Abstract
Pseudomonas aeruginosa can employ many distinct mechanisms of resistance to aminoglycoside antibiotics; however, in cystic fibrosis patients, more than 90% of aminoglycoside-resistant P. aeruginosa isolates are of the impermeability phenotype. The precise molecular mechanisms that produce aminoglycoside impermeability-type resistance are yet to be elucidated. A subtractive hybridization technique was used to reveal gene expression differences between PAO1 and isogenic, spontaneous aminoglycoside-resistant mutants of the impermeability phenotype. Among the many genes found to be up-regulated in these laboratory mutants were the amrAB genes encoding a recently discovered efflux system. The amrAB genes appear to be the same as the recently described mexXY genes; however, the resistance profile that we see in P. aeruginosa is very different from that described for Escherichia coli with mexXY. Direct evidence for AmrAB involvement in aminoglycoside resistance was provided by the deletion of amrB in the PAO1-derived laboratory mutant, which resulted in the restoration of aminoglycoside sensitivity to a level nearly identical to that of the parent strain. Furthermore, transcription of the amrAB genes was shown to be up-regulated in P. aeruginosa clinical isolates displaying the impermeability phenotype compared to a genotypically matched sensitive clinical isolate from the same patient. This suggests the possibility that AmrAB-mediated efflux is a clinically relevant mechanism of aminoglycoside resistance. Although it is unlikely that hyperexpression of AmrAB is the sole mechanism conferring the impermeability phenotype, we believe that the Amr efflux system can contribute to a complex interaction of molecular events resulting in the aminoglycoside impermeability-type resistance phenotype.
Resistance to aminoglycosides in Pseudomonas aeruginosa is usually mediated either by specific enzymatic modification of the drug or by an undefined mechanism that has commonly been referred to as impermeability resistance. Aminoglycoside impermeability-type resistance (AGIR) was originally described for clinical isolates of P. aeruginosa with strains exhibiting diminished uptake of gentamicin in energy-dependent phases I and II and no detectable acetylation or adenylylation activity and having ribosomes that were sensitive to the inhibitory effect of aminoglycosides (3). AGIR strains are now commonly characterized as panaminoglycoside resistant in the absence of modifying enzymes, and the characterization of strains exhibiting this phenotype can be inferred from the pattern of sensitivity to aminoglycosides by a disk diffusion-based assay referred to as the aminoglycoside resistance profile (AGRP) (43). Extensive surveys of aminoglycoside resistance in clinical isolates have established the prevalence of the AGIR phenotype in Pseudomonas isolates (24, 25, 34, 38, 44). In general, these studies found that among clinical strains of P. aeruginosa, impermeability-type resistance was the single most common mechanism even though it was identified less frequently than that caused by modifying enzymes as a whole. In contrast, impermeability resistance predominates (>90%) among P. aeruginosa isolates from patients with cystic fibrosis (CF) (23).
While much has been learned since the original characterization of AGIR strains, the specific mechanism(s) involved in this type of resistance remains unclear. Impermeability-type resistance has been circumstantially linked with changes in outer membrane composition of P. aeruginosa, including alterations in the structure of lipopolysaccharide (6, 15, 49), overexpression of outer membrane protein (OMP) OprH (30, 50), and changes in the electron transport chain (4, 5). In this study we employed a PCR-based, subtractive hybridization technique, representational difference analysis (RDA) (20), to examine differential gene expression associated with the AGIR phenotype in P. aeruginosa. These analyses led to the identification of many genes that may be differentially expressed in AGIR strains. One such genomic region, the amrAB locus, was observed to be up-regulated in AGIR strains, including clinical isolates. These genes encode a P. aeruginosa transporter belonging to the resistance, nodulation, and cell division (RND) family of efflux systems (39) and appear to be the same as the recently described mexXY genes from P. aeruginosa. However, our data shows that the effects of this efflux system in P. aeruginosa are quite different from those described when these genes were expressed in Escherichia coli (26). A knockout of this putative efflux system in an AGIR strain restored sensitivity to aminoglycosides in this mutant. Although these data provide a direct line of evidence supporting a role for AmrAB in the AGIR phenotype, the large number of other genes identified in the RDA analyses supports the hypothesis that impermeability-type resistance to aminoglycosides in P. aeruginosa is more complex. It is likely that mutations in multiple loci are necessary to achieve high-level resistance while maintaining strain viability. In addition, regulatory mutations that affect the up-regulation of the amrAB locus may also result in expression level changes of other P. aeruginosa loci that contribute to the AGIR phenotype.
MATERIALS AND METHODS
Bacterial strains and plasmids.
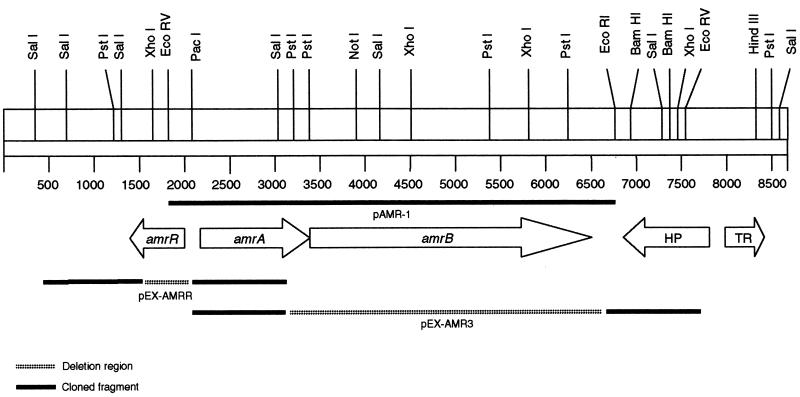
The bacterial strains and plasmids used in this study are described in Table 1. P. aeruginosa PAO1 was used to generate the spontaneous mutant 2547 by plating 108 cells from an overnight culture onto Mueller-Hinton agar (BBL) containing 4 μg of tobramycin per ml. Colonies that appeared after 48 h at 37°C were subsequently characterized for susceptibility to other aminoglycosides. Mutant 2548 was generated in a similar manner by plating strain 2547 onto media containing 32 μg of tobramycin per ml. Biochemical profiles were determined by the API-20NE strip test (Biomerieux), and strains were tested for isogenicity by pulsed-field gel electrophoresis (PFGE) (12). Strain 3579 was constructed by utilizing the pEX-AMRR plasmid to generate a 540-bp deletion within the open reading frame (ORF) of amrR (Table 1; Fig. 1). Strain 3580 was constructed in a similar manner utilizing the pEX-AMR3 plasmid for deletion of a 3.5-kb fragment containing the 3′ terminus of the amrA gene and a majority of the amrB gene (Table 1; Fig. 1). Construction of recombinant plasmids and generation of unmarked deletions are described in detail below. Plasmid pAMR-1 (Fig. 1) was constructed by subcloning a 4.9-kb EcoRV-EcoRI fragment, carrying the amrAB genes, from a cosmid library into pUCP20. The plasmid construction was carried out in Escherichia coli DH5α prior to transformation into P. aeruginosa by electroporation (9).
TABLE 1.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild type | P. Phibbs, S. Lory, University of Washington |
| PAO200 | PAO1 with unmarked Δ(mexAB-oprM) | H. Schweitzer (41) |
| 2547 | Spontaneous tobramycin-resistant derivative of PAO1 | This study |
| 2548 | Spontaneous tobramycin-resistant derivative of 2547 | This study |
| 3579 | PAO1 with unmarked Δ(amrR) | This study |
| 3580 | 2547 with unmarked Δ(amrB) | This study |
| 3582 | PAO1(pAMR-1) | This study |
| 3583 | PAO1(pUCP20) plasmid control strain | This study |
| 3737 | 3579(pXZL34) | This study |
| 913 | CF clinical isolate | CHMC |
| 1030 | CF clinical isolate | CHMC |
| 1085 | CF clinical isolate | CHMC |
| 1104 | CF clinical isolate | CHMC |
| 1151 | CF clinical isolate | CHMC |
| 1168 | CF clinical isolate | CHMC |
| 1200 | CF clinical isolate | CHMC |
| 1249 | CF clinical isolate | CHMC |
| 1250 | CF clinical isolate | CHMC |
| 1365 | CF clinical isolate | CHMC |
| 1452 | CF clinical isolate | CHMC |
| 1457 | CF clinical isolate | CHMC |
| 1520 | CF clinical isolate | CHMC |
| E. coli | ||
| DH5α | F− f80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK−, mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Gibco-BRL |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) ψ 80dlacZΔ M15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU λ− rpsL nupG | Gibco-BRL |
| S17.1λpir | thi pro hsdR recA Tra+ | P. Phibbs, S. Lory, University of Washington |
| SM10 | Kmr; mobilizer strain (thi-1 thr leu tonA lacY supE recA::RP4-2Tc::Mu) | H. Schweizer, Colorado State University (42) |
| Plasmids | ||
| pGEM-T | Apr; T-tailed cloning vector | Promega Corp. |
| pUCP20 | Apr Cbr, ori1600+; broad-host-range cloning vector (GenBank no. U07165) | H. Schweizer (47) |
| pAMR-1 | pUCP20 derivative carrying the amrAB genes on a 4.9-kb EcoRV-EcoRI fragment | This study |
| pXZL34 | pVLT31 derivative carrying the oprM gene under the control of an IPTG-inducible promoter | R. Hancock, University of British Columbia (48) |
| pEX18T | Apr Cbr; sacB+ oriT+; gene replacement vector (GenBank no. AF004910) | H. Schweizer, Colorado State University (42) |
| pPS858 | Apr Gmr; source of Gmr-GFP fragment flanked by FRT sites | H. Schweizer (17) |
| pFLP2 | Apr Cbr; source of yeast Flp recombinase (GenBank no. AF048702) | H. Schweizer (17) |
| pEX-AMRR | pEX18T derivative used for the deletion of the amrR gene | This study |
| pEX-AMR3 | pEX18T derivative used for the deletion of the amrAB genes | This study |
Apr, ampicillin resistant; Cbr, carbenicillin resistant; Gmr, gentamicin resistant; Kmr, kanamycin resistant; CHMC, Children’s Hospital and Medical Center, Seattle, Washington; GFP, green fluorescent protein structural gene; FRT, yeast Flp site-specific recombinase recognition sequence.
FIG. 1.
Physical map of the amr locus in P. aeruginosa. Arrows designate complete ORF. The region downstream of amrB contains two ORFs, one (HP) with homology (54% similar) to yjiK, encoding a hypothetical protein from E. coli, and the other (TR) possessing homology (74% similar to merR from Archaeoglobus fulgidus) to the MerR family of bacterial response regulators. See Materials and Methods for a detailed description of the representative plasmids.
Growth media and susceptibility testing.
E. coli strains were cultivated in Lennox L broth or agar (Gibco-BRL). P. aeruginosa strains were maintained on blood agar (Remel) or L agar and propagated in cation-adjusted Mueller-Hinton broth (BBL) unless otherwise noted. Growth curves were determined by dilution (1:50) of overnight cultures in fresh Mueller-Hinton broth and growth at 37°C in a shaking incubator (250 rpm). Vogel-Bonner (VB) medium (46) was used for selective isolation of P. aeruginosa and supplemented with 5% sucrose for negative selection of strains carrying the sacB gene. Strains containing the pXZL34 plasmid were maintained on L agar containing tetracycline and 0.05 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (48). Antibiotics at various concentrations were used for selection, as follows: for E. coli, ampicillin (100 μg/ml), gentamicin (10 μg/ml), and tetracycline (10 μg/ml); and for P. aeruginosa, carbenicillin (500 μg/ml), gentamicin (200 μg/ml unless otherwise indicated), and tetracycline (100 μg/ml). All antibiotics were supplied by Sigma Chemical Co. (St. Louis, Mo.). MICs were determined by microbroth dilution according to National Committee for Clinical Laboratory Standards guidelines (29). Disk susceptibility determinations were also done according to National Committee for Clinical Laboratory Standards guidelines (28). AGRP assay results were interpreted as described by Shaw et al. (43).
RDA.
The protocol described by Lisitsyn et al. (20) was modified for use with bacterial cDNA as a means of analyzing gene expression. Total bacterial RNA (3 μg), isolated from mid-log-phase cultures, was DNase treated and converted to double-stranded cDNA. As a positive control, MS2 bacteriophage RNA was spiked into a background of PAO1 RNA at 100 copies per cell equivalent and used as a tester against PAO1. The first strand of cDNA was synthesized in a 20-μl random-primed reverse transcription reaction mixture containing 1× first-strand buffer (50 mM Tris-HCl [pH 8.3], 40 mM KCl, 6 mM MgCl2; Gibco-BRL), 100 ng of primer [2:1:1 mix of N6 (SN)3, and (NS)3, where S = G or C] 0.5 mM deoxynucleoside triphosphates (dNTPs), 10 mM dithiothreitol, 5% dimethyl sulfoxide (DMSO), and 200 U of reverse transcriptase (SuperScript II; Gibco-BRL). The second strand was synthesized in a 150-μl polymerization reaction mixture containing 1× second-strand buffer [20 mM Tris-HCl (pH 6.9), 4.6 mM MgCl2, 90 mM KCl, 0.15 mM β-NAD+, 10 mM (NH4)2SO4], 0.2 mM dNTPs, 40 U of E. coli DNA polymerase I, 10 U of E. coli DNA ligase, 2 U of ribonuclease H (all enzymes obtained from Gibco-BRL). All adapter sequences and PCR conditions were as described by Lisitsyn et al. (20), with exceptions as noted. Purified cDNA was digested with DpnII and ligated with oligonucleotide (R-Bgl) adapters. For all ligation reactions, 100 ng of cDNA and 0.5 nmol of each adapter were combined in a T4 DNA ligase reaction (Boehringer Mannheim) at 16°C for 4 h. PCR amplification primed off the terminal adapter sequences was used to produce driver and tester amplicons for subsequent hybridization and selective amplification. Amplicons were purified by extraction with equal volumes of phenol (pH 8.0) and phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol) and precipitated with isopropanol by using tRNA as the carrier. All subsequent PCRs and digests were purified using QiaQuick PCR purification columns (Qiagen) as per the manufacturer’s instructions. R-Bgl adapters were removed by digestion with DpnII, and only tester amplicons were ligated with new (J-Bgl) adapters. A 100-ng amount of the adapterless driver amplicon and 0.1 ng of the newly ligated tester amplicon were denatured and hybridized in a 50-μl phenol emulsion containing the following: 1.5 M sodium thiocyanate, 120 mM phosphate buffer (equimolar mono- and dibasic phosphate), 10 mM EDTA, and 12% (vol/vol) unbuffered phenol. Initial denaturation at 100°C for 10 min was followed by three cycles of hybridization (15 min at 25°C) and denaturation (2 min at 65°C) and a final hybridization (15 min at 25°C). A driver-only reaction was included as a negative control for each hybridization and selective amplification. Hybridizations were extracted with 100 μl of chloroform and concentrated to 20 μl over a QiaQuick column. Selective amplifications of the tester-tester hybrid DNA were performed by two rounds of PCR, with priming from the adapter sequence and an intervening mung bean nuclease digest to remove any remaining single-stranded DNA. Primary amplification of the purified hybridization reaction was done in a 50-μl volume as follows: 1× PCR buffer [67 mM Tris-HCl (pH 8.8), 4 mM MgCl2, 16 mM (NH4)2SO4, 10 mM β-mercaptoethanol, 100 μg of bovine serum albumin per ml], 0.5 mM dNTPs, 0.2 nmol of primer (same 24-bp oligonucleotide adapter to which the tester was ligated), 5% DMSO, and 2 U of Taq polymerase. Reactions were hot-started in the absence of primer and incubated for 10 min at 72°C in order to fill in target adapter ends. Primer was added, and reactions were amplified for 15 cycles of 1 min at 95°C and 3 min at 72°C and then finished with 10 min at 72°C. Purified reaction products were digested with mung bean nuclease (New England Biolabs) for 30 min at 30°C and purified again. Secondary amplification, scaled to a 100-μl reaction volume, was as described for the primary PCR, but the fill-in incubation prior to primer addition was omitted. The resulting difference products were subjected to two more rounds of hybridization and selective amplification, with N-Bgl adapters used for the second round and J-Bgl adapters used for the third round, as described above. Third-round difference products were cloned into pGEM-T and transformed into competent E. coli DH10B cells. Colony PCR was used to amplify the inserted difference product with primers directed to the flanking vector sequence. PCR products were sequenced and analyzed by Basic Local Alignment Search Tool (BLAST) (2) queries of nonredundant public databases, including nucleotide databases (GenBank, EMBL, DDBJ, and PDB) and protein databases (GenBank CDS translations, PDB, SwissProt, PIR, and PRF), and queries at the National Center for Biotechnology Information (29a) as well as by comparison to P. aeruginosa sequences from the Pseudomonas Genome Project.
RT-PCR.
A 1-μg sample of DNase-treated RNA was converted to single-stranded cDNA by reverse transcription with the reverse primers specific to the gene of interest. Specific primer pairs were as follows: for amrA, amrA-F1 (5′-CATCAGCGAACGCGAGTACACCGAAGCG-3′) and amrA-R1 (5′-CACGTAGATCGGATCGATCTGCTCGACGC-3′); for amrB, amrB-F1 (5′-CTGGGTGATCTCCCTGCTGATCGTGCTC-3′) and amrB-R2 (5′-ACTCGACGATCTTCAGGCGGTTCTGCAC-3′); and for rpsL, rpsL-For (5′-GCAACTATCAACCAGCTGGTG-3′) and rpsL-Rev (5′-GCTGTGCTCTTGCAGGTTGTG-3′). Primers specific to the constitutively expressed gene rpsL were used as a positive control. Reverse transcription reaction conditions were as follows: 1× first-strand buffer (50 mM Tris-HCl [pH 8.3], 40 mM KCl, 6 mM MgCl2; Gibco-BRL), 10 mM ditheothreitol, 2 pmol of reverse primer per target transcript, 0.5 mM dNTPs, 5% DMSO, 22 U of anti-RNase (Ambion), and 200 U of reverse transcriptase (Gibco-BRL). The same reaction omitting the reverse transcriptase (RT) was used as a negative control. RT reaction mixtures were serially diluted 1:3 in 10 mM Tris-HCl (pH 8.0), and 5 μl of each dilution was used as the template for PCR amplification with the following: 1× PCR buffer [67 mM Tris-HCl (pH 8.8), 4 mM MgCl2, 16 mM (NH4)2SO4, 10 mM β-mercaptoethanol, 100 μg of bovine serum albumin per ml], 0.2 mM dNTPs, 10 pmol of each primer (forward and reverse), 5% DMSO, and 2.5 U of Taq polymerase (Boehringer Mannheim). Reaction mixtures were incubated for 90 s at 95°C and amplified for 35 cycles of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C and finished with 10 min at 72°C. A 10-μl volume of each reaction product was examined by 1.5% (wt/vol) agarose gel electrophoresis.
Generation of unmarked deletions.
Unmarked deletions of the amrR gene in PAO1 and the amrB gene in strain 2547 were generated according to the method described by Schweizer (41). DNA homologous to sequence flanking the region of deletion was amplified by PCR. Primers 5′ for-amrR (5′-CGTCGCGGGTTTTCTGGGATCCCTCTTTGG-3′) and 5′ rev-amrR (5′-GCAGGAATTCGCGATGCGGATTGCGGAAC-3′) generated a 1-kb fragment with BamHI and EcoRI restriction sites introduced on the respective ends and flanking the 5′ end of amrR. Similarly, primers 3′ for-amrR (5′-GGAAAGCTTGGTGGCGAGGAAGGCATTGG-3′) and 3′ rev-amrR (5′-GCCTCTAGAGCCTGCGCAGTTCTCCCTAC-3′) generated a 1.1-kb fragment with HindIII and XbaI restriction sites introduced on the respective ends and flanking the 3′ end of amrR. The 5′ and 3′ ends flanking the amrAB deletion region were generated with the same respective restriction sites by using primers 5′ for-amr3 (5′-CGTCGCGGGTTTTCTGGAATTCCTCTTTGG-3′), 5′ rev-amr3 (5′-AGCAATTCGGGATCCGGATTGCGGAACAG-3′), 3′ for-amr3 (5′-CAAGCCTGATGCCTCTAGAGAAACTCTCGC-3′), and 3′ rev-amr3 (5′-GAATCTGGTTCAAGCTTGAGCAGCGCTACG-3′). PCR fragments were amplified with 3 cycles at an annealing temperature of 55°C and 31 cycles at an annealing temperature of 60°C. Fragments were digested with the appropriate enzymes and 5′ and 3′ fragments were cloned sequentially into plasmid pEX18T. The Gmr-GFP cassette from pPS858 was isolated as a BamHI fragment and cloned directly into pEX18T containing flanking ends for amrR. Due to BamHI restriction sites in the amrAB 3′ fragment, the Gmr-GFP cassette and XbaI-digested pEX18T containing flanking ends for amrAB were treated with Klenow fragment and blunt-end ligated. All plasmids were constructed and propagated in E. coli DH5α. The final constructs were introduced into the E. coli donor strain, S17.1λpir, by chemical transformation and conjugally transferred to P. aeruginosa. Transconjugates were selected and propagated on L agar containing gentamicin. Secondary selection on L agar with carbenicillin was used to identify double-crossover events of the Gmr and Cbs phenotypes. In the case of pEX-AMR3, primary selection yielded only colonies which were Gmr and Cbr, or single-crossover events. One of these strains was grown overnight in L broth containing gentamicin. Culture dilutions (10−2 and 10−4; volume, 100 μl) were plated on VB plus 5% sucrose agar containing gentamicin in order to select for the loss of the sacB gene. Colonies arising after 48 h were screened for carbenicillin resistance and propagated on L agar containing gentamicin (100 μg/ml). E. coli SM10 was used to mobilize the plasmid pFLP2, which contains the yeast Flp recombinase, into Gmr and Cbs strains of P. aeruginosa by conjugal transfer. Primary selection for P. aeruginosa carrying the pFLP2 plasmid was done on VB agar containing carbenicillin. Plasmids were cured by streaking colonies onto VB agar plus 5% sucrose and subsequently were confirmed to be Gms and Cbs. Both strains 3579 and 3580 were confirmed to contain the proper deletions by comparing them to appropriate parent and single-crossover and double-crossover event and strains by Southern blot analyses (45).
Outer membrane SDS-PAGE gels.
OMPs of P. aeruginosa were prepared by the method of Poxton et al. (37). Bacterial cells were grown to mid- to late-log phase. Cell pellets were resuspended in 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid) buffer (pH 7.4). Cells were broken by ultrasonic disruption, and unbroken cells were removed by low-speed centrifugation (10,000 × g for 10 min). Membranes were subsequently pelleted by high-speed centrifugation (100,000 × g for 30 min), resuspended in 10 mM HEPES with 2% (wt/vol) sodium N-lauroyl sarcosine, and incubated at room temperature for 1 h. Insoluble membrane proteins were pelleted by high-speed centrifugation (100,000 × g for 30 min) and resuspended in distilled water. Samples were assayed for total protein using the DC Protein Assay (Bio-Rad) as per the manufacturer’s instructions. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on a 12% acrylamide/bis-acrylamide (37.5:1) SDS slab gel and analyzed by staining with GELCODE Blue Stain Reagent (Pierce).
Southern blot hybridizations.
SalI-digested genomic DNA (1 μg) was run on a 0.7% agarose gel. DNA was blotted onto positively charged nylon membranes overnight (Nytran Plus; Schleicher & Schuell) by standard alkaline transfer (40). Dry membranes were cross-linked with UV light and probed with random-primed [α-32P]dCTP-labeled DNA. Mutants were confirmed by probing blots with the PCR fragments that were cloned into the recombination vectors. The deletion mutants were confirmed to be unmarked by probing with the Gmr-GFP cassette.
Nucleotide sequence accession numbers.
The sequences of the amrR and amrAB genes have been deposited with GenBank under the accession no. AF147719. Other relevant genes can be referenced with the following GenBank accession no.: AF073776 (mexGH [P. aeruginosa]), AB015853 (mexXY [P. aeruginosa]), and AF072887 (amrAB [Burkholderia pseudomallei]).
RESULTS
Characterization of spontaneous tobramycin-resistant mutants.
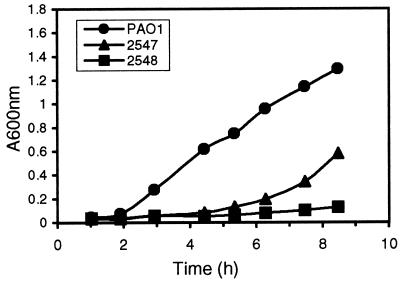
Two spontaneous AGIR mutants were isolated in a stepwise fashion from strain PAO1. The mutants, designated 2547 and 2548, are isogenic to PAO1 as determined by PFGE (data not shown), and they displayed a panaminoglycoside- resistant phenotype, which is characteristic of AGIR strains, in the AGRP assay (Table 2). Strain 2547 showed an intermediate-level aminoglycoside impermeability phenotype, whereas 2548, with no zone of inhibition to any of the tested aminoglycosides, showed a high-level impermeability phenotype. In addition to enhanced aminoglycoside resistance the mutants exhibited other altered phenotypic characteristics compared to PAO1. Neither of the mutants were able to hydrolyze urea, and both displayed impaired growth in culture with rich media (Fig. 2).
TABLE 2.
Aminoglycoside resistance phenotypes
| Strain | Patient and/or relevant genotypeb | AGRP zone diameter (mm)a
|
Disc AGRP Phenotype | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Apra | Forta | 21562 | 21561 | Gent | Tobra | Amk | Isep | Net | 22591 | Kan | Neo | |||
| PAO1 | Wild type | 20 | 20 | 17 | 16 | 16 | 21 | 21 | 20 | 18 | 18 | 8 | 14 | Sensitive |
| 2547 | Isogenic PAO1 mutant | 10 | 6 | 7 | 7 | 6 | 10 | 6 | 7 | 8 | 9 | 6 | 6 | Permeability |
| 2548 | Isogenic 2547 mutant | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | Permeability |
| 1168 | 40-03, 22-7 | 20 | 19 | 16 | 16 | 16 | 20 | 17 | 18 | 16 | 17 | 6 | 12 | Sensitive |
| 1250 | 40-03, 22-7 | 19 | 8 | 6 | 6 | 10 | 19 | 12 | 13 | 11 | 16 | 6 | 6 | Permeability |
| 1249 | 40-03, 22-7 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | Permeability |
| 1151 | 59-01, 11-1 | 27 | 22 | 20 | 20 | 21 | 27 | 23 | 22 | 22 | 23 | 6 | 6 | Sensitive |
| 1030 | 59-01, 10-1 | 8 | 6 | 6 | 9 | 6 | 18 | 6 | 10 | 11 | 12 | 6 | 6 | Permeability |
| 1200 | 59-01, 10-1 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | Permeability |
| 913 | 48-06, 7-1 | 29 | 29 | 28 | 28 | 25 | 29 | 30 | 30 | 30 | 25 | 10 | 18 | Sensitive |
| 1085 | 48-07, 7-1 | 27 | 28 | 25 | 26 | 22 | 26 | 24 | 24 | 24 | 22 | 9 | 16 | Sensitive |
| 1365 | 47-17, 4-1 | 30 | 21 | 22 | 22 | 23 | 31 | 28 | 27 | 27 | 26 | 6 | 16 | Sensitive |
| 1457 | 01-09, ND | 26 | 20 | 21 | 21 | 21 | 27 | 24 | 25 | 22 | 22 | 11 | 16 | Sensitive |
| 1104 | 50-03, 24-4 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | Permeability |
| 1452 | 67-03, ND | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | Permeability |
| 1520 | 53-11, ND | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | Permeability |
Disk diameter is 6 mm. A value of 6 mm indicates no zone of inhibition. Apra, apramycin; Forta, fortamicin; Gent, gentamicin; Tobra, tobramycin; Amk, amikacin; Isep, isepamicin; Net, netilmicin; Kan, kanamycin; Neo, neomycin.
Genotypes for the clinical isolates were determined by randomly amplified polymorphic DNA fingerprinting as described by Bukanov et al. (7). ND, not determined.
FIG. 2.
Comparison of the growth rates of PAO1 and spontaneous tobramycin-resistant mutants.
Identification of the amr locus.
RDA subtractions were designed to identify genes that are up-regulated in the mutants 2547 and 2548 in comparison to strain PAO1. Three subtractions were done with an excess of PAO1-derived cDNA as the driver. Tester cDNA was derived from in vitro cultures of strains 2547, 2548, and 2548 grown in the presence of 16 μg of tobramycin per ml. An average of 200 difference products per subtraction were sequenced, and approximately 30 open reading frames (ORFs) were identified from each subtraction. One of these ORFs which was identified in all three experiments was designated amrB based on strong homology (79% similarity of amino acid sequences) to a previously identified efflux pump component that affects aminoglycoside and macrolide resistance in B. pseudomallei (27). BLAST analysis of the Pseudomonas genome revealed that amrB was located downstream of another gene, designated amrA, which has homology to the membrane fusion proteins of the RND-type transporters. Homology of the predicted amino acid sequence of AmrB and genomic organization of the amrAB genes (Fig. 1) indicate that AmrAB belongs to the RND family of transporters; however, unlike the previously characterized Mex pumps, no OMP was identified downstream of amrB. More recently, the sequence of the amrAB genes was found to be highly similar to two other GenBank sequences from PAO1, mexGH (1) and mexXY (26). While the deduced amino acid sequences of amrAB and mexXY are only 84 to 89% similar, respectively, the restriction pattern published by Mine et al. (26) is the same as that deduced for the amrAB genes. In addition, BLAST analysis of the PAO1 genome (presently more than 99% complete) revealed no other matches for the mexXY or mexGH genes that were perfect or better than amrAB. Therefore, it seems likely that amrAB, mexXY, and presumably mexGH are the same genes.
AmrR transcriptional regulation of amrAB.
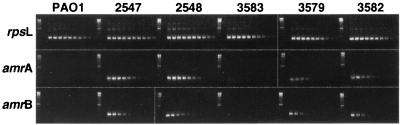
The region located upstream of amrA contains an ORF, designated amrR, belonging to the TetR family of bacterial response regulators (Fig. 1). To determine whether this putative regulator was involved in the transcription of amrAB, an unmarked deletion of amrR was constructed in strain PAO1. The deletion was generated using a gentamicin resistance, green fluorescent protein selectable marker cassette that is flanked by yeast Flp recombinase target sites (17). Generation of an unmarked deletion within a gene by a Flp-based system has been demonstrated in other organisms and shown to cause no polar effects on downstream transcription (8, 21). The resulting strain, 3579, was shown by RT-PCR to transcribe the amrA gene at least 15-fold and the amrB gene at least 12-fold compared to the parental strain PAO1 (Fig. 3). The transcription levels in strain 3579 were approximately equivalent to those in a strain carrying amrAB on a multicopy plasmid, strain 3582, but are still approximately sixfold less than those of the spontaneous mutants 2547 and 2548 (Fig. 3). It has been shown previously that other P. aeruginosa strains that overexpress Mex pumps have mutations in their respective transcriptional regulatory genes (18, 35, 36). However, sequencing of the amrR gene in both 2547 and 2548 revealed no mutations in either the putative 5′ promoter region or the amrR coding sequence compared to wild-type PAO1. Therefore, although it is apparent that amrR can negatively regulate transcription of amrAB, loss of this regulatory function does not seem to be the cause of the up-regulation observed in mutants 2547 and 2548. However, we recently identified an rplA mutation in strains 2547 and 2548 that theoretically would result in a truncated form of ribosomal protein L1. Complementation of mutant 2547 with a wild-type copy of the rplA gene restored aminoglycoside sensitivity, growth rate, and amrAB transcription levels back to PAO1 levels but did not complement the urease defect (16).
FIG. 3.
1.5% agarose gels of RT-PCR products showing relative expression of the genes rpsL (positive control), amrA, and amrB. For each set the two left-most lanes show a 100-bp DNA ladder and an RT-negative template (negative control), respectively, and the other lanes show an RT-positive template (serial 1:3 dilutions).
Deletion of amrAB from strain 2547 results in a reduction in MICs of aminoglycosides.
To determine whether the increased transcription of amrAB contributes to the AGIR phenotype of the laboratory mutants, an unmarked deletion of amrB was constructed in strain 2547 by the same system described above. Strain 3580 (2547ΔamrB) shows MIC values returned to within a fourfold difference of wild-type (PAO1) levels for four of the six aminoglycosides tested (Table 3). Both kanamycin and tobramycin also showed reduced MIC values compared to strain 2547, but in these cases the changes were less than or equal to fourfold. Strains 2547 and 3580 did not show differences in the MICs of any of the other nonaminoglycoside drugs tested, and in addition, the deletion of amrB in strain 3580 did not complement the other phenotypic characteristics, slow growth and lack of urease activity, seen in strain 2547 compared to PAO1.
TABLE 3.
Susceptibilities of P. aeruginosa strains to various antibioticsa
| Strain | Relevant genotype | MIC (μg/ml)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Apra | Gent | Amk | Kan | Neo | Tobra | Nor | Oflox | Cipro | Lom | Erm | Clar | Clin | Tet | ||
| PAO1 | Wild type | 16 | 2 | 4 | 128 | 16 | 0.25 | 2 | 2 | 0.5 | 4 | >32 | >8 | >32 | 32 |
| 3583 | PAO1(pUCP20) | 16 | 4 | 8 | 128 | 16 | 0.5 | 2 | 2 | 0.25 | 4 | >32 | >8 | >32 | 32 |
| 3582 | PAO1(pAMR-1) | 16 | 2 | 8 | 256 | 64 | 1 | 4 | 4 | 1 | 8 | >32 | >8 | >32 | 32 |
| 3579 | PAO1Δ(amrR) | 32 | 4 | 8 | 128 | 32 | 0.5 | 2 | 4 | 0.5 | 8 | >32 | >8 | >32 | 32 |
| PAO200 | PAO1Δ(mexAB-oprM) | 2 | <1 | 2 | 32 | 2 | 0.25 | <0.06 | <0.06 | <0.015 | 0.13 | 32 | 2 | >32 | 0.5 |
| 2547 | PAO1 mutant | 256 | 32 | 128 | >512 | 256 | 16 | 4 | 4 | 1 | 8 | >32 | >8 | >32 | 32 |
| 3580 | 2547Δ(amrB) | 16 | 2 | 16 | 256 | 16 | 4 | 2 | 1 | 0.5 | 2 | >32 | >8 | >32 | 16 |
| 2548 | 2547 mutant | >512 | 128 | 256 | >512 | >512 | 64 | 4 | 4 | 1 | 8 | >32 | >8 | >32 | 32 |
Apra, apramycin; Gent, gentamicin; Amk, amikacin; Kan, kanamycin; Neo, neomycin; Tobra, tobramycin; Nor, norfloxacin; Oflox, ofloxacin; Cipro, ciprofloxacin; Lom, lomefloxacin; Erm, erythromycin; Clar, clarithromycin; Clin, clindamycin; Tet, tetracycline.
Up-regulation of amrAB alone does not alter MICs of aminoglycosides for strain PAO1.
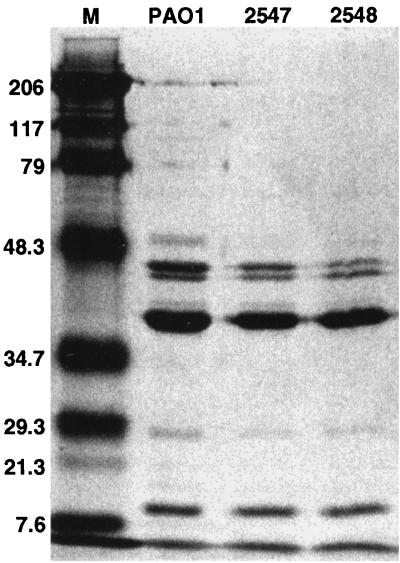
Despite the overexpression of amrAB in strains 3579 and 3582, the MICs for these strains did not change significantly relative to those of PAO1 (Table 3). Since MexXY has been shown to associate with OprM in E. coli (26), we investigated the possibility that OprM might contribute to an increase of aminoglycoside resistance in strain 3579, which overexpresses amrAB (mexXY) as a result of an unmarked deletion of the amrR gene. Transformation of strain 3579 with the plasmid pXZL34, which has been shown previously to express OprM under the control of an IPTG-inducible promoter (48), resulted in strain 3737. Growth of strain 3737 in 0.05 mM IPTG did not impact susceptibility to any of the antibiotics tested (data not shown). In addition, we analyzed outer membrane preparations made from strains PAO1, 2547, and 2548 by SDS-PAGE to look for differences in the levels of the OprM. These gels revealed a dramatic decrease in the amount of OprM present in strains 2547 and 2548 compared to PAO1 (Fig. 4), indicating that OprM is unlikely to be the outer membrane component associated with this efflux system.
FIG. 4.
SDS-PAGE analysis of OMPs. The OprM protein, known to be expressed in wild-type P. aeruginosa, runs in the 50-kDa monomeric form with the inclusion of β-mercaptoethanol in the sample buffer (48). The molecular weight standards (M) are listed to the left in kilodaltons.
amrB transcription is increased in CF clinical isolates that display impermeability-type aminoglycoside resistance.
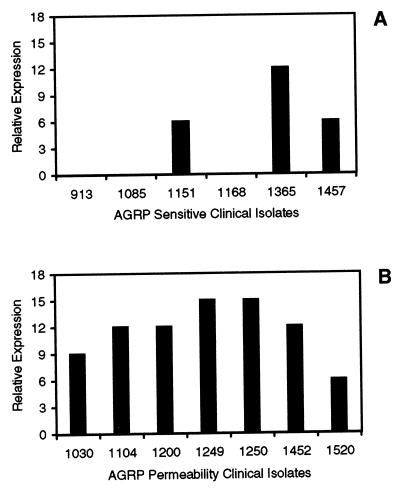
In order to address the possible clinical relevance of the Amr system, we used RT-PCR to examine amrB gene expression level differences among clinical isolates with varying AGRP phenotypes. Transcription of amrB can be detected in clinical isolates of P. aeruginosa that display an impermeability phenotype at levels that are at least 6- to 15-fold greater than that of strain PAO1 (Fig. 5), and gene expression is generally higher in the AGIR strains (Fig. 5A) compared to those clinical isolates with a sensitive phenotype (Fig. 5B). In particular, the isogenic clinical isolates 1168, 1250, and 1249 have AGRP phenotypes (Table 2) and amrB RT-PCR results that mimic directly those of PAO1 and the mutants 2547 and 2548. A trend towards increased amrB transcription was not observed in strains that produce aminoglycoside-modifying enzymes or have multiple AGRP phenotypes (data not shown).
FIG. 5.
Graphs representing expression of amrB in P. aeruginosa clinical isolates having the sensitivity (A) or permeability (B) phenotype from patients with CF relative to that in strain PAO1 as determined by RT-PCR. Relative expression levels are plotted as the number of 1:3 dilutions showing amplification on a 1.5% agarose gel multiplied by three. PAO1 had no detectable amplification, so the relative expression value represents at least that fold increase in expression of amrB by a particular strain compared to PAO1.
DISCUSSION
Drug efflux is a prevalent mechanism of antibiotic resistance in P. aeruginosa. Until recently this mechanism of resistance had been seemingly limited to quinolones, β-lactams, tetracycline and chloramphenicol (14, 31). However, new evidence for B. pseudomallei now indicates an even broader potential for efflux-mediated acquired and intrinsic antibiotic resistance. The recently described B. pseudomallei genes, amrAB, have been shown to contribute to the intrinsic resistance of this organism to both macrolides and aminoglycosides (27).
Aminoglycoside impermeability-type resistance is poorly understood despite the fact that it is the predominant manifestation of aminoglycoside resistance in P. aeruginosa isolates infecting the lungs of CF patients (23). In this study we show that spontaneous mutants of PAO1 displaying the AGIR phenotype also display up-regulation of the amrAB genes. The deletion of the amrB gene resulted in the reversal of the panaminoglycoside resistance phenotype, and in contrast to the recent work describing expression of the mexXY genes in E. coli (26), our data did not show any change in the MICs of a number of quinolones and macrolides for strain 3580 compared to those for strain 2547. It was shown recently that the frequency of emergence of fluoroquinolone resistance in a triple deletion strain of PAO1 (ΔmexAB-oprM::Cm ΔmexEF-oprN::ΩHg ΔmexCD-oprJ::Gm) was essentially undetectable (22). These data would argue against the role of an additional efflux system actively contributing to quinolone resistance in P. aeruginosa. Using disk susceptibility assays we did detect small differences in the zones of inhibition for lomefloxacin, norfloxacin, and ofloxacin for strains PAO1, 2547 and 3580 (Table 4). However, the comparison of these subtle differences with the changes seen in the MICs of quinolones for strain PAO200 (ΔmexAB-oprM) (Table 3) leads us to believe that the observed quinolone effect is more likely to be the result of regulatory cross talk with other efflux systems rather than an affinity of AmrAB for quinolones. Recent data demonstrating coordinated regulation of expression for the MexAB-OprM, MexCD-OprJ, and MexEF-OprN efflux pumps (19) supports the possibility that altering the expression of a single pump may have downstream effects on any number of other efflux systems. In fact the apparent decrease seen for OprM in the mutants 2547 and 2548 compared to PAO1 would be consistent with the quinolone disk resistance data noted above and support a hypothesis of coordinated regulation between MexAB-OprM and the AmrAB efflux systems.
TABLE 4.
Kirby-Bauer disk diffusion assay for selected quinolones
| Strain | Relevant genotype | Zone diameter (mm)a
|
||||
|---|---|---|---|---|---|---|
| Nor | Oflox | Cipro | Lom | Nal | ||
| PAO1 | Wild type | 28 | 20 | 31 | 23 | 7 |
| 2547 | PAO1 mutant | 20 | 12 | 27 | 13 | 7 |
| 2548 | 2547 mutant | 19 | 10 | 26 | 11 | 7 |
| 3580 | 2547Δ(amrB) | 28 | 18 | 36 | 24 | 7 |
Disk diameter is 6 mm. Disk content (in μg/ml) and resistance and sensitivity ranges (R and S, respectively; also in μg/ml) for the drugs tested were as follows: for norfloxacin (Nor), disk content = 10, R ≤ 12, and S ≥ 17; for ofloxacin (Oflox), disk content = 5, R ≤ 12, and S ≥ 16; for ciprofloxacin (Cipro), disk content = 5, R ≤ 15, and S ≥ 21; for lomefloxacin (Lom), disk content = 10, R ≤ 18, and S ≥ 22; and for nalidixic acid (Nal), disk content = 30, R ≤ 13, and S ≥ 19.
Although it is clear from our studies that amrAB can play a role in aminoglycoside resistance, up-regulation of the amrAB genes alone in wild-type P. aeruginosa (PAO1) did not affect susceptibility to aminoglycosides or any of the other antibiotics tested, including quinolones and macrolides. This is not unexpected since RND-type systems are known to require three proteins for active efflux of drugs from the cytoplasm, an inner membrane RND-transport protein, a periplasmic membrane fusion protein, and an OMP (11, 31–33). It is likely that the lack of enhanced resistance displayed by the above strains is due to the absence of corresponding expression of an appropriate OMP. Recent observations by Mine et al. suggest that OprM could be the proper accessory protein for AmrAB (26). Nonetheless, SDS-PAGE gels of outer membrane preparations revealed that very little OprM is even present in the mutant 2547. Our results suggest that the OMP that acts in conjunction with AmrAB in affecting aminoglycoside resistance in P. aeruginosa is unlikely to be OprM but instead some other yet to be identified protein. There are many potential choices for an OMP that will function with the AmrAB pump. Analysis of the newly sequenced PAO1 genome has identified ten potential RND-type drug efflux loci. Four of these loci, including the Amr locus, lack downstream OMPs. Additional phylogenetic analyses have identified three large families of OMPs, each consisting of 18 to 27 members, and numerous smaller families in the genome (13). One large family is most closely related to those proteins previously characterized as having a role in antimicrobial efflux and contains a subfamily that is most related to OprM, OprN, and OprJ. Comparisons of trees generated using these OMPs and the corresponding proposed cytoplasmic membrane and pump components suggests that in some cases there has been a shuffling of the OMP genes relative to the periplasmic and cytoplasmic membrane components of the efflux machinery (13). This is reminiscent of a previous report on other efflux OMPs that suggests that the OMP components evolve independently of the other components (33). The genome analysis reveals a complex set of efflux systems and corresponding OMPs and highlights the need for additional understanding of how these systems may be interrelated and regulated.
Despite the fact that AGIR has been observed in the clinic for decades, the molecular mechanisms of this phenotype are still not well understood. The RDA experiments presented in this study identified a large number of genes that are potentially involved in this type of aminoglycoside resistance and suggest that the AGIR phenotype is multifaceted. While we have shown here that the AmrAB efflux system can contribute to aminoglycoside resistance, it seems that efflux may be only one of the factors that can contribute to this phenotype in spontaneously occurring mutants of P. aeruginosa. The ability to generate stepwise mutants resistant to increasing levels of aminoglycosides (i.e., strain 2548) and the up-regulation of the amrAB genes in a one-step, low-level resistant mutant, strain 2547, in addition to other phenotypic differences, such as reduced growth rate and lack of urease activity, also support the theory that AGIR is a cumulative, multifactorial process. While we have shown here that the amrR gene can negatively regulate transcription of amrAB, the mutation in 2547 resulting in up-regulation at the amr locus does not seem to be related to the amrR gene. Alternate regulatory pathways for the currently characterized Mex pumps are just starting to surface. For instance, a recent publication has demonstrated growth-phase regulation for the mexAB-oprM system and shown that this regulation is not dependent on MexR (10). The rplA mutation we recently identified in strains 2547 and 2548 and its link to aminoglycoside resistance require more study. However, this mutation may affect some global regulatory function that results in deregulation and hyperexpression of amrAB as well as many other genes. In any case, it seems that the functional regulation of this pump is quite complex and the identification of other regulatory systems affecting the expression and function of the amr locus could shed light on the pleiotropic effects noted above.
The association between the AGIR phenotype and hyperexpression of the AmrAB efflux system was initially observed in a laboratory-generated mutant of PAO1. However, the potential relevance of this system to the AGIR observed in clinical isolates is also supported by RT-PCR data demonstrating substantially increased transcription of amrAB in clinical strains displaying the AGIR phenotype when compared to a genotypically matched sensitive isolate (e.g., 1168, 1250, and 1249). Because amrAB up-regulation alone does not appear to be sufficient to affect aminoglycoside resistance and because amrAB hyperexpression may not be universally necessary for AGIR one would not expect a direct correlation between amrB expression and resistance phenotype in all strains. However, a general trend of higher-level amrB expression was observed in clinical isolates displaying the AGIR phenotype compared to sensitive isolates. These studies support the hypothesis that the AGIR phenotype is the result of a complex interaction of molecular events and suggest the possibility that multiple molecular scenarios may result in similar AGIR phenotypes. The latter point may contribute to the difficulty in understanding the exact mechanism of this type of resistance. Ultimately, understanding the complicated and potentially diverse manifestations of impermeability resistance may require looking beyond aminoglycoside resistance to other phenotypic differences among this population and perhaps in the process redefine the nature of impermeability resistance. In conclusion, we believe efflux of aminoglycosides by the AmrAB system to be one of the contributing factors in the multifactorial AGIR phenotype, but further study will be necessary to understand the intricate aspects of this type of resistance in P. aeruginosa.
ACKNOWLEDGMENTS
We thank Herbert P. Schweizer at Colorado State University for providing the bacterial strains and plasmids used for construction of the unmarked deletions, Robert E. W. Hancock at the University of British Columbia for providing the pXZL34 plasmid in addition to his knowledgeable insights, Paul Phibbs and Steve Lory from the University of Washington, Seattle, for the PAO1 strain, Maynard Olson at the University of Washington for the P. aeruginosa cosmid library, Romesh Gautom at the Washington State Department of Health, Seattle, for PFGE, L. Goltry and E. Tolentino for oligonucleotide synthesis and DNA sequencing efforts, and M. Lagrou and S. Mizoguchi for informatics support.
REFERENCES
- 1.Aires, J. R., and P. Plesiat. 1998. mexGH, a new multidrug active efflux operon in Pseudomonas aeruginosa GenBank accession no. AF073776. (Unpublished [direct submission to GenBank].)
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan L E, Haraphongse R, Van den Elzen H M. Gentamicin resistance in clinical-isolates of Pseudomonas aeruginosa associated with diminished gentamicin accumulation and no detectable enzymatic modification. J Antibiot. 1976;29:743–753. doi: 10.7164/antibiotics.29.743. [DOI] [PubMed] [Google Scholar]
- 4.Bryan L E, Kwan S. Aminoglycoside-resistant mutants of Pseudomonas aeruginosa deficient in cytochrome d, nitrite reductase, and aerobic transport. Antimicrob Agents Chemother. 1981;19:958–964. doi: 10.1128/aac.19.6.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryan L E, Nicas T, Holloway B W, Crowther C. Aminoglycoside-resistant mutation of Pseudomonas aeruginosa defective in cytochrome c552 and nitrate reductase. Antimicrob Agents Chemother. 1980;17:71–79. doi: 10.1128/aac.17.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryan L E, O’Hara K, Wong S. Lipopolysaccharide changes in impermeability-type aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1984;26:250–255. doi: 10.1128/aac.26.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukanov N, Ravi V N, Miller D, Srivastava K, Berg D E. Pseudomonas aeruginosa corneal ulcer isolates distinguished using the arbitrarily primed PCR DNA fingerprinting method. Curr Eye Res. 1994;13:783–790. doi: 10.3109/02713689409025132. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 9.Enderle P J, Farwell M A. Electroporation of freshly plated Escherichia coli and Pseudomonas aeruginosa cells. BioTechniques. 1998;25:954–958. doi: 10.2144/98256bm05. [DOI] [PubMed] [Google Scholar]
- 10.Evans K, Poole K. The MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa is growth-phase regulated. FEMS Microbiol Lett. 1999;173:35–39. doi: 10.1111/j.1574-6968.1999.tb13481.x. [DOI] [PubMed] [Google Scholar]
- 11.Fralick J A. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gautom R K. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol. 1997;35:2977–2980. doi: 10.1128/jcm.35.11.2977-2980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock, R. E. Personal communication.
- 14.Hancock R E. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin Infect Dis. 1998;27(Suppl. 1):S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa M, Kobayashi I, Saika T, Nishida M. Drug-resistance patterns of clinical isolates of Pseudomonas aeruginosa with reference to their lipopolysaccharide compositions. Chemotherapy. 1997;43:323–331. doi: 10.1159/000239585. [DOI] [PubMed] [Google Scholar]
- 16.Hickey, M. J. Unpublished data.
- 17.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 18.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechere J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 19.Li, X. Z., and K. Poole. Inverse relationship between MexAB-OprM expression and expression of MexCD-OprJ and MexEF-OprN multidrug efflux systems of Pseudomonas aeruginosa, abstr. 21. In Program and abstracts of the ASM Conference on Pseudomonas ’99: Biotechnology and Pathogenesis. American Society for Microbiology, Washington, D.C.
- 20.Lisitsyn N, Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd A M, Davis R W. Functional expression of the yeast FLP/FRT site-specific recombination system in Nicotiana tabacum. Mol Gen Genet. 1994;242:653–657. doi: 10.1007/BF00283419. [DOI] [PubMed] [Google Scholar]
- 22.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren M S, Boyer E, Chamberland S, Lee V J. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:1340–1346. doi: 10.1128/aac.43.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLeod, D. L., L. E. Nelson, R. M. Shawar, B. B. Lin, L. G. Lockwood, J. E. Dirks, G. H. Miller, J. L. Burns, and R. L. Garber. Aminoglycoside resistance mechanisms for Pseudomonas aeruginosa strains in cystic fibrosis patients are unchanged by chronic treatment with inhaled tobramycin. Submitted for publication. [DOI] [PubMed]
- 24.Miller G H, Sabatelli F J, Naples L, Hare R S, Shaw K J. Resistance to aminoglycosides in Pseudomonas. Aminoglycoside Resistance Study Groups. Trends Microbiol. 1994;2:347–353. [PubMed] [Google Scholar]
- 25.Miller G H, Sabatelli F J, Naples L, Hare R S, Shaw K J. The most frequently occurring aminoglycoside resistance mechanisms—combined results of surveys in eight regions of the world. The Aminoglycoside Resistance Study Groups. J Chemother. 1995;7(Suppl. 2):17–30. [PubMed] [Google Scholar]
- 26.Mine T, Morita Y, Kataoka A, Mizushima T, Tsuchiya T. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:415–417. doi: 10.1128/aac.43.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore R A, DeShazer D, Reckseidler S, Weissman A, Woods D E. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests, 6th ed. Document M2-A6. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, Eighth informational supplement. Document M100-S8. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 29a.National Center for Biotechnology Information Website. 13 October 1999, revision date. [Online.] National Institutes of Health, Bethesda, Md. http: //www.ncbi.nim.nih.gov. [27 October 1999, last date accessed.]
- 30.Nicas T I, Hancock R E. Outer membrane protein H1 of Pseudomonas aeruginosa: involvement in adaptive and mutational resistance to ethylenediaminetetraacetate, polymyxin B, and gentamicin. J Bacteriol. 1980;143:872–878. doi: 10.1128/jb.143.2.872-878.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikaido H. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin Infect Dis. 1998;27(Suppl. 1):S32–S41. doi: 10.1086/514920. [DOI] [PubMed] [Google Scholar]
- 32.Ocaktan A, Yoneyama H, Nakae T. Use of fluorescence probes to monitor function of the subunit proteins of the MexA-MexB-oprM drug extrusion machinery in Pseudomonas aeruginosa. J Biol Chem. 1997;272:21964–21969. doi: 10.1074/jbc.272.35.21964. [DOI] [PubMed] [Google Scholar]
- 33.Paulsen I T, Park J H, Choi P S, Saier M H., Jr A family of gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from gram-negative bacteria. FEMS Microbiol Lett. 1997;156:1–8. doi: 10.1111/j.1574-6968.1997.tb12697.x. [DOI] [PubMed] [Google Scholar]
- 34.Phillips I, King A, Shannon K. Prevalence and mechanisms of aminoglycoside resistance. A ten-year study. Am J Med. 1986;80:48–55. doi: 10.1016/0002-9343(86)90479-1. [DOI] [PubMed] [Google Scholar]
- 35.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 36.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poxton I R, Bell G T, Berdlay G R. The association on SDS-polyacrylamide gels of lipopolysaccharide and outer membrane proteins of Pseudomonas aeruginosa as revealed by monoclonal antibodies and Western blotting. FEMS Microbiol Lett. 1985;27:247–251. [Google Scholar]
- 38.Price K E, Kresel P A, Farchione L A, Siskin S B, Karpow S A. Epidemiological studies of aminoglycoside resistance in the U.S.A. J Antimicrob Chemother. 1981;8(Suppl. A):89–105. doi: 10.1093/jac/8.suppl_a.89. [DOI] [PubMed] [Google Scholar]
- 39.Saier M H, Jr, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11:841–847. doi: 10.1111/j.1365-2958.1994.tb00362.x. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schweizer H P. Intrinsic resistance to inhibitors of fatty acid biosynthesis in Pseudomonas aeruginosa is due to efflux: application of a novel technique for generation of unmarked chromosomal mutations for the study of efflux systems. Antimicrob Agents Chemother. 1998;42:394–398. doi: 10.1128/aac.42.2.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 43.Shaw K J, Hare R S, Sabatelli F J, Rizzo M, Cramer C A, Naples L, Kocsi S, Munayyer H, Mann P, Miller G H, et al. Correlation between aminoglycoside resistance profiles and DNA hybridization of clinical isolates. Antimicrob Agents Chemother. 1991;35:2253–2261. doi: 10.1128/aac.35.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shimizu K, Kumada T, Hsieh W-C, Chung H-Y, Chong Y, Hare R S, Miller G H, Sabatelli F J, Howard J. Comparison of aminoglycoside resistance patterns in Japan, Formosa, and Korea, Chile, and the United States. Antimicrob Agents Chemother. 1985;28:282–288. doi: 10.1128/aac.28.2.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 46.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 47.West S E, Schweizer H P, Dall C, Sample A K, Runyen-Janecky L J. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene. 1994;148:81–86. doi: 10.1016/0378-1119(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 48.Wong K K, Poole K, Gotoh N, Hancock R E. Influence of OprM expression on multiple antibiotic resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2009–2012. doi: 10.1128/aac.41.9.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoneyama H, Sato K, Nakae T. Aminoglycoside resistance in Pseudomonas aeruginosa due to outer membrane stabilization. Chemotherapy. 1991;37:239–245. doi: 10.1159/000238861. [DOI] [PubMed] [Google Scholar]
- 50.Young M L, Bains M, Bell A, Hancock R E. Role of Pseudomonas aeruginosa outer membrane protein OprH in polymyxin and gentamicin resistance: isolation of an OprH-deficient mutant by gene replacement techniques. Antimicrob Agents Chemother. 1992;36:2566–2568. doi: 10.1128/aac.36.11.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]