Abstract
Lamivudine population pharmacokinetics were investigated by using nonlinear mixed-effect modelling (NONMEM) analysis of data from 394 human immunodeficiency virus (HIV)-infected patients treated with lamivudine (150 to 300 mg every 12 h) in two large, phase III clinical efficacy-safety trials, NUCA3001 and NUCA3002. Analyses of 1,477 serum lamivudine concentration determinations showed that population estimates for lamivudine oral clearance (CL/F; 25.1 liters/h) and volume of distribution (V/F; 128 liters) were similar to values previously reported for HIV-infected patients in phase I pharmacokinetic studies. Lamivudine CL/F was significantly influenced by the covariates creatinine clearance and weight and not affected by age, Centers for Disease Control and Prevention (CDC) classification, CD4+ cell count, HIV type 1 (HIV-1) RNA PCR, or gender and race when CL/F was corrected for differences in patient weight. The population estimate for lamivudine V/F was not significantly influenced by the covariates gender, race, age, weight, renal function, HIV-1 RNA PCR, or CDC classification and CD4+ cell count when creatinine clearance was included with CL/F in the model. Lamivudine disposition was significantly influenced by renal function. However, as only three patients had an estimated creatinine clearance of <60 ml/min, dosage adjustments for patients with impaired renal function should not be determined based on the population parameters derived in this analysis.
The reverse transcriptase inhibitor lamivudine, administered in combination with zidovudine and a protease inhibitor, has proven effective in improving surrogate markers of human immunodeficiency virus (HIV) disease and delaying disease progression and death in the treatment of HIV-infected patients (7, 8). Although the pharmacokinetics of lamivudine have been assessed in several phase I and phase I/II dose-ranging studies, each of which involved small numbers of HIV type 1 (HIV-1)-infected patients (4 to 12 patients per study) (1, 10, 11, 13, 14, 16, 17, 19, 21), no study to date has evaluated lamivudine population pharmacokinetics. Population pharmacokinetic studies are important because they address the demographic or disease-related factors in the targeted treatment population that may significantly alter the pharmacokinetics of a particular therapeutic agent (18, 20). Nonlinear mixed-effect modeling (NONMEM), as implemented in the NONMEM program (3), has been applied previously to estimate the population pharmacokinetic parameter means, interindividual variances, and intraindividual (residual) variances of the reverse transcriptase inhibitors zidovudine and didanosine (6, 15). The primary objectives for the present study were to (i) define the population pharmacokinetics of lamivudine from sparse sampling obtained during two large, phase III clinical trials, NUCA3001 and NUCA3002, and (ii) identify the influence of gender, race, renal function, weight, age, and surrogate markers of HIV disease on lamivudine pharmacokinetics.
(The results of this population pharmacokinetic analysis were previously presented in part [13a, 22].)
The study designs for NUCA3001 and NUCA3002 have been described previously (2, 5). NUCA3001 evaluated the clinical efficacy and safety of 150 mg of lamivudine given twice daily with 200 mg of zidovudine three times daily (n = 92), 300 mg of lamivudine given twice daily (n = 87), 300 mg of lamivudine given twice daily with 200 mg of zidovudine given three times daily (n = 94), and 200 mg of zidovudine given three times daily (n = 93) for 52 weeks to 366 antiretroviral therapy-naive patients (≤4 weeks of previous zidovudine treatment) at 26 sites (5). NUCA3002 evaluated the clinical efficacy and safety of 150 mg of lamivudine given twice daily with 200 mg of zidovudine given three times daily (n = 84), 300 mg of lamivudine given twice daily with 200 mg of zidovudine given three times daily (n = 84), and 0.75 mg of zalcitabine given three times daily with 200 mg of zidovudine given three times daily (n = 86) for 52 weeks to 254 zidovudine-experienced patients (≥6 months of previous zidovudine treatment) at 21 sites (2). In both clinical trials, lamivudine doses were given twice daily, as two 75-mg tablets (for the 150-mg regimen) or as one 300-mg tablet of Epivir (Glaxo Wellcome Inc., Research Triangle Park, N.C.), at approximately 8 am and 4 pm.
At the week 2 or 4 clinic visit, blood samples were collected for pharmacokinetic evaluations. Selected centers participated in intensive pharmacokinetic evaluations (six blood samples per visit) in which an indwelling catheter, maintained via saline flush, was placed in a forearm vein for blood sampling. A 2-ml blood sample was drawn and discarded, a 10-ml predose sample was drawn, and then 10-ml samples were drawn at 0.5, 1, 2.5, 3.5, and 8 h postdose. Following collection of the 8-h sample, the second study drug dose was administered and the regular dosing schedule was resumed. The remaining sites participated in limited pharmacokinetic evaluations (two blood samples per visit) at the week 2 and 4 clinic visits. Patients were instructed to refrain from taking their morning dose of the study drug until they arrived at the clinic and a 10-ml sample of blood was collected. If a patient had taken the last dose of the study drug during the 6 h preceding collection of the first sample, a second blood sample was drawn at least 1 h after the first and the usual dosing schedule was resumed. If the last study drug dose was taken more than 6 h before collection of the first blood sample, the study drug was to be administered immediately and a second blood sample was to be taken at least 1 h later. Study drug administration was to be restarted with the evening dose on the day of the clinic visit.
Blood samples were evaluated for serum lamivudine concentrations, which were determined by methods previously reported (9). For NUCA3001, the serum assay for lamivudine was linear over the calibration range of 3 to 5,000 ng/ml (r ≥ 0.996) (9). The interday percent coefficient of variation (%CV) for the assay was 14.7% at 6 ng/ml, 3.6% at 150 ng/ml, and 0.9% at 3,500 ng/ml. For NUCA3002, the serum assay for lamivudine was linear over the calibration range of 3 to 5,000 ng/ml (r ≥ 0.997). The interday %CV for the assay was 15.2% at 6 ng/ml and 4.8% at 3,500 ng/ml.
Data from NUCA3001 and NUCA3002 were combined for pharmacostatistical analysis. Patients were included in this analysis if they had an adequate dosing history (including time of dose administration prior to the time when the serum drug concentration was determined) and measurable serum lamivudine concentrations (above the limit of quantitation of the assay). Creatinine clearance was chosen as the most convenient clinically appropriate measure of renal function, since it is typically used for dose adjustments. Gender, age, weight, serum creatinine, CD4+ cell count, HIV-1 RNA PCR, and Centers for Disease Control and Prevention (CDC) classification were generally obtained during the patient’s second visit after entry into the study. If no value was reported for that date, the next available value for the patient was used, if obtainable.
Lamivudine pharmacokinetic data were fitted to a one-compartment open model with first-order elimination (ADVAN2 TRANS2). A proportional-error model was used to describe the interindividual variability in pharmacokinetic parameters and residual variability in the model. Population pharmacokinetics and Bayesian parameter estimates were based on the intense and limited sampling described above. NONMEM techniques (software package NONMEM, version 4, level 2) (3) were used to develop a pharmacostatistical model to describe lamivudine population pharmacokinetics after oral administration and to define the influence of specific covariates (gender, race, age, weight, renal function, and surrogate markers of HIV disease) on lamivudine disposition. The first-order conditional estimation process with an interaction option was employed in NONMEM analyses. Serum drug concentration and time data were used to estimate oral lamivudine clearance (CL/F), apparent volume of distribution (V/F), and absorption rate constant (ka). A covariance step was executed in each run to obtain the standard errors of the estimate, covariance matrix, and correlation matrix. To assess model fit, scatterplots were created which included predicted versus observed concentrations (PRED versus CONC = DV with unity line), residual and weighted residuals versus time (RES WRES versus TIME), and residual and weighted residuals versus observed and predicted concentrations (RES WRES versus CONC PRED).
The model-building process employed a two-step approach to determine the inclusion of covariates. Initially, a basic (base) pharmacokinetic model was fitted to the data. Individual Bayesian parameter estimates were obtained from this model. Scatterplots and summary plots of individual Bayesian estimates and covariates were used to screen for appropriate covariates to include in the pharmacostatistical model. Likely candidate covariates were then added to the pharmacostatistical model. The influence of each covariate on clearance and volume was examined separately and compared to the base model (no covariates). Covariates that significantly improved the objective function were further examined in combinations as follows: weight and race on CL/F, weight and gender on CL/F, race and creatinine clearance on CL/F, creatinine clearance and CL/F on CD4+ cell count and V/F, and creatinine clearance and CL/F on CDC classification and V/F.
The criteria for accepting a NONMEM model estimation included the following: (i) convergence of the objective function (“successful termination” statement by the NONMEM program), (ii) standard error of estimates not larger than half of the estimates, (iii) number of significant digits >3, (iv) termination of the covariance step without warning messages, and (v) correlations between model parameters of <0.95. The likelihood ratio was used to assess whether the difference in objective function between the base model and the full (more complex) model statistically improved the fit of the model to the data. A decrease in objective function (Δ) of >7.88 (P = 0.005, based on the likelihood ratio, which is approximately χ2 distributed), compared to the base model, was considered significant. A decrease in Δ of >10.60 was considered significant when the model contained two additional parameters. A superior model also was expected to reduce the intersubject variance terms and/or the residual error term.
Statistical analyses (SAS version 6.10) were also used to evaluate potential variables that may influence lamivudine pharmacokinetic parameters obtained from the base model. Stepwise regression analyses were performed to determine relationships between continuous variables (weight, creatinine clearance [4], age, number of CD4+ cells per cubic millimeter, and number of HIV-1 RNA PCR copies per milliliter) and lamivudine CL/F and V/F. Analysis of variance was used to assess the influence of categorical data (gender, race, CDC classification, CD4+ cell count category, and HIV-1 RNA PCR category) on lamivudine CL/F and V/F. Statistically significant (P < 0.05) interactions were examined in the NONMEM model for evaluation of goodness of fit.
Of 1,721 measurable lamivudine concentrations available, 1,477 with adequate dosing and sample time documentation were included in the lamivudine population pharmacokinetic analysis. Of the 441 patients who received lamivudine in the two clinical studies, 394 were included in the analysis (245 from NUCA3001 and 149 from NUCA3002). The patient population was predominantly (87%) male and had a mean age of 35.7 years, a mean weight of 74.2 kg, a mean calculated creatinine clearance of 97.4 ml/min, a mean CD4+ cell count of 306/mm3, and a mean HIV-1 RNA PCR number of 43,723 copies/ml (Table 1).
TABLE 1.
Demographics of patients who participated in NUCA3001 and NUCA3002 and had adequate dosing and sample time documentation
| Parameter | NUCA3001 | NUCA3002 | Combined |
|---|---|---|---|
| No. of evaluable patients | 245 | 149 | 394 |
| No. (%) of: | |||
| Males | 213 (87) | 129 (87) | 342 (87) |
| Females | 32 (13) | 20 (13) | 52 (13) |
| Age (yr) | |||
| Mean | 34.9 | 36.9 | 35.7 |
| Range | 19–61 | 22–64 | 19–64 |
| Wt (kg) | |||
| Mean | 75.3 | 72.4 | 74.2 |
| Range | 47.2–138.5 | 37.2–115.5 | 37.2–138.5 |
| No. (%) that were: | |||
| Caucasian | 152 (62) | 92 (62) | 244 (62) |
| Black | 38 (16) | 20 (13) | 58 (15) |
| Hispanic | 52 (21) | 35 (24) | 87 (22) |
| Other | 3 (1) | 2 (1) | 5 (1) |
| Serum creatinine (mg/dl) | |||
| Mean | 1.11 | 1.09 | 1.1 |
| Range | 0.6–1.6 | 0.6–1.7 | 0.6–1.7 |
| No. (%) CDC category: | |||
| A | 170 (69) | 73 (49) | 243 (62) |
| B | 65 (27) | 60 (40) | 125 (32) |
| C | 10 (4) | 16 (11) | 26 (6) |
| No. (%) with CD4+ cell count (cells/mm3) of: | |||
| <100 | 0 (0) | 12 (8) | 12 (3) |
| 101–200 | 17 (7) | 50 (34) | 67 (17) |
| 201–500 | 196 (80) | 87 (58) | 283 (72) |
| >500 | 32 (13) | 0 (0) | 32 (8) |
| No. (%) with PCR count (copies/ml)a of: | |||
| <50,000 | 244 (99.6) | 77 (52) | 309 (81) |
| >50,000 | 1 (0.4) | 72 (48) | 73 (19) |
| No. (%) receiving: | |||
| Lamivudine alone | 82 (33) | 0 (0) | 82 (21) |
| Lamivudine + zidovudine | 163 (67) | 149 (100) | 312 (79) |
For PCR category only, n was 237 in NUCA3001, 145 in NUCA3002, and 382 for the combined studies.
In stepwise regression analyses performed between CL/F and V/F against age, weight, and creatinine clearance, the following relationships achieved statistical significance: CL/F versus creatinine clearance (P = 0.0001; r2 = 0.0761), CL/F versus weight (P = 0.0265; r2 = 0.0881), and V/F versus weight (P = 0.0001; r2 = 0.5006). In stepwise regression analyses performed between CL/F and V/F against gender, race, CDC classification, CD4+ cell count, and HIV-1 RNA PCR, the following categorical analyses achieved statistical significance: CL/F versus race (P = 0.0307; not significant when CL/F was normalized for weight [P = 0.1270]), weight-normalized V/F versus race (P = 0.0017), CL/F versus gender (P = 0.0221; not significant when CL/F was normalized for weight [P = 0.8538]), V/F and gender (P = 0.0451), and weight-normalized CL/F and HIV-1 RNA PCR (P = 0.0139).
NONMEM results were consistent with stepwise regression and analysis of variance results. No differences were observed with the following models when compared with the base model: V/F versus gender, V/F versus weight, V/F versus race, V/F versus creatinine clearance, V/F and CL/F versus age, V/F and CL/F versus CD4+ cell count, CL/F versus CDC classification, and V/F and CL/F versus HIV-1 RNA PCR. Weight significantly influenced the population estimate of lamivudine CL/F (Δ = −24.818). Less remarkable differences in objective function were found when gender (Δ = −8.945) and race (Δ = −9.171) were included in the model as covariates of CL/F when compared to CL/F normalized for weight. No differences were observed in weight-normalized lamivudine CL/F between Caucasians and members of other races (Δ = −0.879). Differences in CL/F due to gender were observed. However, these were redundant when weight was included in the model (weight-normalized lamivudine CL/F between men and women, Δ = −0.112). CDC classification significantly influenced the population estimate for V/F (Δ = −16.828), but CD4+ cell count, a more sensitive measure of HIV disease, was less remarkable (Δ = −8.441). When creatinine clearance was combined with CDC classification or CD4+ cell count as covariates of CL/F and V/F, respectively, no significant differences were observed (Δ = −8.27 and Δ = −6.513 compared to the creatinine clearance model, respectively).
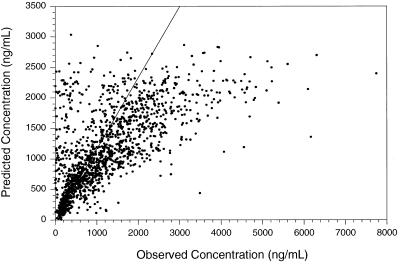
Depending on the model, population pharmacokinetic parameter estimates varied between 4.07 and 26.7 liters/h for CL/F, −55 and 150 liters for V/F, and 3.80 and 5.48 h−1 for ka. The final (superior) model estimated CL/F at 25.1 liters/h, V/F at 128 liters, and ka at 4.65 h−1 (Table 2). As for residual error, ɛ was 0.163 (40.4%), with a standard error (ɛ) of 0.012. An adequate correlation was observed in the final model between predicted and observed serum lamivudine concentrations (Fig. 1), weighted residuals and predicted concentrations, and weighted residuals and time profiles. Lamivudine disposition is best described by a two-compartment model. Attempts to fit a two-compartment model with first-order absorption either failed to converge or failed to converge to a meaningful result, and such a model was not identifiable by using the available data. All possible combinations of additive- or proportional-error models for interpatient variability on pharmacokinetic parameters and error model on concentration were examined. The only compartmental pharmacokinetic model to successfully fit the data was a one-compartment model with first-order absorption. It is known that lamivudine absorption is rapid, with a time to maximum drug concentration in serum of 1 to 1.5 h. Blood samples were generally collected 1 h or later after dosing. Thus, there were insufficient data to describe the absorption process.
TABLE 2.
Lamivudine final model parameter estimates
| Parameter | Population estimate | 95% CIa | % CV | SE (θ)b | ηc | SE (η) |
|---|---|---|---|---|---|---|
| CL/F (liters/h),d θ1 | 25.1 | 23.8–26.4 | 35.2 | 0.683 | 0.124 | 0.0183 |
| V/F (liters),e θ2 | 128 | 117–139 | 31.1 | 5.72 | 0.0965 | 0.0278 |
| ka (h−1),f θ3 | 4.65 | 2.18–7.12 | 1.26 | |||
| θ4 | 0.468 | 0.233–0.703 | 0.120 |
CI, confidence interval.
θ, fixed effects.
η, random effects between individuals.
If male, CL/F = [θ1 · (140 − age/serum creatinine · 100)θ4 EXPη2] · weight/70; if female, CL/F = {θ1 · [(140 − age/serum creatinine · 100) · 0.85]θ4 · Expη2} · weight/70.
V/F = θ2 · EXPη2.
ka = θ3. Error model: F · EXP(ɛ1).
FIG. 1.
Correlation between predicted and observed serum lamivudine concentrations in the final model.
Renal function as a covariate greatly improved the fit of the model to the data. A significant difference in objective function was found when creatinine clearance was included in the model as a covariate of CL/F (Δ = −39.182). The model was defined as follows: CL/F (liters per hour) = 4.07 · (creatinine clearance/100) + 21 · (weight/70), %CV = 38.7%, V/F = 161 liters (%CV = 36.7%), and ka = 6.39 h−1 (residual error, 39.4%). Since the 95% confidence intervals for CL/F included zero for this model, an exponential model was examined. The exponential model with creatinine clearance as a covariate of lamivudine clearance was the final (superior) model (Δ = −34.6).
Overall, the results of this study indicate that population estimates for key lamivudine pharmacokinetic parameters, as derived by NONMEM analysis, are consistent with values reported in earlier, small-scale, phase I pharmacokinetic studies employing traditional, intensive, serial blood sampling (10, 11, 13, 14, 16, 17, 19, 21). According to the final model, population estimates for lamivudine CL/F (25.1 liters/h) and V/F (128 liters [1.8 liters/kg for a 70-kg person]) were within the ranges observed by van Leeuwen et al. (19) and Yuen et al. (21) in patients with asymptomatic HIV infection (CL/F = 15.4 to 28 liters/h; V/F = 1.0 to 2.2 liters/kg), assuming an absolute bioavailability of 82%. A lamivudine ka has not been reported previously in phase I pharmacokinetic studies. The large magnitude of the population estimate for ka derived from the final model (4.65 h−1) was expected because lamivudine is known to be absorbed rapidly, with a time to maximum serum drug concentration of approximately 1 h (19, 21).
This is the first study to assess the population pharmacokinetics of lamivudine. Serum drug concentration data from more patients (n = 394) were evaluated in this study than in comparable population pharmacokinetic studies of zidovudine (72 patients) (6) and didanosine (69 patients) (15). The large population allowed an exploration of possible relationships between various covariates (gender, weight, race, age, renal function, and surrogate markers of HIV disease) and lamivudine pharmacokinetics that would not have been possible in a traditional small-scale pharmacokinetic study.
The final model indicated that renal function was the most significant covariate. This was not surprising, since Heald et al. (10) and Johnson et al. (12) previously demonstrated a significant relationship between impaired renal function and lamivudine disposition after single-dose (300 mg) oral administration of lamivudine. Decreased renal lamivudine clearance, as is evident in renally impaired patients, is associated with increased lamivudine area under the concentration-time curve and maximum concentration of drug in serum, and this necessitates dosage modification according to creatinine clearance (e.g., a 50% reduction in the daily lamivudine dose in patients with creatinine clearances below 50 ml/min). The final model in this population pharmacokinetic study similarly predicted that the lamivudine area under the concentration-time curve estimates following administration of a 150-mg dose increases as renal impairment worsens. One limitation of our study was that most of the patient population evaluated had normal renal function; indeed, only three patients had an estimated creatinine clearance of <60 ml/min. In view of this, the relationship between creatinine clearance and lamivudine CL/F identified in this population pharmacokinetic analysis should not be used as a guide for dose modification.
Note that the population estimate of lamivudine CL/F was not significantly influenced by weight-corrected gender and race. In a previous study by Moore et al. (13), healthy female subjects were found to have a lower lamivudine CL/F (liters per hour) than male subjects but, as in the study presented here, correction for weight eliminated any gender differences in this parameter (measured in liters per hour per kilogram). Age alone did not significantly affect either the lamivudine CL/F or V/F. It should be kept in mind, though, that the age range of the patients studied (19 to 64 years) may limit this interpretation. The lack of influence of gender and age on drug disposition in this study of lamivudine is comparable to that reported in the population pharmacokinetic study of another reverse transcriptase inhibitor, didanosine. Surrogate markers of HIV disease (CD4+ cell count, HIV-1 RNA PCR, and CDC classification) did not influence lamivudine disposition. However, interpretation of these findings may be limited since only 12 patients had CD4+ cell counts of <100/mm3, indicating that most of the patients were not severely immunosuppressed.
In conclusion, the results of this study indicated that population estimates for lamivudine pharmacokinetic parameters were generally in agreement with values derived from earlier, small-scale, phase I pharmacokinetic studies. Lamivudine disposition was significantly influenced by renal function and was not affected by weight-corrected gender and race. Lamivudine dose adjustments in patients with renal impairment should be based on information in the product label.
Acknowledgments
We thank the many clinical investigators, patients, and other people who were associated with the NUCA3001 and NUCA3002 clinical trials.
REFERENCES
- 1.Angel J B, Hussey E K, Hall S T, Donn K H, Morris D M, McCormack J P, Montaner J S G, Ruedy J. Pharmacokinetics of 3TC (GR109714X) administered with and without food to HIV-infected patients. Drug Investig. 1993;6:70–74. [Google Scholar]
- 2.Bartlett J A, Benoit S L, Johnson V A, Quinn J B, Sepulveda G E, Ehmann W C, Tsoukas C, Fallon M A, Self P L, Rubin M for The North American HIV Working Party. Lamivudine plus zidovudine compared with zalcitabine plus zidovudine in patients with HIV infection. Ann Intern Med. 1996;125:161–172. doi: 10.7326/0003-4819-125-3-199608010-00001. [DOI] [PubMed] [Google Scholar]
- 3.Beal S L, Sheiner L B. NONMEM user’s guide. San Francisco: University of California at San Francisco; 1989. [Google Scholar]
- 4.Cockcroft D W, Gault M H. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16:31–41. doi: 10.1159/000180580. [DOI] [PubMed] [Google Scholar]
- 5.Eron J J, Benoit S L, Jemsek J, MacArthur R D, Santana J, Quinn J B, Kuritzkes D R, Fallon M A, Rubin M for The North American HIV Working Party. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. N Engl J Med. 1995;333:1662–1669. doi: 10.1056/NEJM199512213332502. [DOI] [PubMed] [Google Scholar]
- 6.Gitterman S R, Drusano G L, Egorin M J, Standiford H C The Veterans Administration Cooperative Studies Group. Population pharmacokinetics of zidovudine. Clin Pharmacol Ther. 1990;48:161–167. doi: 10.1038/clpt.1990.131. [DOI] [PubMed] [Google Scholar]
- 7.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez G, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 8.Hammer S M, Squires K E, Hughes M D, Grimes J M, Demeter L M, Currier J S, Eron J J, Jr, Feinberg J E, Balfour H H, Jr, Deyton L R, Chodakewitz J A, Fischl M A for The AIDS Clinical Trials Group 320 Study Team. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N Engl J Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 9.Harker A J, Evans G L, Hawley A E, Morris D M. High-performance liquid chromatographic assay for 2′-deoxy-3′-thiacytidine in human serum. J Chromatogr B. 1994;657:227–232. doi: 10.1016/0378-4347(94)80092-8. [DOI] [PubMed] [Google Scholar]
- 10.Heald A E, Hsyu P H, Yuen G J, Robinson P, Mydlow P, Bartlett J A. Pharmacokinetics of lamivudine in human immunodeficiency virus-infected patients with renal dysfunction. Antimicrob Agents Chemother. 1996;40:1514–1519. doi: 10.1128/aac.40.6.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horton C M, Yuen G, Mikolich D M, Fisher A, Rana K, Mydlow P, Dudley M N. Pharmacokinetics (PK) of oral lamivudine administered alone and with oral zidovudine (ZDV) in asymptomatic patients with human immunodeficiency virus (HIV) infection. Clin Pharmacol Ther. 1994;55:198. . (Abstract.) [Google Scholar]
- 12.Johnson M A, Moore K H P, Yuen G J, Bye A, Pakes G E. A review of the clinical pharmacokinetics of lamivudine. Clin Pharmacokinet. 1999;36:1–26. doi: 10.2165/00003088-199936010-00004. [DOI] [PubMed] [Google Scholar]
- 13.Moore K H P, Shaw S, Laurent A L, Lloyd P, Duncan B, Morris D M, O’Mara M J, Pakes G E. Lamivudine/zidovudine as a combined formulation tablet: bioequivalence compared with lamivudine and zidovudine administered concurrently and the effect of food on absorption. J Clin Pharmacol. 1999;39:593–605. doi: 10.1177/00912709922008209. [DOI] [PubMed] [Google Scholar]
- 13a.Moore K H P, Yuen G J, Hussey E K. Abstract book of the National Conference on Women and HIV. 1997. Analysis of potential differences in lamivudine (3TC) disposition using population pharmacokinetics from two phase III clinical trials in HIV-infected patients, abstr. P1.54; pp. 168–169. [Google Scholar]
- 14.Moore K H P, Yuen G J, Raasch R H, Eron J J, Martin D, Mydlow P K, Hussey E K. Pharmacokinetics of lamivudine administered alone and with trimethoprim-sulfamethoxazole. Clin Pharmacol Ther. 1996;59:550–558. doi: 10.1016/S0009-9236(96)90183-6. [DOI] [PubMed] [Google Scholar]
- 15.Pai S M, Shukla U A, Grasela T H, Knupp C A, Dolin R, Valentine F T, McLaren C, Liebman H A, Martin R R, Pittman K A, Barbhaiya R H. Population pharmacokinetic analysis of didanosine (2′,3′-dideoxyinosine) plasma concentrations obtained in phase I clinical trials in patients with AIDS or AIDS-related complex. J Clin Pharmacol. 1992;32:242–247. doi: 10.1002/j.1552-4604.1992.tb03832.x. [DOI] [PubMed] [Google Scholar]
- 16.Pluda J M, Cooley T P, Montaner J S G, Shay L E, Reinhalter N E, Warthan S N, Ruedy J, Hirst H M, Vicary C A, Quinn J B, Yuen G J, Wainberg M A, Rubin M, Yarchoan R. A phase I/II study of 2′-deoxy-3′-thiacytidine (lamivudine) in patients with advanced human immunodeficiency virus infection. J Infect Dis. 1995;171:1438–1447. doi: 10.1093/infdis/171.6.1438. [DOI] [PubMed] [Google Scholar]
- 17.Rana K Z, Horton C M, Yuen G J, Pivarnick P E, Mikolich D M, Fisher A E, Mydlow P K, Dudley M N. Program and abstracts of the 34th Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C.: American Society for Microbiology; 1994. Effect of lamivudine on zidovudine pharmacokinetics in asymptomatic HIV-1-infected individuals, abstr. A62; p. 83. [Google Scholar]
- 18.Sheiner L B, Beal S L. Evaluation of methods for estimating population pharmacokinetic parameters. III. Monoexponential model; routine clinical pharmacokinetic data. J Pharmacokinet Biopharm. 1983;11:303–309. doi: 10.1007/BF01061870. [DOI] [PubMed] [Google Scholar]
- 19.van Leeuwen R, Lange J M A, Hussey E K, Donn K H, Hall S T, Harker A J, Jonker P, Danner S A. The safety and pharmacokinetics of a reverse transcriptase inhibitor, 3TC, in patients with HIV infection: a phase I study. AIDS. 1992;6:1471–1475. doi: 10.1097/00002030-199212000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Whiting B, Kelman A W, Grevel J. Population pharmacokinetics: theory and clinical application. Clin Pharmacokinet. 1986;11:387–401. doi: 10.2165/00003088-198611050-00004. [DOI] [PubMed] [Google Scholar]
- 21.Yuen G J, Morris D M, Mydlow P K, Haidar S, Hall S T, Hussey E K. Pharmacokinetics, absolute bioavailability, and absorption characteristics of lamivudine. J Clin Pharmacol. 1995;35:1174–1180. doi: 10.1002/j.1552-4604.1995.tb04043.x. [DOI] [PubMed] [Google Scholar]
- 22.Yuen G Y, Benoit S, Rubin M, et al. Population pharmacokinetics of lamivudine in HIV-infected patients: results from two phase III clinical trials. Clin Pharmacol Ther. 1996;59:144. . (Abstract.) [Google Scholar]