Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative pathogen of pandemic coronavirus disease 2019 (COVID-19). So far, no approved therapy has been developed to halt the spread of the pathogen, and unfortunately, the strategies for developing a new therapy will require a long time and very extensive resources. Therefore, drug repurposing has emerged as an ideal strategy toward a smart, versatile, quick way to confine the lethal disease. In this endeavor, natural products have been an untapped source for new drugs. This review represents the confederated experience of multidisciplinary researchers of 99 articles using several databases: Google Scholar, Science Direct, MEDLINE, Web of Science, Scopus, and PubMed. To establish the hypothesis, a Bayesian perspective of a systematic review was used to outline evidence synthesis. Our docking documentation of 69 compounds and future research agenda assumptions were directed toward finding an effective and economic anti-COVID-19 treatment from natural products. Glucosinolate, flavones, and sulfated nitrogenous compounds demonstrate direct anti-SARS-CoV-2 activity through inhibition protease enzymes and may be considered potential candidates against coronavirus. These findings could be a starting point to initiate an integrative study that may encompass interested scientists and research institutes to test the hypothesis in vitro, in vivo, and in clinics after satisfying all ethical requirements.
Keywords: SARS-CoV-2, SARS-CoV, MERS-CoV, sinigrin, Prolixin, systematic review
Background
Coronaviruses are a large family of pleomorphic RNA viruses that cause respiratory tract diseases. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the third highly pathogenic and large-scale pandemic beta-coronavirus, after the severe acute respiratory syndrome coronavirus (SARS-CoV) in 2003 that spread throughout in China and the Middle East respiratory syndrome coronavirus (MERS-CoV) that was identified in Saudi Arabia in 2012.1 On March 11, 2020, the World Health Organization (WHO) officially described the new coronavirus disease 2019 (COVID-19) outbreak as a global epidemic disorder.2 SARS-CoV-2 is characterized by mutations in nsp1, nsp3, nsp15, and gene S, which are associated with its epidemic behavior.3 The clinical manifestations of infected cases are varied, from asymptomatic to intensive care hospitalization states with pneumonia disorder.4 Reports have indicated that SARS-CoV-2 has the same cell entry receptor angiotensin-converting enzyme 2 (ACE2) and pathological features as acute respiratory distress syndromes (ARDSs) like SARS-CoV.5 This study aimed to find the similarity between SARS-CoV, MERS-CoV, and SARS-CoV-2 in genomic variation, pathogenesis, and epidemiology, in addition to discussing the theories of SARS-CoV-2 origins and its clinical manifestation, based on insights from COVID-19 genome sequencing and pathogenesis. The research builds a hypothesis and tries to answer a question: Can natural products assist during the COVID-19 crisis?
Method
The following electronic databases were searched: Science Direct, Google Scholar, Web of Science, Scopus, PubMed, and MEDLINE. A literature search was performed using the following key terms: CoV-19/COVID-19/SARS-CoV-2, SARS-CoV, World Health Organization (WHO)/reports, genomic sequence/genetic/origin, mutated protein, difference SARS/MERS/SARS-CoV-2, epidemiology, replication/pathogenesis, transmission, genetic susceptibility/factors affecting, clinical manifestation/clinical radiology/diagnosis, in silico/docking, treatment/FDA, plant remedies/complementary medicine/medicinal plants/natural product/extract/Chinese medicine, severe respiratory syndrome coronavirus/SARS-CoV/plant, Middle East respi-ratory syndrome coronavirus (MERS-CoV)/plant, plant affect pneumonia virus/RSV/HRV/HMPV/HPV, interleukin6/IL-6/clinical/COVID-19, IL-6/secondary metabolite/compound, review/systematic review.
The screening of search outputs was performed in three stages ( Fig. 1):
Stage 1: Collecting research and review articles concerning SARS-CoV-2 and statistically monitoring the virus’s spread worldwide according to WHO reports.
Stage 2: Searching for natural products of medicinal plants that have previously proved to be a potential antiviral against SARS-CoV, MERS-CoV, and pneumonia virus, in addition to phytoconstituents that inhibit interleukin-6 (IL-6; pathogenesis marker of respiratory failure).
Stage 3: Practical point/hypothesis via virtual screening and docking studies.6
Figure 1.
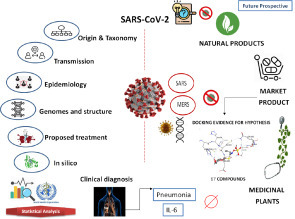
Graphical abstract of systematic review using screening mode design.
Data Analysis
WHO reports for 10-day intervals, from January 20, to June 1, 2020, were entered into Statistical Software Packages (SPSS software version 25; IBM Corp., Armonk, NY) to test the difference between groups using the Kruskal-Wallis test; the results were considered significant at p < 0.05. Descriptive and cumulative statistical methods, including percentage and frequency, were used to present confirmed, infected, and recovered cases and deaths in different regions and countries. Graphs were made using Microsoft Excel (2016; Redmond, WA). Charts and tables were used to present medicinal plant studies. In silico and docking simulation was performed using Molecular Operating Environment (MOE) version 2014.09 (Chemical Computing Group Inc., Montreal, QC, Canada).
Results
Structures of Coronaviruses and Genomic Variation with SARS-CoV-2
Coronaviruses are classified as the largest species of RNA viruses (26–32 kb) as they are about 125 nm in diameter.1 There are four types of viruses: alpha, beta, gamma, and delta. HCoV-229E and HCoV-NL63 are examples of the alpha type, which are responsible for one-third of the common colds, as they use different host proteins as receptors. SARS-CoV, MERS-CoV, and SARS-CoV-2 are examples of a beta-coronavirus; they have the same genomic structure as coronaviruses7 and contain an enveloped, single, positive-stranded genome that encodes four major viral structural proteins (spike [S], envelope [E], membrane [M], and nucleocapsid [N] proteins 3–5, that follow the characteristic gene order, 5′-replicase [rep gene], spike [S], envelope [E], membrane [M], and nucleocapsid [N]-3′) with short untranslated regions at both termini. The S protein binds to the host cell receptor and allows its entrance; therefore, it is considered to be the main target of therapy.8
It is composed of two functional subunits, S1 for binding to the host receptor and S2 for membrane fusion. The cleavage of S1/S2 subunits occurs by one or more host proteases. Moreover, the activation of the SARS-CoV S protein needs additional cleavage by the endosomal cysteine protease cathepsin L and trypsin-like serine protease. Few enzyme inhibitors affecting these proteins have shown anticoronavirus activity in vitro.9
The nonstructural protein is encoded by the rep gene, which represents about two-thirds of the genome at the 5′ end. Open reading frame 1a/b (ORF1a/b) is partially overlapped with the 5′-terminal end, which encodes the large replicase polyproteins 1a (pp1a) and 1ab (pp1ab). Moreover, papain-like cysteine protease (PLpro) and 3C-like serine protease (3CLpro) cleave pp1a and pp1ab to give nonstructural proteins, involving RNA-dependent RNA polymerase (RdRp) and helicase (Hel), which are considered the major enzymes during the process of transcription and replication of coronaviruses. The structural proteins (S, E, M, and N) are encoded by the 3′ one-third of the coronavirus genome, which is important for binding the virus to the receptor host cell assembly of the virion.10
Modification of the spike glycoprotein by homologous recombination is a characteristic difference between SARS-CoV-2 and SARS-CoV,11 in addition to the absence of the 8a protein and the change in the number of amino acids in 8b and 3c proteins.12 Also, the spike glycoprotein of SARS-CoV-2 has composed a combination of bat SARS-CoV and undetected beta-CoV.13 On the other hand, a fluorescence study has proved that SARS-CoV-2 utilizes the ACE2 cell receptor and the mechanism of the entrance to host cell as SARS-CoV.14 The binding affinity of SARS-CoV-2 for ACE2 is increased by the single N501T mutation in the spike protein.15
ACE2 is the main cellular receptor for SARS-CoV-2; the virus binds to these receptors by its spike protein, which facilitates its entrance to the host cell, duplicates genomic materials, and synthesizes many different required proteins using the cellular machinery. After that, it produces new virions from the cell surface.16 Moreover, after the entrance of SARS-CoV-2 through the ACE2, it subsequently downregulates ACE2 expression causing the unchallenged angiotensin II accumulation and local RAAS (renin–angiotensin–aldosterone system) activation, which deranges homeostasis, triggers inflammation, and induces tissue injury in lungs and other organs.17 , 18
Pathogenesis
Life Cycle of SARS-CoV and MERS-CoV in Host Cells
CoVs enter the host cell by exploiting two pathways: the endosomal and cell surface nonendosomal pathways. Low pH and pH-dependent endosomal cysteine protease cathepsins assist the membrane fusion and enosomal CoV cell entry and support endosomal cell entry of coronaviruses.19 Host proteases such as transmembrane protease serine 2 (TMPRSS2) and TMPRSS11D (airway trypsin-like protease) cleave S into the S1 and S2 subunits to activate S for the entrance of cell surface nonendosomal virus at the plasma membrane. Inhibitors of these proteases can prevent this proteolytic cleavage and partially block cell entry. MERS-CoV is additionally activated by furin, a serine endoprotease that has been involved in the processing of fusion proteins and the entrance of the cells of other RNA viruses, including HIV, avian influenza A/H5N1 virus, Ebola virus, Marburg virus, and flaviviruses.20 The Furin is included in MERS-CoV S1/S2 cleavage during exit from the infected cell. Monotherapy and/or combinatorial treatment with inhibitors of host proteases concerned with the various cell entry pathways are potent as anticoronaviruses.21 Table 1 illustrates the major differences between SARS-CoV, MERS-CoV, and SARS-CoV-2.
Table 1.
Differences between SARS, MERS, and SARS-CoV-2.
| SARS-CoV16,31,32 | MERS-CoV31, 32, 33 | SARS-CoV-216,31,34,35 | |
|---|---|---|---|
| First occurrence | November 16, 2002, in Foshan, Guangdong | September 2012 | December 7, 2019, in Wuhan, Hubei |
| Virus type | RNA virus | ||
| Species pathogen | Beta-coronavirus | ||
| Intermediate host | Paguma larvata | Dromedary camel | Pangolin, mink, bat (possible) |
| Definitive host | Rhinolophus sinicus | Tylonycteris pachypus and Pipistrellus abramus | Rhinolophus sinicus (possible) |
| Predominant receptor | Human ACE2 | Human DPP4 (or CD26) | Human ACE2 |
| Total DNA sequence length of pathogen | 29,751 | 30,119 | 29,903 |
| Characteristic gene order | 50-replicase ORF1ab, spike (S), envelope (E), membrane (M), and nucleocapsid (N)-30 | 5′-Replicase ORF1ab-S-envelope (E)-membrane (M)-nucleocapsid (N)-3′; ORF3ab, ORF6, ORF7ab, ORF8, ORF9ab, and ORF10 | |
| Transmission rate | More than MERS-CoV but less than SARS-CoV-2 | Less than SARS-CoV and SARS-CoV-2 | More rapid than MERS-CoV and SARS-CoV |
| Male–female patient ratio | 1.0:1.2 | — | 2.7:1.0 |
| Clinical symptoms | Fever, cough, myalgia, dyspnea, and diarrhea | High fever, chill, cough, shortness of breath, chest pain, headache, myalgia, sore throat, arthralgia, abdominal pain, anorexia, vomiting, and severe diarrhea (some cases) | Fever, fatigue, and dry cough |
| Propagation mode | Droplets or close contacts, fomites, fecal transmission, and handling of animals having the virus | Droplets or close contact with infected dromedary camels | Human to human/spread through respiratory droplets from coughs or sneezes; handling of animals having the virus |
| Diagnostic methods | real-time PCR (RT-PCR), rRT-PCR, RT-LAMP, rRT-LAMP, coronavirus detection kit | Chest radiography, electron microscope, immunofluorescence microscopy, cell culture, enzyme-linked immunosorbent assay (ELISA), and RT-PCR | RT-PCR, rRT-PCR, RT-LAMP, rRT-LAMP, coronavirus detection kit |
| Treatment | Glucocorticoid and interferon | Interferon, lopinavir/ritonavir, cyclosporin A, chloroquine/hydrochloroquine | Lopinavir/ritonavir, chloroquine |
| Mortality | 9.6% | 40.0% | 2%–7% |
| Number of deaths worldwide | 916 | 800 | 379,941 and rising daily (WHO website, June 3, 2020) |
rRT, reverse transcription loop-mediated isothermal amplification; rRT-LAMP, real-time reverse transcription LAMP assay.
Endosomal Pathway of SARS-CoV and MERS-CoV
The S proteins of SARS and MERS bind to two main receptors: cellular receptor ACE2 and cellular receptor dipeptidyl peptidase 4 (DPP4), respectively. After the entrance of the virus into the host cell, the viral RNA is released in the cytoplasm. ORF1a and ORF1ab are translated to produce pp1a and pp1ab, which are then cleaved by the proteases encoded by ORF1a to give 16 nonstructural proteins (RNA replicase–transcriptase complex). The performed complex leads to the production of negative-sense (–) RNA by both replication and transcription. Full-length (–) RNA copies of the genome are produced during replication and used as a template for full-length positive-sense (+) RNA genomes. A subset of seven to nine subgenomic RNAs, included those encoding all structural proteins, is produced by discontinuous transcription.7 , 22
Virion Assembly and Release
Virion assembly occurs in the endoplasmic reticulum–Golgi intermediate compartment (ERGIC) and is coordinated by the M protein. Incorporation of viral proteins into coronavirus virions relies on protein trafficking to protein–protein interactions at the ERGIC. The M, E, and some S proteins have intracellular trafficking signals that are important in their targeting to accumulation beside the budding site.23 , 24 The M protein is essential, and homotypic M-M interactions through multiple contact sites are required to drive virus-like particle (VLP) and coronavirus assembly.25 , 26 Also, the integration of E, S, and ribonucleoproteins (RNPs) with virions occurs by heterotypic interactions with M proteins at the budding site.27 , 28
Life Cycle of SARS-CoV-2
The virus life cycle starts when S protein binds to the cellular receptor ACE2. After binding, modification in the S protein allows fusion between the viral envelope and the cell membrane through the endosomal pathway. After that, RNA is released into the host cell. Genome RNA is then translated into viral replicase pp1a and pp1ab, which are cleaved into small products by viral proteinases. Groups of subgenomic mRNAs are formed by the polymerase throughout discontinuous transcription and finally translated into related viral proteins. Following that, viral proteins and genome RNA are aggregated into virions in the ER and Golgi and then transported by vesicles and released out of the cell.29
Pathogenic Mechanism of SARS-CoV-2
SARS-CoV-2 and Pneumonia
The viral infection has the ability to produce an excessive immune reaction in the host; in some patients, a “cytokine storm” reaction takes place and leads to extensive tissue damage. IL-6, which is known as the protagonist of this storm, can be produced by activated leukocytes. It allows the differentiation of B lymphocytes and the growth of some groups of cells, and inhibits the growth of others. Additionally, it can stimulate the production of acute-phase proteins and plays a vital role in thermoregulation, bone maintenance, and the functionality of the central nervous system. Moreover, the IL-6 level rises during inflammatory diseases, infections, autoimmune disorders, cardiovascular diseases, and some cases of cancer. It is also involved in the pathogenesis of the cytokine release syndrome (CRS), which is characterized by fever and multiple organ dysfunction.30
Inhibition of Human Heme Metabolism
Preprint research has reported that viruses target hemoglobin and attack porphyrins where the viral ORF8 and surface glycoproteins form a porphyrin complex. At the same time, ORF1ab, ORF10, and ORF3a proteins could coordinate an attack on the heme on the 1-beta chain of hemoglobin to dissociate the iron porphyrin. This results in a decreasing the number of hemoglobin carrying oxygen and carbon dioxide. Also, the released iron led to the production of reactive oxygen species (ROS); therefore, the lung cells will have vigorous inflammation and damage.36
Theories of SARS-CoV-2 Origins
In reviewing the SARS-CoV-2 genomic data sequence, two theories explain the origin of the virus. According to Andersen et al,37 the viral genesis depends on natural selection in a nonhuman animal host before zoonotic transfer. Another belief depends on natural selection in humans following a zoonosis transfer; the author reported that analysis of the public genome sequence proved that there is no evidence the virus was made in a laboratory or otherwise engineered.
Several reports record a variable virus origin interpretation, some of which have indicated that most of the coronaviruses in humans are derived from a bat reservoir.1 , 38, 39, 40 Only one study states that the virus matches with pangolin as a host carrier,1 while another report shows a similarity with a bovine sequence with a 90% genome identity.40 , 41
Interestingly, upon searching using the SARS term, a reported invention of a novel strain of SARS coronavirus was found. The patent concerned isolation of the virus strain, with diagnostic reagents and vaccine applications thereof. The virus genome was in the form of complementary DNA, a serine codon reported in positions 23220–23222 of protein S or positions 25298–25300 of the ORF3 gene. There was a glycine codon, and an alanine codon in positions 7918–7920 of FORT la or a serine codon in positions 26857–26859 of the protein M gene. The protein S was a membrane glycoprotein (200–220 kDa) that is in the “spike” emerging from the surface of the viral envelope.42
The genome sequences of SARS-CoV-2 show ≥70%–79% identity to SARS-CoV.1 , 35 , 43 Additionally, Nezhad et al.44 indicated that the structural analysis homology percentage is 96% for the envelope and 89.6% for the nucleotide. These findings are consistent with those of Tahir et al.40 On the other side, Xiaolu et al.35 signify that the virus is similar to MERS-CoV, with 50% similarity, which is in contrast to the investigation of Tahir,40 who indicated an 87% resemblance relationship between the two viruses. From the authors’ point of view, there is a bit of conflict in the virus origin reports; thus, this research area requires more attention by genomic scientists.
Epidemiology
According to the observed data from the early outbreak in mainland China during January 10–24, 2020, the tendency of incidence largely follows exponential growth, and the mean basic reproduction number (R 0) was estimated to range from 2.24 to 3.58, associated with a two- to eightfold increase in the reported rate. Another estimation based on the data from December 31, 2019, to January 28, 2020, suggested similar findings, with R 0 = 2.68 and the epidemic doubling time being 6.4 days. The current estimate of the mean incubation period for COVID-19 is 6.4 days, ranging from 2.1 to 11.1 days, with potential asymptomatic transmission. However, the situation is evolving and further updated data are required to confirm these estimations.45 , 46
Factors Affecting the Prevalence of COVID-19
Sex
In a study of 140 patients in China using single-cell sequencing, it was found that the expression of ACE2 is more predominant in Asian men than women, which might be the reason for the higher prevalence of COVID-19 in men. The data indicate that there might be a sex predisposition to the virus.47
Smoking
Because ACE2 has been identified as a receptor for COVID-19, studies have reported that smoking affects the expression pattern of ACE2 in the respiratory tract, causing differences in susceptibility to the virus.34 Another investigation considered that sex predisposition to SARS-CoV-2 with men might be related to the higher rate of smoking in men than in women in China. Additionally, susceptibility was significantly higher in current smokers of Asian ethnicity than in Asian nonsmokers. In a study by Cai and colleagues (11.8% of smokers had nonsevere disease vs 16.9% of smokers with severe disease), a tendency toward an association between smoking and severity of disease was observed, but it was not significant.47
Chronic Diseases
The most distinctive comorbidities of nonsurvivors from a group of 52 intensive care unit states were cerebrovascular diseases (22%), diabetes (16.2%–22%), hypertension (19.30%–23.7%), and coronary heart disease (5.8%).48
Environmental Factors
Oliveiros et al. reported that the doubling time correlates positively with temperature and inversely with humidity, suggesting a decrease in the rate of virus progression with the arrival of spring and summer in the northern hemisphere. A 20 °C increase is expected to delay the doubling time by 1.8 days. Those variables explain 18% of the variation in disease doubling time; the remaining 82% may be related to containment measures, general health policies, population density, transportation, or cultural aspects.49
The intolerance of coronaviruses to heat is due to the presence of a lipid bilayer that can easily be damaged by high temperatures; also, the transmission of these viruses can be affected by humidity in the air.50 Moreover, SARS-CoV-2, like other respiratory viruses, involves aerial transmissions of respiratory droplets that expose the virus to external environmental conditions. SARS-CoV-2 and SARS-CoV can be viable on surfaces for more than 5 days at temperatures of 11–25 °C and relative humidity of 40%–50%; then it drastically loses its viability as temperatures and humidity increase.50 Cold temperatures and low humidity can increase the half-life and viability of the virus, the stabilization of the droplet, and its propagation in the nasal mucosa.51
Genetic Susceptibility of the Different Populations to Viral (SARS-CoV-2) Infection
To compare the genomic characteristics of ACE2 among different populations, coding region variants in ACE2 and expression quantitative trait loci (eQTL) were systematically analyzed. The findings indicated that no direct evidence was identified to support the existence of coronavirus S protein binding-resistant ACE2 mutants. The data of variant distribution and allele frequency (AF) may contribute to further investigations of ACE2, including its roles in acute lung injury and lung function. The East Asian populations have much higher AFs in the eQTL variants associated with higher ACE2 expression in tissues, which may suggest different susceptibilities or responses to COVID-19/SARS-CoV-2 compared with different populations under similar conditions.52
Statistical Analysis of WHO Reports
The differences between the total infected SARS-CoV-2 cases and the number of people who died in different regions (Western Pacific, Eastern Mediterranean, Southeast Asia, African, European, and America regions) according to WHO reports were statistically interpreted from January 20 to May 30. The total number of confirmed cases (Kruskal-Wallis H = 34.339, df = 5, p < 0.001), the total death numbers (Kruskal-Wallis H = 31.839, df = 5, p < 0.001), and case fatality rates (Kruskal-Wallis H = 16.601, df = 5, p = 0.005) throughout the study period were recorded.
The African, Southeast Asia, Eastern Mediterranean, and Western Pacific regions represent a significantly lower number of reported infected cases than the European regions and regions of America. However, the fatality rate did not directly correlate with the confirmed cases of such regions. Hence, the regions of America and Western Pacific regions showed the least fatal cases worldwide (2.75% and 3.43%, respectively). The bars ( Fig. 2) show that the European region represents a higher total number of confirmed and fatality rates worldwide (7.55%). The Eastern Mediterranean region represents the second-highest fatality rate, although it represents the fourth level of reported cases worldwide.
Figure 2.
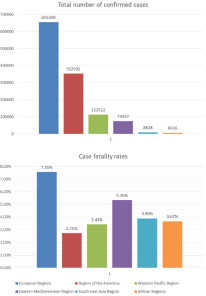
The number of confirmed cases (Kruskal-Wallis H = 34.339, df = 5, p < 0.001) and fatality rates (Kruskal-Wallis H = 16.601, df = 5, p = 0.005) of different worldwide regions distributed according to WHO reports, represented throughout the study period.
From the previous statistical data, we concluded that different factors may contribute to the variation in fatality rate between world regions; for example, genetic susceptibility and lack of widespread systematic testing may be considered the main sources of discrepancy in COVID-19-infected cases. Other predisposing elements are the ratio of men to women infected; complex cases with comorbid disease, especially immunocompromised states; elderly persons who are COVID positive; people’s state of immunity, lifestyle, and diet; hospitalization efficiency protocols, and the readiness of respiratory ventilation systems.
The line graphs compare countries in the same region, to represent the most affected (top 10) countries with reported cases of infection and death. The European region ( Fig. 3A), Spain, Italy, and the United Kingdom, showed a high number of confirmed cases and mortality rates. The Western Pacific region, China, represents a high number of confirmed cases and deaths, with a plateau of patient cases at the end of February. The number of fatalities demonstrated a fluctuation at the end of April; this represents an increase for a short time interval and then reaches a steady state again ( Fig. 3B). The Southeast Asia region, India and Indonesia, recorded a rise of positive cases of the virus with a remarkably significant number between the cases of the two countries (Fig. 3C). In the Eastern Mediterranean region, Iran demonstrated a higher number of COVID-19 patients (about 150,000 cases) and most deaths in the region (more than 7500 persons died) (Fig. 3D). In the region of the Americas, the increase in both infected and death instances recorded by the United States skyrocketed compared with other countries in the same region (Fig. 3E). In the African region, South Africa, followed by Algeria, had the most cases. While Algeria had a significant number of deaths at the end of March, compared with other countries in the region, South Africa achieved the same level by the end of May (Fig. 3F).
Figure 4.
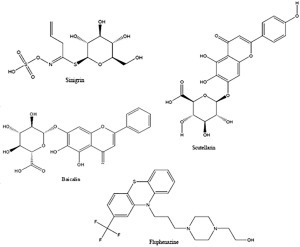
Top 4 hit compounds under docking investigation.
Figure 3.
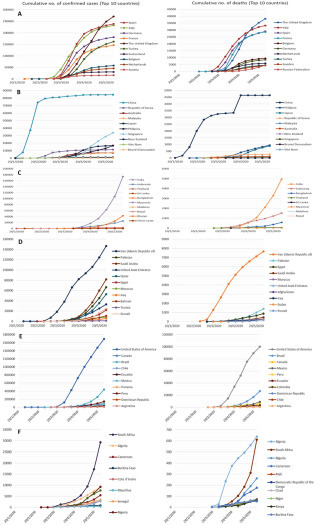
The number of confirmed cases and deaths reported in the (A) European region, (B) Western Pacific region, (C) Southeast Asia region, (D) Eastern Mediterranean region, (E) region of the Americas, and (F) African region according to WHO reports.
Statistical Analysis of Egypt WHO Reports
The infected cases, death numbers, and fatality rate in Egypt compared with the same parameters for the rest of the world were statistically analyzed. The numbers of confirmed cases and deaths in Egypt are in alignment with the world average until March 10 and possesses almost the same level until the end of April, followed by dramatic uplifting. The world case fatality rate was 3%–4% until March 19, which represents a cross section with the line for the death rate in Egypt. Afterward, the mortality rate jumped to an apex on April 19 with a percentage of 7.5, followed by a decline. Afterward, the confirmed case and death numbers surged until the start of June. Continuation in recording cases should be taken into consideration to confirm the passing of the critical phase.
Clinical Manifestation
COVID-19 patients suffer from common symptoms of typical respiratory diseases and other manifestations, like fatigue, diarrhea, and myalgia.53 Clinical radiology is used to distinguish SARS-CoV-2 from other viral pneumonia. Chest computed tomography (CT) records a high specificity but moderate sensitivity. The detailed imaging features can evaluate the severity and extent of disease and differentiate between four clinical types (early, common, critical, and fatal). The imaging features of the early stage include multifocal patchy shadows or ground-glass opacities located in the lung periphery, subpleural area, and both lower lobes on chest CT scans. The long-axis lesion is mostly parallel to the pleura. Additionally, interlobular septal thickening and intralobular interstitial thickening are manifested. After progression of the disease, an enlarged and increased density of the lesions is observable as consolidated lesions with the air bronchogram sign. Increased density of opacity in the whole lung (white lung) and ground-glass opacities can be completely absorbed by some consolidation lesions that leave fibrotic stripes or subpleural reticulation as a critical sign. In time, multiple lobular involvement of lesion expansions is detected at the disease is exacerbated.54, 55, 56, 57
Future Prospective: Role of Natural Compounds against COVID-19
Virus mutation is an evolutionary mechanism for adaptation, and the scientific community faces the challenge of finding preventive vaccines and efficient antiviral therapies.58 The pandemic state of COVID-19 has yielded a globally heavy toll. Researchers are still actively testing various strategies, including new and repurposed drugs, for finding a solution. Hence, there is clear room for the intervention of alternative and complementary medicine as a therapy for SARS-CoV-2 patients, inconsistent with the WHO report, where there is no specific medicine for COVID-19 to date.59
Plant and microbial natural products could represent a new prospect for coronavirus treatments.60 , 61 Medicinal plants are considered a highly attractive strategy for the economic production of a treatment,62 and have also been used by ancient nations against viral respiratory infections. The secondary metabolites of plants are characterized by novelty and potency as bioactive biological agents.63 It is relevant to note that the current clinical guideline in China is to use conventional medicine previously reported to treat SARS and MERS patients. More than 85% of SARS-CoV-2-infected patients in China are receiving traditional Chinese medicine (TCM) treatments, such as Astragali radix, Glycyrrhizae radix, Saposhnikoviae radix, Atractylodis rhizoma, and Lonicerae japonicae. These plants were recorded as the most frequent plants commonly used in preventive formulae for COVID-19.59
Previous reports have indicated the similarity between SARS-CoV-2 and SARS-CoV in genomic sequence (70%–96% in the envelope and nucleotide segment) and pathogenesis pathways1 , 10 , 40 , 44 and proposed a resemblance with the MERS-CoV human virus by 50%.35 Based on these factors, there is a general hypothesis that anti-SARS-CoV and MERS-CoV natural products would be potential ammunition to fight the lethal virus.
SARS-CoV-2 infects the upper and lower respiratory tract and causes mild or highly acute respiratory syndrome with a consequent release of pro-inflammatory cytokines (IL1β and IL-6). Supportive care to ameliorate a patient’s symptoms is the logical option at this time. The pathogenesis of the virus includes binding to the toll-like receptor (TLR) that results in the release of pro-IL-1β, which is a mediator of lung inflammation, fever, and fibrosis. The suppression of IL-1β through IL-37 and IL-38 may have a therapeutic effect, as this strategy has previously inhibited inflammation during viral infections.64 , 65
As a result of the progression of the clinical and radiological speculation regarding severe acute respiratory syndrome (SARS-CoV-2), pneumonia states have been indicated,66 , 67 so we postulate that plants previously used to treat pneumonia disease could help to relieve the symptoms. In Table 2 is a review list of plants that act against different pneumonia viruses: human metapneumo virus (HMPV), human parainfluenza virus (HPIV), respiratory syncytial virus (RSV), and human rhinovirus (HRV).
Table 2.
Reported Plants Affecting Pneumonia Viruses.
| Virus | Plants | Reference |
|---|---|---|
| HMPV | Aspidosperma tometosum, Gaylussacia brasiliensis, Virola sebifera, Arrabideaa chica | 58 |
| HPIV | Dahlia, Glycine max (L.) Merr, Phaeseolus aureus Roxb, Cossypium herbaceum, Allium sativa | 68 |
| Mistletoe (Viscum album L. spp. album) | 69 | |
| Ipomopsis aggregate, Rhus succedanea, Garcinia multiflora, Sanicula europea | 70 | |
| RSV | Alchornea cordifolia, Alchornea floribunda, Nauclea latifolia | 71 |
| Plantago asiatica, Clerodendrum trichotomum | 72 | |
| Ballota glandulosissima | 73 | |
| Allium sativa, Markhamia lutea | 68 | |
| Potentilla arguta, Sambucus racemose, Eleutherococcus senticosus, Myrcianthes ciplatensis, Rhus succedanea, Garcinia multiflora | 70 | |
| HRV | Plectranthus cylindraceus, Pogostemon cablin, Heterotheca grandiflora, Aglaia andamanica, Euodia glabra, Larrea triden tata, Bollata glandulosissima | 73 |
| Pelargonium sidoides radix | 74 | |
| Chrysosplenium tosaense, Calendula arvensis, Eriobotrya japonica Lindl., Urginea scilla steinh | 68 | |
| Woodfordia fruticose | 75 |
From all previous concepts, that is why the literature search and review strategy are based on three different scopes: natural products affecting SARS-CoV and MERS-CoV, compounds able to inhibit IL-6, and medicinal plants that relieve pneumonia.
Natural Products Affecting SARS-CoV
Artemisia annua, Pyrrosia lingua, and Lindera aggregata have been reported to display antiviral activity against SARS-CoV in screening analysis using the Vero cell.76 Houttuynia cordata water extract inhibits 3CL protease enzyme and blocks RNA polymerase activity.77 Also, leukocytic IFN-α, ribavirin, lopinavir, and rimantadine have been indicated as a prophylaxis or curative agent against the virus.78 In the investigation of the activities of 15 plants against viruses, lectins compounds such as sugar N-acetyl-specific agglutinins and nonspecific agglutinin showed a potent anti-SARS-CoV activity, in particular the mannose type.79 Moreover, tryptanthrin, myricetin, scutellarin, baicalin, glycyrrhizin, aloe-emodin, hesperetin, sinigrin, lycorine, and amentoflavone are isolated natural compounds that possess activity against SARS-CoV with different action mechanisms.
Natural Products Affecting MERS-CoV
Park et al.80 demonstrated the inhibitory activity of polyphenol compounds derived from Broussonetia papyrifera (broussochalcone B, broussochalcone A, 4-hydroxyisolonchocarpin, papyriflavonol A, 3′-[3-methylbut-2-enyl]-3′,4,7-trihydroxyflavane, kazinol A, kazinol B, broussoflavan A, kazinol F, and kazinol J) against 3-chymotrypsin-like and papain-like MERS coronavirus cysteine proteases. The prenylated quercetin derivative papyriflavonol A was the most potent compound against papain-like proteases. Furthermore, another natural compound having antiviral activity is resveratrol, which is found in grape skin and seeds, where it inhibits MERS-CoV infection and prolongs cellular survival after virus infection through different pathways: (1) reducing the viral RNA expression, (2) blocking the NF-κB pathway, (3) decreasing nucleocapsid (N) protein essential for virus replication, and (4) inhibiting caspase 3 cleavage and apoptosis induced by virus infection. It also decreases the production of nitric oxide in tissue, thereby reducing tissue inflammation.81 Homoharringtonine (HHT) is a pharmaceutical class of natural alkaloid ester obtained from plant, and Cephalotaxus harringtonii is marketed as an omacetaxine mepesuccinate against MERS-CoV. Moreover, natural products like trigocherin, jatropane esters, and harringtonine have shown potent antiviral effects on the MERS-RNA viruses.
Natural products such as alkaloids, flavonoids, phenolics, and terpenoids are reported to inhibit SARS-CoV and also the pathogenic elevation of IL-6 ( Table 3). Moreover, current market products are available as MERS-CoV and SARS-CoV inhibitors ( Table 4) according to Jadav et al.82
Table 3.
Natural Product Candidates Could Be Used against SARS-CoV-2.
| Compound | Mechanisms | Reference |
|---|---|---|
| Natural products affect SARS-CoV | ||
| Tryptanthrin | RdRp and PLP2 inhibitor Moderates the viral RNA genome synthesis and progeny virus production |
83 |
| Myricetin | Interacts with ATP/ADP binding pocket of Hel protein Interacts with critical residues of the ATPase domain (N265, Y269, and R443) |
84 |
| Scutellarin, baicalin | Conjugate with chemokines and interfere with their capacity to activate cellular receptors CCR5 and CXCR4 | 78 |
| Glycyrrhizin | Induces nitrous oxide synthase, which inhibits virus replication | 78, 85, 86 |
| Aloe emodin, hesperetin sinigrin |
Inhibit cleavage activity of the 3CLpro | 83 |
| Lycorine | Unclear how this compound interacts with expressed viral proteins and antigens | 76 |
| Amentoflavone | SARS-CoV 3CL protease inhibitor | 58 |
| Sugar N-acetyl glucosamine and agglutinins |
Bind to the ACE2 receptor, resulting in the inhibition of SARS-CoV fusion with the target cell | 79 |
| Preparation Echinaforce | Interacts directly with viral envelope proteins | 87 |
| Natural agents affect IL-6 | ||
| Tocilizumab | Inhibits the binding of IL-6 to its receptor | 88 |
| Epigallocatechin-3-gallate | Blocks IL-6 synthesis in IL-1, inhibits p38 pathways, inhibits IL-6–induced apoptosis | 88 |
| Curcumin | Inhibits the production of pro-inflammatory cytokines and reduces IL-6/IL-6-soluble receptor (sIL-6R)-induced STAT3 and ERK phosphorylation | 89 |
| Celastrol | Reduces levels of IL-6 and IL-1β by inhibition of STAT3 phosphorylation to block the IL-6 receptor signaling pathway | 90 |
| Statins | Inhibits the enzyme HMG-CoA reductase and JAK/STAT3 signaling pathway for IL-6-mediated inflammation | 91 |
| Bisphosphonate | Inhibits the enzyme FPP synthase and the JAK/STAT3 signaling pathway for IL-6-mediated inflammation; reduces sIL-6R serum levels | |
| Polyphenolic compounds | Hinder JAK/STAT3 signaling pathway for IL-6-mediated inflammation | |
| Genistein | Decreases IL-6 production | |
| Sophoricoside | Arrests IL-6 bioactivity | |
| Isoflavones | Inhibit production of IL-6 | |
| Eriodictyol | Inhibits expression of inflammatory cytokines TNF-α, IL-6, and IL-1β | 92 |
| Luteolin | Reduces TNF-α, KC, ICAM-1, and SOD; activates MAPK and NF-κB | |
| Quercetin | Diminishes TNF-α, IL1-β, IL-5, and IL-6; NO and COX-2 | |
| Kaempferol | Reduces inflammatory cells; activates MAPK and NF-κB pathways | |
| Mitraphylline | Lowers IL-1α, IL-1β, IL-17, TNF-α, IL-6, and IL-8 | |
| Asperuloside | Depletes TNF-α, IL-1β, and IL-6 levels | |
| Callicarpa japonica | Reduces cytokine IL-6 | |
| Sakuranetin | Minimizes eosinophils, TNF-α, IL-5, IL-1β, M-CSF, and RANTES; inhibits NF-κB in lung, MMP-9-positive, and MMP-12-positive cells; increases TIMP-1 expression | |
| Apigenin | Inhibits eosinophil infiltration in lung tissue and IL-6 | |
| Herbal formula PM014 | Lowers IL-6 levels | |
| Punica granatum | Depletes eosinophils and cytokines IL-1β and IL-5 | |
| Peganum harmale | Inhibits the production of both IL-6 and TNF-α | 93 |
| Baicalin | Decreases the induction of IL-1, IL-6, TNF-α, and IFN-γ | |
| Kaempferia parviflora | Lowers phosphorylation of STAT3, Akt, and the expression of Mcl-1 in response to exogenous IL-6 stimulation | 94 |
Table 4.
Current Available Inhibitors against MERS-CoV* and SARS-CoV.82
| Drug Classification | Drugs | Probable Mode of Action |
|---|---|---|
| Neurotransmitter inhibitor | Clomipramine HCl, chlorphenoxamine HCl, astemizole promethazine HCl, fluphenazine HCl, thiothixene, fluspirilene, benztropine mesylate | Clathrin-mediated endocytosis inhibitors |
| Antibiotic agent | Anisomycin | Protein processing inhibitor |
| Antibacterial agent | Emetine 2HCl·H2O, cycloheximide | Unknown protein synthesis inhibitor |
| Anticancer agents, antiparasitic agent/antimalarial | Dasatinib, imatinib, mesylate, chloroquine, 2H3PO4 mefloquine hydroxychloroquine SO4, amodiaquine 2HCl·H2O | Inhibition of viral replication |
| Estrogen receptor inhibitor | Toremifene citrate, tamoxifen citrate | Viral entry inhibitor |
| Nucleoside analog | Gemcitabine HCl | DNA metabolism inhibitor |
| Cytoskeleton inhibitors | Nocodazole | Microtubule depolymerization |
| Ion channel inhibitors | Monensin, salinomycin Na | Blocks the formation of virus particle and eruption |
Evidence-Based Hypothesis Using Molecular Docking Study
Our docked library was collected based on the aforementioned data hypothesis of 67 tested candidates, including 24 natural compounds, 22 market products previously reported to act against SARS-CoV and MERS-CoV, and 14 natural compounds previously indicated for inhibition of IL-6. In addition, some recommended treatments for SARS-CoV-2-infected patients were used as reference drugs (seven products). The molecular docking trial has been focused on one of the most promising potential target proteins of SARS-CoV-2: the main protease (Mpro or 3CLpro), which hinders enzyme activity, blocks viral replication, and disturbs the viral life cycle.95
Among all the natural compounds, the docking results showed that the top 3 hits are sinigrin, scutellarin, and baicalin based on their docking score (S), number of amino acids that interact, and binding pattern to fit the main protease binding pocket ( Table 5, Fig. 4).
Table 5.
Docking Energy Scores (S) for the Collected Database (in kcal/mol).
| Compound | Docking Score | Compound | Docking Score |
|---|---|---|---|
| Natural Compounds | Market Products | ||
| Natural compounds affecting SARS-CoV | Market products affecting SARS-CoV and MERS-CoV | ||
| Scutellarin | –15.29 | Fluphenazine | –11.75 |
| Sinigrin | –14.2 | Toremifene | –11.72 |
| Hesperetin | –14.19 | Amodiaquine | –11.57 |
| Myricetin | –13.92 | Mefloquine | –11.42 |
| Baicalin | –12.87 | Monensin | –11.26 |
| Aurantiamide | –11.24 | Tamoxifen | –11.15 |
| Lycorine | –10.1 | Thiothixene | –10.71 |
| Glycyrrhizin | –9.49 | Dasatinib | –10.65 |
| Aloe-emodin | –9.37 | Resveratrol | –10.41 |
| Tryptanthrin | –9.17 | Chlorophenoxamine | –10.34 |
| Natural compounds affecting MERS-CoV | Nocodazole | –10.19 | |
| Kazinol F | –12.70 | Imatinib | –9.56 |
| Kazinol A | –12.57 | Anisomycin | –9.50 |
| 4,7-Trihydroxyflavane | –12.41 | Cycloheximidine | –9.26 |
| Hydroxyisolonchocarpin | –12.25 | Chlomipramine | –9.24 |
| Harringtonine | –12.15 | Benzotropine | –9.24 |
| Homoharringtonine | –11.93 | Astemizole | –8.51 |
| Kazinol B | –11.84 | Emetine | –8.46 |
| Broussoflavan A | –11.71 | Fluspirilene | –8.45 |
| Broussochalcone B | –11.58 | ||
| Broussochalcone A | –11.43 | Promethazine | –8.06 |
| Papyriflavonol A | –11.32 | Salinomycin | –7.93 |
| Resveratrol | –10.79 | Hydroxychloroquine | –7.90 |
| Gemcitabine | –7.79 | ||
| Trigocherin | –8.38 | Standard reference regimen | |
| Natural compounds affecting IL-6 | Chloroquine | –9.26 | |
| Eriodictyol | –14.72 | Remdesivir | –8.99 |
| Quercetin | –14.35 | Oseltamivir | –8.88 |
| Kaempferol | –14.13 | Ribavirin | –8.69 |
| Epigallocatechin-3-gallate | –13.55 | Lopinavir | –8.64 |
| Luteolin | –13.30 | Ritonavir | –8.55 |
| Statins | –12.38 | Favipiravir | –6.85 |
| Celastrol | –11.98 | ||
| Apigenin | –11.81 | ||
| Mitraphylline | –11.76 | ||
| Genistein | –11.39 | ||
| Asperuloside | –11.26 | ||
| Sophoricoside | –10.46 | ||
| Curcumin | –9.11 | ||
| Sakuranetin | –8.69 | ||
As shown in Figure 5, the first hit, sinigrin, mediates a network of hydrogen bonding interactions with most of the essential amino acids within the active site—Gly143, His163, Glu166, Gln189, and Thr190—in addition to a hydrophobic interaction with the His41 hot residue. Extra hydrogen bonding interactions formed between sinigrin and Gln192, Cys145, and Pro168 might stabilize its binding within the active site. 2D and 3D analysis of the second hit, scutellarin, showed its binding through hydrogen bonding with Phe140, Cys145, His163, and Glu166. Moreover, a hydrophobic interaction was observed with Met165. The third hit, baicalin, interacts through hydrogen bonding with Phe140, Cys145, His163, Glu166, Gln189, and Gln192, and a Pi–H hydrogen interaction with Glu166.
Figure 5.
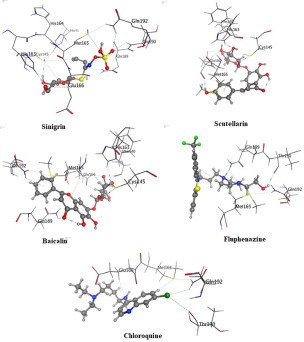
3D representations of the most promising compounds with the main protease active site of SARS-CoV-2.
The docking score, which represents the final score for binding energy, could be used to compare the binding affinity of different ligands to the same protein. Our three hits, sinigrin, scutellarin, and baicalin, attained high docking scores of –14.20, –15.29, and –14.75 kcal/mol, respectively. The docking scores of other natural compounds are illustrated in Table 5.
Twenty-four different pharmaceutical products previously reported to act against SARS-CoV and MERS-CoV were docked within the main protease binding site. The docking scores of the top candidates ranged from –7.79 kcal/mol to –11.75 kcal/mol (Table 5). Analysis of these market product binding modes showed that fluphenazine revealed the best binding pattern and the highest docking score (–11.75 kcal/mol). As shown in Figure 5, fluphenazine could mediate hydrogen bonding interactions with Met165, Gln189, Thr 190, and Gln192.
Chloroquine, Remdesivir, oseltamivir, ribavirin, lopinavir, ritonavir, and favipiravir, which are drugs recommended to be received by SARS-CoV-2-infected patients,10 were also docked within the enzyme binding site as a reference. Chloroquine (docking score = –9.26 kcal/mol) showed a better binding pattern between those recommended medicines. It could bind through hydrogen bonding interactions with Met165, Thr190, and Gln192 residues and hydrophobic interaction with Glu166. Other recommended agents—Remdesivir, oseltamivir, ribavirin, lopinavir, ritonavir, and favipiravir—showed docking scores of –8.99, –8.88, –8.69, –8.64, –8.55, and –6.85 kcal/mol, respectively.
In a comparison of fluphenazine and chloroquine with sinigrin, sinigrin showed an excellent binding pattern with the SARS-CoV-2 main proteinase active site, as illustrated in Figure 5. Moreover, it attained a promising docking score compared with fluphenazine and all the SARS-CoV-2-recommended regimens. Sinigrin is one of the previously reported natural compounds that affect SARS-CoV;83 its docking result reveal it to be a hot candidate for trial against SARS-CoV-2’s main proteinase enzyme inhibition.
Computational Approaches to Estimate Solubility and Permeability of Sinigrin
The bioavailability of the sinigrin compound was assessed using the Molispiration online property calculation toolkit, in particular, compliance with Lipinski’s “rule of five,” which describes molecular properties important for a drug’s pharmacokinetics (ADME) in the human body. Sinigrin obeyed Lipinski’s rule,96 showing no violations: Molecular weight (M.wt) = 359.38 Da; number of hydrogen bond donors (nOHNH) = 5; number of hydrogen bond acceptors (nON) = 10, and calculated octanol/water partition coefficient (c log P) = –3.63. So, it is considered a promising drug candidate.
Practical Points
Drug Research and Development against COVID-19
Even though drugs such as ribavirin, lopinavir, favipiravir, Remdesivir, hydroxychloroquine, Arbidol, and Camostat mesylate should be taken into consideration as an urgent measure, we ought to consider the following practical facts about COVID-19:
-
•
SARS-CoV-2 is an RNA beta-coronavirus with mutations in nsp1, nsp3, nsp15, and gene S that are associated with its epidemic behavior. COVID-19 has seven major target proteins (protease, Hel, hemagglutinin, esterase, membrane, envelope, and spike proteins).
-
•
Activation of SARS-CoV (S) protein resulted from sequential cleavage by the endosomal cysteine protease cathepsin L, and another trypsin-like serine protease and drug target could both be essential for virus inhibition.
-
•
Origin sequence and structural analyses of SARS-CoV-2 revealed virus similarity with ≥70%–96% homology with SARS-CoV and 50%–87% homology with MERS-CoV, which prompted the proposal, from the docking analysis results, that sinigrin and fluphenazine (sulfated nitrogenous compounds) may be considered promising COVID-19 protease inhibitors.
Fluphenazine (brand name Prolixin) is an antipsychotic medication used to treat schizophrenia and psychotic symptoms; it has previously reported activity as an SARS-CoV and MERS-CoV inhibitor. Currently, fluphenazine is not recommended for COVID-2 as a management regimen and may be considered a relatively safe candidate with minor common side effects, like lethargy, dizziness, nausea, loss of appetite, sweating, and dry mouth.97 Sinigrin is an allyl-glucosinolate present in plants of the Brassicaceae family, such as broccoli and brussels sprouts, and Brassica nigra seeds (mustard seeds).98 The docking score indicated that it is almost twice as potent as products from the reference regimen (Remdesivir, oseltamivir, ribavirin, lopinavir, ritonavir, and favipiravir).
-
•
Statistics from WHO reports for confirmed cases and deaths have revealed that temperature may affect the virus. This investigation is aligned with previously reported data that showed the consequences of different environmental states on coronavirus viability.49, 50, 51 On the other hand, countries in the southern hemisphere with high temperatures and low humidity have recorded an increasing number of infected cases, which may be attributed to the formation of virus aerosol particles and their accumulation in a large diameter as a result of lower humidity. This phenomenon could lead to increasing the risk of transmission via aerosol particles.
-
•
To prevent contagion between individuals, it is advisable to avoid close contact with anyone showing symptoms of respiratory illness, such as coughing and sneezing. Wearing masks and gloves is helpful and could decrease the spread of infection.
-
•
Although the rate of death is relatively low in comparison with old SARS-CoV, the high rate of transmission leads to pandemic categorization of the virus by the WHO, which consequently may lead to many economic disasters in the event that no treatment or vaccine is found for a long time.
-
•
Ground-glass opacities, glass nodules, pleural effusion, mediastinal lymphadenopathy on CT scan, and x-ray white lung are the main features of clinical radiology for COVID-19.
Proposed Research Agenda
-
•
Further research on all aspects of the disease is needed to elaborate on the mode of infection (especially concerning the rate of asymptomatic patients), different genetic susceptibilities, virus genomic sequence, and beneficial treatments.
-
•
The high prevalence of COVID-19 in some nations compared with others may inform the relationship between people’s genetic makeup and disease susceptibility.
-
•
Plant or microbial natural products such as glucosinolate, flavones, and sulfated nitrogenous compounds could be promising for future research against the coronavirus family.
Complementary Treatment Candidates
-
•
Punica granatum (pomegranate), Peganum harmale (harmel), Eriobotrya japonica Lindl. (loquat), Urginea Scilla steinh (red squill), Artemisia annua (anuual wormwood), Allium porrum (leek), Allium ursinum (wild garlic), Allium sativum (garlic), Colocasia esculenta (cocoyam), Phragmites australis (common reed), Morus nigra (blackberry), Vitis vinifera (grape seeds), Glycine max (L.) Merr (soybean), Plantago asiatica (Chinese plantain), Clerodendrum trichotomum (peanut butter tree), Markhamia lutea (Nile tulip), and Eleutherococcus senticosus (Siberian ginseng) may be contemplated as source materials for scientific and clinical studies, as they could help in the development of cheap, effective, protective, complementary medicine in the African region. Therefore, we invite curious researchers to carry out detailed studies on nation medicinal plants and food.
-
•
The herbal formula PM01492 and preparation Echinaforce87 could be helpful remedies against COVID-19. Clinical trials are recommended on the bases of previous art.
Drugs Need Further In Vitro, In Vivo, and In-Clinic Studies
-
•
Supplemental market products such as fluphenazine, toremifene, amodiaquine, mefloquine, monensin, and tamoxifen may demonstrate supportive effects against COVID-19. Future clinical studies are recommended to investigate their possible role in the prevention and treatment of SARS-CoV-2 respiratory viral infections.
-
•
Our obtained preliminary results from docking analysis revealed that sinigrin, scutellarin, and baicalin could have direct anti-SARS-CoV-2 activity through inhibition protease enzymes and may be considered potential candidates against COVID-19. Interestingly, the ADMET results of sinigrin showed good bioavailability.
General Recommendation
-
•
Additional studies should also test the possibility of combining available recommended treatments with other natural agents or with repurposed standard therapeutics, as a multitarget therapy could help in reducing the risk of generating drug-resistant viruses.
-
•
The authors wish to expand this research area not only for scientific soundness but also for potential druggability.99
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to thank Dr. Mohamed A. Mekkawy (Health Services Sector, Ministry of Interior, High Institute of Public Health, and Alexandria University, Egypt) for his guidance in the statistical analysis of data.
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
Footnotes
Supplemental material is available online with this article.
Supplemental Material
Supplemental Material
References
- 1.Singhal T. A Review of Coronavirus Disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khan M., Kazmi S., Bashir A., et al. COVID-19 Infection: Origin, Transmission, and Characteristics of Human Coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen F., Yu H., Guo J., et al. Identification of the Hyper-Variable Genomic Hotspot for the Novel Coronavirus SARS-CoV-2. J. Infect. 2020;80:671–693. doi: 10.1016/j.jinf.2020.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sahin A.R., Erdogran A., Agaoglu P.M., et al. 2019 Novel Coronavirus (Covid-19) Outbreak: A Review of the Current Literature. Eurasian J. Med. Oncol. 2020;4:1–7. [Google Scholar]
- 5.Jiang F., Deng L., Zhang L., et al. Review of the Clinical Characteristics of Coronavirus Disease 2019 (COVID-19) J. Gen. Intern. Med. 2020;35:1545–1549. doi: 10.1007/s11606-020-05762-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petticrew M. Time to Rethink the Systematic Review Catechism? Moving from ‘What Works’ to ‘What Happens’. Syst. Rev. 2015;4(36):1–6. doi: 10.1186/s13643-015-0027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song Z., Xu Y., Bao L., et al. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L., He Y., Zhou Y., et al. The Spike Protein of SARS-CoV—A Target for Vaccine and Therapeutic Development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehr R., Perlman S. Coronaviruses: An Overview of Their Replication and Pathogenesis in Coronaviruses. Methods Protoc. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y., Islam S., Wang J., et al. Traditional Chinese Medicine in the Treatment of Patients Infected with 2019-New Coronavirus (SARS-CoV-2): A Review and Perspective. Int. J. Biol. Sci. 2020;16:1708–1717. doi: 10.7150/ijbs.45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shajahan A., Supekar N. T., Gleinich A. S.; et al. Deducing the N- and O-Glycosylation Profile of the Spike Protein of Novel Coronavirus SARS-CoV-2. Glycobiology2020. DOI: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed]
- 12.Lu R., Zhao X., Li J., et al. Genomic Characterisation and Epidemiology of 2019 Novel Coronavirus: Implications for Virus Origins and Receptor Binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y., Liu Q., Guo D. Emerging Coronaviruses: Genome Structure, Replication, and Pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hui D.S., Azhar E., Madani T., et al. The Continuing 2019-nCoV Epidemic Threat of Novel Coronaviruses to Global Health—The Latest 2019 Novel Coronavirus Outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;92:418–423. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan Y., Shang J., Graham R.; et al. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol.2020. DOI: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed]
- 16.Belouzard S., Millet J.K., Licitra B.N., et al. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vinod N. The Agent and Host Factors in Covid-19: Exploring Pathogenesis and Therapeutic Implications. Biomed. J. Sci. Tech. Res. 2020;27:20669–20681. [Google Scholar]
- 18.Shajahan A., Archer-hartmann S. A., Supekar N. T.; et al. Comprehensive Characterization of N- and O-Glycosylation of SARS-CoV-2 Human Receptor Angiotensin Converting Enzyme 2. bioRxiv2020. DOI: 10.1101/2020.05.01.071688. [DOI] [PMC free article] [PubMed]
- 19.Gierer S., Bertram S., Kaup F., et al. The Spike Protein of the Emerging Betacoronavirus EMC Uses a Novel Coronavirus Receptor for Entry, Can Be Activated by TMPRSS2, and Is Targeted by Neutralizing Antibodies. J. Virol. 2013;87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mille K., Whittaker R. Host Cell Entry of Middle East Respiratory Syndrome Coronavirus after Two-Step, Furin-Mediated Activation of the Spike Protein. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simmons G., Zmora P., Gierer S., et al. Proteolytic Activation of the SARS-Coronavirus Spike Protein: Cutting Enzymes at the Cutting Edge of Antiviral Research. Antiviral Res. 2013;100:605–614. doi: 10.1016/j.antiviral.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang C., Zheng W., Huang X., et al. Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J. Proteome Res. 2020;19:1351–1360. doi: 10.1021/acs.jproteome.0c00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lontok E., Corse E., Machamer E. Intracellular Targeting Signals Contribute to Localization of Coronavirus Spike Proteins Near the Virus Assembly Site. J. Virol. 2004;78:5913–5922. doi: 10.1128/JVI.78.11.5913-5922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Machamer E., Rose K. A Specific Transmembrane Domain of a Coronavirus E1 Glycoprotein Is Required for its Retention in the Golgi Region. J. Cell Biol. 1987;105:1205–1214. doi: 10.1083/jcb.105.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Haan M., Kuo L., Masters S., et al. Coronavirus Particle Assembly: Primary Structure Requirements of the Membrane Protein. J. Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Haan M., Vennema H., Rottier M. Assembly of the Coronavirus Envelope: Homotypic Interactions between the M Proteins. J. Virol. 2000;74:4967–4978. doi: 10.1128/jvi.74.11.4967-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masters P.S., Kuo L., Ye R., et al. Genetic and Molecular Biological Analysis of Protein-Protein Interactions in Coronavirus Assembly. Adv. Exp. Med. Biol. 2006;581:163–173. doi: 10.1007/978-0-387-33012-9_29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Haan M., Rottier M. Molecular Interactions in the Assembly of Coronaviruses. Adv. Virus Res. 2005;64:165–230. doi: 10.1016/S0065-3527(05)64006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shereen A., Khan S., Kazmi A., et al. COVID-19 Infection: Origin, Transmission, and Characteristics of Human Coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cascella M., Rajnik M., Cuomo A., et al. StatPearls [Internet] StatPearls Publishing; Petersburg, FL: 2020. Features, Evaluation and Treatment Coronavirus (COVID-19) [PubMed] [Google Scholar]
- 31.Peeri N., Shrestha N., Rahman M.S., et al. The SARS, MERS and Novel Coronavirus (COVID-19) Epidemics, the Newest and Biggest Global Health Threats: What Lessons Have We Learned? Int. J. Epidemiol. 2020;49:717–726. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hilgenfeld R., Peiris M. From SARS to MERS, 10 Years of Research on Highly Pathogenic Human Coronaviruses. Antiviral Res. 2013;100:286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shrwani J., Noureldin M., Dafalla M. MERS-CoV Incidence in the Kingdom of Saudi Arabia and Worldwide: General Review Article. J. Egypt. Soc. Parasitol. 2017;47:467–478. [Google Scholar]
- 34.Wang Y., Chen Y., Qin Q. Unique Epidemiological and Clinical Features of the Emerging 2019 Novel Coronavirus Pneumonia (COVID-19) Implicate Special Control Measures. J. Med. Virol. 2020;92:568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang X., Wu C., Li X., et al. On the Origin and Continuing Evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu W., Li H. COVID-19: Attacks the 1-Beta Chain of Hemoglobin and Captures the Porphyrin to Inhibit Human Heme Metabolism. ChemRxiv. 2020 [Google Scholar]
- 37.Andersen G., Rambaut A., Lipkin I., et al. The Proximal Origin of SARS-CoV-2. Nat. Med. 2020;89:44–48. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu B., Ge X., Shi Z., et al. Bat Origin of Human Coronaviruses. Virol. J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramphul K., Mejias G. Coronavirus Disease: A Review of a New Threat to Public Health. Cureus. 2020;12:e7276. doi: 10.7759/cureus.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tahir M., Alqahtani M., Alamri A.; et al. Structural Basis of SARS-CoV-2 3CL Pro and Anti-COVID-19 Drug Discovery from Medicinal Plants. J. Pharm. Anal.2020. DOI: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed]
- 41.Chan F., Kok K., Zhu Z., et al. Genomic Characterization of the 2019 Novel Human-Pathogenic Coronavirus Isolated from a Patient with Atypical Pneumonia after Visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Werf S., Nicolas E., Bernadette C.; et al. Novel Strain of SARS-Associated Coronavirus and Applications Thereof. Google Patents. 2010. https://patents.google.com/patent/US20070128224A1/en.
- 43.Guo R., Cao D., Hong S., et al. The Origin, Transmission and Clinical Therapies on Coronavirus Disease 2019 (COVID-19) Outbreak—An Update On the Status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nezhad S., Mosaddeghi P., Negahdaripour M.; et al. Therapeutic Approaches for COVID-19 Based on the Dynamics of Interferon-Mediated Immune Responses. https://www.preprints.org/manuscript/202003.0206/v1 (accessed Aug 3, 2020).
- 45.Zu Y., Jiang M.D., Xu P., et al. Coronavirus Disease 2019 (COVID-19): A Perspective from China. Radiology. 2020;2019:200490. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai C., Shih P., Ko C., et al. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Coronavirus Disease-2019 (COVID-19): The Epidemic and the Challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai H. Sex Difference and Smoking Predisposition in Patients with COVID-19. Lancet Respir. Med. 2020;8:e20. doi: 10.1016/S2213-2600(20)30117-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang L., Karakiulakis G., Roth M. Are Patients with Hypertension and Diabetes Mellitus at Increased Risk for COVID-19 Infection? Lancet Respir. Med. 2020;8:e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oliveiros B., Caramelo L., Ferreira C. Role of Temperature and Humidity in the Modulation of the Doubling Time of COVID-19 Cases. medRxiv. 2020. DOI: 10.1101/2020.03.05.20031872.
- 50.Mohammad S., Parham H., Augustin V.; et al. Spread of SARS-CoV-2 Coronavirus Likely to be Constrained by Climate. medRxiv. 2020. DOI: 10.1101/2020.03.12.20034728.
- 51.Sajadi M., Habibzadeh P., Vintzileos A.; et al. Temperature, Humidity, and Latitude Analysis to Predict Potential Spread and Seasonality for COVID-19. https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3550308 (accessed Aug 3, 2020). [DOI] [PMC free article] [PubMed]
- 52.Cao Y., Li L., Feng Z., et al. Comparative Genetic Analysis of the Novel Coronavirus (2019-nCoV/SARS-CoV-2) Receptor ACE2 in Different Populations. Cell Discov. 2020;6:11. doi: 10.1038/s41421-020-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q., Guan X., Wu P., et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen H., Guo J., Wang C., et al. Clinical Characteristics and Intrauterine Vertical Transmission Potential of COVID-19 Infection in Nine Pregnant Women: A Retrospective Review of Medical Records. Lancet. 2020;395:809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dai W., Zhang H., Yu J., et al. CT Imaging and Differential Diagnosis of COVID-19. Can. Assoc. Radiol. J. 2020;71:195–200. doi: 10.1177/0846537120913033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y., Xia L. Coronavirus Disease 2019 (COVID-19): Role of Chest CT in Diagnosis and Management. AJR Am. J. Roentgenol. 2020;214:1–7. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 57.Yoon S.H., Lee K.H., Kim J.Y., et al. Chest Radiographic and CT Findings of the 2019 Novel Coronavirus Disease (COVID-19): Analysis of Nine Patients Treated in Korea. Korean J. Radiol. 2020;21:494–500. doi: 10.3348/kjr.2020.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin T., Hsu C., Lin C. Antiviral Natural Products and Herbal Medicines. J. Tradit. Complement. Med. 2014;4:24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luo H., Tang Q., Shang Y., et al. Can Chinese Medicine Be Used for Prevention of Corona Virus Disease 2019 (COVID-19)? A Review of Historical Classics, Research Evidence and Current Prevention Programs. Chin. J. Integr. Med. 2020;26:243–250. doi: 10.1007/s11655-020-3192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sayed A.M., Alhadrami H.A., El-Gendy A.O., et al. Microbial Natural Products as Potential Inhibitors of SARS-CoV-2 Main Protease (Mpro) Microorganisms. 2020;8:970. doi: 10.3390/microorganisms8070970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mohana N.C., Rao H.Y., Rakshith D., et al. Omics Based Approach for Biodiscovery of Microbial Natural Products in Antibiotic Resistance Era. J. Genet. Eng. Biotechnol. 2018;16(1):1–8. doi: 10.1016/j.jgeb.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mousa L. Prevention and Treatment of Influenza, Influenza-Like Illness, and Common Cold by Herbal, Complementary, and Natural Therapies. J. Evid. Based Complementary Altern. Med. 2017;22:166–174. doi: 10.1177/2156587216641831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mushtaq S., Abbasi B.H., Uzair B., et al. Natural Products as Reservoirs of Novel Therapeutic Agents. EXCLI J. 2018;17:420. doi: 10.17179/excli2018-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conti P., Ronconi G., Caraffa A., et al. Induction of Pro-Inflammatory Cytokines (IL-1 and IL-6) and Lung Inflammation by Coronavirus-19 (COVI-19 or SARS-CoV-2): Anti-Inflammatory Strategies. J. Biol. Regul. Homeost. Agents. 2020;34(2):1. doi: 10.23812/CONTI-E. [DOI] [PubMed] [Google Scholar]
- 65.Kim K.S., Jung H., Shin I.K., et al. Induction of Interleukin-1 Beta (IL-1β) Is a Critical Component of Lung Inflammation during Influenza A (H1N1) Virus Infection. J. Med. Virol. 2015;87:1104–1112. doi: 10.1002/jmv.24138. [DOI] [PubMed] [Google Scholar]
- 66.Lin L., Lu L., Cao W., et al. Hypothesis for Potential Pathogenesis of SARS-CoV-2 Infection—A Review of Immune Changes in Patients with Viral Pneumonia. Emerg. Microbes Infect. 2020;9:727–732. doi: 10.1080/22221751.2020.1746199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woo Y., Lau P., Chu M., et al. Characterization and Complete Genome Sequence of a Novel Coronavirus, Coronavirus HKU1, from Patients with Pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez M. Antiviral Activity of Compounds Isolated from Plants. Pharm. Biol. 2003;41:107–157. [Google Scholar]
- 69.Karagöz A., Önay E., Arda N., et al. Antiviral Potency of Mistletoe (Viscum album ssp. album) Extracts against Human Parainfluenza Virus Type 2 in Vero Cells. Phyther. Res. 2003;17:560–562. doi: 10.1002/ptr.1163. [DOI] [PubMed] [Google Scholar]
- 70.Jassim A., Naji A. Novel Antiviral Agents: A Medicinal Plant Perspective. J. Appl. Microbiol. 2003;95:412–427. doi: 10.1046/j.1365-2672.2003.02026.x. [DOI] [PubMed] [Google Scholar]
- 71.Odimegwu D., Esimone C. In Vitro Antiviral Activity of Nauclea latifolia Root Bark Extract against the Respiratory Syncytial Virus. Eur. J. Med. Plants. 2018;22:1–7. [Google Scholar]
- 72.Chathuranga K., Kim M.S., Lee H.C., et al. Anti-Respiratory Syncytial Virus Activity of Plantago asiatica and Clerodendrum trichotomum Extracts In Vitro and In Vivo. Viruses. 2019;11:604. doi: 10.3390/v11070604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Doğan H., Duman R. Evaluation of the Antiviral Activity of Ballota glandulosissima Hub.-Mor. & Patzak Extract against Respiratory Syncytial Virus (RSV) Trak. Univ. J. Nat. Sci. 2019;20:121–127. [Google Scholar]
- 74.Roth M., Fang L., Stolz D., et al. Pelargonium sidoides Radix Extract EPs 7630 Reduces Rhinovirus Infection through Modulation of Viral Binding Proteins on Human Bronchial Epithelial Cells. PLoS One. 2019;14:1–18. doi: 10.1371/journal.pone.0210702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi J., Song H., Bhatt R., et al. Anti-Human Rhinovirus Activity of Gallic Acid Possessing Antioxidant Capacity. Phyther. Res. 2010;24:1292–1296. doi: 10.1002/ptr.3101. [DOI] [PubMed] [Google Scholar]
- 76.Li S.Y., Chen C., Zhang H.Q., et al. Identification of Natural Compounds with Antiviral Activities against SARS-Associated Coronavirus. Antiviral Res. 2005;67:18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lau K.M., Lee K.M., Koon C.M., et al. Immunomodulatory and Anti-SARS Activities of Houttuynia cordata. J. Ethnopharmacol. 2008;118:79–85. doi: 10.1016/j.jep.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen F., Chan K.H., Jiang Y., et al. In Vitro Susceptibility of 10 Clinical Isolates of SARS Coronavirus to Selected Antiviral Compounds. J. Clin. Virol. 2004;31:69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Keyaerts E., Vijgen L., Pannecouque C., et al. Plant Lectins Are Potent Inhibitors of Coronaviruses by Interfering with Two Targets in the Viral Replication Cycle. Antiviral Res. 2007;75:179–187. doi: 10.1016/j.antiviral.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Park J.Y., Yuk H.J., Ryu H.W., et al. Evaluation of Polyphenols from Broussonetia papyrifera as Coronavirus Protease Inhibitors. J. Enzyme Inhib. Med. Chem. 2017;32:504–512. doi: 10.1080/14756366.2016.1265519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin S.C., Ho C.T., Chuo W.H., et al. Effective Inhibition of MERS-CoV Infection by Resveratrol. BMC Infect. Dis. 2017;17:1–10. doi: 10.1186/s12879-017-2253-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jadav S., Ganta M., Kumar A., et al. The Updates on Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Epidemiology, Pathogenesis, Viral Genome and Currently Available Drugs. J. Pharm. Chem. 2016;3:10. [Google Scholar]
- 83.Lin C.W., Tsai F.J., Tsai C.H., et al. Anti-SARS Coronavirus 3C-Like Protease Effects of Isatis indigotica Root and Plant-Derived Phenolic Compounds. Antiviral Res. 2005;68:36–42. doi: 10.1016/j.antiviral.2005.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu M.S., Lee J., Lee J.M., et al. Identification of Myricetin and Scutellarein as Novel Chemical Inhibitors of the SARS Coronavirus Helicase, nsP13. Bioorganic Med. Chem. Lett. 2012;22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cristina F., Michael E., Rea K., et al. Antiviral Effects of Glycyrrhiza species. Phyther. Res. 2008;22:141–148. doi: 10.1002/ptr.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cinatl J., Morgenstern B., Bauer G., et al. Glycyrrhizin, an Active Component of Liquorice Roots, and Replication of SARS-Associated Coronavirus Progression of Cerebral White Matter Lesions: 6-Year Results of the Austrian Stroke Prevention Study. Lancet. 2003;361:2045–2046. doi: 10.1016/S0140-6736(03)13615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Signer J., Jonsdottir H. R., Albrich W. C.; et al. In Vitro Antiviral Activity of Echinaforce®, an Echinacea Purpurea Preparation, against Common Cold Coronavirus 229E and Highly Pathogenic MERS-CoV and SARS-CoV. Virol. J.2020. DOI: 10.21203/rs.2.24724/v1. [DOI] [PMC free article] [PubMed]
- 88.Ahmed S., Marotte H., Kwan K., et al. Epigallocatechin-3-Gallate Inhibits IL-6 Synthesis and Suppresses Transsignaling by Enhancing Soluble gp130 Production. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14692–14697. doi: 10.1073/pnas.0802675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jiang P., Yang C., Elliott L., et al. Down-Regulation of Expression of Interleukin-6 and Its Receptor Results in Growth Inhibition of MCF-7 Breast Cancer cells. Anticancer Res. 2011;31:2899–2906. [PubMed] [Google Scholar]
- 90.Venkatesha H., Dudics S., Astry B., et al. Control of Autoimmune Inflammation by Celastrol, a Natural Triterpenoid. Pathog. Dis. 2016;74:1–12. doi: 10.1093/femspd/ftw059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Omoigui S. The Interleukin-6 Inflammation Pathway from Cholesterol to Aging—Role of Statins, Bisphosphonates and Plant Polyphenols in Aging and age-Related Diseases. Immun. Ageing. 2007;4:1–22. doi: 10.1186/1742-4933-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Santana F., Pinheiro M., Mernak B., et al. Evidences of Herbal Medicine-Derived Natural Products Effects in Inflammatory Lung Diseases. Mediators Inflamm. 2016;2016:1–14. doi: 10.1155/2016/2348968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hadieh B., Said O., Masalh M., et al. Anti-Inflammatory Effects of Herbal-Derived Factors Are Mediated by Down Regulation of Pro-Inflammatory Cytokines. 2nd Conf. Biotechnol. Appl. Palest. 2010;2:1–12. [Google Scholar]
- 94.Suradej B., Sookkhee S., Panyakaew J., et al. Kaempferia parviflora Extract Inhibits STAT3 Activation and Interleukin-6 Production in HeLa Cervical Cancer Cells. Int. J. Mol. Sci. 2019;20:4226. doi: 10.3390/ijms20174226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang L., Sun X., Curth U., et al. Crystal Structure of SARS-CoV-2 Main Protease Provides a Basis for Design of Improved α-Ketoamide Inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lipinski A., Lombardo F., Dominy W., et al. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2012;64:4–17. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 97.Abbas F., Rajab T., Alsamarrai O., et al. Fluphenazine Decanoate (Timing of Administration) for People with Schizophrenia. Cochrane Database Syst. Rev. 2017;10:1–15. [Google Scholar]
- 98.Mazumder A., Dwivedi A., Plessis D. Sinigrin and Its Therapeutic Benefits. Molecules. 2016;21:416. doi: 10.3390/molecules21040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Badria F., Abou-Mohamed G., El-Mowafy A., et al. Mirazid: A New Schistosomicidal Drug. Pharm. Biol. 2001;39:127–131. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material


