Abstract
Introduction
To describe the protocol and findings of the instrumental validation of three imaging‐based biomarker kits selected by the MarkVCID consortium: free water (FW) and peak width of skeletonized mean diffusivity (PSMD), both derived from diffusion tensor imaging (DTI), and white matter hyperintensity (WMH) volume derived from fluid attenuation inversion recovery and T1‐weighted imaging.
Methods
The instrumental validation of imaging‐based biomarker kits included inter‐rater reliability among participating sites, test–retest repeatability, and inter‐scanner reproducibility across three types of magnetic resonance imaging (MRI) scanners using intra‐class correlation coefficients (ICC).
Results
The three biomarkers demonstrated excellent inter‐rater reliability (ICC >0.94, P‐values < .001), very high agreement between test and retest sessions (ICC >0.98, P‐values < .001), and were extremely consistent across the three scanners (ICC >0.98, P‐values < .001).
Discussion
The three biomarker kits demonstrated very high inter‐rater reliability, test–retest repeatability, and inter‐scanner reproducibility, offering robust biomarkers suitable for future multi‐site observational studies and clinical trials in the context of vascular cognitive impairment and dementia (VCID).
Keywords: biomarker, diffusion tensor imaging, free water, magnetic resonance imaging, peak‐width skeletonized mean diffusivity, small vessel disease, vascular contributions to cognitive impairment and dementia, VCID, white matter hyperintensity, white matter injury
1. BACKGROUND
Vascular contributions to cognitive impairment and dementia (VCID), along with Alzheimer's disease (AD) and mixed pathologies, are predominant contributors to cognitive decline and increased risk of dementia. 1 , 2 Although efforts have been deployed to develop biomarkers of VCID for use in larger scale, multicenter studies including clinical trials, the lack of technical validation, including estimates of repeatability and reproducibility, disqualifies these biomarkers for use in clinical trials by regulatory agencies. 3 In recent efforts to improve early identification, staging, and prediction of risk of persons with small vessel disease (SVD) at risk for VCID for inclusion in clinical trials, the US National Institutes of Health (NIH) National Institute on Neurological Disorders and Stroke (NINDS) funded the MarkVCID consortium (https://markvcid.partners.org/) to identify and validate fluid‐ and imaging‐based biomarkers for the small vessel diseases associated with VCID.
The MarkVCID consortium has developed standardized multi‐site protocols for participant enrollment, clinical and cognitive testing, handling of fluid samples, 4 and acquisition of neuroimaging data. 4 , 5 Furthermore, the consortium has also selected a panel of 11 fully specified biomarker protocols (or “biomarker kits”) for the validation of their instrumental performance (reliability across users, sites, and time points) and clinical significance (association with clinically meaningful aspects of VCID including, for example, cognitive function or dementia rating scales).
The MarkVCID consortium selected seven imaging‐based biomarker kits as part of this process. The present paper describes the results from the instrumental validation for three of these kits: free water fraction (FW) and peak‐width of skeletonized mean diffusivity (PSMD), both measures derived from diffusion tensor imaging (DTI) data, and white matter hyperintensity (WMH) burden derived from T1‐weighted and fluid attenuation inversion recovery (FLAIR) imaging data. FW refers to the extracellular water content contained in a voxel of the white matter (WM) tissue; reflects the amount of water molecules that are relatively unrestricted by their local microenvironment; 6 and is promoted by neuroinflammation, which increases interstitial extraneuronal space. 7 FW has been associated with vascular risk factors, including arterial stiffness and blood pressure in adult individuals 8 and with a network of inflammatory biomarkers in older individuals. 9 FW has also been reported to correlate with cognition status and its trajectory in older individuals, including cognitively normal, mild cognitive impairment, and AD individuals. 10 , 11 PSMD represents the peak‐width distribution of mean diffusivity (MD) between the 5th and 95th percentile of skeletonized FA WM tracts, with higher values indicating greater water dispersion, which has been shown to correlate with general WM microstructural damage. 12 PSMD explains a substantial proportion of variance in processing speed, the cognitive domain predominantly affected in SVD, 13 as well as visuospatial performance and general cognitive ability. 14 WMH is considered one of the paradigmatic markers of cerebrovascular disease. Multiple cross‐sectional studies, including population‐based studies, have found significant associations between age and cerebrovascular risk factors—particularly hypertension—and WMH burden. 15 , 16 Prospective longitudinal studies also show associations between WMH burden and memory and executive function decline, 17 future risk of stroke, mild cognitive impairment, dementia, and death. 18
This study reports the protocol and findings for the instrumental validation of the FW, PSMD, and WMH kits. For the purposes of MarkVCID imaging‐based biomarkers, instrumental validation is operationally defined as follows: (1) inter‐rater reliability (differences between raters at different sites analyzing the same magnetic resonance imaging [MRI] dataset), (2) test–retest repeatability (differences between two scans obtained for the same individual and MRI scanner within 14 days), and (3) inter‐scanner reproducibility (differences across different MRI scanners in the same group of individuals).
2. METHODS
The data used in this study were acquired as part of the MarkVCID consortium. 5 MarkVCID is a consortium of seven sites: Johns Hopkins University School of Medicine (JHU); Rush University Medical Center/Illinois Institute of Technology (RUSH); Universities of California San Francisco, Davis, and Los Angeles (UCSF/UCD/UCLA); University of Kentucky (UKY); University of New Mexico Health Sciences Center (UNM); University of Southern California (USC) and the University of Texas Health Science Center at San Antonio (UTHSCSA, operating as part of the Cohorts for Heart and Aging Research in Genomic Epidemiology [CHARGE] consortium site); and a central coordinating center (Massachusetts General Hospital) working with NINDS and the National Institute on Aging (NIA) under cooperative agreements. 3T scanners used by the different sites included two Siemens systems (TIM Trio and Prisma) and one Philips system (Achieva). The participants included in this study were in the age range of 53 to 78 years. We excluded participants with unstable major medical illness, major primary psychiatric disorder, prevalent stroke at the MRI assessment, or other neurological disorders that might confound the assessment of brain volumes.
2.1. MRI acquisition protocol
Protocols for MarkVCID sequences, including DTI, 3D T1‐weighted, and FLAIR, have been previously described 19 . DTI sequences are used to derive FW and PSMD. To balance accuracy and scan time, the MarkVCID DTI protocol uses a single‐shell (b = 1000 s/mm2), 40‐direction diffusion sequence with a voxel size of 2.0 × 2.0 ×2.0 mm3 and six b = 0 s/mm2. The reverse polarity data were used to estimate and correct image distortions in the DTI data.
FLAIR and T1‐weighted MRI are included to evaluate WMH of the brain. A high‐resolution 3D FLAIR with a sagittal‐plane acquisition and a voxel size of 1.0 × 1.0 × 1.0 mm3 is used. The three‐dimensional T1‐weighted multi‐echo magnetization‐prepared rapid‐acquisition‐of‐gradient‐echo (ME‐MPRAGE) uses a multi‐echo version of MPRAGE with a sagittal‐plane acquisition, four echoes, and a voxel size of 1.0 × 1.0 × 1.0 mm3.
Imaging parameters for MarkVCID sequences on different MRI models (Philips, Siemens Trio, and Prisma) have been previously described. 5 Although none of the consortium's prospective enrollment sites use a General Electric (GE) model MRI, additional sequence parameters were developed for the GE (750W) scanner 5 to enhance inter‐scanner generalizability. The most substantial difference in GE data, in terms of acquisition parameters and compared to data obtained with Philips, Siemens Trio, and Siemens Prisma systems, resides in the GE default use of zero‐filling to expand the image matrix size in the phase‐encoded direction.
RESEARCH IN CONTEXT
Systematic Review: This article describes the instrumental validation of three of the imaging‐based biomarkers selected by the MarkVCID consortium. To assess the reliability, repeatability, and reproducibility properties of the biomarkers, the group reviewed existing publicly available methodological papers. References to these sources are appropriately cited.
Interpretation: Our results indicate that free water fraction (FW), peak‐width of skeletonized mean diffusivity (PSMD), and white matter hyperintensity (WMH) measures are highly reliable among raters, reproducible between test and retest sessions, and consistent across different magnetic resonance imaging (MRI) scanners, offering promising avenues for future multi‐site observational studies and clinical trials in the context of vascular contributions to cognitive impairment and dementia (VCID).
Future Directions: The next step consists, for each kit, to evaluate the kit clinical validation by investigating associations between kit measures with clinically meaningful aspects of VCID including, for example, cognitive function and relevant co‐morbidities.
HIGHLIGHTS
Methods for three imaging‐based biomarkers of small vessel brain disease.
Formal process of imaging‐based biomarker qualification for clinical trial.
Inter‐rater reliability, test–retest repeatability, and inter‐scanner reproducibility.
The institutional review boards at all participating institutions approved this study, and subjects or their legal representatives gave written informed consent.
2.2. MRI processing
Participating sites preprocessed DTI datasets using FSL software tools 20 including correction for eddy current‐induced distortions and participants’ head movements. The brain was masked using the BET tool and fractional anisotropy (FA), MD, axial diffusivity (AD), and radial diffusivity (RD) maps generated using DTIFIT (FMRIB software library; http://fsl.fmrib.ox.ac.uk/fsl/fslwiki). Although presented together in this study, FW and PSMD kits are two independent tools but may use common processing steps.
2.2.1. FW kit
The kit requires, as inputs, a 4D DTI volume (corrected for eddy current distortion), a brain mask, and the b‐vector and b‐values files. The model considers two co‐existing compartments per voxel: one compartment is a FW compartment, which models isotropic diffusion with a diffusion coefficient of water at body temperature (37°C) fixed to 3 × 10−3 mm2/s. 21 The FW fraction is expected to predominantly highlight water molecules in the extracellular space. The second compartment is the tissue compartment, which accounts for all other molecules, that is, all intra‐ and extracellular molecules that are hindered or restricted by tissue membranes. 6 The script contains the following steps: (1) the tissue compartment is modeled by a diffusion tensor characterizing the “tissue” molecules, as well as the fractional volume of the FW compartment in each voxel, resulting in the FW fraction map; (2) the individual FA map obtained from DTIFIT is linearly and non‐linearly registered to the standard FSL FA template space (FMRIB 1‐mm FA template) using linear and nonlinear transformations, (3) the resulting transformation parameters are applied to the FW map, (4) a WM mask is defined by thresholding the FSL FA template at a value of 0.3 to reduce cerebrospinal fluid (CSF) partial volume contamination, 22 (5) an overall measure of mean FW is computed by superimposing the WM mask onto the individual coregistered FW fraction map and averaging values within these WM voxels and (6) an overall measure of mean FW fraction is stored in a text file.
2.2.2. Peak width of skeletonized mean diffusivity (PSMD) kit
The kit follows the PSMD method previously described 12 . Briefly, it requires FA, MD, RD, and AD maps. The script procedure includes the following steps: (1) the FA volume is linearly and non‐linearly registered to the standard space FMRIB FSL 1‐mm FA template; (2) a WM skeleton is created using the standard Tract‐Based Spatial Statistics (TBSS) 23 pipeline available in FSL; (3) subject's FA data are then projected onto the skeleton, which is derived from the standard space template thresholded at an lower‐bound FA value of 0.2 to exclude predominantly non‐WM voxels; 22 (4) MD volume is projected onto the mean FA skeleton using the FA‐derived projection parameters and further thresholded with a template skeleton mask to reduce cerebrospinal fluid (CSF) partial volume contamination; (5) PSMD is calculated as the difference between the 95th and 5th percentiles of the voxel‐based MD values within the subject's MD skeleton; and (6) PSMD is stored in a text file.
2.2.3. WMH kit
The WMH kit requires only three inputs: high‐resolution 3D T1‐weighted image, a raw FLAIR image, and a binary brain mask. The algorithm will return a four‐component (CSF, gray matter, WM, and WMH) grayscale segmented image volume in the native space of the 3D T1‐weighted volume along with a segmented mask of WMH. A step‐by‐step analytic plan has been described previously. 24 It includes (1) linear co‐registration of 3D T1‐weighted image to the FLAIR image, (2) removal of non‐brain elements from the FLAIR image using 3D T1‐weighted brain mask, (3) image intensity normalization of the FLAIR image, (4) non‐linear warping of 3D T1‐weighted brain image to a minimal deformation template, 25 (5) non‐linear deformation of the FLAIR volume to the atlas using the registration parameters for the 3D T1‐weighted volume, (6) application of Bayesian segmentation to both 3D T1‐weighted and the FLAIR volumes, (7) creation of four‐tissue segmentation volume, (8) reverse transformation of three‐tissue segmented volume into 3D T1‐weighted native space, (9) reverse transformation of WMH segmented volume into FLAIR native space, (10) reverse transformation of four‐tissue segmented volume into 3D T1‐weighted native space, (11) output of these volumes into the directory from which the program is launched.
Each of these imaging‐biomarker kits includes a protocol, the script, and instructions; these are available on the MarkVCID website (https://markvcid.partners.org/consortium‐protocols‐resources). FW, PSMD, and WMH processing takes ≈ 4, 10, and 90 minutes, respectively, with a standard desktop computer.
2.3. Datasets
2.3.1. Inter‐rater reliability
The objective of this analysis was to investigate whether different raters (individual MarkVCID site staff) reported consistent FW, PSMD, and WMH measures from the same sample. This sample consisted of 20 participants imaged with the full MarkVCID MRI protocol and selected to cover the full range of SVD severities based on their WMH burden quartiles 24 . DTI sequences, as well as FLAIR/T1 scans, were distributed to each participating site (see Table S1 in supporting information). One individual DTI acquisition had poor image quality and was excluded from the FW and PSMD inter‐rater analyses (see Figure SA in supporting information). Participating sites analyzed these data and shared outputs with the kit lead site, resulting in 7 sites × 19 subjects FW and PSMD measurements and 7 sites × 20 subjects WMH measures.
2.3.2. Test–retest repeatability
To evaluate FW, PSMD, and WMH test–retest repeatability measurements, each actively enrolling participating site recruited a subset of individuals to return for a second MRI using the same scanner and protocol within 1 to 14 days after their initial MRI. For the FW and PSMD kits, repeated DTI datasets were obtained for five (UKY) and six (UCD/UCSF/UCLA, UNM, USC, UTHSCSA, JHU, UKY, and RUSH) individuals. For the WMH kit, repeated FLAIR/T1 datasets were obtained for six individuals for the seven sites. Sites computed FW, PSMD, and WMH metrics for their own test–retest sample and shared outputs with the lead kit site, resulting in a total sample size of 82 FW and PSMD measurements ([6 sites × 6 subjects + 1 site × 5 subjects] × 2 exams) and 84 WMH measures ([7 sites × 6 subjects] × 2 exams).
2.3.3. Inter‐scanner reproducibility
Inter‐scanner reproducibility was assessed by a series of cross‐model MRI scans acquired on 20 individuals, stratified to include 10 with no‐to‐low SVD burden and 10 with moderate‐to‐high SVD assessed by Fazekas Scale 26 scores on previously obtained MRI scans. 5 Each participant was scanned on three MarkVCID sites’ MRI scanners, including Philips Achieva, Siemens Trio, and Siemens Prisma. For the DTI‐derived kits, four individuals were excluded for the following reasons: one individual did not undergo the exam on the Philips machine, the reverse polarity acquisition for another individual (Siemens Trio) was missing, one dataset (Siemens Trio) included only 10 diffusion directions instead of 40, and acquisition for one dataset had poor quality (Siemens Prisma, see Figure SB), resulting in a final sample size of 48 FW and PSMD measurements (3 scanners × 16 subjects). For the WMH kit, one individual did not undergo the Philips MRI exam and was therefore excluded, resulting in a final sample size of 57 WMH measures (3 scanners × 19 subjects).
Finally, although GE data used interpolated resolution for all its sequences, individuals participating in the inter‐scanner reproducibility study underwent an additional MRI exam on a GE scanner (see MRI acquisition section) to enhance generalizability of the kit inter‐scanner reproducibility.
The maximum time between the first and last scans on the four scanners for the same participant was 15 weeks. Data from the inter‐scanner reproducibility scans were transferred to the lead site for each biomarker kit, where they were processed and analyzed.
2.4. Statistical analyses
2.4.1. Inter‐rater reliability
To establish inter‐rater reliability of FW, PSMD, and WMH measurements, we computed intraclass correlation coefficient 27 (ICC) between sites using a two‐way (same raters) random‐effects model (same participants) with single measure and absolute agreement form, noted ICCAA and calculated as follows:
where MSR is the mean square for rows (i.e., participants), MSE the mean square error, MSC the mean square for columns (i.e., raters), n the number of measures, and k the number of raters. ICCAA estimates agreement between measures without allowing systematic error. Pairwise ICCAA for each pair of sites were also computed.
2.4.2. Test–retest repeatability
To evaluate test–retest repeatability of FW, PSMD, and WMH measurements, we computed ICCAA between test and retest FW, PSMD, and WMH measures using a two‐way random‐effects model with an absolute agreement form as described above.
2.4.3. Inter‐scanner reproducibility
Inter‐scanner reproducibility of FW, PSMD, and WMH measures among the three scanners was evaluated using consistency ICC (ICCC), which considers systematic error between measurements. It uses a two‐way random effects model with a consistency form, using single measures and calculated as follows:
Pairwise ICCC between each pair of scanners were also computed.
2.4.4. Generalizability of inter‐scanner reproducibility: GE (750W) scanner
We finally computed pairwise ICCC among FW, PSMD, and WMH measures generated from GE system with measures generated from the three scanners.
For visualization of outcome measures obtained from different sites (inter‐rater study), sessions (test–retest study), and scanners (inter‐scanner study), a Bland‐Altman 28 plot and scatterplot were used. The terms poor, moderate, good, and excellent were used to refer to ICC values < 0.5, between 0.5 and 0.75, between 0.75 and 0.9, and > 0.90, respectively. 29
3. RESULTS
3.1. Inter‐rater reliability
3.1.1. FW
Figure 1 illustrates measures of FW for 19 individuals according to each of the seven participating sites. Overall ICCAA between measures generated by the different sites was found to be 0.997, P < .001 (confidence interval [CI]: [0.993; 0.999]) indicating excellent reliability, that is, a very strong agreement, between the different raters (i.e., sites) when using the FW kit on the same DTI datasets. Pairwise ICCAA for each pair of sites ranged from 0.986 to 1 (P values < .001, see Table 1).
FIGURE 1.
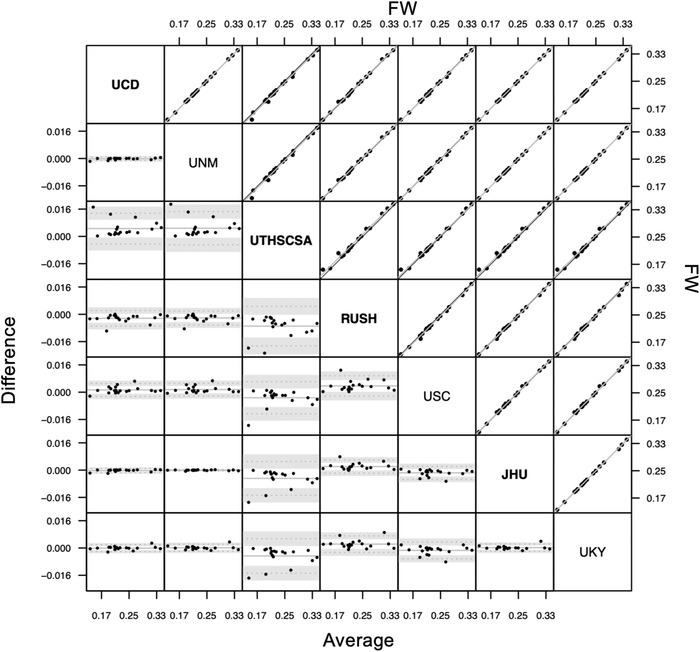
Bland‐Altman plot (lower triangle panel) and scatterplot (upper triangle panel) with identity and linear regression line (black and gray, respectively) of free water (FW) measures obtained from different sites (inter‐rater reliability study) in n = 19 participants. Bold or plain font for site's name indicate that sites used similar preprocessing methods (see Discussion section). JHU, Johns Hopkins University School of Medicine; RUSH, Rush University Medical Center/Illinois Institute of Technology; UCD, University of California Davis; UKY, University of Kentucky; UNM, University of New Mexico Health Sciences Center; USC, University of Southern California; UTHSCSA, University of Texas Health Science Center at San Antonio
TABLE 1.
Intra‐class coefficients of FW, PSMD, and WMH measures between sites (inter‐rater reliability study), sessions (test–retest repeatability study) and scanners (inter‐scanner reproducibility study)
| FW | PSMD | WMH | ||
|---|---|---|---|---|
| Inter‐rater | UCD‐UNM | 1 ([1; 1], P < .001) | 0.906 ([0.732; 0.965], P < .001) | 0.988 ([0.963; 0.996], P < .001) |
| UCD‐UTHSCSA | 0.993 ([0.933; 0.998], P < .001) | 0.991 ([0.977; 0.997], P < .001) | 0.944 ([0.863; 0.977], P < .001) | |
| UCD‐RUSH | 0.998 ([0.981; 1], P < .001) | 0.999 ([0.998; 1], P < .001) | 0.987 ([0.958; 0.995], P < .001) | |
| UCD‐USC | 0.999 ([0.996; 1], P < .001) | 0.907 ([0.709; 0.967], P < .001) | 0.989 ([0.97; 0.996], P < .001) | |
| UCD‐UKY | 1 ([1; 1], P < .001) | 0.908 ([0.746; 0.965], P < .001) | 0.989 ([0.968; 0.996], P < .001) | |
| UNM‐UTHSCSA | 0.992 ([0.932; 0.998], P < .001) | 0.914 ([0.735; 0.969], P < .001) | 0.979 ([0.949; 0.992], P < .001) | |
| UNM‐RUSH | 0.999 ([0.984; 1], P < .001) | 0.905 ([0.729; 0.965], P < .001) | 0.967 ([0.919; 0.987], P < .001) | |
| UNM‐USC | 0.999 ([0.994; 1], P < .001) | 0.992 ([0.981; 0.997], P < .001) | 1 ([0.999; 1], P < .001) | |
| UNM‐UKY | 1 ([1; 1], P < .001) | 0.998 ([0.995; 0.999], P < .001) | 0.999 ([0.997; 1], P < .001) | |
| UTHSCSA‐RUSH | 0.986 ([0.804; 0.997], P < .001) | 0.993 ([0.982; 0.997], P < .001) | 1 ([0.999; 1], P < .001) | |
| UTHSCSA‐USC | 0.994 ([0.977; 0.998], P < .001) | 0.914 ([0.705; 0.97], P < .001) | 0.966 ([0.918; 0.986], P < .001) | |
| UTHSCSA‐UKY | 0.992 ([0.941; 0.998], P < .001) | 0.914 ([0.75; 0.968], P < .001) | 0.968 ([0.922; 0.987], P < .001) | |
| RUSH‐USC | 0.996 ([0.934; 0.999], P < .001) | 0.904 ([0.703; 0.966], P < .001) | 0.973 ([0.935; 0.989], p < 0.001) | |
| RUSH‐UKY | 0.998 ([0.985; 0.999], P < .001) | 0.908 ([0.744; 0.965], P < .001) | 0.969 ([0.926; 0.988], P < .001) | |
| USC‐UKY | 0.999 ([0.996; 1], P < .001) | 0.988 ([0.97; 0.995], P < .001) | 0.907 ([0.781; 0.962], P < .001) | |
| UCD‐JHU | 1 ([1; 1], P < .001) | 0.999 ([0.998; 1], P < .001) | 0.999 ([0.997; 1], P < .001) | |
| UNM‐JHU | 1 ([1; 1], P < .001) | 0.909 ([0.742; 0.966], P < .001) | 1 ([0.999; 1], P < .001) | |
| UTHSCSA‐JHU | 0.992 ([0.929; 0.998], P < .001) | 0.994 ([0.985; 0.998], P < .001) | 0.966 ([0.917; 0.986], P < .001) | |
| RUSH‐JHU | 0.999 ([0.984; 1], P < .001) | 0.999 ([0.998; 1], P < .001) | 0.999 ([0.999; 1], P < .001) | |
| USC‐JHU | 0.999 ([0.993; 1], P < .001) | 0.911 ([0.72; 0.968], P < .001) | 0.966 ([0.918; 0.986], P < .001) | |
| UKY‐JHU | 1 ([0.999; 1], P < .001) | 0.91 ([0.754; 0.966], P < .001) | 0.967 ([0.919; 0.987], P < .001) | |
| Test–retest | 0.995 ([0.99; 0.997], P < .001) | 0.986 ([0.975; 0.993], P < .001) | 0.985 ([0.972; 0.992], P < .001) | |
| Inter‐scanner | Philips‐Siemens_Prisma | 0.977 ([0.936; 0.992], P < .001) | 0.968 ([0.912; 0.989], P < .001) | 0.966 ([0.914; 0.987], P < .001) |
| Philips‐Siemens_Trio | 0.945 ([0.85; 0.98], P < .001) | 0.956 ([0.878; 0.984], P < .001) | 0.973 ([0.931; 0.989], P < .001) | |
| Siemens_Prisma‐Siemens_Trio | 0.958 ([0.883; 0.985], P < .001) | 0.942 ([0.843; 0.979], P < .001) | 0.981 ([0.952; 0.993], P < .001) | |
| Philips‐GE | 0.966 ([0.905; 0.988], P < .001) | 0.956 ([0.879; 0.984], P < .001) | 0.713 ([0.394; 0.879], P < .001) | |
| Siemens_Prisma‐GE | 0.992 ([0.978; 0.997], P < .001) | 0.956 ([0.879; 0.984], P < .001) | 0.634 ([0.265; 0.841], P = .0013) | |
| Siemens_Trio‐GE | 0.949 ([0.861; 0.982], P < .001) | 0.919 ([0.784; 0.971], P < .001) | 0.716 ([0.4; 0.88], P < .001) | |
Abbreviations: FW, free water; GE, General Electric; JHU, Johns Hopkins University School of Medicine; PMSD, peak width skeletonized mean diffusivity; RUSH, Rush University Medical Center/Illinois Institute of Technology; UCD, University of California Davis; UKY, University of Kentucky; UNM, University of New Mexico Health Sciences Center; USC, University of Southern California; UTHSCSA, University of Texas Health Science Center at San Antonio; WMH, white matter hyperintensity.
3.1.2. PSMD
Figure 2 illustrates measures of PSMD for 19 individuals according to each of the seven participating sites. Overall ICCAA between measures generated by the different sites was found to be 0.945, P < .001 (CI: [0.897; 0.976]) indicating excellent reliability between the different raters. Pairwise ICCAA for each pair of sites ranged from 0.904 to 0.999 (P values < .001, see Table 1).
FIGURE 2.
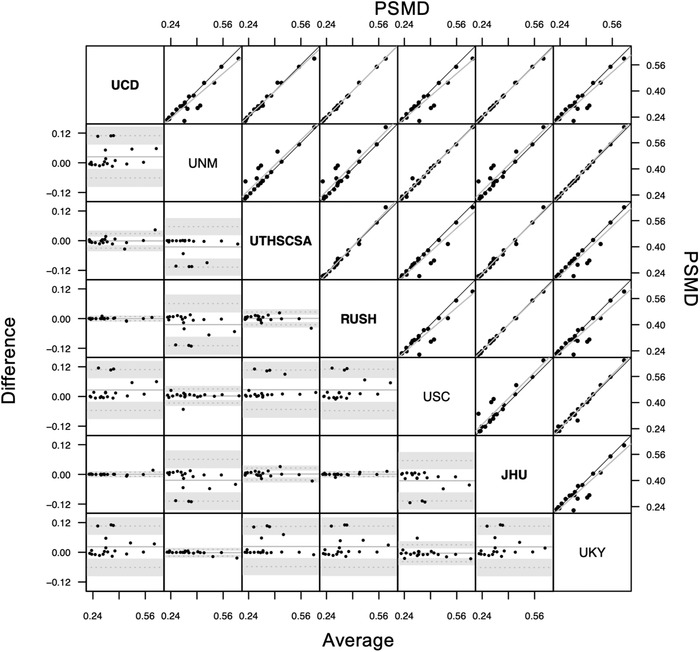
Bland‐Altman plot (lower triangle panel) and scatterplot (upper triangle panel) with identity and linear regression line (black and gray, respectively) of peak width skeletonized mean diffusivity (PSMD) measures obtained from different sites (inter‐rater reliability study) in n = 19 participants. Bold or plain font for site's name indicate that sites used similar preprocessing methods (see Discussion section). JHU, Johns Hopkins University School of Medicine; RUSH, Rush University Medical Center/Illinois Institute of Technology; UCD, University of California Davis; UKY, University of Kentucky; UNM, University of New Mexico Health Sciences Center; USC, University of Southern California; UTHSCSA, University of Texas Health Science Center at San Antonio
3.1.3. WMH
Figure 3 illustrates measures of WMH for 20 individuals according to each of the seven participating sites. Overall ICCAA between measures generated by the different sites was found to be 0.978, P < 0.001 (CI: [0.959; 0.990]) indicating excellent reliability between the different sites. Pairwise ICCAA for each pair of sites ranged from 0.944 to 1 (P values < .001, see Table 1).
FIGURE 3.
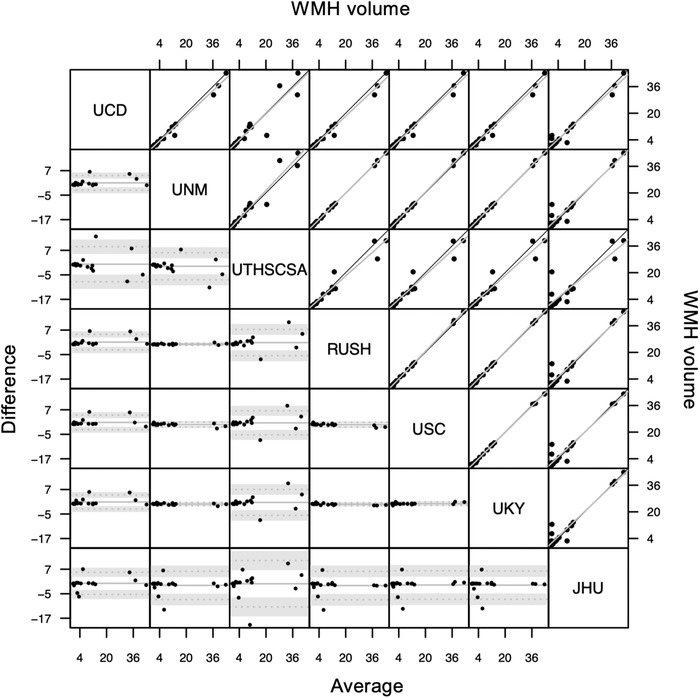
Bland‐Altman plot (lower triangle panel) and scatterplot (upper triangle panel) with identity and linear regression line (black and gray, respectively) of white matter hyperintensity (WMH) volumes obtained from different sites (inter‐rater reliability study) in n = 20 participants. JHU, Johns Hopkins University School of Medicine; RUSH, Rush University Medical Center/Illinois Institute of Technology; UCD, University of California Davis; UKY, University of Kentucky; UNM, University of New Mexico Health Sciences Center; USC, University of Southern California; UTHSCSA, University of Texas Health Science Center at San Antonio
3.2. Test–retest repeatability
3.2.1. FW
Figure 4A illustrates test and retest FW measures. ICCAA between test and retest measures was found to be 0.995, P < .001 (CI: [0.99; 0.997]) indicating an excellent agreement between test and retest measurements of FW.
FIGURE 4.
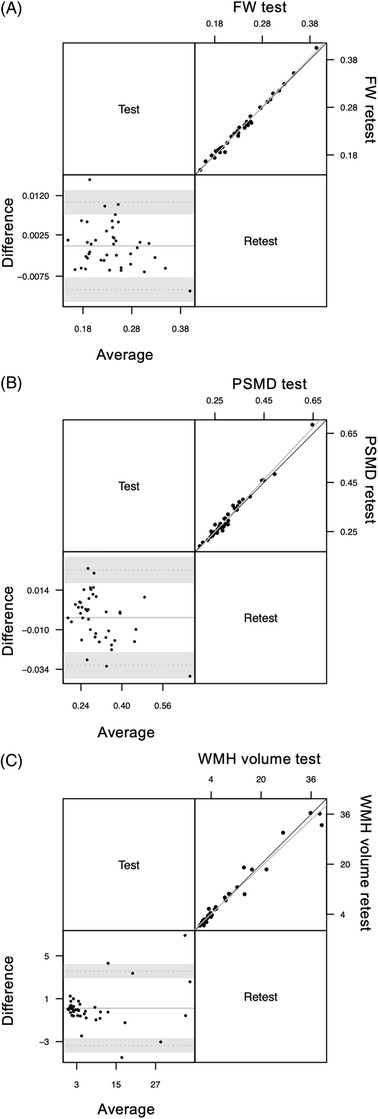
Bland‐Altman plot (lower triangle panel) and scatterplot (upper triangle panel) with identity and linear regression line (black and gray, respectively) of (A) free water (FW; n = 82 participants), (B) peak width skeletonized mean diffusivity (PSMD; n = 82 participants), and (C) white matter hyperintensity (WMH; n = 48 participants) volumes obtained at two different sessions (test–retest repeatability study)
3.2.2. PSMD
Figure 4B illustrates test–retest PSMD measures. ICCAA between test and retest measures was found to be 0.986, P < .001 (CI: [0.974; 0.993]) indicating excellent agreement between test and retest measurements of PSMD.
3.2.3. WMH
Figure 4C illustrates test and retest WMH measures. ICCAA for these measures was found to be 0.985, P < .001 (CI: [0.972; 0.992]) indicating excellent agreement between test and retest measurements of WMH.
3.3. Inter‐scanner reproducibility
3.3.1. FW
Figure 5A illustrates individual measures of FW for 16 individuals according to the three different scanners (Philips, Siemens Prisma, and Siemens Trio). Overall ICCC between FW measures from the three different scanners was found to be 0.96, P < .001 (CI: [0.991; 0.985]). Pairwise ICCC (see Table 1) were all significant: 0.945 ([0.85; 0.98], P < .001) for Philips‐Siemens Trio, 0.977 ([0.936; 0.992], P < .001) for Philips‐Siemens Prisma, and 0.958 ([0.883; 0.985], P < .001) for Siemens Trio‐Siemens Prisma (see Table 1), indicating excellent reproducibility.
FIGURE 5.
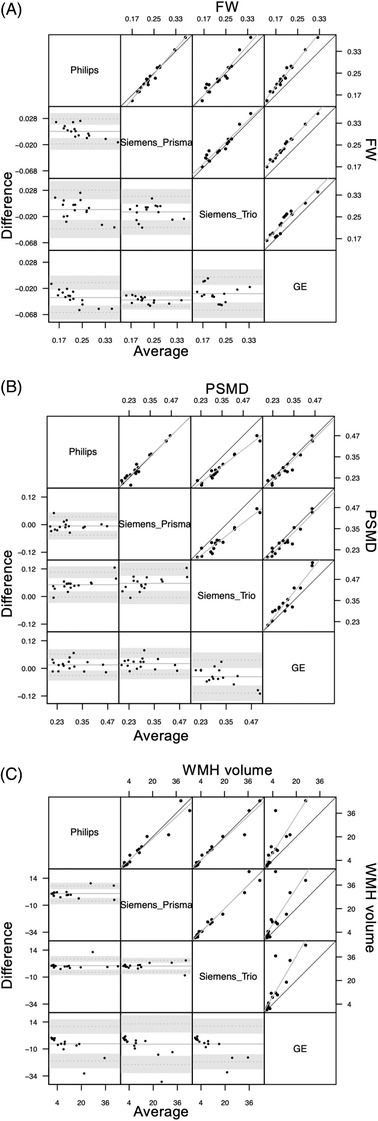
Bland‐Altman plot (lower triangle panel) and scatterplot (upper triangle panel) with identity and linear regression line (black and gray, respectively) of (A) free water (FW), (B) peak width skeletonized mean diffusivity (PSMD), and (C) white matter hyperintensity (WMH) volumes obtained at two different sessions (inter‐scanner reproducibility study) in n = 20 participants. Gray background is used for comparisons that involve General Electric interpolated data
3.3.2. PSMD
Figure 5B illustrates individual measures of PSMD for 16 individuals according to the three different scanners (Philips, Siemens Prisma, and Siemens Trio). Overall ICCC between PSMD measures from the three different scanners was found to be 0.954, P < .001 (CI: [0.899; 0.982]). Pairwise ICCC were all significant: 0.956 ([0.878; 0.984], P < .001) for Philips‐Siemens Trio, 0.968 ([0.912; 0.989], P < .001) for Philips‐Siemens Prisma, and 0.942 ([0.843; 0.979], P < .001) for Siemens Trio‐Siemens Prisma (see Table 1), indicating excellent reproducibility.
3.3.3. WMH
Figure 5C illustrates individual measures of WMH for 19 individuals according to the three different scanners. Overall ICCC between WMH measures generated using images from the three different scanners was found to be 0.974, P < .001 (CI: [0.944; 0.989]). Pairwise ICCC between WMH measures from the three scanners were all significant: 0.973 ([0.931; 0.989], P < .001) for Philips‐Siemens Trio, 0.966 ([0.914; 0.987], P < .001) for Philips‐Siemens Prisma, and 0.981 ([0.952; 0.993], P < .001) for Siemens Trio‐Siemens Prisma (see Table 1), indicating excellent reproducibility.
3.4. Generalizability to other scanners: GE analysis
3.4.1. FW
Pairwise ICCC between FW measures from a GE scanner with other scanners was found to be 0.992 ([0.978; 0.997], P < .001) with Siemens Prisma, 0.949 ([0.861; 0.982], P < .001) with Siemens Trio, and 0.966 ([0.905; 0.988], P < .001) with Philips (see Table 1). These results suggest that, even with interpolated resolution images, FW measurement reproducibility remains excellent.
3.4.2. PSMD
Pairwise ICCC between PSMD measures from a GE scanner with other scanners was found to be 0.956 ([0.879; 0.984], P < .001) with Siemens Prisma, 0.919 ([0.784; 0.971], P < .001) with Siemens Trio, and 0.956 ([0.879; 0.984], P < .001) with Philips Achieva (see Table 1). These results suggest that, even with interpolated resolution images, PSMD measurement reproducibility remains excellent.
3.4.3. WMH
Pairwise ICCC between WMH measures from a GE scanner with other scanners was found to be 0.634 ([0.265; 0.841], P = .0013) with the Siemens Prisma, 0.716 ([0.4; 0.88], P < .001) with the Siemens Trio machine, and 0.713 ([0.394; 0.879], P < 0.001) with Philips (see Table 1). These results suggest moderate reproducibility of WMH measurement compared to data acquired with GE using sequence parameters that were not optimized for the MarkVCID protocol.
4. DISCUSSION
Here we describe the protocols and instrumental validation findings of three of the imaging‐based biomarker kits selected by the MarkVCID consortium: FW, PSMD, and WMH. Protocol for DTI, FLAIR, and T1‐weighted image acquisitions used by these fully automated kits have been published 5 and FW, PSMD, and WMH kits, including script and instructions, are available on the MarkVCID website (https://markvcid.partners.org/consortium‐protocols‐resources). The instrumental validation protocol for imaging‐based biomarker kits was prespecified and included three studies: inter‐rater reliability, test–retest repeatability, and inter‐scanner reproducibility. This aspect is critically important to estimate measurement variability in multicenter studies and is part, along with the clinical validation and feasibility of implementation of a biomarker, of the framework for neuroimaging biomarker development as proposed by the Harmonizing Brain Imaging Methods for Vascular Contributions to Neurodegeneration (HARNESS) initiative. 3 While partial aspects of marker validation, such as repeatability and reproducibility, have already been evaluated in the context of VCID, 30 to our knowledge, this study is the first to cover the entire, formal process of imaging‐based biomarker qualification. 30 We discuss the instrumental validation findings for each biomarker below.
4.1. Inter‐rater reliability
Inter‐rater reliability evaluates differences between values obtained for MRI biomarker kits by raters at different sites analyzing the same MRI data. Each kit included a protocol, video tutorial, and example data that allowed investigators and staff from the participating sites to train on their own and to apply the kits to the 20 datasets shared by the MarkVCID coordinating center. ICC of absolute agreement of biomarkers between raters (i.e., participating sites), for MRI scanners present in the consortium, was above 0.94 for the three kits (see Results section) reflecting excellent inter‐rater reliability for FW, PSMD, and WMH kits. Because FW, PSMD, and WMH are generated by automatic algorithms, we expected that variability introduced by human interaction to be negligible. Potential variability may originate from the input files used by these kits: pre‐processed DTI data (i.e., data corrected for eddy current and brain masked) and brain mask for the FW and PSMD kits and T1 and FLAIR raw volumes and brain mask for the WMH kit. Although the inter‐rater reliability for PSMD was excellent, follow‐up discussions with participating sites revealed that a source of heterogeneity was that, whereas most images were collected at a 2 mm × 2 mm × 2 mm resolution, data sets from the GE machine were interpolated to a 1 mm × 1 mm × 2 mm resolution at the scanner. Some sites down‐sampled the data to 2 mm × 2 mm × 2 mm resolution before using the kit (UCSF, RUSH, JHU, and UTHSCSA). The sites that did not down‐sample the data had slightly higher PSMD values (see Figure 2). When only considering site pairs with identical pre‐processing, overall ICCAA was 0.996 (P < .001, [0.992; 0.998]) among UCSF, RUSH, JHU, and UTHSCSA and 0.993 (P < .001, [0.985; 0.997]) among UKY, UNM, and USC. Interestingly, FW was not found to be susceptible to whether data were down‐sampled or not during preprocessing (see Figure 1). Although preprocessing of DTI images is not part of the FW or PSMD kit, strict uniformity across sites can be obtained if the processing is encapsulated within a container. This step would also ensure that all sites use the same software versions. Although the use of a container will be considered in future kit versions, at present users are encouraged to carefully harmonize preprocessing steps, as well as check the quality of input images, including the presence of artefacts or the generation of the brain mask.
4.2. Test–retest repeatability
Test–retest repeatability assesses the degree of similarity between results for MRI biomarkers computed using scans of the same individual obtained on the same MRI scanner but on different days separated by a short time interval. Test–retest reliability of measurements is essential to establish what part of that variability observed in longitudinal measures is not related to short‐term scan‐to‐scan variations or non–kit‐related factors. Several factors can impact repeated intra‐subject measurements of MRI‐derived measures: imaging acquisition factors such as precise placement in the scanner, fluctuations in scanner function or hardware performance, or non–disease‐related participant physiological state variability 31 . FW, PSMD, and WMH kits all demonstrated excellent test–retest repeatability, with ICC of absolute agreement all above 0.98 (see Table 1), indicating that these measures remained unchanged when measured twice in a short period (<2 weeks). These findings establish that variations in FW, PSMD, and WMH, when used in a longitudinal design and with respect to a standardized imaging protocol, truly reflect a disease‐state change and are not due to the day‐to‐day variation in either the MRI scanner or the patient's physiological state.
4.3. Inter‐scanner reproducibility
Inter‐scanner reproducibility reflects the degree of similarity between results for candidate MRI‐based biomarkers using scans of the same individual obtained on different MRI scanners. Defining a harmonized MRI protocol that results in equivalent image contrast is a first step toward reducing inter‐scanner variability. Several other factors may introduce variability including scanner image‐reconstruction software, the inability to modify certain sequence parameters, and differences in the algorithms used to calculate output metrics (e.g., FA, MD). Using harmonized imaging protocols, FW, PSMD, and WMH measures were found to be highly consistent among Philips Achieva, Siemens Trio, and Siemens Prisma systems, with ICC each > 0.94, and therefore, offer the ability to pool data across multiple sites implementing the MarkVCID protocol across these scanners without increasing the heterogeneity of the imaging measurements. Comparing these measures to those obtained with a GE scanner, with an in‐plane resolution interpolation, revealed that pairwise ICC of consistency remained excellent for DTI‐derived kits. Interpolation appeared to impact WMH estimation, highlighting the importance of using optimized protocol parameters 5 for this biomarker. 5 Importantly, the selection of sequence parameters used for the GE machine occurred before imaging‐based biomarker kits were proposed. These parameters were originally developed by another study (Injury & Traumatic Stress, InTRUST study) and were developed to optimize harmonization, among Siemens, Philips, and GE vendors, of gray/white contrast necessary for FreeSurfer segmentations. Future work is needed to investigate whether suppressing in‐plane resolution interpolation on GE machines may increase WMH inter‐scanner repeatability.
FW, PSMD, and WMH kits have several strengths. They were proposed based on prior literature for their endpoint role in imaging studies evidencing association between these measures and SVD and cognition. 8 , 10 , 11 , 13 , 14 , 17 , 32 , 33 , 34 The kits are publicly available on markvcid.partners.org website. Finally, training documents, example data sets, and video tutorials are also provided. There are, however, several limitations to these three imaging‐based biomarker kits. First, although we demonstrated that FW fraction, PSMD, and WMH burden were reliable in the test–retest analysis, the present study does not address whether these measures are sensitive biomarkers of WM change over time. Previous works support that changes in FW fraction, PSMD, and WMH burden may be detectable over time. 10 , 17 , 32 Further longitudinal studies are needed to characterize the smallest detectable change for each biomarker. Second, this study did not address whether FW, PSMD, and WMH measures are associated with cognition measures. This question constitutes the second phase of the biomarker kit validation, referred as the clinical validation, that will investigate in separate publications the associations of kit biomarker measures with clinically meaningful aspects of VCID including, for example, cognitive function and relevant co‐morbidities as defined in their pre‐specified hypotheses (see Table S1). Third, we did not investigate the pairwise association between FW, PSMD and WMH measures. Although presented together in this paper, each imaging‐based biomarker kit is independent. Each kit aims to reflect a specific component of VCID and future users, depending of their study design (age, clinical diagnosis, sample size), may choose to only focus on specific kits. Finally, although each kit is entirely automatic and can be run using a single command line, these scripts require preprocessed images: a brain mask and a DTI 4D volume corrected for eddy current distortions for the FW and PSMD kits and a brain mask for the WMH kit.
In summary, the present study reports findings from the instrumental validation of three imaging‐based biomarkers of SVD selected by MarkVCID consortium: FW, PSMD, and WMH kits. The three kits demonstrated excellent inter‐rater reliability, test–retest repeatability, and inter‐scanner reproducibility. These findings, along with results from the ongoing clinical validation of imaging‐based biomarker kits, further support the potential role of these biomarker kits beyond imaging studies as an endpoint for future multi‐site observational studies and clinical trials in the context of VCID.
CONFLICTS OF INTEREST
Dr. Pauline Maillard is a consultant for Boston University, outside of the scope of the current work. Dr. Hanzhang Lu has no conflict of interest to report. Dr. Konstantinos Arfanakis is on the advisory board for External Advisory Committee, NIH‐funded clinical research study titled “Determinants of Incident Stroke Cognitive Outcomes and Vascular Effects on RecoverY Network (DISCOVERY).” Dr. Brian T. Gold has no conflict of interest to report. Dr. Christopher E. Bauer has no conflict of interest to report. Dr. Valentinos Zachariou has no conflict of interest to report. Dr. Lara Stables has no conflict of interest to report. Dr. Danny J.J. Wang has no conflict of interest to report. Dr. Kay Jann has no conflict of interest to report. Dr. Sudha Seshadri is a consultant for Biogen. Dr. Marco Duering reports honoraria for lectures from Pfizer (payment to Dr. Duering), Bayer (payment to Dr. Duering,) and Sanofi Genzyme (payment to institution). is a consultant for Roche Pharma (payment to institution) and Hovid Berhad, and participates on the Executive committee of the VAS‐COG Society. Laura J. Hillmer has no conflict of interest to report. Dr. Gary A. Rosenberg has no conflict of interest to report. Dr. Haykel Snoussi has no conflict of interest to report. Dr. Farshid Sepehrband has a patent pending for Mapping Perivascular Space using magnetic resonance imaging. Dr. Mohamad Habes is a consultant for Biogen on ARIA. Baljeet Singh has no conflict of interest to report. Dr. Joel H. Kramer has received royalties from Pearson, Inc. Dr. Roderick A. Corriveau has no conflict of interest to report. Herpreet Singh has no conflict of interest to report. Kristin Schwab has no conflict of interest to report. Dr. Karl G. Helmer has no conflict of interest to report. Dr. Steven M. Greenberg is a consultant for Roche (payment to Dr. Greenberg) and Washington University/IQVIA (payment to Dr. Greenberg), Bayer (payment to Dr. Greenberg), Biogen (payment to Dr. Greenberg), and receives royalties or licenses from Up‐To‐Date (payment to Dr. Greenberg). Dr. Charles DeCarli is a consultant to Novartis on a safety trial for heart failure. Dr. Arvind Caprihan has no conflict of interest to report. Dr. Claudia L. Satizabal has received support for attending meetings and/or travel from the NIH (payment to Dr. Satizabal).
AUTHOR CONTRIBUTIONS
Dr. Konstantinos Arfanakis : R01AG064233 (support to institution); R01AG052200 (support to institution). Dr. Danny Wang: R01EB028297 (support to institution); R01NS114382 (support to institution). Dr. Kay Jann: 1R01AG066711‐01 (support to institution). Dr. Farshid: BRAIN Initiative (support to institution). Dr. Joel Kramer: NIH funding (support to institution). Dr. Karl Helmer: NIH (support to institution), Boston Scientific (support to institution), Minoryx (support to institution). Dr. Steven Greenberg: NIH (support to Dr. Greenberg). Dr. Charles DeCarli: NIH funding (support to institution). Dr. Claudia Satizabal: TARCC/State of Texas 2020‐58‐81‐CR (support to Dr. Satizabal and to institution).
Supporting information
Supporting material
ACKNOWLEDGMENTS
U24NS100591; UH3NS100599; UH3NS100588; UH3NS100608; UH3NS100605; UH3NS100606; UH3NS100598; UH3NS100614; P30 AG 010129; 5UH2NS100614‐02. The authors thank: Bruce Fischl, Department of Radiology, Massachusetts General Hospital; Arnold M. Evia, Rush Alzheimer's Disease Center, Rush University Medical Center and Nazanin Makkinejad, Department of Biomedical Engineering, Illinois Institute of Technology; Xin Li and Yang Li, Department of Radiology, Johns Hopkins University School of Medicine; Samantha Ma, Departments of Neurology and Radiology, Keck School of Medicine, University of Southern California; Elyas Fadaee, Glenn Biggs Institute for Alzheimer's and Neurodegenerative Diseases, University of Texas Health San Antonio; Andrew Warren, Mitchell Horn, and Vanessa Gonzalez, Department of Neurology, Massachusetts General Hospital; and Linda McGavern from NIH/NINDS.
Maillard P, Lu H, Arfanakis K, et al., Instrumental validation of free water, peak‐width of skeletonized mean diffusivity, and white matter hyperintensities: MarkVCID neuroimaging kits. Alzheimer's Dement. 2022;14:e12261. 10.1002/dad2.12261
Arvind Caprihan, Charles DeCarli, and Claudia L. Satizabal equally contributed to this study.
REFERENCES
- 1. Kapasi A, DeCarli C, Schneider JA Impact of multiple pathologies on the threshold for clinically overt dementia. Acta Neuropathologica. 2017;134(2):171–186. 10.1007/s00401-017-1717-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Decarli C. Vascular factors in dementia: an overview. J Neurol Sci. 2004;226:19‐23. [DOI] [PubMed] [Google Scholar]
- 3. Smith EE, Biessels GJ, De Guio F, et al. Harmonizing brain magnetic resonance imaging methods for vascular contributions to neurodegeneration. Alzheimers Dement (Amst). 2019;11:191‐204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilcock D, Jicha G, Blacker D, et al. MarkVCID cerebral small vessel consortium: I. Enrollment, clinical, fluid protocols. Alzheimer's & Dementia. 2021;17(4):704–715. 10.1002/alz.12215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lu H, Kashani AH, Arfanakis K, et al. MarkVCID cerebral small vessel consortium: II. Neuroimaging protocols. Alzheimer's & Dementia. 2021;17(4):716–725. 10.1002/alz.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pasternak O, Sochen N, Gur Y, Intrator N, Assaf Y. Free water elimination and mapping from diffusion MRI. Magn Reson Med. 2009;62:717‐730. [DOI] [PubMed] [Google Scholar]
- 7. Wang Y, Wang Q, Haldar JP, et al. Quantification of increased cellularity during inflammatory demyelination. Brain. 2011;134:3590‐3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maillard P, Mitchell GF, Himali JJ, et al. Aortic Stiffness, Increased White Matter Free Water, and Altered Microstructural Integrity. Stroke. 2017;48(6):1567–1573. 10.1161/strokeaha.116.016321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Altendahl M, Maillard P, Harvey D, et al. An IL‐18‐centered inflammatory network as a biomarker for cerebral white matter injury. PLoS One. 2020;15:e0227835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maillard P, Fletcher E, Singh B, et al. Cerebral white matter free water: a sensitive biomarker of cognition and function. Neurology. 2019;92:e2221‐e2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Duering M, Finsterwalder S, Baykara E, et al. Free water determines diffusion alterations and clinical status in cerebral small vessel disease. Alzheimers Dement. 2018;14:764‐774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baykara E, Gesierich B, Adam R, et al. A novel imaging marker for small vessel disease based on skeletonization of white matter tracts and diffusion histograms. Ann Neurol. 2016;80:581‐592. [DOI] [PubMed] [Google Scholar]
- 13. Vinciguerra C, Giorgio A, Zhang J, et al. Peak width of skeletonized mean diffusivity (PSMD) and cognitive functions in relapsing‐remitting multiple sclerosis. Brain Imaging and Behavior. 2021;15(4):2228–2233. 10.1007/s11682-020-00394-4 [DOI] [PubMed] [Google Scholar]
- 14. Deary IJ, Ritchie SJ, Muñoz Maniega S, et al. Brain Peak Width of Skeletonized Mean Diffusivity (PSMD) and cognitive function in later life. Front Psychiatry. 2019;10:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeCarli C, Murphy DG, Tranh M, et al. The effect of white matter hyperintensity volume on brain structure, cognitive performance, and cerebral metabolism of glucose in 51 healthy adults. Neurology. 1995;45:2077‐2084. [DOI] [PubMed] [Google Scholar]
- 16. Jeerakathil T, Wolf PA, Beiser A, et al. Stroke risk profile predicts white matter hyperintensity volume: the Framingham Study. Stroke. 2004;35:1857‐1861. [DOI] [PubMed] [Google Scholar]
- 17. Maillard P, Carmichael O, Fletcher E, Reed B, Mungas D, DeCarli C. Coevolution of white matter hyperintensities and cognition in the elderly. Neurology. 2012;79:442‐448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Debette S, Beiser A, DeCarli C, et al. Association of MRI markers of vascular brain injury with incident stroke, mild cognitive impairment, dementia, and mortality: the Framingham Offspring Study. Stroke. 2010;41:600‐606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu H, Kashani AH, Arfanakis K, et al. MarkVCID cerebral small vessel consortium: II. Neuroimaging protocols. Alzheimer's & Dementia. 2021;17(4):716–725. 10.1002/alz.12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage. 2012;62:782‐790. [DOI] [PubMed] [Google Scholar]
- 21. Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magn Reson Med. 1996;36:893‐906. [DOI] [PubMed] [Google Scholar]
- 22. Smith SM, Kindlmann G, Jbabdi S. In: Johansen‐Berg H, Behrens TEJ, eds. Cross‐Subject Comparison of Local Diffusion MRI Parameters. Diffusion MRI (Second Edition) (2nd ed.). San Diego: Academic Press; 2014:209‐239. editors. 10.1016/B978-0-12-396460-1.00010-X [DOI] [Google Scholar]
- 23. Smith SM, Jenkinson M, Johansen‐Berg H, et al. Tract‐based spatial statistics: voxelwise analysis of multi‐subject diffusion data. NeuroImage. 2006;31:1487‐1505. [DOI] [PubMed] [Google Scholar]
- 24. DeCarli C, Fletcher E, Ramey V, Harvey D, Jagust WJ. Anatomical mapping of white matter hyperintensities (WMH): exploring the relationships between periventricular WMH, deep WMH, and total WMH burden. Stroke. 2005;36:50‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kochunov P, Lancaster JL, Thompson P, et al. Regional spatial normalization: toward an optimal target. J Comput Assist Tomogr. 2001;25:805‐816. [DOI] [PubMed] [Google Scholar]
- 26. Wahlund LO, Barkhof F, Fazekas F, et al. A new rating scale for age‐related white matter changes applicable to MRI and CT. Stroke. 2001;32:1318‐1322. [DOI] [PubMed] [Google Scholar]
- 27. Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychol Bull. 1979;86:420‐428. [DOI] [PubMed] [Google Scholar]
- 28. Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307‐310. [PubMed] [Google Scholar]
- 29. Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15:155‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Konieczny MJ, Dewenter A, Ter Telgte A, et al. Multi‐shell diffusion MRI models for white matter characterization in cerebral small vessel disease. Neurology. 2021;96:e698‐e708. [DOI] [PubMed] [Google Scholar]
- 31. Boukadi M, Marcotte K, Bedetti C, et al. Test‐retest reliability of diffusion measures extracted along white matter language fiber bundles using HARDI‐based tractography. Front Neurosci. 2018;12:1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beaudet G, Tsuchida A, Petit L, et al. Age‐related changes of Peak Width Skeletonized Mean Diffusivity (PSMD) across the adult lifespan: a multi‐cohort study. Front Psychiatry. 2020;11:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lockhart SN, Mayda AB, Roach AE, et al. Episodic memory function is associated with multiple measures of white matter integrity in cognitive aging. Front Hum Neurosci. 2012;6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seiler S, Fletcher E, Hassan‐Ali K, et al. Cerebral tract integrity relates to white matter hyperintensities, cortex volume, and cognition. Neurobiol Aging. 2018;72:14‐22. Epub Aug 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting material


