Summary
Background
Clonal haematopoiesis driven by mutations in DNMT3A or TET2 has recently been identified as a new risk factor for cardiovascular disease. Experimental studies suggest that these mutations may enhance inflammation which accelerates the disease progression. We aim to investigate the prevalence of mutations in DNMT3A and TET2 and their association with prognosis of patients with ST-segment elevation myocardial infarction (STEMI).
Methods
Targeted deep sequencing for DNMT3A and TET2 and inflammatory cytokines (IL-1β, IL-6, TNF-α, INF-γ) were analyzed in 485 patients with STEMI. Major adverse cardiac events (MACE) was a composite of death, myocardial infarction, stroke, or hospitalization due to heart failure.
Findings
Patients carrying DNMT3A- or TET2-CH-driver mutations with a variant allele frequency (VAF) ≥2% were found in 12.4% (60 of 485) of STEMI patients and experienced an increased incidence of the death (30.9% vs 15.5%, P = 0.001) and MACE (44.5% vs 21.8%, P < 0.001) compared to those who did not, during a median follow up of 3.0 (interquartile range: 2.4–3.4) years. After adjusting for confounders, mutation remained an independent predictor of death (HR = 1.967, 95% CI 1.103–3.507, P = 0.022) and MACE (HR = 1.833, 95% CI 1.154–2.912, P = 0.010). Concentrations of plasma IL-1β (P = 0.010) and IL-6 (P = 0.011) were significantly elevated in DNMT3A/TET2 VAF≥2% group.
Interpretation
DNMT3A- or TET2-CH-driver mutations with a VAF≥2% were observed in over 10% STEMI patients, and were significantly associated with poorer prognosis, which might be explained by higher levels of inflammatory cytokines in mutations carriers.
Funding
National Natural Science Foundation of China; National Key R&D Program of China.
Keywords: Clonal hematopoiesis, Myocardial infarction, DNMT3A/TET2, Prognosis, Inflammation
Research in context.
Evidence before this study
Clonal hematopoiesis of indeterminate potential has been found to be significantly associated with a high risk of coronary heart disease. The most commonly mutated genes, DNMT3A and TET2, were experimentally shown to regulate the inflammatory response of circulating leukocytes, increase release of inflammatory cytokines and accelerate atherosclerosis and heart failure development in mice. Recent studies using targeted sequencing showed that DNMT3A- and TET2-CH-driver mutations were associated with profoundly impaired long-term survival and increased disease progression in patients with chronic heart failure, or aortic valve stenosis undergoing transfemoral aortic valve implantation. However, no studies quantified the prevalence and assessed the prognostic significance of DNMT3A- and TET2-CH-driver mutations in patients with ST-segment elevation myocardial infarction.
Added value of this study
We report the prevalence and prognosis of STEMI patients with CH-driver mutations. The main findings of our study are as followed: (1) DNMT3A- and TET2-CH-driver mutations with a VAF≥2% are not infrequent (12.4%) in patients with STEMI. (2) the presence of DNMT3A- and TET2-CH-driver mutations are associated with profoundly worse outcomes following STEMI. (3) patients carrying DNMT3A- or TET2-CH-driver mutations with a VAF≥2% have significantly higher levels of IL-1β and IL-6.
Implications of all the available evidence
Our study has revealed that STEMI patients carrying DNMT3A- or TET2-CH-driver mutations had worse outcomes and higher levels of inflammatory cytokines. These findings suggest that increased inflammatory status may be worthy of consideration as a new therapeutic target for these patients.
Alt-text: Unlabelled box
Introduction
Clonal hematopoiesis of indeterminate potential (CHIP), defined as the presence of expanded somatic blood cell clones in individuals in the absence of other hematological abnormalities, has been found to be significantly associated with a high risk of coronary heart disease (CHD).1, 2, 3, 4, 5 The most commonly mutated genes in CHIP are the DNA methyltransferase DNMT3A and the DNA demethylase TET2, both of which were experimentally shown to regulate the inflammatory response of circulating leukocytes, increase release of inflammatory cytokines (including IL-1β, IL-6, TNF-α and INF-γ) and accelerate atherosclerosis and heart failure (HF) development in mice.5, 6, 7, 8, 9, 10, 11 Recent studies12, 13, 14, 15 using targeted sequencing showed that DNMT3A- and TET2-CH-driver mutations were associated with profoundly impaired long-term survival and increased disease progression in patients with chronic HF, or aortic valve stenosis undergoing transfemoral aortic valve implantation. Despite use of various therapies, recurrent ischemic events or HF continue to occur in some patients post-myocardial infarction (MI), which are considered to be associated with chronic inflammation.16, 17, 18, 19, 20 Therefore, we hypothesized that patients with ST-segment elevation myocardial infarction (STEMI) and carrying DNMT3A- and TET2-CH-driver mutations had higher level of pro-inflammatory cytokines and experienced poorer prognosis, compared to those that did not.
However, to date, no studies have quantified the prevalence and assessed the prognostic significance of DNMT3A- and TET2-CH-driver mutations in patients with STEMI. In addition, the mean sequencing depth of whole exome sequencing (WES) used in the previous studies1, 2, 3, 4 analyzing the relationship of CHIP-driver mutations and CHD was relatively low (about 84×) compared to that of targeted deep sequencing (about 4000×). Therefore, our study used hybridization capture-based targeted deep sequencing to analyze the prevalence of DNMT3A- and TET2-CH-driver mutations in patients with STEMI and to associate their presence with clinical outcomes.
Methods
Study population and study design
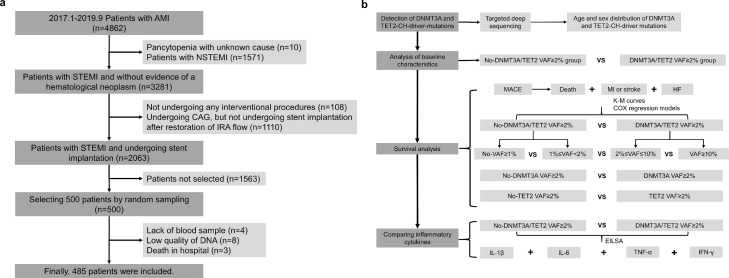
This was a post hoc analysis of patients enrolled in a prospective, observational cohort study (ClinicalTrials.gov ID: NCT03297164). In this cohort, 4862 patients presented with acute myocardial infarction were recruited between January 2017 and September 2019 and were followed up prospectively through September 2021. In the present analysis, only STEMI patients were selected using simple random sampling Figure 1.a details the process of patient selection. At baseline, blood samples were collected from the arterial access site during interventional procedures and separated into whole blood, plasma, serum and white blood cells, and then stored at -80 ℃. Table S1 shows that baseline characteristics of the sampling cohort were comparable to those of the total cohort, and the age distribution between the sampling and total cohort was similar (Fig. S1). Finally, 485 patients were included for final analysis. Fifteen patients were excluded from the analysis due to the following reasons: 4 subjects were excluded due to insufficient blood samples, and 8 subjects were excluded due to an inadequate concentration of DNA isolated from their blood samples, and 3 subjects were excluded due to death in hospital. STEMI was defined as continuous chest pain for > 30 min, ST-segment elevation > 0.1 mV in at least two contiguous leads, or new left bundle-branch block on the 12-lead electrocardiogram (ECG), and elevated cardiac markers (troponin T/I or creatine kinase-MB). Timely reperfusion was defined as first medical contact (FMC) -to-needle ≤30 min for patients undergoing thrombolytic therapy or FMC-to-wire crossing ≤120 min for patients undergoing primary percutaneous coronary intervention (PCI). Family history of CHD was defined as an immediate relative receiving a diagnosis of having CHD before age 60 years. The study design was shown in Figure 1b.
Figure 1.
Workflow diagram and study design. Workflow diagram (a) showing selection of 485 STEMI patients undergoing stent implantation from the total cohort and study design (b) showing the overview of methods used in the study. AMI = myocardial infarction; CAG = coronary angiogram; NSTEMI = non-ST-segment elevation myocardial infarction; IRA = infarct related artery.
Study endpoints
The major adverse cardiac events (MACE) was a composite of all-cause death, recurrent nonfatal MI, nonfatal stroke, or hospitalization due to HF. Detailed definitions of all endpoints are provided in Table S2. After discharge, all patients were followed at 1 month, 6 months, 1 year, and yearly thereafter up to 4 years by phone call or interview. Unscheduled follow-up was carried out if needed.
Targeted deep sequencing
Targeted sequencing was performed by iGenetect Ltd (Beijing, China). In brief, a custom panel comprising 51 CHIP genes (including DNMT3A and TET2; Table S3) commonly mutated in hematologic malignancy was designed to detect the presence of DNMT3A and TET2-clonal hematopoiesis (CH)-driver mutations in patients. DNA was isolated from deep-frozen samples of peripheral blood and the DNA concentration was determined using the Qubit dsDNA HS Assay Kit (Life Technologies) on a Qubit Fluorometer (Life Technologies). The patients’ libraries were generated with the Nextera Flex for enrichment kit (Illumina, San Diego, CA, USA), and sequences for 51 genes were enriched with the iGenetect hybridization capture of DNA libraries protocol and customized probes (iGenetect Ltd, Beijing, China). The libraries were subsequently sequenced on an Illumina NovaSeq 6000. The mean of target mean depth of all 485 samples was 4037×, and the minimum of target mean depth was 2062×. The sequence reads were aligned to the hg19 human reference genome, and variants calling was performed using GATK, and variant annotation was performed using ANNOVAR.
Given that the sensitivity of next-generation sequencing (NGS)21,22 is in the region of variant allele frequency (VAF) 1%, only variants with a VAF≥1% were considered. VAF was calculated by using the formula VAF=alternate reads/(reference + alternate reads). Common single-nucleotide polymorphisms with a minor allele frequency of at least 1% in either the 1000 Genome Project, ESP6500, or ExAC, gnomAD databases were excluded, and synonymous, non-frameshift insertion/deletion variants were also excluded. In addition, variants occurring in more than 5% of the patients in our cohort were considered as technical artifacts and excluded, and variants with fewer than 1000 reads were also filtered out. Furthermore, variants with a VAF of 0.4 to 0.6 or ≥0.9 were considered as potential germline variants and excluded. Finally, the variants were validated according to the previous literature or the Catalogue of Somatic Mutations in Cancer (COSMIC) and ClinVar.
Enzyme-linked immunosorbent assay (ELISA) of inflammatory cytokines
Selected inflammatory cytokines, plasma IL-1β, IL-6, TNF-α and INF-γ were tested by ELISA (ELISA array kit, Boster, Wuhan, China) as per standard manufacturer's protocol. Plasma for ELISA and whole blood for targeted deep sequencing were from the same blood sample.
Statistical analysis
The Kolmogorov–Smirnov test was used as a distribution normality test. Continuous variables were reported as mean ± standard deviation for normally distributed data or as median (25th–75th percentiles) for non-normally distributed data. Independent sample t-test or Mann-Whitney U test was performed for comparison of continuous variables between groups. Categorical data were shown as counts (proportions) and were compared with the χ2 test or Fisher exact test as appropriate. Logistic regression model was used to adjust for age when we analyzed whether female was associated with the presence of DNMT3A- or TET2-CH driver mutations independently.
For survival analysis, time-to-first event curves were generated by Kaplan-Meier analysis, and compared using the log-rank test. In addition, univariate and multivariable Cox proportional-hazards regression models were used to estimate hazard ratios (HRs) and 95% confidence intervals (CI). Adjustments were made for baseline difference and potentially confounding prognostic factors. In adjusted model 1, age, sex, diabetes mellitus, dyslipidemia, hypertension, smoking status were adjusted for, and in adjusted model 2, besides those factors, log-transformed N-terminal pro-B-type natriuretic peptide (NT-proBNP), hyper-sensitivity C-reactive protein (hs-CRP) and left ventricular ejection fraction (LVEF) were also adjusted for. Cox proportional hazards assumptions were tested using Schoenfeld residuals. VAF 2% is typically used as the threshold to define CHIP,12,13,15,23 but for sensitivity analysis, 1 or 5% was also set as the threshold in the COX regression models.
Statistical significance was assumed at p less than 0.05, and all reported p values are 2-sided. Statistical analysis was performed with SPSS (Version 22.0) and Stata (Version 16.0).
Ethics
All procedures were performed according to the Declaration of Helsinki. This study was approved by the Ethics Committee of the 2nd Affiliated Hospital of Harbin Medical University (KY2017-249), and all patients provided written informed consent.
Role of funders
The funders have no role in study design; collection, management, analysis, and interpretation of data; writing of the report.
Results
Detection of CH-driver mutations
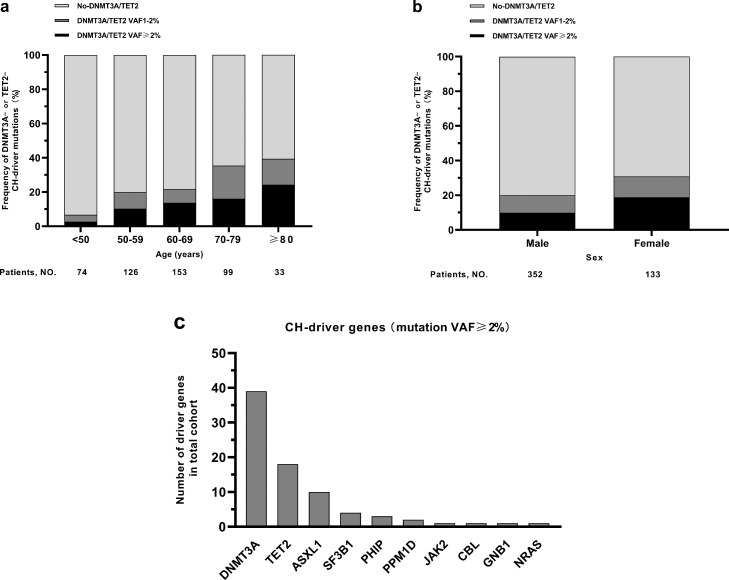
Targeted deep sequencing was performed in a total of 485 patients with STEMI to detect DNMT3A- or TET2-CH-driver mutations in their peripheral blood samples. 60 of 485 patients (12.4%) carried DNMT3A- (n = 43) and/or TET2- (n = 28) CH-driver mutations (Table S4) with a VAF≥2%, with 6 patients (1.2%) harboring mutations in both genes. Furthermore, a VAF≥1% was identified in 111 of 485 patients (22.9%) carrying DNMT3A- (n = 90) and/or TET2- (n = 52) CH-driver mutations, with 17 patients (3.5%) harboring mutations in both genes. Fig. S2 showed the distributions of VAFs of all the DNMT3A- and TET2- CH-driver mutations
The patients had a median age of 62 years (range 25–91), and 133 subjects (27.4%) were female. As shown in Figure 2a, the prevalence of DNMT3A- or TET2-CH-driver mutations with a VAF≥1% in the present cohort with STEMI increased with age from 6.8% (5 out of 74) in the age group of 25–49 years, to 39.4% (13 out of 33) at age 80–91 years. The mutations with a VAF≥1% showed a higher incidence in females (30.8%) than that in males (19.9%) (Figure 2b). After adjusted for age in the multivariate logistic regression analysis, female was on longer associated with the presence of mutations with a VAF≥1% (Unadjusted Model: odds ratio (OR) = 1.795, 95% CI 1.143-2.820, P = 0.011; Adjusted Model: OR = 1.483, 95% CI 0.932–2.359, P = 0.096). Furthermore, when we considered mutations in DNMT3A or TET2 separately, the age or sex distribution showed similar patterns (Fig. S3).
Figure 2.
Distribution of driver mutations and driver genes. Distribution of DNMT3A- or TET2-CH-driver mutations in patients (n = 485) according to age (a) and sex (b). Distribution of driver genes (c).
Besides 43 DNMT3A- and 28 TET2-driver mutations with a VAF≥2%, we also found 31 other genes driver mutations (Table S4) with a VAF≥2%, and all the 102 driver mutations with a VAF≥2% occurred in 80 patients. If a patient simultaneously carried two or more CH-driver mutations in different genes, we considered that the gene with the highest VAF was the driver gene here. As shown in Figure 2c, the driver mutations were identified in the genes DNMT3A (39 patients), TET2 (18 patients), ASXL1 (10 patients), SF3B1 (4 patients), PHIP (3 patients), PPM1D (2 patients), JAK2, CBL, GNB1, NRAS (1 patient each). 63 of 80 patients carrying driver mutations with a VAF≥2% had a driver mutation in only a single gene, whereas 17 patients had 2 ∼ 4 driver mutations simultaneously.
Baseline characteristics
Typically, a subject carrying CH-driver mutations with a VAF≥2% were considered as CHIP carriers.12,13,15,23 So patients were divided into DNMT3A/TET2 VAF≥2% group (carrying DNMT3A- and/or TET2-CH-driver mutations with a VAF≥2%) and no-DNMT3A/TET2 VAF≥2% group (carrying neither DNMT3A- nor TET2-CH-driver mutations with a VAF≥2%) without consideration of the other CHIP genes. The no-DNMT3A/TET2 VAF≥2% group including 20 patients carrying other CHIP mutations with a VAF≥2% (other-CHIP VAF≥2% group) and 405 patients without any CHIP mutations with a VAF≥2% (no-CHIP VAF≥2% group). Baseline characteristics of DNMT3A/TET2 VAF≥2% group and no-DNMT3A/TET2 VAF≥2% group was summarized Table 1.
Table 1.
Baseline characteristics of the study cohort.
| Variables | Overall (n = 485) | No-DNMT3A/TET2VAF≥2% (n = 425) | DNMT3A/TET2VAF≥2% (n = 60) | P-value |
|---|---|---|---|---|
| Age, years | 61.8 ± 12.0 | 61.1 ± 12.0 | 66.5 ± 10.7 | 0.001 |
| Age <50 years, n (%) | 74 (15.3) | 72 (16.9) | 2 (3.3) | 0.006 |
| Female, n (%) | 133 (27.4) | 108 (25.4) | 25 (41.7) | 0.008 |
| Coronary risk factors | ||||
| Hypertension, n (%) | 224 (46.2) | 192 (45.2) | 32 (53.3) | 0.24 |
| Dyslipidemia, n (%) | 321 (66.2) | 287 (67.5) | 34 (56.7) | 0.096 |
| Diabetes mellitus, n (%) | 110 (22.7) | 90 (21.2) | 20 (33.3) | 0.035 |
| Cigarette smoking | 0.81 | |||
| Non-smoker, n (%) | 162 (33.4) | 142 (33.4) | 20 (33.3) | |
| Former smoker, n (%) | 68 (14.0) | 58 (13.6) | 10 (16.7) | |
| Current smoker, n (%) | 255 (52.6) | 225 (52.9) | 30 (50.0) | |
| BMI, kg/m2 | 24.5 (22.5–26.8) | 24.6 (22.5–26.9) | 24.2 (22.4–26.5) | 0.37 |
| CKD, n (%) | 27 (5.6) | 25 (5.9) | 2 (3.3) | 0.61 |
| Family history of CHD, n (%) | 71 (14.6) | 60 (14.1) | 11 (18.3) | 0.39 |
| Previous history | ||||
| Prior MI, n (%) | 11 (2.3) | 9 (2.1) | 2 (3.3) | 0.90 |
| Prior PCI, n (%) | 17 (3.5) | 15 (3.5) | 2 (3.3) | 1.00 |
| LVEF, % | 58 (51–61) | 58 (51–61) | 57 (49–62) | 0.73 |
| Laboratory data | ||||
| hs-CRP, mg/l | 6.39 (2.43–12.12) | 6.27 (2.37–12.00) | 7.87 (3.22–13.25) | 0.13 |
| NT-proBNP, pg/ml | 226 (110–776) | 220 (109–692) | 426 (125–1774) | 0.093 |
| Peak Troponin I, ng/ml | 54 (23–131) | 55 (23–125) | 50 (21–154) | 0.78 |
| White blood count, 103/μl | 11.5 ± 4.0 | 11.5 ± 4.0 | 11.6 ± 3.9 | 0.84 |
| Haemoglobin, g/dl | 14.4 ± 2.0 | 14.4 ± 2.0 | 14.0 ± 1.9 | 0.13 |
| Platelet count, 103/μl | 233 ± 67 | 234 ± 67 | 226 ± 67 | 0.43 |
| TC, mg/dl | 179 ± 41 | 179 ± 42 | 173 ± 37 | 0.23 |
| Triglyceride, mg/dl | 123 (88–176) | 125 (89–178) | 109 (81–158) | 0.21 |
| LDL-C, mg/dl | 110 ± 36 | 111 ± 36 | 104 ± 32 | 0.15 |
| HDL-C, mg/dl | 49 ± 12 | 48.6 ± 12.5 | 48.9 ± 12.4 | 0.85 |
| HbA1c, % | 5.8 (5.5–6.2) | 5.8 (5.4–6.1) | 5.9 (5.6–6.9) | 0.031 |
| Creatinine, mg/dl | 1.03 (0.86–1.18) | 1.03 (0.87–1.18) | 1.03 (0.81–1.16) | 0.17 |
| Procedural characteristics | ||||
| Extent of coronary artery disease | 0.69 | |||
| 1-vessel disease, n (%) | 36 (7.4) | 33 (7.8) | 3 (5.0) | |
| 2-vessel disease, n (%) | 90 (18.6) | 81 (19.1) | 9 (15.0) | |
| 3-vessel disease, n (%) | 359 (74.0) | 311 (73.2) | 48 (80.0) | |
| SYNTAX score | 27.6 ± 11.4 | 27.5 ± 11.6 | 28.5 ± 10.2 | 0.55 |
| Total ischemic time, hour | 5.0 (3.0–7.5) | 5.0 (3.0–7.5) | 5.0 (3.5–7.0) | 0.67 |
| Pre-hospital fibrinolysis, n (%) | 20 (4.1) | 17 (4.0) | 3 (5.0) | 0.99 |
| Type of PCI | ||||
| Primary PCI, n (%) | 438 (90.3) | 383 (90.1) | 55 (91.7) | 0.65 |
| Rescue PCI, n (%) | 5 (1.0) | 4 (0.9) | 1 (1.7) | |
| Delayed PCI, n (%) | 42 (8.7) | 38 (8.9) | 4 (6.7) | |
| Timely reperfusion, n (%) | 412 (84.9) | 361 (84.9) | 51 (85.0) | 0.99 |
| Thrombus aspiration, n (%) | 130 (26.8) | 115 (27.1) | 15 (25.0) | 0.74 |
| Number of stents | 1.29 ± 0.58 | 1.29 ± 0.59 | 1.28 ± 0.52 | 0.89 |
| Mean stent size | 3.13 ± 0.40 | 3.13 ± 0.41 | 3.08 ± 0.39 | 0.35 |
| Total stent length | 35.3 ± 17.8 | 35.2 ± 17.9 | 36.1 ± 17.6 | 0.72 |
| Medication at discharge | ||||
| Aspirin (%) | 479 (98.8) | 420 (98.8) | 59 (98.3) | 0.55 |
| Clopidogrel (%) | 178 (36.7) | 154 (36.2) | 24 (40.0) | 0.57 |
| Ticagrelor (%) | 303 (62.5) | 268 (63.1) | 35 (58.3) | 0.48 |
| Statins (%) | 479 (98.8) | 420 (98.8) | 59 (98.3) | 0.55 |
| β-blocker (%) | 323 (66.6) | 279 (65.6) | 44 (73.3) | 0.24 |
| ACEI (%) | 269 (55.5) | 236 (55.5) | 33 (55.0) | 0.94 |
| ARB (%) | 9 (1.9) | 8 (1.9) | 1 (1.7) | 1.00 |
ACEI = angiotensin-converting enzyme inhibitor; ARB = angiotensin receptor blocker; BMI = body mass index; CHD = coronary heart disease; CKD = chronic kidney disease; HbA1c = Hemoglobin A1C; HDL-C = high-density lipoprotein cholesterol; hs-CRP = hyper-sensitivity C-reactive protein; LDL-C = low-density lipoprotein cholesterol; LVEF = left ventricular ejection fraction; MI = myocardial infarction; NT-proBNP = N-terminal pro brain natriuretic peptide; PCI = percutaneous coronary intervention; SYNTAX = Synergy Between Percutaneous Coronary Intervention With Taxus and Cardiac Surgery; TC = total cholesterol.
As illustrated, patients in the DNMT3A/TET2 VAF≥2% group were significantly older, more female and diabetes mellitus, and had higher serum NT-proBNP and HbA1c levels than those in the no-DNMT3A/TET2-VAF≥2% group. The incidence of other cardiovascular risk factors, including hyperlipidemia, hypertension, chronic kidney disease, smoking and family history of CHD, were not significantly different between the two groups. In addition, the extent of coronary artery disease as assessed by SYNTAX score, total ischemic time, and LVEF showed no difference between the two groups at baseline.
Prognostic significance of DNMT3A- and TET2-CH-driver mutations
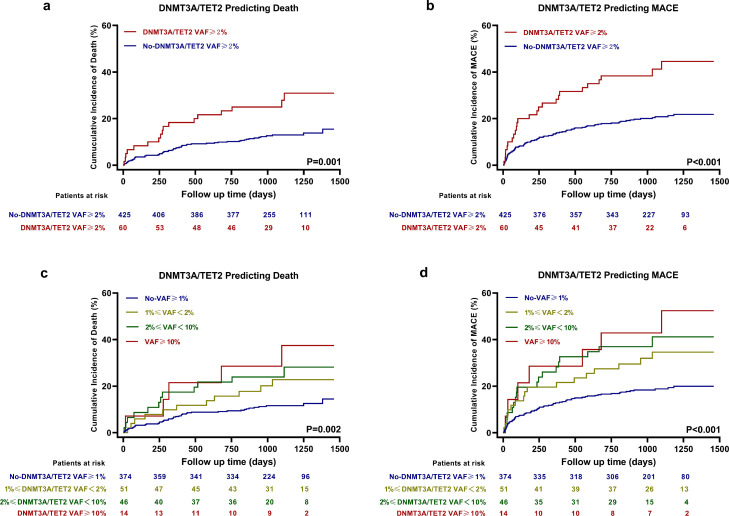
During a median follow-up of 3.0 years (interquartile range: 2.4–3.4 years), 72 patients died, 41 experienced recurrent nonfatal MI, 8 experienced nonfatal stroke and 23 were readmitted due to HF. All-cause death occurred in 17 patients (30.9%) in the DNMT3A/TET2 VAF≥2% group as compared with 55 patients (15.5%) in the no-DNMT3A/TET2 VAF≥2% group, and MACE occurred in 25 patients (44.5%) in the DNMT3A/TET2 VAF≥2% group as compared with 88 patients (21.8%) in the no-DNMT3A/TET2 VAF≥2% group.
Figure 3a and b illustrated the Kaplan-Meier time-to-first event curves for the two groups and demonstrated that patients in the DNMT3A/TET2 VAF≥2% group experienced a significantly worse clinical outcome for all-cause death (Log-rank test, P = 0.001) and MACE (Log-rank test, P < 0.001). Additionally, patients in the DNMT3A/TET2 VAF≥2% group had higher rates of recurrent nonfatal MI and stroke, and hospitalization due to HF (Fig. S3). Using Cox proportional-hazards regression models, the presence of DNMT3A- or TET2-CH-driver mutations with a VAF≥2% showed significant association with all-cause death (Unadjusted Model: HR = 2.442, 95% CI 1.417–4.208, P = 0.001; Adjusted Model 1: HR = 2.053, 95% CI 1.165–3.618, P = 0.013; Adjusted Model 2: HR = 1.967, 95% CI 1.103–3.507, P = 0.022; Table 2), and with MACE (Unadjusted Model: HR = 2.319, 95% CI 1.487–3.618, P < 0.001; Adjusted Model 1: HR = 1.909, 95% CI 1.203–3.031, P = 0.006; Adjusted Model 2: HR = 1.833, 95% CI 1.154–2.912, P = 0.010; Table 2).
Figure 3.
Kaplan-Meier time-to-first event curves for death and MACE. Patients in the DNMT3A/TET2 VAF≥2% group (n = 60) had higher incidence of death (a) and MACE (b) compared to the no-DNMT3A/TET2 VAF≥2% group (n = 425). Further, we divided our cohort into subjects without a VAF≥1% (n = 374); with a VAF of 1, 2% (n = 51); a VAF of 2–10% (n = 46); and a VAF≥10% (n = 14), and found that the higher VAF was associated with poorer outcome (c,d). P-values are for log-rank tests.
Table 2.
Univariate and multivariable COX regression models of DNMT3A/TET2 VAF≥2% group compared to No- DNMT3A/TET2 VAF≥2% group.
| Event | Unadjusted Model |
Adjusted Model 1 |
Adjusted Model 2 |
|||
|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| MACE | 2.319 (1.487–3.618) | <0.001 | 1.909 (1.203–3.031) | 0.006 | 1.833 (1.154-2.912) | 0.010 |
| All-cause Death | 2.442 (1.417–4.208) | 0.001 | 2.053 (1.165–3.618) | 0.013 | 1.967 (1.103-3.507) | 0.022 |
| Nonfatal MI or Stroke | 2.821 (1.463–5.440) | 0.002 | 2.551 (1.277–5.099) | 0.008 | 2.563 (1.283-5.117) | 0.008 |
| Hospitalization due to HF | 3.417 (1.405–8.308) | 0.007 | 2.295 (0.895–5.887) | 0.084 | 2.297 (0.875-6.027) | 0.091 |
Adjusted Model 1: adjusting for age, sex, hypertension, dyslipidemia, diabetes mellitus, smoking status.
Adjusted Model 2: adjusting for age, sex, hypertension, dyslipidemia, diabetes mellitus, smoking status, LVEF, ln_NT-proBNP, hs-CRP.
HF = heart failure; HR = hazard ratio; hs-CRP = hyper-sensitivity C-reactive protein; LVEF = left ventricular ejection fraction; MACE = major adverse cardic events; MI = myocardial infarction; NT-proBNP = N-terminal pro brain natriuretic peptide.
For sensitivity analysis, according to the VAFs of DNMT3A or TET2-CH driver mutations, we also set 1 or 5% as the threshold to group the patients, and Table S5 showed that patients with a VAF no less than the threshold were associated with worse outcomes in Cox proportional-hazards regression models. Further, our STEMI cohort was divided into patients without a VAF≥1%; with a VAF of 1, 2%; a VAF of 2–10%; and a VAF≥10%. In survival analysis, Figure 3c and d and Table S6 showed that the highest VAF group experienced the worst outcomes.
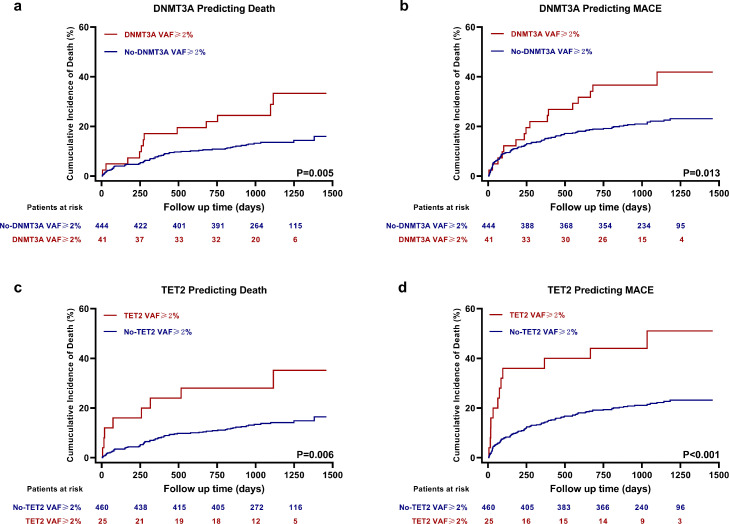
Furthermore, we analyzed Kaplan–Meier survival curves and Cox proportional-hazards regression models separately for patients carrying either DNMT3A- or TET2-CH-driver mutations with a VAF≥2% (patients were classified as DNMT3A VAF≥2% group or no-DNMT3A VAF≥2% group without consideration of the other CHIP genes; so was TET2), and found that both genes mutations were associated with worse outcomes (Figure 4; Table S7).
Figure 4.
Kaplan-Meier Time-to-First Event Curves of DNMT3A or TET2 Separately for Death and MACE. No matter patients with DNMT3A- (a, b) (no-DNMT3A VAF≥2% group, n = 444; DNMT3A VAF≥2% group, n = 41) or TET2- (c,d) (no-TET2 VAF≥2% group, n = 460; TET2 VAF≥2% group, n = 25) CH-driver mutations with a VAF≥2% had higher incidence of death and MACE. P-values are for log-rank tests.
Comparisons of DNMT3A/TET2 VAF≥2% group, other-CHIP VAF≥2% group and no-CHIP VAF≥2% group were also performed by Kaplan–Meier survival curves and Cox proportional-hazards regression models (Fig. S5; Table S8). Compared to the no-CHIP VAF≥2% group, DNMT3A/TET2 VAF≥2% group experienced worse outcomes. However, there was no difference between other-CHIP VAF≥2% group and no-CHIP VAF≥2% group.
Inflammatory cytokines
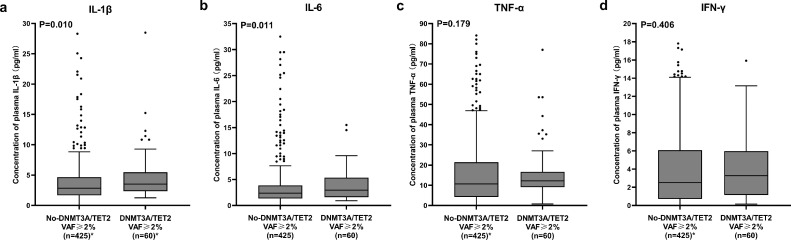
As shown in Figure 5, plasma concentrations of IL-1β [Mann-Whitney test, 3.52 (2.34–5.43) vs 2.83 (1.68–4.63) pg/ml, P = 0.010] and IL-6 [Mann-Whitney test, 2.94 (1.62–5.38) vs 2.38 (1.35–3.87) pg/ml, P = 0.011] in the DNMT3A/TET2 VAF≥2% group were significantly higher than those observed in the no-DNMT3A/TET2 VAF≥2% group, while plasma concentrations of TNF-α [Mann-Whitney test, 12.21 (9.16–16.60) vs 10.69 (4.27–21.35) pg/ml, P = 0.179] and IFN-γ [Mann-Whitney test, 3.29 (1.18–5.78) vs 2.52 (0.73–6.05) pg/ml, P = 0.406] showed no significant difference. After Bonferroni correction to account for multiple testing with a P-value of 0.0125, the difference in concentrations of IL-1β and IL-6 remained statistically significant.
Figure 5.
Concentrations of Inflammatory Cytokines. Box plots of plasma IL-1β (a), IL-6 (b), TNF-α (c) and INF-γ (d) in the no-DNMT3A/TET2 VAF≥2% group (n = 425) and DNMT3A/TET2 VAF≥2% group (n = 60). Box plots represented the median, interquartile (box) and 1.5 interquartile range (whiskers). P-values are for Mann-Whitney tests. *There are 3 points presenting IL-1β, 2 points presenting TNF-α, and 3 points presenting IFN-γ concentrations in the no-DNMT3A/TET2 VAF≥2% group, and 2 points presenting IL-1β concentrations in the DNMT3A/TET2 VAF≥2% group beyond the upper limit of the concentrations in the plot.
Fig. S6 showed the comparisons of cyokines concentrations of DNMT3A VAF≥2% group, TET2 VAF≥2% group and no-DNMT3A/TET2 VAF≥2% group (5 of 6 patients carrying both genes mutations with a VAF≥2% were classified as DNMT3A VAF≥2% group here, since the VAFs of DNMT3A were higher than those of TET2). The IL-1β and IL-6 levels in DNTMT3A VAF≥2% group or TET2 VAF≥2% group tended to be higher than those in no-DNMT3A/TET2 VAF≥2% group.
Discussion
This study investigated the prevalence and prognostic significance of DNMT3A- and TET2-CH-driver mutations in patients with STEMI by targeted deep sequencing. The main findings of our study are: (1) DNMT3A- and TET2-CH-driver mutations with a VAF≥2% are not infrequent (12.4%) in patients with STEMI. (2) the presence of DNMT3A- and TET2-CH-driver mutations are associated with profoundly worse outcomes following STEMI. (3) patients carrying DNMT3A- or TET2-CH-driver mutations with a VAF≥2% have significantly higher levels of IL-1β, and IL-6, which might contribute to disease progression and worse clinical outcomes.
The association of CHIP and atherosclerotic cardiovascular disease was preliminarily described using whole exome sequencing with mean sequencing depth of approximately 84×, which cannot detect SNP mutations with a VAF<3.5% or indel mutations with a VAF<7%, resulting in a comparatively low prevalence of mutations. However, CH-driver mutations with a VAF≥1% can be fully identified by targeted deep sequencing performed in our study, and we found that DNMT3A- and TET2-CH-driver mutations were not infrequent (VAF≥2%, 12.4%; VAF≥1%, 22.9%) in STEMI patients. Consistent with previous studies,1,2,12,14 the prevalence of mutations increased with patients age. Besides, women were observed to have a higher prevalence of mutations. However, given that women presented at an older age than men (mean age 65.9 years vs 60.2 years) and that the association did not remain significant after adjusting for age, the gender difference in mutation frequency might be associated with the difference in age. We also found that DNMT3A- or TET2-CH mutation carriers had higher prevalence of diabetes and higher levels of HbA1c, which is in agreement with previous studies1,10 revealing that diabetes is associated with CHIP mutations in 22 population-based cohorts and that somatic TET2 mutations contribute to insulin resistance in mouse models.
Dorsheimer et al.,12 who investigated the relationship between the outcome of chronic ischemic HF and CHIP, revealed that in patients harboring DNMT3A or TET2 mutations, most deaths were related to progression of HF and arrhythmia. However, besides HF events, in the present study focusing on STEMI patients, DNMT3A- or TET2-CH-driver mutations were also associated with ischemic events, including fatal or nonfatal MI, and ischemic stroke. Previous studies6, 7, 8, 9 and our data revealed that mutations in either DNMT3A or TET2 were associated with the activation of the inflammasome complex, or increased gene expression of inflammatory cytokines, such as IL-1β and IL-6. Therefore, a possible driver for recurrent ischemic events relates to plaque events associated with untreated “vulnerable” non-culprit lesions or accelerated in-stent neoatherosclerosis exposed to a high inflammatory state. It is logical to propose that the increased inflammation associated with DNMT3A- and TET2 mutations may serve as a new therapeutic target to prevent the potential adverse events post-MI.
Typically, 2% VAF have been used as the threshold to define CHIP,12,13,15,23 and indeed, we demonstrated that STEMI patients carrying DNMT3A- or TET2-CH-driver mutations with a VAF≥2% were associated with worse outcomes and higher levels of plasma IL-β and IL-6. However, in our cohort, patients carrying DNMT3A- or TET2-CH-driver mutations with a VAF of 1-2% also experienced more MACE, compared to patients without a VAF≥1%, which was consistent with the previous studies revealing that there was a dose-response association between clone size and death in HF patients12 and that optimized VAF cut-off values of DNMT3A- and TET-CH-driver mutations for prediction of death were 1.15% and 0.73%, respectively.14 So, more studies are needed to identify disease-specific threshold levels of VAF in cardiovascular diseases.
In our study, we found that patients in DNMT3A/TET2 VAF≥2% group had higher plasma concentrations of IL-1β and IL-6 compared to those in no-DNMT3A/TET2 VAF≥2% group, but levels of TNF-α, INF-γ and hs-CRP showed no difference between the two groups. Previous cohort studies showed contradictory results about hs-CRP. Some3,12, 13, 14 found no significant difference in hs-CRP levels between CHIP carriers and non-CHIP carriers, while only a larger single-centre study24 consisting of patients with coronary artery disease revealed that CHIP carriers had 21% higher hs-CRP levels compared with their noncarrier counterparts. Besides, in a study using single-cell RNA–sequencing found that patients carrying DNMT3A or TET2 mutations displayed increased expression of IL-1β and IL-6. Therefore, we considered that plasma levels of IL-1β or IL-6, rather than hs-CRP, could serve as the possible biomarker to help identify carriers of DNMT3A- or TET2- mutations.
The limitations of our study are as follows: firstly, this was a single center study. However, the present study investigating the role of CHIP in this subset of population included the largest number of STEMI patients. Secondly, according to the sensitivity of targeted deep sequencing performed in our study, only mutations with a VAF≥1% were considered. Therefore, the impact of mutations with a VAF<1% on prognosis is unclear. However, with a median of 3.0 years of follow-up, the incidence of MACE in patients without a VAF≥1% was relatively low. Thus, it is reasonable to consider that hazard ratio of DNMT3A/TET2-CH-driver mutations with a VAF<1% is limited. Thirdly, considering the definitive evidence from experiments6, 7, 8, 9 that DNMT3A and TET2 were associated with inflammation and comparatively low prevalence of other CH-driver mutations, we mainly focused on mutations in DNMT3A and TET2. Therefore, the impacts of mutations in other CHIP genes need to be further investigated.
In conclusions, DNMT3A- or TET2-CH-driver mutations with a VAF≥2% were observed in over 10% STEMI patients, and were significantly associated with poorer prognosis, which may be explained by higher levels of inflammatory cytokines in mutations carriers.
Data sharing statement
CHIP-driver mutations are available in supplementary data, and the raw sequencing data have been deposited in NCBI Sequence Read Archive (SRA) database (BioProject accession: PRJNA807074).
Contributors
Conceptualization, Haibo Jia, Bo Yu, Shengfang Wang; Methodology, Shengfang Wang, Xing Luo, Xiaoyi Bao, Ying Lv, Minghao Liu, Ming Zeng, Ji Li; Investigation, Shengfang Wang, Sining Hu, Xing Luo, Chen Zhao, Xi Chen; Writing Original Draft, Shengfang Wang; Review, Haibo Jia, Amanda Unsworth, Sarah Jones, Thomas W. Johnson, Stephen J. White; Funding acquisition, Haibo Jia, Bo Yu. Haibo Jia and Bo Yu have verified the underlying data. All authors read and approved the final version of the manuscript.
Declaration of interests
All authors report to have nothing to disclose.
Acknowledgements
We gratefully acknowledge the participants who provided biological samples and data for this analysis. This work was supported by National Natural Science Foundation of China (Grant Nos.: 81722025, 82061130223, and 81827806); and National Key R&D Program of China (Grant Nos.: 2016YFC1301101 and 2016YFC1301103).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103964.
Contributor Information
Haibo Jia, Email: jhb101180@163.com.
Bo Yu, Email: yubodr@163.com.
Appendix. Supplementary materials
References
- 1.Jaiswal S., Fontanillas P., Flannick J., et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaiswal S., Natarajan P., Silver A.J., et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bick A.G., Pirruccello J.P., Griffin G.K., et al. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation. 2020;141:124–131. doi: 10.1161/CIRCULATIONAHA.119.044362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Honigberg M.C., Zekavat S.M., Niroula A., et al. Premature menopause, clonal hematopoiesis, and coronary artery disease in postmenopausal women. Circulation. 2021;143:410–423. doi: 10.1161/CIRCULATIONAHA.120.051775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaiswal S., Libby P. Clonal haematopoiesis: connecting ageing and inflammation in cardiovascular disease. Nat Rev Cardiol. 2020;17:137–144. doi: 10.1038/s41569-019-0247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuster J.J., MacLauchlan S., Zuriaga M.A., et al. Clonal hematopoiesis associated with tet2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355:842–847. doi: 10.1126/science.aag1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sano S., Oshima K., Wang Y., Katanasaka Y., Sano M., Walsh K. Crispr-mediated gene editing to assess the roles of tet2 and dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ Res. 2018;123:335–341. doi: 10.1161/CIRCRESAHA.118.313225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sano S., Oshima K., Wang Y., et al. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the il-1beta/nlrp3 inflammasome. J Am Coll Cardiol. 2018;71:875–886. doi: 10.1016/j.jacc.2017.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abplanalp W.T., Mas-Peiro S., Cremer S., John D., Dimmeler S., Zeiher A.M. Association of clonal hematopoiesis of indeterminate potential with inflammatory gene expression in patients with severe degenerative aortic valve stenosis or chronic postischemic heart failure. JAMA Cardiol. 2020;5:1170–1175. doi: 10.1001/jamacardio.2020.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuster J.J., Zuriaga M.A., Zorita V., et al. Tet2-loss-of-function-driven clonal hematopoiesis exacerbates experimental insulin resistance in aging and obesity. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Sano S., Yura Y., et al. Tet2-mediated clonal hematopoiesis in nonconditioned mice accelerates age-associated cardiac dysfunction. JCI Insight. 2020;5 doi: 10.1172/jci.insight.135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorsheimer L., Assmus B., Rasper T., et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol. 2019;4:25–33. doi: 10.1001/jamacardio.2018.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mas-Peiro S., Hoffmann J., Fichtlscherer S., et al. Clonal haematopoiesis in patients with degenerative aortic valve stenosis undergoing transcatheter aortic valve implantation. Eur Heart J. 2020;41:933–939. doi: 10.1093/eurheartj/ehz591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assmus B., Cremer S., Kirschbaum K., et al. Clonal haematopoiesis in chronic ischaemic heart failure: prognostic role of clone size for dnmt3a- and tet2-driver gene mutations. Eur Heart J. 2021;42:257–265. doi: 10.1093/eurheartj/ehaa845. [DOI] [PubMed] [Google Scholar]
- 15.Pascual-Figal D.A., Bayes-Genis A., Diez-Diez M., et al. Clonal hematopoiesis and risk of progression of heart failure with reduced left ventricular ejection fraction. J Am Coll Cardiol. 2021;77:1747–1759. doi: 10.1016/j.jacc.2021.02.028. [DOI] [PubMed] [Google Scholar]
- 16.Ridker P.M., Everett B.M., Thuren T., et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377:1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 17.Tardif J.C., Kouz S., Waters D.D., et al. Efficacy and safety of low-dose colchicine after myocardial infarction. N Engl J Med. 2019;381:2497–2505. doi: 10.1056/NEJMoa1912388. [DOI] [PubMed] [Google Scholar]
- 18.Broch K., Anstensrud A.K., Woxholt S., et al. Randomized trial of interleukin-6 receptor inhibition in patients with acute st-segment elevation myocardial infarction. J Am Coll Cardiol. 2021;77:1845–1855. doi: 10.1016/j.jacc.2021.02.049. [DOI] [PubMed] [Google Scholar]
- 19.Abbate A., Toldo S., Marchetti C., Kron J., Van Tassell B.W., Dinarello CA. Interleukin-1 and the inflammasome as therapeutic targets in cardiovascular disease. Circ Res. 2020;126:1260–1280. doi: 10.1161/CIRCRESAHA.120.315937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lawler P.R., Bhatt D.L., Godoy L.C., et al. Targeting cardiovascular inflammation: next steps in clinical translation. Eur Heart J. 2021;42:113–131. doi: 10.1093/eurheartj/ehaa099. [DOI] [PubMed] [Google Scholar]
- 21.Jennings L.J., Arcila M.E., Corless C., et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the association for molecular pathology and college of american pathologists. J Mol Diagn. 2017;19:341–365. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salk J.J., Schmitt M.W., Loeb LA. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat Rev Genet. 2018;19:269–285. doi: 10.1038/nrg.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steensma D.P., Bejar R., Jaiswal S., et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busque L., Sun M., Buscarlet M., et al. High-sensitivity c-reactive protein is associated with clonal hematopoiesis of indeterminate potential. Blood Adv. 2020;4:2430–2438. doi: 10.1182/bloodadvances.2019000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.