Abstract
In this study, the mitochondrial genome of Thinopyrum obtusiflorum was sequenced, assembled, and annotated. The complete circular mitogenome of Th. obtusiflorum is 390,725 bp in length and the overall A + T content of mitogenome is 55.61%. It harbors 33 protein-coding genes (PCGs), 21 transfer RNA genes (tRNAs), six ribosomal RNA genes (rRNAs), and 20 simple sequence repeats (SSRs). Phylogenetic analysis indicates that Th. obtusiflorum is a sister to the clade including Aegilops speltoides, Triticum aestivum, and Triticum aestivum cultivar Chinese Yumai in the Triticeae.
Keywords: Thinopyrum obtusiflorum, mitochondrial genome, Poaceae, phylogenetic analysis
Thinopyrum obtusiflorum (DC.) Banfi (≡Thinopyrum ponticum (Podp.) Barkworth & D.R.Dewey, 1985, 2n = 10× =70) is a perennial allodecaploid species in the Triticeae (Poaceae), mainly native to European and Near Asian East (Banfi 2018; Tiryaki et al. 2021). It is known to possess many elite genes for wheat improvement, such as those for wheat leaf rust resistance (Sepsi et al. 2008), stem rust resistance (Mago et al. 2019), stripe rust resistance (Wang et al. 2020), powdery mildew resistance (He et al. 2017), Fusarium head blight (FHB) resistance (Forte et al. 2014), and tolerance to abiotic stress, such as cold, drought and salinity (Friebe et al. 1996; Linc et al. 2012). All of these characters, coupled with its high cross-compatibility with wheat genome, make this species an important donor of elite genes for improving the genetic diversity of cultivated wheat.
The fresh leaves of Th. obtusiflorum were collected from the Botanical Garden of Center of Wheat Research, Xinxiang, Henan, China (113°88′E 35°30′N). The voucher specimen was deposited at the Herbarium of Henan Institute of Science and Technology, Xinxiang, China (Zhengang Ru; rzgh58@163.com) under the voucher number XM-W027. Total genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) method. Sequencing was carried out on the Illumina NovaSeq 6000 platform and PacBio Sequel platform. The Th. obtusiflorum mitogenome sequences were assembled with the NovoPlasty v.4.3.1 (Dierckxsens et al. 2017), SPAdes-3.13.0 package (Antipov et al. 2016) and Canu v2.1.1 package (Koren et al. 2017), and then annotated using the online GeSeq tool (Tillich et al. 2017) with default parameters to predict protein-coding genes (PCGs), transfer RNA (tRNA) genes, and ribosomal RNA (rRNA) genes. In addition, the simple sequence repeats (SSRs) were detected in the mitogenome using MISA (MIcroSAtellite Identification Tool) (Beier et al. 2017).
The entire mitogenome of Th. obtusiflorum (GenBank accession number: OK120846) is a closed circular double-stranded structure of 390,725 bp in length with base compositions of 27.62% A, 21.96% G, 22.43% C, 27.99% T, exhibiting A + T bias (55.61%). The mitogenome encodes a typical 60 genes, including 33 PCGs, 21 tRNA genes, and six rRNA genes. The cumulative length of the 33 PCGs is 31,404 bp, and the average length of a single-gene is 952 bp, which accounts for 8.04% of the complete genome. The cumulative length of the tRNA genes is 1590 bp, with an average gene length of 75 bp, accounting for 0.41% of the complete genome. The lengths of rRNA genes are 1977 bp (rrn18), 3467 bp (rrn26), and 122 bp (rrn5), which together account for 0.09% of the complete genome. The cumulative length of 20 SSR loci identified is 212 bp which comprises approximately 0.05% of the genome length.
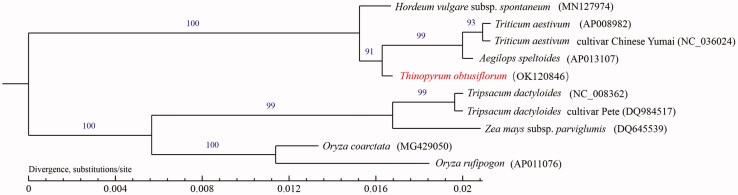
To investigate the evolutionary relationship of Th. obtusiflorum, the mitogenome of the other nine species was downloaded from GenBank (https://www.ncbi.nlm.nih.gov/genbank/). Phylogenetic tree was constructed based on functional protein-coding sequences (CDS), first aligned them with the online tool MUSCLE under the default setting (Madeira et al. 2019), and then performed the maximum-likelihood (ML) phylogenetic analysis using PHYML 3.0 (Guindon et al. 2010). The results of phylogenetic analysis supported a monophyletic relationship of Th. obtusiflorum to Aegilops speltoides, Triticum aestivum, and Triticum aestivum cultivar Chinese Yumai (Figure 1). This study might be helpful for promoting its utilization of distant hybridization in genetic improvement of wheat.
Figure 1.
Maximum-likelihood phylogenetic tree based on complete mitogenome of Thinopyrum obtusiflorum and other nine species of Poaceae. Numbers above the lines represent ML bootstrap values.
Authors contributions
Conception and design: Xiaojun Wu, Xigui Hu, Tiezhu Hu, and Zhengang Ru; analysis and interpretation of the data: Xiangdong Chen, Jinlong Zhang, and Cuicui Ren; the drafting of the paper, revising it critically for intellectual content: Xiaojun Wu, Lintong Song, Fang Fang, and Na Dong; all authors have approved the final version of the manuscript and all authors agree to be accountable for all aspects of the work.
Funding Statement
This work was supported by the Major Science and Technology Project of Henan Province [Grant No. 201300110800]; the Training Plan for Young Backbone Teachers in Colleges and Universities in Henan Province [Grant No. 2021GGJS122]; the Post Expert of Wheat Industry System in Henan Province [Grant No. S2010-01-G09], and the Science and Technology Climbing Plan of Henan Institute of Science and Technology [Project Code: 105020221015/007].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, reference number OK120846. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA782017, SAMN23314009, and SRP347059, respectively.
References
- Antipov D, Korobeynikov A, McLean J, Pevzner P.. 2016. HYBRIDSPADES: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 32(7):1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfi E. 2018. A survey of the Elymus L. s. l. species complex (Triticeae, Poaceae) in Italy: taxa and nothotaxa, new combinations and identification key. Nat Hist Sci. 5(2):57–64. [Google Scholar]
- Beier S, Thiel T, Münch T, Scholz U, Mascher M.. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forte P, Virili M, Kuzmanović L, Moscetti I, Gennaro A, D’Ovidio R, Ceoloni C.. 2014. A novel assembly of Thinopyrum obtusiflorum genes into the durum wheat genome: pyramiding Fusarium head blight resistance onto recombinant lines previously engineered for other beneficial traits from the same alien species. Mol Breed. 34(4):1701–1716. [Google Scholar]
- Friebe B, Jiang J, Raupp WJ, McIntosh RA, Gill BS.. 1996. Characterization of wheat–alien translocations conferring resistance to diseases and pests: current status. Euphytica. 91(1):59–87. [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- He F, Bao Y, Qi X, Ma Y, Li X, Wang H.. 2017. Molecular cytogenetic identification of a wheat–Thinopyrum ponticum translocation line resistant to powdery mildew. J Genet. 96(1):165–169. [DOI] [PubMed] [Google Scholar]
- Koren S, Walenz B, Berlin K, Miller J, Bergman N, Phillippy A.. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linc G, Sepsi A, Molnár-Láng M.. 2012. A FISH karyotype to study chromosome polymorphisms for the Elytrigia elongata E genome. Cytogenet Genome Res. 136(2):138–144. [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47(W1):W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mago R, Zhang P, Xia X, Zhang J, Hoxha S, Lagudah E, Graner A, Dundas I.. 2019. Transfer of stem rust resistance gene SrB from Thinopyrum obtusiflorum into wheat and development of a closely linked PCR-based marker. Theor Appl Genet. 132(2):371–382. [DOI] [PubMed] [Google Scholar]
- Sepsi A, Molnár I, Szalay D, Molnár-Láng M.. 2008. Characterization of a leaf rust-resistant wheat–Thinopyrum obtusiflorum partial amphiploid BE-1, using sequential multicolor GISH and FISH. Theor Appl Genet. 116(6):825–834. [DOI] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones E, Fischer A, Bock R, Greiner S.. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiryaki I, Karaoğlu G, Yücel G, Tuna M.. 2021. Assessment of Thinopyrum obtusiflorum (Podp.) Barkworth & DR Dewey accessions using universal rice primers and molecular cytogenetics. Genet Resour Crop Evol. 68(5):1875–1888. [Google Scholar]
- Wang Y, Cao Q, Zhang J, Wang S, Chen C, Wang C, Zhang H, Wang Y, Ji W.. 2020. Cytogenetic analysis and molecular marker development for a new wheat–Thinopyrum obtusiflorum 1Js (1D) disomic substitution line with resistance to stripe rust and powdery mildew. Front Plant Sci. 11:1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, reference number OK120846. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA782017, SAMN23314009, and SRP347059, respectively.