Abstract
Objective:
To demonstrate the diagnostic ability of label-free, point-scanning, fiber-based Fluorescence Lifetime Imaging (FLIm) as a means of intraoperative guidance during oral and oropharyngeal cancer removal surgery.
Methods:
FLIm point-measurements acquired from 53 patients (n = 67893 pre-resection in vivo, n = 89695 post-resection ex vivo) undergoing oral or oropharyngeal cancer removal surgery were used for analysis. Discrimination of healthy tissue and cancer was investigated using various FLIm-derived parameter sets and classifiers (Support Vector Machine, Random Forests, CNN). Classifier out-put for the acquired set of point-measurements was visualized through an interpolation-based approach to generate a probabilistic heatmap of cancer within the surgical field. Classifier output for dysplasia at the resection margins was also investigated.
Results:
Statistically significant change (P < 0.01) between healthy and cancer was observed in vivo for the acquired FLIm signal parameters (e.g., average lifetime) linked with metabolic activity. Superior classification was achieved at the tissue region level using the Random Forests method (ROC-AUC: 0.88). Classifier output for dysplasia (% probability of cancer) was observed to lie between that of cancer and healthy tissue, highlighting FLIm’s ability to distinguish various conditions.
Conclusion:
The developed approach demonstrates the potential of FLIm for fast, reliable intraoperative margin assessment without the need for contrast agents.
Significance:
Fiber-based FLIm has the potential to be used as a diagnostic tool during cancer resection surgery, including Transoral Robotic Surgery (TORS), helping ensure complete resections and improve the survival rate of oral and oropharyngeal cancer patients.
Keywords: Machine learning, medical robotics, surgical guidance/navigation
I. INTRODUCTION
ORAL and oropharyngeal cancer jointly represent 3.0% of all new cancer cases arising in the United States [1]. These pathologies fall within the wider category of head and neck (H&N) cancer [2]. Accurate cancer margin assessment (also referred to as margin delineation) prior to surgical resection is the key factor influencing the long-term survival of oral and oropharyngeal cancer patients, mitigating local reoccurrence due to residual cancer [3]. A cancer margin, as defined by the NIH National Cancer Institute, is “the edge or border of the tissue removed in cancer surgery.” [4]. If assessed correctly, this border surrounds the cancerous tissue as well as a rim of normal tissue in order to subsequently confirm a successful resection. Margin assessment is highly challenging in the context of H&N cancer due to the relatively complex anatomy of the H&N regions and the associated risk of compromising functional and aesthetical features with the resection of additional tissue [5]. Another challenge faced during margin assessment is the occurrence of dysplasia (abnormal, potentially pre-cancerous cells) within the analyzed epithelial tissue [6], leading to a “gray area” between healthy and cancerous tissue. Current approaches for performing margin assessment in H&N cancer include white-light visualization, tactile feedback and frozen section histopathology (an invasive and time-consuming method with the inherent potential for sampling error) [3], [7]. The development of real-time, non-invasive guidance tools can lead to more accurate, faster and more consistent margin assessments [3], particularly during procedures in which direct tactile feedback from tissue is not possible, such as Transoral Robotic Surgery (TORS) [8]–[10]. Accurate discrimination of tissue conditions (cancer, healthy) is a necessary first step in the development of a tool for margin assessment guidance and that is the focus of this work.
Various technologies have been investigated for their utility in margin assessment, including Raman Spectroscopy [11], [12], Optical Coherence Tomography (OCT) [13], [14], and intensity-based fluorescence imaging (with an exogenous contrast agent) [15], [16]. While promising, each of these modalities present certain limitations (e.g., time-consuming analysis, administration of a contrast agent, controlled lighting environment), which has impacted their clinical adoption [3].
Conversely, tissue autofluorescence has been identified as a viable source of non-invasive, real-time, endogenous, diagnostic contrast which does not require the administration of an exogenous contrast agent [17], [18]. Time-resolved autofluorescence techniques such as FLIm (Fluorescence Lifetime Imaging) have been shown to detect variation in the molecular composition of tissue (matrix proteins, metabolic co-factors) [19], allowing for the discrimination of normal and malignant tissue to be observed for various pathologies including oral cancer [15], [19]–[23]. In a recent study, a custom-built fiber-based FLIm unit [24] integrated with the da Vinci Surgical Si System (Intuitive Surgical Inc.) was used during TORS for interrogation of oropharyngeal cancer in 10 patients [25]. Time-resolved (average lifetime, decay dynamics) and spectral (intensity ratio) FLIm parameters derived from three fluorescence emission spectral bands provided patient-level contrast between point measurements acquired at normal and cancerous regions within the surgical field. However, the specific FLIm parameter (or combination of parameters) required for discrimination of cancer and healthy varied between patients due to tissue heterogeneity (e.g., variable thickness of the epithelial layer, ulcerations within tumor mass, etc). Thus, extending this method to provide a generalized diagnostic model through machine learning is a logical next step in the development of FLIm-based tissue discrimination for tasks such as cancer margin assessment.
The method of feature extraction and choice of classification approach for FLIm-based tissue discrimination are two related challenges that need to be addressed together. Previous studies have relied on various sets of hand-engineered FLIm signal parameters (i.e., calculated or derived from the raw signal) to develop classifiers [26], [27]. In contrast, data-driven feature extraction methods (deep learning) may offer advantages such as task-specific feature learning from fluorescence decay waveforms. Deep learning has been shown to lead to superior classification performance in various medical applications [28], [29]. While deep learning methods have previously been used for the deconvolution of raw fluorescence signals in low photon scenarios [30], to our knowledge this technique has not been investigated for fluorescence decay based tissue classification.
Additional challenges for FLIm-based delineation of H&N cancer margins include biochemical differences between anatomical locations (e.g., tongue, tonsil), changes in surgical environment (e.g., pre- and post-excision), or the presence of nuanced tissue conditions such as dysplasia. Thus, understanding which machine learning approaches can account for changes in experimental conditions is important in designing future large-scale studies.
The goals of this study are as follows: (i) to identify the key sources of FLIm-based diagnostic contrast (time-resolved, spectral properties) that can generalize across patients and anatomies in the context of oral and oropharyngeal cancer; (ii) to investigate which combination of feature extraction and classification method (hand-engineered vs data-driven) leads to superior tissue classification performance during oral and oropharyngeal cancer resection surgery in various imaging contexts (in vivo, ex vivo); (iii) to develop a visualization method that can be integrated into a surgical workflow to provide diagnostic contrast for real-time margin evaluation; and (iv) to compare classifier output for healthy tissue, dysplasia at the excision margins, and cancer to observe how FLIm captures the gradient between healthy tissue and cancer.
II. METHOD
A. FLIm System
A custom-built, fiber-based, point-scanning FLIm system [24] was used to acquire data for this study. This system was designed for real-time intraoperative imaging in which FLIm data is augmented onto the surgical field-of-view (FOV) observed by white-light imaging cameras. For in vivo intraoperative imaging, this system was integrated into two distinct surgical approaches:(i) the da Vinci surgical system equipped with an integrated camera [20] and, (ii) a non-robotic approach which combines a hand-held fiber probe (Omniguide Laser Handpiece) and endoscopic camera (Stryker). For ex vivo imaging, surgically excised tissue specimens were scanned using a hand-held fiberoptic probe and scientific camera (Chameleon3, Point Grey). Acquired FLIm data was augmented onto the surgical console in real time to ensure a complete scan [20]. In brief, tissue autofluorescence was excited with a 355 nm (<600 ps FWHM) pulsed laser (micro Q-switched laser, 120 Hz repetition rate, Teem Photonics, France) delivered through a 365 μm core diameter multimode optical fiber (Thorlabs Inc, numerical aperture 0.22). Various instruments and workflows were used for data acquisition. For robotic surgery cases using the da Vinci Si robot (N = 20) the fiber probe was inserted into a fiber introducer instrument. For robotic surgery cases using the da Vinci SP robot (N = 16) the fiber probe was held using one of the robot’s grasper instruments. For non-robotic surgery cases (N = 17) the fiber probe was held by hand by the surgeon. The same fiber optic used for excitation was also used to collect autofluorescence from the scanned tissue region. The fiber’s proximal end was coupled to a Wavelength Selection Module (WSM) which features a set of four dichroic mirrors and bandpass filters (CH1: 390 20 nm; CH2: 470 ± 14 nm; CH3: 542 ± 25 nm; and CH4: 629 ± 26.5 nm) used to spectrally resolve the autofluorescence signal. These four spectral channels were selected based on the autofluorescence emission maxima of specific endogenous fluorophores previously reported as the main contributors to autofluorescence emission, specifically collagen, NADH, FAD, and porphyrins respectively [19]. CH1, CH2 and, CH3 were used for analysis in this study due to the very low signal intensity observed in CH4. The optical signal from each spectral band was time-multiplexed onto a single microchannel plate photomultiplier tube (MCP-PMT, R3809U-50, 45 ps FWHM, Hamamatsu, Japan), amplified (AM-1607–3000, Miteq Inc., USA), and time-resolved by a high sampling frequency digitizer (12.5 GS/s, 3 GHz, 8-bit, 512 Mbytes, PXIe-5185, National Instruments, Austin, TX, USA). Signal-to-noise ratio (SNR) was calculated for each spectral channel for each digitized point measurement.
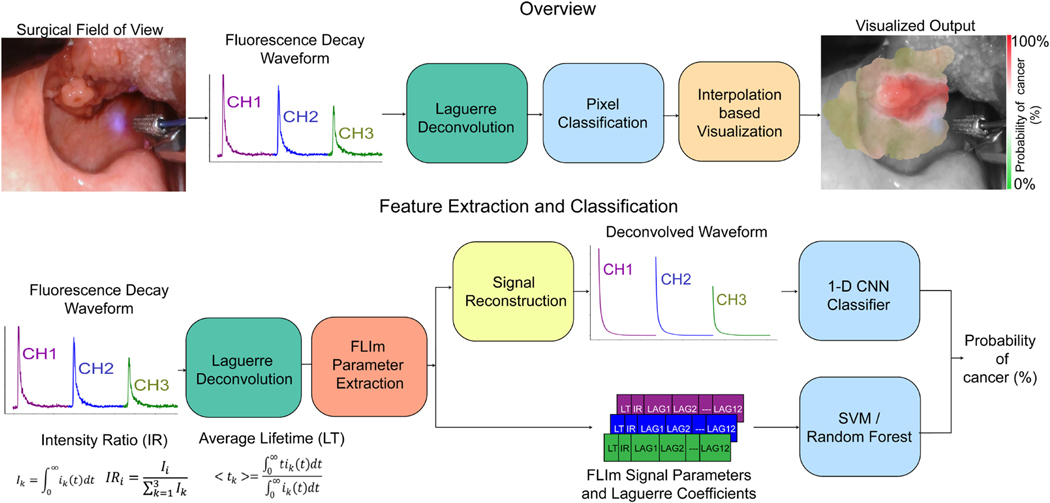
The lateral resolution of the system is determined by the illumination spot size and collection geometry, which is improved when the probe is closer to tissue. A background subtraction step was performed on the acquired waveform using a probe back-ground signal acquired at the beginning of each clinical case. A Laguerre expansion based deconvolution [31] was performed on the acquired signal using the system Impulse Response Function (IRF), producing a fluorescence decay waveform for each spectral channel (following reconstruction) and 12 Laguerre coefficients per channel (see Fig. 1).
Fig. 1.
(Top) Overview of the FLIm-based tissue classification methods used for intraoperative cancer margin assessment. For clinical data collection, a custom-built Fluorescence Lifetime Imaging (FLIm) point-scanning system was integrated with either a robotic surgery platform (da Vinci) or an endoscopic camera. Multi-channel fluorescence decay waveforms were acquired and localized within the white light image of the specimen using the aiming beam (blue). Deconvolution and classification were performed before classifier output was visualized using a distance and signal-to-noise ratio (SNR) based interpolation method. (Bottom) Feature extraction and classification process: A Laguerre expansion based deconvolution was performed using the system impulse response function (IRF) producing a set of 12 Laguerre coefficients per channel. Feature extraction and classifier training was performed for three distinct classification approaches including Support Vector Machine (SVM) and Random Forests (RF) classifiers (both trained on extracted FLIm parameters and Laguerre coefficients) as well as a 1-D CNN classifier trained on fluorescence decay waveforms. The deconvolved waveforms were reconstructed using Laguerre basis functions and the associated set of 12 coefficients computed for each channel.
To localize each FLIm point measurement, a 445 nm continuous-wave aiming beam (TECBL-50G-440-USB, World Star Tech, Canada) was employed, as described earlier [24], [32]. This aiming beam was integrated into the optical path of the WSM and delivered to the specimen through the same optical path used to induce tissue autofluorescence. The aiming beam was localized within a 2-D white light image of the surgical field, which is captured via the integrated camera (see Fig. 1). More specifically, the center of the beam for a given measurement was localized by transforming the image to the HSV color space, thresholding the hue and saturation channels, and performing a series of morphological operations as described previously [32]. Block-matching based motion tracking was retrospectively applied to overcome any errors due to tissue motion encountered during acquisition and to ensure that all point measurement locations are corrected with respect to a desired reference frame in the video sequence which was used for ground truth annotation of the specimen.
B. Data Acquisition, Histological Coregistration and Preprocessing
Fifty three patients undergoing upper aerodigestive oncologic surgery at the University of California Davis Medical Center were recruited after determining their eligibility for the research procedure. Research was performed under Institutional Review Board (IRB) approval and with patient’s informed consent. During a procedure, the surgeon (DGF, ACB, AFB, MA) identified the tissue areas of interest based on preoperative planning. FLIm point measurements were then acquired by scanning the fiber probe over this region as well as peripheral healthy tissue for comparison. FLIm data was collected during two stages of surgical resection (i) in vivo before surgical resection and (ii) ex vivo on the surgically excised specimen.
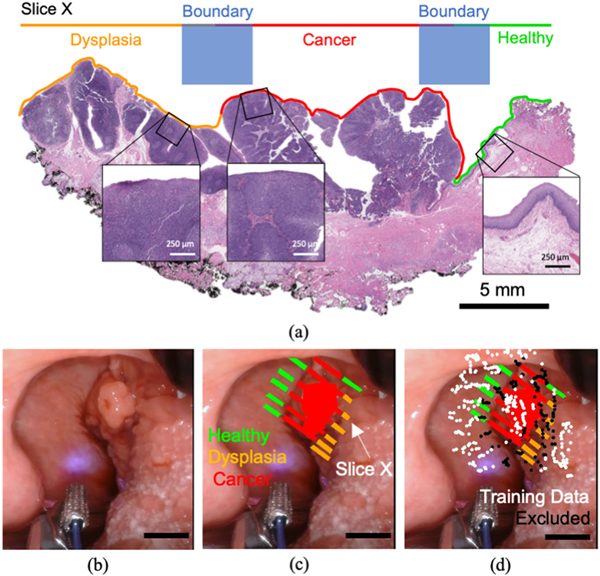
Immediately following the ex vivo scan, the resected specimen was sent for sectioning and histopathology staining (Hematoxylin and Eosin (H&E) and P16 immunostaining when required). The fixed sections were placed on slides and annotated (Fig. 2(a)) for healthy, dysplasia and cancerous regions by a pathologist (RFG). These histology slide annotations were then used to provide labeling of different tissue regions within the white light images of each ex vivo specimen (Fig. 2(c)). This labeling was performed by localizing each histology slide within the ex vivo image using gross sectioning cut locations and morphological landmarks. This set of annotations was then translated to the in vivo images of the specimen by accounting for sample orientation information (obtained from clinical notes taken by the research personnel, surgeons, and pathologist) and the matching of morphological features visible in the surgical field as well as on the excised specimen. Two tissue classes, “healthy” and “cancer”, were considered for classification purposes, while regions of dysplasia (abnormal, possibly pre-cancerous cells) marked at the resected margins were analyzed separately. All white light images were acquired at 720 × 1280 pixel resolution. A pixel/mm scale was derived from the known dimensions of surgical instruments visible in the surgical field. Measured specimen dimensions found in the associated tissue grossing report were used to validate this estimated scale.
Fig. 2.
Tissue annotation and training data selection process. (a) The ground truth for classifier training was derived directly from histopathology via H+E staining. Each annotated slice was coregistered with white light images of the specimen (in vivo and ex vivo), (b) Surgical FOV for in vivo imaging of a given specimen and (c) Corresponding registration of pathology (in vivo). Homogenous regions of a single tissue label (healthy, cancer, dysplasia) were annotated in a region-based fashion. (d) Point measurements centered at a boundary between disparate tissue conditions, specifically within a 15-pixel radius of multiple tissue labels (approx. 2.0 mm), were excluded from classifier training due to their ambiguous ground truth. The scale bar for white light images corresponds to 5 mm.
To account for potential errors in ground truth coregistration (e.g., in vivo motion artifacts, non-uniform shrinkage of tissue after excision, tissue condition boundaries) point measurements centered at a boundary between distinct tissue conditions (i.e., healthy and cancer), specifically within a 15px radius of multiple tissue labels (approx. 2 mm), were excluded from classifier training due to their ambiguous ground truth (Fig. 2(d)). This 2 mm radius was selected to ensure no intermediate tissue conditions (e.g., healthy-cancer tissue interface) at the margins were included when training a binary classifier. Measurements acquired at heterogenous tissue label boundaries were however included when generating a heatmap visualization to qualitatively evaluate classifier output. Data-points located more than 2 mm from an annotated pixel, coregistered with dysplasia or acquired with a signal-to-noise ratio (SNR) lower than 30 dB for any of the three channels were excluded from model training and validation. This SNR threshold was selected as a trade-off between good signal quality and an overly aggressive removal of training data which would limit the robustness of a trained classifier. FLIm data-points for which a correct aiming beam localization and registration was not possible were also excluded from analysis.
C. Classifiers Trained Using Time-resolved and Spectral FLIm Parameters
Support Vector Machine (SVM) [33] and Random Forests (RF) [34] classifiers were trained using hand-engineered FLIm features from three spectral channels. For each channel, average lifetime and spectral intensity ratio parameter values were combined with 12 Laguerre coefficients calculated during the deconvolution process (Fig. 1) [20]. These coefficients capture the decay dynamics of the fluorescence waveform. This results in a total of 42 features per point measurement. Various subsets of this feature vector were investigated. A Radial Basis Function (RBF) kernel was employed for SVM training with the regularization parameter (i.e., C parameter) set to 1.0 as is commonly employed in the field [35], [36]. This C value was not experimented with in this study. The RF model uses 100 decision trees/estimators, each with a max depth of 10. A random forest ensemble size of 100 was selected as the classifier performance reached an upper limit and did not increase further beyond this ensemble size. Class-level weights were applied during training to mitigate class imbalance problems. Equations for average fluorescence lifetime (LT) and spectral intensity ratio (IR) are presented in Fig. 1, with ik referring to the deconvolved decay waveform for a given channel k. A comprehensive description of the deconvolution and parameter extraction process is described in a prior work [31].
D. CNN-Based Feature Extraction and Classification
A 6-layer 1-D convolutional neural network (CNN), consisting of four convolutional layers (12, 24, 48 and 64 kernels respectively) and two fully connected layers (32 and 2 neurons), was trained to classify a given multi-channel fluorescence decay waveform. All 1-D convolutional kernels were 3 units in length. Max pooling (stride: 2, kernel length: 3) was performed following each convolutional layer. A rectified linear unit (ReLU) activation [37] was applied following each network layer (apart from the last). Average pooling was then performed across the entire signal length prior to the first fully connected layer, producing a flattened 64-D input vector. The final fully connected layer was followed by a softmax activation to produce the classification output. An intensity scaling factor was applied to each channel within a given decay waveform based on the relative differences in signal intensity (area under the curve of the original waveform). This ensures only relative differences in spectral intensity are used for classification, rather than the absolute intensity which is largely determined by probe to tissue distance.
Classifier training was performed using Stochastic Gradient Descent (SGD) with backpropagation [38] for 20,000 iterations with a momentum of 0.9, a learning rate of 0.001, a batch size of 64 and an L2 weight decay of 5e-4. This CNN training configuration was employed as it lead to smooth convergence during training. Cross-entropy loss, given in equation 1, was used as the objective function. Weighted batch sampling was performed during model training in order to address any class imbalance within the training data.
| (1) |
Due to the limited training data available, CNN pre-training was performed for a related regression task, average fluorescence lifetime estimation, on synthetic data before fine-tuning the model for classification. 100,000 synthetic 3-channel fluorescence decay measurements were generated through the convolution of synthetic multi-exponential decay waveforms with a measured instrument impulse response function (IRF) as described previously [31]. Each synthetic waveform has a random number of exponential components (between 1 and 6) [31]. Random channel level weights (summing to 1.0) were applied to simulate relative difference in intensity between spectral bands. This dataset was used to pre-train the CNN model for average lifetime estimation using the same configuration as classifier training. An alternate final fully connected layer with 3 neurons was included to perform average lifetime estimation and is removed prior to classifier training. Mean squared error was minimized to train this regression task. Performing CNN classifier training in this study without pre-training often results in the model failing to converge and ultimately very poor performance.
The proposed classification method was developed using the PyTorch numerical library [39]. Network optimization was accelerated using an Nvidia GeForce GTX 1050 GPU (Graphics Processing Unit).
E. Visualization Approach
A classification heatmap was generated for the FLIm scan of a given specimen through a combination of inverse distance weighted interpolation [40] and SNR weighting to aggregate nearby cancer probability scores. Any pixel location within a 4 mm radius of at least 5 data-point centers was included in the heatmap to focus the visualization on the more densely scanned regions. For each heatmap pixel xi, the N cancer classification scores centered within a 4 mm radius were aggregated together using equations 2–4.
| (2) |
| (3) |
| (4) |
N is the number of data-points within the 4 mm radius a given pixel location xi, fi corresponds to the predicted probability of cancer for data-point i, di is the Euclidean distance between the influencing data-point i and the pixel of interest xi. si is the set of channel level SNR values recorded for the influencing data-point i. P is the weighting exponent. Greater values of P assign a greater influence to the data-points closest to the pixel of interest (x, y) as well as data-points with higher SNR. A P alue of 1.0 was used in this study as it results in smooth heatmaps which do not highlight small local variations related to noise. After performing this aggregation for an entire specimen scan, the generated heatmap was colorized using an RGB colormap which interpolates between green (0 255 0), white (255 255 255) and red (255 0 0), with green corresponding to 0.0% predicted cancer probability and red corresponding to 100%. The heatmap was then overlaid onto the white light image of the specimen. An example of this visualization output is shown in Fig. 1 and Fig. 6. The developed visualization method was implemented in the Python programming language and employs the OpenCV image processing library [41].
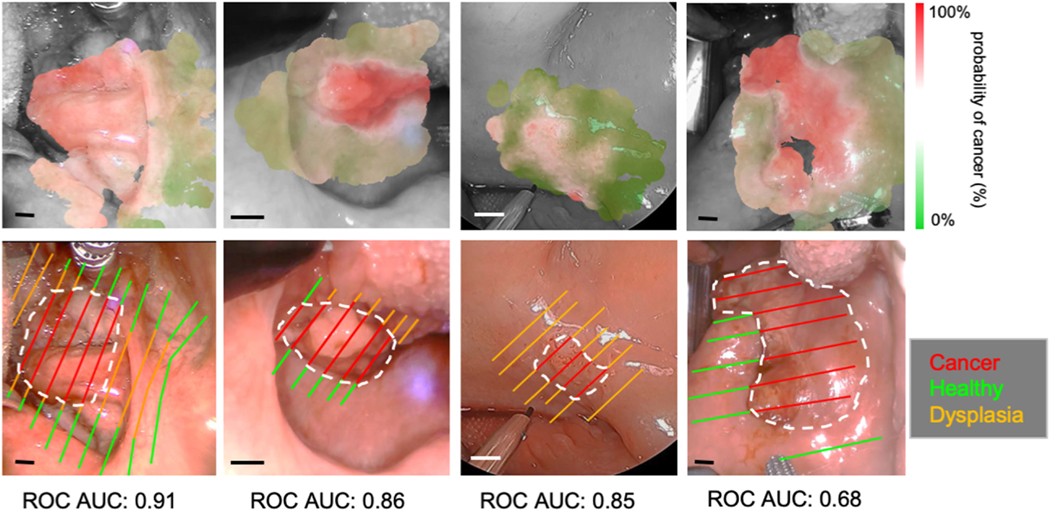
Fig. 6.
Classification visualizations and histological slice annotations for four in vivo patient scans (RF method used). Only labels derived directly from histology are shown with cancer outlined in white. Additional labels for homogeneous regions (healthy/cancer only) were also employed for model training. Clear deliniation of cancer regions is observed, even for patients with a lower AUC score. This is due partially to the presence of dysplasia which is omitted from training/validation as well as a small margin of error with histological coregistration. The scale bar corresponds to 5 mm in all cases.
F. Evaluation Metrics
Classifier evaluation was performed at the tissue region level to evaluate the diagnostic capability of the method and then at the point-measurement level to assess the capability for margin assessment over an entire tissue surface. A tissue region in this context refers an entire cancer or healthy region within the surgical field for a given patient. Both evaluations were performed via a leave-one-out cross-validation.
For tissue region evaluation, a mean probability of cancer was calculated for all point-measurements acquired within a given tissue region producing an overall binary prediction (cancer or healthy). These region level predictions were then used to compute region-level sensitivity, specificity and receiver operator characteristic area-under-the-curve (ROC-AUC) for the entire dataset using equations 5, 6 and 7. An ROC curve based threshold selection step was performed to find the optimal trade-off of sensitivity and specificity for region-level evaluation. TP, FN, TN and FP correspond to the number of true positives, false negatives, true negatives, and false positives respectively. TPR and FPR refer to the true positive rate and false positive rate as a function of decision threshold.
| (5) |
| (6) |
| (7) |
For point-measurement level evaluation, sensitivity, specificity and ROC-AUC were computed for a given patient using all acquired point-measurements before an evenly weighted mean and 95% confidence interval were calculated across all patients. ROC-AUC was only computed at the point-measurement level for cases in which at least 100 data points from healthy and 100 from cancer were acquired in order to prevent misleading patient level scores caused by highly imbalanced data. Sensitivity and specificity were calculated using all patients in which cancer or healthy point measurements were acquired respectively.
G. Statistical Analysis
The Shapiro-Wilk normality test [42] was used to assess whether parametric statistical tests should be used for the FLIm data acquired in this study. Application of this test demonstrated a non-normal distribution at the patient and inter-patient level, therefore the non-parametric Mann-Whitney U test [43] was performed when comparing FLIm parameters for distinct tissue conditions (healthy, cancer).
III. EXPERIMENTAL RESULTS
A. Dataset and Histopathology Breakdown
Table I presents a breakdown of the in vivo and ex vivo FLIm datasets used for analysis in terms of acquired pixels/point measurements following preprocessing. This dataset has the following anatomical breakdown: 20 tonsil, 23 tongue, 1 glossotonsillar sulcus, 1 floor of the mouth, 1 palate, 1 pharynx, 1 left posterior maxilla, 1 gingiva, 1 lip and 3 patient with unknown primary cancer (multiple locations imaged). An in vivo scan was not available for 3 patients due to either acquisition or coregistration challenges; ex vivo scans were however acquired for these cases.
TABLE I.
COMPOSITION OF ORAL AND OROPHARYNGEAL CANCER DATASET
| Imaging Context | Healthy (pixels) | Dysplasia (pixels) | Cancer (pixels) |
|---|---|---|---|
|
| |||
| in vivo (N=50) | 27904 | 16329 | 23660 |
| ex vivo (N=53) | 21396 | 26135 | 32164 |
For classification purposes, the dataset was split into two general classes, “healthy” and “cancer”, with a range of conditions found in each class. Healthy epithelium is made up of a variety of features, including epithelium of varying thickness as well as keratinized epithelium, stratified epithelium and ulcerated tissue. The cancer class consists of invasive, basaloid, verrucous and ulcerated squamous cell carcinoma (SCC). In certain cases tissue from only one of the two general classes (“healthy” and “cancer”) was imaged, either due to prominent dysplasia within the imaged tissue region or a given tumor being identified as benign in histology and deemed “healthy”. Dysplasia at the resection margins was identified in 44 cases. Certain anatomies and conditions (floor of the mouth, pharynx, posterior maxilla, gingiva, ulceration, filiform papillae, lip, palate, excessive bleeding) occur in single patients resulting in these patients being omitted from classifier cross-validation experiments. The ability of the trained classifier to generalize to these cases was then investigated.
B. Univariate Statistics
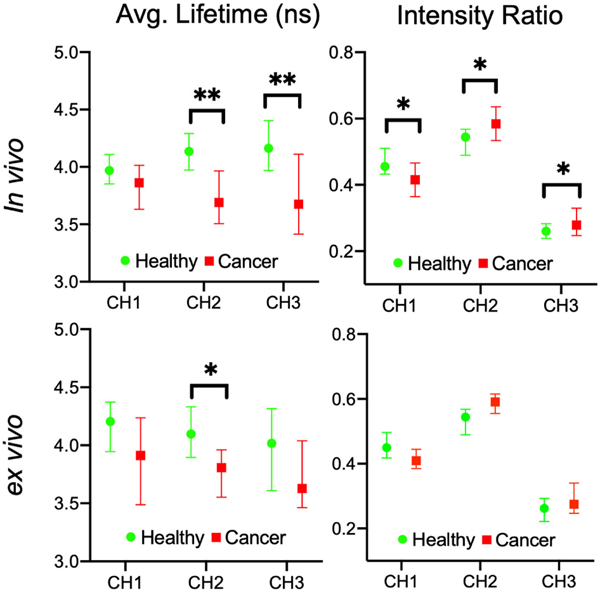
Fig. 3 compares median lifetime and intensity ratio values for cancer and healthy data for all anatomies (in vivo and ex vivo) for the three spectral channels. Statistical significance was investigated using the Mann-Whitney U test, with a greater number of significant parameters observed for in vivo compared to ex vivo, suggesting the additional variation caused by removing tissue from the body (cauterization, loss of blood flow influencing metabolism) has limited the contrast between conditions. The key sources of in vivo contrast include CH2 lifetime and CH3 lifetime. Overall, cancerous tissue presented shorter lifetime values compared to the surrounding healthy tissue. A spectral shift was also observed in cancer (in vivo and ex vivo), with increased CH2 and CH3 intensity relative to CH1.
Fig. 3.
Average fluorescence lifetime and spectral intensity ratio changes between healthy tissue and cancer observed for (a) in vivo and (b) ex vivo FLIm. All anatomies imaged in this study are included while dysplasia point measurements were omitted. Median parameter values were taken from each patient for healthy and cancer, with a 95% confidence interval calculated. The Mann-Whitney U test (*p<0.05 **p<0.01) was applied, with a greater number of significant FLIm parameters observed for in vivo compared to ex vivo, particularly CH2 and CH3 lifetime.
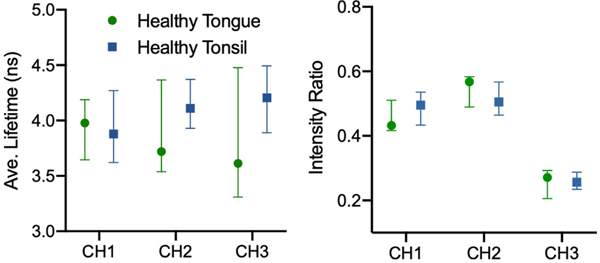
Fig. 4 presents the difference in median FLIm parameter values between healthy tongue and tonsil patients imaged in vivo. Greater variation between anatomies is observed in CH3 lifetime for healthy tissue relative to CH2 suggesting CH2 may provide more robust diagnostic contrast for intraoperative scanning. An increase in fluorescence lifetime with wavelength was observed for healthy tonsil, while the opposite was seen for healthy tongue, suggesting the biochemical differences between these anatomies can be highlighted with FLIm. An ex vivo comparison of healthy tissue by anatomy was not presented due to the inferior contrast observed in this context and given that intraoperative in vivo scanning is the focus of this study. However, this result is included in the supplementary materials (S1).
Fig. 4.
Average fluorescence lifetime and spectral intensity ratio medians (with 95% confidence interval) for healthy tongue and tonsil specimens (in vivo). Higher variability was observed in CH3 lifetime as well as CH1 and CH2 intensity ratio, suggesting these parameters do not generalize well across anatomies due to differences in biochemistry captured by FLIm.
C. Region-Level Classifier Evaluation
Table II presents region-level cancer vs. healthy classification performance of the hand-engineered feature methods (SVM, RF) and deep learning methods (1-D CNN) for in vivo and ex vivo scans, respectively. All three spectral channels were used for training. This leave-one-out cross validation experiment was limited to tongue and tonsil patients only. Classification of in vivo scans was observed to be noticeably superior to ex vivo, while the Random Forests method lead to superior region-level discrimination with an in vivo ROC-AUC of 0.88, sensitivity of 86% and specificity of 87%.
TABLE II.
REGION-LEVEL HEALTHY VS. CANCER CLASSIFICATION PERFORMANCE FOR TONGUE AND TONSIL FLIM SCANS
| Method | in vivo | ex vivo | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Sens.(%) N=30 | Spec.(%) N=39 | ROC-AUC | Sens.(%) N=34 | Spec.(%) N=33 | ROC-AUC | |
|
| ||||||
| 1-D CNN | 79 | 76 | 0.78 | 72 | 66 | 0.69 |
| SVM | 78 | 75 | 0.77 | 72 | 69 | 0.68 |
| RF | 86 | 87 | 0.88 | 78 | 72 | 0.74 |
D. Point-Measurement Level Classifier Evaluation
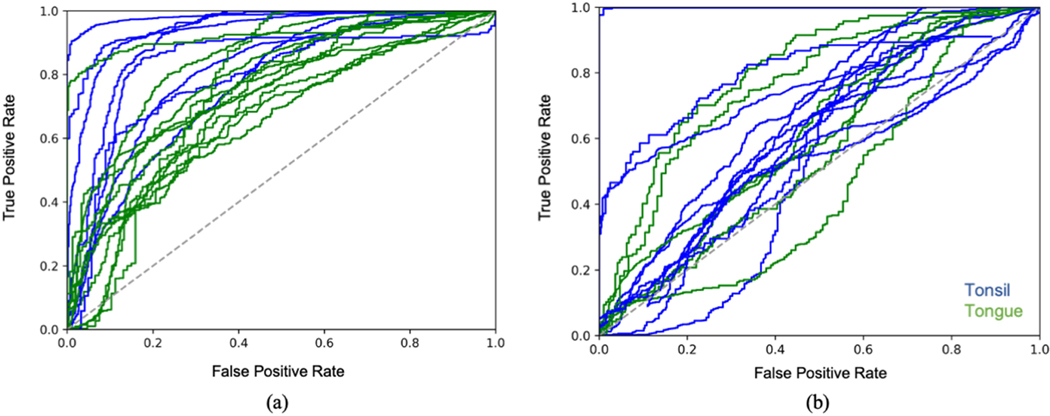
Tables III and IV compare point-measurement level cancer vs. healthy classification performance of the hand-engineered feature methods (SVM, RF) and deep learning methods (1-D CNN) for in vivo and ex vivo scans, respectively. This experiment was limited to tongue and tonsil patients only. The Random Forests method was observed to produce the strongest classification performance for both in vivo and ex vivo, even compared to the 1-D CNN, achieving a mean AUC of 0.79 ± 0.04 and 0.65 ± 0.08 respectively. Fig. 5 presents the ROC curves for in vivo and ex vivo classification. Performance is notably superior for in vivo scans compared to ex vivo, which is consistent with the statistical results reported in the previous subsection. The remainder of the results will focus on in vivo imaging given that this is the target application of the method.
TABLE III.
POINT-MEASUREMENT LEVEL HEALTHY VS. CANCER CLASSIFICATION PERFORMANCE FOR IN VIVO TONGUE AND TONSIL FLIM SCANS (±95% CI)
| Method | ROC-AUC | Sensitivity(%) (n=18291) | Specificity(%) (n=23012) |
|---|---|---|---|
|
| |||
| 1-D CNN | 0.70±0.03 | 62±17 | 67±10 |
| SVM | 0.71±0.07 | 59±13 | 69±10 |
| RF | 0.79±0.04 | 72±11 | 69±10 |
TABLE IV.
POINT-MEASUREMENT LEVEL HEALTHY VS. CANCER CLASSIFICATION PERFORMANCE FOR EX VIVO TONGUE AND TONSIL FLIM SCANS (±95% CI)
| Method | ROC-AUC | Sensitivity(%) (n=26512) | Specificity(%) (n=17526) |
|---|---|---|---|
|
| |||
| 1-D CNN | 0.61±0.08 | 60±11 | 60±9 |
| SVM | 0.60±0.07 | 59±13 | 62±9 |
| RF | 0.65±0.09 | 67±11 | 60±10 |
Fig. 5.
ROC curves for healthy/cancer classification of (a) in vivo and (b) ex vivo FLIm scans at the point-measurement level. Each curve is color coded by anatomy. Only tongue and tonsil patients for which 100 healthy and 100 cancer point measurements were acquired was included in this analysis. The Random Forests classification method (using all three spectral channels) was used in all cases. Superior classification performance was observed for in vivo specimens compared to ex vivo as well as for in vivo tonsil cases relative to in vivo tongue.
E. Contribution of FLIm Parameters to Classification Performance
Table V compares in vivo point-measurement level classification performance for various subsets of the 42 FLIm parameters used. Time-resolved parameters, and in particular the Laguerre coefficients from CH2, were observed to contribute the most towards discrimination, with only a marginal decrease in mean AUC compared to using all FLIm parameters (0.78 ± 0.7 vs. 0.79 ± 0.04). This is consistent with the statistical results from earlier which highlighted CH2 time-resolved parameters as a robust source of contrast. Laguerre coefficients provide a more granular representation of fluorescence decay dynamics than the concise average lifetime representation, leading to superior classification performance.
TABLE V.
FEATURE SET COMPARISON FOR IN VIVO TONGUE AND TONSIL FLIM SCANS USING RANDOM FORESTS (±95% CI)
| Features | ROC-AUC | Sensitivity(%) (n=18291) | Specificity(%) (n=23012) |
|---|---|---|---|
|
| |||
| All FLIm Parameters | 0.79±0.04 | 72±11 | 69±10 |
| Time-resolved Only | 0.79±0.04 | 71±15 | 70±9 |
| Spectral Intensity Only | 0.60±0.07 | 54±16 | 60±12 |
| Time-resolved CHI Only | 0.72±0.04 | 63±12 | 68±9 |
| Time-resolved CH2 Only | 0.77±0.06 | 71±13 | 69±10 |
| Time-resolved CH3 Only | 0.74±0.04 | 71±12 | 61±12 |
| LAG CH2 Only | 0.78±0.07 | 71±15 | 70±10 |
F. Generalization to Unseen Anatomies and Conditions
Table VI presents the AUC score observed when a Random Forests classifier trained on in vivo tongue and tonsil patients with commonly occurring conditions was used to classify previously unseen anatomies and conditions. Discrimination performance similar to that of tongue and tonsil patients (AUC > 0.65, specificity > 0.60) was observed for unseen anatomies (pharynx, lip, palate, floor of mouth, gingiva) as well as unknown primary cases in which several anatomies were scanned. In certain cases only healthy tissue was imaged resulting in specificity being presented. The posterior maxilla and glossotonsillar sulcus cases were only imaged ex vivo. The presence of prominent ulceration on the tissue also does not appear to result in a noticeable decrease in performance. A situation where discrimination is challenging is the occurrence of excessive tissue bleeding within the scanned region. This is due to the absorption of UV excitation light by blood, which attenuates the signal and influences calculated FLIm parameter values. Significant bleeding within the scanned region only occurred in 1/53 patients enrolled in this study. The substantial presence of filiform papillae on one of the tongue specimens results in very poor discrimination, however this patient was noted to be very old (82 years) relative to the median age (65) at the time of surgery. This may lead to additional confounding factors relative to the other cases which were not captured by histopathology. Classification performance for ex vivo imaging of these one-off cases was observed to be inferior and is included in the supplementary materials (S2).
TABLE VI.
TISSUE CLASSIFICATION PERFORMANCE FOR ONE-OFF ANATOMIES AND CONDITIONS (IN VIVO) USING RANDOM FORESTS
| Anatomy/Condition | ROC-AUC |
|---|---|
|
| |
| Excessive Bleeding | 0.51 |
| Ulceration | 0.73 |
| Pharynx | 0.78 |
| Floor of Mouth | Specificity: 0.66 |
| Gingiva | 0.85 |
| Lip | Specificity : 0.75 |
| Palate | 0.86 |
| Filiform Papillae | 0.44 |
| Unknown Primary A | 0.68 |
| Unknown Primary B | Specificity: 0.67 |
| Unknown Primary C | Specificity: 0.95 |
G. Comparison of AUC Score and Visualized Output
Fig. 6 presents the visualized classifier output and AUC score for four in vivo patients. The Random Forests classifier method with all 42 FLIm parameters was employed. In all cases there is clear contrast between the cancer and heathy regions, however, there is no clearly observable relationship with the quality of the associated visualization and the point-measurement level AUC score. This is partially due to the presence of dysplasia which is included in visualization but not in cross-validation. Even for the worst performing of the four (AUC: 0.68) the cancer and healthy regions are clearly delineated. This reduced AUC performance may be related to minor (<1 mm) coregistration errors when labelling the tissue as well as variation in fiber-tissue distance and angle during acquisition. A qualitative evaluation by a physician will be required to ultimately determine the utility of this tool for surgical guidance.
H. Comparison of Classifier Output for Healthy Tissue, Dysplasia and Cancer
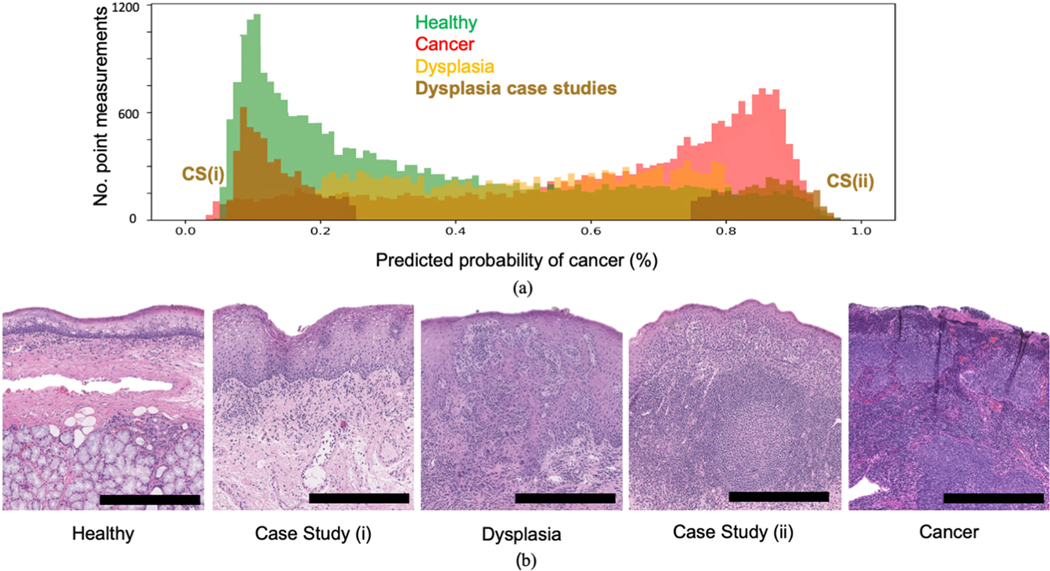
Fig. 7 presents the distribution of classifier output (probability of cancer) for healthy, dysplasia and cancer point measurements taken from all in vivo tongue and tonsil patients. Histology examples of the various conditions are also highlighted. This output was recorded during cross-validation using the Random Forests method (all 3 channels used). Two patients with tissue registered to dysplasia were investigated as case studies due to their predicted cancer probability being consistently low (case study (i)) and high (case study (ii)) respectively. Under the pathologist’s guidance (RGE), closer review showed that the patient in case study (i) contained tissue which was hyperplasic and deemed to be in an earlier stage than the typical dysplasia observed in the dataset. The patient in case study (ii) had significantly higher proportions of lymphatic tissue within the penetration depth of the laser (<250 μm) relative to the typical dysplasia case, likely skewing the classifier to predict high cancer probability. The remaining dysplasia cases consist of probability values spread across the probability distribution suggesting a range of dysplasia gradings are present in the dataset. These results suggest classifier output for FLIm point measurements can potentially be linked with dysplasia grading and various sub-conditions.
Fig. 7.
Distribution of classifier output (% probability of cancer) for in vivo point measurements registered with healthy, dysplasia and cancer from tongue and tonsil specimens. (a) Histograms of classier output for the three conditions including two patient case studies of dysplasia. The tissue condition for each case study was independently verified by a pathologist to prevent bias. The trained classifier predicts low probability of cancer for case study (i) (deemed to be hyperplasia) and high probability of cancer for case study (ii) (shown to be lymphatic tissue). Dysplasia point measurements from other patients are evenly spread across the probability scale. This suggests classifier output for FLIm point measurements can be linked with dysplasia grading. (b) H&E examples showing the gradient between healthy, dysplasia and cancer including the two case study patients. The scale bar in all cases corresponds to 0.5 mm.
I. Effect of Ensemble Learning
Table VII presents the effect of varying the number of estimators/decision trees on in vivo point-measurement level classification performance for tongue and tonsil patients (three spectral channels used). An increased number of RF estimators results in improved generalization, with superior performance to CNN and SVM achieved with just 10 estimators. This increase however plateaus at 100 estimators. These results suggest ensemble learning methods, enabled by the rapid training time of decision trees, leads to reduced overfitting of FLIm data compared to the regularization methods used for CNN and SVM training.
TABLE VII.
THE EFFECT OF VARYING THE NUMBER OF RF ESTIMATORS ON IN VIVO TONGUE AND TONSIL CLASSIFICATION (±95% CI)
| Method | ROC-AUC | Sensitivity(%) (n=18291) | Specificity(%) (n=23012) |
|---|---|---|---|
|
| |||
| RF (200 trees) | 0.79±0.06 | 72±16 | 70±11 |
| RF (100 trees) | 0.79±0.04 | 72±11 | 69±10 |
| RF (10 trees) | 0.77±0.05 | 70±14 | 68±11 |
| RF (1 tree) | 0.68±0.05 | 59±13 | 64±9 |
IV. DISCUSSION
This study highlights FLIm’s potential for performing intraoperative discrimination of tissue types (cancer, healthy) through a combination of machine learning and visualization. Strong diagnostic contrast across various anatomies of the oral cavity and oropharynx was observed with a region-level ROC-AUC of 0.88, sensitivity of 86% and specificity of 87% achieved for intraoperative in vivo scans. For the more challenging point-measurement level in vivo evaluation, an ROC-AUC of 0.79 ± 0.04, a sensitivity of 72 ± 11% and a specificity of 69 ± 10% is observed. The free-hand scanning method employed allows for local diagnostic contrast to be obtained for a desired region of interest within the surgical field. The strong discrimination observed using FLIm highlights the potential for this device to be utilized for label-free margin assessment in a larger study in the future.
The use of a Random Forests classifier results in the strongest discrimination when compared to SVM and 1-D CNN methods. FLIm based tissue classification using Random Forests has previously been observed to achieve strong classification performance for ex vivo breast tissue [27], suggesting this method for modelling/classification is well suited to this data type. FLIm data acquired in a clinical setting is observed to have high variability due to inherent patient-level differences and local heterogeneity within tissue, increasing the likelihood of overfitting. The ensemble approach of Random Forests demonstrates superior generalization to the regularization methods of CNN and SVM. Performing a similar ensemble method using an SVM or CNN is possible [44], [45] but would be incredibly demanding in terms of training time on this dataset, with a single SVM/CNN classifier in this study taking at least 10x longer to train than a full random forest (made up of 100 decision tree classifiers).With robust, real-time classification a requirement of this application, a more in-depth investigation of efficient ensemble classification methods for FLIm will be carried out in a future study.
The key source of diagnostic contrast observed in this study is the time-resolved FLIm parameters acquired from CH2 (470 ± 14 nm), specifically the 12 Laguerre coefficients. This result highlights the value of capturing time-resolved fluorescence decay dynamics, especially given the noticeably inferior classification performance observed using purely spectral intensity parameters. CH2 targets the emission maxima of the metabolic co-factor NADH (Nicotinamide Adenine Dinucleotide (reduced form)) and while there are likely several other fluorophores contributing to the contrast observed in tissue, this spectral band and the biochemical properties it relates to (i.e., the ratio of bound and unbound NADH [19], the Warburg Effect [46]) provide a robust source of diagnostic contrast in oral and Oropharyngeal cancer.
in vivo tissue discrimination is shown to be far superior to ex vivo, both in terms of univariate statistics and classifier performance. The effects of tumor excision (cauterization, loss of blood supply, changes to protein expression) [47]–[50] introduce additional biochemical variability, and in particular metabolic changes, which limits diagnostic contrast. These effects can also vary with resection time during a procedure. The effects of cauterization on FLIm has been demonstrated in an earlier study [47]. Given that the key source of in vivo contrast for these anatomies is CH2 and that this band is linked closely with the metabolic activity, it is not surprising that a loss of oxygenation impacts this source of contrast greatly. Other intrinsic sources of patient-level variation include distinct anatomies, patient age, smoking history and distinct cancer phenotypes. These factors may explain the variation in classification performance between patients. However, none of these factors have the same limiting effect on diagnostic contrast as tumor excision and loss of blood flow. This result suggests that studies of this type which rely on metabolic activity must be performed in vivo and therefore intraoperatively.
It is important to note the distinction between region-level evaluation and point-measurement level evaluation of the developed FLIm method, particularly when comparing to other imaging methods employed in the assessment of head and neck tumour margins. A region-level evaluation, performed through the aggregation of point-measurements, was included to allow for more direct comparisons with other imaging approaches in terms of diagnostic capability. Gao et al. [51] use near-infrared (NIR) fluorescence imaging and a molecular probe (panitumumab-IRDye800CW) to detect cancer in oral and Oropharyngeal tissue specimens, with a sensitivity of 92 ± 2.7% and a specificity of 91 ± 1.5% achieved through a similar region-level evaluation of tissue slices. This performance was achieved with a panitumumab-IRDye800CW dosage of 1.0 mg/kg. Using a lower dosage of 0.5 mg/kg, however, specificity falls to 78 ± 10.3% [51]. The label-free FLIm method reported here achieves competitive diagnostic performance without an exogenous molecular probe or the controlled lighting environment needed for NIR. This lack of restrictions increases the feasibility of FLIm for intraoperative use.
For the task of intraoperative cancer margin assessment the entire tissue surface must be accurately classified, resulting in the need for a point-measurement level evaluation where any individual misclassifications are penalized. This more challenging validation results in a decrease in the various performance metrics (ROC-AUC, sensitivity and specificity) due to individual point-measurement classification errors which must be overcome. Possible sources of such error within a given scan include tissue heterogeneity, variation in the distance and angle of the fiber probe and minor coregistration errors.
One of the key challenges of this study is the generation of accurate tissue condition labels for the in vivo imaging data. This relies on the coregistration of histopathology annotations with the excised specimen, followed by the coregistration of the excised specimen with the interrogated region in the surgical field. Challenges for this last step include differences in sample orientation and limited presence of morphological landmarks easily identified in the white light video as well as grossing pictures. This is further complicated by the changing size, orientation and shape of the specimen once excised and has resulted in some in vivo cases being rejected due to the inability to generate accurate labels. The use of image analysis based registration tools may lead to more consistent and accelerated data labelling. The use of such tools does however present additional challenges (i.e., the possibility of mismatched landmarks). In an earlier study using the same FLIm instrument, ex vivo breast lumpectomy specimens were imaged using a raster scanning scheme (rather than free hand scanning) and annotated using a marker-based coregistration method resulting in very high cancer classification performance (sensitivity and specificity > 97%) [26]. While FLIm-based breast tissue classification may rely on a different source of diagnostic contrast less affected by excision, the standardized annotation and imaging approach are likely key contributors to the strong performance observed.
Other limitations include the varying fiber-tissue distance, leading to variable lateral resolution and excitation-collection efficiency across the field of view. This is one of the down-sides of the free-hand scanning approach employed. A greater fiber-tissue distance can result in fluorescence contributions from multiple adjacent tissue locations being collected and influencing classifier output. A potential approach to overcome this local variation, is the addition of a classification refinement step whereby a given point-measurement prediction is updated based on neighbouring point-measurement predictions. In future studies, we will evaluate such a step as it can mitigate local errors and produce a more robust classification method. Furthermore, an optimized design of the FLIm instrument itself (currently under development) is expected to improve SNR and to generate more consistent signal in future clinical cases.
The developed visualization approach shows great promise as a diagnostic aid, producing smooth probability heatmaps in which one-off misclassifications are mitigated and the distinct tissue regions are clearly visible. However, the level of classifier uncertainty needs to be communicated to the surgeon to allow for informed decision making. A Bayesian machine learning approach [52] could be used to visualize classifier uncertainty levels (e.g., via color transparency level).
When investigating classifier output for dysplasia a wide range of cancer probability scores were observed, suggesting a range of tissue conditions are present within the dysplasia group. These results suggest that FLIm can potentially capture the gradient between healthy and cancerous tissue. With a large enough dataset and more detailed grading of dysplasia there is also the possibility of developing an ordinal regression model which can predict dysplasia grading using FLIm parameters and allows for the detection of potentially pre-cancerous cells.
CONCLUSION
In this work the diagnostic capability of label-free, fiber-based Fluorescence Lifetime Imaging (FLIm) during oral and Oropharyngeal cancer surgery was highlighted. Robust classification of healthy tissue and cancer was observed during intraoperative scans, with the key source of contrast coming from time-resolved FLIm parameters linked with metabolic activity. However, inferior contrast was observed for post-resection imaging of specimens. This lack of contrast for excised specimens suggests metabolic activity is a key contributor to the FLIm-based discrimination observed in these anatomies, highlighting the need for imaging studies which rely on biochemical changes to be performed in vivo. Ensemble methods, specifically in the form of the Random Forests approach, result in superior generalization across patients and tissue conditions, even when compared to CNN-based classification. This highlights the high variability of clinical FLIm data and the need for robust classification methods which prevent overfitting. The developed visualization approach produces smooth cancer probability maps within the surgical field that can mitigate local classification errors and highlight the distinct regions. Classifier output observed for dysplasia at the excision margins suggests that FLIm can capture the gradient between healthy and cancerous tissue, and potentially identify absnormal or pre-cancerous cells. Overall, the results of this study highlight the potential for label-free FLIm to be utilized clinically for margin assessment in a future study.
Supplementary Material
ACKNOWLEDGMENT
The authors would like to thank Angela Beliveau for her assistance as our clinical coordinator, Dr. Jakob Unger and Dr. Joao Lagarto for their contributions to the aiming beam segmentation and control software, Xiangnan Zhou for improving the stability of the system software, Silvia Noble Anbunesan and Dr. Lisanne de Boer for assisting in the acquisition of clinical data, Takanori Fukazawa for identifying motion correction methods and Athena K. Tam for helping in the preparation of data for analysis. We would like to thank Jonathan Sorger for his assistance in the design of the updated fiber probe used with the da Vinci SP robot. The authors declare no conflict of interest for the research.
This work was supported by the National Institutes of Health under Grant R01 CA187427 in collaboration with Intuitive Surgical, Inc.
Contributor Information
Mark Marsden, Department of Biomedical Engineering, University of California, Davis..
Brent W. Weyers, Department of Biomedical Engineering, University of California, Davis..
Julien Bec, Department of Biomedical Engineering, University of California, Davis..
Tianchen Sun, Department of Computer Science, University of California, Davis..
Regina F. Gandour-Edwards, Department of Pathology and Laboratory Medicine, University of California, Davis.
Andrew C. Birkeland, Department of Otolaryngology, University of California, Davis.
Marianne Abouyared, Department of Otolaryngology, University of California, Davis..
Arnaud F. Bewley, Department of Otolaryngology, University of California, Davis..
D. Gregory Farwell, Department of Otolaryngology, University of California, Davis, CA 95817 USA.
Laura Marcu, Department of Biomedical Engineering, University of California, Davis, CA 95616 USA.
REFERENCES
- [1].“National cancer institute: Statistics on oral and oropharyngeal cancer,” Accessed: Mar. 1, 2020. [Online]. Available: https://seer.cancer.gov/statfacts/html/oralcav.html
- [2].Weatherspoon DJ et al. , “Oral cavity and oropharyngeal cancer incidence trends and disparities in the United States: 2000–2010,” Cancer Epidemiol., vol. 39, no. 4, pp. 497–504, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Eldeeb H. et al. , “The effect of the surgical margins on the outcome of patients with head and neck squamous cell carcinoma: Single institution experience,” Cancer Biol. Med, vol. 9, no. 1, pp. 29–33, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].“National Cancer Institute definition of a Cancer Margin,” 2020[Online]. Available: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/margi
- [5].Mishra A. et al. , “Defining optimum surgical margins in buccoalveolar squamous cell carcinoma,” Eur. J. Surgical Oncol, vol. 45, no. 6, pp. 1033–1038, 2019. [DOI] [PubMed] [Google Scholar]
- [6].Shah AK, “Postoperative pathologic assessment of surgical margins in oral cancer: A contemporary review,” J. Oral Maxillofacial Pathol.: JOMFP, vol. 22, no. 1, pp. 78–85, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jaafar H, “Intra-operative frozen section consultation: Concepts, applications and limitations,” Malaysian J. Medical Sci.: MJMS, vol. 13, no. 1, pp. 4–12, 2006. [PMC free article] [PubMed] [Google Scholar]
- [8].Yee S, “Transoral robotic surgery,” AORN J., vol. 105, no. 1, pp. 73–84, 2017. [DOI] [PubMed] [Google Scholar]
- [9].Okamura AM, “Haptic feedback in robot-assisted minimally invasive surgery,” Current Opinion Urol., vol. 19, no. 1, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wedmid A. et al. , “Future perspectives in robotic surgery,” BJU Int., vol. 108, no. 6b, pp. 1028–1036, 2011. [DOI] [PubMed] [Google Scholar]
- [11].Kong K. et al. , “Towards intra-operative diagnosis of tumours during breast conserving surgery by selective-sampling Raman micro-spectroscopy,” Phys. Med. Biol., vol. 59, no. 20, pp. 6141–6152, 2014. [DOI] [PubMed] [Google Scholar]
- [12].Guze K. et al. , “Pilot study: Raman spectroscopy in differentiating premalignant and malignant oral lesions from normal mucosa and benign lesions in humans,” Head Neck, vol. 37, no. 4, pp. 511–517, 2015. [DOI] [PubMed] [Google Scholar]
- [13].Nguyen FT et al. , “Intraoperative evaluation of breast tumor margins with optical coherence tomography,” Cancer Res., vol. 69, no. 22, pp. 8790–8796, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Volgger V. et al. , “Evaluation of optical coherence tomography to discriminate lesions of the upper aerodigestive tract,” Head Neck, vol. 35, no. 11, pp. 1558–1566, 2013. [DOI] [PubMed] [Google Scholar]
- [15].Butte PV et al. , “Intraoperative delineation of primary brain tumors using time-resolved fluorescence spectroscopy,” J. Biomed. Opt., vol. 15, no. 2, 2010, Art. no. 27008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gao RW et al. , “Safety of panitumumab-IRDye800CW and cetuximab-IRDye800CW for fluorescence-guided surgical navigation in head and neck cancers,” Theranostics, vol. 8, no. 9, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Marcu L, “Fluorescence lifetime techniques in medical applications,” Ann. Biomed. Eng., vol. 40, no. 2, pp. 304–331, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pavlova I. et al. , “Understanding the biological basis of autofluorescence imaging for oral cancer detection: High-resolution fluorescence microscopy in viable tissue,” Clin. Cancer Res., vol. 14, no. 8, pp. 2396–2404, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Marcu L. et al. , Fluorescence Lifetime Spectroscopy and Imaging: Principles and Applications in Biomedical Diagnostics - Chapter 3: Tissue Fluorophores and Their Spectroscopic Characteristics. Boca Raton, FL, USA: CRC Press, 2014. [Google Scholar]
- [20].Gorpas D. et al. , “Autofluorescence lifetime augmented reality as a means for real-time robotic surgery guidance in human patients,” Sci. Rep., vol. 9, no. 1, pp. 1–9, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Alfonso-Garcia A. et al. , “Real-time augmented reality for delineation of surgical margins during neurosurgery using autofluorescence lifetime contrast,” J. Biophotonics, vol. 13, 2020, Paper e201900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sun Y. et al. , “Endoscopic fluorescence lifetime imaging for in vivo intraoperative diagnosis of oral carcinoma,” Microscopy Microanalysis, vol. 19, no. 4, pp. 791–798, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jo JA et al. , “Autofluorescence lifetime endoscopy for early detection of oral dysplasia and cancer,” in Latin America Optics and Photonics Conference. Washington, D.C., USA: Optical Society of America, 2018. [Google Scholar]
- [24].Yankelevich DR et al. , “Design and evaluation of a device for fast multispectral time-resolved fluorescence spectroscopy and imaging,” Rev. Sci. Instrum., vol. 85, no. 3, 2014, Art. no. 34303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Weyers BW et al. , “Fluorescence lifetime imaging (FLIm) for intraoperative cancer delineation in transoral robotic surgery (TORS),” Transl. Biophotonics, vol. 1, 2019, Paper e201900017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Phipps JE et al. , “Automated detection of breast cancer in resected specimens with fluorescence lifetime imaging,” Phys. Med. Biol., vol. 63, no. 1, 2017, Art. no. 15003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Unger J. et al. , “Real-time diagnosis and visualization of tumor margins in excised breast specimens using fluorescence lifetime imaging and machine learning,” Biomed. Opt. Express, vol. 11, no. 3, pp. 1216–1230, Mar. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Akkus Z. et al. , “Deep learning for brain MRI segmentation: State of the art and future directions,” J. Digit. Imag., vol. 30, no. 4, pp. 449–459, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Grewal M. et al. , “Radnet: Radiologist level accuracy using deep learning for hemorrhage detection in ct scans,” in Proc. IEEE 15th Int. Symp. Biomed. Imag., 2018, pp. 281–284. [Google Scholar]
- [30].Yao R. et al. , “Fluorescence lifetime imaging with compressive sensing through deep convolutional neural network,” in Optical Tomography and Spectroscopy. Washington, D.C., USA: Optical Society of America, 2018. [Google Scholar]
- [31].Liu J. et al. , “A novel method for fast and robust estimation of fluorescence decay dynamics using constrained least-squares deconvolution with Laguerre expansion,” Phys. Med. Biol., vol. 57, no. 4, pp. 843–865, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Gorpas D. et al. , “Real-time visualization of tissue surface biochemical features derived from fluorescence lifetime measurements,” IEEE Trans. Med. Imag., vol. 35, no. 8, pp. 1802–1811, Aug. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cortes C. and Vapnik V, “Support-vector networks,” Mach. Learn., vol. 20, no. 3, pp. 273–297, 1995. [Google Scholar]
- [34].Breiman L, “Random forests,” Mach. Learn., vol. 45, no. 1, pp. 5–32, 2001. [Google Scholar]
- [35].Pawar S, “Web-based application for accurately classifying cancer type from microarray gene expression data using a support vector machine (svm) learning algorithm,” in Proc. Int. Work-Conf. Bioinformatics Biomed. Eng., 2019, pp. 149–154. [Google Scholar]
- [36].Petrova NV and Wu CH, “Prediction of catalytic residues using support vector machine with selected protein sequence and structural properties,” BMC Bioinformatics, vol. 7, no. 1, pp. 312–324, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hahnloser RHR et al. , “Digital selection and analogue amplification coexist in a cortex-inspired silicon circuit,” Nature, vol. 405, no. 6789, pp. 947–951, 2000. [DOI] [PubMed] [Google Scholar]
- [38].Hecht-Nielsen R, “Theory of the backpropagation neural network,” in Neural Networks for Perception. Amsterdam, The Netherlands: Elsevier, 1992, pp. 65–93. [Google Scholar]
- [39].Paszke A. et al. , “Pytorch: An imperative style, high-performance deep learning library,” in Proc. Adv. Neural Inf. Process. Syst., (NeurIPS), 2019, pp. 8026–8037. [Google Scholar]
- [40].Shepard D, “A two-dimensional interpolation function for irregularly-spaced data,” in Proc. 1968 23rd ACM Nat. Conf., 1968, pp. 517–524. [Google Scholar]
- [41].Bradski G, “The OpenCV Library,” Dr. Dobb’s J. Softw. Tools, vol. 25, pp. 120–125, 2000. [Google Scholar]
- [42].Shapiro SS and Wilk MB, “An analysis of variance test for normality (complete samples),” Biometrika, vol. 52, nos. 3/4, pp. 591–611, 1965. [Google Scholar]
- [43].Mann HB and Whitney DR, “On a test of whether one of two random variables is stochastically larger than the other,” Annals Math. Statistics, vol. 18, pp. 50–60, 1947. [Google Scholar]
- [44].Kim H-C et al. , “Support vector machine ensemble with bagging,” in Proc. Int. Workshop Support Vector Mach., 2002, pp. 397–408. [Google Scholar]
- [45].Perez F. et al. , “Solo or ensemble? Choosing a CNN architecture for melanoma classification,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. Workshops, 2019, pp. 2775–2783. [Google Scholar]
- [46].Warburg O, “On the origin of cancer cells,” Science, vol. 123, no. 3191, pp. 309–314, 1956. [DOI] [PubMed] [Google Scholar]
- [47].Lagarto JL et al. , “Electrocautery effects on fluorescence lifetime measurements: An in vivo study in the oral cavity,” J. Photochemistry Photo bio. B: Biol., vol. 185, pp. 90–99, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Juhl H, “Preanalytical aspects: a neglected issue,” Scandinavian J. Clin. Lab. Investigation, vol. 70, no. sup242, pp. 63–65, 2010. [DOI] [PubMed] [Google Scholar]
- [49].Martinsen OG and Grimnes S, “Postexcision changes and the death process,” in Bioimpedance and Bioelectricity Basics, 3rd ed., New York, NY, USA: Academic, 2015, ch. 4, pp. 77–118. [Google Scholar]
- [50].David KA et al. , “Surgical procedures and postsurgical tissue processing significantly affect expression of genes and EGFR-pathway proteins in colorectal cancer tissue,” Oncotarget, vol. 5, no. 22, 2014, Art. no. 11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gao RW et al. , “Determination of tumor margins with surgical specimen mapping using near-infrared fluorescence,” Cancer Res., vol. 78, no. 17, pp. 5144–5154, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kendall A. and Gal Y, “What uncertainties do we need in bayesian deep learning for computer vision?” in Proc. Adv. Neural Inf. Process. Syst., 2017, pp. 5574–5584. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.