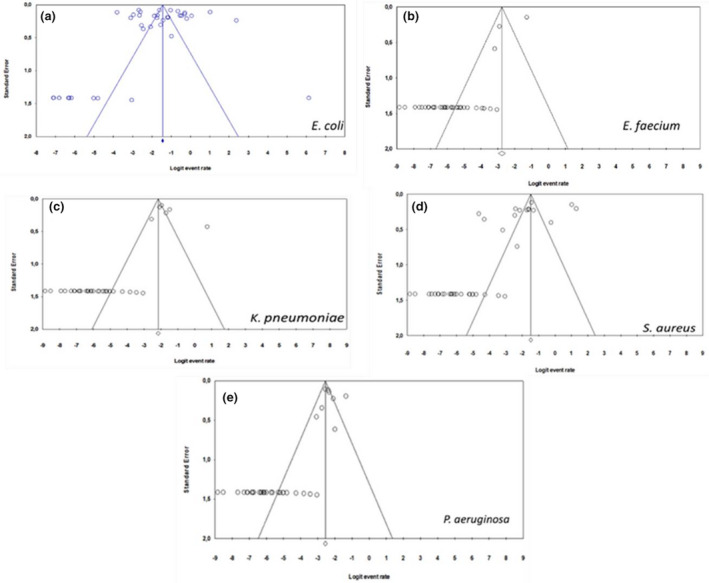
Funnel plots for studies investigating: (a) E. coli, (b) E. faecium, (c) K. pneumoniae, (d) S. aureus and (e) P. aeruginosa
4. DISCUSSION
This study undertook a systematic review and meta‐analysis to estimate the pooled prevalence of the occurrence of pathogenic bacterial species of medical and veterinary importance carried by houseflies globally. The number of pathogens reported by the studies for qualitative synthesis varied greatly with most reporting on pathogenic and virulent bacterial strains. Thus, the pathogenic Gram‐negative bacteria were by far the most frequent with most studies as compared to Gram‐positive bacteria. Different bacterial species found on houseflies depend on the location where the insects are captured (Cervelin et al., 2018). This study has also proved that houseflies are in abundance in different settings and geographical locations. These findings are in accordance with the review by Issa (2019) where they indicated that houseflies live closely with humans and domestic animals and often found in areas of human activities such as restaurants, hospitals, food centres, food markets, fish markets and slaughterhouses. With the available information to date, there is evidence that houseflies are moderate to high in various human and animal settings harbouring clinically and veterinary relevant bacterial species in abundance (Gupta et al., 2012).
M. domestica is most frequently associated with zoonoses worldwide (Wales et al., 2010). Through history, housefly colonization by bacteria has been reported previously by other studies including the study by Moorhead and Weisser (1946) where they recorded and proved the survival of Staphylococcus strains in the gut and excreta of houseflies. Additionally, other studies also documented that M. domestica is becoming heavily colonized by various bacterial genera including Staphylococci, Streptococci and Salmonellae (Mawak & Okulose, 2006). Thus, houseflies have been implicated as vectors or transporters of various human pathogens, including Vibrio cholerae, Enterobacteriaceae pathogens, S. aureus and Pseudomonas spp. (Ahmed et al., 2013; Davari et al., 2010).
The most frequent reported bacterial species by various studies were the following: (a) E. coli, the bacterium that causes nosocomial infections and bloody diarrhoea and haemorrhagic colitis in humans (Blaak et al., 2014; Iwasa et al., 1999; Poudel et al., 2019; Wales et al., 2010); (b) E. faecium, the most important nosocomial pathogens of humans, causing urinary tract infections, bloodstream infections, endocarditis and wound infections (Hammerum, 2012; Macovei et al., 2008; Selleck et al., 2019 ); (c) K. pneumoniae, which cause infections of the respiratory tract and urinary tract, as well as post‐operative wound infections (Blaak et al., 2014; Ranjbar et al., 2016); (d) P. aeruginosa, cause disease in humans and other animals (Banjo et al., 2005); and (e) S. aureus which is capable of causing human illness and food poisoning (Banjo et al., 2005).
Antimicrobial resistance results in a serious public health problem, for it weakens the treatment efficacy against bacterial infections and this poses greater health risks (Russell et al., 2018; Wei et al., 2013). Some studies have reported some bacterial isolates which are resistant to significantly more than one of the commonly used antibiotics tested (Akter et al., 2020; Hemmatinezhad et al., 2015; Nazari et al., 2017; Nwankwo et al., 2020; Odentoyin et al., 2020; Rahuma et al., 2005). According to Russell et al. (2018), dipterans that associate with livestock, livestock waste products and cadavers have the potential to acquire livestock‐associated antibiotic resistant bacteria and transmit them to humans. In the year 2017, the WHO made a list of antibiotic‐resistant priority pathogens which present a great threat to humans (WHO, 2017). Gram‐negative bacterial pathogens including E. coli, K. pneumoniae, P. aeruginosa and Gram‐positive E. faecium and S. aureus were the most listed under the critical and high categories with regard to urgent need of new antibiotics (WHO, 2017). Owing to the distinctive structure amongst the Gram‐negative and Gram‐Positive bacteria, the Gram‐negative bacteria appear to be more resistant to antibiotics and cause significant morbidity and mortality worldwide (Breijyeh et al., 2020).
Release of antibiotic resistance bacteria expressing antibiotic resistance genes into the environment creates an opportunity for other insects to acquire livestock‐associated antibiotic‐resistant bacteria (Russell et al., 2018). Some studies in this review have reported on the presence of resistance genes including mobile colistin, tetracycline and extended‐spectrum β‐lactamase gene, from bacterial isolates identified from houseflies. This raises a very great concern since colistin is considered a last‐resort antimicrobial for treating patients infected with multidrug‐resistant Gram‐negative bacteria (Zhang et al., 2018). The spreading of the plasmid encoded extended‐spectrum β‐lactamase gene which can confer resistance to third generation cephalosporins is the major problem because third generation cephalosporins are clinically important antimicrobials in human and veterinary medicine (Usui et al., 2013). A review by Onwugamba et al. (2018) on the role of flies in the spread of antimicrobial resistance also revealed that ‘filth flies’ are colonized by clinically important antimicrobial resistant bacteria, including extended‐spectrum beta‐lactamase, carbapenemase‐producing, and colistin‐resistant bacteria. The presence of these resistant genes from the bacterial isolates simply indicates that houseflies have the capacity to significantly disseminate bacteria carrying antibiotic resistant genes from various environmental settings to humans (Zhang et al., 2018).
In this study, we observed high variance in the number of studies conducted on bacterial pathogens harboured by houseflies per continent. Thus, Asia had the highest number of eligible studies followed by America, Africa and Europe, respectively. The large number of studies in Asia can be due to the fact that, houseflies originate from Central Asia (Sanchez‐Arroyo & Capinera, 2015). However, they are now common in all climates ranging from tropical regions to temperate climates (Khamesipour et al., 2018; Sanchez‐Arroyo & Capinera, 2015). Hence there's a large number of studies observed from other continents including America (Alves et al., 2018; Burrus et al., 2017; Macovei et al., 2008; Macovei & Zurek, 2006; Olsen & Hammack., 2000; Poudel et al., 2019, Africa (Brits et al., 2016; Ibrahim et al., 2018; Nwankwo et al., 2020; Songe et al., 2016) and Europe (Blaak et al., 2014; Davies et al., 2014; Rosef & Kapperud, 1983; Solà‐Ginés et al., 2015).
Culturing and plating conventional microbiological methods are still regarded as a gold standard, as they enable the identification of various bacterial species effectively (Kotsilkov et al., 2015). However, the combination of both microbiological culturing and molecular methods is increasingly adopted (Langley et al., 2015). Due to the use of different pathogen detection methods (PCR and culture including selective media and/or broth enrichment), the bacterial carrier rates from flies must be cautiously compared (Onwugamba et al., 2018). Most of the studies in this review used selective culture media to identify predominantly pathogenic bacteria or specific species. However, medium preferences and growth condition optima of individual bacterial species may influence the observation of species diversity and the abundance of individual bacteria isolated (Brits et al., 2016). The combination of both culture and molecular techniques has shown to improve the identification and isolation rate of bacterial species. However, the advanced method including PCR is costly (Khamesipour et al., 2018). A review by Adzeity et al. (2013) showed that studies using molecular methods (PCR and/or sequences) yield more pathogens compared to studies using standard cultural methods. However, Gupta et al. (2012) showed that there are benefits and limitations concerning the use of either approach for diagnosis.
Notably, all studies included in this systematic review and meta‐analysis were conducted differently, as some studies investigated large number of samples while others investigated different settings and utilizing various diagnostic techniques. These parameters resulted in the high heterogeneity observed in this study. According to Oppong et al. (2020), heterogeneity could partially be attributed to large variation in sampling and diagnostic techniques applied for the detection of the bacterial pathogens. This is because different techniques differ in their detection sensitivity, specificity, as well as positive and negative predictive values. There was no statistical significance at the study level, but we observed statistical significance at the meta‐analysis. Substantial heterogeneity was observed in studies included in our analyses which can largely be attributed to variation in sample size, sampling method and analysis of data. Some studies presented small sample size resulting in effect estimates with wide confidence intervals.
This study has considerable number of strengths including (a) this study summarizes currently existing knowledge and helps in identifying areas that lack adequate evidence, thereby producing new research questions. (b) We have checked the overall consistency of the searching process, study choice and inclusion/exclusion criteria. All the duplicated records were removed. The authors independently examined the titles, abstracts and full‐text article. This was done in order to negate any possibility of errors. (c) The application of a consistent reviewing approach and extracting relevant data from the available published studies which allowed to reliably determine the pathogenic bacteria isolated from houseflies by various studies around the world and how frequent these studies report on the antibiotic susceptibility profiles and resistant genes in common bacterial pathogens in different countries (d) exclusion of duplicate studies and studies that are not specific with genus and species of the fly, studies using laboratory bred houseflies while including only studies with genus and species confirmed (e) and considering studies with samples collected from different human and animal settings.
Several limitations have been identified which included the following: (a) only journal papers published in English language were considered. This was due to lacking resources for translation of papers that might have been published in other languages. (b) Few studies have failed to describe the sample size despite the importance of considering the sample size in all investigational studies. Sample size calculation is ethically important and form part of the early stages of leading an epidemiological, clinical or laboratory studies (Faber & Fonesca, 2014). (c) Furthermore, some countries had more research reports compared to others.
In conclusion, this systematic review and meta‐analysis has provided global overview on the carriage of significant bacterial pathogens including antibiotic resistant bacteria identified in houseflies from different regions in the world. Houseflies are in abundance and found almost in every human and animal habitation and this pose a serious threat to public health. Most of the studies in this review predominantly identified pathogenic bacteria including E. coli, E. faecium, K. pneumoniae, P. aeruginosa and S. aureus. Few studies presented data on antibiotic susceptibility profile and resistant genes from housefly, and this is of great concern, as flies have the potential to rapidly and widely disseminate antimicrobial resistant genes. Studies in this review were found to employ different diagnostic methods including culture and PCR or the combination of both techniques. Further studies with special focus on antibiotic susceptibility profile and resistant genes from bacteria identified from houseflies will provide more insight global threat to human and animal health (Zhang et al., 2018). Future systematic reviews may also include the following: (a) data from different life cycles of housefly including eggs, larvae and pupa (b) data from culture‐independent studies that allow the identification of the cultivable and non‐culturable fractions of bacteria present in houseflies. The findings of this study provide more insight on the carriage of significant bacterial pathogens of distinct taxonomic identities by M. domestica from different geographical locations across the world. Continuous investigation is required to completely comprehend the significance of medically and veterinary important pathogenic bacteria harboured by houseflies. Also, further investigation is needed to quantify the contamination level in order to analyse the risk of bacterial contamination and dissemination by houseflies.
CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest in relation to this review paper.
AUTHOR CONTRIBUTION
Maropeng Charles Monyama: Conceptualization; Data curation; Formal analysis; Investigation; Writing‐original draft; Writing‐review & editing. Emmanuel ThankGod Onyiche: Methodology; Validation; Writing‐review & editing. Moeti Oriel Taioe: Validation; Writing‐review & editing. Jane Nkhebenyane: Validation; Writing‐review & editing. Oriel Matlhahane Molifi Thekisoe: Conceptualization; Supervision; Validation; Writing‐review & editing.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/vms3.496.
Supporting information
Table S1–Figure S1‐S4
ACKNOWLEDGEMENT
This study was supported financially by the University of South Africa (AQIP Scholarship), North West University (NWU) Postgraduate student bursary and National Research Foundation (NRF) Thuthuka grant (Grant number 117980). The Grant holder acknowledges that opinions, findings and conclusions or recommendations expressed in any publication generated by the NRF supported research is that of the authors, and that the NRF accepts no liability whatsoever in this regard.
Monyama MC, Onyiche ET, Taioe MO, Nkhebenyane JS, Thekisoe OMM. Bacterial pathogens identified from houseflies in different human and animal settings: A systematic review and meta‐analysis. Vet Med Sci. 2022;8:827–844. 10.1002/vms3.496
REFERENCES
- Adzeity, F. , Huda, N. , Rusul, G. , & Ali, R. (2013). Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. Biotechnology, 3, 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, A. S. , Ahmed, K. M. , & Salih, S. S. (2013). Isolation and identification of Bacterial isolates from houseflies in Sulaymaniyah city. Journal of Engineering and Technology, 31, 24–33. [Google Scholar]
- Akter, S. , Sabuj, A. A. M. , Haque, Z. F. , Rahman, M. T. , Kafi, M. A. , & Saha, S. (2020). Detection of antibiotic‐resistant bacteria and their resistance genes from houseflies. Veterinary World, 13(2), 266–274. 10.14202/vetworld.2020.266-274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves, T. S. , Lara, G. H. B. , Maluta, R. P. , Ribeiro, M. G. , & da Silva Leite, D. (2018). Carrier flies of multidrug‐resistant Escherichia coli as potential dissemination agent in dairy farm environment. Science of the Total Environment, 633, 1345–1351. 10.1016/j.scitotenv.2018.03.304 [DOI] [PubMed] [Google Scholar]
- Awache, I. , & Farouk, A. A. (2016). Bacteria and fungi associated with houseflies collected from cafeteria and food Centres in Sokoto. Fuw Trends in Science & Technology Journal, 1(1),123–125. [Google Scholar]
- Baker, S. Z. , Atiyae, Q. M. , & Khairallah, M. S. (2018). Isolation and identification of some species of bacterial pathogens from Musca Domestica and test their susceptibility against antibiotics. Tikrit Journal of Pure Science, 23(9), 20–27. 10.25130/tjps.23.2018.145 [DOI] [Google Scholar]
- Banjo, A. D. , Lawal, O. A. , & Adeduji, O. O. (2005). Bacteria and fungi isolated from housefly (Musca domestica L.) larvae. African Journal of Biotechnology, 4(8), 780–784. [Google Scholar]
- Barreiro, C. , Albano, H. , Silva, J. , & Teixeira, P. (2013). Role of flies as vectors of foodborne pathogens in rural areas. ISRN Microbiology, 2013, 1–7. 10.1155/2013/718780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaak, H. , Hamidjaja, R. A. , Van Hoek, A. H. A. M. , De Heer, L. , De Roda Husman, A. M. , & Schets, F. M. (2014). Detection of extended‐spectrum beta‐lactamase (ESBL)‐producing Escherichia coli on flies at poultry farms. Applied Environmental Microbiology, 80, 239–246. 10.1128/AEM.02616-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouamamaa, L. , Sorlozano, A. , Laglaoui, A. , Lebbadi, M. , Aarab, A. , & Gutierrez, J . (2010). Antibiotic resistance patterns of bacterial strains isolated from Periplaneta americana and Musca domestica in Tangier, Morocco. Journal of Infections Developing Countries, 4(4), 194–201. [DOI] [PubMed] [Google Scholar]
- Breijyeh, Z. , Jubeh, B. , & Karaman, R. (2020). Resistance of Gram‐negative bacteria to current antibacterial agents and approaches to solve it. Molecules, 25(6), 1340. 10.3390/molecules25061340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brits, D. , Brooks, M. , & Villet, M. H. (2016). Diversity of bacteria isolated from the flies Musca domestica (Muscidae) and Chrysomya megacephala (Calliphoridae) with emphasis on vectored pathogens. African Entomology, 24(2), 365–375. [Google Scholar]
- Burrus, R. G. , Hogsette, J. A. , Kaufman, P. E. , Maruniak, J. E. , Simonne, A. H. , & Mai, V. (2017). Prevalence of Escherichia coli O157:H7 from house flies (Diptera: Muscidae) and dairy samples in north central Florida. Journal of Medical Entomology, 54(3), 733–741. [DOI] [PubMed] [Google Scholar]
- Cervelin, V. , Fongaro, G. , Pastore, J. B. , Engel, F. , Reimers, M. A. , & Viancelli, A. (2018). Enterobacteria associated with houseflies (Musca domestica) as an infection risk indicator in swine production farms. Acta Tropica, 185, 13–17. [DOI] [PubMed] [Google Scholar]
- Chaiwong, T. , Srivoramas, T. , Sueabsamran, P. , Sukontason, K. , Sanford, M. R. , & Sukontason, K. L. (2014). The blow fly, Chrysomya megacephala, and the house fly, Musca domestica, as mechanical vectors of pathogenic bacteria in Northeast Thailand. Tropical Biomedicine, 31(2), 336–346. [PubMed] [Google Scholar]
- Choo, L. C. , Saleha, A. A. , Wai, S. S. , & Fauziah, N. (2011). Isolation of Campylobacter and Salmonella from houseflies (Musca domestica) in a university campus and a poultry farm in Selangor, Malaysia. Tropical Biomedicine, 28(1), 16–20. [PubMed] [Google Scholar]
- Davari, B. , Kalantar, E. , Zahirnia, A. , & Moosa‐Kazemi, S. H. (2010). Frequency of resistance and susceptible bacteria isolated from houseflies. Iran Journal of Arthropod‐Borne Disease, 4, 50–55. [PMC free article] [PubMed] [Google Scholar]
- Davies, M. P. , Hilton, A. C. , & Anderson, M. (2014). Isolation and characterization of bacteria associated with Musca domestica (Diptera: muscidae) in hospitals. Proceedings of the Eighth International Conference on Urban Pests Gabi Müller, Reiner Pospischil and William H Robinson. OOK‐Press Kft., H‐8200 Veszprém, Papái ut 37/a, Hungary. [Google Scholar]
- Doud, C. W. , Scott, H. M. , & Zurek, L. (2014). Role of house flies in the ecology of Enterococcus faecalis from wastewater treatment facilities. Microbial Ecology, 67(2), 380–391. 10.1007/s00248-013-0337-6 [DOI] [PubMed] [Google Scholar]
- Faber, J. , & Fonesca, L. M. (2014). How sample size influences research outcomes. Dental Press Journal of Orthodontics, 19(4), 27–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotedar, R. , Banarjee, U. , Samantray, J. C. , & Shriniwas, S. S. (1992). Vector potential of the hospital house flies with special reference to Klebsiella species. Epidemiology and Infection, 109, 143–147. [PMC free article] [PubMed] [Google Scholar]
- Fotedar, R. , Banerjee, U. , Singh, S. , & Verma, A. K. (1992). The house fly (Musca domestica) as a carrier of pathogenic microorganisms in a hospital environment. Journal of Hospital Infections, 20, 209–215. 10.1016/0195-6701(92)90089-5 [DOI] [PubMed] [Google Scholar]
- Ghalehnoo, M. R. (2015). Housefly (Musca domestica) as harrier of enterotoxigenic Staphylococcus aureus in broiler farms in Iran: Is it important for public health? International Journal of Enteric Pathogens, 3(3), e25688. 10.17795/ijep25688 [DOI] [Google Scholar]
- Gupta, A. K. , Nayduch, D. , Verma, P. , Shah, B. , Ghate, H. V. , Patole, M. S. , & Shouche, Y. S. (2012). Phylogenetic characterization of bacteria in the gut of houseflies (Musca domestica L.). FEMS Microbiological Ecology, 79, 581–593. 10.1111/j.1574-6941.2011.01248.x [DOI] [PubMed] [Google Scholar]
- Hammerum, A. M. (2012). Enterococci of animal origin and their significance for public health. European Society of Clinical Microbiology and Infectious Diseases, 18, 619–625. [DOI] [PubMed] [Google Scholar]
- Hemmatinezhad, B. , Ommi, D. , Taktaz‐Hafshejani, T. , & Khamesipour, F. (2015). Molecular detection and antimicrobial resistance of Pseudomonas aeruginosa from houseflies (Musca domestica) in Iran. Journal of Venomous Animals Toxins, 21, 18. 10.1186/s40409-015-0021-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins, J. P. T. , & Thompson, S. G. (2002). Quantifying heterogeneity in meta‐analysis. Statistics in Medicine, 21, 1539e1558. [DOI] [PubMed] [Google Scholar]
- Ibrahim, A. W. , Ajiboye, T. O. , Akande, T. A. , & Anibaba, O. O. (2018). Isolation and identification of pathogenic microorganisms from houseflies. Global Journal of Science Frontier Research, 18(1), 57–64. [Google Scholar]
- Iqbal, W. , Malik, M. F. , Sarwar, M. K. , Azam, I. , Iram, N. , & Rashda, A. (2014). Role of housefly (Musca domestica, Diptera; Muscidae) as a disease vector; a review. Journal of Entomology and Zoology Studies, 2(2), 159–163. [Google Scholar]
- Issa, H. (2019). Musca domestica acts as transport vector hosts. Issa Bulletin of the National Research Centre, 43, 1–5. 10.1186/s42269-019-0111-0 [DOI] [Google Scholar]
- Iwasa, M. , Makino, S. , Asakura, H. , Kobori, H. , & Morimoto, Y. (1999). Detection of Escherichia coli 0157–H7 from Musca domestica (Diptera: Muscidae) at a cattle farm in Japan. Journal of Medical Entomology, 36, 108–112. [DOI] [PubMed] [Google Scholar]
- Kababian, M. , Mozaffari, E. , Akbarzadeh, K. , Kordshouli, R. S. , Saghafipour, A. , & Shams, S. (2020). Identification of bacteria contaminating Musca domestica (Diptera: Muscidae) collected from animal husbandries. Shiraz E‐Med Journal, 21(4), e92018. 10.5812/semj.92018 [DOI] [Google Scholar]
- Khamesipour, F. , Lankarani, K. B. , Honarvar, B. , & Kwenti, T. E. (2018). A systematic review of human pathogens carried by the housefly (Musca domestica L.). BMC Public Health, 18, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsilkov, K. , Popova, C. , Boyanova, L. , Setchanova, L. , & Mitov, I. (2015). Comparison of culture method and real‐time PCR for detection of putative period on to pathogenic bacteria in deep periodontal pockets. Biotechnology and Biotechnological Equipment, 29(5), 996–1002. 10.1080/13102818.2015.1058188 [DOI] [Google Scholar]
- Langley, G. , Besser, J. , Iwamoto, M. , Lessa, F. C. , Cronguist, A. , Skoff, T. H. , Chaves, S. , Boxrud, D. , Pinner, R. W. , & Harrison, L. H. (2015). Effect of culture‐dependent diagnostic tests on future emerging infections program surveillance. Emerging Infectious Disease, 21(9), 1582–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei, L. , Miles, B. , & Zurek, L. (2008). The potential of house flies to contaminate ready to eat food with antibiotic resistant enterococci. Journal of Food Protection, 71, 432–439. 10.4315/0362-028X-71.2.435 [DOI] [PubMed] [Google Scholar]
- Macovei, L. , & Zurek, L. (2006). Ecology of antibiotic resistance genes: Characterization of enterococci from houseflies collected in food settings. Applied Environmental Microbiology, 72, 4028–4035. 10.1128/AEM.00034-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, A. , Singh, N. , & Satya, S. (2007). Housefly (Musca domestica): A review of control strategies for a challenging pest. Journal of Environmental Science and Health, 42, 453–469. [DOI] [PubMed] [Google Scholar]
- Manandhar, R. , & Gokhale, S. (2017). Are houseflies still important vector of gastrointestinal infections? Journal of Bacteriology and Parasitology, 8, 4. 10.4172/2155-9597.1000318 [DOI] [Google Scholar]
- Mawak, J. D. , & Olukose, O. J. (2006). Vector potential of houseflies (Musca domestica) for pathogenic organisms in Jos, Nigeria. Journal of Pest Disease and Vector Management, 7, 418–423. [Google Scholar]
- Mohammed, A. N. , Abdel‐Latef, G. K. , Abdel‐Azeem, N. M. , & El‐Dakhly, K. M. (2016). Ecological study on antimicrobial‐resistant zoonotic bacteria transmitted by flies in cattle farms. Parasitology Research, 115, 3889–3896. 10.1007/s00436-016-5154-7 [DOI] [PubMed] [Google Scholar]
- Moher, D. , Shamseer, L. , Clarke, M. , Ghersi, D. , Liberati, A. , Petticrew, M. , & Stewart, L. A. (2015). Preferred reporting items for systematic review and meta‐analysis protocols (PRISMA‐P) statement. Systematic Reviews, 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead, S. , & Weiser, H. H. (1946). The survival of Staphylococcal food poisoning strain in the gut and excreta of the houseflies. Journal of Milk Technology, 9, 253–259. [Google Scholar]
- Moriya, K. , Fujibayashi, T. , Yoshihara, T. , Matsuda, A. , Sumi, N. , Umezaki, N. , Kurahashi, H. , Agui, N. , Wada, A. , & Watanabe, H. (1999). Verotoxin‐producing Escherichia coli O157: H7 carried by the housefly in Japan. Medical and Veterinary Entomology, 13, 214–216. 10.1046/j.1365-2915.1999.00161.x [DOI] [PubMed] [Google Scholar]
- Nazari, M. , Mahrabi, T. , Hosseini, S. M. , & Alikhani, M. Y. (2017). Bacterial contamination of adult houseflies (Musca domestica) and sensitivity of these bacteria to various antibiotics, captured from Hamadan City. Iran. Journal of Clinical Diagnostic Research, 11, DC04‐DC07. 10.7860/JCDR/2017/23939.9720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupane, S. , Nayduch, D. , & Zurek, L. (2019). House flies (Musca domestica) pose a risk of carriage and transmission of bacterial pathogens associated with bovine respiratory disease (BRD). Insects, 10, 1–7. 10.3390/insects10100358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nmorsi, O. P. G. , Agbozele, G. , & Ukwandu, N. C. D. (2007). Some aspects of epidemiology of filth flies: Musca domestica, Musca domestica vicina, Drosophilia melanogaster and associated bacteria pathogens in Ekpoma, Nigeria. Vector Borne Zoonotic Disease, 7, 107–117. [DOI] [PubMed] [Google Scholar]
- Nwankwo, E. O. , Ekemezie, C. L. , & Adeyemo, S. (2020). Evaluation of microbial flora of the external surface of housefly (Musca domestica) in Umuahia Metropolis, Abia State, Southeast Nigeria. Calabar Journal of Health Sciences, 3(1), 9–15. 10.25259/CJHS_5_2019 [DOI] [Google Scholar]
- Odentoyin, B. , Adeola, B. , & Olaniran, O. (2020). Frequency and Antimicrobial Resistance Patterns of Bacterial Species Isolated from the Body Surface of the Housefly (Musca domestica) in Akure, Ondo State. Nigeria. Journal of Arthropod‐Borne Disease, 14(1), 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, A. R. , & Hammack, T. S . (2000). Isolation of Salmonella spp. from the housefly, Musca domestica L., and the dump fly, Hydrotaea aenescens (Wiedemann) (Diptera: Muscidae), at Cagedlaer house. Journal of Food Protocol, 63, 958–960. [DOI] [PubMed] [Google Scholar]
- Onwugamba, F. C. , Fitzgerald, J. R. , Rochon, K. , Guadabassi, L. , & Alabi, A. (2018). The role of ‘filth flies’ in the spread of antimicrobial resistance. Travel Medicine and Infectious Disease, 22, 8–17. [DOI] [PubMed] [Google Scholar]
- Oppong, T. B. , Yang, H. , Amponsem‐Boateng, C. , Kyere, E. K. D. , Abdulai, T. , Duan, G. , & Opolot, G. (2020). Enteric pathogens associated with gastroenteritis among children under 5 years in sub‐Saharan Africa: A systematic review and meta‐analysis. Epidemiology and Infection, 148, 1–9. 10.1017/S0950268820000618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, R. , Dzialo, M. C. , Spaepen, S. , Nsabimana, D. , Gielens, K. , Devriese, H. , Crauwels, S. , Tito, R. Y. , Raes, J. , Lievens, B. , & Verstrepen, K. L. (2019). Microbial communities of the house fly Musca domestica vary with geographical location and habitat. Microbiome, 7, 147. 10.1186/s40168-019-0748-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlenz, T. D. , Zavadilova, K. , Ghosh, A. , & Zurek, L. (2018). Prevalence of Shiga‐toxigenic Escherichia coli in house flies (Diptera: Muscidae) in an urban environment. Journal of Medical Entomology, 55(2), 436–439. 10.1093/jme/tjx225 [DOI] [PubMed] [Google Scholar]
- Poudel, A. , Hathcock, T. , Butaye, P. , Kang, Y. , Price, S. , Macklin, K. , Walz, P. , Cattley, R. , Kalalah, A. , Adekanmbi, F. , & Wang, C. (2019). Multidrug‐Resistant Escherichia coli, Klebsiella pneumoniae and Staphylococcus spp. in Houseflies and Blowflies from farms and their environmental settings. International Journal of Environmental Research and Public Health, 16, 3583. 10.3390/ijerph16193583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri‐Giri, R. , Ghosh, A. , Thomson, J. L. , & Zurek, L. (2017). House flies in the confined cattle environment carry non‐O157 Shiga toxin‐producing Escherichia coli . Journal of Medical Entomology, 54, 726–732. 10.1093/jme/tjw240 [DOI] [PubMed] [Google Scholar]
- Rahuma, N. , Ghenghesh, K. S. , Ben‐Aissa, R. , & Elamaari, A. (2005). Carriage by the housefly (Musca domestica) of multiple antibiotic‐resistant bacteria that are potentially pathogenic to humans, in hospital and other urban environments in Misurata, Libya. Annals of Tropical Medicine Parasitology, 99, 795–802. [DOI] [PubMed] [Google Scholar]
- Ranjbar, R. , Izadi, M. , Hafshejani, T. T. , & Khamesipour, F. (2016). Molecular detection and antimicrobial resistance of Klebsiella pneumoniae from house flies (Musca domestica) in kitchens, farms, hospitals and slaughterhouses. Journal of Infections and Public Health, 9, 499–505. 10.1016/j.jiph.2015.12 [DOI] [PubMed] [Google Scholar]
- Rosef, O. , & Kapperud, G. (1983). House flies (Musca domestica) as possible vectors of Campylobacter fetus subsp. jejuni . Applied Environmental Microbiology, 45, 381–383. 10.1128/AEM.45.2.381-383.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, S. M. , Chittick, V. A. , Sangweme, D. T. , & Lampert, E. C. (2018). Potential of saprophage Diptera to acquire culturable livestock‐associated antibiotic‐resistant bacteria. Zoonoses Public Health, 65, e216–e221. 10.1111/zph.12431 [DOI] [PubMed] [Google Scholar]
- Sanchez‐Arroyo, H. , & Capinera, J. L. (2015). Housefly, Musca domestica Linnaeus (insecta: Diptera: Muscidae). IFAS Extension University of, Florida.EENY‐048. [Google Scholar]
- Selleck, E. M. , van Tynel, D. , & Gilmore, M. S. (2019). Pathogenicity of Enterococci. Microbiology . Spectrum, 7(4), 10.1128/microbiolspec.GPP3-0053-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobur, M. A. , Levy, S. , Haque, Z. F. , Nahar, A. , Zaman, S. B. , & Rahman, M. T. (2019). Emergence of colistin‐resistant Escherichia coli in poultry, house flies and pond water in Mymensingh, Bangladesh. Journal of Advanced Veterinary and Animal Research, 6(1), 50–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solà‐Ginés, M. , González‐López, J. J. , Cameron‐Veas, K. , Piedra‐Carrasco, N. , Cerdà‐Cuéllar, M. , & Migura‐Garcia, L. (2015). Houseflies (Musca domestica) as vectors for extended spectrum β‐lactamase‐producing Escherichia coli on Spanish broiler farms. Applied Environmental Microbiology, 81, 3604–3611. 10.1128/AEM.04252-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songe, M. , Hangombe, B. , Knight‐Jones, T. , & Grace, D. (2016). Antimicrobial resistant enteropathogenic Escherichia coli and Salmonella spp. in houseflies infesting fish in food markets in Zambia. International Journal of Environmental Research . Public Health, 14, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalanski, A. L. , Owens, C. B. , Mckay, T. , & Steelman, C. D. (2004). Detection of Campylobacter and Escherichia coli O157:H7 from filth flies by polymerase chain reaction. Medical and Veterinary Entomology, 18, 241–246. 10.1111/j.0269-283X.2004.00502.x [DOI] [PubMed] [Google Scholar]
- Tsangaan, A. , Kanuka, H. , & Okado, K. (2015). Study of pathogenic bacteria detected in fly samples using universal primer‐multiplex PCR. Journal of Agricultural Sciences, 15(2), 27–32. [Google Scholar]
- Usui, M. , Iwasa, T. , Fukuda, A. , Sato, T. , Okubo, T. , & Tamura, Y. (2013). The role of flies in spreading the extended‐Spectrum β‐lactamase gene from cattle. Microbial Drug Resistant, 19, 415–420. 10.1089/mdr.2012.0251 [DOI] [PubMed] [Google Scholar]
- Usui, M. , Shirakawa, T. , Fukuda, A. , & Tamura, Y. (2015). The role of flies in disseminating plasmids with antimicrobial‐resistance genes between farms. Microbial Drug Resistance, 21, 562–569. 10.1089/mdr.2015.0033 [DOI] [PubMed] [Google Scholar]
- Vaziriannzadeh, B. , Solary, S. S. , Radhar, M. , Hajhassein, R. , & Mendinejad, M. (2008). Identification of bacteria with possible transmitted by Musca domestica (Diptera: Muscidae) in the region of Ahvaz, SW Iran. Journal of Microbiology, 1(1), 28–31. [Google Scholar]
- Wales, A. D. , Carrique‐Mas, J. J. , Rankin, M. , Bell, B. , Thind, B. B. , & Davies, R. H. (2010). Review of the carriage of zoonotic bacteria by arthropods with special reference to Salmonella in mites, flies and litter beetles. Zoonoses Public Health, 57, 299–314. [DOI] [PubMed] [Google Scholar]
- Wei, T. , Hu, J. , Miyanga, K. , & Tanji, Y. (2013). Comparative analysis of bacterial community and antibiotic resistant strains in different developmental stages of the housefly (Musca domestica). Applied Microbiology and Biotechnology, 97, 1775–1783. 10.1007/s00253-012-4024-1 [DOI] [PubMed] [Google Scholar]
- WHO . (1997). Vector control ‐ methods for use by individuals and communities. https://pdf4pro.com/view/chapter‐6‐house‐ies‐who‐world‐health‐organization‐4e9a57.html. Accessed January 15, 2020. [Google Scholar]
- WHO . (2017). Media Centre. News Release. WHO publishes list of bacteria for which new antibiotics are urgently needed. Available from: http://www.who.int/mediacentre/news/releases/2017/bacteria‐antibiotics‐needed/en/. Accessed March 5, 2020. [Google Scholar]
- Yalli, A. A. , Sambo, S. , Lawal, H. M. , & Tukur, U. (2017). Study of bacteria on the body surfaces of house flies (Musca Domestica) in some homes within Sokoto Metropolis. Journal of Advancement in Medical and Life Sciences, 5(4). ISSN: 2348–294X. [Google Scholar]
- Yosboonruang, A. , Kiddee, A. , Boonduang, C. , & Pibalpakdee, P. (2019). Integron expression in multidrug‐resistant Escherichia coli isolated from house flies within the hospital. Walailak Journal of Science and Technology, 16(5), 319–327. [Google Scholar]
- Zhang, J. , Wang, J. , Chen, L. , Yassin, A. K. , Kelly, P. , Butaye, P. , Li, J. , Gong, J. , Cattley, R. , Qi, K. , & Wang, S. (2018). Housefly (Musca domestica) and blow fly (Protophormia terraenovae) as vectors of bacteria carrying colistin resistance genes. Applied and Environmental Microbiology, 84, e01736‐17. 10.1128/AEM.01736-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek, L. , Schal, C. , & Watson, D. W. (2000). Diversity and contribution of the intestinal bacterial community to the development of Musca domestica (Diptera: Muscidae) larvae. Journal of Medical Entomology, 37(6), 924–928. 10.1603/0022-2585-37.6.924 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1–Figure S1‐S4


