Abstract
Antibacterial activities of gatifloxacin (AM1155), a new C-8-methoxy fluoroquinolone, and two structurally related compounds, AM1121 and ciprofloxacin, were studied with an isogenic set of ten quinolone-resistant, gyrA (gyrase) mutants of Escherichia coli. To compare the effect of each mutation on resistance, the mutant responses were normalized to those of wild-type cells. Alleles exhibiting the most resistance to growth inhibition mapped in α-helix 4, which is thought to lie on a GyrA dimer surface that interacts with DNA. The C-8-methoxy group lowered the resistance due to these mutations more than it lowered resistance arising from several gyrA alleles located outside α-helix 4. These data are consistent with α-helix 4 being a distinct portion of the quinolone-binding site of GyrA. A helix change to proline behaved more like nonhelix alleles, indicating that helix perturbation differs from the other changes at helix residues. Addition of a parC (topoisomerase IV) resistance allele revealed that the C-8-methoxy group also facilitated attack of topoisomerase IV. When lethal effects were measured at a constant multiple of the minimum inhibitory concentration for each fluoroquinolone to normalize for differences in bacteriostatic action, gatifloxacin was more potent than the C-8-H compounds, both in the presence and absence of protein synthesis (an exception was observed when alanine was substituted for aspartic acid at position 82). Collectively, these data show that the C-8-methoxy group contributes to the enhanced activity of gatifloxacin against resistant gyrase and wild-type topoisomerase IV.
Fluoroquinolones are potent antibacterial agents that poison cells by trapping DNA gyrase and DNA topoisomerase IV on chromosomes as quinolone-enzyme-DNA complexes (reviewed in references 4 and 5). Occasionally the compounds are rendered ineffective through the development of bacterial resistance (reviewed in reference 15). To identify fluoroquinolones that more avidly attack resistant bacteria, gyrA (gyrase) mutants have been used to screen new fluoroquinolone derivatives. By this test, particularly potent compounds contain a halogen or methoxy group at the C-8 position (2, 7, 12, 23–25). Avid attack of resistant mutants allows the C-8-methoxy fluoroquinolones to restrict the selection of resistant mutants more effectively than C-8-H derivatives (3). Why the C-8-methoxy fluoroquinolones are particularly active against gyrase (25) and topoisomerase IV mutants (24) is not known.
One way to examine the action of C-8-methoxy fluoroquinolones is to compare the effectiveness of structural variants against a series of resistant gyrase mutants. Since the crystal structure of the breakage-reunion fragment of the GyrA protein from Escherichia coli has been determined (14), a starting point exists for spatially orienting resistance alleles. When the GyrA dimer is modeled, two helices (one α-helix 4 on each GyrA monomer) are found on a surface where DNA is thought to interact. The most commonly selected resistance alleles map on the same surface of α-helix 4. Whether the C-8-methoxy group preferentially enhances attack of mutant gyrase encoded by particular alleles has not been studied.
In the present work we examined the effect of fluoroquinolone structure by comparing the action of gatifloxacin (C-8-methoxy), AM1121 (C-8-H), and ciprofloxacin (C-8-H) against a set of ten quinolone-resistant gyrA mutants of E. coli. Data were normalized to the wild-type value to cancel differences among the compounds with respect to potencies against wild-type strains, and a parC quinolone-resistance allele was included in some mutant strains to minimize effects due to topoisomerase IV. Under these conditions, the highly resistant alleles in α-helix 4 were more sensitive to the presence of a C-8-methoxy group than mutations mapping outside the helix. An exception to the pattern was a helix mutation that placed a proline between the major resistance positions (amino acids 83 and 87). This mutant behaved more like those outside the helix. The helix-disrupting proline substitution probably perturbed the helix as a whole, while the other helix mutations modified portions.
During formation of quinolone-gyrase-DNA complexes, gyrase undergoes a conformational change and DNA is broken (8, 9). Release of double-strand DNA breaks is thought to be the lethal event (1). To estimate lethal effects separately from complex formation, survival of the E. coli mutants was measured at a constant multiple of the minimum inhibitory concentration (MIC) for the three fluoroquinolones. The relative resistance contributed by the alleles differed for lethal and bacteriostatic effects, emphasizing that blocking growth and killing cells are separate events. For most of the mutants, gatifloxacin was more lethal than the two C-8-H compounds. Thus, a C-8-methoxy moiety improves fluoroquinolone action against gyrase at both the bacteriostatic and bactericidal levels.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains, listed in Table 1, were derivatives of E. coli K-12. The gyrA mutations were obtained from a variety of sources (Table 1), and P1-mediated transduction was used to place them in the DM4100 background. For transduction, we first introduced into each gyrA (Nalr) strain zfa-3145::Tn10Kan or zeg::Tn10 that cotransduced with gyrA at frequencies of 70 and 50%, respectively. The gyrA mutations were then transduced into DM4100 by selection for Kanr or Tetr with scoring for resistance to nalidixic acid (10 or 20 μg/ml, depending on the allele). Each gyrA mutant was also introduced into a derivative of DM4100 (KD1373) containing the S80L parC quinolone resistance allele. For this construction, zeg::Tn10 was introduced into each gyrA (Nalr) mutant. P1-mediated transduction, with selection for tetracycline resistance followed by scoring for nalidixic acid resistance, was then used to introduce each gyrA allele into the parC mutant (in gyrA+ cells, the parC allele confers no quinolone resistance).
TABLE 1.
Bacterial strains
| Strain | Relevant genotypea | Source or reference |
|---|---|---|
| DM4100 | Wild type | 20 |
| KD66 | DM4100 gyrA (Nalr S83L) | 17 |
| KD1366 | gyrA (Nalr S83L) parC (Cipr S80L)b | 25 |
| KD1373 | DM4100 parC (Cipr S80L) | 25 |
| KD1525 | gyrA (Nalr D82A) parC (Cipr S80L) | This work, as described in reference 25 |
| KD1721 | DM4100 gyrA (Nalr A51V) | This work, by P1-mediated transduction from T/r (6) |
| KD1911 | DM4100 gyrA (Nalr A67S) | This work, by P1-mediated transduction from P-10 (22) |
| KD1915 | DM4100 gyrA (Nalr G81C) | This work, by P1-mediated transduction from N-97 (22) |
| KD1909 | DM4100 gyrA (Nalr S83W) | This work, by P1-mediated transduction from P-18 (22) |
| KD1913 | DM4100 gyrA (Nalr D87N) | This work, by P1-mediated transduction from N-113 (22) |
| KD1917 | DM4100 gyrA (Nalr Q106H) | This work, by P1-mediated transduction from N-89 (22) |
| KD1957 | gyrA (Nalr A51V) parC (Cipr S80L) | This work, by P1-mediated transduction from KD1721 zeg::Tn10c into KD1373 |
| KD1959 | gyrA (Nalr A67S) parC (Cipr S80L) | This work, by P1-mediated transduction from KD1911 zeg::Tn10 into KD1373 |
| KD1961 | gyrA (Nalr G81C) parC (Cipr S80L) | This work, by P1-mediated transduction from KD1915 zeg::Tn10 into KD1373 |
| KD1963 | gyrA (Nalr S83W) parC (Cipr S80L) | This work, by P1-mediated transduction from KD1909 zeg::Tn10 into KD1373 |
| KD1965 | gyrA (Nalr A84P) parC (Cipr S80L) | This work, by P1-mediated transduction from KD1975 zeg::Tn10 into KD1373 |
| KD1967 | gyrA (Nalr D87Y) parC (Cipr S80L) | This work, by P1-mediated transduction from KD1977 zeg::Tn10 into KD1373 |
| KD1969 | gyrA (Nalr Q106H) parC (Cipr S80L) | This work, by P1-mediated transduction from KD1917 zeg::Tn10 into KD1373 |
| KD1971 | gyrA (Nalr D87N) parC (Cipr S80L) | This work, by P1-mediated transduction from KD1913 zeg::Tn10 into KD1373 |
| KD1973 | DM4100 gyrA (Nalr D82A) | This work, by P1-mediated transduction from KD1525 into DM4100 (25) |
| KD1975 | DM4100 gyrA (Nalr A84P) | This work, by P1-mediated transduction from P-5 (22) |
| KD1977 | DM4100 gyrA (Nalr D87Y) | This work, by P1-mediated transduction from KD1677d (M. Friedman) |
Amino acid abbreviations: A, alanine; D, aspartic acid; C, cysteine; H, histidine; G, glycine; L, leucine; N, asparagine; P, proline; Q, glutamine; S, serine; V, valine; W, tryptophan; Y, tyrosine. The first letter represents the wild-type amino acid at the position indicated by the number. The second letter indicates the identity of the amino acid which was substituted for the original at that position.
By itself this parC allele confers no resistance because topoisomerase IV is a secondary target.
zeg::Tn10 encodes a tetracycline resistance gene that is 50% cotransducible with gyrA.
Obtained by selection on agar containing nalidixic acid (20 μg/ml).
Fluoroquinolones and measurement of potency.
Ciprofloxacin was purchased from Miles Pharmaceutical Co.; gatifloxacin (C-8-methoxy; AM1155) and its C-8-H control AM1121 were obtained from Bristol-Myers Squibb. Stock solutions (10 mg of fluoroquinolone per ml in 0.1 M NaOH) were stored at −70°C.
The MIC at which 99% of the isolates tested were inhibited (MIC99) was determined for each fluoroquinolone by spotting 10-μl aliquots of diluted, late log-phase cultures on Luria-Bertani (LB) agar plates (13) containing various concentrations of the fluoroquinolone. After overnight incubation at 37°C, the number of colonies appearing on the plates was counted and plotted against the fluoroquinolone concentration. The MIC99 of the fluoroquinolone was determined by interpolation.
Bactericidal effects of the fluoroquinolones were measured by incubating exponentially growing cultures with the fluoroquinolones in the presence or absence of chloramphenicol (20 μg/ml) for 2 h. Aliquots were then removed, diluted, and plated on drug-free LB agar for determination of viable cell numbers.
RESULTS
Bacteriostatic effect of fluoroquinolone C-8-methoxy group against resistant gyrase.
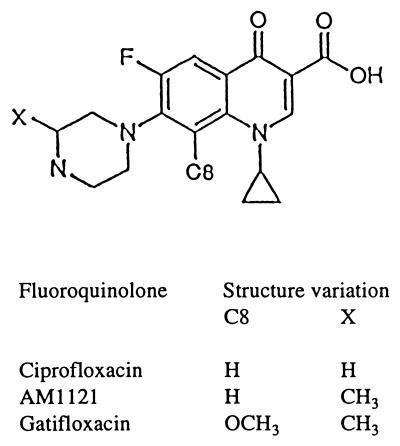
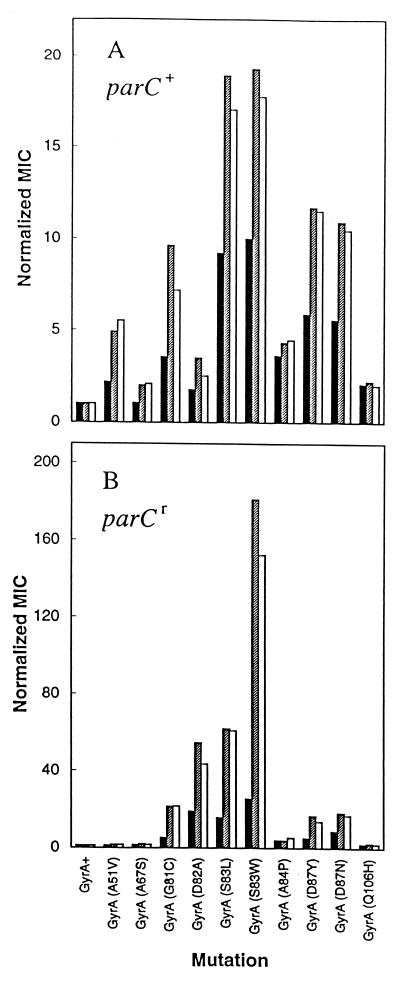
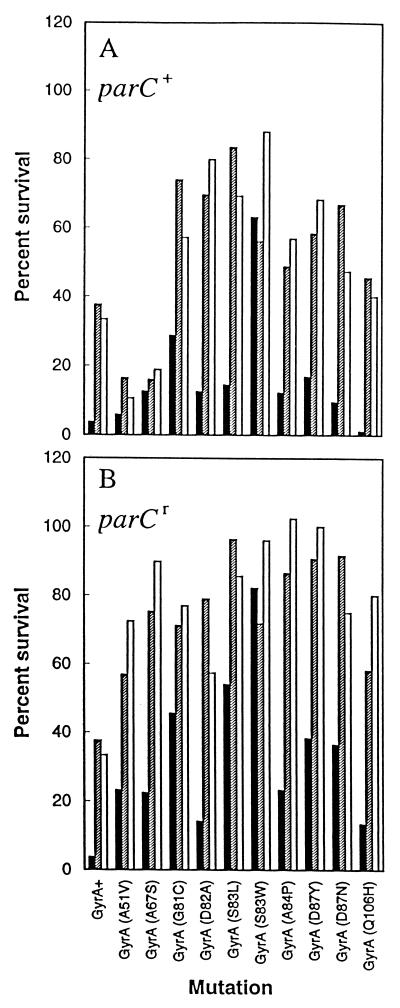
To examine the effects of fluoroquinolone structure on the attack of mutant gyrase, we obtained a variety of E. coli gyrA mutants and prepared an isogenic collection by P1-mediated transduction (Table 1). We then measured the ability of gatifloxacin, AM1121, and ciprofloxacin (Fig. 1) to block the growth of mutant and wild-type strains through determination of MIC99s. Data analysis included normalization of mutant MICs to wild-type values to minimize possible differences among the compounds with respect to nontopoisomerase factors such as drug uptake, efflux, etc. As shown in Fig. 2A, the effect of individual resistance alleles varied considerately, with mutations at codons 83 and 87 in gyrA conferring the most resistance. By this assay gatifloxacin was more active than its C-8-H derivative (AM1121) against all mutations except A84P and Q106H, which showed little difference. Ciprofloxacin exhibited the same general pattern of activity as AM1121 (Fig. 2A).
FIG. 1.
Fluoroquinolone structures.
FIG. 2.
Bacteriostatic action of fluoroquinolones against resistant mutants. Isogenic strains of E. coli containing the indicated alleles of gyrA (A) or gyrA parC (B) were applied to LB agar plates containing various concentrations of the fluoroquinolones for determination of the MIC99s. The MIC99 for each mutant was then divided by the MIC99 of wild-type or parC (S80L) cells to generate the normalized MIC99 (the MIC99s with wild-type cells were 0.038, 0.013, and 0.011 μg/ml for gatifloxacin [C-8-methoxy; solid bars], AM1121 [C-8-H; shaded bars], and ciprofloxacin [C-8-H; open bars], respectively. The data shown are the averages of two determinations.
In E. coli, topoisomerase IV is attacked after gyrase has become resistant (1, 11, 16). Thus some of the effects described above could have arisen from differences in fluoroquinolone action against topoisomerase IV. To minimize these effects we transduced each gyrA allele into an E. coli strain that carried a quinolone-resistance allele of parC (S80L), which alone confers no resistance. The MIC99 of each of the three fluoroquinolones was then determined for each double mutant and normalized to the MIC99 obtained with the gyrA+ parC (S80L) strain to estimate the increase in resistance due to each gyrA allele. Introduction of resistant topoisomerase IV changed the relative resistance contributed by the alleles (compare Fig. 2A and B), indicating that wild-type topoisomerase IV can be a quinolone target when gyrase is resistant. Gatifloxacin was more active than its C-8-H control or ciprofloxacin (Fig. 2B). As described above, mutants carrying the A84P and Q106H alleles showed little difference among the compounds. With respect to absolute values of MIC, gatifloxacin was 60% more effective than ciprofloxacin for the most resistant mutant (S83W).
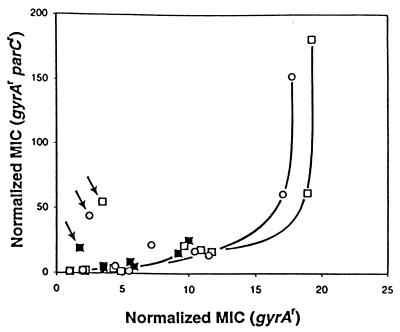
For each of the three quinolones, the effect of introducing a parC resistance allele into gyrA mutants is shown in Fig. 3. In Fig. 3, the abscissa represents the protective effect of the various gyrA mutations in the presence of a wild-type parC allele; the ordinate represents their protective effect in the presence of a parC resistance allele. Resistance due to the less protective gyrA mutations was unaffected by the parC resistance allele, presumably because mutant gyrase was more susceptible than wild-type topoisomerase IV. The curves sharply increase at the point where wild-type topoisomerase IV becomes more susceptible than mutant gyrase—at that point, the parC resistance allele contributes to resistance. The magnitude of protection provided by the parC resistance allele should depend on how well each fluoroquinolone attacks resistant gyrase, since mutant gyrase was the more sensitive target in the double mutant, as indicated by selection of a triple mutant in which the third mutation mapped in the quinolone resistance-determining region of GyrA (data not shown). By this criterion gatifloxacin was more potent than the two other compounds (the parC resistance allele protected least against gatifloxacin [Fig. 3]).
FIG. 3.
Relationship between bacteriostatic activities of fluoroquinolones against gyrA mutants and gyrA parC mutants. The MIC99s for gyrA (Fig. 2A) and gyrA parC (Fig. 2B) mutants, normalized to the MIC99s of wild-type and parC mutant cells, respectively, as described in the legend for Fig. 2, were plotted such that each data point represents one gyrA allele with wild-type and mutant parC alleles. Symbols: ■, gatifloxacin; □, AM1121; and ○, ciprofloxacin. Arrows indicate values for the D82A gyrA allele.
The point (gyrA allele) in Fig. 3 at which the parC resistance allele began to be protective, i.e., the point where parC-dependent resistance increased sharply, depended on how effectively each compound attacked wild-type topoisomerase IV. Thus, gatifloxacin was more active against wild-type topoisomerase IV than AM1121 or ciprofloxacin. The mutation at position 82 showed anomalous behavior (Fig. 3): for each fluoroquinolone tested, the addition of a parC allele was exceptionally protective (see Discussion).
To assess the effect of the C-8-methoxy group on resistance due to each gyrA allele, we divided the normalized MIC99 for gatifloxacin by that for AM1121, obtained with gyrA parC double mutants to minimize the contribution of topoisomerase IV. As shown in Fig. 4, the C-8-methoxy group facilitated attack of alleles in the core of the quinolone-resistance-determining region (positions 81-87). Those lying outside the core were affected little by the methoxy group (Fig. 4). The A84P allele, an α-helix 4 mutation, was unresponsive to the methoxy group (Fig. 4). Thus, specific changes in fluoroquinolone structure elicit effects that depend on GyrA structure.
FIG. 4.
Effect of fluoroquinolone structure on bacteriostatic activity against resistant gyrase. Normalized MIC99s of gyrA parC mutants (Fig. 2B) are expressed as gatifloxacin/AM1121 MIC99 ratios. Filled bars represent alleles located in α-helix 4 according to reference 14; open bars represent alleles outside α-helix 4. The values shown are the averages of two independent determinations. Error bars indicate the upper ranges of the determinations.
Bactericidal effects of fluoroquinolone C-8-methoxy group against resistant gyrase.
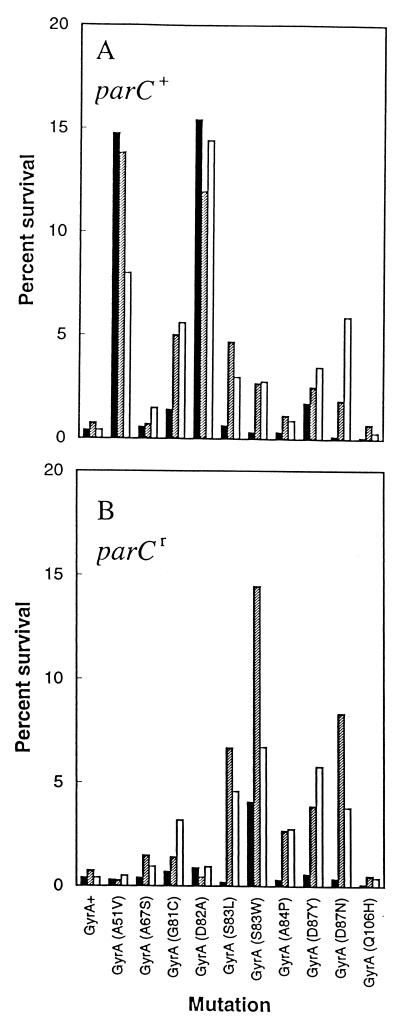
To compare lethal activity among the three compounds for the set of mutants, we measured survival rates at specific multiples of the MIC99s to minimize differences in bacteriostatic activity, which probably reflects formation of quinolone-gyrase-DNA complexes (4). Pilot experiments (data not shown) indicated that the three compounds killed wild-type (strain DM4100) and mutant (strains KD66 and KD1366) cells extensively within 2 h at fluoroquinolone concentrations 10 times higher than the MIC99s. Under these conditions the paradoxical increase in survival rates at high concentrations (reviewed in reference 19) was not observed. Gatifloxacin killed a greater fraction of cells than its C-8-H derivative or ciprofloxacin (Fig. 5A) for all mutants except strains containing A51V and D82A gyrase mutations. By introducing a parC resistance allele we minimized attack of topoisomerase IV, and again gatifloxacin was the most lethal of the three compounds (Fig. 5B). The A51V and D82A alleles proved to be exceptions, with AM1121 showing more activity than gatifloxacin. With some mutants ciprofloxacin was more lethal than AM1121, while the reverse was true with others. Survival after treatment with gatifloxacin was only 2 to 50% of that with AM1121 and 10 to 60% of that with ciprofloxacin.
FIG. 5.
Bactericidal action of fluoroquinolones against resistant mutants. The survival rates of the indicated mutant strains of E. coli were determined following a 2-h incubation in gatifloxacin (filled bars), AM1121 (shaded bars), or ciprofloxacin (open bars) at 10 times the MIC99 for each compound. (A) gyrA mutant strains; (B) gyrA parC double mutants. The data shown are the averages of two determinations.
Effect of chloramphenicol on bactericidal activity of fluoroquinolones.
Fluoroquinolones cause two types of lethal activity, one that requires protein synthesis and one that does not (reviewed in reference 5). With single gyrA mutants gatifloxacin was from 2- to 40-fold more lethal than AM1121 or ciprofloxacin when chloramphenicol was included to block protein synthesis (Fig. 6A). Exceptions were seen with the A67S and S83W alleles; for these there was little difference among the compounds. When the gyrA parC double mutants were examined, gatifloxacin was about twice as effective as the two other compounds, except in the case of the S83W mutation where not much difference was seen (Fig. 6B). Thus, the C-8-methoxy group improved lethal action against most of the gyrA mutants, as was the case when inhibition of growth was measured.
FIG. 6.
Bactericidal action of fluoroquinolones against resistant gyrA and gyrA parC mutants in the presence of chloramphenicol. The survival rates of mutants were determined following a 2-h incubation in chloramphenicol plus gatifloxacin (filled bars), AM1121 (shaded bars), or ciprofloxacin (open bars) at 10 times the respective MIC99s (A) gyrA mutant strains; (B) gyrA parC double mutant strains. The data shown are the averages of two determinations.
DISCUSSION
The work described above compared gatifloxacin, a C-8-methoxy fluoroquinolone, with two structurally related C-8-H compounds for the ability to attack a wide variety of resistant gyrA mutants of E. coli. Since C-8-methoxy and C-8-H compounds are not equally active against wild-type gyrase or wild-type E. coli (18, 21, 25), we normalized all mutant MICs to wild-type values. In some experiments we also included a resistant parC allele to minimize the contribution of a secondary target, DNA topoisomerase IV. For 9 out of 10 gyrA alleles, gatifloxacin exhibited more normalized bacteriostatic activity than the two C-8-H compounds (Fig. 2). Differences caused by the introduction of the parC resistance allele (compare Fig. 2A and B) indicate that wild-type topoisomerase IV can become a target when the gyrase allele provides sufficient resistance. The least protective gyrA mutant with which resistant parC had a substantial effect reflected the susceptibility of wild-type topoisomerase IV to each compound. That value was lowest for gatifloxacin (Fig. 3), which indicated that the C-8-methoxy group enhances attack of wild-type topoisomerase IV. Once gyrase became resistant enough to make wild-type topoisomerase IV the main target, the extent of protection afforded by the parC mutation depended on the gyrA allele and the fluoroquinolone. The least protection was observed with gatifloxacin (Fig. 3).
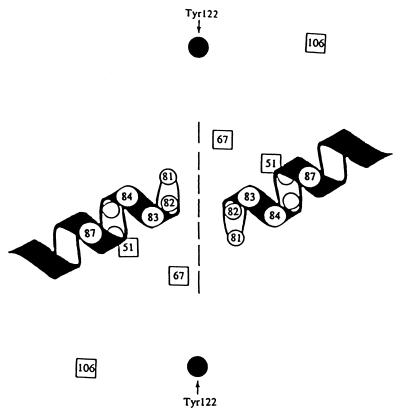
The response of individual mutations to the presence of a C-8-methoxy group can be rationalized by separating the alleles into two categories, those that lie in α-helix 4 of the GyrA protein and those that do not (Fig. 7). For mutations mapping in the helix, resistance to bacteriostatic action was decreased substantially by a C-8-methoxy group, while those mapping outside the helix were relatively insensitive to the group (Fig. 4). These data, plus the observation that mutations in α-helix-4 confer the highest levels of resistance, suggest that α-helix-4 may be part of a quinolone-binding site. Substitution of Leu or Trp for Ser-83 and Asn or Tyr for Asp-87 may confer quinolone resistance by making the microenvironment of α-helix 4 less electron-rich and less able to bind quinolone. C-8-methoxy or C-8-halogenated fluoroquinolones may reduce resistance by partially restoring electron richness to the region.
FIG. 7.
Relative positions of resistance alleles in GyrA dimer. The numbers indicate amino acid positions in the E. coli GyrA protein for resistance mutations and for the active center tyrosine (solid circles). The resistance alleles that reside within α-helix 4 are shown as open circles; residues outside the helix that confer resistance are shown as open squares. The dashed line approximates the interface of the two GyrA subunits. DNA is predicted to lay across the protein at an angle from upper left to lower right such that the two helices fit in the major groove of DNA (14).
Substitution of proline for alanine at codon 84 generated a helix mutant that behaved like the nonhelix alleles (Fig. 4). Unlike other substitutions that confer resistance, a proline is expected to disrupt helix structure. Thus, it is not surprising that the effect of A84P differs from the mutations that probably change local affinity for a quinolone. Perturbation of the helix between positions 83 and 87 might alter the alignment between these putative quinolone-binding sites. According to this idea, placing a proline at position 85 or 86, two positions that lie on the bottom surface of the helix and are not generally associated with quinolone resistance, should have the same effect as seen at position 84.
Comparison of lethal action revealed that gatifloxacin was superior to the two C-8-H compounds for most of the gyrA mutants examined even when cells were treated with chloramphenicol. An exception was observed when tryptophan was substituted for serine at codon 83: little difference was seen between gatifloxacin and AM1121 (Fig. 6). These observations should contribute to a better understanding of quinolonegyrase-DNA complexes when more structural information becomes available.
Resistance associated with mutation of amino acid 82 of GyrA exhibited several unusual features. First, position 82 gyrase mutations arise rarely, if at all, unless topoisomerase IV is also resistant (25). Second, introduction of parC-mediated resistance into a D82A gyrase mutant rendered the double mutant exceptionally resistant when bacteriostatic action was measured (Fig. 3). Third, when lethal activity was measured, the D82A allele was among the more resistant alleles until a parC resistance allele was added (Fig. 5). These observations, which are still unexplained, may reflect special features of this amino acid due to its position at the GyrA dimer interface.
The results described above have several practical implications. First, enhanced ability to block mutant growth and kill mutant cells is expected to restrict the selection of resistant mutants, as observed with fluoroquinolones having structures similar to gatifloxacin (2, 3). Second, the C-8-methoxy group improves activity against gyrA parC double mutants, as revealed by comparison of gatifloxacin and AM1121. Consequently, a concentration should exist at which wild-type cells would need to acquire three mutations to express resistance. For E. coli that concentration is about 1 μg/ml for the most resistant mutant we examined. This value is within the range of serum concentrations normally achieved by fluoroquinolones (10). If the serum concentration is kept above that level, no mutant would be selected (see reference 3). Finally, the C-8-methoxy group improved the attack of topoisomerase IV (as shown by a comparison of results for gatifloxacin and AM1121 [Fig. 3]), which should make gatifloxacin better able to attack species in which topoisomerase IV is the primary quinolone target. Thus, compounds such as gatifloxacin are likely to have widespread application.
ACKNOWLEDGMENTS
We thank M. Gennaro and S. Kayman for critical reading of the manuscript and S. M. Friedman and S. Nakamura for providing mutant strains.
This work was supported by grants from NIH (AI 35257) and Bristol-Meyers Squibb.
REFERENCES
- 1.Chen C-R, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- 2.Dong Y, Xu C, Zhao X, Domagala J, Drlica K. Fluoroquinolone action against mycobacteria: effects of C8 substituents on bacterial growth, survival, and resistance. Antimicrob Agents Chemother. 1998;42:2978–2984. doi: 10.1128/aac.42.11.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dong Y, Zhao X, Domagala J, Drlica K. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1756–1758. doi: 10.1128/aac.43.7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drlica K. Mechanism of fluoroquinolone action. Curr Opin Microbiol. 1999;2:504–508. doi: 10.1016/s1369-5274(99)00008-9. [DOI] [PubMed] [Google Scholar]
- 5.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman S M, Malik M, Drlica K. DNA supercoiling in a thermotolerant mutant of Escherichia coli. Mol Gen Genet. 1995;248:417–422. doi: 10.1007/BF02191641. [DOI] [PubMed] [Google Scholar]
- 7.Ito T, Matsumoto M, Nishino T. Improved bactericidal activity of Q-35 against quinolone-resistant staphylococci. Antimicrob Agents Chemother. 1995;39:1522–1525. doi: 10.1128/aac.39.7.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kampranis S, Maxwell A. Conformational changes in DNA gyrase revealed by limited proteolysis. J Biol Chem. 1998;273:22606–22614. doi: 10.1074/jbc.273.35.22606. [DOI] [PubMed] [Google Scholar]
- 9.Kampranis S, Maxwell A. The DNA gyrase-quinolone complex, ATP hydrolysis and the mechanism of DNA cleavage. J Biol Chem. 1998;273:22615–22626. doi: 10.1074/jbc.273.35.22615. [DOI] [PubMed] [Google Scholar]
- 10.Karabalut N, Drusano G L. Pharmacokinetics of the quinolone antimicrobial agents. In: Hooper D C, Wolfson J S, editors. Quinolone antimicrobial agents. Washington, D.C.: American Society for Microbiology; 1993. pp. 195–223. [Google Scholar]
- 11.Khodursky A B, Zechiedrich E L, Cozzarelli N R. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitamura A, Hoshino K, Kimura Y, Hayakawa I, Sato K. Contribution of the C-8 substituent of DU-6859a, a new potent fluoroquinolone, to its activity against DNA gyrase mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1467–1471. doi: 10.1128/aac.39.7.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. p. 466. [Google Scholar]
- 14.Morais-Cabral J H, Jackson A P, Smith C V, Shikotra N, Maxwell A, Liddington R C. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 15.Nakamura S. Mechanisms of quinolone resistance. J Infect Chemother. 1997;3:128–138. [Google Scholar]
- 16.Ng E Y, Trucksis M, Hooper D C. Quinolone resistance mutations in topoisomerase IV: relationship to the flqA locus and genetic evidence that topoisomerase IV is the primary target and DNA gyrase is the secondary target of fluoroquinolones in Staphylococcus aureus. Antimicrob Agents Chemother. 1996;40:1881–1888. doi: 10.1128/aac.40.8.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pruss G, Franco R, Chevalier S, Manes S, Drlica K. Effects of DNA gyrase inhibitors in Escherichia coli topoisomerase I mutants. J Bacteriol. 1986;168:276–282. doi: 10.1128/jb.168.1.276-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schedletzky H, Wiedemann B, Heisig P. The effect of moxifloxacin on its target topoisomerases from Escherichia coli and Staphylococcus aureus. J Antimicrob Chemother. 1999;43(Suppl. B):31–37. doi: 10.1093/jac/43.suppl_2.31. [DOI] [PubMed] [Google Scholar]
- 19.Smith J. The mode of action of 4-quinolones and possible mechanisms of resistance. J Antimicrob Chemother. 1986;18(Suppl. D):21–29. doi: 10.1093/jac/18.supplement_d.21. [DOI] [PubMed] [Google Scholar]
- 20.Sternglanz R, DiNardo S, Voelkel K A, Nishimura Y, Hirota Y, Becherer A K, Zumstein L, Wang J C. Mutations in the gene coding for Escherichia coli DNA topoisomerase I affecting transcription and transposition. Proc Natl Acad Sci USA. 1981;78:2747–2751. doi: 10.1073/pnas.78.5.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takei M, Fukuda H, Yasue T, Hosaka M, Oomori Y. Inhibitory activities of gatifloxacin (AM-1155), a newly developed fluoroquinolone, against bacterial and mammalian type II topoisomerases. Antimicrob Agents Chemother. 1998;42:2678–2681. doi: 10.1128/aac.42.10.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao B-Y, Pine R, Domagala J, Drlica K. Fluoroquinolone action against clinical isolates of Mycobacterium tuberculosis: effects of a C8-methoxyl group on survival in liquid media and in human macrophages. Antimicrob Agents Chemother. 1999;43:661–666. doi: 10.1128/aac.43.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao X, Wang J-Y, Xu C, Dong Y, Zhou J, Domagala J, Drlica K. Killing of Staphylococcus aureus by C-8-methoxy fluoroquinolones. Antimicrob Agents Chemother. 1998;42:956–958. doi: 10.1128/aac.42.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao X, Xu C, Domagala J, Drlica K. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc Natl Acad Sci USA. 1997;94:13991–13996. doi: 10.1073/pnas.94.25.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]