ABSTRACT
Clostridioides difficile is the most prevalent pathogen of nosocomial diarrhea. In the United States, over 450,000 cases of C. difficile infection (CDI), responsible for more than 29,000 deaths, are reported annually in recent years. Because of the emergence of hypervirulent strains and strains less susceptible to vancomycin and fidaxomicin, new therapeutics other than antibiotics are urgently needed. The gut microbiome serves as one of the first-line defenses against C. difficile colonization. The use of antibiotics causes gut microbiota dysbiosis and shifts the status from colonization resistance to infection. Hence, novel CDI biotherapeutics capable of reconstituting normal gut microbiota have become a focus of drug development in this field.
KEYWORDS: Clostridioides difficile, microbiota, biotherapeutics, dysbiosis, reconstitution
1. Introduction
In 1935, Ivan C. Hall et al. isolated a new bacterial species from the feces of healthy newborn infants. The bacteria discovered in this study was named Bacillus difficilis due to the difficulty encountered in its isolation and culture.1 It was classified as Clostridium difficile in the 1970s, and now Clostridioides difficile.1,2 Hall et al.’s study is the first documented case of C. difficile in human intestinal microbes. Later, more studies reported the isolation of C. difficile from healthy infants and asymptomatic adults which endowed the bacterium with the role of normal intestinal commensal. In fact, C. difficile colonization is found in up to 15% of healthy adults, and its prevalence is even higher in hospitalized patients and residents of long-term care facilities.3–5 In the 1970s, as the surge of antibiotic-associated colitis increased, toxin-producing C. difficile was eventually identified as one of the major pathogens responsible for the disease.6 Two exotoxins, toxin A and B (TcdA and TcdB), are its main virulence factors that can disrupt the architecture of the intestine and induce severe inflammation.7,8 C. difficile has been listed as one of the top antibiotic resistance threats to public health by the Center for Disease Control and Prevention (CDC).9 The clinical spectrum of CDI can vary widely, ranging from asymptomatic colonization of the gastrointestinal (GI) tract to severe disease leading to toxic megacolon or intestinal perforation. Based on CDC guidelines, asymptomatic colonization does not require any treatment. Symptomatic CDI often occurs after antibiotic treatment and long-term hospitalization. It is believed that the perturbation of homeostasis of gut microbes by antibiotic treatment is highly correlated to the infection.
The human GI tract is naturally a huge reservoir of microbes in which trillions of microorganisms inhabit. This group of microorganisms is called gut microbiota (Box 1). The interaction between microbiome (Box 1) and host is absolutely a hotspot of research in recent years. It has aroused extensive attention and been widely studied not only in infectious diseases but also in other disorders.10,11 As technological advances and in-depth understanding of the gut microbiome have increased in the past decade, research has revealed that host-residential bacteria interaction may influence the formation of host neurological systems, nutrition metabolization, and host immune responses.9–12 In addition, recent clinical studies demonstrated that the gut microbiota may affect the efficacy of oncology medications in patients.13,14,15 Although under normal circumstances the host and gut microbiome are mutually beneficial, dysbiosis (Table 1) of the gut microbiota has been associated with many diseases.10–12 Recently, significant progress has been made in developing live biotherapeutics that help to restore the normal gut microbiota and treat CDI. In this review, we briefly summarize the role of gut microbiota in the pathogenesis of CDI and discuss the current development of live biotherapeutics against CDI.
Table 1.
Glossary
| TERMS | DEFINITION |
|---|---|
| MICROBIOTA | Microorganisms, composed of bacteria, fungi, virus, protozoa and archaea, inhabiting a defined environment. |
| MICROBIOME | Generally, microbiota, its genes, gene p roducts and activities in niches in a habitat. |
| METABOLOME | A large array of small molecule metabolites produced by microbiota into the inhabited environment during the metabolism of food and xenobiotics. In the case of the gut metabolome, metabolites from both microbiota and hosts should be included. |
| DYSBIOSIS | There is no consensus that defines dysbiosis despite a high frequency of usage in microbiome studies. It is often described as a state, in which alterations to the microbiota of hosts and its functional components may be correlated with undermined host immunity and increasing susceptibility to diseases, for example blooming of C. difficile. Dysbiosis usually features: i) impaired microbial diversity; ii) loss of beneficial commensal bacteria; iii) thriving of pathogens. |
2. Gut microbiota and C. difficile infection
C. difficile survives in harsh environments in the form of spores that are highly tolerant to heat, oxygen, ultraviolet light, common disinfectants, and antibiotics.16 Ubiquitous spores in the environment, especially in hospitals and healthcare facilities, are the leading source of C. difficile transmission via the oral-fecal route. Healthy individuals upon oral ingestion of C. difficile spores may not develop any sign of disease but shed spores and bacterial debris in their feces, which was defined as colonization by Crobach et al.17 Colonization does not necessarily proceed to symptomatic infection. In fact, an intrinsic homeostasis in the gut microbial niche allows a resistance to C. difficile colonization and infection (Figure 1). Within the niche, the microorganisms benefit each other but competitively restrain the colonization of opportunistic pathogens at the same time. ‘Good’ bacteria build up an ‘unfavorable’ environment in which C. difficile are not able to expand and thrive. Although persistent colonization as asymptomatic carriage can last for months, as long as the homeostasis of the gut microbiome is maintained, infection may not occur.5,18 Based on the current knowledge, gut resident commensals contribute to homeostasis conferring resistance to symptomatic infection in three major mechanisms including nutrition competition, production of inhibitory metabolites and secretion of bactericidal molecules.18–22 These mechanisms have been well discussed in other reviews23,24,25 and therefore will not be discussed in this review.
Figure 1.
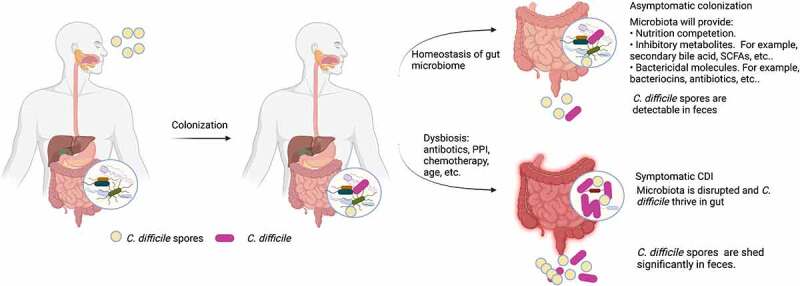
The role of gut microbiota in CDI development.
CDI often occurs upon disruption of the homeostasis of gut microbiota (Figure 1). Loss of the total number of gut microorganisms and diversity of microbiota have been frequently reported in CDI patients.26,27 Perturbation of gut microbiota will consequently lead to an imbalanced living environment, including altered metabolome, pH, and epithelial mucus, of which opportunistic pathogens like C. difficile will take advantage to expand.27–29 Once C. difficile dominates, it will produce two exotoxins that will disrupt the intestinal epithelium and cause inflammation.7,8 Meta-analysis showed that carriers of toxigenic strains are at a higher risk for the development of an infection compared to non-colonized patients.26 Therefore, colonization with toxigenic C. difficile and dysbiosis are prerequisites of CDI. Direct perturbation of microbiota is mainly induced by antibiotics and proton pump inhibitors (PPIs), while some chemotherapies and advanced age may also affect the intestinal microbiome components.31–33 Among them, antibiotic treatment is still the leading risk of primary and recurrent CDI.
Since gut dysbiosis leads to the loss of C. difficile colonization resistance, therapeutic strategies aiming to restore a normal gut microbiota have gained attention in the past decade and achieved some remarkable success. These strategies are pleiotropic as they not only restore the microbiome structure but also recover its biofunctions. When the gut residential microbe community recovers, the biofunctions that produce a normal metabolome will be re-established and shift to C. difficile colonization resistance as described earlier in this review. To date, these strategies mainly include full fecal microbiota transplantation (FMT) and precise fecal bacterial/probiotic strain(s) engraftment. Strategies directly targeting C. difficile are also emerging. In addition, compounds, such as ribaxamase and prebiotics, have been investigated for their ability to manipulate the gut microbes to defend against CDI.34–36
In next sections, we will summarize the interventions using live microorganisms to treat CDI that directly or indirectly restore the homeostasis of the gut microbiome, including their mechanisms, preclinical and clinical progress, and benefits versus risks.
3. FMT and its derivatives
FMT treatment for diarrhea can trace its history back to 1700 years ago by a Chinese physician.37 The first FMT used to treat CDI-related pseudomembranous colitis was recorded in 1958.38 Not until the past decade, have FMT and its derivatives been extensively studied and widely applied in patients with CDI, particularly those with recurrent CDI. Numerous pre-clinical and clinical studies have demonstrated the promising efficacy of FMT and its derivatives in treating CDI, although differences exist between the regimens (Table 2).
Table 2.
Key features of FMT and its derivatives
| SOURCE | CULTIVATED | COMPOSED STRAINS | DOSAGE OF EACH COMPOSITION | |
|---|---|---|---|---|
| TRADITIONAL FMT | HDa | No | Unknown | Unknown |
| DEFINED FMT | HD/CPb | Yes | Designated bacteria mixture | Designated |
| FMT SPORES | HD | No | Firmicutes species | Unknown |
a: healthy donors; b: FMT cured CDI patients.
3.1. FMT
Utilizing a dynamic computer-controlled in vitro model of the human colon, a very recent study has dissected how FMT rescues the gut microbiome from antibiotic therapy-induced loss of microbial diversity and abnormal bioactivities.30 Shortly after FMT application, the fermentation activity, measured by pH and redox potential, gas production and short-chain fatty acids (SCFA) production, quickly recovered to baseline. It also shortened the recovery time of the bacterial profile at both diversity and richness levels.30 Similar to C. difficile treatment, restoration of normal colonic microbial ecology by FMT restores bile acid metabolism and normal bile acid composition in the colon, producing an unfavorable environment for C. difficile spore germination and allowing clinical recovery of recurrent CDI patients.39,40 Jillian R.-M. Brow et al. further found that the ratio of inflammatory to non-inflammatory fatty acids decreased although the total fatty acid levels were restored in patients’ gut.40 Patients post-FMT developed similar microbial structure as donors’ for a certain period.41 Another clinical study also showed that FMT dramatically reduced the abundance of antibiotic-resistant bacteria in the 2 months after administration, suggesting fecal antibiotic resistance gene carriage decreased in direct relationship to the degree to which donor microbiota was engrafted.42
The above findings and many more have established FMT as an evidence-supported treatment option for recurrent CDI. According to the 2017 IDSA/SHEA clinical practice guidelines, FMT is recommended for patients with multiple recurrences of CDI.43 The development of FMT therapeutics is riding on the crest of a wave. Most of the registered clinical trials in the United States targeting gut microbiome to treat CDI or recurrent CDI are FMT (Table 3). Several whole-stool FMT products in the pipeline including RBX2660, VE303, CP101, and RBX7455 have been in clinical trials. Among them, RBX2660 is the only one now in clinical trial phase 3. As a commercial FMT regimen, RBX2660 showed favorable clinical efficacy and safety results in its phase 2 clinical trials.44,45
Table 3.
Registered clinical trials use live microorganisms as interventions to treat CDI in the United States
| NCT Number | Status | Conditions | Intervention | Commercial name | Phase | Ref. |
|---|---|---|---|---|---|---|
| NCT04090346 | Enrolling by invitation | Recurrent CDI | FMT | Phase 4 | ||
| NCT03973697 | Recruiting | Recurrent CDI | FMT | Phase 2 | ||
| NCT03970200 | Recruiting | Severe CDI | FMT | Phase 2 | ||
| NCT03931941 | Recruiting | Recurrent CDI | FMT | RBX2660 | Phase 3 | |
| NCT03829475 | Recruiting | IBD/CDI | Bezlotoxumab and/or FMT | Phase 2 | ||
| NCT03795233 | Suspended | CDI | FMT | Phase 1&2 | ||
| NCT03788434 | Recruiting | Recurrent CDI | FMT | VE303 | Phase 2 | |
| NCT03621657 | Completed (has results) | CDI | FMT | Phase 2 | ||
| NCT03617445 | Suspended | Recurrent CDI | FMT | Phase 2 | ||
| NCT03548051 | Terminated | CDI | FMT | Phase 1&2 | ||
| NCT03497806 | Active, not recruiting | CDI and recurrent CDI | FMT | CP101 | Phase 2 | |
| NCT03298048 | Terminated Has Results | Recurrent CDI | FMT | Phase 2 | ||
| NCT03268213 | Active, not recruiting | CDI/UC/indeterminate colitis | FMT | Early Phase 1 | ||
| NCT03244644 | Completed | Recurrent CDI | FMT | RBX2660 | Phase 3 | |
| NCT03183141 | Recruiting | Recurrent CDI | FBSa | SER-109 | Phase 3 | |
| NCT03183128 | Completed | Recurrent CDI | FBS | SER-109 | Phase 3 | 131 |
| NCT03110133 | Completed | Recurrent CDI | FMT | CP101 | Phase 2 | |
| NCT03106844 | Completed | IBD/CDI | FMT | Phase 1&2 | ||
| NCT03005379 | Recruiting | Recurrent CDI | FMT | Phase 2&3 | ||
| NCT02981316 | Completed | Recurrent CDI | FMT | RBX7455 | Phase 1 | 132 |
| NCT02589964 | Terminated | CDI | Probiotic | Florajen-3 | Phase 1 | |
| NCT02589847 | Completed | Recurrent CDI | FMT | RBX2660 | Phase 2 | |
| NCT02465463 | Completed | CDI | FMT | Phase 1&2 | ||
| NCT02437487 | Completed | Recurrent CDI | FBS | SER-109 | Phase 2 | 56 |
| NCT02423967 | Completed | Recurrent CDI | FMT | Phase 1 | ||
| NCT02403622 | Terminated | Recurrent CDI | FMT | Phase 2 | ||
| NCT02343328 | Terminated | Recurrent CDI | FMT | Phase 1 | ||
| NCT02299570 | Completed | Recurrent CDI | FMT | RBX2660 | Phase 2 | 44,45 |
| NCT02269150 | Active, not recruiting | Prophylaxis of CDI | FMT | Phase 2 | 133 | |
| NCT02255305 | Recruiting | Recurrent CDI | FMT | Phase 2 | ||
| NCT02134392 | Recruiting | Recurrent CDI | FMT | Phase 1 | ||
| NCT02127398 | Recruiting | Recurrent CDI | FMT | Phase 2 | ||
| NCT01972334 | Completed | Recurrent CDI | FMT | Phase 2 | ||
| NCT01925417 | Completed | Recurrent CDI | FMT | RBX2660 | Phase 2 | 42,134 |
| NCT01914731 | Completed | Recurrent CDI | FMT | Phase 1 | 135 | |
| NCT01868373 | Enrolling by invitation | Primary and recurrent CDI | Defined FMT | Phase 1 | ||
| NCT01704937 | Completed | Recurrent CDI | FMT | Phase 1 | 136 | |
| NCT01680874 | Completed | Primary and recurrent CDI | Probioticb | Phase 2 | 137,138 | |
| NCT01259726 | Completed | Prevention of recurrent CDI | NTCD | VP20621 | Phase 2 | 92 |
| NCT01202630 | Suspended | Recurrent CDI | Probioticc | Bio-K, CL1285 | phase 3 | |
| NCT02127814 | Completed | AAD, CDI | Probioticd | NAa | ||
| NCT03562741 | Recruiting | Recurrent CDI | FMT | NA | ||
| NCT02830542 | Unknown | Recurrent CDI | FBS | SER-262 | Phase 1 | |
| NCT02636517 | Active, not recruiting | CDI, IBD, UC, Crohn’s Disease | FMT | NA | ||
| NCT02557685 | Unknown | CDI | FMT | Phase 2 | ||
| NCT02326636 | Completed | CDI | FMT | NA | ||
| NCT02076438 | Terminated | AAD, CDI | Probiotice | Culturelle | NA | |
| NCT01973465 | Unknown | CDI | FMT | NA | ||
| NCT01905709 | Recruiting | CDI | FMT | NA | ||
| NCT01873872 | Unknown | CDI | Probiotics | Theralac & Culturelle | NA | |
| NCT01703494 | Unknown | CDI | FMT | Phase 2 | 41 | |
| NCT01077245 | Withdrawn | CDI | Probiotic CBM588 | MIYA-BM | Phase 2 |
a: FBS = Fecal bacterial spores; NA = Not applicable. b: equal amounts of Lactobacillus acidophilus NCFM® (ATCC 700396), Lactobacillus paracasei Lpc-37 (ATCC SD5275), Bifidobacterium lactis Bi-07 (ATCC SC5220), and Bifidobacterium lactis Bl-04 (ATCC SD5219). c: Lactobacillus acidophilus CL1285® and Lactobacillus casei. d: Lactobacillus reuteri. e: Lactobacillus Rhamnosus GG
Although highly efficacious, FMT is associated with risks, such as heterogeneity among donors, batches and preparations, and unknown long-term impact post FMT treatment.46 The major risks of FMT include: 1) Side effects, such as constipation, diarrhea, bloating, etc., often occurred after transplantation; 2) It may transfer potential pathogens. Although the donors have been screened for recognized pathogens, uncertain pathogens may still exist. The screening procedure for donors was updated by the United States Food and Drug Administration (FDA) in 2019 because of cases of invasive infection caused by extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in two immunocompromised patients after FMT;47 3) Disorders or diseases other than infectious disease may be transferred to the recipients. Most of the clinical trial studies only have a 6-month follow-up period, and no data so far have demonstrated the long-term impact of FMT. As we discussed in the beginning, based on current knowledge, the gut microbiome plays a vital role in many aspects of human physiological functions. Therefore, it is hard to predict whether the donors may transfer their own characteristics to the recipients, for instance, obesity, diabetes, colon carcinoma, neurological disorders, etc.
3.2. Defined FMT
The defined FMT is intrinsically a precise engraftment with a designated fecal bacterial composition. As an alternative to FMT, the defined FMT has multiple advantages. First, the bacterial composition is designable and controllable. The absence and presence of certain bacterial strains are frequently and consistently observed in CDI patients pre- and post-therapy. Thus, replenishment of those bacteria that defined a healthy state may inhibit CDI in vivo. Tvede and Rask-Madsen initiated the proof-of-concept study in which they treated 5 CDI patients through rectal administration with a mixture of equal volume of 10 bacterial cultures isolated from the feces of healthy donors.48 The strains were chosen based on their characteristics as probiotics, their inhibitory effects to C. difficile in vitro, and absence in CDI patients. Similarly, Cammarota et al. introduced a 15-strain consortium derived from successful engraftments in patients cured by FMT.49 For both studies, in response to the treatment procedure, patients were promptly relieved of CDI symptoms. Second, the defined FMT can lower the risk of antibiotic resistance. Petrof and colleagues adopted a regimen possessing a broader spectrum of intestinal microbes (33 strains).50 Unlike Tvede’s and Cammarota’s, the strains in Petrof’s study were prepared to mimic fecal microbiota. The selected strains (except Bifidobacterium spp.) were formulated in an inferred relative ratio based on the MetaREP metagenomic database of stool sample datasets from healthy donors. The authors also considered antibiotic susceptibility and safety, while attempting to generate as much taxonomic diversity as possible. If any selected organism was suspected to be resistant to antibiotics, then it would be ruled out of the mixture. Third, the regimen is reproducible, low cost, stable and can be quality controlled. Variations of the FMT products widely exist between individual donors. Even the same donor may not produce identical microbiota at different times. Therefore, reproducibility is one of the biggest pitfalls for FMT products. However, the defined FMT is a synthetic bacterial mixture cultured in vitro that can be easily produced and genetically monitored. For example, in Petrof’s study, all purified candidates were identified by 16S rRNA sequencing to guarantee the genomic background. Since all bacteria are culturable in vitro, the time and cost required for donor screening will be saved. Fourth, an absence of viruses and other pathogens in the administered mixture can be ensured, thereby improving patient safety.50
Despite these advantages, challenges also exist. In the United States, only one registered ongoing clinical trial using defined FMT as intervention was found on the Clinical trials website (Table 1). In Europe, the result of a randomized controlled clinical trial (RCT) was released recently. In this study, a total of 98 participants with recurrent CDI were enrolled, which presents the largest trial on this topic to date. The treatment did not show a superior efficacy compared to conventional FMT (cure rate 52% vs 76%), although it was comparable to vancomycin treatment (cure rate 52% vs 45%).51 The regimen of 12 well-characterized bacterial strains, selected only based on previous experiences, was a simple equal volume mixture of bacterial candidates. Nowadays, as the development of metagenomic and culturomic techniques progresses, more bacterial strains have been isolated from gut microbiota and identified to be closely related to CDI dysbiosis.52,53 Species composition and interactions are both important determinants of the C. difficile inhibition phenotype, as a simple cocktail of selected strains may not achieve ideal inhibitory outcomes but oppositely may assist C. difficile growth.53 Therefore, regarding the interaction of bacteria with each other in the mix and with other members of gut commensals of hosts, a personalized bacterial cocktail may be required to treat individuals.
3.3. Fecal bacterial spores
Another FMT derived strategy is the mixture of fecal bacterial spores. SER-109, a pipeline product of Seres Therapeutics, Inc., is approaching the market. The company has released inspiring primary endpoint results from its phase 3 trial using SER-109 to treat recurrent CDI. The patients had a significantly lower recurrence rate of 11.1% in the SER-109 treatment group versus 41.3% in the placebo group 8 weeks after treatment.54 The drug is a consortium of spores of multiple Firmicutes species fractionated from the stools of healthy human donors and encapsulated.55,56 Firmicutes play an important role in colonization resistance.57 The SER-109 was manufactured with a sterilization process to reduce potential pathogens and fecal matter and debris. The entire spore ecology of SER-109 is more physiologic than defined FMT as all healthy donors’ spores were represented without depleting or enriching for specific spore species based on sequencing and microbiology studies. However, SER-109 had failed to achieve its primary endpoint in phase 2 in 2016. The chief scientist of the company revealed two important modifications for their phase 3 study: detection method for C. difficile and dosage of SER-109. Instead of amplifying C. difficile genes, ELISA for detecting toxins was performed to differentiate active infection and asymptomatic colonization, implying that the microbiota niche shifted back to a balanced state of colonization resistance after treatment.58 The dosage of the drug was also emphasized to play a critical role in its efficacy.58
4. Probiotics
The current consensus definition of probiotics is “live strains of strictly selected microorganisms that, when administered in adequate amounts, confer a health benefit on the host” by an expert panel of the International Scientific Association for Probiotics and Prebiotics (ISAPP) since October 2013.59 According to the consensus, probiotics should be well-known strains that provide health benefit in single or multiple mechanisms. Hence, the defined FMTs described in a previous section of this review indeed meet the criteria of a probiotic.59 Since the initial aim of defined FMTs was to mimic fecal microbiota, we categorized them as FMT derivatives. In fact, as the in-depth study of FMT advances, the gut commensal will be a source of next-generation probiotics.60 The wide range of the probiotic definition will surely encourage innovation in the field. Although not all mechanisms of probiotics have been confirmed in humans, diverse mechanisms are likely to drive probiotic benefits to host health, such as production of antimicrobial products, cross-feeding the resident commensal, direct interaction with immune cells, etc.61 In this section, we will summarize the non-FMT probiotic-related interventions in CDI prevention.
4.1. Traditional probiotics
The traditional probiotics, such as strains of Lactobacillus, Bifidobacterium, and Saccharomyces have been suggested as dietary supplements for CDI.62 The mechanisms of probiotics against CDI are similar to FMT. A consortium of probiotics, including five Lactobacilli strains, two Bifidobacterium standard strains, and Bifidobacterium infantis obstructs the proliferation of C. difficile through affecting the diversity of gut microbiota and regulating SCFA production, eventually attenuating C. difficile colonization.63 Lactobacillus and Bifidobacterium species have also been shown to colonize the intestine regardless of concurrent antibiotic use, competing with C. difficile for nutrition.64,65 Saccharomyces boulardii was reported to lessen antibiotic induced microbiota shifts.66,67,68 In addition, S. boulardii produces a protease capable of digesting C. difficile toxins, which are etiologies of the disease, and modulates a host of inflammatory signaling pathways to inhibit toxin-induced inflammation.76–79
The traditional probiotics have a long history as safe and effective diet supplements or drugs with health benefit. A major outbreak, associated with the C. difficile NAP1/027/BI strain in the hospital Pierre-Le Gardeur (PLGH) in Quebec, promoted the use of probiotic Bio-K+, containing three Lactobacillus strains, to every adult inpatient on antibiotics in this hospital. The data collected by the Ministry of Health in Quebec showed no episodes of Lactobacillus bacteremia during the entire 10-year experience and that CDI incidents were lower in this hospital.70,71 Although in particular immunocompromised individual treatment with S. boulardii should be avoided or well managed due to fungemia concerns,72,73 it has been used to prevent and treat diarrheal diseases such as antibiotics associated with diarrhea (AAD) for many decades.74,75 Hence, the overall safety of using traditional probiotics to treat CDI is not a major concern. Moreover, the probiotic products can be easily and economically prepared and given to patients daily as yogurt, drinks, cheese, or capsules.64,85–87,88,89 Unlike FMT bacteria, the probiotics are frequently used as prophylactics or adjuvant therapy to reduce the risk of AAD and/or primary CDI in many clinical trials across the world. Inconsistent conclusions drawn from these clinical trials may be the reason that probiotics are not recommended for CDI treatment and prevention by current clinical practice guidelines. However, a meta regression analysis emphasized the efficacy of probiotics in reducing CDI incidence in a high-risk population.79 In this study, a total of 19 published RCTs, comprising 6261 subjects and using different regimens of four probiotic species (Lactobacillus, Saccharomyces, Bifidobacterium, and Streptococcus), were analyzed. The authors found that the timing of probiotic administration was critical to prevent CDI. Future research is still needed to focus on optimal probiotic dose, species, and formulation since no superior regimens were concluded in this study.
4.2. Newly emerging commensal probiotics
Given the broad definition of probiotics, several commensal bacteria may now be categorized as probiotics since they have been frequently demonstrated to play important roles in combatting CDI. Most of them are in the preclinical stage of development. In order for the emerging probiotics to assert benefits in CDI patients, more clinical studies are needed to examine criteria, such as dose responses, biological plausibility, replication of findings, etc.59 Here are some examples of the newly identified probiotic candidates.
Clostridium scindens was identified as a probiotic candidate from commensals to treat CDI through a subtle workflow described by Buffie and colleagues.80 They first analyzed the microbiota composition in mice after different antibiotic regimens and identified 11 strains that were strongly associated with C. difficile colonization resistance. Then, an inference-modeling approach was applied to samples of hospitalized patients in parallel with the murine samples. C. scindens was identified as the strongest correlated bacteria to enhance resistance to CDI in both human and murine CDI. Further study indicated that C. scindens mediated inhibition was bile acid dependent.
CBM588 is a probiotic consisting of Clostridium butyricum, a bacterium that produces a robust amount of butyrate. It has been used as a live biotherapeutic probiotic to treat CDI patients in a phase 2 clinical trial, although the trial was eventually suspended due to lack of enrollment. Preclinical studies showed that C. butyricum activated neutrophils and Th1 and Th17 cells to elicit the protective effects against CDI.81
Li et al. investigated the role of Bacteroides thetaiotaomicron in defending against CDI in a mouse model. Similar to the FMT control, mice administered B. thetaiotaomicron had fewer copies of C. difficile as well as less colonic inflammation. Meanwhile, both FMT and B. thetaiotaomicron improved the gut microbiota composition and reversed the CDI-induced change in bile acid composition, suggesting that B. thetaiotaomicron is a good probiotic candidate to combat CDI.82 The cell-wall associated glycans of B. thetaiotaomicron were shown to suppress the production of the glycosylated toxins of C. difficile in vitro.83 As a cephalosporinase-producing anaerobe, the pre-colonized B. thetaiotaomicron produced β-lactamase enzymes to inactivate intraintestinal β-Lactam antibiotics during antibiotic treatment, providing protection for commensal recovery and preventing overgrowth of C. difficile in a mouse model.84
5. Non-toxigenic C. difficile spores
Nontoxigenic C. difficile (NTCD) strains that lack the genes for active toxin production are frequently found in the hospital environment and colonize hospitalized patients, although patients are usually asymptomatic for CDI. One hospital study found that asymptomatic colonized patients had less chance of developing active CDI,85 implying pre-colonization with NTCD would be a potential treatment to protect hospitalized patients from recurrent CDI. Several preclinical studies have demonstrated the efficacy of NTCD or low-virulent C. difficile to prevent CDI.95–99 The mechanism might be that a certain population of NTCD occupies the living space and outcompetes the invading strains. Recently, Lesile et al. revealed that consumption of glycine by the first colonized strain of C. difficile would decrease germination of the second lethal strain, consequently limiting colonization by the lethal one.90
VP20621, a commercial product of Viropharma Inc, is an oral liquid drug containing the spores of NTCD strain M3 and has completed phase 2 clinical trial. The released results indicated that the drug was quite safe and tolerable as patients took daily dosages ranging from 104 to 108 CFU for 7 or 14 days and only had similar mild side effects.91,92 NTCD-M3 was isolated from human patients.87 In its phase 2 trial, the average fecal colonization rate of NTCD-M3 was 69% (71% with 107 spores/d and 63% with 104 spores/d). Recurrence of CDI occurred in 13 (30%) of 43 placebo patients and in 14 (11%) of 125 NTCD-M3 patients. The NTCD-M3 colonization became completely undetectable after week 22 of follow-up, implying the restoration of the normal microbiota, which may then provide protection against subsequent CDI. Notably, the dosage of 107 spores/d for 7 days had a lower recurrence than the dosage of 107 spores/d for 14 days. The gut microbiome profile alteration between pre- and post-spore treatment needs further investigation to dissect how NTCD competes against invasive C. difficile and restores the microbiota.
At the time of writing, NTCD-M3 is the only biotherapeutic using single-species bacteria that has demonstrated efficacy in reducing recurrent CDI in the clinic. Compared to FMT-related strategies, the NTCD-M3 has a clearer genetic background. Preparation is reproducible and low cost. However, there are a couple of concerns about widespread clinical use, such as antibiotic resistance gene transfer and toxin gene acquisition.93 Horizontal gene transfer occurs both between bacteria of the same species and between different species through variant mechanisms.94–96,C. difficile strains are known to be resistant to a wide spectrum of antibiotics. Extensive use of a highly antibiotic resistant strain of NTCD would increase the risk of spreading antibiotic resistance to other bacteria in the gastrointestinal tract. Meanwhile, the co-colonized toxigenic C. difficile strain may convert the NTCD to a toxin producer strain by horizontal gene transfer.97
6. Bacteriophages
Bacteriophages are viruses that can infect and replicate within their host bacteria and eventually lyse their hosts. Although the use of C. difficile-specific bacteriophages does not directly target gut microbiota, this approach precisely targets C. difficile without using antibiotics. To date, no treatment against CDI using bacteriophages is in clinical practice, although they have been widely used in humans in European countries to treat other infections.98 Several preclinical studies have demonstrated bacteriophages and phage-derived products as potential therapeutics to inhibit C. difficile growth and toxin production both in vivo and in vitro.99–102 Although these results are promising, there are several drawbacks with the current bacteriophage strategies. All the C. difficile phages with complete genomes in the public database are temperate phages, which can be replicated either by the lytic or the lysogenic cycle.103 When the temperate phage goes into a lysogenic cycle, it becomes integrated into the host genome to replicate with the host chromosome or replicate within the host as a plasmid.103 The lysogenic cycle creates phage resistance, leaving the host C. difficile strain tolerant to the phage mediated eradication. Genetically engineered phages that are deficient of lysogen-related genes seemed to be a solution. Selle et al. developed an engineered C. difficile phage carrying a self-targeting CRISPR-Cas array to directly target the bacterial chromosome.104 This recombinant phage was demonstrated to be more efficient at killing C. difficile both in vitro and in vivo while lysogens still accumulated 24 hours post infection. To avoid lysogeny formation, the authors removed a region of the genome encoding the cI repressor and integrase gene (wtPhage Δlys and crPhage Δly). Although no lysogeny from in vitro infection with wtPhage Δlys and crPhage Δly was detected, lysogens appeared in feces of mice treated with each, suggesting other unknown mechanisms may exist to drive the lysogenic replication cycle in C. difficile phages. Unfortunately, this particular characteristic of phages may play an important role in horizontal gene transfer that contributes to bacterial evolution and may generate superbugs. Phages transferring antibiotic resistance to their host strains have been demonstrated. Goh et al. found that phage phiC2 was able to promote the transfer of the transposon Tn6215, which encodes erythromycin resistance, between C. difficile strains.105,106 Furthermore, C. difficile toxins may also originate from phages. The complete functional binary toxin locus was identified in the genome of phage phiSemix9P1.106 Besides, use of phage may risk promoting biofilm since LuxS encoding enzyme mediated induction of prophages likely contributes to C. difficile biofilm structure.107 Finally, the host specificity as its advantage is also a disadvantage for phage therapy. There is not a universal phage regimen that can kill all C. difficile variants. Hence, a personalized bacteriophage regimen may be necessary for CDI treatment.108 C. difficile isolation and sequencing may be needed prior to treatment administration to determine which phage or phage combination may be applicable to the specific patient.
Overall, bacteriophage therapy is a potential candidate to treat CDI that avoids antibiotic usage and disturbs the gut microbiome. More knowledge about the phage-C. difficile interaction is needed to safely translate this therapeutic concept to human patients.
7. Recombinant live biotherapeutic products
According to the FDA-updated guidance in 2016, recombinant live biotherapeutic products (LBPs) are composed of genetically modified microorganisms with the purposeful addition, deletion, or modification of genetic material.109 The genetic modifications can empower additional functions of LBPs besides their intrinsic effects on gut microbiota, and are therefore potentially more promising in fighting against CDI. Although to date, no recombinant LBPs have been approved by the FDA for human diseases, many are in development to target a range of diseases including metabolic disorders, inflammatory bowel disease (IBD), colorectal cancer, and infectious diseases.
7.1. Using probiotic bacteria as chassis
Probiotic bacteria such as E. coli Nissle 1917, Lactobacillus spp., Salmonella Typhi, etc., are often used as the chassis of the recombinant LBPs since genetic toolboxes in those bacteria have been well established.110,111 For example, Synlogic has developed a series of recombinant LBPs using E. coli Nissle 1917 as chassis to treat metabolic disorders.112 Another in the pipeline product of Precigen Actobio in the clinical trial phase 1b/2 for type-1 diabetes adopted Lactococcus lactis as chassis.113 In the context of CDI, two lactic acid bacterial strains, Lactobacillus casei and Lactobacillus acidophilus, engineered to display C. difficile surface layer protein A (SlpA) were proposed by Vedantam et al. to compete with C. difficile for epithelial adherence.114 The proposed strains were demonstrated to protect hamsters and piglets from death or diarrhea post infection of C. difficile strain 630. SlpA is the most abundant component of C. difficile cell wall and mediates the attachment of the bacteria to mucus layer of host intestine that may contribute to colonization.115 Vaccine studies have shown SlpA as a candidate to elicit protective immune responses against CDI in mice but not hamster models.116,117 However, C. difficile has a high level of variability of SlpA between strains.118 Therefore, it is risky for therapeutic development to target one of the SlpA variants solely.
7.2. Using probiotic yeast as chassis
Probiotic yeast S. boulardii has not been broadly used as a live vector for the delivery of therapeutic proteins until 2020 since the first report in 2013. 119–123 As a probiotic, the treatment efficacy of S. boulardii has already been widely assessed in various gut disorders. As an LBP chassis, S. boulardii’s lack of sporulation and stable colonization in the gut may further ease safety concerns.124 The eukaryotic yeast is unlikely to transfer antibiotic resistant genes. In addition, S. boulardii is tolerant to the acidic environment and grows well at 37°C.125 Therefore, S. boulardii is an attractive vehicle to deliver therapeutics in the gut. Recent genomic sequencing studies uncovered the similarities between Saccharomyces cerevisiae and S. boulardii that expands the genetic toolbox to engineer S. boulardii, paving the way to producing heterologous proteins in S. boulardii.126 Several studies have generated auxotrophic strains of S. boulardii and demonstrated the expression of heterologous proteins by this probiotic yeast.127,128 Chen et al. recently reported a rationally designed S. boulardii strain (Sb-ABAB) that has superior prophylactic and therapeutic efficacy in mouse models of primary and recurrent CDI.122 The concept of this design was to utilize a well-documented non-colonizing probiotic to constitutively deliver therapeutic antibodies in situ to combat intestinal colonized pathogens, such as C. difficile. Chen et al.’s report is the first recombinant LBP to use probiotic yeast as chassis to deliver therapeutic antibodies. S. boulardii may also be able to deliver other therapeutics, including bacteriocin, cytokines, peptides, and small molecular drugs.120–123 In order to improve the yield of target cargoes, the components of the gene cassettes, such as signal peptides, promotors/terminators, selection markers, etc., and maintenance of the exogenous gene of interests as plasmids or chromosomal integration have been modified in those studies. Among them, Chen et al. and Liu et al. have demonstrated the in vivo activity of the yeast-secreted heterologous proteins after passing through animal GI tract.121,122
Particularly to Chen and colleagues’ study, Sb-ABAB carries a plasmid harboring a nanobody-encoding gene with an uracil auxotrophic selection marker. It is also the first recombinant LBP to target C. difficile toxins. Several advantages exist in this immunotherapy-based recombinant LBP. First, multiple-effects-in-one confers this engineered S. boulardii a powerful candidate to battle C. difficile. Clearly, the administration of this engineered Sb-ABAB more efficiently protected the host from diarrhea and weight loss and enhanced survival compared to control vehicles. In addition to the probiotic effects described above, the delivered cargo, a tandem tetra-specific VHH antibody named ABAB, neutralized C. difficile toxins TcdA and TcdB simultaneously and efficiently. Although S. boulardii was shown to secrete an enzyme that degrades C. difficile toxins,69 ABAB is more precise and potent, targeting the toxins directly. Further analysis indicated that the secreted endogenous ABAB was stable in vivo and sufficient to decrease intestinal toxin accumulation, accompanied by a decrement of host intestinal damage and inflammation. Dramatically fewer C. difficile colonies were recovered from feces of mice administered Sb-ABAB, implying that colonization resistance was increased. Further exploration is needed to dissect the impact of Sb-ABAB on regulating the entire microbiome. Secondly, it is a less costly way to deliver therapeutic antibodies to protect high-risk patients from CDI. Antibody drugs are currently one of the most popular but pricy therapeutics in the market. Bezlotoxumab (commercial name Zinplava) is a human monoclonal antibody against C. difficile TcdB. It is the first approved antibody by the FDA for the prevention of recurrent CDI. The average wholesale price of Bezlotoxumab is $4560 per vial.129 The cost of antibody drugs is partially due to the complexity of manufacture. By contrast, S. boulardii is economical to manufacture through fermentation. Hence, utilizing the live surrogate to deliver therapeutic antibodies in a real-time fashion will reduce the manufacturing process and save costs. Thirdly, it will simplify the regimen with concurrent administration with antibiotics. Unlike other probiotic bacteria, S. boulardii is not suppressed by vancomycin. Therefore, when used as a prophylactic, the engineered S. boulardii can be taken with antibiotics in a way that will be easy to follow by high-risk patients. Finally, the clear genetic and safety background of S. boulardii will accelerate its translation to human patients.
There are also limitations to S. boulardii based recombinant LBPs. The dissemination of synthetic DNA material is a general concern of the FDA of all genetically modified LBPs, although the current Sb-ABAB adopted an auxotrophic selection marker to avoid spreading antibiotic resistance. The glycosylation profile of yeast produced proteins is different from human proteins and that may interfere with the function of therapeutics in humans, hence possibly shrinking the pool of candidates.130 Despite many efforts, the yield of target cargoes is still a limitation since none of the studies discussed earlier demonstrated an abundant secretion amount.
8. Conclusions
Due to the wide usage of antibiotics, antibiotic resistance has become a challenge in clinics to treat infectious diseases. C. difficile is one of the representative superbugs that is hard to treat with antibiotics. As knowledge grows, the role of gut microbiota in human health turns more transparent and gains more attention. Over the past decade, numerous studies have provided evidence to support that homeostasis of gut microbiome will shield hosts from invasion by opportunistic pathogens. Thus, ideas about reconstituting the disturbed microbiome to treat CDI are emerging. Precise and generic intestinal engraftments with various microbiota have been widely explored. Remarkable milestones have been achieved in CDI treatment using microbiota, although the long-term impact on human health is unknown. Genetically modified probiotics that specifically target C. difficile pathogenesis provide a brand-new direction for the treatment of this antibiotic resistant superbug. Despite a short history, microbial therapies in the C. difficile field open up a new era in drug development targeting gut disorders. Meanwhile, criteria about using microbiota are also in urgent need to unify the application in patients.
Supplementary Material
Acknowledgments
This work is funded by grants R01AI132207-01 (NIH/NIAID), U19 AI142725-01 (NIAID), R01AI148270-01 (NIAID), R01AI148357-01 (NIAID).
Funding Statement
This work was supported by the grants from National Institute of Health.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Hall IC, Denver T, Denver T, Denver T, Denver T.. Intestinal flora in new-born infants: with a description of a new pathogenic anaerobe, Bacillus difficilis. American Journal of Dieseases of Children. 1935;49(2):390–18. doi: 10.1001/archpedi.1935.01970020105010. [DOI] [Google Scholar]
- 2.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. Reclassification of clostridium difficile as Clostridioides difficile (hall and O’Toole 1935) prévot 1938. Anaerobe. 2016;40:95–99. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 3.Zacharioudakis IM, Zervou FN, Pliakos EE, Ziakas PD, Mylonakis E. Colonization with toxinogenic C. difficile upon hospital admission, and risk of infection: a systematic review and meta-analysis. American Journal of Gastroenterology. 2015;110(3):381–390. doi: 10.1038/ajg.2015.22. [DOI] [PubMed] [Google Scholar]
- 4.Orraine L, Yne K, Arny IW, Mir A, Amar Q, Elly PK. Asymptomatic carriage of clostridium difficile and serum levels of IgG antibody against Toxin A. N Engl J Med. 2000;342(6):390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 5.Ozaki E, Kato H, Kita H, Karasawa T, Maegawa T, Koino Y, Matsumoto K, Takada T, Nomoto K, Tanaka R, et al. Clostridium difficile colonization in healthy adults: transient colonization and correlation with enterococcal colonization. J Med Microbiol. 2004;53(2):167–172. doi: 10.1099/jmm.0.05376-0. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett JG, Chang T, Gurwith M, Gorbach SL, Onderdonk AB. Antibiotic-associated pseudomembranous colitis due to toxin-producing clostridia. New England Journal of Medicine. 1978;298(10):531–534. doi: 10.1056/NEJM197803092981003. [DOI] [PubMed] [Google Scholar]
- 7.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467(7316):711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Feng H, Mobley H. Pathogenic effects of glucosyltransferase from Clostridium difficile toxins. Pathog Dis. 2016;74(4):ftw024. doi: 10.1093/femspd/ftw024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Antibiotic resistance threats in the United States, 2019. Atlanta, Georgia: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention. 2019. 10.15620/cdc:82532. [DOI] [Google Scholar]
- 10.Hwang I, Park YJ, Kim YR, Kim YN, Ka S, Lee HY, Seong JK, Seok YJ, Kim JB. Alteration of gut microbiota by vancomycin and bacitracin improves insulin resistance via glucagon-like peptide 1 in diet-induced obesity. FASEB Journal. 2015;29(6):2397–2411. doi: 10.1096/fj.14-265983. [DOI] [PubMed] [Google Scholar]
- 11.Frye RE, Rose S, Chacko J, Wynne R, Bennuri SC, Slattery JC, Tippett M, Delhey L, Melnyk S, Kahler SG, et al. Modulation of mitochondrial function by the microbiome metabolite propionic acid in autism and control cell lines. Transl Psychiatry. 2016;6(10):e927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bates NA, Li A, Fan T, Cutcliffe MP, Dagenet CB, Sleiman KC, Ma H, Tahsin S, Garrett CS, Altemus J, et al. Gut commensal segmented filamentous bacteria fine-tune T follicular regulatory cells to modify the severity of systemic autoimmune arthritis. The Journal of Immunology. 2021;206(5):941–952. doi: 10.4049/jimmunol.2000663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogbonnaya ES, Clarke G, Shanahan F, Dinan TG, Cryan JF, O’Leary OF. Adult hippocampal neurogenesis is regulated by the microbiome. Biol Psychiatry. 2015;78(4):e7–9. doi: 10.1016/j.biopsych.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 14.Routy B, le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 15.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente † D, Hoffman K, Wei SC, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol. 2014;22(7):406–416. doi: 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crobach MJT, Vernon JJ, Loo VG, Kong LY, Péchiné S, Wilcox MH, Kuijper EJ. Understanding clostridium difficile colonization. Clin Microbiol Rev. 2018;31(2):e00021–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato H, Kita H, Karasawa T, Maegawa T, Koino Y, Takakuwa ÃH, Saikai T, Kobayashi ÃK, Yamagishi T, Nakamura S. Colonisation and transmission of Clostridium diffcile in healthy individuals examined by PCR ribotyping and pulsed-field gel electrophoresis. J Med Microbiol. 2001;50(8):720–727. doi: 10.1099/0022-1317-50-8-720. [DOI] [PubMed] [Google Scholar]
- 19.Lopez CA, McNeely TP, Nurmakova K, Beavers WN, Skaar EP. Clostridioides difficile proline fermentation in response to commensal clostridia. Anaerobe. 2020;63:102210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weingarden AR, Chen C, Zhang N, Graiziger CT, Dosa PI, Steer CJ, Shaughnessy MK, Johnson JR, Sadowsky MJ, Khoruts A. Ursodeoxycholic acid inhibits clostridium difficile spore germination and vegetative growth, and prevents the recurrence of ileal pouchitis associated with the infection. J Clin Gastroenterol. 2016;50(8):624–630. doi: 10.1097/MCG.0000000000000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDonald JAK, Mullish BH, Pechlivanis A, Liu Z, Brignardello J, Kao D, Holmes E, Li J, Clarke TB, Thursz MR, et al. Inhibiting growth of Clostridioides difficile by restoring valerate, produced by the intestinal microbiota. Gastroenterology. 2018;155(5):1495–1507.e15. doi: 10.1053/j.gastro.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rea MC, Sit CS, Clayton E, O’Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, Hill C. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proceedings of the National Academy of Sciences of the United States of America 2010; 107:9352–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatziioanou D, Gherghisan-Filip C, Saalbach G, Horn N, Wegmann U, Duncan SH, Flint HJ, Mayer MJ, Narbad A. Discovery of a novel lantibiotic nisin O from Blautia obeum A2-162, isolated from the human gastrointestinal tract. Microbiology (United Kingdom). 2017;163:1292–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ducarmon QR, Zwittink RD, Hornung BVH, van Schaik W, Young VB, Kuijper EJ. Gut microbiota and colonization resistance against bacterial enteric infection. Microbiology and Molecular Biology Reviews. 2019;83(3):e00007–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reed AD, Theriot CM. Contribution of inhibitory metabolites and competition for nutrients to colonization resistance against clostridioides difficile by commensal clostridium. Microorganisms. 2021;9(2):1–14. doi: 10.3390/microorganisms9020371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernández M, de Frutos M, Rodríguez-Lázaro D, López-Urrutia L, Quijada NM, Eiros JM, Fung K, Ip M. Fecal microbiota of toxigenic clostridioides difficile-associated diarrhea. Front Microbiol. 2019;10:10. doi: 10.3389/fmicb.2019.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berkell M, Mysara M, Xavier BB, van Werkhoven CH, Monsieurs P, Lammens C, Ducher A, Vehreschild MJGT, Goossens H, de Gunzburg J, et al. Microbiota-based markers predictive of development of Clostridioides difficile infection. Nat Commun. 2021;12(1):12. doi: 10.1038/s41467-020-20168-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engevik MA, Beth Yacyshyn M, Engevik KA, Wang J, Darien B, Hassett DJ, Yacyshyn BR, Worrell RT. Human Clostridium difficile infection: altered mucus production and composition. Am J Physiol Gastrointest Liver Physiol. 2015;308(6):510–524. doi: 10.1152/ajpgi.00091.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fletcher JR, Erwin S, Lanzas C, Theriot CM. Shifts in the gut metabolome and Clostridium difficile transcriptome throughout colonization and infection in a mouse model. mSphere 2018; 3. [DOI] [PMC free article] [PubMed]
- 30.Verdier C, Denis S, Gasc C, Boucinha L, Uriot O, Delmas D, Dore J, le Camus C, Schwintner C, Blanquet‐diot S. An oral fmt capsule as efficient as an enema for microbiota reconstruction following disruption by antibiotics, as assessed in an in vitro human gut model. Microorganisms. 2021;9(2):1–24. doi: 10.3390/microorganisms9020358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milani C, Ticinesi A, Gerritsen J, Nouvenne A, Andrea Lugli G, Mancabelli L, Turroni F, Duranti S, Mangifesta M, Viappiani A, et al. Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci Rep. 2016;6(1):6. doi: 10.1038/s41598-016-0015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rea MC, O’Sullivan O, Shanahan F, O’Toole PW, Stanton C, Ross RP, Hill C. Clostridium difficile carriage in elderly subjects and associated changes in the intestinal microbiota. J Clin Microbiol. 2012;50(3):867–875. doi: 10.1128/JCM.05176-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galloway-Peña JR, Shi Y, Peterson CB, Sahasrabhojane P, Gopalakrishnan V, Brumlow CE, Daver NG, Alfayez M, Boddu PC, Khan MAW, et al. Gut microbiome signatures are predictive of infectious risk following induction therapy for acute myeloid leukemia. Clinical Infectious Diseases. 2020;71(1):63–71. doi: 10.1093/cid/ciz777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kokai-Kun JF, Roberts T, Coughlin O, Le C, Whalen H, Stevenson R, Wacher VJ, Sliman J. Use of ribaxamase (SYN-004), a β-lactamase, to prevent Clostridium difficile infection in β-lactam-treated patients: a double-blind, phase 2b, randomised placebo-controlled trial. Lancet Infect Dis. 2019;19(5):487–496. doi: 10.1016/S1473-3099(18)30731-X. [DOI] [PubMed] [Google Scholar]
- 35.Maltz C, Miskovitz PF, Hajifathalian K. Lactulose may reduce Clostridium difficile -related diarrhea among patients receiving antibiotics. JGH Open. 2020;4(6):1088–1090. doi: 10.1002/jgh3.12390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rätsep M, Kõljalg S, Sepp E, Smidt I, Truusalu K, Songisepp E, Stsepetova J, Naaber P, Mikelsaar RH, Mikelsaar M. A combination of the probiotic and prebiotic product can prevent the germination of Clostridium difficile spores and infection. Anaerobe. 2017;47:94–103. doi: 10.1016/j.anaerobe.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Zhang F, Luo W, Shi Y, Fan Z, Ji G. Should we standardize the 1,700-year-old fecal microbiota transplantation? American Journal of Gastroenterology. 2012;107(11):1755. doi: 10.1038/ajg.2012.251. [DOI] [PubMed] [Google Scholar]
- 38.Eiseman B, Silen W, Bascom GS, Kauvar AJ. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery. 1958;44(5):854–9. [PubMed] [Google Scholar]
- 39.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. J Physiol Gastrointest Liver Physiol. 2014;306(4):310–319. doi: 10.1152/ajpgi.00282.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown JRM, Flemer B, Joyce SA, Zulquernain A, Sheehan D, Shanahan F, O’Toole PW. Changes in microbiota composition, bile and fatty acid metabolism, in successful faecal microbiota transplantation for Clostridioides difficile infection. BMC Gastroenterol. 2018;18(1):18. doi: 10.1186/s12876-018-0748-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly CR, Khoruts A, Staley C, Sadowsky MJ, Abd M, Alani M, Bakow B, Curran P, McKenney J, Tisch A, et al. Effect of fecal microbiota transplantation on recurrence in multiply recurrent clostridium difficile infection. Ann Intern Med. 2016;165(9):609–616. doi: 10.7326/M16-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langdon A, Schwartz DJ, Bulow C, Sun X, Hink T, Reske KA, Jones C, Burnham CAD, Dubberke ER, Dantas G. Microbiota restoration reduces antibiotic-resistant bacteria gut colonization in patients with recurrent Clostridioides difficile infection from the open-label PUNCH CD study. Genome Med. 2021;13(1):13. doi: 10.1186/s13073-021-00828-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma J, Erik Dubberke, Dubberke E. Current management of Clostridioides difficile infection in adults a summary of recommendations from the 2017 IDSA:SHEA clinical practice guideline. Polish Archives of Internal Medicine. 2019;129(3):189–198. doi: 10.20452/pamw.4377. [DOI] [PubMed] [Google Scholar]
- 44.Kwak S, Choi J, Hink T, Reske KA, Blount K, Jones C, Bost MH, Sun X, Burnham CAD, Dubberke ER, et al. Impact of investigational microbiota therapeutic RBX2660 on the gut microbiome and resistome revealed by a placebo-controlled clinical trial. Microbiome. 2020;8(1):8. doi: 10.1186/s40168-019-0776-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubberke ER, Lee CH, Orenstein R, Khanna S, Hecht G, Gerding DN. Results from a randomized, placebo-controlled clinical trial of a RBX2660 - A microbiota-based drug for the prevention of Recurrent Clostridium difficile infection. Clinical Infectious Diseases. 2018;67(8):1198–1204. doi: 10.1093/cid/ciy259. [DOI] [PubMed] [Google Scholar]
- 46.Baktash A, Terveer EM, Zwittink RD, Hornung BVH, Corver J, Kuijper EJ, Smits WK. Mechanistic insights in the success of fecal microbiota transplants for the treatment of Clostridium difficile infections. Front Microbiol. 2018;9. doi: 10.3389/fmicb.2018.01242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, Turbett S, Chung RT, Chen Y-B, Hohmann EL, et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. New England Journal of Medicine. 2019;381(21):2043–2050. doi: 10.1056/NEJMoa1910437. [DOI] [PubMed] [Google Scholar]
- 48.Tvede M, Rask-Madsen J. Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. The Lancet. 1989;333(8648):1156–1160. doi: 10.1016/S0140-6736(89)92749-9. [DOI] [PubMed] [Google Scholar]
- 49.Cammarota G, Ianiro G, Masucci L, Quaranta G, Paroni Sterbini F, Simonelli C, Maletesta L, Gasbarrini A, Sanguinetti M. a culturomics-based synthetic microbiota consortium, derived from successful bacterial engrafters, is a safe and effective treatment for recurrent clostridium difficile infection: a pilot study. United European Gastroenterology Journal. 2018;6:A70–A70. [Google Scholar]
- 50.Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, Daigneault MC, Brown EM, Schroeter K, Allen-Vercoe E. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: “RePOOPulating” the gut. Microbiome. 2013;1(1):1. doi: 10.1186/2049-2618-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rode AA, Chehri M, Krogsgaard LR, Heno KK, Svendsen AT, Ribberholt I, Helms M, Engberg J, Schønning K, Tvede M, et al. Randomised clinical trial: a 12-strain bacterial mixture versus faecal microbiota transplantation versus vancomycin for recurrent Clostridioides difficile infections. Aliment Pharmacol Ther. 2021;53(9):999–1009. doi: 10.1111/apt.16309. [DOI] [PubMed] [Google Scholar]
- 52.Amrane S, Hocquart M, Afouda P, Kuete E, Pham TPT, Dione N, Ngom II, Valles C, Bachar D, Raoult D, et al. Metagenomic and culturomic analysis of gut microbiota dysbiosis during Clostridium difficile infection. Sci Rep. 2019;9(1). doi: 10.1038/s41598-019-49189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghimire S, Roy C, Wongkuna S, Antony L, Maji A, Keena MC, Foley A, Scaria J. Identification of Clostridioides difficile -inhibiting gut commensals using culturomics. Phenotyping, and Combinatorial Community Assembly mSystems. 2020;5(1):e00620–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seres therapeutics announces positive topline results from SER-109 phase 3 ECOSPOR III study in recurrent C. difficile infection. Business Wire. 2020. Available from: https://www.businesswire.com/news/home/20200810005194/en/ 10 August. [Google Scholar]
- 55.Khanna S, Pardi DS, Kelly CR, Kraft CS, Dhere T, Henn MR, Lombardo MJ, Vulic M, Ohsumi T, Winkler J, et al. A novel microbiome therapeutic increases gut microbial diversity and prevents recurrent Clostridium difficile infection. Journal of Infectious Diseases. 2016;214(2):173–181. doi: 10.1093/infdis/jiv766. [DOI] [PubMed] [Google Scholar]
- 56.McGovern BH, Ford CB, Henn MR, Pardi DS, Khanna S, Hohmann EL, O’Brien EJ, Desjardins CA, Bernardo P, Wortman JR, et al. SER-109, an investigational microbiome drug to reduce recurrence after Clostridioides difficile infection: lessons learned from a phase 2 trial. Clinical Infectious Diseases. 2021;72(12):2132–2140. doi: 10.1093/cid/ciaa387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Samarkos M, Mastrogianni E, Kampouropoulou O. The role of gut microbiota in Clostridium difficile infection. European Journal of Internal Medicine2018. 2018;50:28–32. doi: 10.1016/j.ejim.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 58.Ratner M. Microbial cocktails raise bar for C. diff. treatments. Nat Biotechnol. 2020;38(12):1366–1367. doi: 10.1038/s41587-020-00765-8. [DOI] [PubMed] [Google Scholar]
- 59.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document: the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11(8):506–514. doi: 10.1038/nrgastro.2014.66. [DOI] [PubMed] [Google Scholar]
- 60.Satokari R. Modulation of gut microbiota for health by current and next-generation probiotics. Nutrients. 2019;11(8):1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16(10):605–616. doi: 10.1038/s41575-019-0173-3. [DOI] [PubMed] [Google Scholar]
- 62.Chamberlain R, Lau C. Probiotics are effective at preventing Clostridium difficile-associated diarrhea: a systematic review and meta-analysis. Int J Gen Med. 2016;9:27–37. doi: 10.2147/IJGM.S98280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, Chu Q, Huang Y, Xiao Y, Song L, Zhu S, Kang Y, Lu S, Xu J, Ren Z. Consortium of probiotics attenuates colonization of Clostridioides difficile. Frontiers in Microbiology 2019; 10. [DOI] [PMC free article] [PubMed]
- 64.Wullt M, Johansson Hagslätt ML, Odenholt I, Berggren A. Lactobacillus plantarum 299v enhances the concentrations of fecal short-chain fatty acids in patients with recurrent Clostridium difficile-associated diarrhea. Dig Dis Sci. 2007;52(9):2082–2086. doi: 10.1007/s10620-006-9123-3. [DOI] [PubMed] [Google Scholar]
- 65.Forssten S, Evans M, Wilson D, Ouwehand A. Influence of a probiotic mixture on antibiotic induced microbiota disturbances. World Journal, of Gastroenterology. 2014;20(33):11878–11885. doi: 10.3748/wjg.v20.i33.11878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kabbani TA, Pallav K, Dowd SE, Villafuerte-Galvez J, Vanga RR, Castillo NE, Hansen J, Dennis M, Leffler DA, Kelly CP. Prospective randomized controlled study on the effects of Saccharomyces boulardii CNCM I-745 and amoxicillin-clavulanate or the combination on the gut microbiota of healthy volunteers. Gut Microbes. 2017;8(1):17–32. doi: 10.1080/19490976.2016.1267890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castagliuolo I, Lamont JT, Nikulasson ST, Pothoulakis C. Saccharomyces boulardii protease inhibits Clostridium difficile toxin a effects in the rat ileum. Infect Immun. 1996;64(12):5225–5232. doi: 10.1128/iai.64.12.5225-5232.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pothoulakis C, Kelly CP, Joshi MA, Gao N, O’keane CJ, Castagliuolo I, Lamont JT. Saccharomyces boulardii inhibits Clostridium difficile toxin A binding and enterotoxicity in rat ileum. Gastroenterology. 1993;104(4):1108–1115. doi: 10.1016/0016-5085(93)90280-P. [DOI] [PubMed] [Google Scholar]
- 69.Castagliuolo I, Riegler MF, Valenick L, Lamont JT, Pothoulakis C, Kozel TR. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins a and b in human colonic mucosa. Infect Immun. 1999;67(1):302–307. doi: 10.1128/IAI.67.1.302-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X, Kokkotou EG, Mustafa N, Bhaskar KR, Sougioultzis S, O’Brien M, Pothoulakis C, Kelly C. Saccharomyces boulardii inhibits ERK1/2 mitogen-activated protein kinase activation both in vitro and in vivo and protects against Clostridium difficile toxin A-induced enteritis. Journal of Biological Chemistry. 2006;281(34):24449–24454. doi: 10.1074/jbc.M605200200. [DOI] [PubMed] [Google Scholar]
- 71.Maziade PJ, Pereira P, Goldstein EJC. A decade of experience in primary prevention of clostridium difficile infection at a community hospital using the probiotic combination lactobacillus acidophilus CL1285, lactobacillus casei LBC80R, and lactobacillus rhamnosus CLR2 (Bio-K+). Clinical Infectious Diseases. 2015;60(suppl_2):S144–7. doi: 10.1093/cid/civ178. [DOI] [PubMed] [Google Scholar]
- 72.Cassone M, Serra P, Mondello F, Girolamo A, Scafetti S, Pistella E, Venditti M. Outbreak of Saccharomyces cerevisiae subtype boulardii fungemia in patients neighboring those treated with a probiotic preparation of the organism. J Clin Microbiol 41 11 . 2003;5340–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, et al. CX3CR1 + mononuclear phagocytes control immunity to intestinal fungi. Science. 2018;360(6387):359. doi: 10.1126/science.aat8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McFarland L. Systematic review and meta-analysis of Saccharomyces boulardii in adult patients. World Journal of Gastroenterology2010. 2010;16(18):2202–2222. doi: 10.3748/wjg.v16.i18.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Szajewska H, Kołodziej M. Systematic review with meta-analysis: saccharomyces boulardii in the prevention of antibiotic-associated diarrhoea. Alimentary Pharmacology and Therapeutics2015. 2015;42(7):793–801. doi: 10.1111/apt.13344. [DOI] [PubMed] [Google Scholar]
- 76.Rajkumar C, Wilks M, Islam J, Ali K, Raftery J, Davies KA, Timeyin J, Cheek E, Cohen J, Wright J, et al. Do probiotics prevent antibiotic-associated diarrhoea? Results of a multicentre randomized placebo-controlled trial. Journal of Hospital Infection. 2020;105(2):280–288. doi: 10.1016/j.jhin.2020.01.018. [DOI] [PubMed] [Google Scholar]
- 77.Lahtinen SJ, Forssten S, Aakko J, Granlund L, Rautonen N, Salminen S, Viitanen M, Ouwehand AC. Probiotic cheese containing Lactobacillus rhamnosus HN001 and Lactobacillus acidophilus NCFM® modifies subpopulations of fecal lactobacilli and Clostridium difficile in the elderly. Age. 2012;34(1):133–143. doi: 10.1007/s11357-011-9208-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wong S, Jamous A, O’Driscoll J, Sekhar R, Weldon M, Yau CY, Hirani SP, Grimble G, Forbes A. A Lactobacillus casei Shirota probiotic drink reduces antibiotic-associated diarrhoea in patients with spinal cord injuries: a randomised controlled trial. British Journal of Nutrition. 2014;111(4):672–678. doi: 10.1017/S0007114513002973. [DOI] [PubMed] [Google Scholar]
- 79.Shen NT, Maw A, Tmanova LL, Pino A, Ancy K, Crawford C, Simon MS, Evans AT. Timely use of probiotics in hospitalized adults prevents Clostridium difficile infection: a systematic review with meta-regression analysis. Gastroenterology. 2017;152(8):1889–1900.e9. doi: 10.1053/j.gastro.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 80.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517(7533):205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hayashi A, Nagao-Kitamoto H, Kitamoto S, Kim CH, Kamada N. The butyrate-producing bacterium Clostridium butyricum suppresses Clostridioides difficile infection via neutrophil- and antimicrobial cytokine–dependent but GPR43/109a-independent mechanisms. The Journal of Immunology. 2021;206(7):1576–1585. doi: 10.4049/jimmunol.2000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li X, Kang Y, Huang Y, Xiao Y, Song L, Lu S, Ren Z. A strain of bacteroides thetaiotaomicron attenuates colonization of Clostridioides difficile and affects intestinal microbiota and bile acids profile in a mouse model. Biomedicine and Pharmacotherapy. 2021;137:111290. doi: 10.1016/j.biopha.2021.111290. [DOI] [PubMed] [Google Scholar]
- 83.Elahi M, Nakayama-Imaohji H, Hashimoto M, Tada A, Yamasaki H, Nagao T, Kuwahara T. The human gut microbe bacteroides thetaiotaomicron suppresses toxin release from clostridium difficile by inhibiting autolysis. Antibiotics. 2021;10(2):1–16. doi: 10.3390/antibiotics10020187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stiefel U, Nerandzic MM, Pultz MJ, Donskeya CJ. Gastrointestinal colonization with a cephalosporinase-producing bacteroides species preserves colonization resistance against vancomycin-resistant enterococcus and Clostridium difficile in cephalosporin-treated mice. Antimicrob Agents Chemother. 2014;58(8):4535–4542. doi: 10.1128/AAC.02782-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet. 1998;351(9103):633–636. doi: 10.1016/S0140-6736(97)08062-8. [DOI] [PubMed] [Google Scholar]
- 86.Borriello SP, Barclay FE. Protection of hamsters against C.difficile ileocaecitis by prior colonisatton with non-pathogenic strains. J Med Microbiol. 1985;19(3):339–350. doi: 10.1099/00222615-19-3-339. [DOI] [PubMed] [Google Scholar]
- 87.Sambol SP, Merrigan MM, Tang JK, Johnson S, Gerding DN. Colonization for the prevention of Clostridium difficile disease in hamsters. Journal of Infectious Diseases. 2002;186(12):1781–1789. doi: 10.1086/345676. [DOI] [PubMed] [Google Scholar]
- 88.Oliveira Júnior CA, Silva ROS, Cruz DSG, Pires IH, Guedes RMC, Faria Lobato FC. The non-toxigenic strain of Clostridioides difficile Z31 can prevent infection by C. difficile in experimental model piglets. Anaerobe. 2019;55:24–28. doi: 10.1016/j.anaerobe.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 89.Etienne-Mesmin L, Chassaing B, Adekunle O, Mattei LM, Bushman FD, Gewirtz AT. Toxin-positive Clostridium difficile latently infect mouse colonies and protect against highly pathogenic C. Difficile Gut. 2018;67(5):860–871. doi: 10.1136/gutjnl-2016-313510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Leslie JL, Jenior ML, Vendrov KC, Standke AK, Barron MR, O’brien TJ, Unverdorben L, Thaprawat P, Bergin IL, Schloss PD, et al. Protection from lethal clostridioides difficile infection via intraspecies competition for cogerminant. mBio 12 2 . 2021;e00522–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Villano SA, Seiberling M, Tatarowicz W, Monnot-Chase E, Gerding DN. Evaluation of an oral suspension of VP20621, spores of nontoxigenic Clostridium difficile strain M3, in healthy subjects. Antimicrob Agents Chemother. 2012;56(10):5224–5229. doi: 10.1128/AAC.00913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gerding DN, Meyer T, Lee C, Cohen SH, Murthy UK, Poirier A, van Schooneveld TC, Pardi DS, Ramos A, Barron MA, et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C difficile infection. JAMA - Journal of the American Medical Association. 2015;313(17):1719–1727. doi: 10.1001/jama.2015.3725. [DOI] [PubMed] [Google Scholar]
- 93.Gerding DN, Sambol SP, Johnson S. Non-toxigenic clostridioides (formerly clostridium) difficile for prevention of C. difficile infection: from bench to bedside back to bench and back to bedside. Front Microbiol. 2018;9. doi: 10.3389/fmicb.2018.01700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McInnes RS, McCallum GE, Lamberte LE, van Schaik W. Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Current Opinion in Microbiology2020. 2020;53:35–43. doi: 10.1016/j.mib.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 95.Peng Z, Jin D, Kim HB, Stratton CW, Wu B, Tang YW, Suna X, Kraft CS. Update on antimicrobial resistance in Clostridium difficile: resistance mechanisms and antimicrobial susceptibility testing. Journal of Clinical Microbiology2017. 2017;55(7):1998–2008. doi: 10.1128/JCM.02250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Spigaglia P. Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Therapeutic Advances in Infectious Disease. 2016;3(1):23–42. doi: 10.1177/2049936115622891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brouwer MSM, Roberts AP, Hussain H, Williams RJ, Allan E, Mullany P. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun. 2013;4:2601. doi: 10.1038/ncomms3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM. Phage treatment of human infections. Bacteriophage. 2011;1(2):66–85. doi: 10.4161/bact.1.2.15845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meader E, Mayer MJ, Steverding D, Carding SR, Narbad A. Evaluation of bacteriophage therapy to control clostridium difficile and toxin production in an invitro human colon model system. Anaerobe. 2013;22:25–30. doi: 10.1016/j.anaerobe.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 100.Meader E, Mayer MJ, Gasson MJ, Steverding D, Carding SR, Narbad A. Bacteriophage treatment significantly reduces viable Clostridium difficile and prevents toxin production in an in vitro model system. Anaerobe. 2010;16(6):549–554. doi: 10.1016/j.anaerobe.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 101.Nale JY, Redgwell TA, Millard A, Clokie MRJ. Efficacy of an optimised bacteriophage cocktail to clear Clostridium difficile in a batch fermentation model. Antibiotics. 2018;7(1):1–15. doi: 10.3390/antibiotics7010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nale JY, Spencer J, Hargreaves KR, Trzepiński P, Douce GR, Clokie MRJ, Clokie MRJ. Bacteriophage combinations significantly reduce Clostridium difficile growth in vitro and proliferation in vivo. Antimicrob Agents Chemother. 2016;60(2):968–981. doi: 10.1128/AAC.01774-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fortier LC. 2018. Bacteriophages contribute to shaping clostridioides (Clostridium) difficile species. Frontiers in Microbiology. 9:2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Selle K, Fletcher JR, Tuson H, Schmitt DS, McMillan L, Vridhambal GS, Rivera AJ, Montgomery SA, Fortier L-C, Barrangou R, et al. In vivo targeting of Clostridioides difficile using phage-delivered CRISPR-Cas3 antimicrobials downloaded from. mBio. 2020;11(2):e00019–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goh S, Hussain H, Chang BJ, Emmett W, Riley T, Mullany P. Phage C2 mediates transduction of Tn6215, encoding erythromycin resistance, between Clostridium difficile strains. mBio. 2013;4(6):e00840–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Riedel T, Wittmann J, Bunk B, Schober I, Spröer C, Gronow S, Overmann J. A Clostridioides difficile bacteriophage genome encodes functional binary toxin-associated genes. J Biotechnol. 2017;250:23–28. doi: 10.1016/j.jbiotec.2017.02.017. [DOI] [PubMed] [Google Scholar]
- 107.Slater RT, Frost LR, Jossi SE, Millard AD, Unnikrishnan M. Clostridioides difficile LuxS mediates inter-bacterial interactions within biofilms. Sci Rep. 2019;9(1). doi: 10.1038/s41598-019-46143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heuler J, Fortier L-C, Sun X. Clostridioides difficile phage biology and application. FEMS Microbiol Rev. 2021;45(5):fuab012. doi: 10.1093/femsre/fuab012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Early Clinical trials with live biotherapeutic products: chemistry, manufacturing, and control information. US Food and Drug Administration. 2016. Available from: http://www.fda.gov/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guida [Google Scholar]
- 110.Ozdemir T, Fedorec AJH, Danino T, Barnes CP. Synthetic biology and engineered live biotherapeutics: toward increasing system complexity. Cell Systems2018. 2018;7(1):5–16. doi: 10.1016/j.cels.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 111.Charbonneau MR, Isabella VM, Li N, Kurtz CB. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat Commun. 2020;11(1):1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kurtz CB, Millet YA, Puurunen MK, Perreault M, Charbonneau MR, Isabella VM, Kotula JW, Antipov E, Dagon Y, Denney WS, et al. An engineered E. coli Nissle improves hyperammonemia and survival in mice and shows dose-dependent exposure in healthy humans. Sci Transl Med 11 475 . 2019;eaau7975. [DOI] [PubMed] [Google Scholar]
- 113.Precigen actobio announces positive topline results from phase 1b study of AG019 actoBioticstm, A novel therapy designed to address the underlying cause of type 1 diabetes. PR Newswire. 2021. Available from: https://www.prnewswire.com/news-releases/precigen-actobio-announces-positive-topline-results-from-phase-1b-study-of-ag019-actobiotics-a-novel-therapy-designed-to-address-the-underlying-cause-of-type-1-diabetes-301108842.html Accessed 10 June 2021. [Google Scholar]
- 114.Vedantam G, Kochanowsky J, Lindsey J, Mallozzi M, Roxas JL, Adamson C, Anwar F, Clark A, Claus-Walker R, Mansoor A, et al. An engineered synthetic biologic protects against Clostridium difficile infection. Front Microbiol. 2018;9. doi: 10.3389/fmicb.2018.02080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Merrigan MM, Venugopal A, Roxas JL, Anwar F, Mallozzi MJ, Roxas BAP, Gerding DN, Viswanathan VK, Vedantam G. Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PloS one. 2013;8(11):e78404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bruxelle J-F, Mizrahi A, Hoys S, Collignon A, Janoir C, Péchiné S. Immunogenic properties of the surface layer precursor of Clostridium difficile and vaccination assays in animal models. Anaerobe. 2016;37:78–84. doi: 10.1016/j.anaerobe.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 117.Ní Eidhin DB, O’Brien JB, McCabe MS, Athié-Morales V, Kelleher DP. Active immunization of hamsters against Clostridium difficile infection using surface-layer protein. FEMS Immunol Med Microbiol. 2008;53(1):52. doi: 10.1111/j.1574-695X.2008.00401.x. [DOI] [PubMed] [Google Scholar]
- 118.Dingle KE, Didelot X, Ansari MA, Eyre DW, Vaughan A, Griffiths D, Clc I, Batty EM, Golubchik T, Bowden R, et al. Recombinational switching of the Clostridium difficile S-layer and a novel glycosylation gene cluster revealed by large-scale whole-genome sequencing. J Infect Dis. 2013;207(4):675–686. doi: 10.1093/infdis/jis734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Michael S, Keubler LM, Smoczek A, Meier M, Gunzer F, Pöhlmann C, Krause-Buchholz U, Hedrich HJ, Bleich A. Quantitative phenotyping of inflammatory bowel disease in the IL-10-deficient mouse by use of noninvasive magnetic resonance imaging. Inflamm Bowel Dis. 2013;19(1):185–193. doi: 10.1002/ibd.23006. [DOI] [PubMed] [Google Scholar]
- 120.Durmusoglu D, Al’Abri IS, Collins SP, Cheng J, Eroglu A, Beisel CL, Crook N. In situ biomanufacturing of small molecules in the mammalian gut by probiotic Saccharomyces boulardii. ACS Synth Biol. 2021;10(5):1039–1052. doi: 10.1021/acssynbio.0c00562. [DOI] [PubMed] [Google Scholar]
- 121.Liu CH, Chang JH, Chang YC, Mou KY. Treatment of murine colitis by Saccharomyces boulardii secreting atrial natriuretic peptide. J Mol Med. 2020;98(12):1675–1687. doi: 10.1007/s00109-020-01987-8. [DOI] [PubMed] [Google Scholar]
- 122.Chen K, Zhu Y, Zhang Y, Hamza T, Yu H, Fleur AS, Galen J, Yang Z, Feng H. A probiotic yeast-based immunotherapy against Clostridioides difficile infection. Sci Transl Med. 2020;12(567):eaax4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li R, Wan X, Takala TM, Saris PEJ. Heterologous expression of the leuconostoc bacteriocin leucocin c in probiotic yeast Saccharomyces boulardii. Probiotics Antimicrob Proteins. 2021;13(1):229–237. doi: 10.1007/s12602-020-09676-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sen S, Mansell TJ. Yeasts as probiotics: mechanisms, outcomes, and future potential. Fungal Genetics and Biology2020. 137:103333. [DOI] [PubMed] [Google Scholar]
- 125.Edwards-Ingram L, Gitsham P, Burton N, Warhurst G, Clarke I, Hoyle D, Oliver SG, Stateva L. Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl Environ Microbiol. 2007;73(8):2458–2467. doi: 10.1128/AEM.02201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Khatri I, Tomar R, Ganesan K, Prasad GS, Subramanian S. Complete genome sequence and comparative genomics of the probiotic yeast Saccharomyces boulardii. Sci Rep. 2017;7(1). doi: 10.1038/s41598-017-00414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu JJ, Kong II, Zhang GC, Jayakody LN, Kim H, Xia PF, Kwak S, Sung BH, Sohn JH, Walukiewicz HE, et al. Metabolic engineering of probiotic Saccharomyces boulardii. Appl Environ Microbiol. 2016;82(8):2280–2287. doi: 10.1128/AEM.00057-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bagherpour G, Ghasemi H, Zand B, Zarei N, Roohvand F, Ardakani EM, Azizi M, Khalaj V. Oral administration of recombinant Saccharomyces boulardii expressing ovalbumin-CPE fusion protein induces antibody response in mice. Front Microbiol. 2018;9. doi: 10.3389/fmicb.2018.00723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yuman L, Pbwilpcibpsmpecp NMMP. Bezlotoxumab (Zinplava) for Clostridium difficile infection. Drug Forecast. 2017;42:735–738. [Google Scholar]
- 130.Wildt S, Gerngross TU. The humanization of N-glycosylation pathways in yeast. Nature Reviews Microbiology2005. 2005;3(2):119–128. doi: 10.1038/nrmicro1087. [DOI] [PubMed] [Google Scholar]
- 131.Feuerstadt P, Louie TJ, Lashner B, Wang EEL, Diao L, Bryant JA, Sims M, Kraft CS, Cohen SH, Berenson CS, et al. SER-109, an oral microbiome therapy for recurrent Clostridioides difficile infection. New England Journal of Medicine. 2022;386(3):220–229. doi: 10.1056/NEJMoa2106516. [DOI] [PubMed] [Google Scholar]
- 132.Khanna S, Pardi DS, Jones C, Shannon WD, Gonzalez C, Blount K. RBX7455, a non-frozen, orally administered investigational live biotherapeutic, is safe, effective, and shifts patients’ microbiomes in a phase 1 study for recurrent clostridioides difficile infections. Clinical Infectious Diseases. 2020;73(7):e1613–e1620. doi: 10.1093/cid/ciaa1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Taur Y, Jenq RR, Ubeda C, van den Brink M, Pamer EG. Role of intestinal microbiota in transplantation outcomes. Best Practice and Research: Clinical Haematology2015. 2015;28(2–3):155–161. doi: 10.1016/j.beha.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Orenstein R, Dubberke E, Hardi R, Kelly C, Ray A, Mullane K, Pardi DS, Ramesh MS, Dubberke ER, Hardi R, et al. Safety and durability of RBX2660 (microbiota suspension) for recurrent Clostridium difficile infection: results of the PUNCH CD study. Clinical Infectious Diseases. 2016;62(5):596–602. doi: 10.1093/cid/civ938. [DOI] [PubMed] [Google Scholar]
- 135.Youngster I, Russell GH, Pindar C, Ziv-Baran T, Sauk J, Hohmann EL. Oral, capsulized, frozen fecal microbiota transplantation for relapsing Clostridium difficile infection. JAMA - Journal of the American Medical Association. 2014;312(17):1772–1778. doi: 10.1001/jama.2014.13875. [DOI] [PubMed] [Google Scholar]
- 136.Youngster I, Sauk J, Pindar C, Wilson RG, Kaplan JL, Smith MB, Alm EJ, Gevers D, Russell GH, Hohmann EL. Fecal microbiota transplant for relapsing clostridium difficile infection using a frozen inoculum from unrelated donors: a randomized, open-label, controlled pilot study. Clinical Infectious Diseases. 2014;58(11):1515–1522. doi: 10.1093/cid/ciu135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Barker AK, Duster M, Valentine S, Hess T, Archbald-Pannone L, Guerrant R, Safdar N. A randomized controlled trial of probiotics for Clostridium difficile infection in adults (PICO). Journal of Antimicrobial Chemotherapy. 2017;72(11):3177–3180. doi: 10.1093/jac/dkx254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Barker A, Duster M, Valentine S, Archbald-Pannone L, Guerrant R, Safdar N. Probiotics for Clostridium difficile infection in adults (PICO): study protocol for a double-blind, randomized controlled trial. Contemp Clin Trials. 2015;44:26–32. doi: 10.1016/j.cct.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


