Abstract
Animal products may play a role in developing and spreading antimicrobial resistance in several ways. On the one hand, residues of antibiotics not adequately used in animal farming can enter the human body via food. However, resistant bacteria may also be present in animal products, which can transfer the antimicrobial resistance genes (ARG) to the bacteria in the consumer’s body by horizontal gene transfer. As previous studies have shown that fermented foods have a meaningful ARG content, it is indicated that such genes may also be present in silage used as mass feed in the cattle sector. In our study, we aspired to answer what ARGs occur in silage and what mobility characteristics they have? For this purpose, we have analyzed bioinformatically 52 freely available deep sequenced silage samples from shotgun metagenome next-generation sequencing. A total of 16 perfect matched ARGs occurred 54 times in the samples. More than half of these ARGs are mobile because they can be linked to integrative mobile genetic elements, prophages or plasmids. Our results point to a neglected but substantial ARG source in the food chain.
Subject terms: Metagenomics, Antimicrobial resistance
Introduction
In intensive cattle farming, silage is an essential component of feed. An average dairy cow consumes 25–27 kg of this forage a day, reaching up to a silage consumption of 12,500 kg per lactation1,2. Silage is most commonly produced from maize or legume plants by the process of anaerobic fermentation. Throughout the fermentation process, fermenting microorganisms, including bacteria, multiply. As a result of this biochemical transformation, the silage is enriched with beneficial nutrients. If bacteria that are involved in the process harbor antimicrobial resistance genes (ARGs), the amount of these genes in the silage will increase in parallel with the bacterial counts. Consequently, silage, as a mass feed may continuously supply the gastrointestinal tract of animals with bacteria carrying ARGs. Bacteria entering the digestive system may come into contact with the host microbiota that facilitates the exchange of bacterial genes (e.g. ARGs) by horizontal gene transfer (HGT). HGT may take place as a result of three different processes: conjugation, transduction and transformation. Except for transformation, by which a bacterium can take up any gene from its environment, the routes of HGT require particular active delivery processes. By conjugation, cell-to-cell contact provides the opportunity for a copy of a plasmid to translocate to a recipient bacterium3. Transduction negates the condition of cell-to-cell contact, as in this case, bacteriophages act as a conduit for shuttling genes among bacteria4. The genetic environment of the genes involved in the transfer significantly influences the efficacy of the latter two HGT processes, i.e., the genes’ mobility. The reason why the mobility characteristics of ARGs involved in silage are worth taking into consideration is the following. If ARGs from silage are transmitted to pathogenic bacteria within an animal’s body, efficacy of antibiotic (AB) treatment may be reduced on the consequent bacterial diseases. In addition, in case of the gut colonization of silage-borne bacteria that carry ARGs, the appearance and enrichment of bacterial ARGs may take place in the animals’ environment after defecation. Decreased efficacy of AB treatments may result in economic loss, and the increased environmental ARG level may have additional veterinary and human health consequences. It is proven in former publications that the number of ARGs in fermented dairy products can increase due to the multiplication of fermenting bacteria5. Nevertheless, the description of this phenomenon cannot be found for silage in the literature. Our study aimed to examine the diversity, bacterial relatedness and mobility potential of ARGs deriving from silage. For this purpose, freely available next-generation sequencing (NGS) shotgun metagenome datasets were analyzed by a unified bioinformatics pipeline.
Results
Based on the taxon classification performed on a database containing complete reference genomes of plants, the most dominant plants in the silage belong to the Medicago genus and most likely to the alfalfa (M. sativa) species. Further results of the analysis of the 52 shotgun metagenomic sequenced samples (Table 2) are summarised in the following sections. After presenting the bacteriome and the identified AGRs (resistome), predictions regarding the mobility potential of ARGs were also resumed based on genetic characteristics that may play a significant role in HGT.
Table 2.
Analyzed samples.
| BioProject | PRJNA495415 | PRJNA764355 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Group | A | B | C | D | E | ||||
| Id | Run | Id | Run | Id | Run | Id | Run | Id | Run |
| 1 | SRR7990583 | 11 | SRR7990582 | 21 | SRR7990580 | 31 | SRR7990581 | 41 | SRR16036389 |
| 2 | SRR7990587 | 12 | SRR7990586 | 22 | SRR7990585 | 32 | SRR7990584 | 42 | SRR16036390 |
| 3 | SRR7990591 | 13 | SRR7990590 | 23 | SRR7990589 | 33 | SRR7990588 | 43 | SRR16036391 |
| 4 | SRR7990592 | 14 | SRR7990593 | 24 | SRR7990594 | 34 | SRR7990595 | 44 | SRR16036392 |
| 5 | SRR7990598 | 15 | SRR7990599 | 25 | SRR7990596 | 35 | SRR7990597 | 45 | SRR16036393 |
| 6 | SRR7990604 | 16 | SRR7990605 | 26 | SRR7990600 | 36 | SRR7990601 | 46 | SRR16036394 |
| 7 | SRR7990608 | 17 | SRR7990609 | 27 | SRR7990602 | 37 | SRR7990603 | 47 | SRR16036395 |
| 8 | SRR7990610 | 18 | SRR7990611 | 28 | SRR7990606 | 38 | SRR7990607 | 48 | SRR16036396 |
| 9 | SRR7990612 | 19 | SRR7990613 | 29 | SRR7990614 | 39 | SRR7990615 | 49 | SRR16036397 |
| 10 | SRR7990616 | 20 | SRR7990617 | 30 | SRR7990618 | 40 | SRR7990619 | 50 | SRR16036398 |
| 51 | SRR16036399 | ||||||||
| 52 | SRR16036400 | ||||||||
The samples of dataset PRJNA495415 were taken on days 0, 7, 14 and 28 were classified in groups A, B, C and D, respectively. All samples from BioProject PRJNA764355 are assigned to group E. Column Run contains the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) run identifiers.
Bacteriome
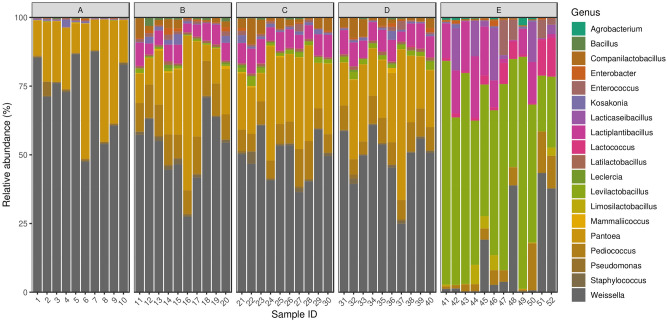
By taxon classification, the number of reads aligning to bacterial genomes varied by samples (median: 20.6 × 106, IQR: 2.9 × 106). The relative abundances of genera that achieved more than 1% of the bacterial hits in any of the samples are shown in Fig. 1.
Figure 1.
Silage core bacteriome. Relative abundances of genera that achieved more than 1% of the bacterial hits in any of the samples. The elements of the PRJNA495415 dataset were taken on days 0, 7, 14 and 28 were classified into groups A, B, C and D, respectively. All items from BioProject PRJNA764355 are assigned to group E.
The dominant bacterial genera (with mean abundance) in descending order were Weissella (45.7%), Pantoea (18.5%), Levilactobacillus (13.5%), Pediococcus (6.7%), Lactiplantibacillus (6.3%), Companilactobacillus (1.7%), Lacticaseibacillus (1.3%), Enterococcus (1.2%), Lactococcus (1%), Kosakonia (0.8%), Staphylococcus (0.6%), Enterobacter (0.5%), Latilactobacillus (0.5%), Bacillus (0.4%), Limosilactobacillus (0.4%), Pseudomonas (0.4%), Leclercia (0.2%), Mammaliicoccus (0.2%), Agrobacterium (0.1%).
Resistome
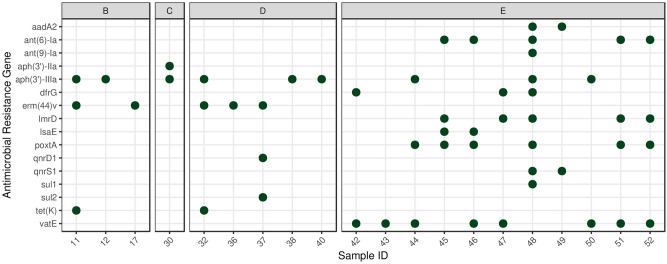
The median length of the filtered contigs harboring ARGs constructed by de novo assembly was 4,204 bp (IQR: 2,832). The number of ARGs found on the contigs ranged from 1 to 2. The identified 16 ARG types appeared 54 times in 20 of the analyzed 52 samples. These ARGs were the following: aadA2, ant(6)-Ia, ant(9)-Ia, aph(3’)-IIa, aph(3’)-IIIa, dfrG, erm(44)v, lmrD, lsaE, poxtA, qnrD1, qnrS1, sul1, sul2, tet(K), vatE (Fig. 2). The resistance mechanism of identified ARGs was the antibiotic inactivation (48.1%), antibiotic target protection (20.3%), antibiotic efflux (13.0%), antibiotic target alteration (9.3%), antibiotic target replacement (9.3%) in descending order of frequency. Table 1 shows the bacterial species to which the ARG harboring contigs were assigned based by the taxon classification. In addition, the table also presents which drug classes are affected by the ARGs.
Figure 2.
Identifed antimicrobial resistance genes (ARGs) by samples. Perfect ARG matches were plotted by samples. The data of the PRJNA495415 taken on days 0, 7, 14 and 28 were classified in groups A, B, C and D, respectively. All samples from BioProject PRJNA764355 were assigned to group E.
Table 1.
Identified ARGs and the drug classes affected by them per bacterial species of origin.
| Bacteria | ARG(s) | Drug class |
|---|---|---|
| Acinetobacter baumannii | aadA2 | Aminoglycoside |
| Amylolactobacillus amylophilus | ant(6)-Ia | Aminoglycoside |
| Bacillus subtilis | aph(3’)-IIa | Aminoglycoside |
| Cronobacter sp. JZ38 | qnrS1 | Fluoroquinolone |
| Enterobacter hormaechei | aadA2, sul1 | Aminoglycoside, sulfonamide |
| Enterococcus faecalis | aph(3’)-IIIa | Aminoglycoside |
| Enterococcus faecium | poxtA | Lincosamide, macrolide, oxazolidinone, phenicol, pleuromutilin, streptogramin, tetracycline |
| Escherichia coli | sul2 | Sulfonamide |
| Gracilibacillus sp. SCU50 | dfrG | Diaminopyrimidine |
| Lacticaseibacillus manihotivorans | ant(6)-Ia | Aminoglycoside |
| Lacticaseibacillus paracasei | poxtA | Lincosamide, macrolide, oxazolidinone, phenicol, pleuromutilin, streptogramin, tetracycline |
| Lactiplantibacillus plantarum | aph(3’)-IIIa, poxtA, vatE | Aminoglycoside, lincosamide, macrolide, oxazolidinone, phenicol, pleuromutilin, streptogramin, tetracycline |
| Lactococcus lactis | lmrD | Lincosamide |
| Levilactobacillus brevis | poxtA | Lincosamide, macrolide, oxazolidinone, phenicol, pleuromutilin, streptogramin, tetracycline |
| Ligilactobacillus acidipiscis | ant(9)-Ia | Aminoglycoside |
| Providencia rettgeri | qnrD1 | Fluoroquinolone |
| Staphylococcus aureus | ant(6)-Ia, tet(K) | Aminoglycoside, tetracycline |
| Staphylococcus carnosus | erm(44)v | Lincosamide, macrolide, streptogramin |
| Staphylococcus pseudoxylosus | erm(44)v | Lincosamide, macrolide, streptogramin |
| Staphylococcus saprophyticus | erm(44)v | Lincosamide, macrolide, streptogramin |
| Streptococcus suis | lsaE | Lincosamide, macrolide, oxazolidinone, phenicol, pleuromutilin, streptogramin, tetracycline |
| Tetragenococcus halophilus | aph(3’)-IIIa | Aminoglycoside |
| Weissella paramesenteroides | ant(6)-Ia | Aminoglycoside |
Mobilome
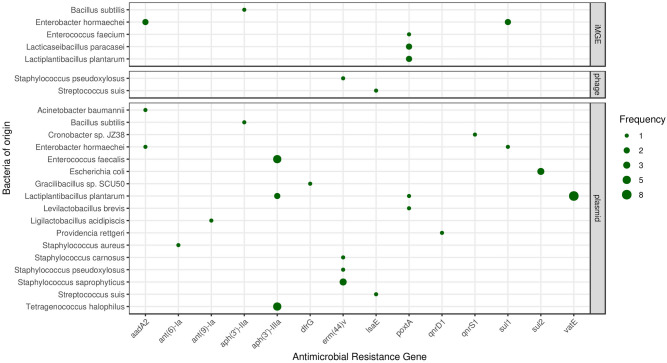
We found a total of 53 ARGs that are assumably mobile. Ten of these ARGs are linked to integrative mobile genetic elements (iMGE). A further two ARGs were detected in prophages and forty-one on plasmids. The frequencies of ARGs associated with iMGEs, phages and plasmids are summarized in Fig. 3 by bacterial species of origin.
Figure 3.
Mobile antimicrobial resistance gene frequency by bacteria of origin. The size of the dots indicates the occurrence frequency of the ARGs flanked by iMGE, positioned in a plasmid or a phage.
Following the distance method proposed by Johansson et al.6, integrated mobile genetic element associated ARGs were detected in five samples (30, 45, 46, 48, 52) and five species (B. subtilis, E. hormaechei, E. faecium, L. paracasei, L. plantarum). B. subtilis associated aph(3ʹ)-IIa in sample 30, and poxtA of E. faecium in sample 45, of L. plantarum in sample 52, and of L. paracasei in sample 46 were detected as iMGE linked gene. Sul1 and aadA2 were detected in E. hormaechei co-existed with integrated mobile elements in sample 48. Two prophage-linked ARGs were identified, the contig harboring erm(44)v classified to S. pseudoxylosus by VirSorter2 was found to be of dsDNA phage origin while the contig of lsaE from S. suis was predicted as ssDNA derived. These phage associated ARGs were detected in sample 37 and sample 45 respectively. Contings with ARGs were predicted to belong to plasmids in 19 samples (Nr. 11, 12, 17, 30, 32, 36, 37, 40, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52).
Discussion
Throughout our study, numerous perfect ARG matches were identified in the metagenome of Medicago silage samples. All but group A of the analyzed subsets had at least one sample containing one or more ARG. Among the PRJNA495415 Bioproject samples, the highest number of ARGs were found in group D. The interpretation of this finding is limited due to the lack of detailed information on the samples. Interestingly, all but one of the PRJNA764355 bioproject samples contained ARGs. Due to the lack of metadata, it is hard to find any reason for this high ARG level. However, one possible cause might be that the PRJNA764355 samples were sequenced deeper and thus contained approximately 1.3 times more reads than the PRJNA495415 samples. It is known from previous studies that deeper sequencing leads to the generation of more complete genes by the de novo assembly5,7.
In the following, our results will be interpreted from a perspective of bacteriological significance, genomic relevance and furthermore, antimicrobial stewardship and possible clinical aspects.
Taking the microbiome into consideration, bacteria that were predicted to harbor the identified ARGs can be classified according to their presence in silage. In the literature, the following bacteria are mentioned to be characteristic for silage: B. subtilis8 E. faecium9 E. coli10, L. plantarum11, L. lactis12, L. brevis13–15, L. acidipiscis12 W. paramesenteroides16. The genera of these species dominate the bacteriome of the samples. The identified Cronobacter sp. JZ3817 may be of plant origin. However, it can be assumed that other species may be present as contaminants of the silage: A. amylophilus, E. hormaechei, E. faecalis, Gracilibacillus sp. SCU50, L. manihotivorans, L. paracasei, P. rettgeri, S. aureus, S. carnosus, S. pseudoxylosus, S. saprophyticus, S. suis, T. halophilus. Nevertheless, some of these bacteria are members of the Lactobacillaceae family, the Leuconostoc or Enterobacter genera. Numerous species of these groups are typical for fermented food and feed components.
From a genomic point of view, the following was found in the literature regarding the co-occurrence of the ARGs identified in our study and the bacteria carrying them. AadA2 encoding an aminoglycoside nucleotidyltransferase has been described in A. baumanni in former publications18,19. ant(6)-Ia, that is an aminoglycoside nucleotidyltransferase gene, appears in many species, including Lactobacillus spp.20. Its species-specific association with A. amylophilus has not been described in any former publications. aph(3’)-IIa, an aminoglycoside phosphotransferase21, to our knowledge, has not been detected in B. subtilis up untill now. QnrS1 encoding a quinolone resistance protein was originally identified in Shigella flexneri22. In line with our results, this gene has recently been mentioned to appear in Cronobacter spp. in a case report23. E. hormaechei deriving aadA2 and sul1, a sulfonamide resistant dihydropteroate synthase gene that is described to appear in Gram-negative bacteria21 have been reported to appear in the genom of Enterobacter spp. and E. hormaechei, respectively in former publications as well24,25. Within the Enterococcus genus, two perfect ARG matches were identified, namely aph(3’)-IIIa in E. faecalis and poxtA in E. faecium. aph(3’)-IIIa is an aminoglycoside phosphotransferase that normally appears in S. aureus21 and Enterococcus spp.26, while poxtA is a gene encoding an ABC-F subfamily (ATP-binding cassette-F) protein that facilitates resistance to tetracycline, phenicol, and oxazolidinone via modification of the bacterial ribosome. First detection of poxtA took place in a methicillin-resistant S. aureus strain21, followed by other bacterial species, including E. faecium27. Sul2, a sulfonamide resistant dihydropteroate synthase of Gram-negative bacteria is commonly described in E. coli21,28. DfrG is a plasmid-encoded dihydrofolate reductase21 that, to our knowledge, has not been described in Gracibacillus spp. up untill now, but has already appeared in the Bacillaceae family29. ant(6)-Ia, an aminoglycoside nucleotidyltransferase gene appears in many species, including Lactobacillus spp.20. Its species-specific association with L. manihotivorans has not been described in any publications. PoxtA that was detected in L. paracasei, L. plantarum and L. brevis in the silage samples, has been described to appear in Lactobacillaceae, namely L. acidophilus, but not in these very species30. Another species that was detected harboring aph(3’)-IIIa in the silage samples was L. plantarum. This finding is in line with the ARG-species match results mentioned in former publications31. Furthermore, L. plantarum was also associated with vatE that encodes an acetyltransferase conferring resistance against streptogramins21. VatE was originally found in E. faecium21 and has since then been identified in Lactobacillaceae32, but not specifically in L. plantarum. L. acidipiscis ant(9)-Ia, an aminoglycoside nucleotidyltransferase gene21 was associated with this genus for first within this study. Gene qnrD1 encoding a quinolone resistance protein that is normally detected in Salmonella enterica21, has already been found in Providencia spp.33 and was attached to P. rettgeri in our study as well. S. aureus could have been associated with two ARGs, ant(6)-Ia and tetK encoding a tetracycline efflux protein, that are both common findings in Staphylococcus spp.34,35. Although, erm(44)v was first detected in the S. saprophyticus36, no literature could be found about the appearance of this gene in S. carnosus or in S. pseudoxylosus species. lsaE encoding another ABC-F subfamily protein conferring resistance to pleuromutilin, lincosamide, and streptogramin A is a common finding in Streptococcus spp.37 and has also been associated with S. suis in previous publications38. Besides the bacterial species mentioned above, aph(3’)-IIIa was also detected in T. halophilus. This ARG is often appears in Enterococcaceae21 but has not yet been written down in this species. Furthermore, to our knowledge, W. paramesenteroides associated ant(6)-Ia has first been detected in this study.
Throughout our study, several ARGs were predicted to be co-occurring with genetic attributes facilitating mobility. The bioinformatic analysis of the mobility characteristics relied upon the identification of three major mobility determination groups, namely iMGEs, phages and plasmids. We found aph(3’)-IIa linked to an integrated mobile genetic element in B. subtilis that is in line with similar findings of E. coli.21 While aadA2 and sul1 have both been described to appear on plasmids in E. hormaechei39, we found them associated with iMGEs. Our finding on iMGE flanked poxtA in E. faecium is in line with the current literature40. We found the same co-occurrence, namely poxtA and an iMGE, in L. paracasei. This phenomenon has not been published in that species to the best of our knowledge. Gene erm(44)v and lsaE were associated with prophages in S. pseudoxylosus and S. suis. While a similar linkage can be found in the literature in connection with erm(44)v41, no details of mobility characteristics are mentioned in a recent report of the latter gene38. All other mobile ARGs were detected on contigs that were predicted to derive from plasmids. In case of aadA2 in A. baumannii21; aadA2 and sul1 in E. hormaechei21; aph(3’)-IIIa in E. faecalis21; sul2 in E. coli21; aph(3’)-IIIa in L. plantarum42; qnrD1 in P. rettgeri21; ant(6)-Ia in S. aureus21 and erm(44)v in S. saprophyticus41 plasmid associations have been formerly described in the literature. To our knowledge, no publications have yet been released on the plasmid occurence of aph(3’)-IIa in B. subtilis, dfrG in Gracilibacillus sp. SCU50, ant(9)-Ia in L. acidipiscis, lsaE in S. suis, qnrS1 in Cronobacter sp. JZ38, erm(44)v in S. carnosus and S. pseudoxylosus. Hao et al. described poxtA embedded in a multi-resistance plasmid with mobile elements flanking in E. fecalis. This gene has been found in a number of Gram-positive bacteria, including enterococci as well, but it has neither been identified in L. plantarum nor L. brevis43. Previous findings confirm the occurrence of vat(E) on plasmids44. Nevertheless, in spite of its frequent presence in enterococci45 there is no evidence of its former plasmid-associated appearance in L. plantarum. We found that gene aph(3’)-IIIa of T. halophilus was encoded on a plasmid that is consistent with the fact that aph(3’)IIIa is often identified on high molecular weight plasmids and chromosomes of the enterococcal species46. Nonetheless, to the best of our knowledge, a description of the aph(3’)-IIIa gene in T. halophilus is a pioneer finding.
The mobility characteristics of the ARGs may not only provide us with information regarding the public health risk that may be associated with the samples, but also point to the possible origins of the genes. Regardless of human intervention, ARGs are present in the microbial communities47. However, antimicrobial use and abuse intensifies the horizontal transfer of ARGs and thus contributes to the spread of AMR. In the animal production sector, the use of antibiotics is common, thus bacteria appearing in the feces and in the surroundings of the animals (e.g. in farm air, on tools, vehicles or other settings related to animals) often harbor bacteria with an advanced ARG set. Silage may get in direct physical contact with these bacteria at the farms and thus get contaminated with a few ARGs. Consequently, the presence of ARGs in the silage samples was well-expected, but the abundance of resistance genes and MGEs may increase due to the application of antibiotics.
Examining further aspects of antimicrobial stewardship and possible clinical relevance, phenotypical manifestations and public health considerations associated with the detected ARGs are both important. Intense antimicrobial use (AMU) can be associated with the headway of AMR, as antibiotic pressure selects for bacteria carrying ARGs that facilitate bacterial survival. Quantifically, the majority of AMU around the globe occurs in agricultural settings48,49. Intensive farming, that serves to fulfill the high global demand for animal proteins relies on an antibiotic infrastructure to treat and prevent disease and occasionally, to increase feed efficacy. In order to maximize economic gains, few countries still apply regulations that facilitate the use of low doses of antibiotics as growth promoters50, while other regions, like the U.S. or Europe, have banned this practice. Nevertheless, besides the treatment of symptomatic infectious diseases, antibiotics are still widely used in the livestock sector for metaphylactic and prophylactic purposes in higher doses51,52. Even though, compared to the poultry and pig production sector, average antibiotic usage has relatively lower rates by cattle53, antimicrobial compounds are often chosen in this species as well. In cattle farming, mastitis is the most predominant reason for the administration of antibiotics by adult cattle, while enteritis and pneumonia is the most common reason for calves54,55. According to various reports and studies from around the world54,56,57 tetracyclines are of inevitable significance in the medication of cattle, while beta-lactams, macrolides, sulfonamides, lincosamides and ionophore antibiotics are also very widely used. Of the European Medicines Agency (EMA) Highest-Priority Critically Important Antibiotics (HPCIAs), namely third and fourth generation cephalosoprines, fluoroquinolones and polymixins, polymixins and fluoroquinolones are the most applied, although their sales rates are still far below the most frequently administered antibiotic groups by livestock species57. In our samples E. faecium, L. paracasei, L. plantarum, L. brevis, S. aureus and S. suis harbored genes, namely poxtA and lsaE that may confer resistance against multiple antibiotic groups, including tetracyclines. Moreover, poxtA was detected in the proximity of iMGEs in L. paracasei and E. faecium and harbored on a plasmid in L. brevis. In line with our findings, Enterococcus species related to cattle were heavily associated with tetracycline resistance by other authors too58. At some species poxtA and lsaE were even predicted to co-occur with more than one MGE type. In the genome of L. plantarum poxtA was predicted to be positioned on a plasmid and associated with an iMGE, while S. suis associated lsaE was located on a plasmid attached to a phage. Such genetic features may contribute to the horizontal transfer of ARGs among bacteria which is of outstanding clinical relevance in the case of such a commonly applied antibiotic group in cattle medicine, as tetracyclines. Perfect matches of genes conferring resistance against other clinically significant antibiotic groups, such as macrolides and sulfonamides were also identified in the genome of E. hormaechei, E. faecium, E. coli, L. paracasei, L. plantarum, L. lactis, L. brevis, S. carnosus, S. pseudoxylosus, S. saprophyticus and S. suis. Of these genes, lsaE, sul1, sul2 and poxtA were even predicted to have enhanced mobility due to their association with multiple MGE groups. The only perfect match for an ARG against HPCIAs, qnrS1, that can confirm resistance against fluoroquinolones, has been detected in a Cronobacter spp. The presence of several ARGs presumably associated with iMGEs in the feed of dairy cows harbors the potential to affect the resident microbiota of the animals. As B. subtilis and E. faecium frequently appear in probiotics for cattle59,60 it is possible that some microorganisms colonize niches in the foregut and proliferate the ARGs they possess. However, even if they cannot reproduce in the ruminal environment, ARGs can still be disseminated through horizontal gene transfer, especially in the presence of antibiotic therapy. Furthermore, ARGs can possibly spread further, to lower gastrointestinal (GI) regions. Fecal microbiota transfer administered to the stomach could restore the microbial population of the colon in human patients61, indicating a high volume of viable bacteria reaching the distal regions. Similar results were found in cattle with rumen microbiota transplantation affecting the microbial population of the hindgut62. If ARGs spread all around the GI tract, serious animal and public health concerns could be raised. Among enteral diseases, salmonellosis is the major indication of antibiotic therapy in dairy cattle. Enhanced antibiotic resistance of these bacteria could contribute to the economic loss from the disease as many strains already exhibit resistance to several antibiotics63. Furthermore, during pathological conditions, like ruminal acidosis, bacteria can translocate to distant locations in the host’s body. Interestingly bacterial translocation was even described in the absence of GI diseases in case of specific microorganisms in humans and rodents64,65.
As a consequence of the colonization and possible ARG proliferation processes, pathologies caused by phenotypically resistant bacteria can induce animal welfare and economic issues. Animals harboring ARGs in their gut can contaminate their environment with ARGs through fecal matter as well as farm workers who get in direct contact with the animals, even consumers of dairy products can be affected, as farm animal-borne bacteria that harbor potentially mobile ARGs66–68 might be distributed by products intended for human consumption. For instance, we have previously found ARGs in raw milk samples provided for human consumption69. Fecal contamination during milking70 is a possible way of ARG transfer into raw milk, however other routes are also possible. In humans and in rodents for instance, maternal mononuclear cells transfer microorganisms to milk during lactation66,67. The possibility for this phenomenon was described in cattle as well68.
ARGs that are transferred to the human body through these routes might decrease the efficacy of antibiotic therapy. In order to gain a deeper insight into the exact role of silage in possible ARG transmission processes, many points still need to be examined and clarified. It would be essential to analyse the colonization success of ARG harboring silage-borne bacteria that enter the body of animals and the extent of ARG transfer of invasive donor bacteria to recipient bacteria living in the gastrointestinal tract. The silage involved in the study is of Medicago origin, and our results are based on data from only two projects. Hence, it would also be necessary to investigate the ARG content of other alfalfa and corn silages.
Antimicrobial resistance is an emerging global threat to public health that both affects agriculture and the healthcare sector. The usage of antibiotics in livestock species exceeds the rate of human applications71. Antibiotic use in food animal medicine is also considered a risk as it may provide an indirect transfer route of antibiotic residual72 ARGs via the food chain73. Even though antimicrobials administered for veterinary use, may exert an undesired effect on the food chain, the presence of ARGs in dairy cattle nutrition research is still underrepresented in the literature. According to our results, microbial mass contained in fermented feeds have other medical risks than transmitting contagious diseases, like listeriosis74. The bacterial content of these mass feeds, that is either, required for the fermentation processes or collected from various sources of contamination on the farms, could play an essential role in the ARG shift through the food chain.
Materials and methods
Data
We searched appropriate datasets in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) repository. In December 2021, only two shotgun metagenomic BioProjects (PRJNA49541575, PRJNA764355) could have been found that had adequate depth for the de novo assembly that our study is based on. The median read count (interquartile range, IQR) of the samples was 26.5 × 106 (3.0 × 106) and 34.7 × 106 (1.5 × 106) in datasets PRJNA495415 and PRJNA764355, respectively. There is limited metadata available of the samples in the NCBI SRA database (Table 2). Nevertheless, it can be assumed from the metadata that the samples of PRJNA495415 were taken at different fermentation periods. Samples were taken on days 0, 7, 14 and 28 were classified in groups A, B, C and D, respectively. Based on metadata of PRJNA764355 samples, no such stratification was possible, so all samples were classified as group E.
Bioinformatic analysis
Quality based filtering and trimming of the raw short reads was performed with TrimGalore (v.0.6.6, https://github.com/FelixKrueger/TrimGalore), setting 20 as a quality threshold. Only reads longer than 50 bp were retained and taxonomically classified using Kraken2 (v2.1.1)76 and a database created (24/03/2021) from the NCBI RefSeq complete archaeal, bacterial, viral and plant genomes. For this taxon assignment the -confidence 0.5 parameter was used to obtain more precise species level hits. The taxon classification data was managed in R77 using functions of the packages phyloseq78 and microbiome79. The preprocessed reads were assembled to contigs with MEGAHIT (v1.2.9)80 using default settings. The contigs were also classified taxonomically with Kraken2 with the same database as above. From the contigs all possible open reading frames (ORFs) were gathered with Prodigal (v2.6.3)81. The protein translated ORFs were aligned to the ARG sequences of the Comprehensive Antibiotic Resistance Database (CARD, v.3.1.3)21,82 by Resistance Gene Identifier (RGI, v5.2.0) with Diamond83. ORFs having a perfect match against the CARD database were exclusively kept for further analysis. Integrative mobile genetic element (iMGE) content of contigs harboring ARG was analyzed with MobileElementFinder (v1.0.3) and its database (v1.0.2)6. Following the distance concept of Johansson et al.6 for each bacterial species, only those with a distance threshold defined within iMGEs and ARGs were considered associated. In the MobileElementFinder database (v1.0.2) for E. hormaechei, the longest composite transposon (cTn) was the Tn3000. In case of this species, its length (11,823 bp) was taken as the cut-off value. For E. faecium, this threshold was the length of the Tn6246 transposon, namely 5,147 bp. As the database neither contains species-level, nor genus-level cTn data for Bacillus, Lactiplantibacillus and Lacticaseibacillus species, a general cut-off value was chosen for the contigs of these species. This value was declared as the median of the longest cTns per species in the database (10,098 bp). The plasmid origin probability of the contigs was estimated by PlasFlow (v.1.1)84 The phage content of the assembled contigs was prediced by VirSorter2 (v2.2.3)85. The findings were filtered for dsDNAphages and ssDNAs. All data management procedures, analyses and plots were performed in R environment (v4.1.0)77.
Acknowledgements
The authors would like to thank the providers of BioProject PRJNA495415 and PRJNA764355. The research was supported by the European Union’s Horizon 2020 research and innovation program under Grant Agreement No. 874735 (VEO) and the Ministry of Innovation and Technology NRDI Office within the framework of the Artificial Intelligence National Laboratory Program MILAB. The authors are thankful to Oz Kilim in the manuscript editing.
Author contributions
N.S. takes responsibility for the integrity of the data and the accuracy of the data analysis. A.G.T., M.P., N.S. and S.Á.N. conceived the concept of the study. A.G.T., M.P., N.S. and S.Á.N. participated in the bioinformatic analysis. A.G.T., K.S., M.P., N.S. and S.Á.N. participated in the drafting of the manuscript. A.G.T., K.S., M.P., N.S. and S.Á.N. carried out the critical revision of the manuscript for important intellectual content. All authors read and approved the final manuscript.
Funding
Open access funding provided by University of Veterinary Medicine.
Data availability
The datasets analysed in the current study are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) repository and can be accessed through the PRJNA495415 and PRJNA764355 BioProject identifiers.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Eastridge M. Major advances in applied dairy cattle nutrition. J. Dairy Sci. 2006;89:1311–1323. doi: 10.3168/jds.S0022-0302(06)72199-3. [DOI] [PubMed] [Google Scholar]
- 2.Driehuis F, Spanjer M, Scholten J, Te Giffel M. Occurrence of mycotoxins in maize, grass and wheat silage for dairy cattle in the Netherlands. Food Addit. Contam. 2008;1:41–50. doi: 10.1080/19393210802236927. [DOI] [PubMed] [Google Scholar]
- 3.Cabezón E, Ripoll-Rozada J, Peña A, De La Cruz F, Arechaga I. Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 2015;39:81–95. doi: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- 4.Goh, S. Clostridium difficile: Methods and Protocols, chap. Phage Transduction, 177–185 (Springer, 2016).
- 5.Tóth AG, et al. A glimpse of antimicrobial resistance gene diversity in kefir and yoghurt. Sci. Rep. 2020;10:22458. doi: 10.1038/s41598-020-80444-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson MH, et al. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021;76:101–109. doi: 10.1093/jac/dkaa390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- 8.McAllister T, et al. Silage review: Using molecular approaches to define the microbial ecology of silage. J. Dairy Sci. 2018;101:4060–4074. doi: 10.3168/jds.2017-13704. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Nishino N. Monitoring the bacterial community of maize silage stored in a bunker silo inoculated with Enterococcus faecium, Lactobacillus plantarum and Lactobacillus buchneri. J. Appl. Microbiol. 2011;110:1561–1570. doi: 10.1111/j.1365-2672.2011.05010.x. [DOI] [PubMed] [Google Scholar]
- 10.Ogunade I, et al. Bacterial diversity and composition of alfalfa silage as analyzed by Illumina MiSeq sequencing: Effects of Escherichia coli O157:H7 and silage additives. J. Dairy Sci. 2018;101:2048–2059. doi: 10.3168/jds.2017-12876. [DOI] [PubMed] [Google Scholar]
- 11.Zhao S, Wang Y, Yang F, Wang Y, Zhang H. Screening a Lactobacillus plantarum strain for good adaption in alfalfa ensiling and demonstrating its improvement of alfalfa silage quality. J. Appl. Microbiol. 2020;129:233–242. doi: 10.1111/jam.14604. [DOI] [PubMed] [Google Scholar]
- 12.Khota W, Pholsen S, Higgs D, Cai Y. Natural lactic acid bacteria population of tropical grasses and their fermentation factor analysis of silage prepared with cellulase and inoculant. J. Dairy Sci. 2016;99:9768–9781. doi: 10.3168/jds.2016-11180. [DOI] [PubMed] [Google Scholar]
- 13.Xu Z, He H, Zhang S, Kong J. Effects of inoculants Lactobacillus brevis and Lactobacillus parafarraginis on the fermentation characteristics and microbial communities of corn stover silage. Sci. Rep. 2017;7:13614. doi: 10.1038/s41598-017-14052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feyereisen M, et al. Comparative genome analysis of the Lactobacillus brevis species. BMC Genom. 2019;20:416. doi: 10.1186/s12864-019-5783-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paradhipta DHV, et al. Dual-purpose inoculants and their effects on corn silage. Microorganisms. 2020;8:765. doi: 10.3390/microorganisms8050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Björkroth KJ, et al. Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov., detected in food and clinical samples. Int. J. Syst. Evol. Microbiol. 2002;52:141–148. doi: 10.1099/00207713-52-1-141. [DOI] [PubMed] [Google Scholar]
- 17.Eida AA, et al. Genome insights of the plant-growth promoting bacterium Cronobacter muytjensii JZ38 with volatilemediated antagonistic activity against Phytophthora infestans. Front. Microbiol. 2020 doi: 10.3389/fmicb.2020.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, et al. Prevalence and characterization of integrons in multidrug resistant Acinetobacter baumannii in Eastern China: A multiple-hospital study. Int. J. Environ. Res. Public Heal. 2015;12:10093–10105. doi: 10.3390/ijerph120810093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mak JK, Kim M-J, Pham J, Tapsall J, White PA. Antibiotic resistance determinants in nosocomial strains of multidrug-resistant Acinetobacter baumannii. J. Antimicrob. Chemoth. 2009;63:47–54. doi: 10.1093/jac/dkn454. [DOI] [PubMed] [Google Scholar]
- 20.Dec M, Nowaczek A, Stepien-Pyśniak D, Wawrzykowski J, Urban-Chmiel R. Identification and antibiotic´ susceptibility of lactobacilli isolated from turkeys. BMC Microbiol. 2018;18:168. doi: 10.1186/s12866-018-1269-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia B, et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hata M, et al. Cloning of a novel gene for quinolone resistance from a transferable plasmid in Shigella flexneri 2b. Antimicrob. Agents Chemother. 2005;49:801–803. doi: 10.1128/AAC.49.2.801-803.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachowska M, Izdebski R, Urbanowicz P, Zabicka D, Królak-Olejnik B. Infection of˙ Cronobacter sakazakii ST1 producing SHV-12 in a premature infant born from triplet pregnancy. Microorganisms. 2021;9:1878. doi: 10.3390/microorganisms9091878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du N, et al. Transmission and characterization of bla NDM-1 in Enterobacter cloacae at a teaching hospital in Yunnan, China. Ann. Clin. Microbiol. Antimicrob. 2017;16:58. doi: 10.1186/s12941-017-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Indugu N, Sharma L, Jackson CR, Singh P. Whole-genome sequence analysis of multidrug-resistant Enterobacter hormaechei isolated from imported retail shrimp. Microbiol. Resour. Announc. 2020;9:e01103–e1120. doi: 10.1128/MRA.01103-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woegerbauer M, Kuffner M, Domingues S, Nielsen KM. Involvement of aph(3)-IIa in the formation of mosaic aminoglycoside resistance genes in natural environments. Front. Microbiol. 2015;6:442. doi: 10.3389/fmicb.2015.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Na S-H, et al. Detection of the phenicol–oxazolidinone resistance gene poxta in Enterococcus faecium and Enterococcus faecalis from food-producing animals during 2008–2018 in Korea. Microorganisms. 2021;8:1839. doi: 10.3390/microorganisms8111839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammerum AM, et al. Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. Int. J. Food Microbiol. 2006;106:235–237. doi: 10.1016/j.ijfoodmicro.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 29.Noor Uddin GM, et al. Identification and antimicrobial resistance of bacteria isolated from probiotic products used in shrimp culture. PLoS ONE. 2015;10:e0132338. doi: 10.1371/journal.pone.0132338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang Z, et al. Comparative genomics and specific functional characteristics analysis of Lactobacillus acidophilus. Microorganisms. 2021;9:1992. doi: 10.3390/microorganisms9091992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rojo-Bezares B, et al. Assessment of antibiotic susceptibility within lactic acid bacteria strains isolated from wine. Int. J. Food Microbiol. 2006;111:234–240. doi: 10.1016/j.ijfoodmicro.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Bischoff KM, Skinner-Nemec KA, Leathers TD. Antimicrobial susceptibility of Lactobacillus species isolated from commercial ethanol plants. J Ind. Microbiol. Biotechnol. 2007;34:739–744. doi: 10.1007/s10295-007-0250-4. [DOI] [PubMed] [Google Scholar]
- 33.Mao Y-C, Chang C-L, Huang Y-C, Su L-H, Lee C-T. Laboratory investigation of a suspected outbreak caused by Providencia stuartii with intermediate resistance to imipenem at a long-term care facility. J. Microbiol. Immunol. Infect. 2018;51:214–219. doi: 10.1016/j.jmii.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Trzcinski K, Cooper BS, Hryniewicz W, Dowson CG. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2000;45:763–770. doi: 10.1093/jac/45.6.763. [DOI] [PubMed] [Google Scholar]
- 35.Hauschild T, et al. Aminoglycosides resistance in clinical isolates of Staphylococcus aureus from a University Hospital in Bialystok, Poland. Folia Histochem. Cytobiol. 2008;46:225–228. doi: 10.2478/v10042-008-0034-3. [DOI] [PubMed] [Google Scholar]
- 36.Strauss C, Hu Y, Coates A, Perreten V. A Novel erm(44) gene variant from a human Staphylococcus saprophyticus isolate confers resistance to macrolides and lincosamides but not streptogramins. Antimicrob. Agents Chemother. 2016;61:e01655–e1716. doi: 10.1128/AAC.01655-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malbruny B, Werno AM, Murdoch DR, Leclercq R, Cattoir V. Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa (C) gene in Streptococcus agalactiae UCN70. Antimicrob. Agents Chemother. 2011;55:1470–1474. doi: 10.1128/AAC.01068-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholson TL, et al. Comparative virulence and genomic analysis of Streptococcus suis Isolates. Front. Microbiol. 2021;11:3563. doi: 10.3389/fmicb.2020.620843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Umeda K, et al. Molecular characterization of bla KHM-1 encoding plasmid in an Enterobacter hormaechei subsp. hoffmannii isolate from blood culture. PLoS ONE. 2020;15:e0227605. doi: 10.1371/journal.pone.0227605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei C-W, et al. Clonal spread and horizontal transfer mediate dissemination of phenicol-oxazolidinone-tetracycline resistance gene poxtA in enterococci isolates from a swine farm in China. Vet. Microbiol. 2021;262:109219. doi: 10.1016/j.vetmic.2021.109219. [DOI] [PubMed] [Google Scholar]
- 41.Wendlandt S, et al. Detection of the macrolide-lincosamide-streptogramin B resistance gene erm (44) and a novel erm variant in staphylococci from aquatic environments. FEMS Microbiol. Ecol. 2015;91:fiv090. doi: 10.1093/femsec/fiv090. [DOI] [PubMed] [Google Scholar]
- 42.Zakaria AS, Kassem MA, El Far MS, Edward EA. Characterization, in-vitro evaluation of probiotic potential and antagonistic activity of selected lactic acid bacteria strains isolated from natural origin against some human pathogens. Bull. Pharm. Sci. 2021;44:225–241. doi: 10.21608/BFSA.2021.174149. [DOI] [Google Scholar]
- 43.Hao W, et al. Analysis of a poxtA-and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J. Antimicrob. Chemother. 2019;74:1771–1775. doi: 10.1093/jac/dkz109. [DOI] [PubMed] [Google Scholar]
- 44.Soltani M, Beighton D, Philpott-Howard J, Woodford N. Mechanisms of resistance to quinupristin-dalfopristin among isolates of Enterococcus faecium from animals, raw meat, and hospital patients in Western Europe. Antimicrob. Agents Chemother. 2000;44:433–436. doi: 10.1128/aac.44.2.433-436.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho S, Barrett JB, Frye JG, Jackson CR. Antimicrobial resistance gene detection and plasmid typing among multidrug resistant enterococci isolated from freshwater environment. Microorganisms. 2020;8:1338. doi: 10.3390/microorganisms8091338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jaimee G, Halami P. Conjugal transfer of aac(6’)Ie-aph(2”)Ia gene from native species and mechanism of regulation and cross resistance in Enterococcus faecalis MCC3063 by real time-PCR. Microb. Pathog. 2017;110:546–553. doi: 10.1016/j.micpath.2017.07.049. [DOI] [PubMed] [Google Scholar]
- 47.Martínez JL. Antibiotics and antibiotic resistance genes in natural environments. Science. 2008;321:365–367. doi: 10.1126/science.1159483. [DOI] [PubMed] [Google Scholar]
- 48.Landers TF, Cohen B, Wittum TE, Larson EL. A review of antibiotic use in food animals: perspective, policy, and potential. Public Heal. Rep. 2012;127:4–22. doi: 10.1177/003335491212700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Boeckel TP, et al. Global trends in antimicrobial resistance in animals in low-and middle-income countries. Science. 2019;365:eaaw1944. doi: 10.1126/science.aaw1944. [DOI] [PubMed] [Google Scholar]
- 50.Founou LL, Founou RC, Essack SY. Antibiotic resistance in the food chain: a developing country-perspective. Front. Microbiol. 2016;7:1881. doi: 10.3389/fmicb.2016.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baptiste KE, Kyvsgaard NC. Do antimicrobial mass medications work? A systematic review and meta-analysis of randomised clinical trials investigating antimicrobial prophylaxis or metaphylaxis against naturally occurring bovine respiratory disease. Pathog. Dis. 2017 doi: 10.1093/femspd/ftx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Word A, Wickersham T, Trubenbach L, Mays G, Sawyer J. Effects of metaphylaxis on production responses and total antimicrobial use in high-risk beef calves. AAS. 2020;36:265–270. doi: 10.15232/aas.2019-01914. [DOI] [Google Scholar]
- 53.Van Boeckel TP, et al. Global trends in antimicrobial use in food animals. PNAS. 2015;112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawant A, Sordillo L, Jayarao B. A survey on antibiotic usage in dairy herds in Pennsylvania. J. Dairy Sci. 2005;88:2991–2999. doi: 10.3168/jds.S0022-0302(05)72979-9. [DOI] [PubMed] [Google Scholar]
- 55.Krömker V, Leimbach S. Mastitis treatment–Reduction in antibiotic usage in dairy cows. Reprod. Domest. Anim. 2017;52:21–29. doi: 10.1111/rda.13032. [DOI] [PubMed] [Google Scholar]
- 56.FDA. Summary report on antimicrobials sold or distributed for use in food-producing animals. U.S. Department of Health and Human Services. https://www.fda.gov/media/154820/download (2020).
- 57.EMA. Sales of veterinary antimicrobial agents in 31 european countries in 2018. EMA/24309/2020. 10.2809/195073 (2020).
- 58.Zaheer R, et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci. Rep. 2020;10:3937. doi: 10.1038/s41598-020-61002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kowalski Z, et al. Performance of Holstein calves fed milk-replacer and starter mixture supplemented with probiotic feed additive. J. Anim. Feed. Sci. 2009;18:399–411. [Google Scholar]
- 60.Mamuad LL, et al. Rumen fermentation and microbial community composition influenced by live Enterococcus faecium supplementation. AMB Expr. 2019;9:123. doi: 10.1186/s13568-019-0848-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cammarota G, Ianiro G, Gasbarrini A. Fecal microbiota transplantation for the treatment of Clostridium difficile infection: A systematic review. J. Clin. Gastroenterol. 2014;48:693–702. doi: 10.1097/MCG.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 62.Ji S, et al. Ecological restoration of antibiotic-disturbed gastrointestinal microbiota in foregut and hindgut of cows. Front. Cell. Infect. Microbiol. 2018;8:79. doi: 10.3389/fcimb.2018.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith G. Antimicrobial decision making for enteric diseases of cattle. Vet. Clin. N. Am. Food Anim. Pract. 2015;31:47–60. doi: 10.1016/j.cvfa.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez AV, Baigorı MD, Alvarez S, Castro GR, Oliver G. Phosphatidylinositol-specific phospholipase C activity in Lactobacillus rhamnosus with capacity to translocate. FEMS Microbiol. Lett. 2001;204:33–38. doi: 10.1111/j.1574-6968.2001.tb10858.x. [DOI] [PubMed] [Google Scholar]
- 65.Yelin I, et al. Genomic and epidemiological evidence of bacterial transmission from probiotic capsule to blood in icu patients. Nat. Med. 2019;25:1728–1732. doi: 10.1038/s41591-019-0626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perez PF, et al. Bacterial imprinting of the neonatal immune system: Lessons from maternal cells? Pediatrics. 2007;119:e724–e732. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 67.Rodríguez JM. The origin of human milk bacteria: Is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 2014;5:779–784. doi: 10.3945/an.114.007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Young W, Hine BC, Wallace OA, Callaghan M, Bibiloni R. Transfer of intestinal bacterial components to mammary secretions in the cow. PeerJ. 2015;3:e888. doi: 10.7717/peerj.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tóth AG, et al. Antimicrobial resistance genes in raw milk for human consumption. Sci. Rep. 2020;10:7464. doi: 10.1038/s41598-020-63675-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oliver SP, Jayarao BM, Almeida RA. Foodborne pathogens in milk and the dairy farm environment: Food safety and public health implications. Foodborne Pathog. Dis. 2005;2:115–129. doi: 10.1089/fpd.2005.2.115. [DOI] [PubMed] [Google Scholar]
- 71.Liu J, et al. The fecal resistome of dairy cattle is associated with diet during nursing. Nat. Commun. 2019;10:4406. doi: 10.1038/s41467-019-12111-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J, Ying G-G, Deng W-J. Antibiotic residues in food: Extraction, analysis, and human health concerns. J. Agric. Food Chem. 2019;67:7569–7586. doi: 10.1021/acs.jafc.9b01334. [DOI] [PubMed] [Google Scholar]
- 73.Pokharel S, Shrestha P, Adhikari B. Antimicrobial use in food animals and human health: Time to implement ‘One Health’approach. Antimicrob. Resist. Infect. Control. 2020;9:181. doi: 10.1186/s13756-020-00847-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raschle S, et al. Environmental dissemination of pathogenic Listeria monocytogenes in flowing surface waters in Switzerland. Sci. Rep. 2021;11:9066. doi: 10.1038/s41598-021-88514-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.You L, et al. Changes in chemical composition, structural and functional microbiome during Alfalfa (Medicago sativa) ensilage with Lactobacillus plantarum PS-8. Anim. Nutr. 2022 doi: 10.1016/j.aninu.2021.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wood DE, Lu J, Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2021).
- 78.McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lahti, L. & Shetty, S. microbiome R package (2012–2019).
- 80.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 81.Hyatt D, et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McArthur AG, et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 84.Krawczyk PS, Lipinski L, Dziembowski A. PlasFlow: Predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res. 2018;46:e35. doi: 10.1093/nar/gkx1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guo J, et al. VirSorter2: A multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome. 2021;9:37. doi: 10.1186/s40168-020-00990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets analysed in the current study are available in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) repository and can be accessed through the PRJNA495415 and PRJNA764355 BioProject identifiers.