Abstract
PCR was used to identify antibiotic resistance determinants in 31 Canadian Salmonella serovar Typhimurium DT104 isolates. Genes encoding resistance to ampicillin (pse1 or blaP1), chloramphenicol (pasppflo-like), streptomycin-spectinomycin (aadA2), sulfonamide (sulI), and tetracycline [tet(G)] were mapped to a 13-kb region of DNA of one isolate. Two copies of sulI were identified and mapped to the 3′ end of either pse1 or aadA2 integrons. The two integrons were separated by the pasppflo-like gene and the tet(G) gene. The kanamycin resistance determinant (aphA-1) was present on a 2.0-MDa plasmid (five isolates) or on the chromosome (three isolates).
Recently in Canada there has been an increase in occurrence of Salmonella serovar Typhimurium DT104 isolates (17, 18). Many of these isolates are resistant to ampicillin (A), chloramphenicol (C), streptomycin (S), sulfonamide (Su), and tetracycline (T), like those found in the United Kingdom (23) and the United States (7, 9), and some are also kanamycin resistant (KANr). This study describes the identification of the resistant genes involved in 31 Canadian isolates of Salmonella serovar Typhimurium DT104 as well as the location and gene order in a selected isolate (96-5227) with the ACSSuT resistance phenotype.
The antimicrobial susceptibilities (15, 20) of the 31 Salmonella serovar Typhimurium DT104 are shown in Table 1, and the PCR primers designed to identify their resistance genes are shown in Table 2. PCR conditions were optimized based on the melting temperature (Tm) of the primers, and long PCR was performed in accordance with the supplier’s protocol (Roche Diagnostics, Laval, Quebec, Canada). All primers were synthesized with a Beckman Oligo 1000M DNA synthesizer, and DNA sequencing was carried out with an ABI Prism 377 DNA sequencer (Applied Biosystems Division of Perkin-Elmer, Foster City, Calif.). DNA sequences were aligned with sequences in the National Center for Biotechnology Information database by using Blast version 2.0.4 (1).
TABLE 1.
Source, antibiograms, antibiotic resistance genes, and plasmid profiles of Canadian Salmonella serovar Typhimurium DT104 isolates
| Source | Total no. of isolates | Antibiograma | Genotype identified | Plasmid size profile (MDa) |
|---|---|---|---|---|
| Human | 6 | ACSSuTr | pse1 pasppflo-likecaadA2 sulI tet(G) | 60 |
| 3 | ACSSuT-KANr | pse1 aadA2 sulI tet(G) aphA-1 | 60, 2.0 | |
| 1 | ACSuTr | pse1 sulI tet(G) | 60 | |
| 1 | SSuTr | tet(A) | 60 | |
| 3 | T-KANr | tet(B) aphA-1 | 60 | |
| 6 | Susceptibleb | 60 | ||
| 1 | Susceptibleb | 60, 2.2, 1.4 | ||
| Nonhuman | 2 | ACSSuTr | pse1 aadA2 sulI tet(G) | 60 |
| 1 | ACSSuT-KANr | pse1 aadA2 sulI tet(G) aphA-1 | 60, 2.0 | |
| 5 | ACSuTr | pse1 sulI tet(G) | 60 | |
| 2 | Susceptibleb | 60 | ||
| Unknown | 14d | ASSu-TMPr | NAf | 60, 50, 4.6, 3.7 |
| 16e | ASSu-KANr | sulI aphA-1 | 60, 3.4, 2.3, 2.0, 1.4 |
TMPr, trimethoprim resistant.
Susceptible to all antibiotics tested.
Putative chloramphenicol resistance determinant (2, 5), identified in strain 96-5227 by the PCR method.
A phage-type-atypical (AT) isolate containing TMP resistance on a self-transmissible 4.6-MDa plasmid was used as a control for the mating experiment.
Phage type PT120.
NA, unable to be determined in this study.
TABLE 2.
PCR primers used in the identification of integrons and resistance genes
| Gene | PCR primer sequences 5′-3′ | Expected size (bp) | GenBank accession no. | Reference strain (plasmid) | Source (reference) |
|---|---|---|---|---|---|
| Integron | 5′-CS, GGC ATC CAA GCA GCA AG; 3′-CS, AAG CAG ACT TGA CCT GA | ?a | M73819 | NAb | P. Roy (11) |
| pse1 | L, AAT GGC AAT CAG CGC TTC CC; R, GGG GCT TGA TGC TCA CTC CA | 586 | Z18955 | Salmonella serovar Newport E1572 | G. Jacoby (10) |
| aadA2 | L, TGT TGG TTA CTG TGG CCG TA; R, GCT GCG AGT TCC ATA GCT TC | 381 | X68227 | Escherichia coli(pSa) | D. Taylor (3) |
| sulI | qacEΔ1, TAG TTG GCG AAG TAA TCG CA; orf5, AGC TTG TGC AGA TAT GCG G; sulI, TGA AGG TTC GAC AGC AC | 1,581 | U49101 | Proteus mirabilis 88071820 | P. Roy (11) |
| aphA-1 | L, TTA TGC CTC TTC CGA CCA TC; R, GAG AAA ACT CAC CGA GGC AG | 489 | U63147 | Escherichia coli JE2571(pHH1457) | D. Taylor (4) |
| intI1 | L, GCC TTG CTG TTC TTC TAC GG; R, GAT GCC TGC TTG TTC TAC GG | 558 | X12870 | NA | P. Roy (11) |
| pasppflo-like | L, CAC GTT GAG CCT CTA TAT GG; R, ATG CAG AAG TAG AAC GCG AC | 868 | AFO71555 | NA | GenBank (5) |
| tet(A) | L, GCT ACA TCC TGC TTG CCT TC; R, CAT AGA TCG CCG TGA AGA GG | 210 | X61367 | Escherichia coli D20-15(pSL18) | S. Levy (14) |
| tet(B) | L, TTG GTT AGG GGC AAG TTT TG; R, GTA ATG GGC CAA TAA CAC CG | 659 | J01830 | Escherichia coli D20-16(pRT11) | S. Levy (13) |
| tet(C) | L, CTT GAG AGC CTT CAA CCC AG; R, ATG GTC GTC ATC TAC CTG CC | 418 | J01749 | Escherichia coli D20-6(pBR322) | S. Levy (13) |
| tet(D) | L, AAA CCA TTA CGG CAT TCT GC; R, GAC CGG ATA CAC CAT CCA TC | 787 | L06798 | Escherichia coli D22-2(pSL106) | S. Levy (13) |
| tet(E) | L, AAA CCA CAT CCT CCA TAC GC; R, AAA TAG GCC ACA ACC GTC AG | 278 | L06940 | Escherichia coli HB101(pSL1504) | S. Levy (12) |
| tet(G) | L, GCT CGG TGG TAT CTC TGC TC; R, AGC AAC AGA ATC GGG AAC AC | 468 | S52437 | Escherichia coli(pJA8122) | T. Aoki (24) |
| tet(G) | L, CAG CTT TCG GAT TCT TAC GG; R, GAT TGG TGA GGC TCG TTA GC | 844 | S52437 | Escherichia coli(pJA8122) | T. Aoki (24) |
| tet(S) | L, CAT AGA CAA GCC GTT GAC C; R, ATG TTT TTG GAA CGC CAG AG | 667 | X92946 | Escherichia coli(pAT451) | M. Roberts (6) |
?, variable size depending on the inserted gene(s).
NA, not available.
To obtain a detailed map and elucidate the order of the ACSSuT resistance phenotype genes, Southern blots of isolate 96-5227 DNA digested with BamHI, HindIII, EcoRI, BglI, XbaI, and corresponding double digests were probed sequentially with amplicons of the identified genes.
Class 1 integrons which have conserved regions (5′CS and 3′CS) often contain antimicrobial resistance gene cassettes (8, 19). PCR amplification using primers 3′-CS and 5′-CS on genomic DNA from all of the DT104 ACSSuT-resistant isolates resulted in 1.0- and 1.2-kb amplicons. DNA sequence analyses (1) of the 1.0-kb amplicon from one selected isolate (96-5227) showed a sequence identical to that of a region on InCg, an integron on Corynebacterium glutamicum plasmid pCG4 (emb|;Y14748), containing aadA2, which encodes streptomycin-spectinomycin resistance (3, 16), while the 1.2-kb amplicon contained sequence with 100% identity to pse1 (blaP1) (carb-2, emb|Z18955, coordinates 35 to 1150), which encodes ampicillin resistance (25). In addition, PCR amplification using intragenic primers (Table 2) identified the presence of pse1 in the 1.2-kb amplicons from 18 (8 ACSSuTr, 4 ACSSuT-KANr, and 6 ACSuTr) isolates and of aadA2 in the 1.0-kb amplicon of 12 (8 ACSSuTr and 4 ACSSuT-KANr) isolates (Table 1). Other studies also found the pse1 and aadA2 genes within 1.0- and 1.2-kb amplicons in DT104 isolates with ACSSuT resistance (5, 20, 21).
Class 1 integrons may contain the qacEΔ1-sulI (sulfonamide resistance)-orf5 region (8, 19). All 18 isolates that had 5′CS and 3′CS sequence produced a PCR amplicon with qacEΔ1 and orf5 primers (Table 2). Partial DNA sequence analysis and PCR using intragenic sulI primers confirmed the presence of the sulfonamide resistance gene in these isolates. Furthermore, these isolates also contained the integrase (intI1) gene sequence as determined by PCR.
PCR using primers specific for tetracycline resistance genes (Table 2) showed that tet(A) was identified in one isolate with an SSuT resistance profile while tet(B) was found in three isolates with T-KANr profiles. In addition, tet(G) was identified in 18 DT104 isolates with ACSSuT resistance, ACSSuT-KANr, and ACSuT resistance profiles. DNA sequence analysis of the tet(G) PCR product from isolate 96-5227 (gb|AF11924) showed 100% identity with published sequences from Salmonella serovar Typhimurium DT104 (gb|AF071555) (5) and Pseudomonas plasmid pPSTG2 (gb|AF133140) and 93% identity (CG instead of GC at coordinates 1351 and 1352 of gb|S52437) with that of Vibrio anguillarum (24).
The chloramphenicol resistance determinant in 96-5227 was identified as the previously described pasppflo-like (flor) gene (2, 5) by using PCR primers (Fig. 1) specific for this gene (5) (gb|AF071555).
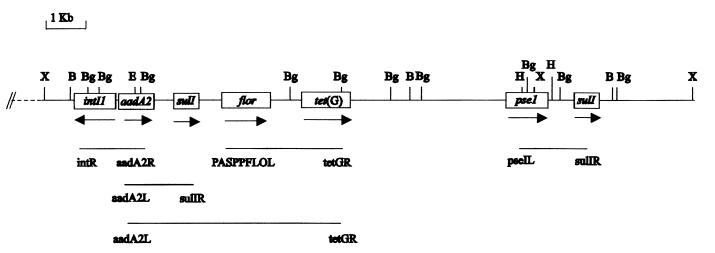
FIG. 1.
Restriction map of Salmonella serovar Typhimurium DT104 multidrug resistance region from isolate 96-5227. The orientations of the genes are indicated by arrows. The respective positions of the antibiotic resistance genes are shown in boxes. Lines below the map indicate the sizes of products obtained by using the primer sets listed. Restriction enzyme abbreviations: X, XbaI; B, BamHI; Bg, BglI; H, HindII.
All of the identified resistance genes as well as the integrase gene were mapped to an 11.8-kb XbaI fragment, and their order is shown in Fig. 1. Although Ridley and Threlfall (20) have suggested that two integron-associated genes in DT104 isolates map to a single 10-kb XbaI chromosomal fragment, our results showed that the pse1 gene, which has an internal XbaI site, actually extends to an adjacent 4.2-kb XbaI fragment (Fig. 1). This difference is probably due to the elution of the 4.2-kb XbaI fragment from their pulsed-field gel electrophoresis gels (20). The relative positions and orientation of genes on the restriction map were confirmed by using various combinations of PCR primers (Fig. 1). The structure of the integron containing aadA2 is similar to the map published by Briggs and Fratamico (5). However, we did not detect the partial integrase gene (groEL-intI1b) (5) in the 5′ region of the pse1 gene by hybridization with the intI1 probe.
It had been reported that the ACSSuT resistance genes in DT104 were located on the chromosome and were not self-transmissible (22, 23). All ACSSuT-resistant isolates in this study harbored only the 60-MDa virulence plasmid which has not been reported to confer antimicrobial resistance (23). Furthermore, this plasmid was also found in susceptible isolates (Table 1). Therefore, it is likely that the ACSSuT resistance determinants are on the chromosome of isolate 96-5227.
Kanamycin resistance of eight isolates, including four with ACSSuT-KANr, was encoded by the aphA-1 gene. The amplicons were verified to contain aphA-1 by AluI and DdeI digestion (gb|U63147). The aphA-1 probe hybridized to a 2-MDa plasmid present in five of the eight isolates (Table 1). This suggests that the other three isolates, containing only the 60-MDa plasmid, have aphA-1 located on the chromosome. Mating experiments showed that the 2-MDa plasmid in a selected Salmonella serovar Typhimurium isolate 97-5025 was not self-transmissible.
In summary, the order, orientation, and location of the ACSSuT resistance genes in the Canadian isolate (96-5227) were similar to those found by others (5, 20, 21). Although the origin of DNA containing the ACSSuT resistance genes remains unknown, our observations together with other published data (5, 20, 21) suggested the possible clonal dissemination of these isolates in the population. The rapid increase of ACSSuT-resistant DT104 isolates in Canada and other countries may suggest that there are other genes in the DNA region which account for the rapid dissemination of this isolate. In an attempt to better understand this unique region of the genome, we have constructed DNA libraries in order to identify other genes and determine the size of the inserted DNA in isolate 96-5227. To this end, sequence analysis and hybridization experiments showed that the insertion site of the unique region is between 24 and 30 kb upstream of the intI1 gene.
Acknowledgments
We thank Rae Bosy and Romeo Hizon for performing the susceptibility testing, G. Wang for the pulsed-field gel electrophoresis, David Woodward for the identification of Salmonella, Rasik Khakhria for phage typing and providing the surveillance data, and Annie Savoie for her technical assistance. We also thank T. Aoki, Miyazaki University, Miyazaki, Japan; D. Taylor, University of Alberta, Edmonton, Alberta, Canada; G. Jacoby, Massachusetts General Hospital, Boston; S. Levy, Tufts University, Boston, Mass.; M. Roberts, University of Washington, Seattle; and P. Roy, Université Laval, Québec, Canada, for providing isolates. We also thank Michael Coulthart, Shaun Tyler, and the staff of the DNA Core Facility, Bureau of Microbiology, Laboratory Centre for Disease Control, for their technical advice and for performing the DNA sequencing of DNA amplicons and the synthesis of oligonucleotides.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcangioli M-A, Leroy-Sétrin S, Martel J-L, Chaslus-Dancla E. A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol Lett. 1999;174:327–332. doi: 10.1111/j.1574-6968.1999.tb13586.x. [DOI] [PubMed] [Google Scholar]
- 3.Bito A, Susani M. Revised analysis of aadA2 gene of plasmid pSa. Antimicrob Agents Chemother. 1994;38:1172–1175. doi: 10.1128/aac.38.5.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley D E, Taylor D E, Cohen D R. Specification of surface mating systems among conjugative drug resistance plasmids in Escherichia coli K-12. J Bacteriol. 1980;143:1400–1470. doi: 10.1128/jb.143.3.1466-1470.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs C E, Fratamico P M. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother. 1999;43:846–849. doi: 10.1128/aac.43.4.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.François B, Charles M, Courvalin P. Conjugative transfer of tet(S) between strains of Enterococcus faecalis is associated with the exchange of large fragments of chromosomal DNA. Microbiology. 1997;143:2145–2154. doi: 10.1099/00221287-143-7-2145. [DOI] [PubMed] [Google Scholar]
- 7.Glynn M K, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo F J. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–1338. doi: 10.1056/NEJM199805073381901. [DOI] [PubMed] [Google Scholar]
- 8.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 9.Hosek G, Leschinsky D D, Irons S, Safranek T J. Multidrug-resistant Salmonella serotype Typhimurium—United States, 1996. Morbid Mortal Weekly Rep. 1997;46:308–310. [PubMed] [Google Scholar]
- 10.Huovinen P, Jacoby G A. Sequence of the PSE-1 β-lactamase gene. Antimicrob Agents Chemother. 1991;35:2428–2430. doi: 10.1128/aac.35.11.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lévesque C, Piché L, Larose C, Roy P H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall B, Morrissey S, Flynn P, Levy S B. A new tetracycline-resistance determinant, class E, isolated from Enterobacteriaceae. Gene. 1986;50:111–117. doi: 10.1016/0378-1119(86)90315-x. [DOI] [PubMed] [Google Scholar]
- 13.Marshall B, Tachibana C, Levy S B. Frequency of tetracycline resistance determinant classes among lactose-fermenting coliforms. Antimicrob Agents Chemother. 1983;24:835–840. doi: 10.1128/aac.24.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendez B, Tachibana C, Levy S B. Heterogeneity of tetracycline resistance determinants. Plasmid. 1980;31:99–108. doi: 10.1016/0147-619x(80)90101-8. [DOI] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. Vol. 17. 1997. , no. 2. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Villanova, Pa. [Google Scholar]
- 16.Nešvera J, Hochmannová J, Pátek M. An integron of class 1 is present on the plasmid pCG4 from Gram-positive bacterium Corynebacterium glutamicum. FEMS Microbiol Lett. 1998;169:391–395. doi: 10.1111/j.1574-6968.1998.tb13345.x. [DOI] [PubMed] [Google Scholar]
- 17.Ng L-K, Khakhria R, Woodward D, Johnson W. National laboratory surveillance of enteric pathogens. Can J Infect Dis. 1997;8:133–136. [Google Scholar]
- 18.Poppe C, Smart N, Khakhria R, Johnson W, Spika J, Prescott J. Salmonella Typhimurium DT104: a virulent and drug-resistant pathogen. Can Vet J. 1998;39:559–565. [PMC free article] [PubMed] [Google Scholar]
- 19.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 20.Ridley A, Threlfall E J. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT104. Microb Drug Resist. 1998;4:113–118. doi: 10.1089/mdr.1998.4.113. [DOI] [PubMed] [Google Scholar]
- 21.Sandvang D, Aarestrup F M, Jensen L B. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 1998;160:37–41. doi: 10.1111/j.1574-6968.1998.tb12887.x. [DOI] [PubMed] [Google Scholar]
- 22.Schmieger H, Schicklmaier P. Transduction of multiple drug resistance of Salmonella enterica serovar typhimurium DT104. FEMS Microbiol Lett. 1999;170:251–256. doi: 10.1111/j.1574-6968.1999.tb13381.x. [DOI] [PubMed] [Google Scholar]
- 23.Threlfall E J, Frost J A, Ward L R, Rowe B. Epidemic in cattle and humans of Salmonella typhimurium DT104 with chromosomally integrated multiple drug resistance. Vet Rec. 1994;134:577. doi: 10.1136/vr.134.22.577. [DOI] [PubMed] [Google Scholar]
- 24.Zhao J, Aoki T. Nucleotide sequence analysis of the class G tetracycline resistance determinant from Vibrio anguillarum. Microbiol Immunol. 1992;36:1051–1060. doi: 10.1111/j.1348-0421.1992.tb02109.x. [DOI] [PubMed] [Google Scholar]
- 25.Zuhlsdorf M T, Wiedemann B. Tn21-specific structures in gram-negative bacteria from clinical isolates. Antimicrob Agents Chemother. 1992;36:1915–1921. doi: 10.1128/aac.36.9.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]