Abstract
CDKL5 Deficiency Disorder (CDD) is a severe encephalopathy characterized by intractable epilepsy, infantile spasms, and cognitive disabilities. The detrimental CNS manifestations and lack of therapeutic interventions represent unmet needs, necessitating identification of CDD-dependent phenotypes for in vitro disease modeling and therapeutic testing. Here, we optimized a high-content assay to quantify cilia in CDKL5-deficient neurons. Our work shows that Cdkl5-knockdown neurons have elongated cilia and uncovers cilium lengthening in hippocampi of Cdkl5 knockout mice. Collectively, our findings identify cilia length alterations under CDKL5 activity loss in vitro and in vivo and reveal elongated cilia as a robust functional phenotype for CDD.
Keywords: CDKL5, Neurodevelopmental disorder, Seizure, Cilia, Autism
CDKL5 Deficiency Disorder (CDD) is an X-linked syndrome caused by mutations in the cyclin-dependent kinase-like 5 (CDKL5) gene and is characterized by severe neurological symptoms such as early-onset of infantile spasms, intellectual disability, intractable epilepsy and hypotonia (Olson et al., 2019). CDKL5 encodes a serine threonine kinase, which is highly expressed and developmentally regulated in the brain, reaching the highest expression in the early postnatal stages (Rusconi et al., 2008). The functional relevance for CDKL5 kinase activity in CDD is underscored by the fact that most of the missense mutations found in CDD patients are concentrated in the catalytic domain (Hector et al., 2017). Studies in rodents have shown that loss of neuronal CDKL5 in the central nervous system (CNS) results in altered dendritic arborization, axonal outgrowth and synaptic plasticity, suggesting a role in neuronal maturation and function (Rusconi et al., 2008). Given its neurodevelopmental regulation and activity-dependent expression, CDKL5 has emerged as a key player in brain plasticity and neurocognitive function (Kilstrup-Nielsen et al., 2012). Accordingly, Cdkl5 mutant mice have impairments in hippocampal-dependent functions, autistic-like behaviors and spontaneous seizures (Wang et al., 2012; Amendola et al., 2014; Wang et al., 2021).
Although little is known regarding the downstream molecular mechanisms affected by CDKL5 gene loss, previous work has indicated an association between CDKL5 and primary cilia (hereafter cilia): i) the homologue of the human CDKL5 gene in Chlamydomonas, long-flagella (LF) gene LF5, encodes a kinase that regulates ciliary length (Tam et al., 2013); ii) in dividing cells CDKL5 localizes to the centrosome, and its loss is associated with defective centrosome functions (Barbiero et al., 2017); iii) the human CDKL5 kinase localizes to cilia and C. elegans mutants expressing disease-linked mutations showed elongated ciliary length (Canning et al., 2018); iv) the C. elegans CDKL-1 is required for the assembly of the proximal and distal segment of cilia [in press: Park et al., Curr Biology (2021)].
Cilia are evolutionarily conserved, microtubule-based membrane extensions protruding from the surface of cells that coordinate extracellular ligand-based signaling and cellular polarity (Lee and Gleeson, 2011). Most ciliopathies are characterized by cilia shortening, resulting in defective ciliary signaling and aberrant neuronal development (Park et al., 2019). Furthermore, altered cilia gene expression has been found as a convergent risk factor for neurological and psychiatric disorders (Migliavacca et al., 2015; Di Nardo et al., 2020a). Cilia lengthening has recently also been observed in developmental disorders associated with severe epilepsy (Wang et al., 2020; Srivastava et al., 2017). Although CDD is not currently classified as a ciliopathy, a contribution of cilial dysfunction to the phenotype is likely. However, the relationship between CDKL5 and cilia in mammalian neurons has not been investigated.
Here, we used a rat hippocampal primary neuronal culture system to test the hypothesis that Cdkl5-knockdown affects cilia. We found that Cdkl5-knockdown neurons have longer cilia and consistent with our in vitro model, elongated cilia were also identified in vivo in Cdkl5 KO mouse brains. Loss of Cdkl5 in rat hippocampal neurons was associated with oxidative stress response and alteration in the Akt/mTOR signaling. Together, our work uncovers that in vitro and in vivo rodent models of CDD have elongated primary cilia and establishes a high-throughput cilia length-based platform that could be used in functional assays for drug discovery and therapeutic testing.
All procedures and animal care were approved by Boston Children’s Hospital Institutional Animal Care and Use Committee (IACUC). Animals were raised and bred in house on a 12 h light/dark cycle with food and water ad libitum. To generate experimental animals, heterozygous CDKL5+/− female mice (Wang et al., 2012; JAX#021967) were mated with C57BL/6 J males. Controls were sex- and age-matched littermates. Rat hippocampal neurons were cultured as previously described (Di Nardo et al., 2020a, Di Nardo et al., 2020b). Lentiviral stocks were prepared as previously described (Di Nardo et al., 2020a). Control shRNA construct against the luciferase gene (ctrl-sh) was previously described (Di Nardo et al., 2009). Cdkl5-sh RNA was from Sigma cat. No.: TRCN0000023097.
Mice were transcardially perfused with saline, followed by phosphate-buffered 4 % paraformaldehyde solution (PFA). Brains were dissected, post-fixed in PFA overnight at 4 °C, cryoprotected in an ascending series of sucrose solutions (10, 20 and 30 %) and frozen in O. C.T. Tissue-Tek (VWR). Coronal slices (40um thick) through the forebrain were sectioned with a cryostat (Leica Microsystems). Brain sections were washed 4 times with Tris Buffered Saline pH 7.4 (TBS), mounted on superfrost slides and dried overnight. The next day, sections were incubated for 2 h at room temperature in blocking buffer (5 %BSA, 0.1 % Triton X-100, 10 % goat serum). Primary antibodies were incubated in 1 % BSA, 0.1 % Triton X-100, at 4 °C for overnight. The day after, sections were washed in TBS buffer followed by incubation with fluorochrome-coupled secondary antibody. Imaging of the Cdkl5 control and mutant hippocampi was performed by imaging 6–9 random regions in the CA1 of the hippocampus. The average percentage of neurons with cilia (NeuN+/ACIII+) was calculated for each of the images. Cilia length measurements were performed by tracing the ACIII stained cilia using the ImageJ Software freehand tool. A threshold for cilia count was set such that only ACIII positive objects that measured longer than 1μm were counted as cilia. All the imaging and the quantification were done in a blinded way. Confocal images were acquired with a Nikon Ultraview Vox Spinning Disk Confocal microscope using 63x oil-immersion objective equipped with Hamamatsu camera.
Protein extracts were prepared as previously described (Di Nardo et al., 2020a). The following antibodies were used: Cdkl5 (Sigma, cat. HPA002847), Cdkl5 (Proteintech cat. 12973-1-AP), p-EB2 S222 (Covalab, cat. pab0132-P), EB2 (Abcam cat. ab4576), GAPDH (Ambion cat. AM4300), β-actin (Cell Signaling, cat. 3700S), pS6 (Cell Signaling cat. 5364), S6 (Santa Cruz cat. sc-74459), p-Akt Ser473 (Cell Signaling cat. 4060), Akt (Cell Signaling cat. 4691), p-GSK Ser9 (Cell Signaling cat. 5558), GSK (Cell Signaling cat. 2456), HO-1 (Proteintech cat. 10701-1-AP), ACIII (Proteintech cat. 19492-1-AP), Arl13b (Proteintech cat. CL488-17711), NeuN (Millipore cat. MAB377), Acetylated tubulin (Sigma cat. T7451), β-Tubulin III (Sigma cat. T8660), γ–tubulin (Millipore cat. MAB 1864), GFP (Thermo Fisher Scientific cat. A10262), GLI1 (Abcam cat. ab273018), Smoothened (Invitrogen cat. PA5-113312), active (non-phospho) β-Catenin (Ser45) (Cell Signaling cat. 19807), Dvl2 (Cell Signaling cat. 3224S). Western blot quantifications were performed by protein normalization using loading controls. Level of phosphorylated proteins was expressed as the ratio of phosphorylated/total level after loading control normalization. GraphPad PRISM was used for statistical quantifications. Significant different values were considered p < 0.05.
Imaging was performed using the ImageXpress Micro Confocal platform (IXM-C from Molecular Devices) available at the Human Neuron Core of the Translational Neuroscience Center (Boston Children’s Hospital). The DAPI channel was used for nuclei identification. When needed, focal planes were adjusted to the best optimal resolution for each channel. Once optimized, Z offsets were kept the same throughout the scans. Imaging of the ciliaHCA was done with a 40x objective on forty-nine fields of view per well (18 % of the well). The nuclei were detected by Hoechst staining at 386 nm emission for 50 ms, the LV-transduced neurons were detected using GFP staining at 485 nm emission for 25 ms, and cilia were detected using ACIII staining at 647 emission for 50 ms or using Arl13b staining at 647 emission for 30 ms using a stack of images at three different focal planes for optimal cilia imaging with a step size of 1 μm/step. Object selection for GFP and ACIII spots identification were filtered using area and shape measurements. Data analysis was done using the MetaExpress Software. After background removal, the cilia were detected using the area measurement as a filter for object identification. A subsequent mask was used to filter the cilia based on the identification of the objects with a minimum length of 1 μm. To identify the transduced neurons, we generated a mask using the GFP staining and used a co-localization module to identify the length of the cilia in the cell bodies of the GFP+ neurons. A threshold for cilia length was set such that only positive objects longer than 1μm were counted as cilia. Additional cilia length cut-offs were set at 2, 3 and 4μm.
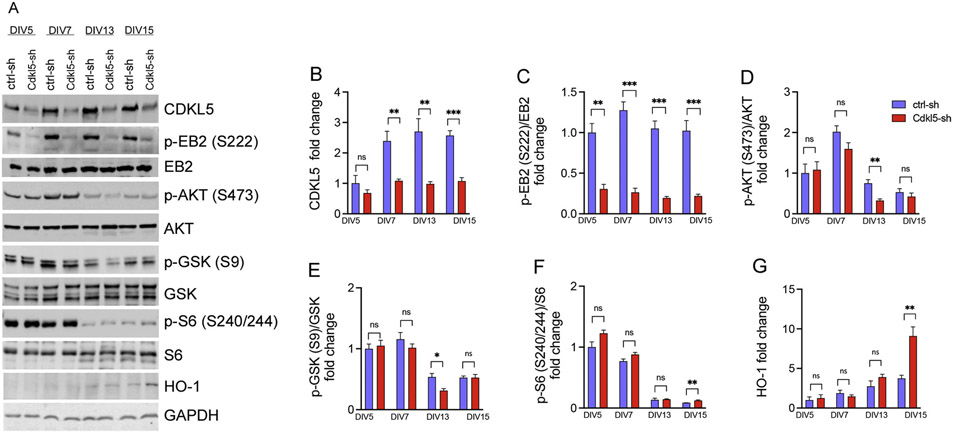
As a CDD in vitro model, we used rat hippocampal neurons transduced with GFP tagged lentiviral vectors (LV) expressing a short hairpin RNA (shRNA) directed against either the Cdkl5 (Cdkl5-sh) or the luciferase gene as control (ctrl-sh). To determine the time course of effects of Cdkl5 gene silencing, we measured protein levels of CDKL5, and its major phosphorylated substrates. Neurons were transduced with ctrl-sh and Cdkl5-sh at day in vitro (DIV) DIV1, and protein knock-down efficiency was assessed at DIV5, DIV7, DIV13 and DIV15 by western blot analysis. We found that seven days in vitro are sufficient to significantly knockdown CDKL5 protein expression (Fig. 1A-B). Notably, phosphorylation of the bona fide CDKL5 substrate EB2 at Ser222 (Baltussen et al., 2018; Munoz et al., 2018) was significantly reduced at all time points we investigated (Fig. 1A, C). Previous work indicated that loss of CDKL5 is associated with a reduction in survival, proliferation and altered mTORC1 signaling cascade (Amendola et al., 2014; Wang et al., 2012). Consistent with these findings, we found that Cdkl5-sh knockdown neurons have a significant reduction in AKT activity, which was associated with reduced AKT-dependent inhibitory phosphorylation of its downstream target GSK at Ser9 but only at DIV13 (Fig. 1A, D-E). Loss of Cdkl5 activity was also associated with a small increase in the phosphorylation of the mTORC1 target S6 at DIV15 but did not affect mTOR activation at other timepoints. Importantly, expression of the heme oxygenase, indicating presence of oxidative stress, was also markedly increased at DIV15 (Abrous et al., 2005) (Fig. 1A, F-G).
Fig. 1. Time course of rat Cdkl5 knockdown in hippocampal neurons.
(A) Representative western blot of protein lysates from rat hippocampal neurons transduced with ctrl-sh and Cdkl5-sh lentiviral vectors in a time course experiment at DIV5, 7, 13 and 15. Quantification of CDKL5 (B), p-EB2 (C), p-AKT (D), p-GSK (E), pS6 (F), HO-1 (G). Data are average fold changes of ctrl-sh neurons. GAPDH was used as loading control for protein level normalization. Phospho-antibodies levels were quantified as the ratio to their respective total antibody after protein normalization to GAPDH. Quantifications are relative to ctrl-sh neurons (n = 4 unpaired Student’s t test, *p < 0.05). Error bars indicate ± SEM.
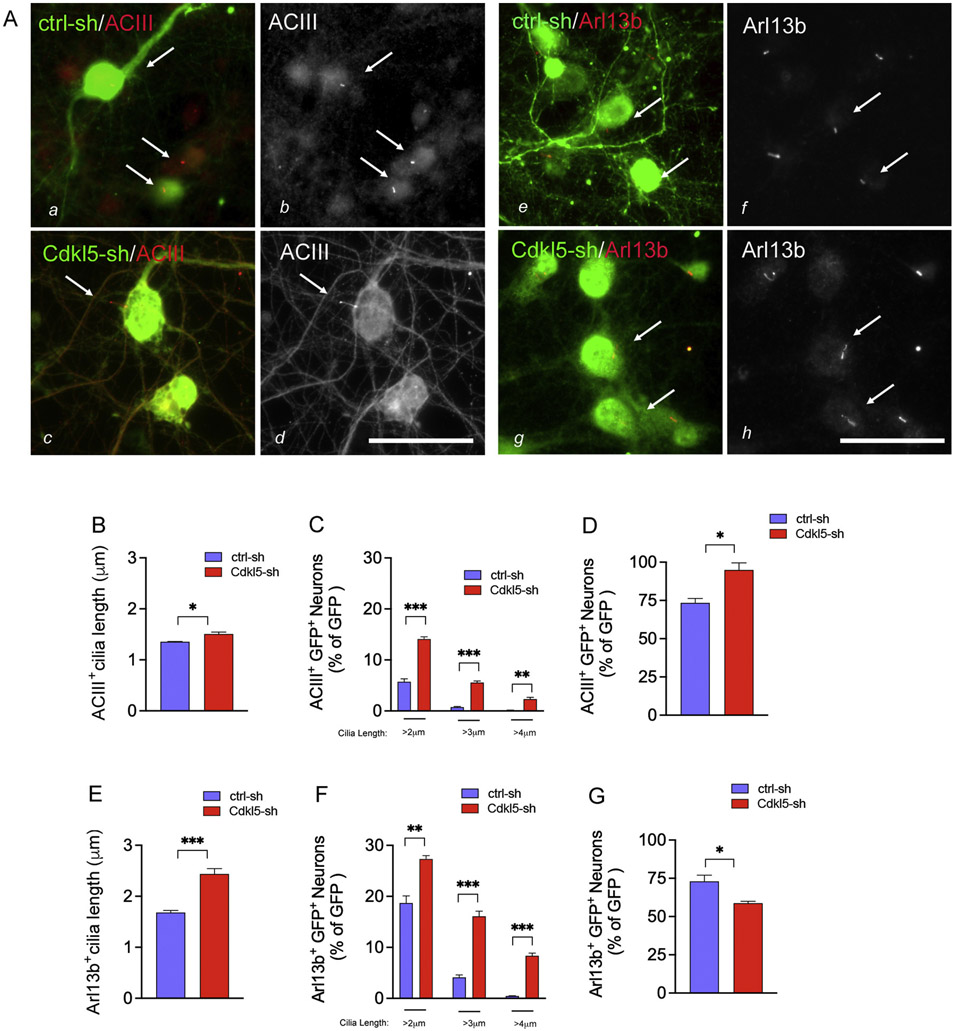
To assess functional consequences of Cdkl5 gene silencing, we imaged cilia in cultured neurons with an assay that we previously developed for unbiased cilia quantification (ciliaHCA) (Di Nardo et al., 2020a, b). We optimized ciliaHCA to measure cilia length (hereafter ciliaLHCA) using the ImageXpress Micro Confocal (XM-C) platform (Molecular Devices) at 13 days in culture (DIV13). Objects that measured longer than 1μm and were immunoreactive to either of the ciliary markers adenylyl cyclase III (ACIII) and the ADP Ribosylation Factor Like GTPase 13b (Arl13b) (Bishop et al., 2007; Chih et al., 2012) were quantified as cilia. Lentiviral transduced neurons were identified by GFP immunoreactivity (Fig. 2A). Compared to controls, Cdkl5-knockdown neurons displayed a significant increase in cilia length for both cilia markers (Fig. 2A-B, E). To increase assay sensitivity and identify optimal threshold for separation between controls and Cdkl5-sh neurons, we assessed cilia length at cut-offs ranging between 2–4 μm. Based on this classification, the most significant cilia length changes occurred with a cut-off between 2–3μm for the ACIII+ cilia and between 3–4μm for the Arl13b+ cilia (Fig. 2C, F). The percentage of transfected neurons in which cilia were identified by ACIII+ was increased, while those with Arl13b+ cilia were decreased with Cdkl5 loss (Fig. 2D, G). Given that ACIII is found more enriched in mature neurons compared to Arl13b, it is possible that these differences might reflect a CDKL5-dependent effect in modulating neuronal maturation (Sterpka and Chen, 2018).
Fig. 2. Loss of neuronal Cdkl5 activity results in elongated ACIII+ and Arl13b+ cilia.
(A) Representative immunofluorescence images of ACIII (a–d) and Arl13b cilia in ctrl-sh and Cdkl5-sh neurons (e–h) fixed at DIV13. Merged images show the GFP+ LV-infected neurons (green in a, c, e, g), the ACIII+ cilia (red in a, c; greyscale in b, d) and the Arl13b+ cilia (red in e, g; greyscale in f, h). White arrows indicate cilia. Bar is 50μm. Quantification of the average length of all cilia in the GFP+ neurons (ACIII+ cilia in B; Arl13b+ cilia in E. Percent of GFP + neurons with cilia above 2, 3 and 4 μm length (ACIII+ cilia in C; Arl13b+ cilia in F). Percent of GFP + neurons with cilia above 1 μm length. Data represent GFP+ ACIII+ (in D) and GFP+ Arl13b+ (in G) neurons as percent of GFP+ cells. Data represent n = 12–24 biological replica/experiment, n = 3 experiments/condition, error bars indicate ± SEM (unpaired t-test *p < 0.05, **p < 0.01, ***p < 0.0005).
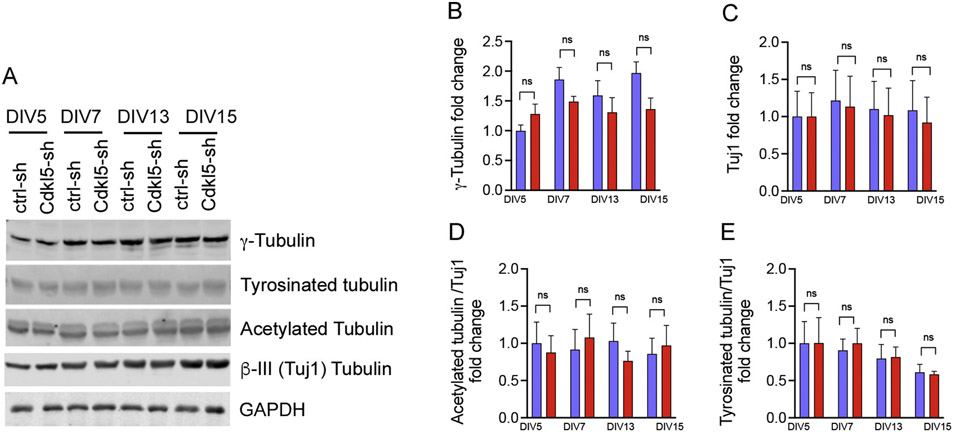
Cilia are microtubule-based organelles originating from the basal body, which is made of gamma tubulin (Werner et al., 2017). Since previous studies identified microtubule-associated proteins as major substrates for CDKL5 kinase (Baltussen et al., 2018), we asked whether there were changes in the levels of tubulin expression level and post-translational modifications. Notably, we found that the cilia length changes seen in the Cdkl5-sh neurons were not associated with alterations in the expression of gamma or beta III (neuronal-specific, Tuj1) tubulin or post-translational modifications such as tyrosination and acetylation (Fig. 3A-E). While the Cdkl5-knockdown neurons did not show overall changes in tubulin modification affecting microtubule stability and plasticity, there was a significant reduction in the phosphorylation of the Cdkl5 substrate, microtubule binding protein EB2 (Fig. 1A, C). However, we cannot exclude the possibility that other tubulin-related perturbations may contributing to the cilia phenotype, as suggested in previous work for other negative kinase regulators of ciliary length (Omori et al., 2010).
Fig. 3. Cdkl5 knockdown neurons display no change in tubulin expression and post-translational modification.
(A) Representative western blot of ctrl-sh and Cdkl5-sh lentiviral vectors in a time course experiment at DIV5, 7, 13 and 15. Quantification of γTubulin (n = 3, in (B), Tuj1 (n = 4, in C), Acetylated tubulin (n = 4, in D), and Tyrosinated tubulin (E n = 3), Data are average fold changes of ctrl-sh neurons. GAPDH was used as loading control for protein level normalization. Quantifications are relative to ctrl-sh neurons (unpaired Student’s ns = not significant, error bars indicate ± SEM).
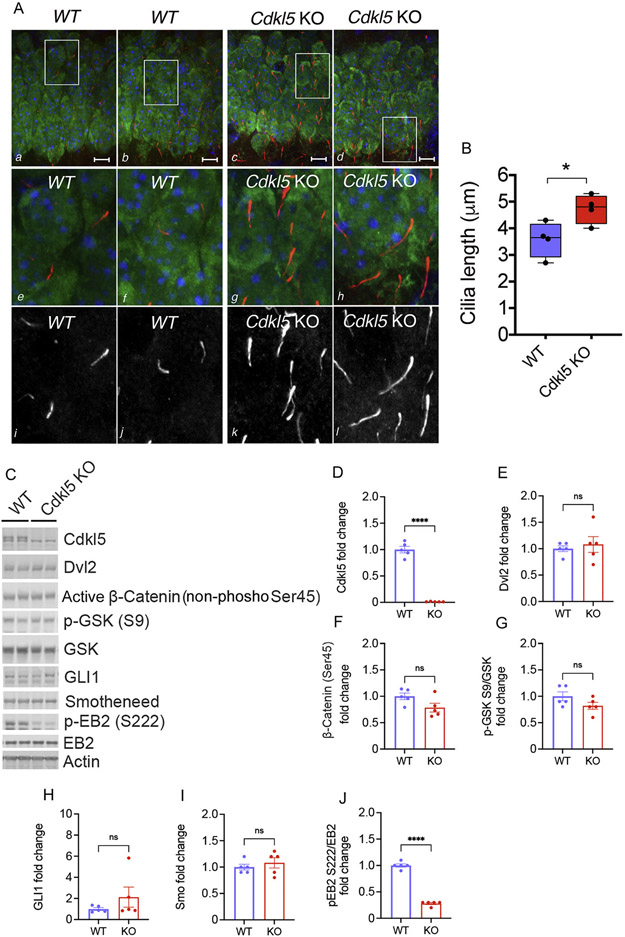
Cilia play a key role in vertebrate tissue homeostasis and function (Gerdes et al., 2009). Mutations in genes required for cilia assembly and/or function result in ciliopathies, which are heterogeneous pediatric disorders affecting several organs with detrimental neurological manifestations in the CNS (Ferkol and Leigh, 2012). To investigate whether changes in cilia length are present in vivo, we monitored ciliation in the Cdkl5 KO mice that have impairments in hippocampal-dependent functions and autistic-like behaviors (Wang et al., 2012). Immunohistochemistry was performed on hippocampal CA1 pyramidal neurons of control and Cdkl5 KO mice. Co-labeling with ACIII and NeuN was used to respectively identify cilia and neurons, nuclei were identified by Hoechst staining. Consistent with the in vitro findings, the Cdkl5 mutant mice showed longer cilia in the hippocampus, further supporting elongated cilia as a robust phenotype related to neuronal CDKL5 loss (Fig. 4A-B). Given the implications of cilia in intracellular signaling pathways, we investigated the effect of Cdkl5-dependent cilia lengthening on sonic hedgehog and Wnt signaling in brain lysates from Cdkl5 KO mice. Our data showed no significant changes in key components of the Wnt signaling pathway, including the Wnt intermediate Dishevelled (Dvl2), downstream target GSK, and active β-catenin (non-phosphorylated at Ser45) (Fig. 4C, E-G). We did not observe changes in components of the sonic hedgehog pathway, including the transcriptional activator GLI1 and G protein-coupled receptor smoothened (Smo) (Fig. 4C, H-I). Although these data show that Cdkl5-dependent cilia lengthening does not affect the expression of these factors, our results do not exclude alterations in their localization or responsiveness to these intracellular signaling mechanisms, as previously reported for other kinases also implicated in cilia-lengthening (Chaya et al., 2014). Future studies are necessary to identify the mechanistic links between CDKL5 loss, lengthening of cilia, and neuronal dysfunction leading to the neurological symptoms observed in CDD.
Fig. 4. Cdkl5 mice have increased cilia length but no changes in components of Wnt and sonic hedgehog signaling pathway.
A. Representative confocal images of the hippocampal CA1 region of WT and Cdkl5 KO mice at P15 stained with the cilia marker ACIII (in red a–h; in greyscale i–l), with the neuronal marker NeuN (in green a–h) and with the nuclei marker Hoechst (in blue a–h). White boxes in a–d represent the regions for zoom in e–l. Scale bar is 10μm. B. Cilia Length quantification. Data are average cilia length (n = 500 average cilia /mouse *p < 0.05 n=4 mice/genotype). C. Representative western blot of protein lysates from brains of control and Cdkl5 KO mice. Quantification of Dvl2 (E n = 5), active β-catenin (non-phosphorylated at S45) (F n = 5), p-GSK (G n = 5), GLI1 (H n = 5), Smo (I n = 5). Loss of Cdkl5 expression and function is shown by reduced Cdkl5 protein level (D n = 5) and EB2 phosphorylation (J n = 5). β-actin was used as loading control for protein level normalization. Phospho-antibodies levels were quantified as the ratio to their respective total antibody after protein normalization to β-actin. Quantifications are relative to ctrl-sh neurons (n = 5 unpaired Student’s t test, ****p < 0.00001, ns = not significant). Error bars indicate ± SEM.
Future studies addressing the interplay between cilia assembly and neuronal activity will help elucidate whether defective cilia length might underlie some of the neuronal alterations seen in these epilepsy disorders. High-throughput functional assays represent powerful preclinical approaches to monitor efficacy of drug screens and gene therapy. The cilia-length based high content assay we have developed represents a valuable platform for unbiased and systematic drug testing in preclinical models of CDD. Finally, given the role of cilia in neuronal function, the establishment of high-content cilia length-based assay in Cdkl5-deficient neurons might also have broader implications in understanding the role of elongated cilia in the neuronal phenotypes associated with CDD.
Acknowledgements
The authors thank the Assistant Director of the Screening Core Facility at the Human Neuron Core of the Translational Neuroscience Center (Boston Children’s Hospital) Lee Barrett for conceptual guidance and methodology support for the cilia high-content imaging. The authors thank Drs. Kellen Winden and Nickesha Anderson for constructive discussions on the project. This study was supported by the Loulou Foundation, Orphan Disease Center, administered by the University of Pennsylvania’s Perelman School of Medicine. M.S. is supported by grant funding from the National Institutes of Health (NIH) (R01NS113591).
Footnotes
Declaration of Competing Interest
Mustafa Sahin reports grant support from Novartis, Biogen, Astellas, Aeovian, Bridgebio, and Aucta. He has served on Scientific Advisory Boards for Novartis, Roche, Regenxbio, and Alkermes.
References
- Abrous DN, Koehl M, Le Moal M, 2005. Adult neurogenesis: from precursors to network and physiology. Physiol. Rev 85, 523–569. [DOI] [PubMed] [Google Scholar]
- Amendola E, Zhan Y, Mattucci C, Castroflorio E, Calcagno E, Fuchs C, Lonetti G, Silingardi D, Vyssotski AL, Farley D, Ciani E, Pizzorusso T, Giustetto M, Gross CT, 2014. Mapping pathological phenotypes in a mouse model of CDKL5 disorder. PLoS One 9, e91613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltussen LL, Negraes PD, Silvestre M, Claxton S, Moeskops M, Christodoulou E, Flynn HR, Snijders AP, Muotri AR, Ultanir SK, 2018. Chemical genetic identification of CDKL5 substrates reveals its role in neuronal microtubule dynamics. EMBO J. 37 (24), e99763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbiero I, Valente D, Chandola C, Magi F, Bergo A, Monteonofrio L, Tramarin M, Fazzari M, Soddu S, Landsberger N, Rinaldo C, Kilstrup-Nielsen C, 2017. CDKL5 localizes at the centrosome and midbody and is required for faithful cell division. Sci. Rep 7, 6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop GA, Berbari NF, Lewis J, Mykytyn K, 2007. Type III adenylyl cyclase localizes to primary cilia throughout the adult mouse brain. J. Comp. Neurol 505, 562–571. [DOI] [PubMed] [Google Scholar]
- Canning P, Park K, Goncalves J, Li C, Howard CJ, Sharpe TD, Holt LJ, Pelletier L, Bullock AN, Leroux MR, 2018. CDKL family kinases have evolved distinct structural features and ciliary function. Cell Rep. 22, 885–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaya T, Omori Y, Kuwahara R, Furukawa T, 2014. ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J. 33, 1227–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Liu P, Chinn Y, Chalouni C, Komuves LG, Hass PE, Sandoval W, Peterson AS, 2012. A ciliopathy complex at the transition zone protects the cilia as a privileged membrane domain. Nat. Cell Biol 14, 61–72. [DOI] [PubMed] [Google Scholar]
- Di Nardo A, Kramvis I, Cho N, Sadowski A, Meikle L, Kwiatkowski DJ, Sahin M, 2009. Tuberous sclerosis complex activity is required to control neuronal stress responses in an mTOR-dependent manner. J. Neurosci 29, 5926–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A, Lenoel I, Winden KD, Ruhmkorf A, Modi ME, Barrett L, Ercan-Herbst E, Venugopal P, Behne R, Lopes CAM, Kleiman RJ, Bettencourt-Dias M, Sahin M, 2020a. Phenotypic screen with TSC-deficient neurons reveals heat-shock machinery as a druggable pathway for mTORC1 and reduced cilia. Cell Rep. 31, 107780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A, Vasquez S, Sahin M, 2020b. A cell-based assay optimized for high-content cilia imaging with primary rat hippocampal neurons. STAR Protoc. 1, 100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferkol TW, Leigh MW, 2012. Ciliopathies: the central role of cilia in a spectrum of pediatric disorders. J. Pediatr 160, 366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N, 2009. The vertebrate primary cilium in development, homeostasis, and disease. Cell 137, 32–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RD, Kalscheuer VM, Hennig F, Leonard H, Downs J, Clarke A, Benke TA, Armstrong J, Pineda M, Bailey MES, Cobb SR, 2017. CDKL5 variants: improving our understanding of a rare neurologic disorder. Neurol. Genet 3, e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilstrup-Nielsen C, Rusconi L, La Montanara P, Ciceri D, Bergo A, Bedogni F, Landsberger N, 2012. What we know and would like to know about CDKL5 and its involvement in epileptic encephalopathy. Neural Plast. 2012, 728267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Gleeson JG, 2011. Cilia in the nervous system: linking cilia function and neurodevelopmental disorders. Curr. Opin. Neurol 24, 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliavacca E, Golzio C, Mannik K, Blumenthal I, Oh EC, Harewood L, Kosmicki JA, Loviglio MN, Giannuzzi G, Hippolyte L, Maillard AM, Alfaiz AA, Van Haelst MM, Andrieux J, Gusella JF, Daly MJ, Beckmann JS, Jacquemont S, Talkowski ME, Katsanis N, Reymond A, 2015. A potential contributory role for ciliary dysfunction in the 16p11.2 600 kb BP4-BP5 pathology. Am. J. Hum. Genet 96, 784–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz IM, Morgan ME, Peltier J, Weiland F, Gregorczyk M, Brown FC, Macartney T, Toth R, Trost M, Rouse J, 2018. Phosphoproteomic screening identifies physiological substrates of the CDKL5 kinase. EMBO J. 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson HE, Demarest ST, Pestana-Knight EM, Swanson LC, Iqbal S, Lal D, Leonard H, Cross JH, Devinsky O, Benke TA, 2019. Cyclin-dependent kinase-like 5 deficiency disorder: clinical review. Pediatr. Neurol 97, 18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori Y, Chaya T, Katoh K, Kajimura N, Sato S, Muraoka K, Ueno S, Koyasu T, Kondo M, Furukawa T, 2010. Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc. Natl. Acad. Sci. U. S. A 107, 22671–22676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Jang HJ, Lee JH, 2019. Roles of primary cilia in the developing brain. Front. Cell. Neurosci 13, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi L, Salvatoni L, Giudici L, Bertani I, Kilstrup-Nielsen C, Broccoli V, Landsberger N, 2008. CDKL5 expression is modulated during neuronal development and its subcellular distribution is tightly regulated by the C-terminal tail. J. Biol. Chem 283, 30101–30111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Ramsbottom SA, Molinari E, Alkanderi S, Filby A, White K, Henry C, Saunier S, Miles CG, Sayer JA, 2017. A human patient-derived cellular model of Joubert syndrome reveals ciliary defects which can be rescued with targeted therapies. Hum. Mol. Genet 26, 4657–4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterpka A, Chen X, 2018. Neuronal and astrocytic primary cilia in the mature brain. Pharmacol. Res 137, 114–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam LW, Ranum PT, Lefebvre PA, 2013. CDKL5 regulates flagellar length and localizes to the base of the flagella in Chlamydomonas. Mol. Biol. Cell 24, 588–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IT, Allen M, Goffin D, Zhu X, Fairless AH, Brodkin ES, Siegel SJ, Marsh ED, Blendy JA, Zhou Z, 2012. Loss of CDKL5 disrupts kinome profile and event-related potentials leading to autistic-like phenotypes in mice. Proc. Natl. Acad. Sci. U. S. A 109, 21516–21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang EJ, Gailey CD, Brautigan DL, Fu Z, 2020. Functional alterations in ciliogenesis-associated kinase 1 (CILK1) that result from mutations linked to juvenile myoclonic epilepsy. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HT, Zhu ZA, Li YY, Lou SS, Yang G, Feng X, Xu W, Huang ZL, Cheng X, Xiong ZQ, 2021. CDKL5 deficiency in forebrain glutamatergic neurons results in recurrent spontaneous seizures. Epilepsia 62, 517–528. [DOI] [PubMed] [Google Scholar]
- Werner S, Pimenta-Marques A, Bettencourt-Dias M, 2017. Maintaining centrosomes and cilia. J. Cell. Sci 130, 3789–3800. [DOI] [PubMed] [Google Scholar]