Abstract
The emergence and spread of artemisinin-resistant Plasmodium falciparum, first in the Greater Mekong Subregion (GMS), and now in East Africa, is a major threat to global malaria elimination ambitions. To investigate the artemisinin resistance mechanism, transcriptome analysis was conducted of 577 P. falciparum isolates collected in the GMS between 2016–2018. A specific artemisinin resistance-associated transcriptional profile was identified that involves a broad but discrete set of biological functions related to proteotoxic stress, host cytoplasm remodelling, and REDOX metabolism. The artemisinin resistance-associated transcriptional profile evolved from initial transcriptional responses of susceptible parasites to artemisinin. The genetic basis for this adapted response is likely to be complex.
Subject terms: Parasite genomics, Predictive markers, Evolutionary genetics
Transcriptomic analysis of isolates from the malaria parasite (Plasmodium falciparum) in the Greater Mekong Subregion of Southeast Asia identifies gene expression patterns that are correlated with resistance to a common anti-malaria drug, artemisinin.
Introduction
Artemisinin-based combination therapies (ACTs) have been critical to the success in reducing the global burden of falciparum malaria between 2000 and 20151. Loss of these drugs to resistance would be a disaster. Historically the Greater Mekong Subregion (GMS) has been the origin of antimalarial drug resistance in P. falciparum. In recent years the emergence and spread of artemisinin resistance and, subsequently, partner drug resistance has led to high failure rates of ACTs in several parts of the GMS2,3. Recently artemisinin resistance has arisen independently in East Africa (parts of Rwanda and Uganda)4. The phenotypic manifestation of artemisinin-resistant P. falciparum infections in vivo is slowed parasite clearance after treatment with artesunate. The slow clearance phenotype, defined by a parasite clearance half-life (PC½) > 5 h, is attributed to loss of sensitivity of P. falciparum to artemisinins during the early stage of the intraerythrocytic developmental cycle (IDC), ring stage5,6 and is causally associated with nonsynonymous mutations in the propeller region of the P. falciparum kelch 13 gene (PfK13)7,8. Artemisinin resistance was first reported in western Cambodia, Pailin province in 2009. Initially, over 20 different PfK13 mutations were associated with the slow parasite clearance phenotype8–10. However, since 2013 the soft sweeps resulting in multiple emergences of PfK13 mutations in the eastern part of the GMS were largely replaced by a selective sweep of a haplotype bearing a single nonsynonymous PfK13 SNP (C580Y)11. This single lineage of artemisinin-resistant P. falciparum spread and expanded through western and northern Cambodia, northeastern Thailand and southern Vietnam, and Lao PDR8,12,13. This was soon joined with molecular markers associated with resistance to the ACT partner drug piperaquine. PfK13-propeller domain mutations in artemisinin-resistant parasite lineages have also emerged independently and spread through Myanmar and western Thailand14. Moreover, PfK13-propeller domain mutations have been reported in Northern India15, and more recent foci include independent emergence in Papua New Guinea16, Rwanda4, Ethiopia17, and other parts of sub-Saharan Africa18.
The molecular mechanism by which the PfK13 mutations confer artemisinin resistance is a subject of intense research using in vitro and in vivo models (reviewed in19–23). Collectively, these studies have proposed the involvement of multiple cellular and metabolic processes in artemisinin resistance including haemoglobin degradation, proteotoxic/unfolded protein stress response, vesicular biogenesis as well as oxidative stress response and mitochondrial functions. Translational suppression mediated by phosphorylation of eIF2α, linked to cell quiescence and slowing of the IDC can also confer artemisinin resistance in vitro24. Missense or loss-of-function alleles of other genes were also shown to contribute to artemisinin resistance in vitro. These include coronin25, falcipain2a/b26, ubiquitin hydrolase (pcubp1)27, and μ-subunit of the AP2 vesicular trafficking complex (pcap2)27. Thus, it appears that artemisinin resistance is mediated by a multifaceted mechanism that results from a concerted action of several metabolic and cellular factors. These may act in different, not mutually exclusive, combinations28. Undoubtedly these mechanisms drive artemisinin resistance of P. falciparum in in vitro conditions in which the parasites are supplied with superfluous amounts of nutrients, kept at uniform temperature, are not targeted by the host’s immune system and other ambient stresses exerted by the host’s environment29. The question now is what are the roles of each of these identified components of artemisinin resistance in natural infections, in vivo.
To investigate this, several genome-wide association studies (GWAS) were conducted. These identified large regions on chromosome 10 and 13, and 1430,31, and subsequently seven nonsynonymous SNPs associated with PfK13 SNPs32. A concurrent longitudinal study of the GMS parasites collected between 2001 and 2014 suggested that additional genes might be associated with artemisinin resistance including an additional kelch protein on chromosome 1033. Collectively, these studies detected genes that could be loosely linked with the artemisinin resistance-implicated biological functions, however, no experimental evidence of their role in the artemisinin resistance clinical phenotype has been reported so far.
Transcriptome-wide association analysis (TWAS) is currently emerging as the method of choice for identifying causative genetic variations of complex traits in a wide range of biological systems ranging from plants34 to human35–37. This is based on the wealth of GWAS studies showing that the vast majority of genetic polymorphisms associated with complex genetic traits (such as genetic diseases) lay within the noncoding regions38. These are typically affecting DNA regulatory sequences and thus gene expression through which the phenotype is manifested39. This likely also applies to P. falciparum as suggested by our earlier study investigating expression quantitative trait loci (eQTL) in the TRACI parasite isolates40. We then carried out transcriptome analyses of the P. falciparum parasites from the Tracking Resistance to Artemisinin Collaboration (TRACI) study conducted in the GMS countries between 2011 and 20138,41. This showed that artemisinin resistance is associated with broad transcriptional changes of many genes, some of which may be linked with inductions of the unfolded protein response (UPR) and with a general deceleration of the IDC. Here we present a transcriptome analysis of parasite isolates from a more recent cohort of P. falciparum natural infections collected in the GMS between 2016 and 2018; 5–7 years after the initial TRACI study8, and after the recent selective sweep of PfK13 C580Y2. We identified a spectrum of transcriptionally correlated genes that likely contribute to artemisinin resistance via their altered transcriptional levels. We termed this the artemisinin resistance-associated transcriptional profile (ARTP) and provide evidence that its constitutive expression may have evolved from the initial transcriptional responses of sensitive P. falciparum parasites to the artemisinins.
Results
Transcriptome of the P. falciparum population in the GMS 2016–2018
The main purpose of this study was to identify specific genes, presumably acting in concordance with the PfK13 mutations, whose expression activity mediates/contributes to the physiological state that enables the parasite cell to withstand the parasiticidal effects of artemisinins. We conducted transcriptome analysis of P. falciparum parasites derived from the blood of patients with uncomplicated P. falciparum infections in 13 field sites across 6 GMS countries (Fig. 1a and Supplementary Table 1). These samples were collected during a large clinical treatment trial (TRACII) carried out from 2016 to 2018 that also characterized the spread of artemisinin resistance in the GMS2. We isolated total P. falciparum RNA from the patients’ blood samples and performed both DNA microarray and Next Generation Sequencing (RNA-seq) analysis42. Overall, we analyzed 577 samples collected at study enrolment to characterize the baseline transcriptomic profiles (baseline, (bl)0 h sample set) (Supplementary Fig. 1a). For 459 (of the 577) patients, samples were also collected 6 h after administering an ACT to characterize the transcriptional response of P. falciparum parasites to the artemisinin in vivo (transcriptional response, (tr)6 h sample set) (Supplementary Fig. 1b). While the DNA microarray was used to generate the transcriptomes for all collected samples, there were sufficient levels of parasite mRNA to perform RNA-seq-based transcriptome analyses for 188 and 159 of the (bl)0 h and (tr)6 h samples respectively42 (Supplementary Fig. 1c, d).
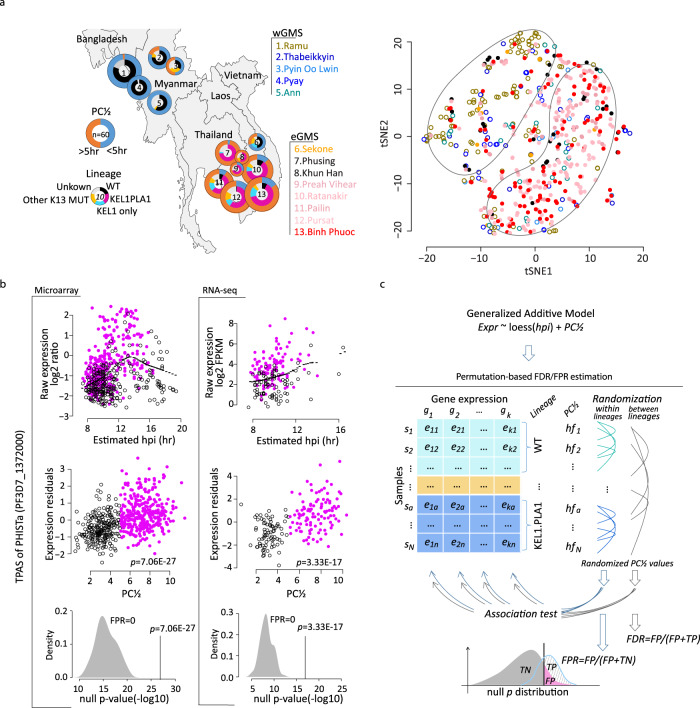
Fig. 1. Transcriptome of TRACII P. falciparum parasite population and a schematic illustration of the TPAS methodology.
a Geographic distribution of all samples used in this study. Pie charts represent the proportion of slow (PC½ > 5 h) and fast (PC½ < 5 h) clearing parasites, or lineages in categories based on PfK13 mutations and plasmepsin II/III copy numbers (WT, KEL1 only, KEL1PLA1, other PfK13 MUT and unknown). On the right, 577 samples before drug treatment are plotted to display two main clusters formed by the eGMS or wGMS parasites geographical distribution using t-SNE algorithm based on the top 2-12 PCs. b TPAS explanation using an example gene of PHISTa (PF3D7_1372000). The expression-hpi/age relationship is shown as the raw expression level (log2 ratio) plotted against the estimated hpi for the microarray data on the left (577 samples) and RNA-seq data on the right (188 samples) with purple dots representing resistant parasites (PC½ > 5 h), black circles for susceptible parasites (PC½ < 5 h) and black dotted lines for the loess curve using all dots. The expression-resistance relationship is represented by the expression residuals plotted against the PC½ for each data set. The density plots at the bottom represent 100 times permutation result within lineages for the FPR calculation. c Workflow of FDR and FPR estimation for the TPAS. The null p distribution was built using permutated resistance status (PC½ values) across parasite samples within lineages for FPR estimation and between lineages for multiple testing correction.
From the (bl)0 h sample set, we established the distribution of the P. falciparum life cycle developmental stages in the peripheral blood including the asexual IDC stages expressed as hours post invasion (hpi) and the fractions of sexual stages (gametocytes)40 (Supplementary Fig. 2 and Supplementary Data 1). The (bl)0 h sample set represented synchronized ring stage parasites between 6 and 20 hpi, with ≤ 5% of the overall parasitemia corresponding to gametocytes. Principle component analysis (PCA) of the (bl)0 h transcriptome revealed parasite hpi composed the major transcriptome difference across samples as it is strongly correlated (Spearman rho = 0.87) with the first principal component (PC1) accounting for up to 32% of the overall transcriptome variation (Supplementary Fig. 3a). A significant non-linear relationship was observed between the hpi and expression (p < 0.001) of most of the studied genes measured by DNA microarrays (70%) and RNA-seq (62%, Supplementary Fig. 4). In contrast to the PC1, none of the subsequent top 2 to 12 PCs (each contributing > 1% of the overall transcriptome variation) correlated with any epidemiological variables (i.e. patient sex, age and time of collection etc.) and molecular biology parameters (i.e., RNA yield, quality assessments etc.) suggesting that the generated dataset was largely unaffected by the methodological approach or epidemiology aspects of the study (Supplementary Fig. 3a). The t-distributed stochastic neighbour embedding (t-SNE) method using the PC2-12 revealed two major parasite groups which separated according to the geographic regions: western GMS (wGMS) and eastern GMS (eGMS) (Fig. 1a). This grouping pattern also corresponds to the prevalence of artemisinin resistance in the eGMS that is demarcated by the expansion of the P. falciparum lineage carrying the PfK13 C580Y mutation (named KEL1 lineage or PfPailin) of which 76% also carried multiple copies of plasmepsin II (KEL1PLA1, Fig. 1a)2. PfK13 wild type parasites from the eGMS did not exhibit any strong association with either group, which further strengthens this model (Supplementary Fig. 3a). Altogether these results suggest that the selective sweep of the PfPailin or KEL1PLA1 lineage(s) in the eGMS over the last 5–7 years43 is mirrored by transcriptional convergence with the eGMS parasites that is distinct from the wGMS. This transcriptome pattern is presumably either contributing to the resistance mechanism and/or alleviating a fitness cost to support its selection.
Transcriptome-phenotype association study of artemisinin resistance
In human genetics, TWAS typically combine GWAS of a studied phenotype on one side and independent datasets linking DNA elements with transcription on the other36,37. By the virtue of being a unicellular organism, in P. falciparum, we can correlate transcriptional differences with the studied phenotype (here artemisinin resistance) directly; the approach we term Transcriptome-Phenotype Association Studies (TPAS) (see “Methods”). First, we conducted TPAS analysis on the (bl)0 h samples in order to identify genes whose steady state mRNA levels correlated with the level of artemisinin resistance represented by PC½. For this we applied a generalized additive model to relate each gene’s expression to the PC½ with a loess function along hpi (example in Fig. 1b). In this model, the expression residuals reflect the relative mRNA abundance unaffected by hpi and thus can be directly correlated to PC½2. To control for multiple testing in correlating between mRNA levels and PC½ values for the whole transcriptome, we calculated the false discovery rate (FDR) for each gene based on 1000 permutations (Fig. 1c). The expression-resistance association could also represent an expression-lineage relationship since the artemisinin resistance status is confounded with K13 lineages that 77% of the susceptible (PC½ < 5 h) samples were from the WT parasites in wGMS and 95% of the resistant (PC½ < 5 h) samples were from the K13 mutant in eGMS (Fig. 1a). Due to this homogeneity, expression-resistance associations will have a lineage effect if we use the entire data set for TPAS analysis or lose power if we test only within sub geographical region (w/eGMS). To overcome this, we estimated false positive rates (FPR) for each gene to control the type I error by repeating the analysis 100 times with randomly generated permutations (Fig. 1c), a method commonly used in human genetics studies44,45. In each permutation, the lineage structure was maintained and PC½ values were randomized amongst the parasites within each lineage. Subsequently FPR was calculated based on the null p distribution reflecting the probability of expression-resistance association caused by expression-lineage relationship. FPR < 0.05 (95% confidence) was applied to define robust expression-resistance associations beyond parasite lineage effect. By this we aimed to eliminate expression-lineage associations and identified transcripts whose expression levels are associated strictly with the PC½. One of the clearest examples was PHISTa (PF3D7_1372000) which displayed a strong association with artemisinin resistance at p = 7.06E-27 (FDR = 0) with FPR = 0 (Fig. 1b, for more examples see Supplementary Fig. 5).
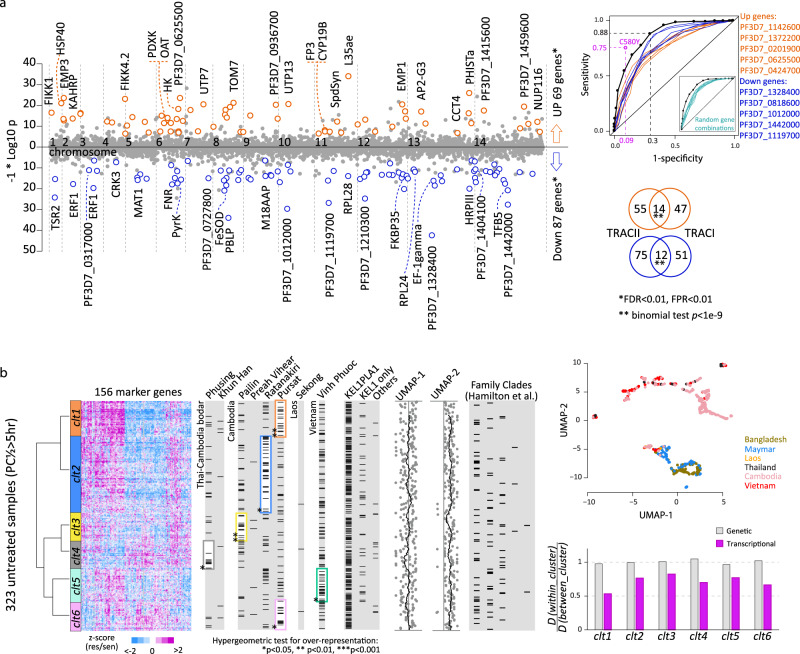
Next, we applied the above TPAS method to the DNA microarray-derived data of 577 (bl)0 h samples and RNA-seq-derived data of 188 (bl)0 hr samples separately. We observed good correlation (Spearman rho = 0.68) between the TPAS results obtained from these two platforms (Supplementary Fig. 6). Merging the two results, we identified 69 upregulated and 87 downregulated transcripts whose levels were significantly associated with artemisinin resistance (FDR < 0.05, corresponding to p < 1e−10 in microarray, p < 1e−6 in RNA-seq, note: these criteria were subsequently used to define the ARTP, Fig. 2a). Out of these, 60S ribosomal protein gene, L35ae (PF3D7_1142600), PHISTa (PF3D7_1372000) erythrocyte membrane protein PfEMP3 (PF3D7_0219000), and PF3D7_1328400, BEM46 (PF3D7_0818600), and PF3D7_1012000 showed the strongest correlations with PC½ for up- and downregulated genes, respectively. Overall, both the upregulated and downregulated genes contribute to a broad but defined set of biological functions previously linked with artemisinin resistance such as protein and REDOX metabolism, digestive vacuole- and mitochondrial-linked biological functions, host cell remodelling etc. (Supplementary Data 2, also see below). When compared to the transcripts identified by the same approach in parasites collected during TRACI (between 2011–2013)41, we observed a significant overlap (binomial test p < 1e−9) in upregulated and downregulated genes, including the PHISTa, KAHRP, FIKK, ATG5, and FKBP35 (for full list see Supplementary Data 2). For further analysis, we term the transcriptional variations of the 156 selected genes (69 upregulated and 87 downregulated) the artemisinin resistance-associated transcriptional profile (ARTP) and investigate further its biological relevance for the putative artemisinin resistance mechanism. We also show that the individual components of the ARTP can be used to predict/diagnose artemisinin resistance either individually or in combinations (Fig. 2a). Specifically, the receiver operating characteristic (ROC) curve combining the 156 ARTP genes was able to distinguish between resistant and susceptible parasites in the studied dataset with a high level of sensitivity (0.88) and specificity (0.7) which is close to the performance of the C580Y mutation (sensitivity = 0.75 and specificity = 0.91).
Fig. 2. Transcriptional resistance markers.
a The scatter plot represents all studied genes along the genomic coordinates on the X-axis with their resistance-association p-values displayed as -log10 p on the Y-axis. Genes passing the threshold (FDR < 0.05, FPR < 0.05) are highlighted in 69 orange circles (upregulation) and 87 blue circles (downregulation). The ROC plot shows the ability to distinguish resistant and susceptible parasites in TRACII for 5 upregulated genes (orange) and 5 downregulated genes (blue) with the best performance. Their gene IDs are shown on the side from top to bottom for the line from left corner to the centre of the plot. The black line with dots represents the distinguishing ability of the combination of all the 156 genes which was calculated as the sum up of expression values of the 69 upregulations with a subtraction of the sum up of the 87 downregulations. It is compared to the roc of randomly selected gene set on the bottom right (see “Methods”). The Venn diagrams show the overlap of TPAS results between the TRACII and TRACI studies. b A heatmap represents the transcriptional profiles clustering for 323 eGMS (bl)0 h samples showing prolonged PC½( > 5 h) based on the 156 resistance markers. The colour (purple to blue) indicates the level of differential expression (upregulation to downregulation) in the resistant parasites. The left dendrogram represents Ward’s clustering result and the colour bars represent the 6 clusters obtained by clustering tree cutting. On the right, samples in each column (marked by black bars) are categorized by their respective sites or lineages, or population structure. Frames mark the overrepresentations of categorized samples in the corresponding clusters. *p < 0.05, **p < 0.01 and ***p < 0.001. The scatter plot represents the population structure of the whole GMS samples, and the bar plot represents the ratio of distance between within-clusters individuals and between-clusters individuals (see “Methods”).
Artemisinin resistance-associated transcriptional profile
Next, we aimed to investigate the utility of the ARTP as a marker of the spread and evolution of artemisinin resistance in the GMS and possibly beyond. Hence, we conducted a clustering analysis to the 323 resistant parasite samples (PC½ > 5 h). Specifically, the Ward’s method was applied for clustering based on the Euclidean distance matrix of similarity of the ARTP formed by the 156 transcript levels in each of the (bl)0 h samples. This approach identified six clusters that displayed a distinct geographical segregation (hypergeometric test p < 0.05) (Fig. 2b). It yielded two main clades with high relatedness between cluster 1 (Pursat) and 2 (Ratanakiri) on one side and the rests of the sites in clusters 3–6 including, Phusing (4) and Binh Phuoc (5) and Pailin (3). The majority of the 323 samples were from KEL1PLA1 lineage (216/323, 67%) with the remainder made up of 36 (11%) from KEL1, 19 (6%) from other lineages, and 52 (17%) from unknown lineages. No lineage bias was observed with any cluster. To further investigate the impact of population-driven genetic factors, population structure of the entire GMS samples was analyzed by PLINK46 and visualized using UMAP47. The UMAP-2 clearly distinguishes the eGMS parasites from the wGMS and UMAP-1 mainly reflects the genetic differences among parasites within each region (Fig. 2b). Either the UMAP-1 or UMAP-2 lack significant correlation to the individual clusters that suggest the genetic differences differentiating the eGMS from wGMS or the population substructure is not a significant confounding factor for the ARTP differential expression. To strengthen the validity of this observation we compared the genetic differences to the transcriptional differences between clusters. The ratio of average distance was calculated between within-cluster individuals and between-clusters individuals genetically and transcriptionally separately (see “Methods”). Here the genetic distance ratios are consistently ~1 across the cluster 1–6 which contrasts the transcriptional distance ratios that were 0.7 in average, ranging from 0.5 to 0.8 (Fig. 2b). This further indicates that the individual clusters are defined by their transcriptional characteristics but indistinguishable genetically. Moreover, no correlation was observed between the ARTP transcriptional clusters and previously reported subpopulations which reflect the founder population effect of the KEL1PLA1 parasites expansion43 (Fig. 2b). Taken together, the results from the UMAP analysis, the genetic/transcriptional distance ratio, and the funder subpopulation associations suggest that the ARTP represents transcriptional re-tuning of at least 156 genes that is largely independent of background genetic differences. Although, the ARTP seems to function mainly in conjunction with the nonsynonymous PfK13 SNP, it is feasible to speculate that this expression profile could drive (at least to some degree) resistance to artemisinins even in an PfK13-independat manner.
In vivo transcriptional response of P. falciparum to ACTs
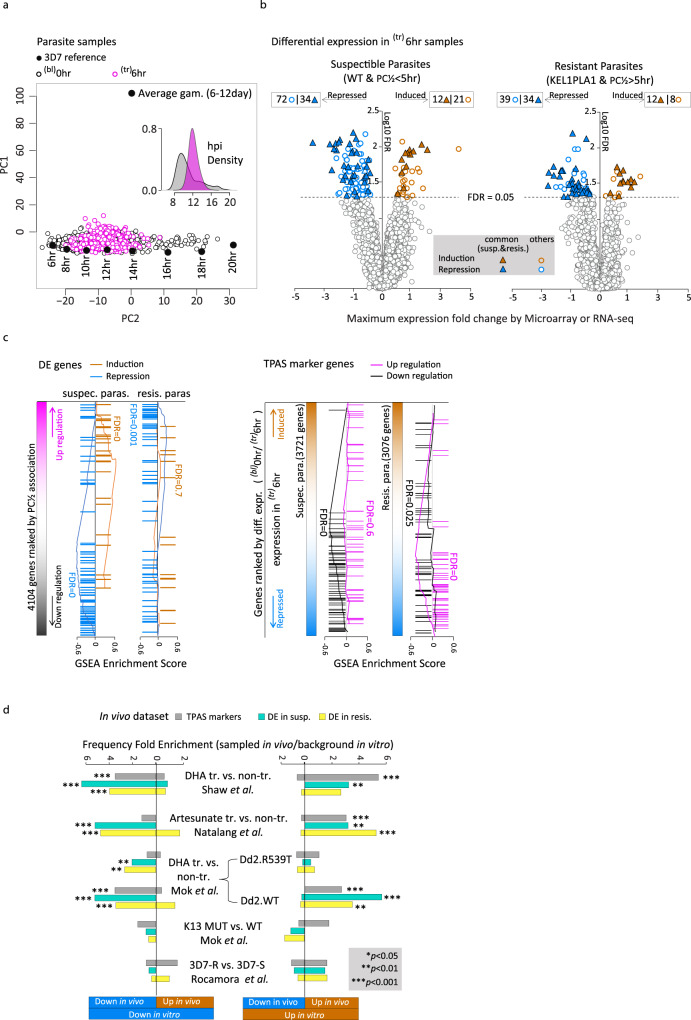
To complement the baseline TPAS, we evaluated the (tr)6 h sample set to assess transcriptional responses of P. falciparum exposed to ACT in vivo. The (tr)6 h parasites exhibited a tighter distribution of the IDC stages compared to those in the (bl)0 h, falling between 8 and 14 hpi with a median ~12 hpi, which might reflect the more rapid loss of mature ring stage parasites because of their higher sensitivity to artemisinins (Fig. 3a and Supplementary Fig. 2) (authors’ note). There was also a reduction (2–5%) in the fraction of gametocytes. Comparative transcriptome analysis between the (bl)0 h and (tr)6 h was conducted to identify transcriptional responses for the resistant (KEL/PLA1 with PC½ > 5 h) and susceptible (WT with PC½ < 5 h) parasites separately (for details see “Methods”). In total, we identified 20 and 73 genes that were induced or repressed, respectively, after 6 h in vivo treatment in the KEL1PLA1/resistant parasites. Similarly, 33 and 106 genes were induced or repressed, respectively, in the WT/susceptible parasites (FDR < 0.05, corresponding p < 1e–14) (Fig. 3b). Highly consistent results were observed between the pairwise and non-pairwise analysis with Pearson Correlation Coefficient of the average expression change (resistant/susceptible) being 0.99 (Supplementary Fig. 7). Given the marked differences in the hpi distributions between the (bl)0 h and (tr)6 h parasite sets (Fig. 3a), the non-pairwise analysis provides additional confidence for the differential genes expression, reaching beyond the individual strain differences. Interestingly, the susceptible and resistant parasites exhibited distinct transcriptional responses to artemisinin with a minimal (albeit statistically significant) overlap including 12 and 34 commonly induced and repressed genes, respectively (Supplementary Data 2). Out of these, six genes were also downregulated in the baseline (bl)0 h sample set while the only upregulated gene in both sample sets was PHISTa (PF3D7_1372000). There was a skewed distribution of the (tr)6 hr induced and repressed genes in the WT/susceptible parasites along the distribution of the transcriptional associations of the (bl)0 h parasites (Fig. 3c). Specifically, the 33 drug-induced genes were ranked significantly towards upregulation in (bl)0 h and the 106 drug-repressed genes towards downregulation (FDR = 0). Contrastingly, the (tr)6 h induced genes in the PfPailin (KEL1PLA1)/resistant parasites show no association with the (bl)0 h baseline ARTP but the (tr)6 h repressed genes exhibit a moderate overlap with upregulated genes in the (bl)0 h baseline ARTP (Fig. 3c). This suggests that the transcriptional response of P. falciparum to 6 h artemisinin exposure in vivo is related to the ARTP which could be its precursor (see “Discussion”).
Fig. 3. In vivo transcriptional response to artemisinins.
a The principal components space of PC1 vs. PC2 was constructed by the PCA on reference transcriptomes of the laboratory strain 3D7 at ring stage and gametocyte stages (average of the day 5th–12th). It was used to visualize transcriptome differences driven by parasite age/hpi or developmental stage (asexual/sexual). The 577 (bl)0 h (black circles) and 459 (tr)6 h (purple circles) samples are projected onto this space to show their age differences. The density plot represents the estimated hpi distribution for (bl)0 hr (grey) and tr)6 hr (purple) parasites. b Volcano plot represents each gene’s association p value of differential expression against the average expression fold change between the (tr)6 h and (bl)0 h parasites for the susceptible group (left, WT with PC½ < 5 h) and the resistant group (right, KELPLA1 with PC½ > 5 h) respectively. c Genes differentially expressed as upregulation/induction (orange) and downregulation/repression (blue) are marked along the rank of their association to PC½ (from TPAS). Markers associated with PC½ positively (purple) or negatively (black) are marked along the rank of their differential expression levels (from b). GSEA was applied to estimate the FDR for ranking bias to either side of upregulation/induction or downregulation/repression. d Bar plots represent significant overlaps between our in vivo study and other independent in vitro studies, In vivo: baseline TPAS analysis ((bl)0 h, 156 resistant markers, grey); post-treatment differentially expressed genes ((tr)6 h/(bl)0 h) in susceptible (turquoise) and resistant parasites (yellow) group, In vitro: transcriptional response to DHA treatment in the K1 rings48; Artesunate treatment in the FCR3 strain49; DHA treatment in the Dd2 R539T or WT stain50; differential expression at the baseline level between the PfK13 MUT and WT stains50 as well as that between lab-derived ART-resistant and ART-sensitive 3D7 ring parasites51. Stars mark intersections having > 3 genes with hypergeometric test p < 0.05. .
There were highly significant gene-by-gene overlaps between the in vivo results and several in vitro studies of transcriptional responses of P. falciparum to artemisinin(s) (Fig. 3d and Supplementary Table 2). This applies to the (bl)0 h TPAS as well as the in vivo transcriptional response of (tr)6 h parasites. There are good agreements between these results and earlier studies of in vitro transcriptional response of P. falciparum K1 strain exposed to dihydroartemisinin (DHA) for 1 h48, the FCR3 strain exposed to artesunate for 3 hours49 and the recently reported transcriptional responses of the Dd2wt strain exposed to DHA for 6 hours50. This is consistent with our observation showing that the basal level ARTP overlaps well with transcriptional responses of the susceptible parasites (Fig. 3c). We also observed a limited overlap with our previously derived in vitro P. falciparum parasites with ring stage-specific artemisinin resistance51 suggesting that additional mechanisms conferring artemisinin resistance exist and could rise in vivo in the future.
Discussion
This study identified at least 156 genes whose altered transcription may contribute significantly to artemisinin resistance. Artemisinin resistance involves a complex array of processes that have been selected to counter the parasiticidal effect of the drug20–23. These processes occur early in the asexual cycle and attenuate ring stage parasite killing. We found marked overlaps between the ARTP and the genes involved in the transcriptional response to the direct action of artemisinin observed in vivo (Fig. 3a, b) and/or in vitro48–51. This suggests that the mechanisms contributing to artemisinin resistance have arisen (at least in part) from the initial transcriptional response of P. falciparum parasites to the direct effect of the drug in which the artemisinin induced transcriptional changes became constitutive. Indeed, there are many overlaps on the gene-by-gene level between gene expression changes in the baseline ARTP and genes induced/suppressed by artemisinins in vivo and in vitro (Supplementary Table 2). In particular, we identified many determinants of protein metabolism including translation, folding, and degradation that were also found to be a part of P. falciparum response to artesunate49, and DHA of both susceptible and resistant parasite lines in vitro48,50,51. Notable examples include gene encoding 60S ribosomal subunits (L24), and elongation factors EF-1-gamma whose proteins products were found to be direct artemisinin targets in the parasite cells52,53. Related to this, we observed significant transcriptional changes for several determinants of protein turnover and protein folding from which DnaJ proteins and the T-complex-protein 1 subunit are likely to be direct protein targets of the artemisinin drugs (Supplementary Table 2). We also found transcriptional suppression of factors involved in REDOX functionalities, some of which are related to mitochondrial functions recently shown to play a key role in artemisinin resistance in vitro50. There was marked transcriptional activity of genes involved in biosynthetic pathways including pyridoxine/polyamine and purine/pyrimidine synthesis and glycolysis. Interestingly several enzymes encoded by these transcripts were also found to be inhibited by artemisinin directly including ornithine aminotransferase (OAT), spermidine synthase, pyruvate kinase, and hexokinase52,53. This also applies to the P. falciparum pyridoxal kinase, PDXK, whose mammalian orthologue, was shown to interact directly and being inhibited by artesunate54. Finally, the ARTP contains strong baseline-level upregulations of several transcripts encoding proteins exported to the host erythrocyte. These are implicated in host cell remodelling and/or host-parasite interactions that are paralleled by drug-induced transcriptional responses (Supplementary Table 2) (see below). Taken together, these observations support a model in which the initially adaptive transcriptional response became constitutively expressed as a result of drug selection, predisposing the parasite to withstand the drug’s parasiticidal effect.
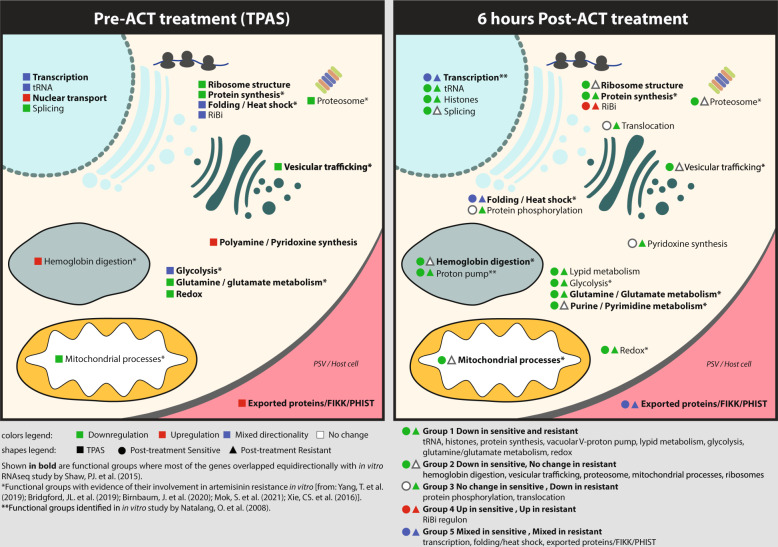
Functional assignments of the transcriptional markers of both the ARTP and in vivo induced artemisinin responses revealed by this study support their role in key biological processes aligned with both PfK13-dependent and independent mechanism(s) of artemisinin resistance suggested by previous in vitro studies (summarized in Fig. 4). First, PfK13, the key causal factor in artemisinin resistance, was shown to localize predominantly to the cytostomes, possibly regulating endocytosis of hemoglobin55. PfK13 mutations lead to a reduced rate of endocytosis and thus hemoglobin digestion, which in turn lessens the bioactivation of artemisinin as a result of lower levels of Fe2+ in the parasite cytoplasm. We observed baseline upregulation of factors involved in hemoglobin degradation and their suppression in sensitive parasites after 6 h in vivo treatment. Second, reductions of the PfK13 protein levels, resulting from the mutations, were also shown to suppress the proteotoxic shock and subsequent cell death normally induced by artemisinins56,57. This suppression can be reversed by proteasome inhibitors in both P. falciparum58 and P. berghei59. We found marked transcriptional suppression of protein synthesis, folding, and turnover including the core subunits of the proteasome (Supplementary Data 2 and Fig. 4). Finally, PfK13 SNP-driven reduction of hemoglobin digestion and thus cytoplasmic amino acid levels can be mitigated by upregulation of nutrient permeable channels (NPC) of the parasitophorous vacuolar membrane60. Here we observed upregulation of at least one of the NPC encoding gene, exp1, that has the potential to alleviate fitness cost caused by the PfK13 mutations. In addition to transcriptional characteristics traced directly to PfK13 function, we observed transcriptional variability of factors involved in transcription, mRNA processing, ribosomal biogenesis (RiBi), translation, translational translocation, and protein transport (Supplementary Table 2 and Supplementary Data 2). This is also consistent with our earlier TPAS study of the TRACI samples where we observed upregulation of the UPR that is canonically related to proteotoxic shock response40,41. Third, PfK13 was also shown to function as an adaptor for the cullin3-RING-E3 ubiquitin ligase targeting a specific set of proteins for proteasome-mediated degradation61. Mutated forms of PfK13 fail to recognize their targets including phosphoinositide 3-kinase (PI3K), which subsequently leads to an increase in PI3P vesicles that alters the rate of hemoglobin endocytosis62. A set of genes involved in vesicular trafficking and the underlying lipid metabolism was also found to be transcriptionally associated with artemisinin resistance in this study (Supplementary Table 2 and Fig. 4). Fourth, PfK13 mutations appear to mediate rewiring of the P. falciparum metabolic programme to higher levels of survival50. This involves mainly induction of mitochondrial processes including damage sensing and oxidative stress response that counteract artemisinin induced oxidative and alkylation activity58. In line with this, induced oxidative stress response was also detected in two independently derived in vitro P. falciparum artemisinin-resistant lines51,63. One of the chief functions of the parasite mitochondria lies in regeneration of ubiquinone, a necessary ECT component in pyrimidine biosynthesis pathways64 which was also shown to be altered by PfK13 mutations50. Here we observed transcriptional variations of multiple REDOX and mitochondria-related factors including purine/pyrimidine synthesis pathway-related markers (OPRT, and HGPRT) suppressed in susceptible parasites but also several downregulated transcripts pertaining to ECT (SDHB, CytC) and TCA-metabolism (Supplementary Data 2). Fifth, several genes encoding proteins exported to the host cell cytoplasm were found amongst the transcriptional resistance markers. These include several members of the FIKK and PHIST gene families that are mostly implicated in host-cell remodelling affecting rigidity and/or cytoadhesion of the infected RBCs65–68. This could suggest that altered host-parasite interactions might contribute to artemisinin resistance; albeit indirectly by reducing parasite clearance by the spleen. Given the functional diversification of the FIKK and PHIST as well as other P. falciparum gene families annotated as exported proteins their direct role in artemisinin resistance mechanisms cannot be ruled out. Several of the exported proteins factors as well as many of the above-mentioned biological pathways have been recently linked with artemisinin resistance in a large-scale P. falciparum piggyBac-transposon mutant screen69. Altogether these observations support the majority of previous studies implicating a wide spectrum of biological functions discovered by in vitro experiments to play specific roles in artemisinin resistance in vivo.
Fig. 4. Functional assignment of resistance markers.
Functional assignments of transcriptional artemisinin resistance markers derived from TPAS analysis at baseline-level ((bl)0 h) and that of genes repressed/induced after 6 h of ACT treatment ((tr)6 h). Colours represent transcriptome directionality for each group. The association with previous independent in vitro studies is also shown.
Since its emergence in 2009, artemisinin resistance exerted strong selection effects on the GMS P. falciparum population due to a continuous drug pressure resulting from intensive high-coverage treatment programmes implemented by all countries in the region11,43,70. There is now a clear distinction between the P. falciparum population in the wGMS (Myanmar, Thai-Myanmar border, and Bangladesh) and eGMS (Cambodia Laos and Southern Vietnam), the latter of which likely provided the supporting genetic background for artemisinin resistance40. Moreover, substantial founder population effects were detected in eGMS also likely as a result of drug selection71. The main example represents the recent selective sweep of the KEL1/PLA1 parasite line throughout eGMS during 2013–2016; prior to the TRACII epidemiological survey11,43. Indeed, the TRACII P. falciparum cohort used in this study reflected both the w/eGMS segregation and KEL1/PLA1 selection sweep. This provided us with the unique opportunity to study artemisinin resistance-associated transitional changes at its origin, but also presented a major challenge due to the complex population structure. To overcome this, we implemented multiple steps during data analysis. In addition of the FPR-based differential gene expression pipeline (Fig. 1b, c), we carried out multiple confounding factor evaluations including tSNE and PCA with the whole transcriptome dataset (Supplementary Fig. 3) and subsequently UMAP, genetic/transcriptional distance ratio, and the Hamilton’s founder subpopulation with the ARTP (Fig. 2b). Notwithstanding the possible weakness of each analytical method, the combination of these provides a strong confidence that identified 156 genes can be linked to putative multifaceted artemisinin resistance mechanism. However, it is feasible to expect that there are more genes contributing to artemisinin resistance that were not uncovered by this work. This is likely due to the challenging nature of the TRACII epidemiological datasets that is characterized by a strong population structure (Fig. 1a) that forced us to develop a highly stringent bioinformatics pipeline to overcome this confounding factor. As a result, we focused on transcriptional changes of individual genes instead of gene groups capturing entire biochemical and cellular pathways that were a subject of our previous TPAS studies with TRACI cohort40,41. In the TRACI dataset, we identified inductions of multiple pathways related to the UPR and REDOX metabolism and suppression of multiple processes facilitating the parasite developmental progression (deceleration of the IDC). These results were reproduced in the TRACII only partially. However, multiple genes related to UPR, REDOX as well as IDC progression identified amongst the ARTP genes signal that these pathways still play a key role in artemisinin resistance. Future GWAS and TPAS using broader epidemiological cohorts will provide deeper insight into the multifaceted artemisinin resistance mechanism relevant to clinical settings.
In contrast to transcriptome-based analyses, GWAS have until now failed to identify such factors making limited observations of direct links between the molecular effectors of artemisinin resistance in vivo; with the exception of the PfK13 SNP32. Although it is still possible that more mutations in the coding regions may contribute to artemisinin resistance they were not detected because of typical sample size limitations of GWAS39; in the context of a highly complex P. falciparum population structure in the GMS71. Our result strongly suggests that many causative mutations might be situated within DNA regulatory elements affecting the regulation of gene expression. It was previously shown that the majority of sequence polymorphisms in P. falciparum occurs in the noncoding regions accumulated particularly around core promoter regions as Insertion/Deletion within Short Tandem Repeats (STR)72. The high AT content (~85%) of the P. falciparum noncoding regions hampered most whole-genome sequencing efforts on field samples done up until now. However, the results of this study make a case for synergizing GWAS and TPAS (combined into eQTL studies) as a potential strategy to monitor the spread and evolution of malaria drug resistance in the future genomic epidemiological surveys.
Methods
Ethics statement
All the samples used in this study were collected with written informed consent from the patients or their legal guardians. All protocols were approved by the appropriate national ethics committees or the Oxford Tropical Research Ethics Committee.
In vivo sample collection
All samples used in this study were collected from patients involved in Tracking Resistance to Artemisinin Collaboration 2 (TRAC2), a multi-site trial that took place between Aug 7, 2015, and Feb 8, 2018. The total number of 1100 patients with uncomplicated Plasmodium falciparum malaria were recruited in eight countries. In the presented study 680 patients’ samples from 6 countries and 13 sites across Southeast Asia Greater Mekong Region were analyzed (Ramu in Bangladesh; Thabeikkyin, Pyay, Pyin Oo Lwin and Ann in Myanmar; Phusing and Khun Han in Thailand; Pailin, Pursat, Ratanakiri, and Preah Vihear in Cambodia; Sekong in Laos; Bin Phuoc in Vietnam). All details regarding samples collection, site locations, inclusion criteria, parasitemia assessment, and given treatments were published previously2. In brief, samples were collected from venous blood of malaria-infected patients at admission to the clinic (Hour 0, 0 h) and 6 h after their respective treatment (Hour 6, 6 h). Depending on their location, patients were randomly assigned different double or triple Artemisinin Combination Therapy (ACT) as follows: Thailand, Cambodia, Vietnam, and Myanmar - dihydroartemisinin-piperaquine or dihydroartemisinin-piperaquine plus mefloquine; Cambodia - artesunate-mefloquine or dihydroartemisinin-piperaquine plus mefloquine; Laos, Myanmar, Bangladesh - artemether-lumefantrine or artemether-lumefantrine plus amodiaquine. Following collection, blood was subsequently depleted of plasma and buffy coat. Hour 0 samples were additionally purified from white blood cells (WBC) using CF11 columns using the Worldwide Antimalarial Resistance Network (WWARN) protocols. 0.2–0.5 mL of packed red blood cells (pRBC) from each sample was homogenized by mixing with 10 volumes of TRIzol (Invitrogen), frozen at −80 °C, and then shipped to Nanyang Technological University, Singapore.
RNA isolation
All clinical samples mixed with TRIzol were supplemented with 5 volumes of chloroform (Merck) and processed as per the manufacturer’s instructions to obtain phase separation. Top (aqueous) phase was mixed 1:1 (v/v) with 100% analytical grade ethanol (Merck) and RNA was extracted using the Direct-Zol-96 extraction kit (Zymo) following the manufacturer’s guide. For this study, we have adopted the RNA extraction kit to a high-throughput EVO 200 robotic platform (Tecan). RNA integrity was assessed using Agilent Bioanalyzer (RIN) (Agilent), concentration was measured using Qubit RNA Broad Range kit (Invitrogen) and RNA contamination was estimated with Nanodrop (Thermo Fisher)42.
cDNA synthesis and microarray hybridization
For each sample, 250 ng of total RNA was used for subsequent reverse transcription reaction followed by 19 rounds of PCR amplification using modified Smart-Seq2 method73. In brief, oligo-dT30 (IDT Asia) was used as a primer to enrich for mRNA and avoid ribosomal rRNA amplification (AAGCAGTGGTATCAACGCAGAGTACT30VN; V = A,C,G; N = A,C,G,C). The amplified product was purified using Ampure XP magnetic beads (Beckman Coulter) on Microlab Nimbus robotic platform (Hamilton) and 100 ng of cDNA was used for subsequent 10 rounds of amplification to generate aminoallyl-coupled cDNA for the hybridizations74,75. 17ul (~5 µg) of each Cy-5-labelled (GE Healthcare) cDNA of patient’s sample and an equal amount of Cy-3-labelled (GE Healthcare) cDNA of the reference pool were then hybridized together on our customized microarray chip using commercially available hybridization platform (Agilent) for 20 h at 70 °C with rotation at 10 rpm. Microarrays were washed and immediately scanned using Power Scanner (Tecan) at 10 µM resolution and with automated photomultiplier tubes gain adjustments to balance the signal intensities between both channels. The reference pool used for microarray was a mixture of 3D7 parasite strain RNA collected every 6 h during 48 h of the full intraerythrocytic developmental cycle.
RNA-sequencing
Purified 1 ng of cDNA was used to generate sequencing libraries using Illumina Nextera XT kit as described in manufacturer’s protocol. Purified cDNA libraries were analyzed on the Bioanalyzer High-Sensitivity DNA chips (Agilent), subsequently pooled (20–24 samples per lane) and sequenced on Illumina HiSeq4000 platform generating 150 bp paired end reads with 110 Gb data output generated per lane.
In vitro culture
Plasmodium falciparum 3D7 strain was obtained from BEI Resources (MRA-102) and maintained in purified human pRBC in RPMI 1640 medium (Gibco) supplemented with Albumax I (Gibco) (0.25%), hypoxanthine (Sigma) (0.1 mM), Sodium bicarbonate (Sigma) (2 g/L), and gentamicin (Gibco) (50 μg/L). Cultures were kept at 37 °C with 5% CO2, 3% O2, and 92% N2. Culture media were replenished every 24 h. Freshly washed pRBC were added to the culture when necessary. Both parasitemia and parasite morphology were assessed by microscopic examination of blood smears stained with Giemsa (Sigma).
Intraerythrocytic asexual developmental cycle reference time course (IDC)
IDC reference time course data were obtained from the study published previously42. In brief, prior to time-point experiment, parasites were double-synchronized with 5% sorbitol solution to achieve a synchrony of +/−6 h and cultured under constant agitation. For a sampling of highly synchronous parasites during the asexual life cycle, the first time point was considered as the TP1 (Time Point 1) when > 95% of early ring stage parasites (approx. 4 hpi) were present in the culture. Starting from TP1, parasites were collected every 2 h for 25 successive time points. The total of 24 time points were used here to build an asexual reference transcriptome generated using two different platforms: microarrays and RNA-seq.
Raw transcriptome of P.falciparum parasites
The transcriptome was generated by quantifying the total RNAs for each parasite sample using two-colour microarray or RNA-seq or both. Additionally, control samples derived from ring stages 3D7 lab strain were quantified together with those clinical samples in microarray method. It was made up of a total of 30 samples for pre-treatment and 27 samples for post-treatment.
The raw data of microarray was acquired using GenePix Pro v6.0 software (Axon Instruments). Then the signal intensities were Loess-normalized within arrays followed by quantile-normalization between samples/arrays using Limma package of R76. Missing values were assigned to weak signal probes showing median foreground intensity less than 1.5-fold of the median background intensity at either Cy5 (sample RNA) or Cy3 (reference pool RNA) channel. Each gene expression was estimated as the average of log2 ratios (Cy5/Cy3) for probes representing it.
For RNA-seq, raw reads were trimmed to remove sequencing adapters, amplification primers, and low-quality bases from 3′ ends. HISAT2 aligner77 was used to perform alignment to the P.falciparum genome downloaded from geneDB database in version of March 2018. BEDTools78 was applied to calculate the read counts for each annotated transcript based on only uniquely mapped reads with pairs in proper orientation. At last, the transcriptional level was estimated for each gene by calculating the Fragments per kilo base per million mapped reads (FPKM) at the gene.
Developmental stage estimation
For each parasite sample, we estimated the asexual age (hours post-invasion, hpi) and the proportion of gametocytes using the method described in Zhu et al’s and Lemieux et al’s works40,79. In brief, the expression value of a gene, Eg, is assumed as a sum of the expression in asexual stage of hpi h, denoted as xg(h), and the average expression of sexual stages (from the 5th to 12th day during gametocytes developing), denoted as zg, mixed in certain proportions. The mixture model is presented in formula as:
| 1 |
where α is the proportion of gametocytes to the total parasite count (sum of sexual and asexual parasites) and the εg is the associated error term. xg(h) is estimated by the reference transcriptome generated during asexual intraerythrocytic cycle in P. falciparum 3D7 strain which has a total of 25 time points with 2 h intervals of 48 h, accessible via GEO accession number GSE149865. To obtain a higher resolution of the asexual reference transcriptome for stages estimation, 24 time points of the data (the 9th time point removed due to its big dissimilarity to others) were interpolated into 240 data points by smooth splines method. zg is estimated by the identical reference gametocyte transcriptome described in Zhu et al’s work9, accessible via GEO accession number GSE121505. In practice, samples were normalized by control samples across batches by adding a scaling factor for each batch before applying the prediction model. The scaling factor was estimated for each batch by calculating the average expression difference between controls and the reference at corresponding ages. Finally, we evaluated the log-likelihood values over a grid of mixtures for varying the gametocyte proportion α, and hpi h, for each sample. The results of estimated hpi and gametocytes proportion were listed in Supplementary Data 1.
Transcriptome filtering
To achieve high quality data for the following analyses, we pruned the sample set according to their transcriptome heterogeneity and discarded samples with low signal intensities. In practice, outlier samples were removed if they were distinct from the majorities with < 10 cohort samples at the similarity threshold for grouping. The threshold was determined by the average similarity of controls to clinical samples using Spearman’s rho. Second, we removed samples displaying extremely low intensities (mode of the intensity < 10) of Cy5 signal on the microarray (Supplementary Fig. 8). In addition, to reduce the prediction errors (if any) affecting on the following study, we removed 2 samples having very high PC½ as 13.1 and 19.3 h (>2 times Median Absolute Deviation to the median), and 24 samples with extremely high gametocytes prediction (>18%, 3times MAD to the median) as most of the parasite samples exhibiting a low even zero proportion of gametocytes. We found all the discarded samples at this end mostly having a low parasitemia or high human content. Finally, with the microarray method, we identified a transcriptome of 4779 genes presented across > 75% of the 577 samples pre-treatment and 4714 genes across 459 samples post-treatment. Among the total of 577 pre-treatment samples and 459 post-treatment samples, 438 samples were paired (collected from the identical patient). With the RNA-seq method, we asked for at least 1 M uniquely mapped reads in a library to call for the transcriptome. Overall, we identified the transcriptome of 4305 genes with > 75% representation for 188 samples pre-treatment and 3923 genes for 159 samples post-treatment for further analyses. The data is accessible in Gene Expression Omnibus (GEO) database via the series accession number GSE149735 for microarray and GSE169520 for RNA-seq.
PCA and population transcriptomic analysis
Principle Component Analysis (PCA) was applied to the (bl)0 h transcriptome data set. We inspected the top 12 PCs for the following association analysis because each of the rest PCs contributed to < 1% of the overall transcriptome variations. The top 12 PCs were tested against all the clinical and technical factors collected during sample processing to assess the potential environmental influence on the population transcriptomic structure. For the factors represented in categorical variables like sampling sites, parasite lineages, and patient’s gender, ANOVA test was used for assessing the statistical significance of associations. For other factors in continues/discrete variables like parasitemia, hpi and patient enrolment time etc., linear regression was used to test the statistical relationship between each factor and each PC with Spearman’s rho calculated for each pair of variables as well. The results are visualized in Supplementary Fig. 3a.
The regression analysis revealed that parasite age (hpi) significantly (p = 7.96e−283) correlated to the top PC (PC1) with showing a high Spearman’s rho as 0.87 in the (bl)0 hr data set by microarray and 0.85 by RNA-seq. PCA of all samples including both (bl)0 h and (tr)6 h resulted in similar high correlations with displaying rho as 0.78 in microarray and 0.76 in RNA-seq (Supplementary Fig. 3b). It indicates that hpi is the major factor distinguishing parasites transcriptome in field. Correlation was also observed between the PC2 and the ratio of parasite to human content; the PC6 and estimated gametocytes proportion but the Spearman’s rho was dropped to 0.53 and 0.52, respectively. These two factors (parasite/human ratio and gametocytes fraction) can interact with other experimental or environmental conditions to drive the minor (compare to hpi) differences in parasites transcriptome. For this study, we considered only hpi as the major factor contributing to gene expression variation across nature parasites and used it as a major predictor variable in modelling gene expression for the further regression analysis. The potential for expression variation caused by other factors, like sampling site and parasite lineage, are analyzed for particular genes associated to resistance in the following studies.
To visualize the parasite population structure in a two-dimensional map, t-distributed stochastic neighbour embedding was applied to the PC2-12 with PC1 excluded to minimize the hpi effects. It was implemented in R with the M3C package80.
The PC space projection
We performed PCA to the reference transcriptome of 3D7 parasite stain at ring stages together with the average transcriptome of 3D7 mature gametocytes (5th–12th day during development). The resulted PC1 clearly distinguishes the sexual and asexual stages, as well as the PC2, reflects the ring-stage parasite development as it obviously separates the 8 ring-stage transcriptomes with 2 h difference in between. With the space of PC1 vs PC2, parasite age can be visualized without bias by transcriptome projection. Therefore, we normalized all the clinical samples to the centre transcriptome derived from the above PCA and rotated it according to the PC1 and PC2 loadings. At last, the 577 pre-treatment samples and 459 post-treatment samples were projected onto the PC space showing an obvious age window shift in the parasites after drug treatment (Fig. 3a).
Population stratification
The SNP information was provided by the Wellcome Sanger Institute. We extracted out a high-quality set of 7009 SNPs for the population stratification analysis here. The filtering criteria are as follow: (1) each polymorphism was covered by at least 10 reads; (2) the genotyping quality score was greater than 30 by GATK; (3) the minor allele count was at least 5 across the genotyped samples we studied, and the minor allele frequency was at least 0.05. (4) Only SNPs showing two alleles with our studied samples are included; (5) only SNPs in coding regions were included; (6) samples with more than 50% high quality SNPs missing were removed from this analysis. At last, 7009 SNPs and 528 isolate samples were used to determine the population stratification using Plink multidimensional scaling function with IBS pairwise distances. UMAP algorithm was applied to visualize the result in Fig. 2b. The UMAP-2 majorly distinguished the eGMS parasites from the wGMS and the UMAP-1 reflected the genetic difference within each region’s parasites.
To measure the genetic differences between the ARTP clusters (Fig. 2b), we constructed the genetic distance matrix using the IBS pairwise distances. And the pair-wise distance was calculated between individuals within each cluster and between clusters. The ratio of the average distance between within-cluster, D(within-cluster), and between-cluster, D(between-cluster) individuals was used to indicate genetic differences between clusters and compare to the ratio calculated from transcriptional distance matrix.
Transcriptome-phenotype association study
Transcriptome-phenotype association study (TPAS) was carried out in order to call for the marker candidates whose mRNA levels were positively or negatively associated with artemisinin resistance. Since parasite age contributed the most to expression variation across clinical samples, we designed a generalized additive model with the age/hpi specified in a loess function to test the expression-resistance association over the dynamic relationship between expression and age. The regression analysis formula for each gene is represented as:
| 2 |
Where E denotes gene expression across samples, f(hpi) denotes the function of hpi which is loess regression here, hf denotes the variable of PC½, β0,1,2 represents the parameters of intercept and slopes to predict and ε is the error terms. Therefore, the resulted p-value of this regression analysis reflects the relationship between expression residuals of age fitting to the half-lives (PC½), alternatively the expression-resistance association independent of age.
With this approach, we tested the expression-resistance association for all the genes, 4779 genes with microarray and 4714 genes with RNA-seq, individually. To correct for the multiple testing, we estimated FDR for each gene by 1000 times permutation constructing a null p distribution based on testing the association of expression to randomized PC½ values (Fig. 1c). In addition, to address the cofounding relation between artemisinin resistance (PC½ > 5 h) and parasite genetic lineage which also largely coincided with the geographical region (w/eGMS), we estimated a FPR value for each gene to control the type I error by 100 times permutations. In each permutation, the lineage structure was maintained and PC½ values were randomized amongst the parasites within each lineage. The resulted null p-values reflect the significance of expression-lineage associations independent of PC½ and the FPR calculated from the null p distribution (Fig. 1b) reflect the probability of expression-resistance association caused by expression-lineage relationship. At last, FPR < 0.05 (95% confidence) was applied to define the robust expression-resistance associations beyond parasite lineage effect. We plotted the expression residuals against the original and randomized PC½ values for PHISTa gene (shown in Fig. 1b) to illustrate the randomization procedure, also for three example genes with significant expression-resistance associations (FDR < 0.05) and different levels of FPR (0.01, 0.53, and 1) together with other two genes with FDR > 0.5 and FPR < 0.01 for reference (Supplementary Fig. 5).
We applied this approach to microarray and RNA-seq data separately. The results agreed to each other with showing a high correlation (Pearson correlation coefficient = 0.68) of the average expression fold change of resistant/susceptible between the two datasets (Supplementary Fig. 6). We merged the markers defined by microarray or RNA-seq excluding 10 genes that displayed conflicting directions of expression changes in resistant parasite in two techniques. Finally, our approach determined 69 genes upregulated and 87 genes downregulated in the resistant parasites at FDR < 0.05 (corresponding p < 1e−10 in microarray, p < 1e−6 in RNA-seq) and FPR < 0.05.
We applied the same TPAS pipeline to the TRACI data and reanalyzed 824 samples collected during 2011–1013. It resulted in 61 expression upregulation and 63 downregulation associated to artemisinin resistance with the identical criteria above (FDR < 0.05 & FPR < 0.05, corresponding p < 8.5e−6). This result significantly overlaps that from TRACII with 14 upregulation and 12 downregulation in common (binomial test p < 1e−9).
ROC curve
ROC curve was applied to access the ability of distinguishing the resistance and susceptible parasites for individual expression markers, combination of markers, and the C580Y mutation. For each gene, the sensitivity and specificity of classifying PC½ into groups of > 5 h or < 5 h was calculated at a varied expression threshold which has 20 data points evenly distributed from the minimum to the maximum expression value observed in the studied parasites. For the combination of markers, we use the measurement as the sum up expression values of the 69 upregulations with a subtraction of the sum up of the 87 downregulations. Then the varied threshold of 20 points was set similarly as the above. Since the combination of multiple genes can always perform better than single, we also generate ROC curve for 10 randomly selected gene sets. Each of the 10 gene sets combined 69 genes randomly selected from genes over-expressed in resistance parasites and 87 random under-expressed genes. The result suggests the combination of the 156 markers having the best performance comparing to each single marker or random genes and it is the most close to the performance of the C580Y mutation (sensitivity = 0.75 and specificity = 0.91, Fig. 2a).
ARTP clustering
To investigate the resistance-associated transcriptome structure, we first defined the artemisinin resistance-associated transcriptional profiles (ARTP) for each parasite sample using the 156 marker genes. To obtain expression levels with hpi effects maximumly reduced, we second extracted out the expression residuals from the formula (2) with the hpi function fitting only for each gene. The expression residuals were normalized for the 323 resistant parasite samples (PC½ > 5 h and from eGMS) against that of 104 susceptible samples (PC½ < 5 h and from wGMS) by calculating z-scores to represent the number of standard deviations by which the expression of the studied gene in resistant parasite was above or below the mean in susceptible parasites. Next, Euclidean distance was applied to the similarity matrix of ARTP to construct the sample distance matrix and the Ward’s method was used to obtain the dendrogram of clustering tree for the 323 resistant parasite samples (Fig. 2b). The six clusters shown in Fig. 2b were defined by the tree cutting at the 1/6 of tree height using the “cutree” function in R stats package.
To measure the transcriptional differences between clusters, we used the Pearson distance of the ARTP to construct the transcriptional distance matrix. And the pair-wise distance was calculated between individuals within each cluster and between clusters. The ratio of the average distance between within-cluster, D(within-cluster), and between-cluster, D(between-cluster) individuals was used to indicate transcriptional differences between clusters and compare to the ratio calculated from a genetic distance matrix.
In vivo transcriptional response measurement
The transcriptional response to artemisinin was inspected for the field P.falciparum parasites by comparing the (tr)6 h sample set to the (bl)0 h sample set. Grouping the (bl)0 h samples by lineage and resistance status level (PC½ greater/smaller than 5 h) revealed two largest groups which are KEL1PLA1 parasites with PC½ > 5 h (37% of the total 577 samples) and PfK13 WT parasites with PC½ < 5 h (29% of 577). Excluding the lineage unknown samples, all the rest groups contain < 6% of the total samples. For this study, we performed the comparative transcriptional analysis specifically for the PfK13 WT samples with PC½ < 5 h (the susceptible parasites) and the KEL1PLA1 samples with PC½ > 5 h (the resistant parasites).
We adjusted the above regression model of formula (2) to better present the data of (tr)6 h and (bl)0 h samples as:
| 3 |
Where treatment indicates patient treatment condition which is pre-treatment ((bl)0 h) or post-treatment ((tr)6 h), and resistance_status indicates PC½ greater/smaller than 5 h. To discover the distinct transcriptional response for resistant and susceptible parasites individually, we performed a comparative analysis for each parasite group (resistant/susceptible). We aimed to define top drug-response genes for each parasite group independent of age and batch effects (due to the collection and transportation issue with the (bl)0 h and (tr)6 h sample set, the treatment condition here unavoidably confounded with the batch of transcriptome measurement). To achieve that, we first extracted the expression residuals from the model (3) with the loess fit only which maximumly removed the expression variations caused by age from the raw data. Second, we conducted Mann–Whitney test on the expression residuals between treatment conditions to compare 216 (bl)0 h to 180 (tr)6 h samples for resistant parasites and 168 (bl)0 h to 130 (tr)6 h samples for susceptible parasites with microarray measurement. To balance the sample size differences, subsampling was applied to 130 samples 100 times per gene to obtain the p-value at 80% confident level. For the multiple test correction, we calculated FDR for each gene using the distribution of 500,000 null p-values generated from expression permutation based on all bl)0 h and (tr)6 h samples. To control the effect of treatment/batch, the structure of the treatment condition was maintained during each time permutation.
We repeated the same analysis with the RNA-seq data. Mann–Whitney test was conducted to compare 67 (bl)0 h to 55 (tr)6 h samples for resistant parasites and 57 (bl)0 h to 46 (tr)6 h samples for susceptible parasites. And the p-values were obtained by 100 subsampling of 46 samples per gene. The result was merged with that from microarray for susceptible and resistance group, respectively, at FDR < 0.05.
By this approach, we identified 20 significantly induced genes and 73 repressed genes upon drug in the KEL1PLA1 resistant parasites and 33 induced genes and 106 repressed genes in the WT susceptible parasites (FDR < 0.05, corresponding p < 1e−14). Significant common response genes were observed between the resistant and susceptible parasite samples which included up to 12 induced and 34 repressed genes (Fig. 3b and Supplementary Data 2).
Statistics and reproducibility
We analyzed 577 samples pre-treatment with 459 samples post-treatment using microarray method and 188 samples pre-treatment with 159 samples post-treatment using RNA-seq method. Due to the nature of this study as a part of the larger clinical trial, no biological replicates were used. In this study, binomial test or hypergeometric test was applied to the enrichment analyses; ANOVA test, Pearson Correlation Coefficient, and linear regression were performed for association or correlation testing between factors. We used FDR and FPR to help control for false positives that resulted in 156 resistance-associated marker genes defined at FDR < 0.05, FPR < 0.05, and the corresponding p < 8.5e−6. For the differential expression analysis, Mann–Whitney test was applied for group comparison. Differentially expressed genes were defined at FDR < 0.05 and the corresponding p < 1e−14. Subsampling method was used to balance sample size difference and the p-values were obtained at 80% confidence level. All details regarding the statistics of this study can be found above in the description of each respective analysis.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We would like to thank all members of the Tracking Resistance to Artemisinin Collaboration for providing access to clinical samples used in this study as well as all patients that have agreed to participate in this TRACII clinical trials. We also would like to thank Sachel Mok for providing the parasite reference pool and Cholrawee Promnarate, Nakarin Aud-Ai for coordinating field specimen collection. This research was funded by Singapore National Medical Research Council grant #NMRC/OFIRG/0040/2017 and Singapore Ministry of education grant #MOE2019-T3-1-007 and #MOE2017-T2-2-030 (S) awarded to Z. Bozdech. Moreover, A.D. coordinated funding for the TRACII epidemiology studies funded by DFID (FCDO): Artemisinin Resistant Malaria Research Programme (TRAC). DFID PO 5408 and Wellcome Trust: MOP Thailand Core award 220211/Z/20/Z and 220211/A/20/Z.
Author contributions
L.Z. performed all the analysis of this study, prepared the figures, and drafted the manuscript. R.V.D.P. and Z.B. designed the study, performed the analysis, and drafted the manuscript; M.K. performed all lab experiments including the design of the experimental protocols, necessary validations, and helped to analyze the data and draft the manuscript and figures; J.T. contributed to the protocol design for lab experiments; S.N. helped to process RNA-Seq data; D.P.K., F.N., H.G., N.P.J.D., N.J.W., and A.M.D. participated in the design and supervised the project and gave vital comments to the Discussion. Tracking Resistance to Artemisinin Collaboration II authors carried out all the field and laboratory work necessary to obtain P. falciparum samples. The manuscript was reviewed by all the authors.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. This article has been peer-reviewed as part of Springer Nature’s Guided Open Access initiative.
Data availability
All data is available in the main text or the supplementary materials or from the corresponding author upon request. All transcriptome data used in this study are available at NCBI’s Gene Expression Omnibus (GEO) database with the series accession number GSE149735 for microarray and GSE169520 for RNA-seq.
Competing interests
Authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lei Zhu, Rob W van der Pluijm, Michal Kucharski
Contributor Information
Arjen M. Dondorp, Email: arjen@tropmedres.ac
Zbynek Bozdech, Email: zbozdech@ntu.edu.sg.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-022-03215-0.
References
- 1.World malaria report 2019. Geneva: World Health Organization; 2019. Licence:CC BY-NC-SA 3.0 IGO.
- 2.van der Pluijm, R. W. et al. Triple artemisinin-based combination therapies versus artemisinin-based combination therapies for uncomplicated Plasmodium falciparum malaria: a multicentre, open-label, randomised clinical trial. Lancet395, P1345–1360 (2020). [DOI] [PMC free article] [PubMed]
- 3.Phyo AP, et al. Declining efficacy of artemisinin combination therapy against P. Falciparum malaria on the Thai-Myanmar border (2003–2013): The role of parasite Genetic Factors. Clin. Infect. Dis. 2016;63:784–791. doi: 10.1093/cid/ciw388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uwimana A, et al. Emergence and clonal expansion of in vitro artemisinin-resistant Plasmodium falciparum kelch13 R561H mutant parasites in Rwanda. Nat. Med. 2020;26:1602–1608. doi: 10.1038/s41591-020-1005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chotivanich K, et al. Laboratory detection of artemisinin-resistant Plasmodium falciparum. Antimicrob. Agents Chemother. 2014;58:3157–3161. doi: 10.1128/AAC.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witkowski B, et al. Reduced artemisinin susceptibility of Plasmodium falciparum ring stages in western Cambodia. Antimicrob. Agents Chemother. 2013;57:914–923. doi: 10.1128/AAC.01868-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ariey F, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashley EA, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WWARN K13 Genotype-Phenotype Study Group. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments-a WWARN individual patient data meta-analysis. BMC Med. 2019;17:1. doi: 10.1186/s12916-018-1207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takala-Harrison, S. et al. Independent emergence of Artemisinin resistance mutations among plasmodium falciparum in Southeast Asia. J. Infect. Dis.211, 670–9 (2014). [DOI] [PMC free article] [PubMed]
- 11.Imwong M, et al. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: A molecular epidemiology observational study. Lancet Infect. Dis. 2017;17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dondorp AM, et al. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Pluijm RW, et al. Determinants of dihydroartemisinin-piperaquine treatment failure in Plasmodium falciparum malaria in Cambodia, Thailand, and Vietnam: A prospective clinical, pharmacological, and genetic study. Lancet Infect. Dis. 2019;19:952–961. doi: 10.1016/S1473-3099(19)30391-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boulle M, et al. Artemisinin-resistant plasmodium falciparum K13 mutant alleles, Thailand-Myanmar border. Emerg. Infect. Dis. 2016;22:1503–1505. doi: 10.3201/eid2208.160004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra N, et al. Emerging polymorphisms in falciparum Kelch 13 gene in Northeastern region of India. Malar. J. 2016;15:583. doi: 10.1186/s12936-016-1636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miotto O, et al. Emergence of artemisinin-resistant Plasmodium falciparum with kelch13 C580Y mutations on the island of New Guinea. PLoS Pathog. 2020;16:e1009133. doi: 10.1371/journal.ppat.1009133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayih AG, et al. A unique Plasmodium falciparum K13 gene mutation in Northwest Ethiopia. Am. J. Trop. Med. Hyg. 2016;94:132–135. doi: 10.4269/ajtmh.15-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamau E, et al. K13-propeller polymorphisms in Plasmodium falciparum parasites from sub-Saharan Africa. J. Infect. Dis. 2015;211:1352–1355. doi: 10.1093/infdis/jiu608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng CL, Fidock DA. Plasmodium falciparum in vitro drug resistance selections and gene editing. Methods Mol. Biol. 2019;2013:123–140. doi: 10.1007/978-1-4939-9550-9_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenthal MR, Ng CL. Plasmodium falciparum artemisinin resistance: The effect of heme, protein damage, and parasite cell stress response. ACS Infect. Dis. 2020;6:1599–1614. doi: 10.1021/acsinfecdis.9b00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talman AM, Clain J, Duval R, Menard R, Ariey F. Artemisinin bioactivity and resistance in malaria parasites. Trends Parasitol. 2019;35:953–963. doi: 10.1016/j.pt.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 22.Tilley L, Straimer J, Gnadig NF, Ralph SA, Fidock DA. Artemisinin action and resistance in plasmodium falciparum. Trends Parasitol. 2016;32:682–696. doi: 10.1016/j.pt.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Xu C, Lun ZR, Meshnick SR. Unpacking ‘artemisinin resistance’. Trends Pharm. Sci. 2017;38:506–511. doi: 10.1016/j.tips.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M, et al. Inhibiting the Plasmodium eIF2alpha kinase PK4 prevents artemisinin-induced latency. Cell Host Microbe. 2017;22:766–776 e4. doi: 10.1016/j.chom.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demas AR, et al. Mutations in Plasmodium falciparum actin-binding protein coronin confer reduced artemisinin susceptibility. Proc. Natl Acad. Sci. USA. 2018;115:12799–12804. doi: 10.1073/pnas.1812317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klonis N, et al. Artemisinin activity against Plasmodium falciparum requires hemoglobin uptake and digestion. Proc. Natl Acad. Sci. USA. 2011;108:11405–11410. doi: 10.1073/pnas.1104063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henrici, R. C., van Schalkwyk, D. A. & Sutherland, C. J. Modification of pfap2mu and pfubp1 markedly reduces ring-stage susceptibility of plasmodium falciparum to artemisinin in vitro. Antimicrob. Agents Chemother.64, e01542–19 (2019). [DOI] [PMC free article] [PubMed]
- 28.Paloque L, Ramadani AP, Mercereau-Puijalon O, Augereau JM, Benoit-Vical F. Plasmodium falciparum: Multifaceted resistance to artemisinins. Malar. J. 2016;15:149. doi: 10.1186/s12936-016-1206-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LeRoux M, Lakshmanan V, Daily JP. Plasmodium falciparum biology: Analysis of in vitro versus in vivo growth conditions. Trends Parasitol. 2009;25:474–481. doi: 10.1016/j.pt.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Cheeseman IH, et al. A major genome region underlying artemisinin resistance in malaria. Science. 2012;336:79–82. doi: 10.1126/science.1215966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takala-Harrison S, et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc. Natl Acad. Sci. USA. 2013;110:240–245. doi: 10.1073/pnas.1211205110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miotto, O. et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat. Genet.47, 226–34 (2015). [DOI] [PMC free article] [PubMed]
- 33.Cerqueira GC, et al. Longitudinal genomic surveillance of Plasmodium falciparum malaria parasites reveals complex genomic architecture of emerging artemisinin resistance. Genome Biol. 2017;18:78. doi: 10.1186/s13059-017-1204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma, Y. et al. Combined transcriptome GWAS and TWAS reveal genetic elements leading to male sterility during high temperature stress in cotton. New Phytol.231, 165–181 (2021). [DOI] [PMC free article] [PubMed]
- 35.Wainberg M, et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamazon ER, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gusev A, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visscher PM, et al. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cano-Gamez E, Trynka G. From GWAS to function: Using functional genomics to identify the mechanisms underlying complex diseases. Front. Genet. 2020;11:424. doi: 10.3389/fgene.2020.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu L, et al. The origins of malaria artemisinin resistance defined by a genetic and transcriptomic background. Nat. Commun. 2018;9:5158. doi: 10.1038/s41467-018-07588-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mok S, et al. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science. 2015;347:431–435. doi: 10.1126/science.1260403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kucharski M, et al. A comprehensive RNA handling and transcriptomics guide for high-throughput processing of Plasmodium blood-stage samples. Malar. J. 2020;19:363. doi: 10.1186/s12936-020-03436-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hamilton WL, et al. Evolution and expansion of multidrug-resistant malaria in southeast Asia: A genomic epidemiology study. Lancet Infect. Dis. 2019;19:943–951. doi: 10.1016/S1473-3099(19)30392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pourhoseingholi MA, Baghestani AR, Vahedi M. How to control confounding effects by statistical analysis. Gastroenterol. Hepatol. Bed Bench. 2012;5:79–83. [PMC free article] [PubMed] [Google Scholar]
- 45.Westra HJ, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat. Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McInnes et al. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw.3, 861, 10.21105/joss.00861 (2018).
- 48.Shaw PJ, et al. Plasmodium parasites mount an arrest response to dihydroartemisinin, as revealed by whole transcriptome shotgun sequencing (RNA-seq) and microarray study. BMC Genomics. 2015;16:830. doi: 10.1186/s12864-015-2040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Natalang O, et al. Dynamic RNA profiling in Plasmodium falciparum synchronized blood stages exposed to lethal doses of artesunate. BMC Genomics. 2008;9:388. doi: 10.1186/1471-2164-9-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mok S, et al. Artemisinin-resistant K13 mutations rewire Plasmodium falciparum’s intra-erythrocytic metabolic program to enhance survival. Nat. Commun. 2021;12:530. doi: 10.1038/s41467-020-20805-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rocamora F, et al. Oxidative stress and protein damage responses mediate artemisinin resistance in malaria parasites. PLoS Pathog. 2018;14:e1006930. doi: 10.1371/journal.ppat.1006930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang J, et al. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat. Commun. 2015;6:10111. doi: 10.1038/ncomms10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ismail HM, et al. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc. Natl Acad. Sci. USA. 2016;113:2080–2085. doi: 10.1073/pnas.1600459113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kasaragod VB, et al. Pyridoxal kinase inhibition by artemisinins down-regulates inhibitory neurotransmission. Proc. Natl Acad. Sci. USA. 2020;117:33235–33245. doi: 10.1073/pnas.2008695117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Birnbaum J, et al. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science. 2020;367:51–59. doi: 10.1126/science.aax4735. [DOI] [PubMed] [Google Scholar]
- 56.Dogovski C, et al. Targeting the cell stress response of Plasmodium falciparum to overcome artemisinin resistance. PLoS Biol. 2015;13:e1002132. doi: 10.1371/journal.pbio.1002132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang T, et al. Decreased K13 abundance reduces hemoglobin catabolism and proteotoxic stress, underpinning artemisinin resistance. Cell Rep. 2019;29:2917–2928 e5. doi: 10.1016/j.celrep.2019.10.095. [DOI] [PubMed] [Google Scholar]
- 58.Bridgford JL, et al. Artemisinin kills malaria parasites by damaging proteins and inhibiting the proteasome. Nat. Commun. 2018;9:3801. doi: 10.1038/s41467-018-06221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simwela, N. V. et al. Plasmodium berghei K13 mutations mediate in vivo artemisinin resistance that is reversed by proteasome inhibition. mBio11, e02312–20 (2020). [DOI] [PMC free article] [PubMed]
- 60.Mesen-Ramirez P, et al. The parasitophorous vacuole nutrient channel is critical for drug access in malaria parasites and modulates the artemisinin resistance fitness cost. Cell Host Microbe. 2021;29:1774–1787 e9. doi: 10.1016/j.chom.2021.11.002. [DOI] [PubMed] [Google Scholar]
- 61.Mbengue A, et al. A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature. 2015;520:683–687. doi: 10.1038/nature14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bhattacharjee S, et al. Remodeling of the malaria parasite and host human red cell by vesicle amplification that induces artemisinin resistance. Blood. 2018;131:1234–1247. doi: 10.1182/blood-2017-11-814665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cui L, et al. Mechanisms of in vitro resistance to dihydroartemisinin in Plasmodium falciparum. Mol. Microbiol. 2012;86:111–128. doi: 10.1111/j.1365-2958.2012.08180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Painter HJ, Morrisey JM, Mather MW, Vaidya AB. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature. 2007;446:88–91. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- 65.Moreira CK, et al. The Plasmodium PHIST and RESA-like protein families of human and rodent malaria parasites. PLoS One. 2016;11:e0152510. doi: 10.1371/journal.pone.0152510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oberli A, et al. Plasmodium falciparum Plasmodium helical interspersed subtelomeric proteins contribute to cytoadherence and anchor P. falciparum erythrocyte membrane protein 1 to the host cell cytoskeleton. Cell Microbiol. 2016;18:1415–1428. doi: 10.1111/cmi.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schneider AG, Mercereau-Puijalon O. A new Apicomplexa-specific protein kinase family: Multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics. 2005;6:30. doi: 10.1186/1471-2164-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: The kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Min Zhanga, C. W. et al. The endosymbiotic origins of the apicoplast link fever-survival and artemisinin-resistance in the malaria parasite. Preprint at bioRxiv10.1101/2020.12.10.419788 (2021). [DOI] [PMC free article] [PubMed]
- 70.Imwong M, et al. Evolution of multidrug resistance in Plasmodium falciparum: A longitudinal study of genetic resistance markers in the greater Mekong subregion. Antimicrob. Agents Chemother. 2021;65:e0112121. doi: 10.1128/AAC.01121-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miotto O, et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat. Genet. 2013;45:648–655. doi: 10.1038/ng.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miles A, et al. Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res. 2016;26:1288–1299. doi: 10.1101/gr.203711.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 74.Bozdech Z, et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc. Natl Acad. Sci. USA. 2008;105:16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mackinnon MJ, et al. Comparative transcriptional and genomic analysis of Plasmodium falciparum field isolates. PLoS Pathog. 2009;5:e1000644. doi: 10.1371/journal.ppat.1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quinlan AR, Hall IM. BEDTools: Aa flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lemieux JE, et al. Statistical estimation of cell-cycle progression and lineage commitment in Plasmodium falciparum reveals a homogeneous pattern of transcription in ex vivo culture. Proc. Natl Acad. Sci. USA. 2009;106:7559–7564. doi: 10.1073/pnas.0811829106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.John CR, et al. M3C: Monte Carlo reference-based consensus clustering. Sci. Rep. 2020;10:1816. doi: 10.1038/s41598-020-58766-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the main text or the supplementary materials or from the corresponding author upon request. All transcriptome data used in this study are available at NCBI’s Gene Expression Omnibus (GEO) database with the series accession number GSE149735 for microarray and GSE169520 for RNA-seq.