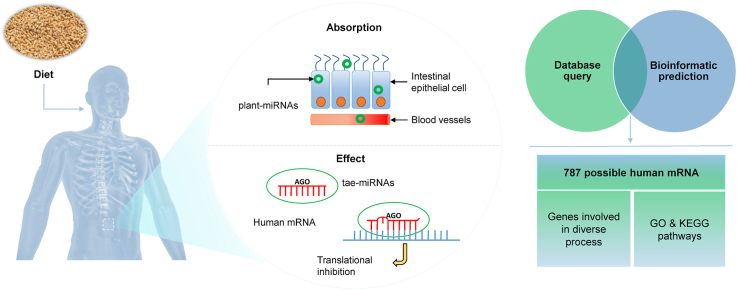
Abstract
Plant-derived miRNAs can be found in the human body after dietary intake, and they can affect post-transcriptional gene regulation in human. It is important to identify targets to determine the possible effects in human genes by using computational approach. In this study, 787 possible mRNAs human targets were predicted by 84 miRNAs of wheat. A total of 14 miRNAs were identified with individual binding to 33 mRNAs associated with schizophrenia, epilepsy, neurodevelopmental disorders, and various cancers, located in the 3′UTR of the mRNA. A functional enrichment was carried out, where the results showed associations to pathways such as dopaminergic synapse (hsa04728), and signaling pathways, significantly associated with the target genes. The prediction of target mRNAs in humans by wheat miRNAs, offer candidates that could facilitate the search and verification, which could be of relevance for future projects and therefor contribute in the therapeutic treatment of various human diseases.
Keywords: microRNA, mRNA, Triticum aestivum, Human, Functional enrichment
Graphical abstract
1. Introduction
The microRNAs (miRNAs) of plants and animals are an abundant class of small non-coding RNAs sequences around ∼22 nucleotides long [1]. They are derived from hairpin precursors, which have remained evolutionarily similar in various species, and have the ability to negatively regulate gene expression at the post-transcriptional level by complementary base binding in various regions of the mRNA (3′UTR, 5′UTR and ORF) and via the RNA-induced silencing complex (RISC) whose binding causes mRNA cleavage or translation inhibition [ [[2], [3], [4]]].
It is expected that 30–60% of the genes in humans are miRNA regulatory targets [5], involved in various developmental processes, molecular, cellular pathways and human disease [6]. When crossing the intestinal barrier after ingestion miRNAs can be packaged in microvesicles, circulate through the human body and be identified in fluids such as human plasma [7] and breast milk [8].
Identifying mammalian gene targets in a timely manner by plant miRNA offers a reliable, effective and economical way to determine the possible effects of interactions given by exogenous molecules, regardless of their origin, on human mRNA [ [[9], [10], [11], [12], [13], [14]]].
Recently, plant miRNAs and their interactions have been actively studied, contributing to the knowledge of coevolution mechanisms between plant miRNAs and human mRNAs. For example, the in silico prediction on the possible regulation of plant miRNA on human mRNA was tested, coincided with an increase in cholesterol-LDL levels in vivo and in vitro, caused by the decrease of LDLRAP1 protein expression, providing the first evidence that food miRNA could bind to a target mRNA and influence the regulation of interspecies expression [15]. Similarly, through bioinformatics analysis, found a number of potential target genes for MIR2911 from the Lonicera japonica plant (honeysuckle) were found in different viruses such as H1N1, H3N2, H5N1 and H7N9. Later, in cultures of cellular lines infected with H1N1 and transfected with synthetic MIR2911, they managed to significantly decrease the viral count because the target genes: PB2 and NS1, are essential for replication. Subsequent functional studies showed that miR2911 inhibited several IAVs (influenza A viruses) including H1N1, H5N1 and H7N9, decreasing mortality in an animal model [16]. Suggesting the first reports that exogenous miRNAs regulate gene expression in endogenous mRNAs similar to mammalian miRNAs and in turn the use of the bioinformatics approach, playing an important role in target identification.
In the present study we have selected the monocotyledonous plant Triticum aestivum (wheat) because it is a common source in human nutrition, we performed an in silico prediction analysis of potential mRNAs target in Homo sapiens (human) by mature miRNAs of wheat, and its relationship to diseases through an enrichment of the possible target genes. In addition, we identified target genes with diverse functions in cellular processes, providing information on regulatory mechanisms given by wheat miRNAs. Also, a functional enrichment has been carried out to explore the participation of the target human mRNAs involved in biological processes and associated pathways, including apelin signaling pathway, fanconi anemia pathway and dopaminergic synapse.
2. Material and methods
2.1. Data collection
The complete set of known mature miRNA sequences, which includes a total of 122 miRNAs from wheat, were downloaded from the registered database (version 22.1) (http://www.mirbase.org) in FASTA format. To reduce errors in prediction and future detection, we applied rigorous filtering for mature miRNAs, i) The mature miRNA should have more than 1400 reads during sequencing reported in its bioinformatic analysis; ii) The miRNA size should be in the range of 19–25 nucleotides and iii) All mature miRNAs should be in the same direction from 5′ to 3'. Following this, 84 miRNAs from wheat were selected as reference data set for the study.
2.2. Prediction of potential targets of miRNAs
The prediction of possible targets on human was carried out by the sequence similarity search tool BLASTn (2.9.0) on its NCBI website (http://www.ncbi.nlm.nih.gov) using the default settings, an E-value threshold of 10; and the search coordinates were set in the nucleotide sequence 2 to 13 of each miRNA. From the RefSeq RNA database the predicted miRNA target sites were identified by aligning the sequences in a complementary way to the miRNA of wheat with human mRNA. RNAhybrid [17] was used to scan the hybridization patterns of the individual miRNAs/mRNA following Zhou Z. et al. methodology with some modifications [16]. i) The seed region had to perfectly match the nucleotide 2 to 7 of the mature miRNA. ii) The minimum free energy (MFE) for the hybridization was below −25 kcal/mol iii) No G-U bonds in the seed region. We also used the MIR168a as a target prediction control [15].
2.3. miRNA homology
We searched for homology using MirCompare between 2042 H. sapiens and 84 T. aestivum matured miRNAs from miRBase was used [18]. The parameters used for this analysis were taken from Minutolo et al., 2018 [19], with modification in the threshold of the seed region according to the guidelines provided by the default settings of the tool. The possible target genes of the resulting human miRNAs with homology to wheat miRNAs as reported by MirCompare were found by an analysis with TargetScan 8.0 algorithms (http://www.targetscan.org).
2.4. Functional enrichment analysis
The functional analysis of the targets, including biological processes was done using clusterProfiler [20], package inside programming language R version 4.0.3 [21]; with only those target sequences with miRNA binding sites in the 5′UTR, 3′UTR and CDS regions. Designating the potential processes with an adjusted p-value lower than 0.05 (FDR <0.05) that involve the predicted mRNA sequences and suggesting the probable influence in the regulation of plant miRNAs in the human organism.
3. Results
3.1. Characteristics of the binding of wheat mature miRNAs with human mRNAs
According to the methodology previously mentioned, the results obtained were similar to the ones described by Zhang et al. 2012 [15]. We selected 84 mature miRNAs from wheat and predict putative targets genes in humans, based on the criteria specified in methods section, which resulted in a total 889 miRNA/target interactions with 787 genes with diverse functions, the characteristics and specific target sequences of the 84 miRNAs analyzed in this study are provided in the (Supplementary Table 1). 70 of these genes had interactions with more than one miRNA and 6 come from miRNAs families, as previously reported by Han R. et al. [22] (Table 1). These miRNAs were divided in two groups; in group one, 14 miRNAs identified with one to four target genes, giving a total of 30 target genes. In group two 70 miRNAs with five or more target genes, giving a total of 757 target genes. The tae-miR9652–3p and tae-miR9652–5p of group one, originated from the same precursor (pre-miRNA) in miRNA biogenesis, have binding sites in the mRNAs of different genes. In the second group the tae-miR9657b-3p and tae-miR9657b5-p, were found as a pair originating from the same pre-miRNA, obtaining 18 targets for miR-3p, and 10 for miR-5p strand, all being different. The miRNAs with the highest number of targets were tae-miR9657c (19 genes), tae-miR9654a (21 genes), tae-miR160 (22 genes), tae-miR396 (23 genes), tae-miR9662a (23 genes), tae-miR9666a (24 genes), tae-miR9664 (28 genes), tae-miR9655 (31 genes), tae-miR9666b-3p (34 genes), tae-miR164 (35 genes).
Table 1.
Potential target genes of 84 wheat miRNAs identified in human.
| miRNA ID | Number of targets | miRNA family | Target Gene |
|---|---|---|---|
| Tae-miR1117 | 9 | TULP4, CHSY3, SMARCC1, C5orf52, DSCAML1, BZW1, UBR5, LOC102724740, ACVR1B | |
| Tae-miR1118 | 7 | ARHGAP32, PRUNE2, XKRX, HOOK3, LOC105372263, ANTXR2, MAP1B | |
| Tae-miR1120b | 1 | RPAP2 | |
| Tae-miR1122a | 6 | SERINC5, ZBTB39, GDNF, SPAG9, ZC3H12C, CDC42BPB, | |
| Tae-miR1122b | 2 | LOC105375144, SPTLC3 | |
| Tae-miR1122c | 1 | PPP2R3A | |
| Tae-miR1123 | 9 | EPO, LOC105375220, ASGR2, HPCAL1, CNFN, COL16A1, IGF2BP2, BEST1, LOC100507516 | |
| Tae-miR1125 | 5 | CHRM3, SLC38A8, RALY, TIMM10, RYR2 | |
| Tae-miR1127a | 4 | MSRB3, IPO13, RRAD, MFAP3 | |
| Tae-miR1127b | 9 | XKR6, STXBP5L, NOL8, ZNF691, KRIT1, RP1L1, ZNF510, SNX13, RGS6 | |
| Tae-miR1128 | 12 | CNTNAP4, LOC105371840, ESR1, RAPGEF6, LOC107986008, TMEM161B-AS1, BRINP1, INTS4, THAP9, ADCY1, RUSC2, KLHL36 | |
| Tae-miR1130b | 2 | PRORY, ASPH | |
| Tae-miR1131 | 7 | SMARCC1, DSCAML1, UBR5, CHSY3, BZW1, TULP4, STXBP5L | |
| Tae-miR1133 | 7 | WDR75, MALT1, RIMKLB, SKAP1, ACAD11, GPR176, ST8SIA4 | |
| Tae-miR1135 | 5 | LOC112268243, LOC105371988, SPACA1, THSD7A, DDX5 | |
| Tae-miR1136 | 11 | OBSCN, DHX37, MRC1, STAB1, LOC105379283, SNORA14B, ZNF174, CALN1, TMEFF2, CALN1, UQCC2 | |
| Tae-miR1137a | 3 | ATP6V1B2, ZFPM2, ATXN1L | |
| Tae-miR1137b | 8 | RBX1, STK35, CREB5, RNF121, KIF27, JAG1, PKHD1, ITGA11 | |
| Tae-miR1138 | 8 | KDM3B, USP35, PRKACB, MAK, ZBED2, TESK2, BMP3, ZSCAN25, | |
| Tae-miR1139 | 5 | ZFHX3, ZC3H8, CDON, MID1, DZIP1L | |
| Tae-miR156 | 14 | CCDC88C, F11R, FAM13A, KIF5A, LOC105369388, MAPK11, ZBTB40, LINCOO312, CCDC15, TMEM201, ADGRD1, CAPN13, TGOLN2, CCNH | |
| Tae-miR159a | 8 | CNOT9, GPR4, TEX13C, LOC105377862, MDH1B, LINC02789, MAN1A2, GPR135, | |
| Tae-miR159b | 8 | miR159 | CNOT9, GPR4, TEX13C, LOC105377862, MDH1B, LINC02789, MAN1A2, GPR135, |
| Tae-miR160 | 22 | miR160 | PCARE, FAM118B, BCAS1, BCAP29, TMEM241, CAMTA1, FMRD8, PSMD7, ASPDH, CRYBB3, PLEKHA6, BTNL9, ANKRD35, KAT5, DAAM2, PARVB, PRKAG2, RTN4RL1, CHST10, RHPN1, GHRL, LOC100996662 |
| Tae-miR164 | 35 | miR164 | FGFRL1, ZNF701, SLC9A9, MTCL1, PPRC1, GREM1, SYNE1, C8orf33, GRM2, PCYT2, GLI3, SNED1, GNAI2, SMARCC2, CSRP1, B3GNT7, KCNA2, LOC107986317, RFPL4B, FSTL4, SLC25A1, RASSF6, KCNH6, APOBEC3B, APOBE3CA, ABCB9, BIRC6, NCAPH, SETD4, SLC6A7, LRRC55, MNT, ACBD4, TRIM41, PBXIP1 |
| Tae-miR167a | 13 | miR167 | PRICKLE2, WASHC2A, TM2D3, CACNA2D4, LOC105375492, NOX5, CBX7, TNFRSF10D, BCL7B, MBD3, NCBP3, SLC9B2, THBS3 |
| Tae-miR167b | 9 | EME1, ZNF20, PLCE1, LOC107986558, CACNA1C-IT1, LOC440292, CAMK2D, GPD1, SNRNP200 | |
| Tae-miR167c | 12 | TM2D3, WASHC2A, CACNA2D4, NCBP3, BCL7B, LOC105375492, MBD3, NOX5, CBX7, TNFRSF10D, SLC9B2, THBS3 | |
| Tae-miR171a | 2 | miR171 | IGF1, C15orf56 |
| Tae-miR171b | 11 | TMEM8A, RNF112, TMEM151A, NAT16, RPAP1, LIMK2, GCC1, LOC107986282, MPRIP, RAPH1, LOC112268024 | |
| Tae-miR1847 | 15 | CRISP2, CYP27A1, ALG3, LAMA5, N4BP1, ART3, ZNF619, UBQLN4, TMEM178B, LMOD1, LGI3, LOC105377347, SH3PXD2A, CRISPLD1, MRM1 | |
| Tae-miR319 | 14 | SCART1, HOXB3, SLC1A4, TIGD7, FAT4, FAM205C, KLC2, KMT5A, ACLY, MUC16, CCDC134, LOC105375153, RAB5B, SUOX | |
| Tae-miR395a | 7 | CLCN5, LOC107984275, VAPA, LOC105373908, CERS4, PPARGC1B, ITGB8 | |
| Tae-miR395b | 11 | miR395 | CLCN5, USP6NL, LINCO2739, FN1, CASP5, WDFY3, CEP63, METRN, UBAP1L, LOC112267986, LYPLAL1 |
| Tae-miR396 | 23 | SLC25A25, CPOG1, CYFIP1, NOTCH2NLB, ZNF814, KDM1A, PELI3, GCDH, ZNF32, ADAMTS18, ZNF776, CL11A1, PTCH2, UNC119B, ZBTB14, LOC107984192, REST, FOXP2, SLITRK9, CDC5L, EFHB, KIF2A, GRIN1 | |
| Tae-miR397 | 5 | PALM2AKAP2, SURF6, BCAR1, C1orf67, RNF175 | |
| Tae-miR398 | 14 | PPM1H, CORO1B, ANKK1, LOC105374534, FBXO10, DNMT1, SLC37A1, PCNX3, LOC105370363, LOC105378740, ZNF691, KIAA0408, SOGA3, PPIP5K2, | |
| Tae-miR399 | 16 | DRICH1, ANKRD36, SET, LINC02142, ARIH2, FAM166C, ZFP2, SCCPDH, ADPRM, LRRIQ1, AOC3, NUP210, CALM1, MANBA, MYLK4, ANKFN1 | |
| Tae-miR408 | 16 | USP43, ATPAF2, PSMD11, ADGRF5, DIAPH3, AGO3, ABCG8, LOC105376475, ZNF518A, EPS15L1, ANXA11, ROS1, RYR3, KCNB2, LOC112268372, KSR1 | |
| Tae-miR444a | 14 | LOC112268280, RUNDC1, LOC105371800, ZC3H4, LOC107986334, CDC25A, ZC3H4, CCDC171, SUPT20HL1, LOC101929566, SYNJ2, TEKT3, FRMD3, YPEL5 | |
| Tae-miR444b | 13 | LOC112268280, RUNDC1, LOC107986334, LOC105371800, ZC3H4, CDC25A, CCDC171, SUPT20HL1, LOC101928566, SYNJ2, TEKT3, FRMD3, YPEL5 | |
| Tae-miR5048 | 5 | CHRM3, TBC1D4, ADAM22, GRM4, LOC105378532 | |
| Tae-miR5049 | 2 | LOC147004, PNPT1 | |
| Tae-miR5050 | 9 | PEG3, IPPK, ITPA, TRIO, TEX30, SCFD2, FOSL2, LINC01226, BCLAF1 | |
| Tae-miR5062 | 9 | ARMC9, YEATS2, LOC107985153, CD2BP2, ABI2, RNF183, CACNA2D4, AHNAK, HS6ST2 | |
| Tae-miR5175 | 16 | SETBP1, RBBBP6, GPRC5D, CHST2, C4orf54, CENPS-CORT, CORT, RNLS, TRIM37, NSL1, LINC01588, SNRNP27, LOC101929270, SUCO, STK31, DNAJC6 | |
| Tae-miR5200 | 5 | LINC02842, LOC101927610, LOC107986016, NRG4, LOC101928421 | |
| Tae-miR531 | 18 | REPIN1, LOC107985320, SLC5A10, GBX2, GPC6, NINL, BACH2, B3GLCT, PLXNB3, DAB2IP, GCK, DCP2, ZNF282, NECTIN4, RFTN1, GPR22, ZNF618, GPR153 | |
| Tae-miR6197 | 3 | EXOC6, ABCD2, ZNF519 | |
| Tae-miR7757 | 2 | RPR6KA2, HDAC8 | |
| Tae-miR9652–3p | 1 | PSMD13 | |
| Tae-miR9652–5p | 1 | POU2F1 | |
| Tae-miR9653b | 15 | ADCK5, SHE, TPST2, CPNE2, FGGY, P4HA2, TPM2, P4HA2, LOC105372821, VPS26C, HES7, ABCG8, RAB37, MTOR. RAB37 | |
| Tae-miR9654a | 21 | DTX2, SGO2, LOC10537819, TBRG1, LOC107984429, NT5DC2, LOC107986811, ELP6, LYRM9, LOC105375206, CFAP61, CHI3L2, GEMIN5, AP3M2, ZSCAN26. FBXO40, BZW2, TNRC6B, SESTD1, EBF2, TBL2, | |
| Tae-miR9654b | 12 | ZNF740, CNF740, CNST, HOXD4, ARHGAP1, SLITRK3, KALRN, UFL1-AS1, SLCO4A1, PCDH8, MLYCD, PDE1A | |
| Tae-miR9655 | 31 | KPNA6, ELMO1, GEMIN5, TOMM20, TRIM71, CCBE1, CMBL, TAX1BP3, FGFR10P, CCR9, ELOVL2, ADH6, ARHGAP33, RNF8, TTC39B, CHD1, HMBOX1, MYADM, PLSCR3, HIPK1, STX7, LOC105372520, LOC105377622, WBP2, NIPAL3, TRPC3, VAMP2, PRRT2, USP12, HOOK3, AP1S3 | |
| Tae-miR9656 | 7 | NPSR1-AS1, DAAM1, LDLRAP1, TRIM72, FAM49B, TRAK1, FAM49B | |
| Tae-miR9657a | 18 | MECR, C17orf80, PCNT, STOX1, MAGI2, MELTF, PITRM1, LASP1, EPHA10, FYCO1, RGS6, SNX29, LZTS3, ZNF395, LINC00473, TTF1, MYO10, CCDC40 | |
| Tae-miR9657b-3p | 17 | C17orf80, MELTF, PCNT, STOX1, MAGI2, LZTS3, PITRM1, EPHA10, RGS6, SMX29, LASP1, TTF1, ZNF395, FYCO1, LONC00473, MYO10, GAD1 | |
| Tae-miR9657b-5p | 10 | MAFB, SPON1, MAF, TBC1D8, MAFA, FSTL4, CEBPB, DMXL1, SCAMP4, ELFN2 | |
| Tae-miR9657c | 19 | MECR, C17orf80, MELTF, PCNT, STOX1. MAGI2, LZTS3, PITRM1, EPHA10, RGS6, SNX29, LASP1, TTF1, ZNF395, FYCO1, LINC00473, MYO10, CCDC40, GAD1 | |
| Tae-miR9662a | 23 | LOC101929698, LRCOL1, LOC105370502, KCNA7, RBCK1, CLDN20, HRH2, TAF5L, MIR155HG, LINC02568, RIBC2, PFKFB2, TOP3A, LOC107986445, MOB3C, CLTRN, JAKMIP2, HIRA, LOC105374262, PCNT, SS18L2, CASP9, ARNTL | |
| Tae-miR9662b | 12 | LOC101929698, PIGV, FAM83H-AS1, LOC105370502, AOX3P-AOX2P, HRH2, LOC105371449, LINC02568, HIRA, LOC107986445, LOC102723784, GNB5 | |
| Tae-miR9663 | 5 | TAF1, ZC3H7B, PSMB4, DHPS, SGF29 | |
| Tae-miR9664 | 28 | IQGAP1, TNXB, RIN3, USP4O, LOC112268055, TNC, SOX30, ABCC1, FAHD2B, ABCC1, FAHD2CP, TNR, MLST8,CLEC3B, CCN1, CASZ1, KRT31, COPG1, NTNG2, CORIN, ZBTB48, CFAP46, TENM3, TELO2, LOC112268408, ALPK1, DENND3, KIAA0586 | |
| Tae-miR9666a | 24 | RAPGED4-AS1, BOC, OVOL1, USH2A, NACA, C8orf34, ANXA11, DOK1, CDH11. MST1R, CSRNP1, MBD1, PRR23D1, STON2, DCHS1, MARC1, WDR46, MBD1, DEFB119, ZFAND5, CDH6, SFMBT1, SEC14L2, DLX5, RAD51AP2 | |
| Tae-miR9666b-3p | 34 | SEC14L2, DLX5, RAD51AP2, POLR2A, YEATS4, CC2D2A, PCDHA6, PCDHA2, PCDHA10, PCDHA4, PCDHA13, IFT22, RAET1G, BSN, PDLIM5, ADAMTS10, KCNIP3, GABBR1, PCDHA8, PDZD2, ATP13A1, RBM6, ARID5B, ARHGAP26, PHF2, PCDHA12, PCDHA5, SGPP2, PCDHA7, ARHGAP26, CLOCK, WDR1, SLC24A1, PEX5L | |
| Tae-miR9668 | 6 | LIN02362, TMEM185B, POLE2, LOC101928985, LOC105376956, SLX4 | |
| Tae-miR9669 | 8 | TOP3A, RXRG, SSH2, ENGASE, LRP6, GAB1, SMUG1, BAHCC1 | |
| Tae-miR9670 | 7 | LOC105372542, NCAPD3, TPM2, GDAP1, SMG7, SCTR, NRG1 | |
| Tae-miR9672a | 12 | FRG2C, DCTN5, BMP8A, PKHD1, PKHD1L1, LOC107987108, NELL2, CEP152, SEC22B, WDFY3, PLXNA4, TACR3 | |
| Tae-miR9672b | 6 | TOX4, LOC102724245, MYO18B, GNL2, APMAP, KIAA1217 | |
| Tae-miR9674a | 18 | ACSS1, FANK1, TJP2, LOC105370291, EIF3A, DYRK1A, ATP9B, ZNF629, PDZRN3, LIPG, RWDD1, PRMT3, TMEM246, ELF4, VPS33A, ERICH1, FAR1, LINGO2 | |
| Tae-miR9674b | 15 | TBX19, DSTYK, TXNDC9, FANCE, NSDHL, GALNT3, TCF4, FAAH2, TOP2B, LOC112268009, TCF4, TSTD2, GCLC, SBF2, LINC03094 | |
| Tae-miR9675 | 5 | BTRC, TMEM135, MRRF, ATRIP, ATRIP-TREX1 | |
| Tae-miR9676 | 3 | TMEM259, GRID1, FOXD1 | |
| Tae-miR9677a | 6 | MAN2A2, RRAS2, OLFML2B, LOC100132249, ABCC8, SLC8B1 | |
| Tae-miR9679 | 11 | DPH7, ABLIM1, ZMIZ1, BCL7B, MBOAT2, CBX5, DRP2, INPP5B, ZNF704, TBC1D12, RELN | |
| Tae-miR9772 | 5 | AAK1, RAP1B, NUCB2, SRGAP2, LOC105374129 | |
| Tae-miR9774 | 6 | CTB-3M24.3, H6PD, OTUD6B, EDNRB, RGS3, EXPH5 | |
| Tae-miR9775 | 3 | MGAT4D, MED27, USP11 | |
| Tae-miR9776 | 5 | OLIG2, HECA, ZBTB11, RNF213, LOC105370091 | |
| Tae-miR9782 | 16 | LOC112267915, RCSD1, RNLS, ZNF184, SAMD12, CYP24A1, GASK1A, LOC105371867, LOC101928819, TRDMT1, LOC105375471, NIPAL2, SRGAP1, ALG10B, CD38, LOC107987129 | |
| Tae-miR9783 | 7 | EIF4EBP2, FGFBP2, DHX8, GINS2, ARPC5, TRPC3, FAM209B |
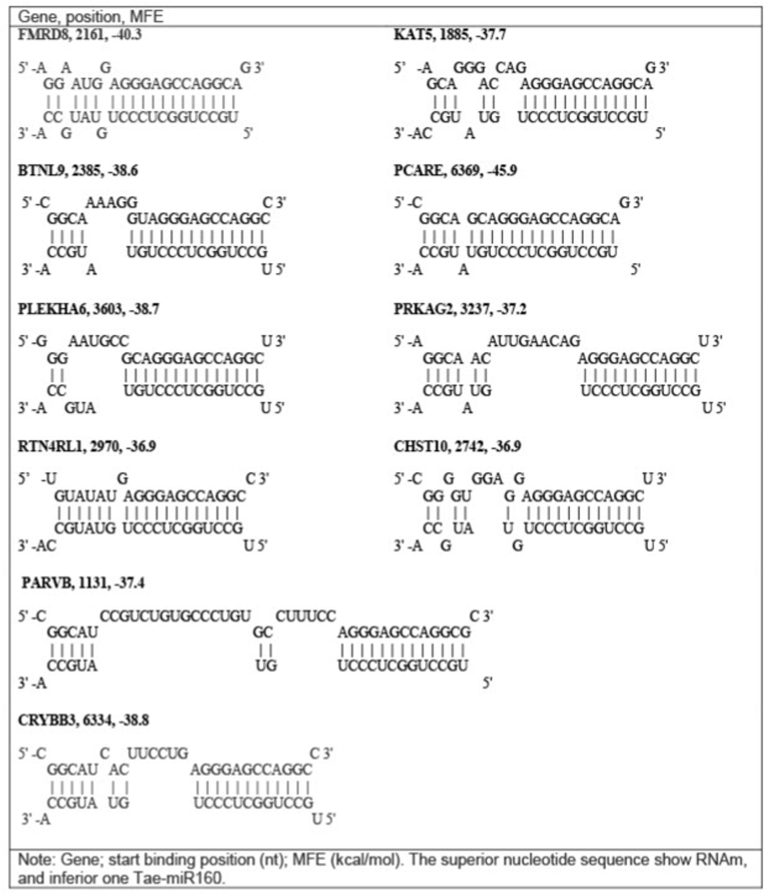
In order to show the targets and MFE for the binding between some wheat miRNAs with human mRNAs, data was filtered to show the targets genes with lower value of MFE and were the sites union are located in 3′UTR binding (Table 2). 10 of the miRNAs show 1 target gene, 2 miRNAs show two targets, and 2 miRNAs show three and six targets, with a MFE between −34.1 and −43 kcal/mol. The tae-miR160 shows 10 target genes with a free energy value between −36.9 and −45.9 kcal/mol.
Table 2.
Individual interaction between wheat miRNAs and human mRNAs at the 3′ UTR site.
|
Gene |
miRNA ID | kcal/mol |
|---|---|---|
| EPO | Tae-MIR1123 | −39.1 |
| RALY | Tae-MIR1125 | −35.3 |
| CNTNAP4 | Tae-MIR1128 | −38.7 |
| CCDC88C | Tae-MIR156 | −34.9 |
| PCARE | Tae-MIR160 | −45.9 |
| FRMD8 | Tae-MIR160 | −40.3 |
| CRYBB3 | Tae-MIR160 | −38.8 |
| PLEKHA6 | Tae-MIR160 | −38.7 |
| BTNL9 | Tae-MIR160 | −38.6 |
| KAT5 | Tae-MIR160 | −37.7 |
| PARVB | Tae-MIR160 | −37.4 |
| PRKAG2 | Tae-MIR160 | −37.2 |
| CHST10 | Tae-MIR160 | −36.9 |
| RTN4RL1 | Tae-MIR160 | −36.9 |
| MTCL1 | Tae-MIR164 | −37.5 |
| GNAI2 | Tae-MIR164 | −35 |
| SMARCC2 | Tae-MIR164 | −34.9 |
| CSRP1 | Tae-MIR164 | −34.3 |
| B3GNT7 | Tae-MIR164 | −34.2 |
| KCNA2 | Tae-MIR164 | −34 |
| NOX5 | Tae-MIR167a | −34.5 |
| CBX7 | Tae-MIR167a | −34.2 |
| BCL7B | Tae-MIR167c | −35 |
| MBD3 | Tae-MIR167c | −34.6 |
| SURF6 | Tae-MIR397 | −34.1 |
| CORO1B | Tae-MIR398 | −36.1 |
| DRICH1 | Tae-MIR399 | −34.8 |
| ATPAF2 | Tae-MIR408 | −36.8 |
| DIAPH3 | Tae-MIR408 | −35.6 |
| ZNF518A | Tae-MIR408 | −34.2 |
| LOC107985320 | Tae-MIR531 | −43 |
| TPM2 | Tae-MIR9653b | −37.4 |
| LRCOL1 | Tae-MIR9662a | −35.4 |
Because genes are nucleotide sequences coding for diffusible elements that participate in all cellular processes, it is important to show information about some human target genes by wheat miRNAs, involved in various biological processes (Supplementary Table 2): RALY - lung cancer, CNTNAP4 - epilepsy, CCDC88C - neurodevelopment, PCARE - pigmentary retinosis, FRMD8 - inflammation, CRIBB33 - cataracts, PLEKHA6 - schizophrenia, BTNL9 - ocular melanoma, KAT5 - anaplastic thyroid cancer, PARBV - oral squamous cell carcinoma, PRKAG2-nonalcoholic steatohepatitis, CHST10-tumor suppressor, RTN4RL1 - nasopharyngeal carcinoma, MTCL1 – cerebellar ataxia, GNAI2 – ovarian cancer, SMARCC2 – embryogenesis and corticogenesis, CSRP1 – lung fibrosis, B3GNT7 – colon cancer, KCNA2 – epileptic encephalopathy, NOX5 – diabetic nephropathy, CBX7 – hepatocellular carcinoma, BCL7B – Wnt signaling, MBD3 – pancreatic cancer, CORO1B - TGFβ1 signaling, ATPAF2 – dementia risk, DIAPH3 – lung adenocarcinoma, and TPM2 – aortic dissection.
Interactions between wheat miRNA and human mRNA can show how efficiently these molecules act. Some tae-miRNA160/mRNAs binding patterns in the 3′UTR of the mRNA, resulting from the bioinformatic tool RNAhybrid are shown in (Fig. 1). Taking into consideration the nucleotide sequence after the seed region of the mature miRNA, in the in silico prediction through the RNAhybrid tool, it is possible to observe the type of interaction between the tae-miRNAs with their targets and provide an approach to the type of silencing like repression and/or degradation through the complementary base pairing. For this reason, when examining the type of binding or complementarity that was performed in each miRNA/mRNA result, in a complete or incomplete complementary binding, it was found that 100% of the 889 predicted tae-miRNA/target interactions, were shown as imperfect hybrids in at least one nucleotide of the complementary mRNA after the seed region.
Fig. 1.
Schemes of the interaction of wheat miRNA nucleotide sequences with human mRNA genes.
3.2. Analysis of human homologs of wheat miRNAs
In the analysis we found homology candidates in 9 human miRNA sequences, with 7 wheat miRNA sequences. Supporting this, we identified target gene predictions of human miRNAs using the TargetScan tool and determined those targets in common predicted in this study as a whole (Table 3).
Table 3.
Potential target genes in common by wheat and human miRNAs.
| Tae-miRNA | Hsa-miRNA | Seed Score | Genes |
|---|---|---|---|
| Tae-miR9772 | Hsa-miR-143-3p | 1 | AAK1 |
| Tae-miR171a | Hsa-miR-8082 | 0.8125 | IGF1 |
| Tae-miR164 | Hsa-miR-139-3p | 0.8125 | ZNF701, FSTL4, RASSF6, LRRC55, MNT |
| Tae-miR9674b-5p | Hsa-miR-4524b-5p | 0.8125 | DSTYK, TOP2B |
| Tae-miR164 | Hsa-miR-5703 | 0.8125 | ZNF701, SLC9A9, GREM1, GLI3, SNED1, SMARCC2, CSRP1, B3GNT7, RASSF6, SLC25A1, NCAPH, LRRC55, MNT, ACBD4, TRIM41, PBXIP1 |
| Tae-miR9666b-3p | Hsa-miR-4259 | 0.8125 | PCDHA6, PCDHA10, PCDHA13, PCDHA2, PCDHA4, PCDHAC2,PCDHA3, RBM33, ARID5B, PCDHA12, PCDHA5, PCDHA7, CLOCK |
| Tae-miR164 | Hsa-miR-765 | 0.8125 | SLC9A9, PPRC1, GREM1, SYNE1, C8orf33, GLI3, SNED1, B3GNT7, FSTL4, KCNH6, LRCC55, ACBD4 |
| Tae-miR5048-5p | Hsa-miR-1205 | 0.8125 | CHRM3, ADAM22, GRM4 |
| Tae-miR395a | Hsa-miR-203a-3p | 0.8125 | VAPA, PPARGC1B |
3.3. Functional enrichment of target genes by wheat miRNAs
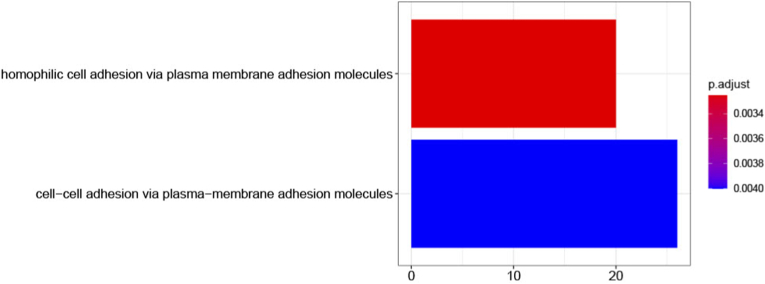
Functional annotation of target genes is an important step in understanding the respective functions. A gene ontology (GO) enrichment analysis was performed in the Kyoto Encyclopedia of Genes and Genomes (KEGG) to identify the functions that miRNAs could have on their predicted targets in humans, where all mRNAs that had binding sites in the prediction were used. From the complete target search data, 670 different genes were used, selected according to probable binding site, including only those in the 5′ UTR, 3′UTR, and CDS region and excluding those in non-coding sequences for functional enrichment analysis. The main GO terms classified in biological process are shown, with significance adjusted p-values (<0.05) indicating that a total of 17 mRNAs enriched the biological process of homophilic cell adhesion (GO:0007156), through adhesion molecules to the plasma membrane, and 22 mRNAs enriched the biological process of cell-cell adhesion through adhesion molecules to the plasma membrane (GO:0098742), by miRNAs known from wheat in the prediction (Fig. 2).
Fig. 2.
Gene ontology analysis of predicted human target genes derived from miRNAs of wheat.
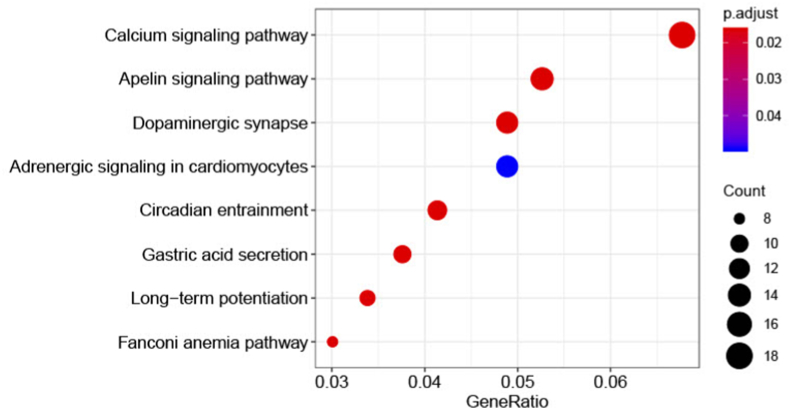
Enrichment analysis of the kEGG pathway showed that the 670 predicted genes of the 84 selected wheat miRNAs were significantly enriched in 8 pathways, with an adjusted p value (<0.05) (Supplementary Table 3). 18 genes were enriched in calcium signaling pathway (hsa04020), 10 genes were enriched in the gastric acid secretion pathway (hsa04971), 14 genes were enriched in the apelin signaling pathway (hsa04371), 9 genes were enriched in the long-term potentiation pathway (hsa04720), 11 genes were enriched in the circadian entrainment pathway (hsa04713), 8 genes were enriched in the fanconi anemia pathway (hsa03460), 13 genes were enriched in the dopaminergic synapse pathway (hsa04728), and 13 genes enriched the adrenergic signaling pathway in cardiomyocytes (hsa04261) (Fig. 3).
Fig. 3.
Pathway enrichment of predicted human target genes derived from miRNAs of wheat in 8 significant KEGG categories.
4. Discussion
Based on the results of this work and the extensive study on the mechanisms of plant miRNAs when entering the circulatory system through the diet, entering the cells to achieve a target and regulate the expression of a gene [15]. Some authors [23,24], suggest the use of available and evaluated tools for predictive analyses, since they recognize the fundamental importance of identifying the target genes in the human genome by exogenous miRNAs, in order to infer their functions and regulatory mechanisms, however they agree that it is convenient to explore and combine predictive tools because individually, they may not reflect the abundance and possible regulatory capacity of miRNAs, improving the speed and accuracy of the targets. In our research we used two search engines, that way we were able to simulate a possible interaction between species. By using BLASTn as the first instance as a search engine for complementary sequences with the updated version of the RefSeq human transcriptome database, we were able to find several targets and their variants, which is important because factors related to expression variation in target genes can provide a solid foundation and more systematic view of the genetic mechanisms that are related to human genetic variation [25].
The results obtained by the BLASTn analysis were filtered in a second computational method using RNAhybrid tool as used successfully used in several studies of cross kingdom miRNAs [16,26,27], which allows us to determine the MFE, characteristics and complementary regions of the miRNA/mRNA interaction, which allowed filtering of those interactions that we consider energetically favorable (MFE-25 kcal/mol). This provided us with a combination of algorithms in the study that allowed us to obtain a total of 787 genes and their variants with different functions as shown in (Supplementary Table 1). In addition, Table S1 shows the transcripts of non-coding RNAs (ncRNAs) that were targeted by the 84 miRNAs, since miRNA targets are not limited to coding transcripts, but can also be found in ncRNAs, indicating that miRNA targeting mechanisms can be complex by binding to a wide variety of targets [28].
A total of target genes for 84 miRNA of wheat, divided into two groups according to the number of targets, and families that each miRNA obtained, since miRNAs with a larger number of target genes should be monitored in human fluids, due to their possible ability to significantly regulate human receptor cells [9]. These targets were located in various regions along the mRNA (5′UTR, CDS, 3′UTR), increasing the area of study on the interactions given by exogenous molecules, it could help understand the role of binding sites in the mRNA with the miRNAs [29]. We also evaluated the type of binding, obtaining 100% of the possible binding as imperfect base-pair by at least 1 base, however the analysis was made with the criterion of perfect binding of the seed region which would suggest that the possible mechanism of regulation of expression according to the reported silencing by mature miRNAs [2] could be translation inhibition and not degrading by cleavage the mRNA, which is sufficient for the miRNAs to suppress their targets [23,30]. In plants, miRNA bind to their targets with high complementarity, causing messenger degradation, however in animals, miRNA recognition in just their seed region is extremely important in target recognition, this complementarity can trigger expression regulation, [31,32]. Even in humans, the seed region of some miRNA can define toxicity or tumor-suppressive for some cancer cell lines [33]. This is due to seed region target recognition mechanisms [34,35], show that an initial search for target sites transitions from being transient to kinetically stable, when target complementarity binds to the full length of the seed region of miRNA [36]. Furthermore, several authors have identified and demonstrated regulation of target gene expression, even when miRNAs have not bound in a fully complementary manner, showing evidence of their function [16,37,38].
In this study we included a homology analysis, identifying 7 wheat-derived miRNAs show homology with 9 human miRNAs, revealing a series of target genes in common, through our prediction. Although plant sequences do not show high homology with human sequences, results obtained by author [39], show that they can perform similar functional activities to endogenous sequences, in a cross-kingdom interaction [19]. Therefore, we consider this dataset provides a solid basis for future projects.
In our mRNA and miRNA interaction analysis we aimed to identify the genes that are the target of multiple miRNAs and their possible health effect through KEGG pathways. We found that the target genes are associated with the calcium signaling pathway, showing participation of the CD38 gene, associated with diseases such as asthma, neoplasia and neuroimmune diseases [40], as well as the ADCY1 gene, involved in insulin secretion, related to diabetes mellitus and its correlation with pancreatic cancer [41].
Among the pathways, three stand out, the pathway of fanconi anemia (hsa03460), circadian entrainment (hsa04713), and dopaminergic synapse (hsa04728), which we have found a relation with a couple of publications [42,43](Gluten-Free Diet for the Treatment of ADHD; Pilot Study) where in a case study they describe the difficulty of finding a diagnosis to a series of neuropsychiatric manifestations, described as “non-celiac gluten sensitivity” (NCGS), where a series of symptoms related to the consumption of gluten-containing foods (wheat) are characterized in people not affected by celiac disease (CD) or wheat allergy (AW).). Attention deficit hyperactivity disorder (ADHD) related to the circadian entrainment pathway [44], and schizophrenia related to the dopaminergic synaptic pathway, are some of the main symptoms related to wheat consumption in case studies, in addition, including a low-ferritin anemia in the clinical picture. Despite attempts to consolidate a possible explanation, this has not been achieved due to lack of information on response mechanisms or genetic components of the disease and therefore the number of people who may be affected is uncertain. In the enrichment results, we have 5 genes, GNB5, GNAI2, PRKACB, CALM1, and CAMK2D as possible targets in common for miRNA regulation of wheat involved in the dopaminergic synapse pathway and circadian entrainment.
Since the report of miRNA plants, regulating gene expression by miR168a in mice and humans [15], the identification of new targets by miRNA is of major importance, due to its high efficiency, low cost, and time saving [[16], [38], [45]]. Since this could be a way to explain negative events in human health, related to some diseases that could risk public health, such as cancer, schizophrenia, and metabolic diseases.
5. Conclusion
Considering that wheat and humans are different species, it is difficult to determine the possible alterations given by exogenous molecules caused by cross-kingdom interactions, however, it is necessary to highlight the importance of the use of bioinformatics tools, for the prediction of possible targets, because this study demonstrates the possible effects of wheat miRNAs in regulating complex disease networks like a schizophrenia, epilepsy, tumor suppressor, inflammation, and diverse type of cancer. Due to the fact that the information collected could facilitate the search and verification, that could be of importance for future projects. In addition, these findings can generate new lines of research in various areas, such as nutrition, medicine, biotechnology, agriculture, etc. It can also show possible miRNAs candidates that can further contribute in therapeutic treatment of various human diseases.
CRediT authorship contribution statement
Daniel Sánchez-Romo: Writing – original draft, Methodology, Investigation. César I. Hernández-Vásquez: Formal analysis. Benito Pereyra-Alférez: Conceptualization, Resources. Jorge H. García-García: Project administration, Writing – review & editing.
Declaration of competing interest
None.
Acknowledgments
Non applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2022.03.002.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Friedman R.C., Farh K.K.-H., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graves P., Zeng Y. Biogenesis of mammalian MicroRNAs: a global view. Genomics, Proteomics Bioinforma. 2012;10:239–245. doi: 10.1016/j.gpb.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee I., Ajay S.S., Jong I.Y., Hyun S.K., Su H.H., Nam H.K., Dhanasekaran S.M., Chinnaiyan A.M., Athey B.D. New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res. 2009;19:1175–1183. doi: 10.1101/gr.089367.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perge P., Nagy Z., Decmann Á., Igaz I., Igaz P. Potential relevance of microRNAs in inter-species epigenetic communication, and implications for disease pathogenesis. RNA Biol. 2017;14:391–401. doi: 10.1080/15476286.2016.1251001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y., Cong L., Lukiw W.J. Plant and animal microRNAs (miRNAs) and their potential for inter-kingdom communication. Cell. Mol. Neurobiol. 2018;38:133–140. doi: 10.1007/s10571-017-0547-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang H., Zhang S., Fu Z., Wang Y., Wang N., Liu Y., Zhao C., Wu J., Hu Y., Zhang J., Chen X., Zen K., Zhang C.Y. Effective detection and quantification of dietetically absorbed plant microRNAs in human plasma. J. Nutr. Biochem. 2015;26:505–512. doi: 10.1016/j.jnutbio.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Stephen B.J., Pareek N., Saeed M., Kausar M.A., Rahman S., Datta M. Xeno-miRNA in maternal-infant immune crosstalk: an aid to disease alleviation. Front. Immunol. 2020;11:1–8. doi: 10.3389/fimmu.2020.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rakhmetullina A., Pyrkova A., Aisina D., Ivashchenko A. In silico prediction of human genes as potential targets for rice miRNAs. Comput. Biol. Chem. 2020;87 doi: 10.1016/j.compbiolchem.2020.107305. [DOI] [PubMed] [Google Scholar]
- 10.Pirim D., Dogan B. In silico identification of putative roles of food-derived xeno-mirs on diet-associated cancer. Nutr. Cancer. 2020;72:481–488. doi: 10.1080/01635581.2019.1670854. [DOI] [PubMed] [Google Scholar]
- 11.Patel M., Mangukia N., Jha N., Gadhavi H., Shah K., Patel S., Mankad A., Pandya H., Rawal R. Computational identification of miRNA and their cross kingdom targets from expressed sequence tags of Ocimum basilicum. Mol. Biol. Rep. 2019;46:2979–2995. doi: 10.1007/s11033-019-04759-x. [DOI] [PubMed] [Google Scholar]
- 12.He J., Chen T., Xi Q., Sun J., Luo J., Li M., Zhang H., Zeng B., Wu J., Zhang Y. Identification of microRNA in Houttuynia cordata Thunb and prediction of cross kingdom functions. ExRNA. 2019;1 doi: 10.1186/s41544-019-0028-7. [DOI] [Google Scholar]
- 13.Kumar D., Kumar S., Ayachit G., Bhairappanavar S.B., Ansari A., Sharma P., Soni S., Das J. Cross-kingdom regulation of putative miRNAs derived from happy tree in cancer pathway: a systems biology approach. Int. J. Mol. Sci. 2017;18:1–21. doi: 10.3390/ijms18061191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gadhavi H., Patel M., Mangukia N., Shah K., Bhadresha K., Patel S.K., Rawal R.M., Pandya H.A. Transcriptome-wide miRNA identification of Bacopa monnieri: a cross-kingdom approach. Plant Signal. Behav. 2020;15 doi: 10.1080/15592324.2019.1699265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Hou D., Chen X., Li D., Zhu L., Zhang Y., Li J., Bian Z., Liang X., Cai X., Yin Y., Wang C., Zhang T., Zhu D., Zhang D., Xu J. 2012. Exogenous Plant MIR168a Specifically Targets Mammalian LDLRAP1 : Evidence of Cross-Kingdom Regulation by microRNA; pp. 107–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Z., Li X., Liu J., Dong L., Chen Q., Liu J., Kong H., Zhang Q., Qi X., Hou D., Zhang L., Zhang G., Liu Y., Zhang Y., Li J., Wang J., Chen X., Wang H., Zhang J., Chen H., Zen K., Zhang C.Y. Honeysuckle-encoded atypical microRNA2911 directly targets influenza A viruses. Cell Res. 2015;25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giegerich R., Rehmsmeier M., Steffen P., Ho M. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604.and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirrò S., Minutolo A., Galgani A., Potestà M., Colizzi V., Montesano C. Bioinformatics prediction and experimental validation of MicroRNAs involved in cross-kingdom interaction. J. Comput. Biol. 2016;23:976–989. doi: 10.1089/CMB.2016.0059. [DOI] [PubMed] [Google Scholar]
- 19.Minutolo A., Potestà M., Gismondi A., Pirrò S., Cirilli M., Gattabria F., Galgani A., Sessa L., Mattei M., Canini A., Muleo R., Colizzi V., Montesano C. Olea europaea small RNA with functional homology to human miR34a in cross-kingdom interaction of anti-tumoral response. Sci. Rep. 2018;8:1–14. doi: 10.1038/s41598-018-30718-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G., Wang L.G., Han Y., He Q.Y. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R core Team R. R Foundation for Statistical Computing; Vienna.: 2018. A Language and Environment for Statistical Computing.https://www.r-project.org/ [Google Scholar]
- 22.Han R., Jian C., Lv J., Yan Y., Chi Q., Li Z., Wang Q., Zhang J., Liu X., Zhao H. Identification and characterization of microRNAs in the flag leaf and developing seed of wheat (Triticum aestivum L.) BMC Genom. 2014;15:289. doi: 10.1186/1471-2164-15-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B., Li J., Cairns M.J. Identifying miRNAs, targets and functions. Briefings Bioinf. 2014;15:1–19. doi: 10.1093/bib/bbs075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Min H., Yoon S. Got target?: computational methods for microRNA target prediction and their extension. Exp. Mol. Med. 2010;42:233–244. doi: 10.3858/emm.2010.42.4.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C., Rennie W.A., Carmack C.S., Kanoria S., Cheng J., Lu J., Ding Y. Effects of genetic variations on microRNA: target interactions. Nucleic Acids Res. 2014;42:9543–9552. doi: 10.1093/nar/gku675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Li Y., Liu Y., Liu H., Wang H., Jin W., Zhang Y., Zhang C., Xu D. Role of plant MicroRNA in cross-species regulatory networks of humans. BMC Syst. Biol. 2016;10:1–10. doi: 10.1186/s12918-016-0292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Y., Wang P., Wang X., Wang Y., Mu Z., Li Q., Fu Y., Xiao J., Li G., Ma Y., Gu Y., Jin L., Ma J., Tang Q., Jiang A., Li X., Li M. Detection of dietetically absorbed maize-derived microRNAs in pigs. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-00488-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helwak A., Kudla G., Dudnakova T., Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu W., Xu Y., Xie X., Wang T., Ko J.H., Zhou T. The role of RNA structure at 5′ untranslated region in microRNA-mediated gene regulation. RNA. 2014;20:1369–1375. doi: 10.1261/rna.044792.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam J.K.W., Chow M.Y.T., Zhang Y., Leung S.W.S. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samad A.F.A., Kamaroddin M.F., Sajad M. Cross-kingdom regulation by plant microRNAs provides novel insight into gene regulation. Adv. Nutr. 2020:1–15. doi: 10.1093/advances/nmaa095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huntzinger E., Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 33.Gao Q.Q., Putzbach W.E., Murmann A.E., Chen S., Sarshad A.A., Peter J.M., Bartom E.T., Hafner M., Peter M.E. 6mer seed toxicity in tumor suppressive microRNAs. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-06526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klum S.M., Chandradoss S.D., Schirle N.T., Joo C., Macrae I.J. Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. EMBO J. 2018;37:75–88. doi: 10.15252/embj.201796474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein M., Chandradoss S.D., Depken M., Joo C. Why Argonaute is needed to make microRNA target search fast and reliable. Semin. Cell Dev. Biol. 2017;65:20–28. doi: 10.1016/j.semcdb.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 36.Chandradoss S.D., Schirle N.T., Szczepaniak M., Macrae I.J., Joo C. A dynamic search process underlies MicroRNA targeting. Cell. 2015;162:96–107. doi: 10.1016/j.cell.2015.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M., Chen T., He J.J., Wu J.H., Luo J.Y., Ye R.S., Xie M.Y., Zhang H.J., Zeng B., Liu J., Xi Q.Y., Jiang Q.Y., Sun J.J., Zhang Y.L. Plant MIR167e-5p inhibits enterocyte proliferation by targeting β-catenin. Cells. 2019;8:1–14. doi: 10.3390/cells8111385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chin A.R., Fong M.Y., Somlo G., Wu J., Swiderski P., Wu X., Wang S.E. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26:217–228. doi: 10.1038/cr.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Avsar B., Zhao Y., Li W., Lukiw W.J. Atropa belladonna expresses a microRNA (aba-miRNA-9497) highly homologous to Homo sapiens miRNA-378 (hsa-miRNA-378); both miRNAs target the 3′-untranslated region (3′-UTR) of the mRNA encoding the neurologically relevant, zinc-finger transcription factor. Cell. Mol. Neurobiol. 2020;40:179–188. doi: 10.1007/s10571-019-00729-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deshpande D.A., Guedes A.G.P., Graeff R., Dogan S., Subramanian S., Walseth T.F., Kannan M.S. Mediators Inflamm; 2018. CD38/cADPR Signaling Pathway in Airway Disease: Regulatory Mechanisms. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pang W., Yao W., Dai X., Zhang A., Hou L., Wang L., Wang Y., Huang X., Meng X., Li L. Pancreatic cancer-derived exosomal microrna-19a induces β-cell dysfunction by targeting adcy1 and epac2. Int. J. Biol. Sci. 2021;17:3622–3633. doi: 10.7150/ijbs.56271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Catassi C., Bai J.C., Bonaz B., Bouma G., Calabrò A., Carroccio A., Castillejo G., Ciacci C., Cristofori F., Dolinsek J., Francavilla R., Elli L., Green P., Holtmeier W., Koehler P., Koletzko S., Meinhold C., Sanders D., Schumann M., Schuppan D., Ullrich R., Vécsei A., Volta U., Zevallos V., Sapone A., Fasano A. Non-celiac gluten sensitivity: the new frontier of gluten related disorders. Nutrients. 2013;5:3839–3853. doi: 10.3390/nu5103839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lionetti E., Leonardi S., Franzonello C., Mancardi M., Ruggieri M., Catassi C. Gluten psychosis: confirmation of a new clinical entity. Nutrients. 2015;7:5532–5539. doi: 10.3390/nu7075235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palm D., Uzoni A., Simon F., Fischer M., Coogan A., Tucha O., Thome J., Faltraco F. Evolutionary conservations, changes of circadian rhythms and their effect on circadian disturbances and therapeutic approaches. Neurosci. Biobehav. Rev. 2021;128:21–34. doi: 10.1016/j.neubiorev.2021.06.007. [DOI] [PubMed] [Google Scholar]
- 45.Wang W., Liu D., Zhang X., Chen D., Cheng Y., Shen F. Plant microRNAs in cross-kingdom regulation of gene expression. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19072007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.