Abstract
Objectives
We systematically reviewed the current literature evaluating the ability of fully-automated deep learning (DL) and semi-automated traditional machine learning (TML) MRI-based artificial intelligence (AI) methods to differentiate clinically significant prostate cancer (csPCa) from indolent PCa (iPCa) and benign conditions.
Methods
We performed a computerised bibliographic search of studies indexed in MEDLINE/PubMed, arXiv, medRxiv, and bioRxiv between 1 January 2016 and 31 July 2021. Two reviewers performed the title/abstract and full-text screening. The remaining papers were screened by four reviewers using the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) for DL studies and Radiomics Quality Score (RQS) for TML studies. Papers that fulfilled the pre-defined screening requirements underwent full CLAIM/RQS evaluation alongside the risk of bias assessment using QUADAS-2, both conducted by the same four reviewers. Standard measures of discrimination were extracted for the developed predictive models.
Results
17/28 papers (five DL and twelve TML) passed the quality screening and were subject to a full CLAIM/RQS/QUADAS-2 assessment, which revealed a substantial study heterogeneity that precluded us from performing quantitative analysis as part of this review. The mean RQS of TML papers was 11/36, and a total of five papers had a high risk of bias. AUCs of DL and TML papers with low risk of bias ranged between 0.80–0.89 and 0.75–0.88, respectively.
Conclusion
We observed comparable performance of the two classes of AI methods and identified a number of common methodological limitations and biases that future studies will need to address to ensure the generalisability of the developed models.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13244-022-01199-3.
Keywords: Prostate cancer, MRI, Artificial intelligence, Deep learning, Machine learning
Key points
Fully-automated and semi-automated MRI-based AI algorithms show comparable performance for differentiating csPCa/iPCa.
DL and TML papers share common methodological limitations discussed in this review.
Consensus on datasets, segmentation, ground truth assessment, and model evaluation are needed.
Background
The introduction of pre-biopsy multiparametric magnetic resonance imaging (mpMRI) has considerably improved the quality of prostate cancer (PCa) diagnosis by reducing the number of unnecessary biopsies and increasing the detection of clinically significant disease compared to the conventional PSA-transrectal ultrasound (TRUS) pathway [1–3]. However, the high dependence of the diagnostic performance of mpMRI on reader experience [4, 5] and image quality [6], coupled with the need to balance the time-consuming delineation of biopsy targets against the increasing pressure on radiology departments [7], limits the population-based delivery of high-quality mpMRI-driven PCa diagnosis.
The recent joint position paper by the European Society of Urogenital Radiology (ESUR) and European Association of Urology (EAU) Section of Urological Imaging (ESUI) has highlighted the importance of developing robust and clinically applicable artificial intelligence (AI) methods for overcoming the aforementioned limitations and facilitating the successful deployment of the mpMRI-driven PCa diagnostic pathway [8] to the community. Importantly, the authors suggest the use of AI as a triage tool to detect and delineate areas suspicious for clinically significant PCa (csPCa), where its accurate differentiation from indolent PCa (iPCa) and benign conditions determines the need for subsequent biopsy and defines the diagnostic accuracy of mpMRI. While several recent systematic [9–12] and narrative [13] reviews have described the performance of AI methods for detecting csPCa on MRI, little is known about the comparative performance of fully-automated and semi-automated approaches when applied to this specific clinical task. The rationale for this comparison is based on several inherent differences between the two approaches. Specifically, fully-automated methods rely on learned deep radiomic features and do not require human input following initial training and validation, which underpins their disruptive potential for significantly reducing the radiologists’ clinical workload. Conversely, semi-automated methods, most commonly based on hand-engineered radiomic features, require manual delineation and image pre-processing that may increase the radiologists’ time while not adding significant diagnostic benefit.
Therefore, the primary objective of this systematic review was to analyse the current literature on fully-automated and semi-automated AI methods to differentiate csPCa from iPCa and benign disease on MRI. In addition, we aimed to both identify and offer prospective solutions to common methodological limitations and biases of the existing studies. Addressing these issues going forward will facilitate the development of robust, generalisable, and clinically applicable MRI-derived AI models for PCa diagnosis.
Materials and methods
To avoid bias, the review protocol was agreed by all authors and registered with PROSPERO (CRD42021270309) before the start of the review process.
Search strategy
Data collection and reporting were conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [14], with a complete PRISMA 2020 checklist presented in Additional file 2: Table S1. We performed a computerised bibliographic search of published and unpublished studies indexed in MEDLINE/PubMed, arXiv, medRxiv, and bioRxiv between 1 January 2016 and 31 July 2021. The full search strategy is summarised in Additional file 1.
Eligibility criteria
The population of interest included treatment-naïve patients who underwent MRI of the prostate that was subsequently processed using either fully-automated or semi-automated AI methods for lesion detection and subsequent binary classification as (a) csPCa or (b) iPCa or benign disease. The performance of AI methods (index test) was referenced against histopathological assessment of MRI target lesions, with csPCa defined as International Society of Urogenital Pathology (ISUP) grade group ≥ 2 disease and iPCa defined as ISUP grade group 1 disease. The outcome measures included the diagnostic performance of AI approaches for differentiating csPCa from iPCa and benign disease measured as an area under the receiving operator characteristic curve (AUC), sensitivity, specificity, accuracy, positive predictive value (PPV), and negative predictive value (NPV). Only studies written in English and presenting original results were included in this review.
Systematic review process
We deployed a three-stage process to identify papers suitable for inclusion in this review using Covidence [15] as a Web-based support tool. In the first stage, a team of two reviewers (N.S., L.R.) independently performed the title and abstract screening to ensure relevance, with conflicts resolved by the third reviewer (T.B.). In the second stage, the same two reviewers screened the full text of each paper for eligibility, with conflicts resolved by the same third reviewer. In the third stage, four reviewers (Team 1, N.S., NMDS; Team 2, L.R., M.Y.) evaluated the quality of the documentation of methodologies in the papers to assess the reproducibility of their results. Papers using fully-automated AI methods based on deep learning (DL) methods were assessed using the Checklist for Artificial Intelligence in Medical Imaging (CLAIM) [16], while studies deploying semi-automated AI approaches relying on traditional machine learning (TML) methods were evaluated using the Radiomics Quality Score (RQS) [17] as detailed in Additional file 1.
Risk of bias assessment
We used the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool [18] to assess the risk of bias and applicability of studies included in this systematic review. In line with the QUADAS-2 guidance, we developed a review-specific protocol on how to assess each signalling question, which is summarised in Additional file 1. QUADAS-2 assessment was conducted by the same two teams of two reviewers, with each paper reviewed independently by the reviewers prior to conflict resolution by consensus of all four reviewers.
Data extraction
The data extraction criteria were agreed prior to the review commencement and then independently extracted by the same reviewer teams. The full list of extracted parameters is presented in Additional file 3, with the key diagnostic performance characteristics being AUC, sensitivity, specificity, accuracy, NPV and PPV for the internal holdout or external test sets (when available).
Data analysis
Given the substantial heterogeneity of patient characteristics, AI algorithms, ground truth assessment methods, and validation strategies used in the diagnostic accuracy studies included in this review, we chose narrative synthesis over meta-analysis of the pooled data to avoid a biased result [19].
Results
Study selection
The study selection process is presented in Fig. 1. Our initial search identified 314 papers, of which 4 were highlighted as duplicates by Covidence and removed by us following manual verification. 60/310 papers had titles or abstract deemed relevant to the review question; of those, 28 were retained for quality review after full-text screening. 12/28 papers deployed fully-automated AI methods based on DL methods and were therefore screened using CLAIM, while 16/28 papers used TML methods to develop semi-automated AI approaches and were assessed using RQS. Of these, 5/12 (42%) DL papers [20–24] and 12/16 (43%) TML papers [25–36] passed the quality screening and were subject to full QUADAS-2 assessment, data extraction, and narrative synthesis.
Fig. 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2020 flow diagram for literature search. csPCa, clinically significant prostate cancer; iPCa, indolent prostate cancer
Quality review
Three out of 12 DL studies (25%) [37–39] that underwent quality screening using CLAIM failed at least three pre-identified mandatory criteria, with 2/12 [40, 41] failing two, and 2/12 [42, 43] failing just one criterion. Four of the seven rejected papers (57%) [37–39, 43] did not describe data processing steps in sufficient detail (Q9), 4/7 [38–40, 42] did not explain the exact method of selecting the final model (Q26), and 3/7 [38, 40, 41] failed to provide enough details on training approach (Q25). Following the subsequent full CLAIM assessment of the remaining five papers, we found that none of them reported the following items: selection of data subsets (Q10), robustness or sensitivity analysis (Q30), validation or testing on external data (Q32), and failure analysis of incorrectly classified cases (Q37). The results of CLAIM quality screening and full assessment are presented in Additional file 1.
One out of 16 TML studies (6%) [44] that underwent quality screening using RQS scored 2/8, 1/16 [45] scored 6/8, and 2/16 [46, 47] scored 7/8, which led to their exclusion from subsequent full RQS assessment. None of the excluded papers had well-documented imaging protocols (Q1) and neither performed multiple segmentations by different radiologists nor conducted robustness analysis of image segmentations to region-of-interest (ROI) morphological perturbations (Q2). The mean RQS of the remaining 12 papers that underwent full assessment was 10.9 ± 2.0 (standard deviation) out of 36 points possible. None of the papers performed phantom studies to detect scanner-dependent features (Q3), reported calibration statistics (Q10), registered a prospective study (Q11), and reported on the cost-effectiveness of the clinical application of the proposed models (Q14). Only one (8%) paper [32] discussed a potential biological correlate for some radiomic features included in the final model (Q7), and only two papers [28, 36] performed external testing of their models (Q12). Furthermore, only six out of 12 (50%) papers [25, 26, 29–32] had image segmentation performed by multiple radiologists or instead assessed the robustness of radiomic features to ROI morphological perturbations (Q2). Eight out of 12 (67%) papers [25–27, 30–32, 34, 35] did not make available any images, code, or feature values used to train the models (Q16), and only 4/12 (33%) papers [30, 31, 34, 36] incorporated non-radiomic features into the multivariable analysis (Q6). The results of RQS screening and full assessment are presented in Additional file 3.
Risk of bias assessment
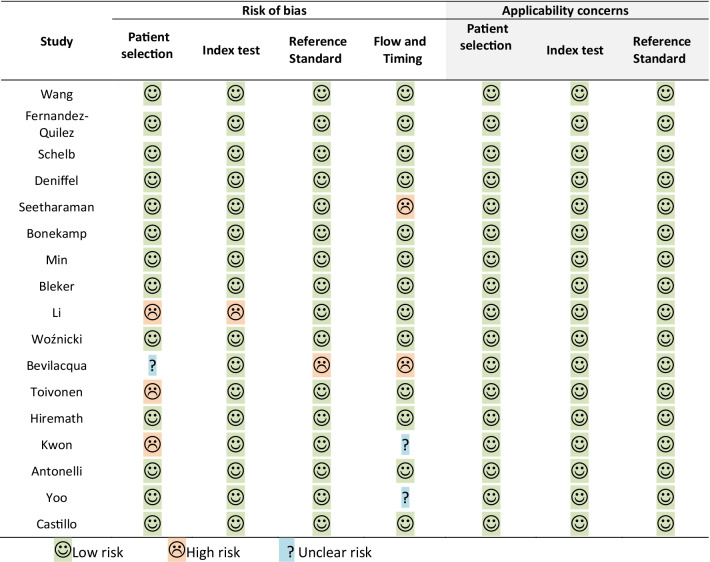
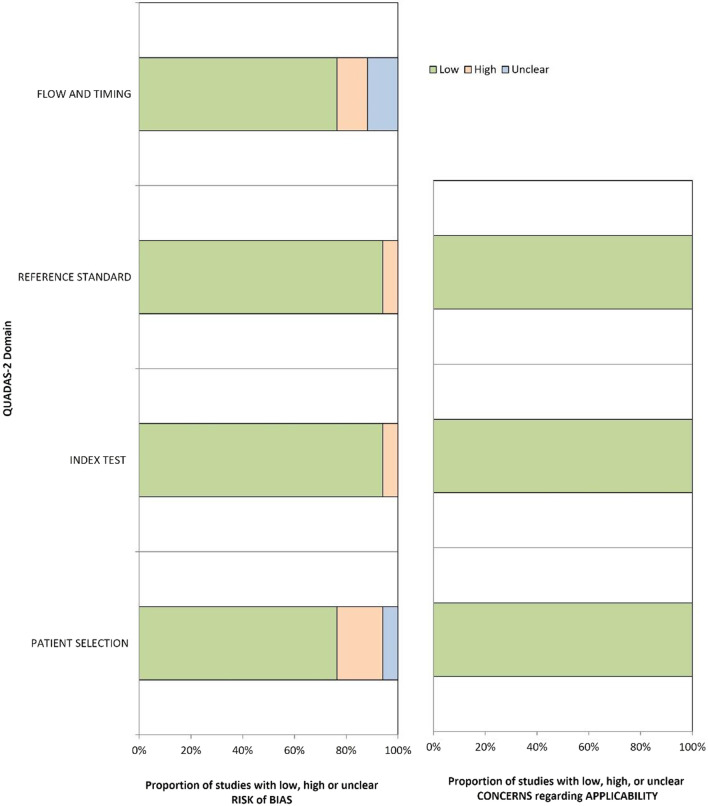
The full results of QUADAS-2 assessment are presented in Additional file 1, with their graphical summary provided in Table 1 and Fig. 2. Overall, 11/17 (65%) [12, 20–23, 25, 26, 29, 31, 34, 36], 1/17 [35], and 5/17 [24, 27, 30, 32, 33] papers had low, unclear, and high risk of bias, respectively. All papers had low applicability concerns. Inappropriate patient selection led to a high risk of bias in 3/5 (60%) studies [27, 30, 33], with two papers containing inappropriate exclusions and one study using a case–control design. One study [30] did not pre-specify a threshold prior to evaluation of the index test performance on the test set. One study [32] used transrectal ultrasound guided (TRUS) biopsy performed six weeks prior to MRI as a reference standard, which introduced a high risk of bias. Two (40%) papers [24, 32] had high risk of bias associated with data flow and timing between the index test (MRI) and reference standard (biopsy), with one paper using both surgical pathology and biopsy results as reference standards, and one paper reporting a six week interval between biopsy and MRI, which was below the recommended threshold of at least six months [48]. The only paper with an unclear risk of bias did not report any information regarding the timing between MRI and biopsy, as well as the specific type of biopsy and whether it was consistent in all patients in the study.
Table 1.
QUADAS-2 risk of bias and applicability concerns
Fig. 2.
Summary QUADAS-2 risk of bias and applicability concerns assessment
Study characteristics
Summary demographic characteristics of patients included in the studies that passed the quality screening are presented in Table 2. Two out of five (40%) DL papers [20, 21] used patient data available as part of the open-source PROSTATEx challenge dataset [49], while the remaining three (60%) studies [22–24] used data from single institutions. Importantly, one paper [24] used radical prostatectomy and targeted biopsy interchangeably in one of its patient cohorts. None of the DL studies reported the time between MRI and biopsy, while all studies performed MRI using a single vendor. The number of readers annotating MR images varied between 1 and 4, with reader experience ranging between 0.5 and 20 years.
Table 2.
Summary demographic characteristics of patients included in the studies selected for narrative synthesis
| Study | Year | Country | No. of patients | Age, years | PSA, ng/mL | Patient population | Bx | MRI vs Bx | Time MRI to Bx | No. of centres /vendors | No. of readers | Reader experience, years |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Studies using deep learning-based fully-automated AI methods | ||||||||||||
| Wang [20] | 2020 | Netherlands | 346 | 66 (48–83) | 13 (1–56) | Clinically suspected | TB | Pre-Bx | NR | 1/1 | 1 | 20 |
| Fernandez-Quilez [21] | 2021 | Netherlands | 200 | 66 (48–83) | 13 (1–56) | Clinically suspected | TB | Pre-Bx | NR | 1/1 | 4 | NR |
| Schelb [22] | 2019 | Germany | 312 | Training: 64 [58–71] Test: 64 [60–69] |
Training: 7.0 [5.0–10.2] Test: 6.9 [5.1–8.9] |
Clinically suspected | TB | Pre-Bx | NR | 1/1 | 2 | 0.5, 10 |
| Deniffel [23] | 2020 | Canada | 499 | Training: 63.8 ± 8.1 Test: 64.4 ± 8.4 |
Training: 7.6 [5.0–10.8]a Test: 7.2 [5.2–11.2] |
Clinically suspected | TB | Pre-Bx | NR | 1/1 | 2 | 15, 3 |
| Seetharaman [24] | 2021 | USA | 424 |
Training: 63.8 (49–76) Test: 65 (38–82) |
Training: 6.8 (3.3–28.6) Test: 7.1 (0.9–63.0) |
Clinically suspected | RP or TB | Pre-Bx or Pre-Op | NR | 1/1 | Unclear | Unclear |
| Studies using traditional machine learning-based semi-automated AI methods | ||||||||||||
| Bonekamp [25] | 2018 | Germany | 316 | 64 [58–71] |
Training: 6.6 [4.9–9.5] Test: 7.5 [5.4–11.0] |
Clinically suspected | TB | Pre-Bx | NR | 1/1 | 2 | 0.5, 8 |
| Min [26] | 2019 | China | 280 |
Training,csPCa: 68.8 ± 8.3Training, iPCa: 71.5 ± 8.4 Test, csPCa: 70.3 ± 7.8 Test, iPCa: 71.6 ± 5.7 |
NRb | Clinically suspected | TB | Pre-Bx | NR | 1/1 | 2 | NR, 20 |
| Kwon [27] | 2018 | Netherlands | 344 | 66 (48–83) | 13 (1–56) | Clinically suspected | TB | Pre-Bx | NR | 1/1 | 2 | > 25 |
| Castillo [28] | 2021 | Netherlands | 107 |
C1: 64 ± 7 C2: N/A C3: N/A |
C1: 12 ± 10 C2: 9 ± 5 C3: 10 ± 8 |
Clinically suspected | RP | Pre-Op | NR | 3/3 | 1 | NR |
| Bleker [29] | 2019 | Netherlands | 206 | 66 (48–83) | 13 (1–56) | Clinically suspected | TB | Pre-Bx | NR | 1/1 | Unclear | Unclear |
| Li [30] | 2020 | China | 381 |
csPCa:75 [68–81] iPCa: 69 [63–75] |
csPCa:49.3 [21.1–83.4 iPCa:9.9 [6.7–15.9] |
Clinically suspected | TB | Pre-Bx | NR | 1/1 | 2 | 3, 9 |
| Woźnicki [31] | 2020 | Germany | 191 | Training: 68 [63–74] Test: 69 [63–72] |
Training:7.6 [5.7–11.0] Test: 8.2 [6.8–11.9] |
Clinically suspected | TB | Pre-Bx | Bx 3 months before MRI | 1/2 | 2 | 7, 7 |
| Bevilacqua [32] | 2021 | Italy | 76 |
csPCa: 66 ± 6.8 iPCa: 65 ± 8.8 |
csPCa: 7.8 ± 7.5 iPCa: 5.3 ± 3.0 |
Biopsy-proven | TB | Post-Bx | Bx 6 weeks before MRI | 1/1 | 2 | 7, 25 |
| Toivonen [33] | 2019 | Finland | 62 | 65 (45–73) | 9.3 (1.3–30) | Biopsy-proven | RP | Pre-Bx | NR | 1/1 | 2 | NR |
| Antonelli [34] | 2019 | UK | 164 | 64 (43–83) | 7.4 (2.5–30.3) | Clinically suspected | TB | Pre-Bx | NR | 1/1 | 1 | 3 |
| Yoo [35] | 2019 | Canada | 427 | NR | NR | Clinically suspected | NR | Pre-Bx | NR | 1/1 | NR | NR |
| Hiremath [36] | 2021 | USA, Netherlands | 592 |
C1: 65.5 (59–72) C2: 63 (59–68) C3: 62 (56–66) C4: 65.5 (62–73) |
C1: 6.6 (0.25–88.2) C2: 6.7 (5–10) C3: 5.7 (4.54–9.58) C4: 7.7 (4.8–11.3) |
Clinically suspected |
RP or SB or TB |
Pre-Bx | NR | 5/3 | 5 |
> 15, > 15, > 15, > 10, > 10 |
Bx, biopsy; C, cohort; MRI, magnetic resonance imaging; NR, not reported; PSA, prostate-specific antigen; RP, radical prostatectomy; SB, systematic biopsy; TB, targeted biopsy
aData missing for 110 cases
bPSA values were reported by subcategories (< 4 ng/mL, 4–10 ng/mL, > 10 ng/mL), see the original reference [26] for further details
Ten out of 12 (83%) TML papers [12, 25, 26, 30–36] utilised non-publicly available institutional datasets, with the remaining 2/12 (17%) studies [27, 29] using the PROSTATEx challenge dataset [49]. In eight (67%) papers [25–27, 29–32, 34], the histopathological ground truth was obtained using targeted biopsy, while two studies [28, 33] relied on radical prostatectomy data, one [36] was a multi-institutional study relying on either biopsy (targeted or systematic) or prostatectomy data in different cohorts, and one [35] did not explicitly report the source of ground truth. Only two (17%) papers [31, 32] reported the time between biopsy and MRI; in these studies, biopsy was performed either three months [31] or six weeks [32] prior to MRI. Nine (75%) studies [25–27, 29, 30, 32–35] had one centre and one vendor each, while the remaining three studies [28, 31, 36] were multi-vendor. The number of readers varied between 1 and 5, with reader experience ranging between 0.5 and more than 25 years.
Predictive modelling characteristics
Summary predictive modelling characteristics of DL papers are presented in Table 3. All studies used different convolutional neural network (CNN) architectures, with 3/5 (60%) studies [20, 23, 24] proposing their own networks and 2 papers using off-the-shelf networks, including VGG16 [21] and U-Net [22]. None of the papers included non-imaging features for the purposes of predictive modelling and conducted external testing of the developed predictive models. All DL papers were designed as a classification task to distinguish csPCa from iPCa and benign lesions. Four (80%) studies [21–24] performed the analysis at the level of the whole prostate, and one study [20] separately analysed peripheral and transition zone lesions. Importantly, none of the DL studies validated their results using external datasets.
Table 3.
Predictive modelling characteristics of studies using deep learning-based fully-automated AI methods
| Study | No. of patients | Training set | Validation set | Test set | Algorithm | MRI input | Image registration | Image segmentation | Outcome | Zone | Analysis | Evaluation strategy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Wang [20] | 346 | 204 | fivefold CV | 142 | CNN (MISN) |
ADC, BVAL, DWI0, DWI1, DWI2, Ktrans, T2WI-Cor, T2WI-Sag, T2WI-Tra |
NR | Open data | csPCa vs iPCa or benign lesions | PZ or TZ | Per lesion | Internal hold-out |
| Fernandez-Quilez [21] | 200 | NRa | NRa | NRa | CNN (VGG16) | T2WI, ADC | NR | Open data | csPCa vs iPCa or benign lesions | WP | Per lesion | Internal hold-out |
| Schelb [22] | 312 | 250 | No | 62 | CNN (U-Net) | T2WI, DWI | SimpleITK, non-rigid Bspline with Mattes mutual information criterion | Automated (U-Net) | csPCa vs iPCa or benign lesions | WP | Per lesion, per patient | Internal hold-out |
| Deniffel [23] | 499 | 324 | 75 | 50b | CNN (3D) | T2WI, ADC, DWI | Static, affine | Manual bounding boxes | csPCa vs iPCa or benign lesions | WP | Per patient | Internal hold-out |
| Seetharaman [24] | 424 | 102 | fivefold CV | 322 | CNN (SPCNet) | T2WI, ADC | Manual | Registration from pathology images | csPCa vs iPCa or benign lesions | WP | Per pixel, per lesion | Internal hold-out |
ADC, apparent diffusion coefficient; CNN, convolutional neural networks; csPCa, clinically significant prostate cancer; CV, cross-validation; DWI, diffusion-weighted imaging; iPCa, indolent prostate cancer; MISN, multi-input selection network; MRI, magnetic resonance imaging; NR, not reported; PZ, peripheral zone; T2WI, T2-weighted imaging; TZ, transition zone; WP, whole prostate
aThe study included 200 patients and 299 lesions, of which 70% were used to train train, 20% to test, 10% to fine-tune the models
bDescribes the calibration cohort
Similar predictive modelling characteristics of TML papers are summarised in Table 4. The three most commonly used ML models included random forests (50% papers), logistic regression (42% papers), and support vector machines (25% papers), with 7/12 studies testing several different models. Imaging features were extracted from apparent diffusion coefficient maps, T2-weighted images, and diffusion-weighted images with different b-values in 12/12 (100%) [25–36], 9/12 [25–31, 33, 36], and 7/12 [25–29, 32, 35] papers, respectively. In contrast to the DL papers, only 7/12 (58%) TML studies [25, 27, 29–31, 35, 36] differentiated csPCa from iPCa and benign lesions, whereas the remaining five studies (42%) [26, 28, 32–34] did not include benign disease, thereby focusing only on distinguishing csPCa from iPCa. Eight (67%) papers [26, 28, 30–33, 35, 36] performed the analysis at the level of the whole prostate, two [27, 34] reported the results for peripheral and transition zone lesions separately, one [25] developed models for the whole prostate as well as peripheral and transition zone lesions, and one [29] included peripheral zone tumours only. Seven (58%) studies [25–27, 29–32] validated their results using internal hold-out, three papers [33–35] used cross-validation, and the remaining two studies [28, 36] used either a mixed hold-out cohort or a fully external hold-out dataset.
Table 4.
Predictive modelling characteristics of studies using traditional machine learning-based semi-automated AI methods
| Study | No. of patients | Training set | Validation set | Test set | Algorithm | MRI input | IR | IS | Discriminative features | No. of features used for training | Outcome | Zone | Analysis | Evaluation strategy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bonekamp [25] | 316 | 183 | NR | 133 | RF | T2WI, ADC, b = 1500 | No | Manual | First-order, volume, shape, texture | NR | csPCa vs iPCa or benign lesions | WP or PZ or TZ | Per lesion and per patient | Internal hold-out |
| Min [26] | 280 | 187 | NR | 93 | LR | T2WI, ADC, b = 1500 | No | Manual | Intensity, shape, texture, wavelet | 9 | csPCa vs iPCa | WP | Per lesion | Internal hold-out |
| Kwon [27] | 344 | 204 | tenfold CV | 140 | CART, RF, LASSO | T2WI, DWI, ADC, DCE | Rigid | No | Intensity | 54 | csPCa vs iPCa or benign lesions | PZ or TZ | Per lesion | Internal hold-out |
| Castillo [28] | 107 | 80% | 20% of training (100 random repeats) | 20% | LR, SVM, RF, NB, LQDA | T2WI, DWI, ADC | HPa | Manual | Shape, local binary patterns, GLCM | NR | csPCa vs iPCa | WP | Per lesion, Per patient | Mixed hold-out |
| Bleker [29] | 206 | 130 | NR | 76 | RF, XGBoost | T2WI, b = 50, b = 400, b = 800, b = 1400, ADC, Ktrans | No | Manual | Intensity, texture | NR | csPCa vs iPCa or benign lesions | PZ | Per lesion | Internal hold-out |
| Li [30] | 381 | 229 | NR | 152 | LR | T2WI, ADC | No | Manual | Intensity, age, PSA, PSAd | 15 | csPCa vs iPCa or benign lesions | WP | Per lesion | Internal hold-out |
| Woźnicki [31] | 191 | 151 | fivefold CV | 40 | LR, SVM, RF, XGBoost, CNN | T2WI, ADC | No | Manual | Intensity, shape, PI-RADS, PSAd, DRE | 15 | csPCa vs iPCa or benign lesions | WP | Per patient | Internal hold-out |
| Bevilacqua [32] | 76 | 48 | threefold CV | 28 | SVM | ADC, b = 2000 | No | Manual | Intensity | 10 | csPCa vs iPCa | WP | Per lesion | Internal hold-out |
| Toivonen [33] | 62 | 62 | LPOCV | N/A | LR | T2WI, ADC, Ktrans, T2 map | No | Manual | Intensity, Sobel, texture | NR | csPCa vs iPCa | WP | Per lesion | LPOCV |
| Antonelli [34] | 164 | 134 | NR | 30 |
PZ: LinR TZ: NB |
ADC, DCE | Rigid | Manual | Texture, PSAd | NR | csPCa vs iPCa | PZ or TZ | Per lesion | fivefold CV |
| Yoo [35] | 427 | 271 | 48 | 108 | CNN, RF | ADC, DWI | No | No | First-order statistics of deep features | 90 | csPCa vs iPCa or benign lesions | WP | Per slice, Per patient | tenfold CV |
| Hiremath [36] | 592 | 368 | threefold CV | 224 | AlexNet or DenseNet and Nomogram | T2WI, ADC | Rigid, affine | Manual | Deep learning imaging predictor, PI-RADS, PSA, gland volume, tumour volume | NR | csPCa vs iPCa or benign lesions | WP | Per patient | External hold-out |
ADC, apparent diffusion coefficient; CART, classification and regression trees; CNN, convolutional neural networks; GLCM, grey level co-occurrence matrix; HP, histopathology; IR, image registration; IS, image segmentation; LASSO, least absolute shrinkage and selection operator; LinR, linear regression; LQDA, linear and quadratic discriminant analysis; LR, logistic regression; NB, naïve Bayes; PI-RADS, prostate imaging-reporting and data system; PSA, prostate-specific antigen; PSAd, prostate-specific antigen density; RF, random forests; SVM, support-vector machines
aHistopathology images registered with T2-weighted images using specialised software
Comparative performance of fully-automated and semi-automated AI methods
Three out of 5 (60%) DL studies [21–23] had clearly defined thresholds at which performance characteristics of the developed models were calculated; these are presented in Table 5. For studies combining peripheral and transition zone lesions for classification [21, 23, 24], the AUCs of the best-performing models reported in the test sets for differentiating csPCa from iPCa and benign disease ranged between 0.80 and 0.89. Importantly, the AUC range changed to 0.85–0.89 when a study by Seetharaman et al. [24] was excluded from the calculation due to its high risk of bias reported on QUADAS-2 assessment (Table 1). In a study by Wang et al. [20], AUCs for peripheral zone and transition zone lesions were 0.89 [0.86–0.93] and 0.97 [0.95–0.98], respectively, and a study by Schelb et al. [22] did not report AUC values. Four (80%) studies [21–24] did not report accuracy of the developed models, while Wang et al. [20] reported accuracy of 0.91 [0.86–0.95] and 0.89 [0.87–0.91] in the peripheral and transition zone lesions, respectively. All studies reported sensitivity and specificity of the proposed models, while only 2/5 (40%) [22, 23] studies presented NPV and PPV, with NPV being higher in both cases (Table 5).
Table 5.
Diagnostic performance of fully-automated and semi-automated AI methods for differentiating between csPCa and iPCa or benign disease
| Study | Threshold | AUC [95% CI] | Accuracy | Sensitivity | Specificity | NPV | PPV |
|---|---|---|---|---|---|---|---|
| Studies using deep learning-based fully-automated AI algorithms | |||||||
| Wang [20] | NR |
PZ: 0.89 [0.86–0.93] TZ: 0.97 [0.95–0.98] |
PZ: 0.91 [0.86–0.95] TZ: 0.89 [0.87–0.91] |
PZ: 0.60 [0.52–0.69] TZ: 1.0 [1.0–1.0] |
PZ: 0.98 [0.95–1.0] TZ: 0.88 [0.82–0.93] |
NR | NR |
| Fernandez-Quilez [21] | 0.5 | 0.89 | NR | 0.85 | 0.94 | NR | NR |
| Schelb [22] | Several for different PI-RADS cut-offs | NR | NR |
PI-RADS ≥ 3: 0.96 PI-RADS ≥ 4: 0.92 |
PI-RADS ≥ 3: 0.31 PI-RADS ≥ 4: 0.47 |
PI-RADS ≥ 3: 0.84 PI-RADS ≥ 4: 0.83 |
PI-RADS ≥ 3: 0.53 PI-RADS ≥ 4: 0.67 |
| Deniffel [23] | Risk of csPCa ≥ 0.2 | 0.85 [0.76–0.97] | NR | 1.0 [1.0–1.0] | 0.52 [0.32–0.68] | 1.0 [1.0–1.0] | 0.56 [0.48–0.66] |
| Seetharamana [24] | NR | 0.80 (per lesion) | NR | 0.70 (per lesion) | 0.77 (per lesion) | NR | NR |
| Studies using traditional machine learning-based semi-automated AI algorithms | |||||||
| Bonekamp [25] | 0.79 |
WP: 0.88 PZ: 0.84 TZ: 0.89 (per lesion) |
NR |
WP: 0.97 (per lesion) |
WP: 0.58 (per lesion) |
NR | NR |
| Min [26] | NR | 0.82 [0.67–0.98] | NR | 0.84 | 0.73 | NR | NR |
| Castillo [28] | NR | 0.75 | NR | 0.88 | 0.63 | NR | NR |
| Bleker [29] | NR | 0.87 [0.75–0.98] | NR | 0.86 | 0.73 | NR | NR |
| Woźnicki [31] | 0.45 | 0.84 [0.6–1.0] | NR | 0.91 [0.81–0.98] | 0.57 [0.38–0.74] | NR | NR |
| Antonelli [34] | Reader SP (training) |
PZ: 0.83 TZ: 0.75 |
NR |
PZ: 90 TZ: 92 |
PZ: 65 TZ: 56 |
NR | NR |
| Hiremath [36] | Maximising accuracy (0.361) | 0.81 [0.76–0.85] | 0.78 | 0.83 | 0.59 | NR | NR |
| Kwona [27] | NR | WP: 0.82 | NR | NR | NR | NR | NR |
| Lia [30] | − 0.42 | 0.98 [0.97–1.00] | 0.90 | 0.95 | 0.87 | 0.97 | 0.82 |
| Bevilacquaa [32] | 0.58 | 0.84 [0.63–0.90] | NR | 0.9 | 0.75 | NR | NR |
| Toivonena [33] | NR | 0.88 [0.92–0.95] | NR | NR | NR | NR | NR |
| Yooa [35] | NR | 0.84 [0.76–0.91] | NR | NR | NR | NR | NR |
AUC, area under the receiver operating characteristic curve; NPV, negative predictive value; NR, not reported; PI-RADS, prostate imaging-reporting and data system; PPV, positive predictive value; PZ, peripheral zone; SP, specificity; TZ, transition zone; WP, whole prostate
aThese papers had either high or unclear risk of bias on QUADAS-2 assessment (see Table 1; Fig. 2)
Six out of 12 (50%) TML studies [25, 30–32, 34, 36] defined specific thresholds for diagnostic performance, with the resulting characteristics summarised in Table 5. The AUCs of the best-performing models for studies combining peripheral and transition zone lesions ranged between 0.75 and 0.98. The AUC range changed to 0.75–0.88 when five papers [27, 30, 32, 33, 35] with high or unclear risk of bias on QUADAS-2 (Table 1) were removed from the calculation. A study by Li et al. [30] (high risk of bias, see Table 1) was one of two papers reporting accuracy of the proposed model (0.90), in addition to a study by Hiremath et al. [36] where it reached 0.78; both studies applied their models to peripheral and transition zone lesions combined. 3/12 (25%) [27, 33, 35] papers did not report sensitivity and specificity of their models, and only one study by Li et al. [30] presented NPV and PPV of their model.
Discussion
This systematic review highlights the intensity of research efforts in developing both fully-automated and semi-automated MRI-derived AI methods for differentiating csPCa from iPCa and benign disease. While formal meta-analysis and direct comparison of the two approaches were not possible due to a substantial heterogeneity of studies included in this review, the narrative synthesis revealed their comparable performance that was marginally higher for fully-automated methods. If common methodological limitations outlined in this review are addressed, future studies will have the potential to make AI-driven expert-level prostate MRI assessment widely accessible and reproducible among multiple centres and readers with different experiences.
In keeping with this report, previous systematic and narrative reviews investigating the diagnostic performance of DL- and TML-based AI methods for PCa diagnosis [9, 11–13] have also highlighted substantial heterogeneity and poor reproducibility of the developed predictive models. While a meta-analysis by Cuocolo et al. [10] showed higher AUC of TML-based models compared to DL-based models, the authors drew the data from all studies included in the qualitative synthesis. Some of these studies had a high risk of bias and showed important differences among their patient populations, ground truth assessment methods, zonal distribution of predictive models, and other potential confounders. In our review, the addition of full CLAIM and RQS quality evaluation to QUADAS-2 assessment highlighted high methodological heterogeneity of both DL- and TML-based studies, which limited the reliability of their quantitative synthesis. The outcomes of qualitative synthesis, however, suggest that DL-based fully-automated AI methods may prove more clinically useful in the long run given their comparable performance to TML-based semi-automated methods. A crucial practical advantage of fully-automated approaches is its potential time-saving effect that is important in the context of an ever-increasing workload in radiology departments. That said, almost all DL papers included in this review still require even minimal manual interaction from the readers, including lesion identification as patches [20] or bounding boxes [22–24], thereby still introducing a known element of interobserver variability. However, a head-to-head comparison of DL- and TML-based AI methods in the same patient cohort presents a highly important area of unmet research need. If addressed, this has the capacity to directly answer the clinical question behind this review.
In this review, a combination of full CLAIM, RQS, and QUADAS-2 assessment revealed several common methodological limitations, some of which are applicable to both DL and TML studies. These common limitations fall into four distinct domains: (1) datasets used for model development, (2) methods used to ensure quality and reproducibility of image segmentation, (3) ground truth assessment methods, (4) strategies used for model evaluation. The following paragraphs summarise the key limitations within each of the four domains, with detailed recommendations for their prospective mitigation provided in Additional file 1.
First, the overwhelming majority of papers included in this review either utilised non-publicly available single-centre datasets or used the same open-source single-centre PROSTATEx challenge dataset [49]. The use of single-centre datasets, both public and private, without external testing presents a critical limitation to the clinical applicability of the developed models. Conversely, the use of a single public dataset without additional data encourages community-wide overfitting that limits the utility of the dataset itself.
Second, nearly half of the studies did not process images segmented by multiple radiologists, thus limiting the generalisability of the developed predictive models due to known interobserver variability even among experts [50–52]. The same applies to the original PROSTATEx dataset [49] that includes lesion coordinates based on the outlines provided by a single reader. While one DL study included in our review [20] used the original single-reader segmentations, another study [21] overcame this limitation by utilising segmentations validated by several readers in a dedicated study by Cuocolo et al. [53]. Even if trained on the same dataset and using the same AI methods, models developed using different segmentations will inevitably differ in their performance, which brings additional layer of heterogeneity to the field.
Third, only 80% of DL and 67% of TML papers used MRI-targeted biopsy specimens as a source of ground truth. The remaining studies either relied on radical prostatectomy data or included mixed patient cohorts where the ground truth was obtained using different methods. While radical prostatectomy specimens offer definitive assessment of lesion morphology, the resulting predictive models will have very limited clinical applicability due to overrepresentation of patients with intermediate-risk disease. If predictive models are trained to differentiate between iPCa and csPCa and therefore help clinicians decide on the need for subsequent biopsy, then MRI-targeted biopsy using cognitive, US/MRI fused, or in-bore approaches present an appropriate standard for ground truth assessment.
Fourth, none of the DL papers and only two TML papers used external testing to assess the generalisability of the developed predictive models [54]. Given the intrinsically low reproducibility and repeatability of MRI-derived radiomic features [55, 56], the lack of robust external testing and prior assessment of feature robustness to scanning parameters present major obstacles to the clinical use of any MRI-based AI algorithms. However, even if external testing becomes the norm, it is also important to avoid common mistakes in reporting standard measures of discrimination that help evaluate model performance. These often include the lack of clearly identified operating points at which they were calculated and confidence intervals that reflect the uncertainty in the estimate. Ideally, the operating points should reflect the expected performance of expert radiologists, with the pooled NPV of 97.1% (95% CI 94.9–98.7%) [2] being the key clinical benchmark that has established mpMRI as a diagnostic test that can effectively rule out csPCa. Importantly, a thorough failure analysis of incorrectly classified cases is key to understanding and communicating diagnostic pitfalls of the developed models, which is paramount to their safe and evidence-based clinical use. Finally, despite pointing out the above pitfalls, we acknowledge the overall high quality of publications in the field of applying AI methods to mpMRI-driven PCa diagnosis. Improving their methodological quality, the next steps will require a consolidated international and multi-institutional effort, the success of which will primarily depend on the quality of data used for training and validating AI algorithms.
This review has several limitations. The introduction of stringent CLAIM and RQS methodological screening led to the exclusion of several high-quality papers published in high-impact journals, such as Journal of Magnetic Resonance Imaging, European Radiology, and Cancers. This approach, which we previously adopted for another review [57], allowed us to only include studies that are reproducible. It is, however, important to acknowledge that the CLAIM requirements are harder to fulfil compared to the RQS ones. We also acknowledge that some relevant studies may not have been included, particularly those published between our search and publication of this review. Due to the considerable heterogeneity of studies, we did not pool the data for a formal comparison of the diagnostic accuracy of fully-automated and semi-automated AI methods. This was, however, compensated by an extensive narrative synthesis that identified common pitfalls and inconsistencies of the included studies that formed the basis of their heterogeneity.
Conclusions
In conclusion, we observed comparable performance of fully-automated and semi-automated MRI-derived AI methods for differentiating csPCa from iPCa and benign disease. In-depth CLAIM and RQS methodological quality assessment of the studies included in this review revealed several important pitfalls that limit clinical applicability and generalisability of the vast majority of the proposed predictive models. These include, but are not limited to, the use of single-centre datasets without external test cohorts, lack of multi-reader image segmentation, use of inappropriate ground truth assessment methods, and insufficient reporting of model evaluation metrics that can inform their interpretability and clinical applicability. Future studies that address these limitations will help to unlock the disruptive potential of AI and harness the benefits of expert-quality mpMRI-driven PCa diagnosis for the wider community.
Supplementary Information
Additional file 1. Master document summarising the review guidance and results of CLAIM, RQS, and QUADAS-2 assessment.
Additional file 2. PRISMA-2020 checklist.
Additional file 3. Supplementary Methods.
Abbreviations
- ADC
Apparent diffusion coefficient
- AI
Artificial intelligence
- AUC
Area under the receiver operating characteristic curve
- CART
Classification and regression trees
- CLAIM
Checklist for Artificial Intelligence in Medical Imaging
- CNN
Convolutional neural network
- csPCa
Clinically significant prostate cancer
- DL
Deep learning
- GLCM
Grey level co-occurrence matrix
- HP
Histopathology
- iPCa
Indolent prostate cancer
- LASSO
Least absolute shrinkage and selection operator
- LinR
Linear regression
- LQDA
Linear and quadratic discriminant analysis
- LR
Logistic regression
- mpMRI
Multiparametric MRI
- NB
Naïve Bayes
- NPV
Negative predictive value
- NR
Not reported
- PCa
Prostate cancer
- PI-RADS
Prostate imaging–reporting and data system
- PPV
Positive predictive value
- PSA
Prostate-specific antigen
- PSAd
Prostate-specific antigen density
- PZ
Peripheral zone
- QUADAS-2
Quality Assessment of Diagnostic Accuracy Studies
- RF
Random forests
- RQS
Radiomics quality score
- SVM
Support-vector machine
- TML
Traditional machine learning
- TZ
Transition zone
- WP
Whole prostate
Authors' contributions
NS formulated the review concept, registered the review, contributed to all stages of the review process, and wrote the first draft of the manuscript. NMDS, MY, and TB contributed to the review registration and all stages of the review process. ES, MR, and LR oversaw the review conceptualisation, registration, and execution. All authors contributed equally to the preparation of the final version of the manuscript. All authors read and approved the final manuscript
Funding
National Institute of Health Research Cambridge Biomedical Research Centre. Award Number: BRC-1215-20014|Recipient: Not applicable. Cancer Research UK. Award Number: C197/A16465|Recipient: Not applicable. Engineering and Physical Sciences Research Council Imaging Centre in Cambridge and Manchester. Award Number: None|Recipient: Not applicable. Cambridge Experimental Cancer Medicine Centre. Award Number: None|Recipient: Not applicable. The Mark Foundation for Cancer Research and Cancer Research UK Cambridge Centre. Award Number: C9685/A25177|Recipient: Not applicable. CRUK National Cancer Imaging Translational Accelerator (NCITA). Award Number: C42780/A27066|Recipient: Not applicable. Wellcome Trust Innovator Award. Award Number: 215733/Z/19/Z. Recipient: Not applicable.
Availability of data and materials
All data generated or analysed using this study are included in this published article and its supplementary information files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
N.M.D.S., M.R., and L.R. are Machine Learning Consultants for Lucida Medical. E.S is the Chief Medical Officer of Lucida Medical.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Michael Roberts and Leonardo Rundo have contributed equally to this work
References
- 1.Drost FJH, Osses D, Nieboer D, et al. Prostate magnetic resonance imaging, with or without magnetic resonance imaging-targeted biopsy, and systematic biopsy for detecting prostate cancer: a cochrane systematic review and meta-analysis. Eur Urol. 2020;77:78–94. doi: 10.1016/j.eururo.2019.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Sathianathen NJ, Omer A, Harriss E, et al. Negative predictive value of multiparametric magnetic resonance imaging in the detection of clinically significant prostate cancer in the prostate imaging reporting and data system era: a systematic review and meta-analysis. Eur Urol. 2020;8(3):402–414. doi: 10.1016/j.eururo.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 3.Kasivisvanathan V, Stabile A, Neves JB, et al. Magnetic resonance imaging-targeted biopsy versus systematic biopsy in the detection of prostate cancer: a systematic review and meta-analysis. Eur Urol. 2019;76:284–303. doi: 10.1016/J.EURURO.2019.04.043. [DOI] [PubMed] [Google Scholar]
- 4.Hansen NL, Koo BC, Gallagher FA, et al. Comparison of initial and tertiary centre second opinion reads of multiparametric magnetic resonance imaging of the prostate prior to repeat biopsy. Eur Radiol. 2017;27:2259–2266. doi: 10.1007/S00330-016-4635-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park KJ, Choi SH, Lee JS, Kim JS, Kim MH. Interreader agreement with prostate imaging reporting and data system version 2 for prostate cancer detection: a systematic review and meta-analysis. J Urol. 2020;204:661–670. doi: 10.1097/JU.0000000000001200. [DOI] [PubMed] [Google Scholar]
- 6.de Rooij M, Israël B, Tummers M, et al. ESUR/ESUI consensus statements on multi-parametric MRI for the detection of clinically significant prostate cancer: quality requirements for image acquisition, interpretation and radiologists’ training. Eur Radiol. 2020 doi: 10.1007/s00330-020-06929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sushentsev N, Caglic I, Sala E, et al. The effect of capped biparametric magnetic resonance imaging slots on weekly prostate cancer imaging workload. Br J Radiol. 2020;93:20190929. doi: 10.1259/bjr.20190929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penzkofer T, Padhani AR, Turkbey B, et al. ESUR/ESUI position paper: developing artificial intelligence for precision diagnosis of prostate cancer using magnetic resonance imaging. Eur Radiol. 2021 doi: 10.1007/S00330-021-08021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanzione A, Gambardella M, Cuocolo R, Ponsiglione A, Romeo V, Imbriaco M, et al. Prostate MRI radiomics: a systematic review and radiomic quality score assessment. Eur J Radiol. 2020;129:109095. doi: 10.1016/j.ejrad.2020.109095. [DOI] [PubMed] [Google Scholar]
- 10.Cuocolo R, Cipullo MB, Stanzione A, et al. Machine learning for the identification of clinically significant prostate cancer on MRI: a meta-analysis. Eur Radiol. 2020;30:6877–6887. doi: 10.1007/S00330-020-07027-W. [DOI] [PubMed] [Google Scholar]
- 11.Syer T, Mehta P, Antonelli M, et al. Artificial intelligence compared to radiologists for the initial diagnosis of prostate cancer on magnetic resonance imaging: a systematic review and recommendations for future studies. Cancers (Basel) 2021 doi: 10.3390/CANCERS13133318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castillo JMT, Arif M, Niessen WJ, Schoots IG, Veenland JF. Automated classification of significant prostate cancer on MRI: a systematic review on the performance of machine learning applications. Cancers (Basel) 2020;12:1–13. doi: 10.3390/CANCERS12061606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Twilt JJ, van Leeuwen KG, Huisman HJ, Fütterer JJ, de Rooij M. Artificial intelligence based algorithms for prostate cancer classification and detection on magnetic resonance imaging: a narrative review. Diagnostics (Basel) 2021 doi: 10.3390/DIAGNOSTICS11060959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021 doi: 10.1136/BMJ.N71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Covidence Systematic Review Software (Veritas Health Innovation, 2021). https://www.covidence.org/. Accessed 14 Oct 2021
- 16.Mongan J, Moy L, Kahn CE. Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiol Artif Intell. 2020;2:e200029. doi: 10.1148/RYAI.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambin P, Leijenaar RTH, Deist TM, et al. (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;1412(14):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 18.QUADAS-2 | Bristol Medical School: Population Health Sciences | University of Bristol. https://www.bristol.ac.uk/population-health-sciences/projects/quadas/quadas-2/. Accessed 14 Oct 2021
- 19.Macaskill P, Gatsonis C, Deeks JJ, Harbord RM, Takwoingi Y (2010) Cochrane handbook for systematic reviews of diagnostic test accuracy chapter 10 analysing and presenting results. Available via http://methods.cochrane.org/sites/methods.cochrane.org.sdt/files/uploads/Chapter%2010%20-%20Version%201.0.pdf
- 20.Wang Y, Wang M. Selecting proper combination of mpMRI sequences for prostate cancer classification using multi-input convolutional neuronal network. Phys Med. 2020;80:92–100. doi: 10.1016/J.EJMP.2020.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez-Quilez A, Eftestøl T, Goodwin M, Kjosavik SR, Oppedal K (2021) Self-transfer learning via patches: a prostate cancer triage approach based on bi-parametric MRI. Med Image Anal
- 22.Schelb P, Kohl S, Radtke JP, et al. Classification of cancer at prostate MRI: deep learning versus clinical PI-RADS assessment. Radiology. 2019;293:607–617. doi: 10.1148/RADIOL.2019190938. [DOI] [PubMed] [Google Scholar]
- 23.Deniffel D, Abraham N, Namdar K, et al. Using decision curve analysis to benchmark performance of a magnetic resonance imaging-based deep learning model for prostate cancer risk assessment. Eur Radiol. 2020;30:6867–6876. doi: 10.1007/S00330-020-07030-1. [DOI] [PubMed] [Google Scholar]
- 24.Seetharaman A, Bhattacharya I, Chen LC, et al. Automated detection of aggressive and indolent prostate cancer on magnetic resonance imaging. Med Phys. 2021;48:2960–2972. doi: 10.1002/MP.14855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonekamp D, Kohl S, Wiesenfarth M, et al. Radiomic machine learning for characterization of prostate lesions with MRI: comparison to ADC values. Radiology. 2018;289:128–137. doi: 10.1148/RADIOL.2018173064. [DOI] [PubMed] [Google Scholar]
- 26.Min X, Li M, Dong D, et al. Multi-parametric MRI-based radiomics signature for discriminating between clinically significant and insignificant prostate cancer: Cross-validation of a machine learning method. Eur J Radiol. 2019;115:16–21. doi: 10.1016/J.EJRAD.2019.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Kwon D, Reis IM, Breto AL, et al. Classification of suspicious lesions on prostate multiparametric MRI using machine learning. J Med Imaging. 2018;5:1. doi: 10.1117/1.JMI.5.3.034502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castillo TJM, Starmans MPA, Arif M, et al. A multi-center, multi-vendor study to evaluate the generalizability of a radiomics model for classifying prostate cancer: high grade vs. low grade. Diagnostics (Basel) 2021;11:369. doi: 10.3390/DIAGNOSTICS11020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bleker J, Kwee TC, Dierckx RA, de Jong IJ, Huisman H, Yakar D. Multiparametric MRI and auto-fixed volume of interest-based radiomics signature for clinically significant peripheral zone prostate cancer. Eur Radiol. 2020;30:1313–1324. doi: 10.1007/S00330-019-06488-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Chen T, Zhao W, et al. Radiomics prediction model for the improved diagnosis of clinically significant prostate cancer on biparametric MRI. Quant Imaging Med Surg. 2020;10:368–379. doi: 10.21037/qims.2019.12.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woźnicki P, Westhoff N, Huber T, et al. Multiparametric MRI for prostate cancer characterization: combined use of radiomics model with PI-RADS and clinical parameters. Cancers (Basel) 2020;12:1–14. doi: 10.3390/CANCERS12071767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bevilacqua A, Mottola M, Ferroni F, Rossi A, Gavelli G, Barone D. The primacy of high B-value 3T-DWI radiomics in the prediction of clinically significant prostate cancer. Diagnostics (Basel) 2021 doi: 10.3390/DIAGNOSTICS11050739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toivonen J, Perez IM, Movahedi P, et al. Radiomics and machine learning of multisequence multiparametric prostate MRI: towards improved non-invasive prostate cancer characterization. PLoS One. 2019 doi: 10.1371/JOURNAL.PONE.0217702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antonelli M, Johnston EW, Dikaios N, et al. Machine learning classifiers can predict Gleason pattern 4 prostate cancer with greater accuracy than experienced radiologists. Eur Radiol. 2019;29:4754–4764. doi: 10.1007/S00330-019-06244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoo S, Gujrathi I, Haider MA, Khalvati F. Prostate cancer detection using deep convolutional neural networks. Sci Rep. 2019 doi: 10.1038/S41598-019-55972-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiremath A, Shiradkar R, Fu P, et al. An integrated nomogram combining deep learning, Prostate Imaging-Reporting and Data System (PI-RADS) scoring, and clinical variables for identification of clinically significant prostate cancer on biparametric MRI: a retrospective multicentre study. Lancet Digit Heal. 2021;3:e445–e454. doi: 10.1016/S2589-7500(21)00082-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanyal J, Banerjee I, Hahn L, Rubin D. An automated two-step pipeline for aggressive prostate lesion detection from multi-parametric MR sequence. AMIA Summits Transl Sci Proc. 2020;2020:552. [PMC free article] [PubMed] [Google Scholar]
- 38.Cao R, Zhong X, Afshari S, et al. Performance of deep learning and genitourinary radiologists in detection of prostate cancer using 3-T multiparametric magnetic resonance imaging. J Magn Reson Imaging. 2021;54:474–483. doi: 10.1002/JMRI.27595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao R, Namdar K, Liu L, Haider MA, Khalvati F. A comprehensive study of data augmentation strategies for prostate cancer detection in diffusion-weighted MRI using convolutional neural networks. J Digit Imaging. 2021;34:862–876. doi: 10.1007/S10278-021-00478-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong X, Cao R, Shakeri S, et al. Deep transfer learning-based prostate cancer classification using 3 Tesla multi-parametric MRI. Abdom Radiol (NY) 2019;44:2030–2039. doi: 10.1007/S00261-018-1824-5. [DOI] [PubMed] [Google Scholar]
- 41.Zabihollahy F, Ukwatta E, Krishna S, Schieda N. Fully automated localization of prostate peripheral zone tumors on apparent diffusion coefficient map MR images using an ensemble learning method. J Magn Reson Imaging. 2020;51:1223–1234. doi: 10.1002/jmri.26913. [DOI] [PubMed] [Google Scholar]
- 42.Aldoj N, Lukas S, Dewey M, Penzkofer T. Semi-automatic classification of prostate cancer on multi-parametric MR imaging using a multi-channel 3D convolutional neural network. Eur Radiol. 2020;30:1243–1253. doi: 10.1007/s00330-019-06417-z. [DOI] [PubMed] [Google Scholar]
- 43.Kohl S, Bonekamp D, Schlemmer H-P et al (2017) Adversarial networks for the detection of aggressive prostate cancer. arXiv
- 44.Winkel DJ, Breit HC, Shi B, Boll DT, Seifert HH, Wetterauer C. Predicting clinically significant prostate cancer from quantitative image features including compressed sensing radial MRI of prostate perfusion using machine learning: comparison with PI-RADS v2 assessment scores. Quant Imaging Med Surg. 2020;10:808–823. doi: 10.21037/qims.2020.03.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen T, Li M, Gu Y, et al. Prostate cancer differentiation and aggressiveness: assessment with a radiomic-based model vs PI-RADS v2. J Magn Reson Imaging. 2019;49:875–884. doi: 10.1002/JMRI.26243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Algohary A, Shiradkar R, Pahwa S, et al. Combination of peri-tumoral and intra-tumoral radiomic features on Bi-parametric MRI accurately stratifies prostate cancer risk: a multi-site study. Cancers (Basel) 2020;12:1–14. doi: 10.3390/CANCERS12082200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giannini V, Mazzetti S, Cappello G, et al. Computer-aided diagnosis improves the detection of clinically significant prostate cancer on multiparametric-MRI: a multi-observer performance study involving inexperienced readers. Diagnostics (Basel) 2021 doi: 10.3390/DIAGNOSTICS11060973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Latifoltojar A, Dikaios N, Ridout A, et al. Evolution of multi-parametric MRI quantitative parameters following transrectal ultrasound-guided biopsy of the prostate. Prostate Cancer Prostatic Dis. 2015;184(18):343–351. doi: 10.1038/pcan.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. Computer-aided detection of prostate cancer in MRI. IEEE Trans Med Imaging. 2014 doi: 10.1109/TMI.2014.2303821. [DOI] [PubMed] [Google Scholar]
- 50.Greer MD, Shih JH, Barrett T, et al. All over the map: an interobserver agreement study of tumor location based on the PI-RADSv2 sector map. J Magn Reson Imaging. 2018;48:482–490. doi: 10.1002/JMRI.25948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montagne S, Hamzaoui D, Allera A, et al. Challenge of prostate MRI segmentation on T2-weighted images: inter-observer variability and impact of prostate morphology. Insights Imaging. 2021 doi: 10.1186/S13244-021-01010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greer MD, Shih JH, Lay N, et al. Interreader variability of prostate imaging reporting and data system version 2 in detecting and assessing prostate cancer lesions at prostate MRI. AJR Am J Roentgenol. 2019;212:1197–1205. doi: 10.2214/AJR.18.20536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cuocolo R, Stanzione A, Castaldo A, De Lucia DR, Imbriaco M. Quality control and whole-gland, zonal and lesion annotations for the PROSTATEx challenge public dataset. Eur J Radiol. 2021 doi: 10.1016/J.EJRAD.2021.109647. [DOI] [PubMed] [Google Scholar]
- 54.Castiglioni I, Rundo L, Codari M, et al. AI applications to medical images: from machine learning to deep learning. Phys Med. 2021;83:9–24. doi: 10.1016/J.EJMP.2021.02.006. [DOI] [PubMed] [Google Scholar]
- 55.Schwier M, van Griethuysen J, Vangel MG, et al. Repeatability of multiparametric prostate MRI radiomics features. Sci Rep. 2019;9:1–16. doi: 10.1038/s41598-019-45766-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J, Steinmann A, Ding Y, et al. Radiomics feature robustness as measured using an MRI phantom. Sci Rep. 2021;111(11):1–14. doi: 10.1038/s41598-021-83593-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roberts M, Driggs D, Thorpe M, et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell. 2021;33(3):199–217. doi: 10.1038/s42256-021-00307-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Master document summarising the review guidance and results of CLAIM, RQS, and QUADAS-2 assessment.
Additional file 2. PRISMA-2020 checklist.
Additional file 3. Supplementary Methods.
Data Availability Statement
All data generated or analysed using this study are included in this published article and its supplementary information files.