Abstract
The ongoing global pandemic of COVID-19, caused by SARS-CoV-2 has killed more than 5.9 million individuals out of ∼43 million confirmed infections. At present, several parts of the world are encountering the 3rd wave. Mass vaccination has been started in several countries but they are less likely to be broadly available for the current pandemic, repurposing of the existing drugs has drawn highest attention for an immediate solution. A recent publication has mapped the physical interactions of SARS-CoV-2 and human proteins by affinity-purification mass spectrometry (AP-MS) and identified 332 high-confidence SARS-CoV-2-human protein-protein interactions (PPIs). Here, we taken a network biology approach and constructed a human protein-protein interaction network (PPIN) with the above SARS-CoV-2 targeted proteins. We utilized a combination of essential network centrality measures and functional properties of the human proteins to identify the critical human targets of SARS-CoV-2. Four human proteins, namely PRKACA, RHOA, CDK5RAP2, and CEP250 have emerged as the best therapeutic targets, of which PRKACA and CEP250 were also found by another group as potential candidates for drug targets in COVID-19. We further found candidate drugs/compounds, such as guanosine triphosphate, remdesivir, adenosine monophosphate, MgATP, and H-89 dihydrochloride that bind the target human proteins. The urgency to prevent the spread of infection and the death of diseased individuals has prompted the search for agents from the pool of approved drugs to repurpose them for COVID-19. Our results indicate that host targeting therapy with the repurposed drugs may be a useful strategy for the treatment of SARS-CoV-2 infection.
Abbreviations: COVID-19, Coronavirus disease-2019; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; 2019-nCoV, 2019 Novel Coronavirus; MERS-CoV, Middle East respiratory syndrome coronavirus; ARDS, Acute Respiratory Distress Syndrome; PPIs, Protein-Protein interactions; AP-MS, Affinity purification-mass spectrometry; PPIN, Protein-Protein interaction network; DC, Degree centrality; CC, Closeness centrality; BC, Betweenness centrality; EC, eigenvector centrality; GO, Gene ontology; MF, Molecular function; CC, Cellular component; BP, Biological process; MgATP, Magnesium ATP; MgADP, Magnesium ADP; PKA, Protein kinase A; AAV2, Adeno-associated virus; HCMV, Human cytomegalovirus; HCV, Hepatitis C virus; HIV-1, Human immunodeficiency virus 1; VSV, vesicular stomatitis virus; ARDS, Adult Respiratory Distress Syndrome; CDK5RAP2, Cyclin dependent kinase 5 regulatory subunit associated protein 2; CNS, Central nervous system; IL-2, Interleukin-2; PD-L1, programmed death-ligand 1
Keywords: COVID-19, Novel coronavirus, SARS-CoV-2, Drug repurposing, RdRP, Network biology, Rank aggregation, Functional annotation
1. Introduction
The ongoing pandemic of Coronavirus disease-2019 (COVID-19) has resulted in over 5.9 million deaths among more than 43 million confirmed cases in over 200 countries and area of territories as on 28th February 2022 (https://www.who.int/emergencies/diseases/novel-coronavirus-2019). The outbreak that started in the Wuhan City of China in December 2019, rapidly engulfed the entire World [1]. Currently, many parts of the world is experiencing of 3rd wave. COVID-19 is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), also called 2019 Novel Coronavirus (2019-nCoV). In the past two decades, two highly pathogenic human coronavirus infections, namely the SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) emerged [2]. Human coronaviruses primarily target the respiratory system that may lead to pneumonia, Acute Respiratory Distress Syndrome (ARDS), multi-organ failure and death. However, the earlier pandemics, despite being more fatal, resulted in much fewer infections and total deaths [3]. The real danger of SARS-CoV-2 lies in its ability to rapidly spread from the infected individuals to susceptible population [4]. Although most patients of COVID-19 are either asymptotic or mildly symptomatic, they may act as reservoirs of infection that pose risk of severe disease in the older populations and patients with co-morbidities [5], [6].
SARS-CoV-2 belongs to the family of single-stranded positive-sense RNA viruses [7]. Whole genome sequencing of SARS-CoV-2 reveals highest sequence identity with SARS-CoV (79.6%) [8]. In absence of the availability of effective antiviral drugs, social distancing, repeated hand washing, isolation/quarantine of potentially infected individuals and containment of high incidence zones have been shown to be the most effective strategies to control the spread of SARS-CoV-2 infection. Mass vaccination has already been introduced in several countries and more than 100 candidate vaccines and nearly 150 potential drug molecules are currently under investigation [9], [10]. While vaccines will provide long-term solutions, they are less likely to be effective for current mutant variants. Developments of new drugs also take years before their clinical efficacy and safety is ensured. The scientific community is looking up to repurposing of the existing drugs as the best immediate hope against COVID-19 [7], [8], [11], [12], [13]. A number of antiviral (favipiravir, remdesivir, lopinavir/ritonavir), antibiotic (doxycycline, azithromycin) and other (for eg, chloroquine) drugs, and were tried with only modest success, but none were universally effective in reducing severity or mortality. Interim data analysis of the WHO Solidarity Trial concluded little or no effect of remdesivir, hydroxychloroquine, lopinavir, and interferon regimens on hospitalized patients with Covid-19, as indicated by overall mortality, initiation of ventilation, and duration of hospital stay [14]. Several studies reported that prophylactic use of hydroxychloroquine might reduce the incidence of severe infections, but others failed to reproduce it. On the other hand, use of hydroxychloroquine to treat symptomatic infection is discouraged because of no clear benefits along with the risk of serious cardiac side effects in severe cases [15], [16]. More drugs need to be urgently investigated for repurposing. However, major bottleneck in this endeavour lies in the lack of understanding about the novel coronavirus and its interactions with the host. Identification of the host proteins that play a critical role in COVID-19 pathogenesis will significantly increase the number of potential drug targets to be repurposed for host-directed therapies.
Network biology approaches were introduced recently to identify the critical host proteins for SARS-CoV-2 infection and their interactor drugs/compounds [8], [17], [18]. However, the key protein sets found in these studies were non– overlapping. In this study, we aimed to identify the critical host targets of COVID-19, using a network biology study. Recently, Gordon et al. identified 67 druggable human proteins from 332 high confidence SARS-CoV-2-human protein-protein interactions (PPIs) [17]. We considered the important network biology and functional annotation properties of the above SARS-CoV-2 targeted human proteins. Drug-protein interactions were integrated into this study for the identification of candidate drug for repurposing.
2. Materials and methods
2.1. Collection of SARS-CoV-2-human Protein-Protein interactions
We obtained SARS-CoV-2-human protein-protein interactions (PPIs) from a recently published literature [17]. The authors identified 332 SARS-CoV-2-human PPIs using affinity purification-mass spectrometry (AP-MS). Furthermore, we have considered interaction between the spike protein of SARS-CoV-2 and angiotensin-converting enzyme 2 (ACE2) of human [19], [20]. We found that the interactors included 26 SARS-CoV-2 and 333 human proteins. We observed that all the human proteins were reviewed and well-annotated in UniProtKB [21]. We then searched for all human PPIs for the above 333 human proteins in the STRING database version 11 [22]. We found 991 human PPIs among 312 human proteins with the combined scores greater than 0.4 and considered them for further analysis. Workflow of the proposed method is shown in Fig. 1 .
Fig. 1.
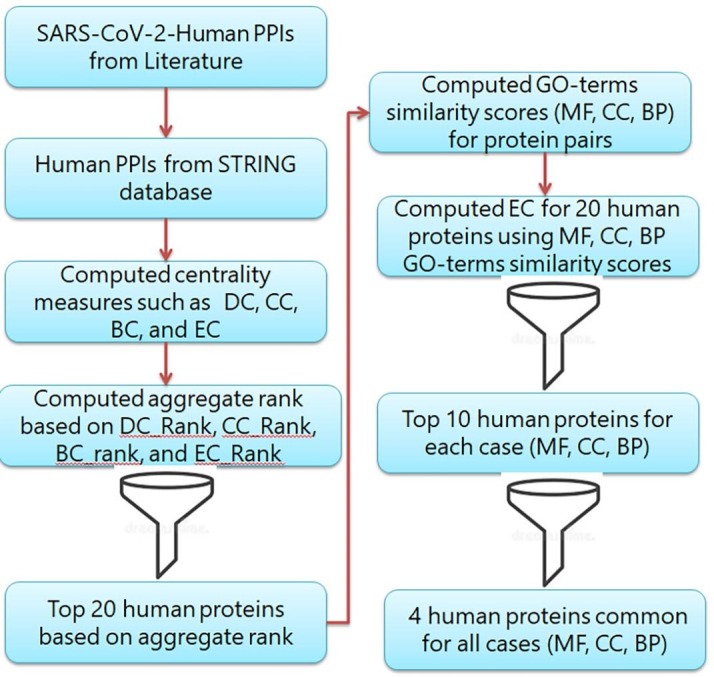
Workflow of the proposed method.
2.2. Network properties
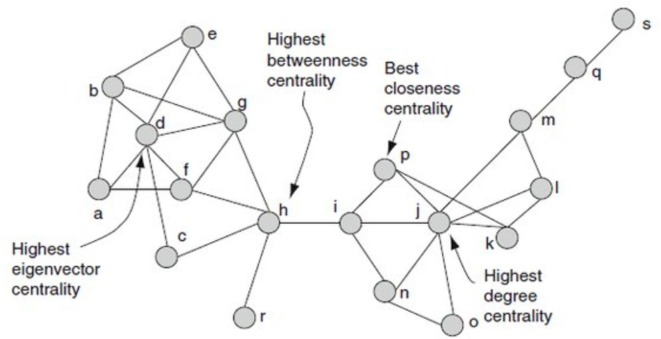
We constructed a network of 991 human PPIs using Cytoscape version 3.9.1 [23]. In this protein–protein interaction network (PPIN), each node indicates a protein and an edge indicates an interaction between the proteins. We integrated combined interaction score between the proteins as an edge weight. A weighted PPIN can be represented as a weighted undirected graph G = (V, E), where V is a set of vertices representing the nodes (proteins) and E is a set of edges representing the connections (PPI) between the nodes. Each edge (i, j) ∊ E is assign with a weight wi,j, which represents the combined interaction score between node i and node j. We calculated decisive network properties, including the degree centrality (DC), closeness centrality (CC), betweenness centrality (BC) and eigenvector centrality (EC) of the edge-weighted network using CytoNCA [24]. A simple example of these centrality measures are shown in Fig. 2 . CytoNCA computes the centrality measures of an edge-weighted network using the following equations.
where, Nu is the node set containing all the neighbors of node u. w(u,v) is the weight of the edge connecting node u and node v.
where, is the weighted distance of the shortest path from node u to node v.
Fig. 2.
An example of four centrality measures is demonstrated in the same network.
is the weight of the edge from node s to node t.
where, p(s,t) is the total number of shortest paths from node s to node t. p(s,u,t) is the number of those paths that pass through u.
where, is the eigenvector corresponding to the largest eigenvalue of the weighted adjacency matrix
2.3. Rank aggregation
We calculated four different rankings of the human proteins in the above network using the DC, CC, BC and EC values. We computed the median ranking score of each protein by an excel formula, instead of exploring each ranking score separately. Subsequently, we sorted the individual human protein in the network using the median ranking score. Top 20 proteins from the candidate pool of 312 human proteins were considered for further analysis.
2.4. Functional similarity
All possible combination pairs of the top 20 proteins were considered for functional similarity. We employed the GOSim R package (version 1.22.0) to compute functional similarity of the protein pairs [25]. The Resnik terms similarity was calculated for all the three domains of gene ontology (GO), viz., the molecular function (MF), cellular component (CC) and biological process (BP) [26].
2.5. Identification of important proteins
Eigenvector and eccentricity centrality are significant centrality measures to identify the influential nodes in a network [27]. For EC calculation, the weighted value of the link of a node is more important than the number of links. The GO similarity score of a protein pair is represented as an edge-weight in the network. In an edge-weighted network, EC of each node was calculated using CytoNCA. Top 20 proteins of all three GO domains (MF, CC, BP) were sorted using EC scores. To find the most influential proteins common to the above domains, we considered the top 10 proteins from each domain.
2.6. Enrichment analysis of pathways and GO
Identification of the critical disease-associated pathways could help drug discovery. Pathway analysis also helps to explore disease mechanisms. The gene ontology biological process (GO BP) analysis assists in understanding the processes that need the support of a protein to become complete. We used “Enrichr: a comprehensive gene set enrichment analysis web server” for the enrichment analysis of Pathways and GO [28]. The pathway with an adjusted P-value < 0.05 was considered as an enriched pathway. We also considered an adjusted P-value < 0.05 for GO BP enrichment analysis.
2.7. Drug-protein interaction network
To identify a drug/compound for the target protein, we searched for drug-protein interactions in the STITCH database [29]. We examined only the experimentally validated and manually curated databases to find reliable drug-protein interactions. The drug/compound was prioritized using a combined score greater than 0.9.
3. Results
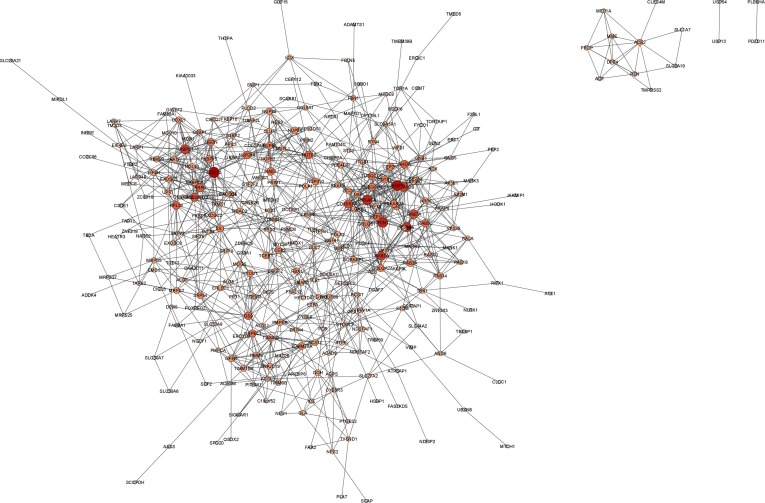
We found a candidate pool of 312 human proteins from 991 human PPIs (Fig. 3 ). The proteins were ranked based on their DC, CC, BC and EC scores, for which we observed the maximum ranks of 312, 311, 245 and 295, respectively (Supplementary Table 1 ). All the centrality measures are important to find the crucial node in a network. Therefore, we sorted the proteins based on the median ranking scores and found a minimum and a maximum score of 3 and 296.5, respectively (Supplementary Table 2 ). Top 20 targets from this candidate protein pool were extracted for functional similarity analysis (Table 1).
Fig. 3.
Human PPIs of SARS-CoV-2 targeted human proteins, visualized by Cytoscape 3.9.1. The human proteins are represented by the nodes. The size and the color of the nodes are different according to their degree.
Table 1.
Top 20 proteins based on the median ranking scores.
| Protein | DC_Rank | CC_Rank | BC_Rank | EC_Rank | Median_Rank |
|---|---|---|---|---|---|
| AKAP9 | 1 | 6 | 18 | 1 | 3.5 |
| PRKACA | 3 | 1 | 8 | 4 | 3.5 |
| PRKAR2B | 4 | 4 | 59 | 2 | 4 |
| RAB1A | 7 | 2 | 4 | 17 | 5.5 |
| RAB8A | 2 | 5 | 26 | 9 | 7 |
| PCNT | 5 | 10 | 43 | 3 | 7.5 |
| CDK5RAP2 | 8 | 7 | 47 | 5 | 7.5 |
| RAB7A | 9 | 8 | 6 | 21 | 8.5 |
| RHOA | 13 | 3 | 5 | 16 | 9 |
| CEP135 | 17 | 15 | 103 | 6 | 16 |
| CNTRL | 19 | 17 | 132 | 7 | 18 |
| DDX10 | 6 | 30 | 7 | 105 | 18.5 |
| PRKAR2A | 26 | 13 | 104 | 13 | 19.5 |
| RPL36 | 10 | 19 | 22 | 113 | 20.5 |
| TUBGCP3 | 20 | 25 | 46 | 11 | 22.5 |
| CEP250 | 21 | 24 | 234 | 8 | 22.5 |
| RAE1 | 16 | 23 | 25 | 81 | 24 |
| NUTF2 | 23 | 26 | 1 | 59 | 24.5 |
| TUBGCP2 | 22 | 31 | 93 | 12 | 26.5 |
| OS9 | 35 | 18 | 2 | 133 | 26.5 |
Table 2.
Eigenvector centrality score of human proteins using GO MF, CC and BP similarity scores.
| Protein |
Eigenvector centrality score |
||
|---|---|---|---|
| MF | CC | BP | |
| AKAP9 | 0.221214 | 0.183526 | 0.247266 |
| CDK5RAP2 | 0.278605 | 0.272746 | 0.256644 |
| CEP135 | 0.107631 | 0.198114 | 0.23159 |
| CEP250 | 0.203105 | 0.273835 | 0.241416 |
| CNTRL | 0.118379 | 0.204541 | 0.234498 |
| DDX10 | 0.270821 | 0.265321 | 0.10762 |
| NUTF2 | 0.14052 | 0.250276 | 0.162841 |
| OS9 | 0.310271 | 0.095256 | 0.170248 |
| PCNT | 0.114475 | 0.17833 | 0.269729 |
| PRKACA | 0.326611 | 0.265322 | 0.240817 |
| PRKAR2A | 0.333194 | 0.262849 | 0.157465 |
| PRKAR2B | 0.333194 | 0.265352 | 0.212394 |
| RAB1A | 0.307002 | 0.249588 | 0.22605 |
| RAB7A | 3.86E-13 | 0.243784 | 0.233122 |
| RAB8A | 2.71E-11 | 0.279244 | 0.248546 |
| RAE1 | 0.199742 | 0.135516 | 0.200893 |
| RHOA | 0.317055 | 0.247685 | 0.244188 |
| RPL36 | 0.037185 | 0.132995 | 0.252692 |
| TUBGCP2 | 0.140298 | 0.164321 | 0.229425 |
| TUBGCP3 | 0.140298 | 0.167172 | 0.233789 |
To understand the functions of the proteins at the molecular level, their functional location in the cell and the biological processes they support, we included all the aspects of the GO annotation and considered different GO domains, viz., MF, CC and BP. For a protein pair, all possible term similarities were calculated. We observed a minimum score of 0, a maximum score of 1 and a mean score of 0.317 for MF terms similarity (Supplementary Table 3 ). CC terms similarity scores were 0.158 and 1 as the minimum and maximum scores, respectively, with a mean of 0.632 (Supplementary Table 4). Finally, the BP terms similarity scores had a mean of 0.547, with the minimum and maximum scores of 0 and 1, respectively (Supplementary Table 5).
Table 3.
Identified drugs/compounds that modulate SARS-CoV-2 interactors in the human cell.
| Protein | Drug/Compound | Experimentally_determined_interaction | Database_annotated | Combined_score | PubChem CID |
|---|---|---|---|---|---|
| PRKACA | pSer | 0.957 | 0.8 | 0.991 | 24779545 |
| PRKACA | A-674563 | 0.942 | 0.8 | 0.988 | 11314340 |
| PRKACA | H-89 dihydroch. | 0.91 | 0.8 | 0.981 | 5702541 |
| PRKACA | balanol | 0.911 | 0.8 | 0.981 | 5287736 |
| PRKACA | A-443654 | 0.912 | 0.8 | 0.981 | 10172943 |
| PRKACA | calcium ions | 0.795 | 0.9 | 0.978 | 271 |
| PRKACA | MgATP | 0.795 | 0.9 | 0.978 | 15126 |
| PRKACA | magnesium | 0.795 | 0.9 | 0.978 | 5462224 |
| PRKACA | MgADP | 0.795 | 0.9 | 0.978 | 30103 |
| PRKACA | manganese | 0.795 | 0.9 | 0.978 | 15551713 |
| PRKACA | adenosine mono. | 0.795 | 0.9 | 0.978 | 6083 |
| PRKACA | phosphate | 0.795 | 0.9 | 0.978 | 1061 |
| RHOA | phosphate | 0.933 | 0.9 | 0.993 | 1061 |
| RHOA | magnesium | 0.852 | 0.9 | 0.984 | 5462224 |
| CDK5RAP2 | MgADP | 0 | 0.9 | 0.9 | 30103 |
| CDK5RAP2 | MgATP | 0 | 0.9 | 0.9 | 15126 |
| CDK5RAP2 | guanosine trip. | 0 | 0.9 | 0.9 | 135398633 |
| CEP250 | guanosine trip. | 0 | 0.9 | 0.9 | 135398633 |
| CEP250 | MgATP | 0 | 0.9 | 0.9 | 15126 |
| CEP250 | MgADP | 0 | 0.9 | 0.9 | 30103 |
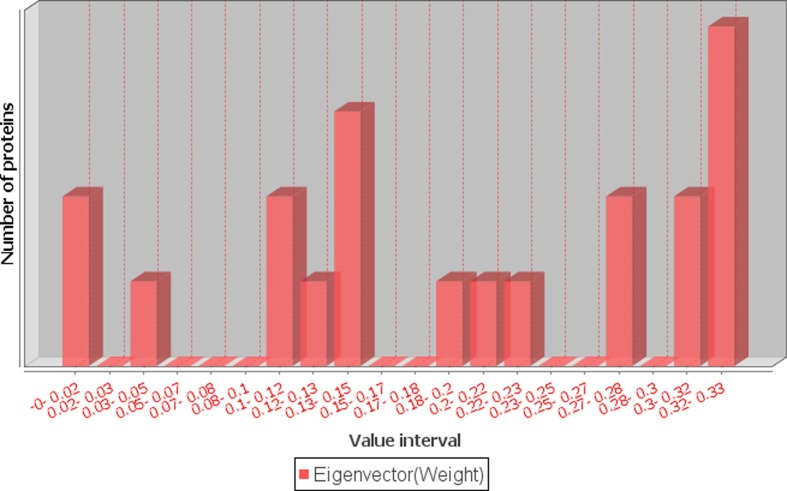
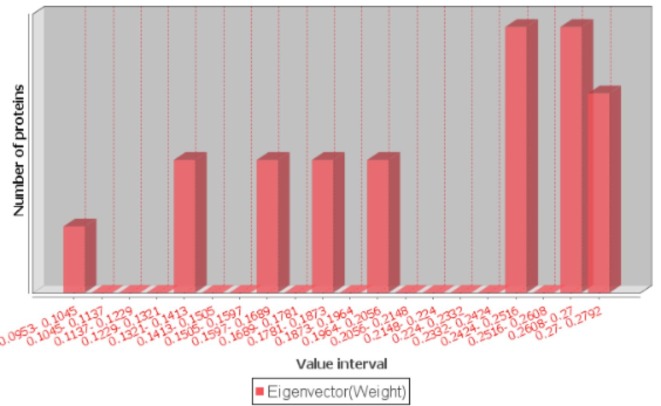
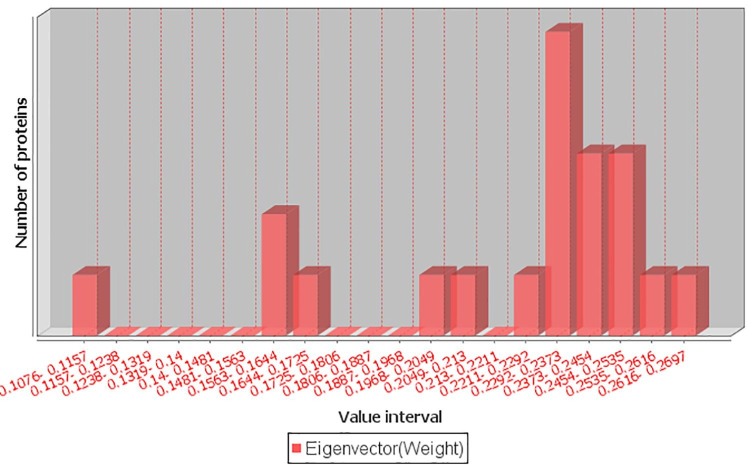
We constructed three networks using the MF, CC and BP similarity scores. For a protein pair, the GO similarity score was treated as the weight of an edge between them. For these networks, only the edge weights (GO similarity scores) were different, while nodes and number of edges, as well as their connections were the same. The edge weights were the main differentiators for eigenvector centrality calculation. As shown in Table 2, the eigenvector centrality scores for MF, CC, and BP ranged between 3.86E-13 and 0.33, 0.09 and 0.28 and 0.11 and 0.27, respectively (Fig. 4 and Supplementary Figure S1 and S2).
Fig. 4.
Distribution of eigenvector centrality measure scores for GO MF.
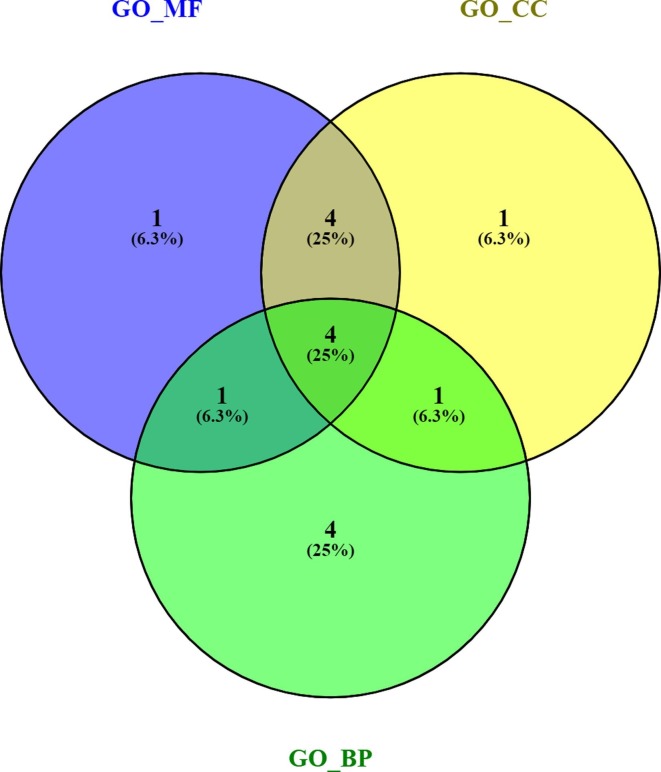
In each case, 10 proteins with the highest eigenvector centrality scores were considered to find the most common influential protein set. Interestingly, we found that four proteins (gene name PRKACA, RHOA, CDK5RAP2 and CEP250) were common to all the cases (Fig. 5 ). The pathway enrichment analysis of these proteins showed vascular smooth muscle contraction, chemokine signaling and viral carcinogenesis pathways etc. (Supplementary Table 6). The gene ontology BP enrichment analysis showed enriched biological processes like ciliary basal body-plasma membrane docking, cell cycle G2/M phase transition, mitotic cell cycle phase transition and cilium assembly (Supplementary Table 7).
Fig. 5.
Venn diagram of eigenvector centrality based top 10 proteins from each GO similarity scores (MF, CC, BP).
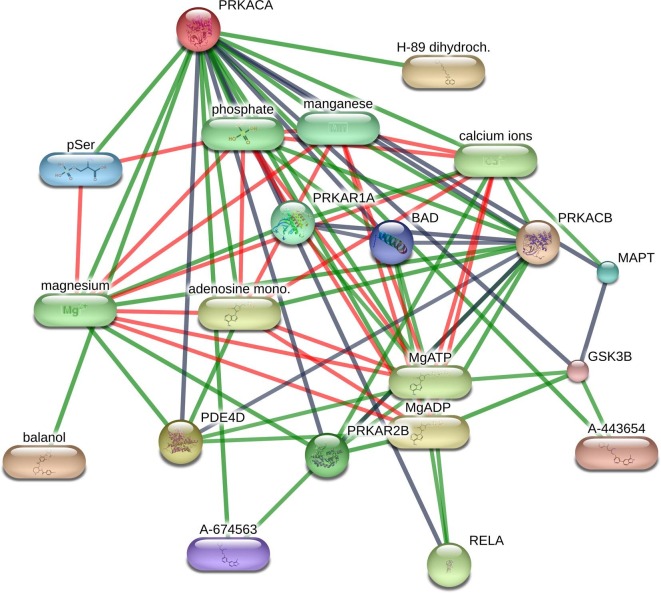
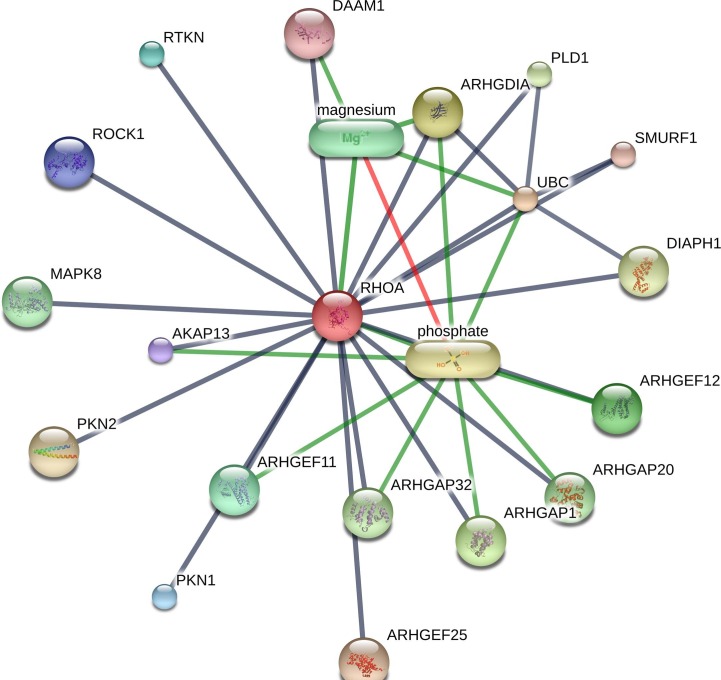
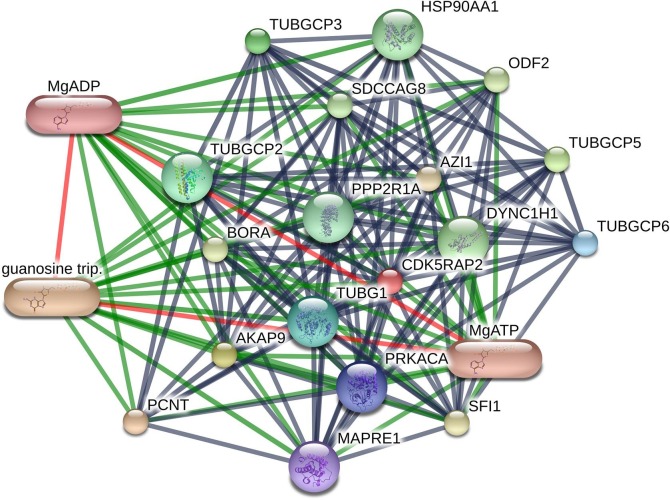
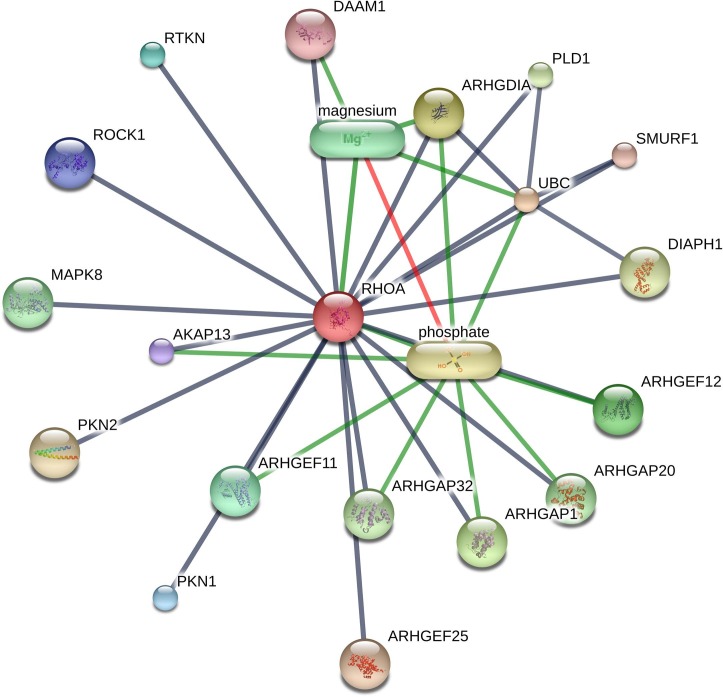
The drug-protein interaction network analysis of the above four proteins showed multiple interactions (Table 3). PRKACA interacted with H-89 dihydrochloride, balanol, MgATP (magnesium ATP), MgADP (magnesium ADP), magnesium, manganese and adenosine monophosphate, etc. (Fig. 6 ). RHOA showed interaction with phosphate andmagnesium (Supplementary Figure S3). Both CDK5RAP2 and CEP250 interacted with MgATP, MgADP and guanosine triphosphate (Supplementary Figure S4-S5).
Fig. 6.
PRKACA protein interactions in STITCH database.
4. Discussion
Knowledge about SARS-CoV-2 targeted human proteins and their interaction network are crucial to understand disease pathogenesis as well as for the diagnosis of COVID-19. Identification of the human targets of SARS-CoV-2 might help the development of an effective treatment. Network biology approaches have demonstrated their effectiveness in the identification of repurposable drugs for various human diseases [8], [30], [31]. Here, we propose a novel network biology approach using the SARS-CoV-2-human PPI and human PPI to identify the critical human targets of SARS-CoV-2 infection. Most of the experimental proteomic studies for SARS-CoV-2–host PPIs are unable to find the interactions between the spike protein and ACE2 [32]. One of the probable reasons might be the expression level of ACE2 is low in most experimental cell lines [33]. However, ACE2 plays a vital role by helping viruses to enter the human cells during disease onset [34]. Therefore, we have considered interactions between the spike protein and ACE2 in this study. The concept of centrality measures, such as DC, CC, BC, and EC have been extensively used to identify the important proteins in the PPIN [35], [36], [37]. However, majority of the network analysis methods have utilized only one or two centrality measures, although it is not clear which one performs the best. We computed four different ranks for each protein, using the DC, CC, BC and EC scores. To find out the key human protein set required for SARS-CoV-2 infection, we opted for a median ranking score instead of considering each ranking method separately. The top 20 proteins from a candidate pool of 312 proteins were considered for the identification of the most crucial human protein set for SARS-CoV-2 infection.
Average term similarity scores of 0.317, 0.632, and 0.547 were found for MF, CC, and BP, respectively, indicating notable functional similarities between the top 20 genes. To identify the most prominent members from the above list, we computed eigenvector centrality, using the MF, CC, and BP terms similarity scores. We found that human proteins PRKACA (UniprotID: P17612), RHOA (UniprotID: P61586), CDK5RAP2 (UniprotID: Q96SN8), and CEP250 (UniprotID: Q9BV73) with high eigenvector centrality were common to all three GO domains. This suggested that these proteins might be the most important targets for SARS-CoV-2 infection. The pathways and GO BP enrichment analysis showed that they target critical pathways and biological processes related to infectious diseases. PRKACA and CEP250 were recently identified as potential therapeutic targets in COVID-19 by Gordon et al. [17]. The authors found that PRKACA and CEP250 interact with nonstructural protein-13 (Nsp13) of SARS-CoV-2. Host interactions with Nsp13 (golgins) may enable dramatic reconfigurations of Golgi trafficking. During disease onset ACE2 plays a vital role for virus entry into the human cells but during the disease progression, PRKACA, RHOA, CDK5RAP2, and CEP250 proteins play a vital role. Moreover, we have employed the proposed methodology on other viruses like hepatitis C virus (HCV) and dengue virus to find essential proteins. We found 3 essential human genes (APOA2, APOA1 and APOB) from a candidate pool of 241 targets for HCV and 3 other genes (RPL5, RPL12 and CALR) from a candidate pool of 244 human genes for the dengue virus [38], [39].
PRKACA is the major catalytic subunit of protein kinase A (PKA), which is ubiquitously present throughout the body, with moderate to high levels of expression in the lung epithelial, vascular endothelial and immune cells. These are the most important cell types involved in the development of lung pathology of COVID-19. Protein kinase A, a serine/threonine kinase and the most important effector of cyclic AMP in the cell has more than 250 substrates that regulate a large number of cellular processes, including cell growth, division and differentiation, metabolism, gene transcription etc. [40]. Major targets of PKA, such as CREB, NF-κB and MAP kinases are critical regulators of pathogenesis of infectious diseases and host immune response [41]. PKA inhibitor (PKI 14–22) suppressed ZIKA virus replication at the post entry stage by interfering with negative-sense RNA synthesis and viral protein translation [42]. Inhibitors of adenylate cyclase and PKA signalling also inhibits epithelial infection of human cytomegalovirus (HCMV), Hepatitis C virus (HCV), and Human immunodeficiency virus 1 (HIV-1) viral replication in CD4 cells [43], [44], [45]. Chloroquine and antiviral drug lopinavir binds to the active site of PKA and have shown inhibitory effects on SARS-CoV-2 and other coronavirus replication in cell culture assays [46], [47]. Additional benefits from PKA inhibition may come from the fact that PKA signalling exacerbates hypoxic injury to cells [41], [48]. Pulmonary thromboembolism has been implicated as one of the major causes of death in COVID-19 and PKA-inducible HIF-1α was shown to increase coagulation factors and thrombus formation [49]. However, the potential beneficial role of PKA, such as T regulatory cell homeostasis [50], prevention of increased endothelial permeability induced by inflammatory mediators [51] and beta adrenoreceptor mediated alveolar liquid clearance [52] should be taken into account before considering therapeutic modulation of PKA in COVID-19.
Rho GTPases are key regulators of actin cytosketon organization and microtubule dynamics. A large number of viruses exploit Rho GTPases to facilitate their entry into the cells (hepatitis C virus, Kaposis Sarcoma Herpes Virus, herpes simplex virus-1, rotavirus and coronaviruses), intracellular replication (herpesvirus, adenovirus, influenza virus) and cell-to-cell spread (influenza virus). Even before the cellular entry, retroviruses, papillomaviruses, herpesviruses, poxviruses, dengue virus and vesicular stomatitis virus (VSV) engage Rho GTPase signaling to facilitate unidirectional movements toward the cell [53], [54], [55], [56]. Recently, Rho kinase inhibitors were proposed as a potential therapy for severe SARS-CoV-2 infection, leading to Adult Respiratory Distress Syndrome (ARDS) and cardiovascular comorbidity [57], [58]. Extensive literature review suggests that activation of RhoA/ROCK signaling pathways causes a burst in inflammation, immune cell migration, cell adhesion to pulmonary endothelium, coagulation, endothelial cell contraction and apoptosis. These may result in endothelium barrier dysfunction and edema, the two hallmarks of ARDS. Additionally, RhoA decreases SARS-CoV-2 receptor ACE2 expression [59].
Cyclin dependent kinase 5 regulatory subunit associated protein 2 (CDK5RAP2) regulates the activation of CDK5, a proline-directed serine/threonine protein kinase. Viruses regulate their replication through the effects on host cell cyclin-dependent kinases (CDKs), thereby modulating cell cycle progression. This is underscored by the inhibition of HSV-1 replication in vitro by Palbociclib, a CDK6 inhibitor and CDK9-targeting alvocidib showing activity against influenza A. Moreover, CDK4/6 inhibitor, abemaciclib, was one of the FDA-approved drugs to display in vitro activity against SARS-CoV-2 [60], [61]. CDK5 was reported to increase programmed death-ligand 1 (PD-L1) expression on tumour cells, which interacted with PD-1 on T cells and impaired anti-tumour immune response [62]. Given that increased PD-1 expression was thought to underlie the immune pathogenesis of COVID-19 [63], it needs to be investigated if CDK5 blocking could be helpful. CDK5 also exerts pro-inflammatory role during infection by regulating neutrophil degranulation and lactoferin secretion and by inhibiting LPS-induced IL-10 production [64]. Activated neutrophils and macrophages play key roles in the lung inflammation of COVID-19. Several reports suggested that early institution of methylprednisolone might help to resolve lung pathology and accelerate recovery while dexamethasone treatment of severe COVID-19 reduce death [65]. A recent study reported that deletion or functional inhibition of CDK5 resulted in the suppression of nitric oxide (NO) generation in the inflammatory macrophages, thus augmenting the anti-inflammatory effects of glucocorticoids [66]. Addition of CDK5 inhibitor might help to reduce the dose of corticosteroids without compromising its therapeutic efficacy.
Drug-protein interactions analysis for the above proteins displayed the most common interactions with MgATP, MgADP, magnesium, and guanosine triphosphate drug/compound. A recent article proposed that the suppresion of guanosine triphosphate might cause reduction of cellular magnesium levels via conversion to MgATP (magnesium ATP). Reduction of magnesium level resulted in blocking of RNA synthesis through RNA dependent RNA Polymerase (RdRP) that might help COVID-19 treatment [67]. Antiviral drug remdesivir also functions by inhibiting RdRP and has shown promise in treating severe COVID-19 infection [68], [69], [70], [71]. Unavailability of experimental study of PPIs limits our analysis for mutant variants. Moreover, we found that most mutations occur in spike protein, resulting in variants with increased binding affinity with human ACE2 receptor and transmissibility and reduced antibody neutralization [72], [73].
5. Conclusion
SARS-CoV-2 targeted human proteins and their intra-species interactions are important to understand the mechanism behind COVID-19. Identification of the critical human interactors of SARS-CoV-2 may aid in the development of host-directed therapies. We found a more influential human protein set relevant for COVID-19 by integrating SARS-CoV-2-human PPIs and interaction dynamics of human PPIs. Our finding suggests that 4 human proteins play a key role in disease development. Studies focused on these proteins may lead to potential biomarkers of SARS-CoV-2 infection. These proteins may be good templates for the development of drugs for COVID-19 and two of these proteins were reported by another study as potential drug targets. The study also indicated candidate repurposed drugs for COVID-19. Further experimental and clinical validations are required to substantiate the findings of this article.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
R.K.B. and S.D. acknowledges the Senior Research Fellowship of Indian Council of Medical Research [No. ISRM/11(39)/2017]. A. M. acknowledges the support received from the research project (Memo No: 355(Sanc.)/ST/P/S&T/6G-10/2018 dt. 08/03/2019) of Dept. of Science & Technology and Biotechnology, Govt. of West Bengal, India.
Funding information
No funding was obtained for this study. Only fellowship was obtained. The funding body did not play any roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Authors’ Contributions
R.K.B., A.M., U.M. and S.D. conceived and designed experiments, R.K.B. executed experiments, R.K.B., A.M., U.M. and S.D. analyzed data and wrote manuscript. All the authors have read and approved the final manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ymeth.2022.03.016.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary figure 1.
Distribution of eigenvector centrality measure scores for GO CC.
Supplementary figure 2.
Distribution of eigenvector centrality measure scores for GO BP.
Supplementary figure 3.
RHOAhuman protein targeted compounds.
Supplementary figure 4.
CDK5RAP2 human protein targeted compounds.
Supplementary figure 5.
CEP250 human protein targeted compounds.
References
- 1.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections-more than just the common cold. JAMA. 2020 doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 3.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson R.M., Heesterbeek H., Klinkenberg D., Hollingsworth T.D. How will country-based mitigation measures influence the course of the COVID-19 epidemic? Lancet. 2020;395(10228):931–934. doi: 10.1016/S0140-6736(20)30567-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55(3) doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singhal T. A Review of Coronavirus Disease-2019 (COVID-19) Indian J. Pediatr. 2020;87(4):281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tufan A., Avanoglu Guler A., Matucci-Cerinic M. COVID-19, immune system response, hyperinflammation and repurposing antirheumatic drugs, Turk. J Med Sci. 2020;50(SI-1):620–632. doi: 10.3906/sag-2004-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullard A. Flooded by the torrent: the COVID-19 drug pipeline. Lancet. 2020;395(10232):1245–1246. doi: 10.1016/S0140-6736(20)30894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thanh Le T., Andreadakis Z., Kumar A., Gomez Roman R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19(5):305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 11.Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14(1):72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Cremades M., Solans B.P., Hughes E., Ernest J.P., Wallender E., Aweeka F., Luetkemeyer A., Savic R.M. Optimizing hydroxychloroquine dosing for patients with COVID-19: An integrative modeling approach for effective drug repurposing. Clin. Pharmacol. Ther. 2020 doi: 10.1002/cpt.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odhar H.A., Ahjel S.W., Albeer A., Hashim A.F., Rayshan A.M., Humadi S.S. Molecular docking and dynamics simulation of FDA approved drugs with the main protease from 2019 novel coronavirus. Bioinformation. 2020;16(3):236–244. doi: 10.6026/97320630016236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium W.H.O.S.T., Pan H., Peto R., Henao-Restrepo A.M., Preziosi M.P., Sathiyamoorthy V., Abdool Karim Q., Alejandria M.M., Hernandez Garcia C., Kieny M.P., Malekzadeh R., Murthy S., Reddy K.S., Roses Periago M., Abi Hanna P., Ader F., Al-Bader A.M., Alhasawi A., Allum E., Alotaibi A., Alvarez-Moreno C.A., Appadoo S., Asiri A., Aukrust P., Barratt-Due A., Bellani S., Branca M., Cappel-Porter H.B.C., Cerrato N., Chow T.S., Como N., Eustace J., Garcia P.J., Godbole S., Gotuzzo E., Griskevicius L., Hamra R., Hassan M., Hassany M., Hutton D., Irmansyah I., Jancoriene L., Kirwan J., Kumar S., Lennon P., Lopardo G., Lydon P., Magrini N., Maguire T., Manevska S., Manuel O., McGinty S., Medina M.T., Mesa Rubio M.L., Miranda-Montoya M.C., Nel J., Nunes E.P., Perola M., Portoles A., Rasmin M.R., Raza A., Rees H., Reges P.P.S., Rogers C.A., Salami K., Salvadori M.I., Sinani N., Sterne J.A.C., Stevanovikj M., Tacconelli E., Tikkinen K.A.O., Trelle S., Zaid H., Rottingen J.A., Swaminathan S. Repurposed Antiviral Drugs for Covid-19 - Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021;384(6):497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbosa J., Kaitis D., Freedman R., Le K., Lin X. Clinical Outcomes of Hydroxychloroquine in Hospitalized Patients with COVID-19: A Quasi-Randomized Compararative Study, In Review at. N. Engl. J. Med. 2020 [Google Scholar]
- 16.M.G.S. Borba, F.D.A. Val, V.S. Sampaio, M.A.A. Alexandre, amp, uacutejo, G.C. Melo, M. Brito, Mour, amp, M.P.G. atildeo, J. Brito Sousa, amp, e. Diego, D.C. Baia-da-Silva, M.V.F. Guerra, L.A. Hajjar, R.C. Pinto, A.A.S. Balieiro, F.G. Naveca, M.S. Xavier, A. Salomão, A.M. Siqueira, A. Schwarzbolt, J.H.R. Croda, M.L. Nogueira, G.A.S. Romero, Q. Bassat, C.J. Fontes, B.C. Albuquerque, C.T. Daniel-Ribeiro, W.M. Monteiro, M.V.G. Lacerda, Chloroquine diphosphate in two different dosages as adjunctive therapy of hospitalized patients with severe respiratory syndrome in the context of coronavirus (SARS-CoV-2) infection: Preliminary safety results of a randomized, double-blinded, phase IIb clinical trial (CloroCovid-19 Study), medRxiv (2020) 2020.04.07.20056424.
- 17.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Huettenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., Garcia-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S. COVID-19: A drug repurposing and biomarker identification by using comprehensive gene-disease associations through protein-protein interaction network analysis. Preprints. 2020 [Google Scholar]
- 19.M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Kruger, T. Herrler, S. Erichsen, T.S. Schiergens, G. Herrler, N.H. Wu, A. Nitsche, M.A. Muller, C. Drosten, S. Pohlmann, SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell 181(2) (2020) 271-280 e8. [DOI] [PMC free article] [PubMed]
- 20.Q. Wang, Y. Zhang, L. Wu, S. Niu, C. Song, Z. Zhang, G. Lu, C. Qiao, Y. Hu, K.Y. Yuen, Q. Wang, H. Zhou, J. Yan, J. Qi, Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2, Cell 181(4) (2020) 894-904 e9. [DOI] [PMC free article] [PubMed]
- 21.UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., Mering C.V. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Y., Li M., Wang J., Pan Y., Wu F.X. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi: 10.1016/j.biosystems.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 25.Frohlich H., Speer N., Poustka A., Beissbarth T. GOSim–an R-package for computation of information theoretic GO similarities between terms and gene products. BMC Bioinf. 2007;8:166. doi: 10.1186/1471-2105-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.P. Resnik, Using Information Content to Evaluate Semantic Similarity in a Taxonomy, Proceedings of the 14th International Joint Conference on Artificial Intelligence, Montreal 1 (1995) 448-453.
- 27.Batool K., Niazi M.A. Towards a methodology for validation of centrality measures in complex networks. PLoS ONE. 2014;9(4) doi: 10.1371/journal.pone.0090283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., McDermott M.G., Monteiro C.D., Gundersen G.W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn M., von Mering C., Campillos M., Jensen L.J., Bork P., Stitch, interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36(Database issue):D684–D688. doi: 10.1093/nar/gkm795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng F., Lu W., Liu C., Fang J., Hou Y., Handy D.E., Wang R., Zhao Y., Yang Y., Huang J., Hill D.E., Vidal M., Eng C., Loscalzo J. A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat. Commun. 2019;10(1):3476. doi: 10.1038/s41467-019-10744-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stolfi P., Manni L., Soligo M., Vergni D., Tieri P. Designing a Network Proximity-Based Drug Repurposing Strategy for COVID-19. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.545089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., Huuskonen S., Laitinen T., Redchuk T., Bogacheva M., Salokas K., Pohner I., Ohman T., Tonduru A.K., Hassinen A., Gawriyski L., Keskitalo S., Vartiainen M.K., Pietiainen V., Poso A., Varjosalo M. SARS-CoV-2-host proteome interactions for antiviral drug discovery. Mol. Syst. Biol. 2021;17(11) doi: 10.15252/msb.202110396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hikmet F., Mear L., Edvinsson A., Micke P., Uhlen M., Lindskog C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020;16(7) doi: 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369(6508):1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozgur A., Vu T., Erkan G., Radev D.R. Identifying gene-disease associations using centrality on a literature mined gene-interaction network. Bioinformatics. 2008;24(13):i277–i285. doi: 10.1093/bioinformatics/btn182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pavlopoulos G.A., Hooper S.D., Sifrim A., Schneider R., Aerts J. Medusa: A tool for exploring and clustering biological networks. BMC research notes. 2011;4:384. doi: 10.1186/1756-0500-4-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavlopoulos G.A., Secrier M., Moschopoulos C.N., Soldatos T.G., Kossida S., Aerts J., Schneider R., Bagos P.G. Using graph theory to analyze biological networks. BioData mining. 2011;4:10. doi: 10.1186/1756-0381-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barman R.K., Mukhopadhyay A., Maulik U., Das S. Identification of critical host targets for HCV infection: a systems biology approach. Trans. Indian Natl. Acad. Eng. 2021;6(3):755–763. [Google Scholar]
- 39.Barman R.K., Sen S., Mukhopadhyay A., Maulik U., Das S. 2020 International Conference on Computer, Electrical & Communication Engineering (ICCECE) 2020. System biology approach to identify critical host genes for dengue infection; pp. 1–5. [Google Scholar]
- 40.Soberg K., Skalhegg B.S. The molecular basis for specificity at the level of the protein kinase a catalytic subunit. Front. Endocrinol. (Lausanne) 2018;9:538. doi: 10.3389/fendo.2018.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skalhegg B.S., Funderud A., Henanger H.H., Hafte T.T., Larsen A.C., Kvissel A.K., Eikvar S., Orstavik S. Protein kinase A (PKA)–a potential target for therapeutic intervention of dysfunctional immune cells. Curr. Drug Targets. 2005;6(6):655–664. doi: 10.2174/1389450054863644. [DOI] [PubMed] [Google Scholar]
- 42.Cheng F., Ramos da Silva S., Huang I.C., Jung J.U., Gao S.J. Suppression of Zika Virus Infection and Replication in Endothelial Cells and Astrocytes by PKA Inhibitor PKI 14–22. J. Virol. 2018;92(4) doi: 10.1128/JVI.02019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.X. E, P. Meraner, P. Lu, J.M. Perreira, A.M. Aker, W.M. McDougall, R. Zhuge, G.C. Chan, R.M. Gerstein, P. Caposio, A.D. Yurochko, A.L. Brass, T.F. Kowalik, OR14I1 is a receptor for the human cytomegalovirus pentameric complex and defines viral epithelial cell tropism, Proc Natl Acad Sci U S A 116(14) (2019) 7043-7052. [DOI] [PMC free article] [PubMed]
- 44.Riva L., Song O.R., Prentoe J., Helle F., L'Homme L., Gattolliat C.H., Vandeputte A., Feneant L., Belouzard S., Baumert T.F., Asselah T., Bukh J., Brodin P., Cocquerel L., Rouille Y., Dubuisson J. Identification of Piperazinylbenzenesulfonamides as New Inhibitors of Claudin-1 Trafficking and Hepatitis C Virus Entry. J. Virol. 2018;92(10) doi: 10.1128/JVI.01982-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh M., Singh P., Vaira D., Torheim E.A., Rahmouni S., Tasken K., Moutschen M. The RIAD peptidomimetic inhibits HIV-1 replication in humanized NSG mice. Eur. J. Clin. Invest. 2014;44(2):146–152. doi: 10.1111/eci.12200. [DOI] [PubMed] [Google Scholar]
- 46.Adeoye A.O., Oso B.J., Olaoye I.F., Tijjani H., Adebayo A.I. Repurposing of chloroquine and some clinically approved antiviral drugs as effective therapeutics to prevent cellular entry and replication of coronavirus. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1765876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B.G., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58(8):4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gozal E., Metz C.J., Dematteis M., Sachleben L.R., Jr., Schurr A., Rane M.J. PKA activity exacerbates hypoxia-induced ROS formation and hypoxic injury in PC-12 cells. Toxicol. Lett. 2017;279:107–114. doi: 10.1016/j.toxlet.2017.07.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bullen J.W., Tchernyshyov I., Holewinski R.J., DeVine L., Wu F., Venkatraman V., Kass D.L., Cole R.N., Van Eyk J., Semenza G.L. Protein kinase A-dependent phosphorylation stimulates the transcriptional activity of hypoxia-inducible factor 1. Sci Signal. 2016;9(430):ra56. doi: 10.1126/scisignal.aaf0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rueda C.M., Jackson C.M., Chougnet C.A. Regulatory T-Cell-Mediated Suppression of Conventional T-Cells and Dendritic Cells by Different cAMP Intracellular Pathways. Front. Immunol. 2016;7:216. doi: 10.3389/fimmu.2016.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiao J., Huang F., Lum H. PKA inhibits RhoA activation: a protection mechanism against endothelial barrier dysfunction. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003;284(6):L972–L980. doi: 10.1152/ajplung.00429.2002. [DOI] [PubMed] [Google Scholar]
- 52.Maron M.B., Folkesson H.G., Stader S.M., Walro J.M. PKA delivery to the distal lung air spaces increases alveolar liquid clearance after isoproterenol-induced alveolar epithelial PKA desensitization. Am. J. Physiol. Lung Cell. Mol. Physiol. 2005;289(2):L349–L354. doi: 10.1152/ajplung.00134.2004. [DOI] [PubMed] [Google Scholar]
- 53.Brazzoli M., Bianchi A., Filippini S., Weiner A., Zhu Q., Pizza M., Crotta S. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J. Virol. 2008;82(17):8316–8329. doi: 10.1128/JVI.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Husain M., Harrod K.S. Enhanced acetylation of alpha-tubulin in influenza A virus infected epithelial cells. FEBS Lett. 2011;585(1):128–132. doi: 10.1016/j.febslet.2010.11.023. [DOI] [PubMed] [Google Scholar]
- 55.Jiang W., Wang Q., Chen S., Gao S., Song L., Liu P., Huang W. Influenza A virus NS1 induces G0/G1 cell cycle arrest by inhibiting the expression and activity of RhoA protein. J. Virol. 2013;87(6):3039–3052. doi: 10.1128/JVI.03176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mercer J., Schelhaas M., Helenius A. Virus entry by endocytosis. Annu. Rev. Biochem. 2010;79:803–833. doi: 10.1146/annurev-biochem-060208-104626. [DOI] [PubMed] [Google Scholar]
- 57.Seccia T.M., Rigato M., Ravarotto V., Calo L.A. ROCK (RhoA/Rho Kinase) in Cardiovascular-Renal Pathophysiology: A Review of New Advancements. J Clin Med. 2020;9(5) doi: 10.3390/jcm9051328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abedi F., Rezaee R., Karimi G. Plausibility of therapeutic effects of Rho kinase inhibitors against Severe Acute Respiratory Syndrome Coronavirus 2 (COVID-19) Pharmacol. Res. 2020;156 doi: 10.1016/j.phrs.2020.104808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calo L.A., Bertoldi G., Davis P.A. Rho kinase inhibitors for SARS-CoV-2 induced acute respiratory distress syndrome: Support from Bartter's and Gitelman's syndrome patients. Pharmacol. Res. 2020;158 doi: 10.1016/j.phrs.2020.104903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma S., Sicinski P. A kinase of many talents: non-neuronal functions of CDK5 in development and disease. Open Biol. 2020;10(1) doi: 10.1098/rsob.190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weisberg E., Parent A., Yang P.L., Sattler M., Liu Q., Liu Q., Wang J., Meng C., Buhrlage S.J., Gray N., Griffin J.D. Repurposing of Kinase Inhibitors for Treatment of COVID-19. Pharm. Res. 2020;37(9):167. doi: 10.1007/s11095-020-02851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shupp A., Casimiro M.C., Pestell R.G. Biological functions of CDK5 and potential CDK5 targeted clinical treatments. Oncotarget. 2017;8(10):17373–17382. doi: 10.18632/oncotarget.14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Na Y.R., Jung D., Gu G.J., Jang A.R., Suh Y.H., Seok S.H. The early synthesis of p35 and activation of CDK5 in LPS-stimulated macrophages suppresses interleukin-10 production. Sci. Signal. 2015;8(404):ra121. doi: 10.1126/scisignal.aab3156. [DOI] [PubMed] [Google Scholar]
- 65.Fadel R., Morrison A.R., Vahia A., Smith Z.R., Chaudhry Z., Bhargava P., Miller J., Kenney R.M., Alangaden G., Ramesh M.S., Force H.-F.-C.-M.-T. Early Short Course Corticosteroids in Hospitalized Patients with COVID-19. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pfander P., Fidan M., Burret U., Lipinski L., Vettorazzi S. Cdk5 Deletion Enhances the Anti-inflammatory Potential of GC-Mediated GR Activation During Inflammation. Front. Immunol. 2019;10:1554. doi: 10.3389/fimmu.2019.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D.m. s., Guanosine as RNA Replication Blocker for Therapeutic Treatment of COVID 19, figshare. Journal contribution. 2020 [Google Scholar]
- 68.Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020;38(4):379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- 69.McCreary E.K., Pogue J.M. Coronavirus Disease 2019 Treatment: A Review of Early and Emerging Options, Open Forum. Infect Dis. 2020;7(4):ofaa105. doi: 10.1093/ofid/ofaa105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sheahan T.P., Sims A.C., Leist S.R., Schafer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11(1):222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tchesnokov E.P., Feng J.Y., Porter D.P., Gotte M. Mechanism of Inhibition of Ebola Virus RNA-Dependent RNA Polymerase by Remdesivir. Viruses. 2019;11(4) doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buratto D., Saxena A., Ji Q., Yang G., Pantano S., Zonta F. Rapid Assessment of Binding Affinity of SARS-COV-2 Spike Protein to the Human Angiotensin-Converting Enzyme 2 Receptor and to Neutralizing Biomolecules Based on Computer Simulations. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.730099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodriguez-Rivas J., Croce G., Muscat M., Weigt M. Epistatic models predict mutable sites in SARS-CoV-2 proteins and epitopes. Proc Natl Acad Sci U S A. 2022;119(4) doi: 10.1073/pnas.2113118119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.