Abstract
The ponA gene of Mycobacterium smegmatis encodes a 95-kDa penicillin binding protein, PBP1, that is similar to PBP1s of Mycobacterium tuberculosis and Mycobacterium leprae. Transposon disruption of ponA in M. smegmatis resulted in a PBP1-deficient mutant that was sensitive to β-lactam antibiotics, was more permeable to glycine, and grew slowly in liquid culture.
Most bacteria have several penicillin binding proteins (PBPs) that catalyze the final steps of peptidoglycan synthesis. In Escherichia coli PBP1a (encoded by ponA) and PBP1b (encoded by ponB) are similar high-molecular-weight (HMW) class A PBPs possessing transglycosylase and transpeptidase activities. E. coli can tolerate inactivation of either ponA or ponB without any significant phenotypic changes, but disruption of both is lethal (13). Both Mycobacterium leprae and Mycobacterium tuberculosis have pairs of genes encoding putative HMW class A PBP1s. In M. tuberculosis the genes are denoted ponA1 and ponA2 (4), while in M. leprae they are denoted ponA (2) and pon1* (8). We identified a ponA gene of Mycobacterium smegmatis that encodes a PBP1 and have characterized a mutant whose ponA was disrupted by transposon mutagenesis.
M. smegmatis mc2155 transposon mutants (3) were grown on media containing azocarmine to screen for mutants that had cell permeability defects. One mutant, designated MUT1, stained intensely and was tested for antibiotic sensitivity. The MICs were determined by an agar dilution method. MUT1 was markedly more sensitive to β-lactam antibiotics than the parental strain; however, it had normal levels of sensitivity to the other antibiotics tested (Table 1).
TABLE 1.
MICs of antimicrobial agents for M. smegmatis mc2155 and mutants MUT1 and MUT2
| Antimicrobial agent | MIC (μg/ml)a for:
|
||
|---|---|---|---|
| mc2155 | MUT1 | MUT2 | |
| Ampicillin | 200–400 | 6.25–25 | 400 |
| Cefamandole | 400–800 | 6.25–12.5 | 400 |
| Cephalothin | 200–800 | 25–50 | 200 |
| Cephaloridine | 200 | 6.25 | 200 |
| Vancomycin | 12.5–25 | 3.13–12.5 | 12.5 |
| Isoniazid | 8 | 8 | 4–8 |
| Ethionamide | 16–128 | 16–128 | 32 |
| Ethambutol | 1–2 | 0.5–2 | 1 |
| Rifampin | 16–32 | 8–32 | 16–32 |
| Cycloserine | 100–200 | 100–200 | 100–200 |
Results are ranges of results from at least two experiments performed in duplicate.
β-Lactam antibiotics covalently bind and inactivate PBPs, resulting in cell death. M. smegmatis is naturally resistant to β-lactam antibiotics. β-Lactam sensitivity in MUT1 may result from increased cell penetration, loss of β-lactamase activity, or alteration of the target PBPs. The β-lactamase activity in cell lysates was determined by monitoring the rate of hydrolysis of the chromogenic cephalosporin nitrocefin (6). Assays were done three times in duplicate. The levels of β-lactamase activity were equivalent in mc2155 (0.291 ± 0.035 μmol/min/mg [mean ± standard deviation]) and MUT1 (0.273 ± 0.035 μmol/min/mg); therefore, β-lactam antibiotic sensitivity was not due to reduced β-lactamase activity.
The permeability of the cell envelopes was determined by measurement of the rate permeation by [14C]glycine into the cell (6, 15). The rate of [14C]glycine uptake by MUT1 (15.33 ± 0.79 nmol/min/mg) was twice the rate of uptake by mc2155 (7.70 ± 0.46 nmol/min/mg), indicating that the mutant was more permeable than the wild type. The increased permeability of the cell envelope of the mutant may contribute to its antibiotic sensitivity by allowing more β-lactam antibiotic to enter the cell.
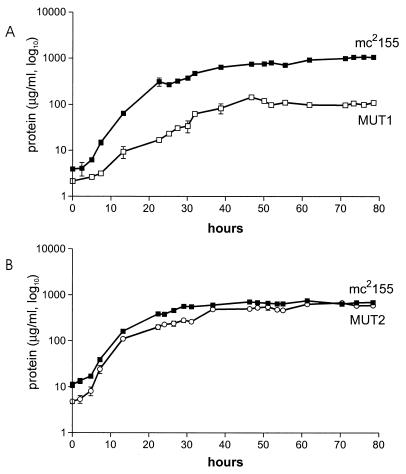
The growth rate of MUT1 was compared to that of the parent strain in several liquid media, including brain heart infusion broth (Oxoid), Middlebrook 7H9 (Difco), Penassay broth (Difco), and Luria-Bertani (LB) broth (Oxoid). The NaCl content of all media was adjusted to 0.17 M NaCl. Growth of the culture was monitored by counting viable cells or measuring biomass accumulation (9). In all media, the mutant MUT1 grew slower than the wild type and failed to reach the same final cell density at stationary phase, suggesting that PBP1 was required for normal growth. Representative data for cultures grown in LB broth are shown (Fig. 1A).
FIG. 1.
Growth in liquid culture of M. smegmatis mc2155 compared to that of MUT1 (A) and MUT2 (B). Bacteria were seeded into LB broth, and cultures were grown with aeration at 39°C for 78 h. Curves were plotted from the total cellular protein content (in micrograms) per milliliter of culture. Each datum point represents the average of two measurements. Error bars show standard deviations of the means.
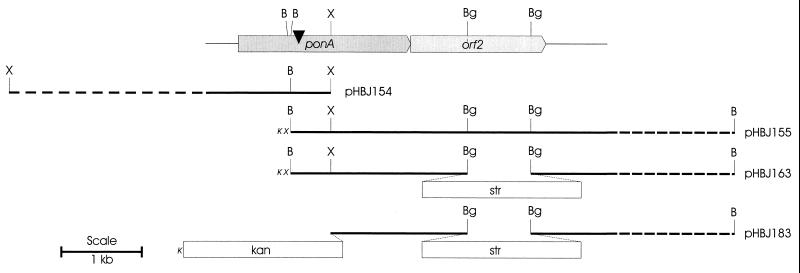
The gene disrupted in MUT1 by Tn611 was determined by inverse-PCR amplification of the chromosomal sequence flanking the site of insertion with the Tn611-specific oligonucleotide primers 5′ GAACCGCTTCGCTGCCTTG and 5′ AACCACCATTTCGCAGCAGC on a genomic DNA template that had been digested with RsaI and then self-ligated. The nucleotide sequence of the PCR product showed identity to M. tuberculosis ponA1 and M. leprae ponA. The PCR product was used as a probe to screen a Lambda Fix II (Stratagene) M. smegmatis genomic library by plaque hybridization (11). Two clones were further analyzed by restriction endonuclease digestion and Southern hybridization with the PCR-generated probe. Two overlapping fragments, a 4-kb XhoI fragment and a 5-kb BamHI fragment, were subcloned into pBluescript SK (Stratagene) for sequence analysis. The resulting plasmids, named pHBJ154 and pHBJ155, were used to derive the complete sequence of ponA and that of a neighboring gene designated orf2 (Fig. 2). A Southern blot of MUT1 genomic DNA, probed with sequences from Tn611, showed that only one copy of Tn611 had inserted into the chromosome (data not shown).
FIG. 2.
Maps of plasmids used in this study are shown in relation to the coding regions of ponA and orf2 in M. smegmatis. The position where Tn611 inserted in ponA in MUT1 is shown with a black triangle. Restriction endonuclease sites are shown for sequenced regions (solid line). B, BamHI; X, XhoI; Bg, BglII. Relevant restriction endonuclease sites within the vector, pBluescript SK, are indicated with small, italicized capital letters as follows: X for XhoI and K for KpnI.
The ponA gene encoded a protein (715 amino acids) with high sequence similarity to the PBP1s of M. tuberculosis ponA1 (83%) and M. leprae ponA (78%). Similarity to the E. coli PBP1a was low and was restricted mainly to motifs that are conserved in HMW class A PBPs (5). An open reading frame (ORF) with a putative coding region of 1,662 bp was adjacent to ponA. The ORF encoded a product of unknown function and was provisionally designated orf2 (Fig. 2). The orf2 gene product had high amino acid sequence similarity to the proteins encoded by CY21D4.14 in M. tuberculosis (80%) and MLCB1913.22c in M. leprae (63%) but was not similar to other sequences in GenBank. The organizations of the ponA and orf2 homologues were conserved in all three of the mycobacterial species. In each case the orf2-like ORF overlapped the 3′ end of ponA.
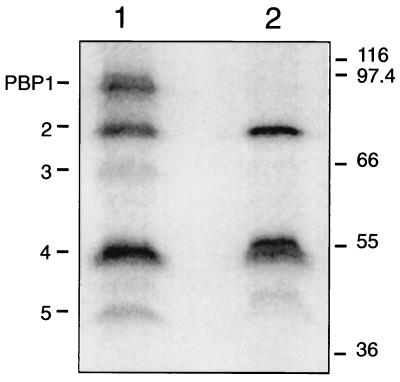
A PBP detection assay was performed to determine if the disruption of ponA altered the PBP profile in M. smegmatis. Membranes were extracted from wild-type and MUT1 cells that had been grown to mid-log phase (1), and the membrane-associated PBPs were labelled with [14C]benzylpenicillin (12). Labelled PBPs were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7) and fluorography. Five major PBPs were detected in the wild type, as was seen previously for this species (1) (Fig. 3). The most intense bands on the fluorogram corresponded to PBP1 (95 kDa), PBP2 (80 kDa), and PBP4 (55 kDa). The ponA mutant completely lacked the 95-kDa PBP1 (Fig. 3). Bands corresponding to PBP3 (66 kDa) and PBP5 (40 kDa) (Fig. 3) were less intense and varied between experiments, possibly due to differences in sample preparation.
FIG. 3.
Penicillin binding assays were performed with M. smegmatis mc2155 (lane 1) and MUT1 (lane 2). Membranes, labelled with [14C]benzylpenicillin, were solublized in 1% sodium lauroyl sarcosinate, and the supernatant was loaded onto sodium dodecyl sulfate-polyacrylamide gels. The gels were dried and subjected to fluorography. The masses of protein markers are shown in kilodaltons on the right, and PBP numbers are on the left.
The Tn611 insertion in ponA could have had polar effects on orf2 and downstream genes. Such effects may contribute to the observed phenotype of MUT1. In order to test for polar effects, another mutant was made by targeted disruption of orf2 as follows. The plasmid pHBJ155, which contains orf2 on a 5-kb BamHI fragment in pBluescript SK (Fig. 2), was digested with BglII to remove a 752-bp fragment from the coding region of orf2. A streptomycin resistance cassette was excised from pUC19Ω (10) by BamHI digestion and ligated into BglII-digested pHBJ155, resulting in pHBJ163 (Fig. 2). A kanamycin resistance marker was excised from pUC4K (14) with SalI and cloned blunt ended into XhoI-cut pHBJ163. The product of this cloning, pHBJ183, had orf2 disrupted by the streptomycin resistance gene and had a kanamycin resistance gene to facilitate screening for double-crossover events into the mycobacterial chromosome (Fig. 2). Wild-type M. smegmatis was transformed with pHBJ183, and some streptomycin-resistant, kanamycin-sensitive transformants were obtained. One such transformant, designated MUT2, had undergone double homologous recombination with pHBJ183 (data not shown), resulting in the disruption of the chromosomal copy of orf2. MUT2 was essentially the same as the wild-type strain with regard to growth rate, salt requirements (Fig. 1B), and antibiotic sensitivity (Table 1). These results indicate that polar effects of Tn611 insertion in ponA did not influence the phenotype observed in MUT1.
In summary, we have identified an ORF, ponA, in M. smegmatis that encodes a 95-kDa protein named PBP1. Insertional inactivation of ponA resulted in a mutant that lacked PBP1, was sensitive to β-lactam antibiotics, and had increased cell wall permeability. The 95-kDa PBP1 must not be essential to M. smegmatis, and other isoenzymes may partially substitute for PBP1 during peptidoglycan synthesis.
Nucleotide sequence accession number.
The nucleotide sequences of ponA and orf2 were submitted to the Genbank, EMBL, and DDBL databases and assigned accession no. AF187306.
Acknowledgments
This work was supported by a project grant from the National Health and Medical Research Council and equipment grants from The Victorian Tuberculosis and Lung Association and the Rebecca L. Cooper Foundation Ltd.
REFERENCES
- 1.Basu J, Chattopadhyay R, Kundu M, Chakrabarti P. Purification and partial characterization of a penicillin-binding protein from Mycobacterium smegmatis. J Bacteriol. 1992;174:4829–4832. doi: 10.1128/jb.174.14.4829-4832.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basu J, Mahapatra S, Kundu M, Mukhopadhyay S, Nguyendisteche M, Dubois P, Joris B, Vanbeeumen J, Cole S T, Chakrabarti P, Ghuysen J M. Identification and overexpression in Escherichia coli of a Mycobacterium leprae gene, pon1, encoding a high-molecular-mass class A penicillin-binding protein, PBP1. J Bacteriol. 1996;178:1707–1711. doi: 10.1128/jb.178.6.1707-1711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billman-Jacobe H, Sloan J, Coppel R L. Analysis of isoniazid-resistant transposon mutants of Mycobacterium smegmatis. FEMS Microbiol Lett. 1996;144:47–52. doi: 10.1111/j.1574-6968.1996.tb08507.x. [DOI] [PubMed] [Google Scholar]
- 4.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornby T, Jagels K, Krogh A, Barrell B G, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 5.Goffin C, Ghuysen J M. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarlier V, Nikaido H. Permeability barrier to hydrophilic solutes in Mycobacterium chelonei. J Bacteriol. 1990;172:1418–1423. doi: 10.1128/jb.172.3.1418-1423.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 8.Lepage S, Dubois P, Ghosh T K, Joris B, Mahapatra S, Kundu M, Basu J, Chakrabarti P, Cole S T, Nguyendisteche M, Ghuysen J M. Dual multimodular class A penicillin-binding proteins in Mycobacterium leprae. J Bacteriol. 1997;179:4627–4630. doi: 10.1128/jb.179.14.4627-4630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyers P R, Bourn W R, Steyn L M, van Helden P D, Beyers A D, Brown G D. Novel method for rapid measurement of growth of mycobacteria in detergent-free media. J Clin Microbiol. 1998;36:2752–2754. doi: 10.1128/jcm.36.9.2752-2754.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 12.Spratt B G. Properties of the penicillin-binding proteins of Escherichia coli K12. Eur J Biochem. 1977;72:341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki H, Nishimura Y, Hirota Y. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci USA. 1978;75:664–668. doi: 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor L A, Rose R E. A correction in the nucleotide sequence of the Tn903 kanamycin resistance determinant in pUC4K. Nucleic Acids Res. 1988;16:358. doi: 10.1093/nar/16.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trias J, Benz R. Permeability of the cell wall of Mycobacterium smegmatis. Mol Microbiol. 1994;14:283–290. doi: 10.1111/j.1365-2958.1994.tb01289.x. [DOI] [PubMed] [Google Scholar]