Abstract
The global pandemic caused by the SARS-CoV-2 infection is a health emergency that needs to be addressed immediately. The international scientific community, following World Health Organization (WHO) indications, launched different trials for testing drugs putatively able to block the SARS-CoV-2 infection or treat the COVID-19 disease symptoms. In parallel, studies devoted to a better understanding of SARS-CoV-2 biology are in the course for designing an effective vaccine. One of the human membrane proteins known to be docked by the virus is angiotensin-converting enzyme 2 (ACE2), proposed to be responsible for viral entry in target cells. Recently, the 3D structure of ACE2 has been obtained, showing its physical interaction with B0AT1 (SLC6A19), a plasma membrane transporter involved in the trafficking of amino acids in cells. The receptor targeted by SARS-CoV-2 is a supercomplex formed by a dimer of ACE2-B0AT1, in which ACE2 binds the viral protein and B0AT1 stabilizes the heterodimer. As a serendipity occurrence, nimesulide was shown to abolish the transport function of B0AT1. Here we suggest including nimesulide in the list of drugs to be tested for the identification of co-adjuvants in the treatment of COVID-19.
Keywords: SARS-CoV-2, nimesulide, COVID-19, drug discovery, SLC6A19, ACE2-B0AT1
The emergency that the human population is currently experiencing calls for rapid identification of drugs to treat the COVID-19 disease caused by SARS-CoV-2. However, the design of novel drugs is a long process that cannot be pursued with immediate results. Indeed, a complete study starts with improvement of knowledge of the virus biology, moving to human target identification and drug design, then performing tests in in vitro and in vivo models, and concluding with human trials; in terms of time, this means more than 5 years. From the depicted scenario, it is evident that alternative, but much faster, paths should be followed, starting from actual knowledge on SARS-CoV-2. This virus belongs to the coronavirus family, is able to infect vertebrates, and is the seventh coronavirus acknowledged to infect humans; the outbreak of SARS-CoV-2, together with that of SARS-CoV and MERS-CoV, is the third passage of a Coronaviridae virus from an animal to humans causing a major epidemic event with severe and life-threatening disease.1 , 2 SARS-CoV-2, like other members of the family, is an enveloped positive-strand RNA virus that spreads quickly among humans. The rapid diffusion of the infections, together with the percentage of deaths, has boosted the efforts of the scientific community to identify the molecular mechanisms of infection. This primary knowledge is crucial for designing effective vaccines and drugs, able to prevent and block or reduce the infections, respectively. The approach proposed by the World Health Organization (WHO), to save time, is to find old approved and safe drugs to actively treat the SARS-CoV-2 infection, as a serendipity effect. In this respect, some drugs have already been highlighted as candidates for a megatrial called Solidarity (“Solidarity” clinical trial for COVID-19 treatments: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments). The first list includes (1) remdesivir, already used against the Ebola virus and able to inhibit the key viral enzyme RNA-dependent RNA polymerase;3 (2) the antimalaric chloroquine and hydroxychloroquine, for their capacity to block SARS-Cov-2 in vitro as well as in a group of COVID-19 patients;4 (3) the anti-HIV complex ritonavir/lopinavir, which has protease activity and has been revealed to be effective with the coronavirus MERS;5 and (4) the complex ritonavir/lopinavir in association with interferon-β, for modulation of the inflammatory response.6 In the meantime, comparative analysis with other known coronaviruses has allowed us to conclude that a known SARS target, angiotensin-converting enzyme 2 (ACE2),7 is one of the ways in which SARS-CoV-2 docking to human cells can occur.8 , 9 This hypothesis was demonstrated by the recently solved structure of the human ACE2, in complex with the viral receptor-binding domain (RBD), at 2.9 Å resolution by cryo-electron microscropy.8 Interestingly, the 3D structure of the human receptor brought to light a more complex architecture in which ACE2 forms a heterodimer with B0AT1 (ACE2-B0AT1). ACE2 consists of an extracellular peptidase domain (PD) and a collectrin-like domain (CLD) that ends with a single transmembrane α-helix. This is the ACE2 moiety interacting with B0AT1 (SLC6A19), an integral membrane protein formed by 12 transmembrane α-helices. B0AT1 belongs to the SLC6 family and is responsible for the sodium-dependent transport of neutral amino acids, mainly in the intestine and kidney.10 The interaction between ACE2 and B0AT1 is well documented; indeed, ACE2 plays a major role in trafficking B0AT1 at the plasma membrane in the intestine but not in the kidney, where collectrin is B0AT1’s chaperone. The complex reported by Yan et al. has a peculiar assembly in which two heterodimers, constituting ACE2-B0AT1, form a homodimeric superstructure (ACE2-B0AT1)2, as shown by Figure 1. The proposed role for B0AT1 is that of stabilizing each single heterodimeric complex ACE2-B0AT1.8 In line with this, the presence of B0AT1 was crucial for solving the 3D structure of the entire complex. Furthermore, the resolution of the (ACE2-B0AT1)2 superstructure disclosed the existence of two different conformations, called “open” and “closed.” The transition between the two states is characterized by coordinated conformational changes occurring, to a major extent, at the level of ACE2 and, to a minor extent, at the level of B0AT1.8 Yan et al. showed that the viral RBD interacts with (ACE2-B0AT1)2 only when the receptor is in the closed state. The docking occurs at the level of the PD of ACE2, similarly to the mechanism already described for the SARS-CoV infection.8 The requirement of the superstructure for viral infection was confirmed by the interaction of two RBDs with (ACE2-B0AT1)2.8 Even though the process of internalization of the virus is still not completely understood, it seems that the protease TMPRSS2 is also involved; this should cleave some residues of ACE2 upon interaction with RBD. Based on the structural data, the authors propose that B0AT1 plays a regulatory role in the receptor complex for ACE2 interaction with RBD. Altogether, the structural data suggest that, even if B0AT1 does not directly interact with the virus, it may play a role in the internalization of the virus, stabilizing and/or participating in the receptor’s conformational changes.8 Therefore, it is not trivial to believe that the receptor complex is a promising drug target for COVID-19. Indeed, ongoing studies aim to select approved drugs able to directly or indirectly target ACE2.9 , 11 Noteworthy, as a serendipity occurrence, we previously described the high-affinity interaction of the commonly used drug nimesulide with the rat isoform of the B0AT1 transporter, which is virtually identical to the human one.12 Biochemical data together with computational analysis showed that nimesulide completely abolishes B0AT1 transport function by reaching a high-affinity pocket accessible from the extracellular milieu, depicted in Figure 1.13 , 14 This previous finding allowed us to envisage the use of nimesulide to interfere with RBD docking on (ACE2-B0AT1)2. It is not guaranteed that nimesulide could truly help in COVID-19 treatment. An open question is the presence of the receptor (ACE2-B0AT1)2 in the lung, which is the tissue mainly damaged by the viral infection. Indeed, as above highlighted, both ACE2 and B0AT1 are highly expressed in the intestine and kidney, even if their direct interaction has been demonstrated only in the intestine.10 On the contrary, expression in the lung can be only inferred from available transcriptomic and proteomic data, which suggest that both ACE2 and B0AT1 are present in the lung at a much lower level than in the intestine (https://www.uniprot.org/). Therefore, even if definitive proof is not available, the existence of (ACE2-B0AT1)2 in the lung can be hypothesized. Interestingly, nimesulide was not listed as one of the potential drugs for treating COVID-19 by a first network-based analysis;11 however, more recent in silico studies included it among potential candidates against COVID-19.15 , 16 Our commentary aims to point out that nimesulide should be added to the list of drugs already under trial for COVID-19 treatment. In this frame, it is important to highlight that the use of nimesulide has been controversial since its first appearance and, in fact, the pharmacological target of nimesulide is still not well defined.17 Nimesulide has never been listed among the allowed drugs in several countries, including the United States, because of its liver toxicity.18 For this reason, nimesulide was initially withdrawn from the market in Europe. However, after reevaluation of the data, hepatotoxicity was revealed to be a rare occurrence, and the European Medicines Agency’s Committee for Medicinal Products for Human Use (CHMP) confirmed that the benefits of nimesulide exceed its risks, allowing its use in Europe. In conclusion, this commentary highlights the presence of a molecular basis, besides the recently published in silico studies, for considering nimesulide as an additional opportunity in the ongoing trials for COVID-19 treatment.
Figure 1.
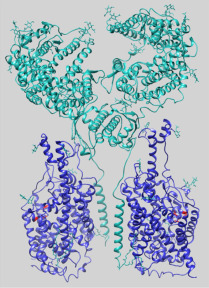
Overall 3D structure of the supercomplex (ACE2-B0AT1)2. The structure (PDB: 6M18) is represented in closed conformation as a ribbon.8 The superstructure is composed of two heterodimers of ACE2-B0AT1; each heterodimer comprises one ACE2 subunit (cyan) and one B0AT1 subunit (blue). In red are shown residues predicted to form the nimesulide binding site on the B0AT1 subunit.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgments
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by PRIN (Progetti di Ricerca di Interesse Nazionale) project no. 2017PAB8EM MIUR (Italian Ministry of Instruction, University and Research) to C.I.
References
- 1.Wu F., Zhao S., Yu B., et al. A New Coronavirus Associated with Human Respiratory Disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Species Severe Acute Respiratory Syndrome-Related Coronavirus: Classifying 2019-nCoV and Naming It SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheahan T.P., Sims A.C., Graham R.L., et al. Broad-Spectrum Antiviral GS-5734 Inhibits Both Epidemic and Zoonotic Coronaviruses. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao J., Tian Z., Yang X. Breakthrough: Chloroquine Phosphate Has Shown Apparent Efficacy in Treatment of COVID-19 Associated Pneumonia in Clinical Studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 5.Chan J.F., Yao Y., Yeung M.L., et al. Treatment with Lopinavir/Ritonavir or Interferon-beta1b Improves Outcome of MERS-CoV Infection in a Nonhuman Primate Model of Common Marmoset. J. Infect. Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arabi Y.M., Asiri A.Y., Assiri A.M., et al. Treatment of Middle East Respiratory Syndrome with a Combination of Lopinavir/Ritonavir and Interferon-beta1b (MIRACLE trial): Statistical Analysis Plan for a Recursive Two-Stage Group Sequential Randomized Controlled Trial. Trials. 2020;21:8. doi: 10.1186/s13063-019-3846-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F., Li W., Farzan M., et al. Structure of SARS Coronavirus Spike Receptor-Binding Domain Complexed with Receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 8.Yan R., Zhang Y., Li Y., et al. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaduganathan M., Vardeny O., Michel T., et al. Renin–Angiotensin–Aldosterone System Inhibitors in Patients with Covid-19. N. Engl. J. Med. 2020;382:1653–1659. doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camargo S.M., Singer D., Makrides V., et al. Tissue-Specific Amino Acid Transporter Partners ACE2 and Collectrin Differentially Interact with Hartnup Mutations. Gastroenterology. 2009;136:872–882. doi: 10.1053/j.gastro.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon D. E., Jang G. M., Bouhaddou M.; et al. A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing. Nature2020. DOI: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed]
- 12.Pochini L., Seidita A., Sensi C., et al. Nimesulide Binding Site in the B0AT1 (SLC6A19) Amino Acid Transporter. Mechanism of Inhibition Revealed by Proteoliposome Transport Assay and Molecular Modelling. Biochem. Pharmacol. 2014;89:422–430. doi: 10.1016/j.bcp.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 13.Broer S. Amino Acid Transporters as Disease Modifiers and Drug Targets. SLAS Discov. 2018;23:303–320. doi: 10.1177/2472555218755629. [DOI] [PubMed] [Google Scholar]
- 14.Yadav A., Shah N., Tiwari P.K., et al. Novel Chemical Scaffolds to Inhibit the Neutral Amino Acid Transporter B(0)AT1 (SLC6A19), a Potential Target to Treat Metabolic Diseases. Front. Pharmacol. 2020;11:140. doi: 10.3389/fphar.2020.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cava C., Bertoli G., Castiglioni I. In Silico Discovery of Candidate Drugs against Covid-19. Viruses. 2020;12:404. doi: 10.3390/v12040404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y., Hou Y., Shen J., et al. Network-Based Drug Repurposing for Novel Coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caiazzo E., Ialenti A., Cicala C. The Relatively Selective Cyclooxygenase-2 Inhibitor Nimesulide: What’s Going On? Eur. J. Pharmacol. 2019;848:105–111. doi: 10.1016/j.ejphar.2019.01.044. [DOI] [PubMed] [Google Scholar]
- 18.Kwon J., Kim S., Yoo H., et al. Nimesulide-Induced Hepatotoxicity: A Systematic Review and Meta-Analysis. PloS One. 2019;14:e0209264. doi: 10.1371/journal.pone.0209264. [DOI] [PMC free article] [PubMed] [Google Scholar]


