Abstract
Objective
Recent studies reported that SLE is characterised by altered interactions between the microbiome and immune system. We performed a meta-analysis of publications on this topic.
Methods
Case–control studies that compared patients with SLE and healthy controls (HCs) and determined the diversity of the gut microbiota and the abundance of different microbes were examined. Stata/MP V.16 was used for the meta-analysis. A Bonferroni correction for multiple tests was used to reduce the likelihood of false-positive results.
Results
We included 11 case–control studies that examined 373 patients with SLE and 1288 HCs. These studies were performed in five countries and nine cities. Compared with HCs, patients with SLE had gut microbiota with lower Shannon-Wiener diversity index (weighted mean difference=−0.22, 95% CI −0.32 to –0.13, p<0.001) and lower Chao1 richness (standardised mean difference (SMD)=−0.62, 95% CI −1.04 to –0.21, p=0.003). Patients with SLE had lower abundance of Ruminococcaceae (SMD = −0.49, 95% CI −0.84 to −0.15, p=0.005), but greater abundance of Enterobacteriaceae (SMD=0.45, 95% CI 0.01 to 0.89, p=0.045) and Enterococcaceae (SMD=0.53, 95% CI 0.05 to 1.01, p=0.03). However, only the results for Ruminococcaceae passed the Bonferroni correction (p=0.0071). The two groups had no significant differences in Lachnospiraceae and Bacteroides (both p>0.05). Patients with SLE who used high doses of glucocorticoids had altered gut microbiota based on the Chao1 species diversity estimator, and hydroxychloroquine use appeared to reduce the abundance of Enterobacteriaceae.
Conclusions
Patients with SLE have imbalanced gut microbiota, with a decrease in beneficial bacteria and an increase in harmful bacteria. Drugs used to treat SLE may also alter the gut microbiota of these patients.
Keywords: systemic lupus erythematosus; autoimmune diseases; autoimmunity; outcome assessment, health care
Key messages.
What is already known about this subject?
SLE is characterised by altered interactions between the microbiome and immune system.
What does this study add?
Ruminococcaceae, Enterococcaceae and Enterobacteriaceae were imbalanced in patients with SLE.
The dosage of glucocorticoids and use of hydroxychloroquine alter the abundance of some gut microbes.
How might this impact on clinical practice or future developments?
This study proposes some factors that may cause discrepant results in the gut microbiota in SLE and provides references for future research.
Introduction
SLE is a chronic and systemic autoimmune disease that has complex clinical manifestations and is characterised by excessive activation of B cells and T cells, production of autoantibodies, and injury of multiple organs and tissues.1 Although there have been improvements in diagnosis and treatment strategies, treatment efficacy remains inadequate due to limited understanding of the aetiology and pathogenesis of SLE. The morbidity and mortality of SLE therefore continue to increase.2 3
Recent studies found that the gut microbiota has a role in the development and regulation of the immune system.4 5 In particular, disruption of the gut microbiota may affect autoimmunity by altering the balance between tolerogenic and inflammatory microbes.6 Therefore, studies on alterations in the gut microbiota of patients with SLE may provide a new avenue for examining the pathogenesis of SLE.
Researchers initially discovered the presence of dysbiosis in the gut microbiota of patients with SLE, in that patients with SLE had a lower ratio of Firmicutes to Bacteroidetes than healthy controls (HCs).7 8 This led to speculation on the underlying connection between SLE and intestinal microbes. One hypothesis is that a disrupted gut barrier leads to a ‘leaky gut’, which allows bacteria to transfer to other tissues, thus activating immune factors and causing systemic autoimmunity.9 The decrease in gut microbial diversity and diminution of Lactobacillaceae may function in this response.10 Another hypothesis is that stimulation of specific molecular pathways by the gut microbiota induces SLE. In support of this later hypothesis, there is evidence that orthologues of the autoantigen Ro60 in the gut microbiota cross-react with human B cells and T cells, thus stimulating autoimmune diseases in a mechanism involving Ruminococcus gnavus.11 Thus, changes in the gut microbiota, the immune system and SLE appear to be related.
Although many studies have examined the association between SLE and gut microbiota, some of the results are still controversial, such as the reported decrease in alpha diversity and changes in the abundance of some gut microbes in patients with SLE. For example, Bellocchi et al12 and WF et al13 reported different results regarding alterations in the Shannon-Wiener diversity index between patients with SLE and HCs. A change in the abundance of Enterobacteriaceae in patients with SLE is also uncertain.12 14 Because the composition of the gut microbiota is dynamic and is affected by genes, diet, geographical region, use of immunosuppressants and other factors,15–20 there could be many explanations for the
differences in these experimental results. In addition, differences in the design of experiments and methods used to study the gut microbiota can also impact the results.21
Thus, we examined the changes in the gut microbiota of patients with SLE and the factors that may affect these changes by performing a meta-analysis of relevant research publications from around the world in an effort to provide an evidence-based approach for follow-up research.
Methods
This meta-analysis was based on the procedures of the Meta-analysis of Observational Studies in Epidemiology group, which provides meta-guidance for observational studies.22 The present study was registered in PROSPERO (International Prospective Record of Systematic Reviews) on 17 May 2021 (CRD42021249607).
Literature search
Two researchers (SX and YQ) conducted the literature search using the following English and Chinese databases: PubMed, Embase, Cochrane, Web of Science, Wanfang Database and Chinese National Knowledge Infrastructure (http://www.cnki.net). Combinations of medical subject headings and synonyms were used to search for studies (online supplemental file 1). The literature search was finalised on 1 March 2021.
lupus-2021-000599supp001.pdf (79.6KB, pdf)
Study selection criteria
All included studies performed research at the population level; examined patients in a case group who were adults diagnosed with SLE based on the American College of Rheumatology (ACR) criteria; compared changes in the gut microbial diversity, or the relative abundance of microbes, in patients with SLE and HCs; examined faecal samples; and had data available for analysis. Studies were excluded if they were animal experiments, provided irrelevant research content, were review articles or failed to provide complete experimental data.
Data extraction
The following characteristics were recorded from the selected studies: (1) first author, year of publication, and country and region where the study was performed; (2) characteristics of the subjects (sex, average age, population, current use of medications and doses); (3) experimental methods (diagnostic criteria, sample size, techniques for assessment of the gut microbiota); (4) alpha diversity of the gut microbiota in patients and HCs; (5) abundance of different gut microbial species, which was expressed as the mean proportion of each microbiota; and (6) results reported as mean and respective SD.
In addition, since beta diversity of the gut microbiota in patients and HCs was depicted with a principal coordinates analysis plot, we were unable to extract data for our study. Therefore, this study did not perform a meta-analysis of beta diversity.
Data extraction and transformation
The original data from each publication were extracted for the meta-analysis and the results are expressed as tables and graphs. Because some publications provided relevant data indirectly, GetData Graph Digitizer V.2.25 (http://getdata-graph-digitizer.com) was used to extract data from plots23–25 using the methods described by Wan et al.26
Quality assessment
Two researchers (SX and YQ) independently extracted relevant data from the included studies. Disagreements were resolved through discussion and consultation with a third researcher (SQ). The nine-star Newcastle-Ottawa Quality Assessment Scale (NOS) for case–control studies was used to assess the quality of all studies.27 This scale considers adequate definition of cases, representativeness of cases, selection of controls, definition of controls, ascertainment of exposure, same method of ascertainment for cases and controls, and non-response rate. The highest possible score was 9. Studies with scores of 6–9 points (high-quality publications) were included in the meta-analysis.
Statistical analysis
The collected or converted data (including the Shannon-Wiener diversity index, Chao1 richness estimator and abundance of specific gut microbes) were collated. When four or more studies examined the same outcome, data were analysed using Stata V.16.0 software and the results were displayed as forest plots. When the data included were continuous variables and the measurement methods were the same, weighted mean differences (WMDs) were used as the effect scale. When there was a large difference in mean or SD among the included studies, standardised mean differences (SMDs) were used as the effect scale.28
Statistical heterogeneity was evaluated using the χ2-based Q statistic test. I2 was used as an index to evaluate heterogeneity and was defined as moderate (25%), large (50%) or extreme (75%).29 Direct meta-analyses were performed using a sample-size weighted random effects model.30 An I2 greater than 25% was considered to indicate statistically significant heterogeneity. A p value below 0.05 was considered significant.31 However, since our study has been tested seven times, a Bonferroni correction was considered. A p value <0.0071 meets the Bonferroni threshold of statistical significance (0.0071=0.05/7). Results with p value >0.0071 were not considered statistically significant.32
Subgroup analyses were performed to determine the effect of population and use of medications. Sensitivity analysis was performed to evaluate the effect of individual studies on the overall results by deleting one study at a time and combining the effect values of the remaining studies.
Due to the limited number of studies ultimately included, standard bias assessment results may not be sufficiently reliable. Nonetheless, publication bias was evaluated by Egger’s test,33 Begg’s test34 and a funnel plot. The Egger’s and Begg’s tests indicated no publication bias when the p value was greater than 0.05, and otherwise indicated publication bias. When the results of Egger’s and Begg’s tests were inconsistent, the results of the Egger’s test were used as the reference value.
Results
Study selection and characteristics
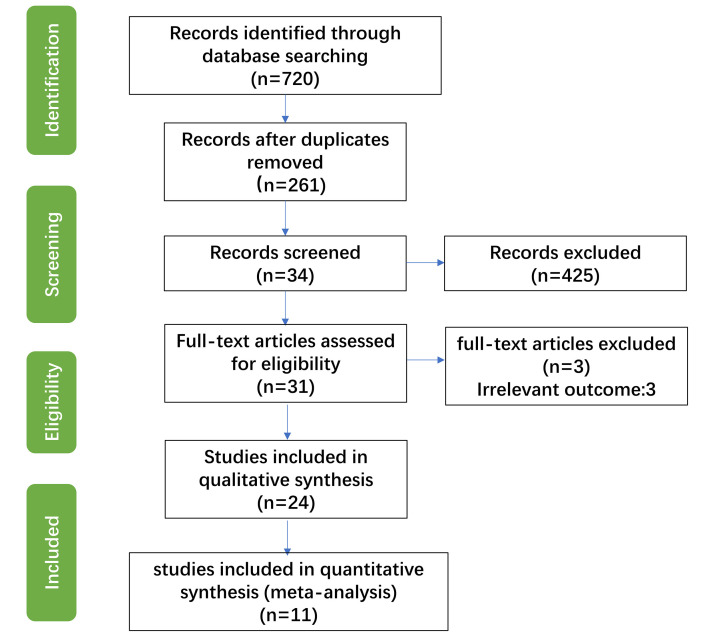
We initially retrieved 720 potentially eligible studies (figure 1). We then deleted duplicate publications using EndNote, and also deleted publications that were unrelated to the research topic, publications that examined animals and review articles. After reading the full text of the remaining publications, we deleted publications that provided incomplete experimental results, unclear experimental procedures or examined faecal samples that were not from humans. The remaining 31 studies reported Shannon-Wiener diversity indexes, Chao1 richness estimators and abundance of some gut microbes. Twenty-four studies remained after deleting eight studies where the data could not be collected, or where the gut microbiota were described as log10 bacteria per gram of faeces or as a linear discriminant analysis score. After quality evaluation, we ultimately included 11 studies in the meta-analysis, 9 English-language studies and 2 Chinese-language studies (table 1).
Figure 1.
Flow chart of selection and inclusion of studies in the meta-analysis.
Table 1.
Characteristics of included studies
| Study | Location | SLE case/HC case | SLE case | Experimental methods | |||
| Country/race | n (female sex, %) | Age, mean±SD | Medication/dosage | Technique | Compositional data analysis methods | Reference panels | |
| Bellocchi et al12 | Italy/not Chinese | 27 (88.9)/27 (74.1) | 47.7±16.6/52.5±10.0 | Prednisone (<10 mg/day)/immunosuppressant/HCQ/statin | 16S rRNA sequencing | OTU | SILVA |
| van der Meulen et al8 | The Netherlands/white/European ethnic background | 35 (94.3)/965 (58.0) | 47.0±14.0/45.0±13.0 | Proton pump inhibitors/NSAIDs/corticosteroids (<7.5 mg/day)/antimalarial | 16S rRNA sequencing | OTU | SILVA |
| Luo et al66 | America/African American (not Caribbean) and Caucasian, non-Hispanic | 14 (71.4)/17 (NA) | 43.2±18.1/NA | HCQ/belimumab/MMF/MTX/rituximab/AZA/tacrolimus | 16S rRNA sequencing | OTU | Greengenes |
| Hevia et al 7 |
Spain/Caucasian origin | 20 (100)/20 (100) | 49.2±10.7/46.9±8.6 | NA | 16S rRNA sequencing | OTU | Greengenes |
| Wei et al14 | China/Chinese | 14 (92.9)/16 (87.5) | 40.7±13.9/38.6±14.5 | NA | 16S rRNA sequencing | OTU | Greengenes |
| He et al63 | China/Chinese | 45 (100)/48 (100) | 46.0±1.8/43.5±2.4 | NA | 16S rRNA sequencing | OTU | SILVA |
| Guo et al55 | China/Chinese | 17 (100)/20 (100) | 34.4±3.4/30.4±1.9 | NA | 16S rRNA sequencing | Feature | Greengenes |
| Li et al67 | China/Chinese | 40 (100)/22 (100) | 37.5±14.2/37.2±14.7 | NA | 16S rRNA sequencing | OTU | RDP |
| Zhu13 | China/Chinese | 32 (93.8)/26 (87.5) | 33.8±14.2/NA | Immunosuppressant/hormone drugs (no record) | 16S rRNA sequencing | OTU | RDP |
| Chen et al52 | China/Beijing | 117 (72.8)/115 (84.3) | 30.8±10.9/32.4±11.3 | NA | Shotgun sequencing | Gene mapping | Immune Epitope Database |
| Sun et al68 | China/Heilongjiang | 12 (NA)/12 (NA) | NA | NA | 16S rRNA sequencing | OTU | RDP |
AZA, Azathioprine; Greengenes, The Greengenes Database; HC, healthy control; HCQ, hydroxychloroquine; HCQ, Hydroxychloroquine; MMF, Mycophenolate Mofetil; MTX, Methotrexate; NA, Not using any medication; NSAIDs, Nonsteroidal Antiinflammatory Drugs; OTU, operational taxonomic unit; OTU, Operational Taxonomic Units; RDp, RibosomalDatabase Project; SILVA, SILVA rRNA database.
These 11 studies examined 373 patients with SLE and 1288 HCs. All 11 studies used human faeces as experimental samples, used diagnostic criteria for SLE from the ACR and were observational case–control studies. According to the NOS, each of the included studies had high quality (NOS score: 6–8; table 2).
Table 2.
Score of studies included in the meta-analysis based on NOS
| Reference | Selection | Comparability | Exposure | Total score | |||||
| Adequate definition of cases | Representativeness of the cases | Selection of controls | Definition of controls | Comparability of cases and controls on the basis of the design or analysis | Ascertainment of exposure | Same method of ascertainment for cases and controls | Non-response rate | ||
| Bellocchi et al12 | ☆ | – | ☆ | ☆ | ☆☆ | – | ☆ | – | 6 |
| van der et al8 | ☆ | ☆ | ☆ | – | ☆☆ | ☆ | ☆ | – | 7 |
| Luo et al66 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | – | 8 |
| Hevia et al7 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆☆ | – | – | 8 |
| Wei et al14 | ☆ | ☆ | ☆ | ☆ | ☆☆ | – | ☆ | – | 7 |
| He et al63 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | 8 | |
| Guo et al55 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | – | 8 |
| Li et al67 | ☆ | ☆ | ☆ | – | ☆☆ | ☆ | ☆ | – | 7 |
| Zhu13 | ☆ | – | ☆ | ☆ | ☆☆ | ☆ | ☆ | – | 7 |
| Chen et al52 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | – | 8 |
| Sun et al68 | ☆ | ☆ | ☆ | ☆ | ☆☆ | ☆ | ☆ | – | 8 |
☆: representative studies meet one criteria ☆☆: representative studies meet two criteria.
NOS, Newcastle-Ottawa Quality Assessment Scale.
Changes in alpha diversity of gut microbes in patients with SLE
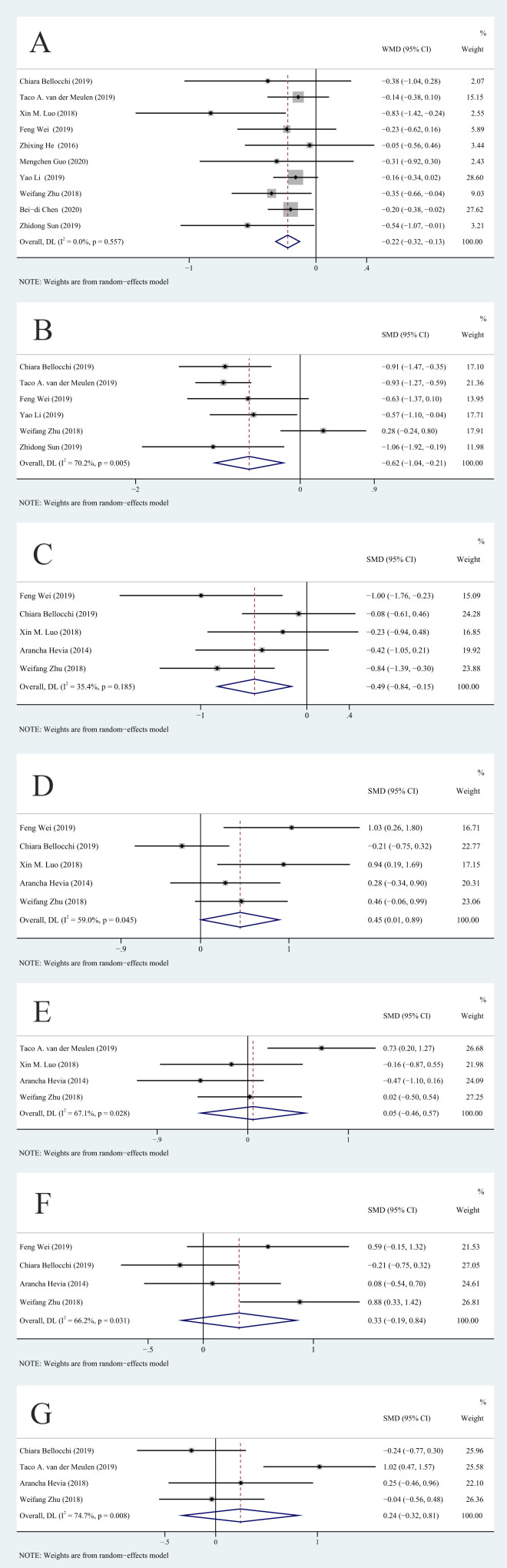
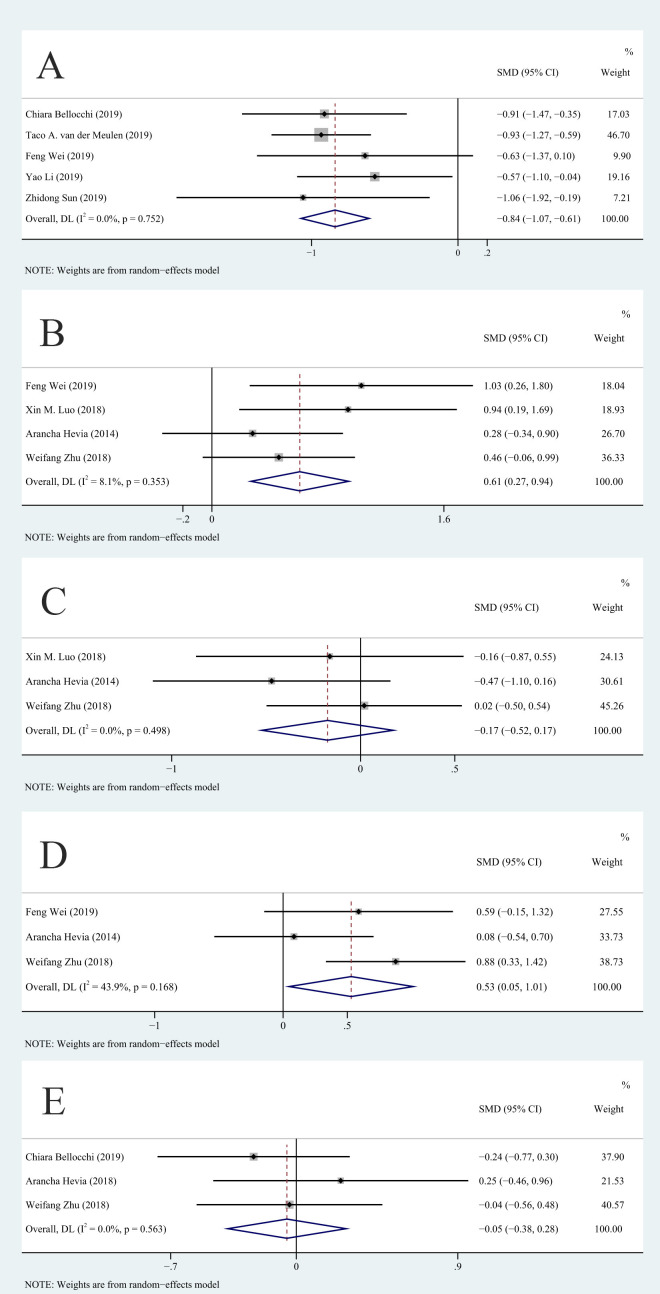
We compared the Shannon-Wiener diversity index and the Chao1 richness estimator for the SLE and HC groups to determine the relationship of SLE with alpha diversity and overall richness of the gut microbiota. Because there was a difference of more than 10 times in the average values of other indicators except for the Shannon-Wiener diversity index, we used SMD as the effect scale. Analysis of the Shannon-Wiener diversity index showed that the studies had no significant heterogeneity (I2=0%, p>0.01) and that the SLE group had a lower diversity of gut microbiota than the HC group (WMD=−0.22, 95% CI −0.32 to –0.13, p<0.001, n=10; figure 2A). There was significant heterogeneity among the studies in the Chao1 richness estimator (I2=70.2%, p<0.01). After sensitivity analysis, we removed the study of Zhu,13 and this eliminated the heterogeneity in Chao1 richness estimator (figure 3A). Analysis of the remaining studies indicated lower richness of the gut microbiota in the SLE group than in the HC group (SMD=−0.84, 95% CI −1.07 to –0.61, p<0.001, n=5; figure 3A).
Figure 2.
Forest plots of alterations in the gut microbiota of patients with SLE versus healthy controls: (A) Shannon-Wiener diversity index, (B) Chao1 richness estimator, (C) Ruminococcaceae, (D) Enterobacteriaceae, (E) Lachnospiraceae, (F) Enterococcaceae and (G) Bacteroides. SMD, standardised mean difference; WMD, weighted mean difference. DL: DerSimonian–Laird.
Figure 3.
Eliminated sources of heterogeneity by sensitivity analysis: (A) Chao1 richness estimator, (B) Enterobacteriaceae, (C) Lachnospiraceae, (D) Enterococcaceae and (E) Bacteroides. SMD, standardised mean difference. DL: DerSimonian–Laird.
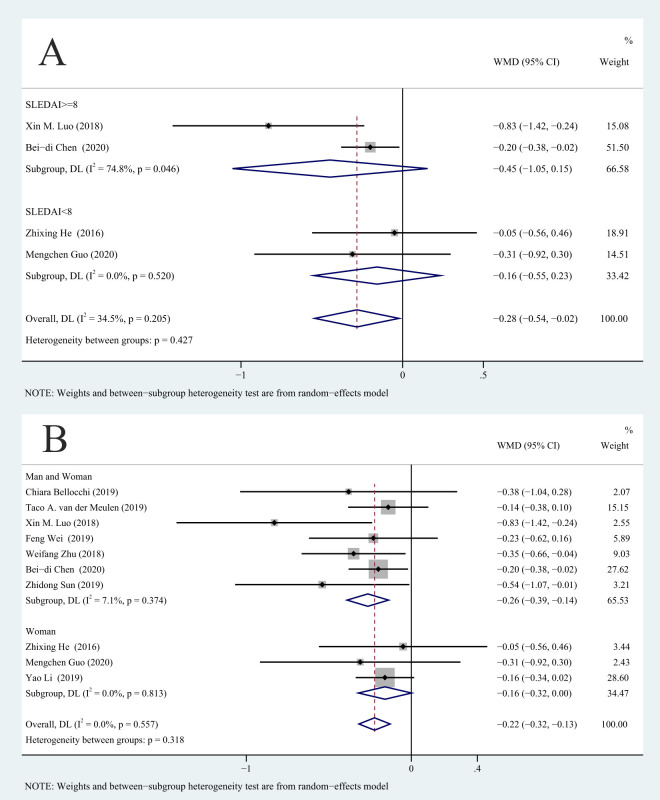
We used the Systemic Lupus Erythematosus Disease Activity Index (SLEDAI)35 to group patients into an active SLE group (SLEDAI ≥8) and an SLE remission group (SLEDAI <8). Subgroup analysis of these two groups indicated no significant difference in the Shannon-Wiener diversity index in the SLE remission and SLE active groups (p>0.05; figure 4A).
Figure 4.
Correlation between SLEDAI, gender and Shannon-Wiener diversity index: (A) SLEDAI and (B) gender. DL: DerSimonian–Laird; SLEDAI, Systemic Lupus Erythematosus Disease Activity Index; WMD, weighted mean difference.
Because the prevalence of SLE is much higher in women than in men, we conducted a subgroup analysis to examine the effect of gender. The results showed that the decrease in the Shannon-Wiener diversity index of male and female patients with SLE together was greater than HCs (WMD=−0.26, 95% −0.39 to –0.14, p<0.001, n=7). However, there was no significant difference between female patients with SLE and HCs (WMD=−0.16, 95% −0.32 to 0.00, p=0.052, n=3; figure 4B).
Changes in specific gut microbes in SLE
We then analysed the different taxa in the gut microbiota of patients with SLE and HCs. The results indicated the SLE group had a decreased abundance of Ruminococcaceae (, (SMD = −0.49, 95% CI = −0.84 to –0.15, p=0.005, n=5; figure 2C) and an increased abundance of Enterobacteriaceae (SMD=0.45, 95% CI 0.01 to 0.89, p=0.045, n=5; figure 2D). However, the results for Enterobacteriaceae were not statistically significant according to the Bonferroni statistical significance threshold (p>0.0071). In addition, the SLE and HC groups had no significant differences in Lachnospiraceae, Enterococcaceae or Bacteroides (all p>0.05).
Due to the significant heterogeneity among the studies, we used sensitivity analysis to evaluate the source of this heterogeneity (online supplemental file 2). After removing the van der Meulen et al study8 and the Bellocchi et al study,12 the heterogeneity in Chao1 richness estimator, Enterobacteriaceae (figure 3B), Lachnospiraceae (figure 3C), Enterococcaceae (figure 3D) and Bacteroides (figure 3E) decreased or disappeared, but the results for Lachnospiraceae (p=0.327) and Bacteroides (p=0.756) were unaffected. However, after sensitivity analysis, the p value for Enterococcaceae indicated a significant difference (SMD=0.53, 95% CI 0.05 to 1.01, I2=43.9%, p=0.03, n=3; figure 3D). In other words, the abundance of Enterococcaceae in the SLE group was greater than in the HC group, but the results did not strictly meet the multiple comparisons criterion (p>0.0071). Removing some of the included studies did not reduce the statistical heterogeneity in Ruminococcaceae. In addition, after sensitivity analysis, the heterogeneity in Enterococcaceae was still higher than 25%.
lupus-2021-000599supp002.pdf (631.7KB, pdf)
We also performed a subgroup analysis with stratification by population and use of medications. The abundance of Ruminococcaceae in patients with SLE was lower than in HCs (SMD=−0.90, 95% CI −1.34 to –0.45, p<0.001; table 3), but this difference was not significant for the non-Chinese subgroup (SMD=−0.22, 95% CI=−0.58 to 0.13, p=0.216; table 3). In the Chinese subgroup, the abundance of Enterococcaceae in patients with SLE was higher than in HCs (SMD=0.77, 95% CI 0.34 to 1.21, p=0.001; table 4), but there were no differences between the non-Chinese subgroups (SMD=−0.08, 95% CI −0.49 to 0.32, p=0.682; table 4). The use of medications had inconsistent effects in these different taxonomic groups.
Table 3.
Results of subgroup analyses of Ruminococcaceae
| Studies (n) | SMD | P value (%) | I2 (%) | P value for heterogeneity | 95% CI | |
| Population | ||||||
| Chinese | 2 | −0.895 | 0.000 | 0.0 | 0.752 | −1.337 to −0.452 |
| Non-Chinese | 2 | −0.223 | 0.216 | 0.0 | 0.714 | −0.576 to 0.130 |
| Take medication or not | ||||||
| Take medication | 3 | −0.394 | 0.019 | 52.6 | 0.121 | −0.889 to 0.100 |
| Do not take medication | 2 | −0.665 | 0.118 | 22.4 | 0.256 | −1.220 to −0.109 |
SMD, standardised mean difference.
Table 4.
Results of subgroup analyses of Enterococcaceae
| Studies (n) | SMD | P value (%) | I2 (%) | P value for heterogeneity | 95% CI | |
| Population | ||||||
| Chinese | 2 | 0.775 | 0.001 | 0.0 | 0.536 | 0.338 to 1.212 |
| Non-Chinese | 3 | −0.085 | 0.682 | 0.0 | 0.480 | −0.490 to 0.320 |
| Take medication or not | ||||||
| Take medication | 2 | 0.326 | 0.541 | 87.2 | 0.005 | −0.734 to 1.398 |
| Do not take medication | 2 | 0.296 | 0.234 | 5.1 | 0.305 | −0.191 to 0.783 |
SMD, standardised mean difference.
Analysis of publication bias
We assessed the risk of publication bias using Egger’s and Begg’s tests and based on the symmetry of a funnel chart (online supplemental file 2, figure S1). The results indicated no evidence of publication bias, indicating the conclusions of the meta-analysis were relatively robust.
Discussion
Our meta-analysis compared the gut microbiota of patients with SLE and HCs and led to three major conclusions. First, patients with SLE had a lower diversity of gut microbiota, and the Shannon-Wiener diversity index was slightly lower in patients with active SLE than in those in remission. Second, analysis of the different taxonomic groups of gut microbes indicated that patients with SLE had a decreased abundance of Ruminococcaceae, increased abundance of Enterobacteriaceae and Enterococcaceae, and no significant changes in Lachnospiraceae and Bacteroides. Third, changes in Ruminococcaceae and Enterococcaceae in patients with SLE were greater in Chinese patients with SLE.
The Chao1 richness estimator is simply an indicator of the overall number of species richness,36 whereas the Shannon-Wiener diversity index is an indicator of the uniformity of the number of different species.37 Our results indicated that both parameters were decreased in the gut microbiota of patients with SLE. As a result, we posit that the gut microbiota of these patients appear to have reduced stability, altered structure and function, and reduced ability to resist changes. Our comparison of patients with SLE in active and remission with HCs indicated no significant effect on the Shannon-Wiener diversity index. This result seems to contradict the findings of Azzouz et al.38 There was significant heterogeneity among the studies in the SLE active groups and a larger sample size may change the result. In addition, we found that the Shannon-Wiener diversity index was lower for all subjects (men and women) than women alone. This preliminary finding suggests a need for further research on the effect of gender on the gut microbiota of patients with SLE.
Our findings of a decreased abundance of Ruminococcaceae in patients with SLE differed from the results of Wen et al.39 Microbes in this family can produce short-chain fatty acids (SCFAs),25 which are considered potential ‘orchestrators’ of the cross-talk between the gut microbiota and the host metabolism.40 A reduction of these microbes may therefore lead to reduced SCFA production and disruptions of human metabolism. Some reports found that SCFAs can inhibit B cell activation-induced cytidine deaminase (AID) and Blimp1 expression, plasma cell differentiation and class-switching by autoantibodies’ class switching; prevent IgG1/IgG2 deposition in the kidney; and prevent lupus skin lesions.41 Therefore, a decreased abundance of species in Ruminococcaceae may be responsible for some of the complications experienced by patients with SLE. In addition, SCFAs are the main energy source of colon cells,42 and these protect the integrity of the small intestinal epithelial cell membrane.43 44 There is also evidence that a decreased in the abundance of Ruminococcaceae may also lead to ‘leaky gut’ (increased intestinal permeability).
Enterobacteriaceae is in the Proteobacteria phylum and is a large family in which many species are pathogens or produce inflammatory reactions. Enterobacteriaceae are among the most common pathogens responsible for abdominal infections, and the production of extended-spectrum beta-lactamases is the main mechanism of their pathogenesis.45 Previous research showed that a high level of S100A8-A9 (a calcium-binding extracellular complex) in the intestines of infants can reduce the abundance of intestinal Enterobacteriaceae through the expansion of Tregs, and thereby promote the healthy development of intestinal microflora.46 The abundance of Enterobacteriaceae is also associated with T cells, and this may be the mechanism by which changes in the abundance of Enterobacteriaceae affect the occurrence and development of SLE. Enterobacteriaceae are also among the main pathogens responsible for pulmonary infections.47 Thus, the increased abundance of Enterobacteriaceae in the gut of patients with SLE may be one of the factors that contribute to multiple infections of the lungs and abdominal cavity in these patients.
Enterococcaceae are generally considered a beneficial microflora because they function in reducing gastrointestinal damage.48 However, species in this family may also contribute to the development of cancer.49 Further studies are needed to determine whether the increase of Enterococcaceae in patients with SLE is a compensatory response to restore gut homeostasis25 50 or whether it leads to tumourigenesis.
However, there is a risk of false-positives in the results for Enterobacteriaceae and Enterococcaceae after correction for multiple comparisons. The risk may be due to insufficient sample size51 as these two groups of gut microbes had the smallest sample sizes of all the positive samples. Therefore, further studies with a larger sample size are necessary to confirm our results.
It is important to consider technical methods used to assess the gut microbiota in the 11 different studies in our meta-analysis. The study by Chen et al52 was the only study to use shotgun sequencing, and all the others relied on 16S rRNA sequencing. This later method provides information on phylogenetic abundance up to the genus level, but has low taxonomic resolution at the species level.53 Notably, the metagenome-wide study of Chen et al52 found an increased level of R. gnavus in patients with SLE. There is evidence that intestinal expansion of this species leads to cross-reaction with lupus anti-double stranded DNA (dsDNA) antibodies and correlated with lupus nephritis.38 Bagavant et al54 also found an increase in Enterococcus gallinarum in patients with SLE and that this was significantly associated with the presence of anti-dsDNA and anti-Sm autoantibodies. However, we cannot infer changes at the level of the species or genus based on changes at the level of the family, as determined by 16S RNA data.
We also examined the effect of medications on the gut microbiota of patients with SLE using sensitivity analysis. Previous research showed that glucocorticoid therapy similarly altered the diversity of operational taxonomic units in patients with SLE and HCs in that it restored the ratio of Firmicutes to Bacteroidetes.55 Our study indicated that medication use may affect the Chao1 index. Guo et al55 reported the gut microbiota was similar in HCs and patients with SLE who received prednisone doses up to 20 mg. None of the studies we included examined patients who received glucocorticoid doses above 10 mg/day, and this may be the reason for our discrepant results. Studies of mice with experimentally induced SLE also support the argument that a high dose of prednisone (10 mg/kg) can change the Shannon-Weiner index and Chao1 estimator of the gut microbiota.56 Another study reported that administration of different doses of corticosteroids to mice with SLE led to changes in different bacterial taxa and alterations in metabolic functions.56 Thus, changes in the gut microbiota of patients with SLE may be used as a reference for clinical adjustment of the glucocorticoid dose because this can be difficult to adjust in the clinic.
Short-term/high-dose or long-term use of hydroxychloroquine (HCQ) can also alter the gut microbiota, resulting in a decreased level in the relative abundance of Firmicutes.57 58 For example, Bellocchi et al12 examined patients with SLE, 70.37% of whom were taking HCQ. HCQ can inhibit the stimulation of Toll-like receptors59 and reduce the level of interleukin 1 and 6, thus achieving anti-inflammatory effects. This previous study found that HCQ combined with hormone therapy may cause a decrease in the abundance of Enterobacteriaceae compared with glucocorticoids or HCQ alone, a possible mechanism by which HCQ can reduce the risk of infection rate in patients with SLE. In addition, HCQ may also cause a decrease in the abundance of Enterococcaceae, which may be one of the mechanisms for its gastrointestinal adverse effects.60
In addition to disease, diet and environment, race and ethnicity can also impact the gut microbiota.61 62 For example, the van der Meulen et al study8 may have been a source of heterogeneity in our meta-analysis due to the different backgrounds of the included population; 28 were white/European, but none of the HCs was white/European. Thus, there was a significant decrease in Ruminococcaceae in the faeces of patients with SLE from Spain, but this phenomenon was not evident in Chinese patients.63 Some other research suggested an influence of genetic background on the gut microbiota. For example, Goodrich et al64 analysed the gut microbiota in 1126 pairs of British twins and found that Christensenellaceae was the most heritable taxon. It is possible that Ruminococcaceae and Enterococcaceae families may also be heritable.
Chen et al52 and and Tomofuji et al53 used a metagenome-wide approach to identify the high abundance of Streptococcaceae and Streptococcus in patients with SLE, and also analysed their results at the level of genes, biological pathways and functional level. However, most of our results were based on studies that used 16S rRNA sequencing, which has limitations. Thus, we could not analyse microbes at the genus level, such as Streptococcus and Veillonella. We anticipate that future research on this topic will increasingly use a metagenome-wide approach. The lack of using compositional data analysis is a limitation of these studies, which may produce errant results and spurious correlations.65 Another limitation is that very little is known about the effects of glucocorticoid dose on the abundance and metabolomics of gut microbiota, so our meta-analysis could not examine this important topic. In addition, we were unable to analyse the changes in the gut microbiota between different genders due to the limitations of the study population. Also, different studies used different reference panels (standard 16S rRNA databases used for alignment), which may lead to different inferences, confounding and heterogeneity.
In conclusion, our meta-analysis summarised the differences in the gut microbiota between patients with SLE and HCs, including the changes in diversity and in the relative abundance of different taxa. Our findings suggest that targeted alteration in the abundance of intestinal flora has potential for the treatment of SLE. In addition, we also found that medication use can affect the gut microbiota.
Acknowledgments
This article was already published as a non-peer reviewed article in Research Square (DOI 10.21203/rs.3.rs-608010/v1).
Footnotes
SX, YQ and SQ contributed equally.
Contributors: Conceptualisation: SX and YQ. Methodology: SQ. Software: SX. Validation: YQ and SQ. Formal analysis: SX. Investigation: RW and YJ. Resources: RW and YJ. Data curation: RW. Writing - original draft preparation: SX. Writing - review and editing: SX and YQ. Visualisation: SQ. Supervision: XD. Project administration: XD. Funding acquisition: XD. All authors have discussed the results and contributed to the writing of the manuscript and approved the final manuscript. XD is responsible for the overall content as guarantor.
Funding: This research was funded by the National Natural Science Foundation of China (no: 81774179 and 81973778).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
Not applicable.
References
- 1.Kiriakidou M, Ching CL. Systemic lupus erythematosus. Ann Intern Med 2020;172:ITC81-ITC96. 10.7326/AITC202006020 [DOI] [PubMed] [Google Scholar]
- 2.Durcan L, O'Dwyer T, Petri M. Management strategies and future directions for systemic lupus erythematosus in adults. Lancet 2019;393:2332–43. 10.1016/S0140-6736(19)30237-5 [DOI] [PubMed] [Google Scholar]
- 3.Jorge AM, Lu N, Zhang Y, et al. Unchanging premature mortality trends in systemic lupus erythematosus: a general population-based study (1999-2014). Rheumatology 2018;57:337–44. 10.1093/rheumatology/kex412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010;107:11971–5. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umesaki Y, Setoyama H, Matsumoto S, et al. Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology 1993;79:32–7. [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 2010;330:1768–73. 10.1126/science.1195568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hevia A, Milani C, López P, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio 2014;5:e01548–01514. 10.1128/mBio.01548-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Meulen TA, Harmsen HJM, Vila AV, et al. Shared gut, but distinct oral microbiota composition in primary Sjögren's syndrome and systemic lupus erythematosus. J Autoimmun 2019;97:77-87. 10.1016/j.jaut.2018.10.009 [DOI] [PubMed] [Google Scholar]
- 9.Kim J-W, Kwok S-K, Choe J-Y, et al. Recent advances in our understanding of the link between the intestinal microbiota and systemic lupus erythematosus. Int J Mol Sci 2019;20:4871. 10.3390/ijms20194871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mu Q, Kirby J, Reilly CM, et al. Leaky gut as a danger signal for autoimmune diseases. Front Immunol 2017;8:598. 10.3389/fimmu.2017.00598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greiling TM, Dehner C, Chen X, et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med 2018;10:eaan2306. 10.1126/scitranslmed.aan2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellocchi C, Fernández-Ochoa Álvaro, Montanelli G, et al. Identification of a shared microbiomic and metabolomic profile in systemic autoimmune diseases. J Clin Med 2019;8:1291. 10.3390/jcm8091291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu WF. Study on the changes of intestinal flora in patients with systemic lupus erythematosus. Zhejiang University 2018. [Google Scholar]
- 14.Wei F, Xu H, Yan C, et al. Changes of intestinal flora in patients with systemic lupus erythematosus in northeast China. PLoS One 2019;14:e0213063. 10.1371/journal.pone.0213063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonder MJ, Kurilshikov A, Tigchelaar EF, et al. The effect of host genetics on the gut microbiome. Nat Genet 2016;48:1407–12. 10.1038/ng.3663 [DOI] [PubMed] [Google Scholar]
- 16.Cuervo A, Hevia A, López P, et al. Association of polyphenols from oranges and apples with specific intestinal microorganisms in systemic lupus erythematosus patients. Nutrients 2015;7:1301–17. 10.3390/nu7021301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma Y, Shi N, Li M, et al. Applications of next-generation sequencing in systemic autoimmune diseases. Genomics Proteomics Bioinformatics 2015;13:242–9. 10.1016/j.gpb.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rojo D, Hevia A, Bargiela R, et al. Ranking the impact of human health disorders on gut metabolism: systemic lupus erythematosus and obesity as study cases. Sci Rep 2015;5:8310. 10.1038/srep08310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature 2012;486:222–7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsokos GC. Systemic lupus erythematosus. N Engl J Med 2011;365:2110–21. 10.1056/NEJMra1100359 [DOI] [PubMed] [Google Scholar]
- 21.Casals-Pascual C, González A, Vázquez-Baeza Y, et al. Microbial diversity in clinical microbiome studies: sample size and statistical power considerations. Gastroenterology 2020;158:1524–8. 10.1053/j.gastro.2019.11.305 [DOI] [PubMed] [Google Scholar]
- 22.Stroup DF, Berlin JA, Morton SC, et al. Meta-Analysis of observational studies in epidemiology: a proposal for reporting. meta-analysis of observational studies in epidemiology (moose) group. JAMA 2000;283:2008–12. 10.1001/jama.283.15.2008 [DOI] [PubMed] [Google Scholar]
- 23.Liao X, Li G, Wang A, et al. Repetitive transcranial magnetic stimulation as an alternative therapy for cognitive impairment in Alzheimer's disease: a meta-analysis. J Alzheimers Dis 2015;48:463–72. 10.3233/JAD-150346 [DOI] [PubMed] [Google Scholar]
- 24.Tang Q, Li G, Liu T, et al. Modulation of interhemispheric activation balance in motor-related areas of stroke patients with motor recovery: systematic review and meta-analysis of fMRI studies. Neurosci Biobehav Rev 2015;57:392–400. 10.1016/j.neubiorev.2015.09.003 [DOI] [PubMed] [Google Scholar]
- 25.Shen T, Yue Y, He T, et al. The association between the gut microbiota and Parkinson's disease, a meta-analysis. Front Aging Neurosci 2021;13:636545. 10.3389/fnagi.2021.636545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan X, Wang W, Liu J, et al. Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol 2014;14:135. 10.1186/1471-2288-14-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. 10.1007/s10654-010-9491-z [DOI] [PubMed] [Google Scholar]
- 28.Bakbergenuly I, Hoaglin DC, Kulinskaya E. Estimation in meta-analyses of mean difference and standardized mean difference. Stat Med 2020;39:171–91. 10.1002/sim.8422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins JPT, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557–60. 10.1136/bmj.327.7414.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chawla N, Anothaisintawee T, Charoenrungrueangchai K, et al. Drug treatment for panic disorder with or without agoraphobia: systematic review and network meta-analysis of randomised controlled trials. BMJ 2022;376:e066084. 10.1136/bmj-2021-066084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chavalarias D, Wallach JD, Li AHT, et al. Evolution of reporting P values in the biomedical literature, 1990-2015. JAMA 2016;315:1141–8. 10.1001/jama.2016.1952 [DOI] [PubMed] [Google Scholar]
- 32.Siegmann E-M, Müller HHO, Luecke C, et al. Association of depression and anxiety disorders with autoimmune thyroiditis: a systematic review and meta-analysis. JAMA Psychiatry 2018;75:577–84. 10.1001/jamapsychiatry.2018.0190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashino Y, Noguchi Y, Fukui T. Systematic evaluation and comparison of statistical tests for publication bias. J Epidemiol 2005;15:235–43. 10.2188/jea.15.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Begg CB, Mazumdar M. Operating characteristics of a RANK correlation test for publication bias. Biometrics 1994;50:1088–101. [PubMed] [Google Scholar]
- 35.Romero-Diaz J, Isenberg D, Ramsey-Goldman R. Measures of adult systemic lupus erythematosus: updated version of British Isles lupus assessment group (bilag 2004), European consensus lupus activity measurements (eclam), systemic lupus activity measure, revised (slam-r), systemic lupus activity questionnaire for population studies (slaq), systemic lupus erythematosus disease activity index 2000 (sledai-2k), and systemic lupus international collaborating clinics/american College of rheumatology damage index (SDI). Arthritis Care Res 2011;63:S37–46. 10.1002/acr.20572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao A. Nonparametric estimation of the number of classes in a population. Scand J Statist 1984;11:265–70. [Google Scholar]
- 37.Chao A, Shen T-J. Nonparametric estimation of shannon’s index of diversity when there are unseen species in sample. Environ Ecol Stat 2003;10:429–43. 10.1023/A:1026096204727 [DOI] [Google Scholar]
- 38.Azzouz D, Omarbekova A, Heguy A, et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis 2019;78:947–56. 10.1136/annrheumdis-2018-214856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wen M, Liu T, Zhao M, et al. Correlation analysis between gut microbiota and metabolites in children with systemic lupus erythematosus. J Immunol Res 2021;2021:5579608. 10.1155/2021/5579608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodríguez-Carrio J, López P, Sánchez B, et al. Intestinal dysbiosis is associated with altered short-chain fatty acids and serum-free fatty acids in systemic lupus erythematosus. Front Immunol 2017;8:23. 10.3389/fimmu.2017.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez HN, Moroney JB, Gan H, et al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun 2020;11:60. 10.1038/s41467-019-13603-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Canfora EE, Jocken JW, Blaak EE. Short-Chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol 2015;11:577–91. 10.1038/nrendo.2015.128 [DOI] [PubMed] [Google Scholar]
- 43.Peng L, Li Z-R, Green RS, et al. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr 2009;139:1619–25. 10.3945/jn.109.104638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elamin EE, Masclee AA, Dekker J, et al. Short-Chain fatty acids activate AMP-activated protein kinase and ameliorate ethanol-induced intestinal barrier dysfunction in Caco-2 cell monolayers. J Nutr 2013;143:1872–81. 10.3945/jn.113.179549 [DOI] [PubMed] [Google Scholar]
- 45.Mureșan MG, Balmoș IA, Badea I, et al. Abdominal sepsis: an update. J Crit Care Med 2018;4:120–5. 10.2478/jccm-2018-0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willers M, Ulas T, Völlger L, et al. S100A8 and S100A9 are important for postnatal development of gut microbiota and immune system in mice and infants. Gastroenterology 2020;159:2130–45. 10.1053/j.gastro.2020.08.019 [DOI] [PubMed] [Google Scholar]
- 47.de Maio Carrillho CMD, Gaudereto JJ, Martins RCR, et al. Colistin-Resistant Enterobacteriaceae infections: clinical and molecular characterization and analysis of in vitro synergy. Diagn Microbiol Infect Dis 2017;87:253–7. 10.1016/j.diagmicrobio.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 48.Guo H, Chou W-C, Lai Y, et al. Multi-Omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science 2020;370:eaay9097. 10.1126/science.aay9097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang P-Y, Yang Y-C, Wang C, et al. Increase in Akkermansiaceae in Gut Microbiota of Prostate Cancer-Bearing Mice. Int J Mol Sci 2021;22. 10.3390/ijms22179626. [Epub ahead of print: 06 Sep 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallen ZD, Appah M, Dean MN, et al. Characterizing dysbiosis of gut microbiome in PD: evidence for overabundance of opportunistic pathogens. NPJ Parkinsons Dis 2020;6:11. 10.1038/s41531-020-0112-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leon AC. Multiplicity-adjusted sample size requirements: a strategy to maintain statistical power with Bonferroni adjustments. J Clin Psychiatry 2004;65:1511–4. [PubMed] [Google Scholar]
- 52.Chen B-di, Jia X-M, Xu J-Y, et al. An autoimmunogenic and proinflammatory profile defined by the gut microbiota of patients with untreated systemic lupus erythematosus. Arthritis Rheumatol 2021;73:232–43. 10.1002/art.41511 [DOI] [PubMed] [Google Scholar]
- 53.Tomofuji Y, Maeda Y, Oguro-Igashira E, et al. Metagenome-wide association study revealed disease-specific landscape of the gut microbiome of systemic lupus erythematosus in Japanese. Ann Rheum Dis 2021;80:1575–83. 10.1136/annrheumdis-2021-220687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bagavant H, Araszkiewicz AM, Ingram JK, et al. Immune Response to Enterococcus gallinarum in Lupus Patients Is Associated With a Subset of Lupus-Associated Autoantibodies. Front Immunol 2021;12:635072. 10.3389/fimmu.2021.635072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo M, Wang H, Xu S, et al. Alteration in gut microbiota is associated with dysregulation of cytokines and glucocorticoid therapy in systemic lupus erythematosus. Gut Microbes 2020;11:1758–73. 10.1080/19490976.2020.1768644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang M, Zhu Z, Lin X, et al. Gut microbiota mediated the therapeutic efficacies and the side effects of prednisone in the treatment of MRL/lpr mice. Arthritis Res Ther 2021;23:240. 10.1186/s13075-021-02620-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan Z-Y, Chang Y, Han N, et al. Short-Term high-dose gavage of hydroxychloroquine changes gut microbiota but not the intestinal integrity and immunological responses in mice. Life Sci 2021;264:118450. 10.1016/j.lfs.2020.118450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Angelakis E, Million M, Kankoe S, et al. Abnormal weight gain and gut microbiota modifications are side effects of long-term doxycycline and hydroxychloroquine treatment. Antimicrob Agents Chemother 2014;58:3342–7. 10.1128/AAC.02437-14 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Kuznik A, Bencina M, Svajger U, et al. Mechanism of endosomal TLR inhibition by antimalarial drugs and imidazoquinolines. J Immunol 2011;186:4794–804. 10.4049/jimmunol.1000702 [DOI] [PubMed] [Google Scholar]
- 60.Matsuda T, Ly NTM, Kambe N, et al. Early cutaneous eruptions after oral hydroxychloroquine in a lupus erythematosus patient: a case report and review of the published work. J Dermatol 2018;45:344–8. 10.1111/1346-8138.14156 [DOI] [PubMed] [Google Scholar]
- 61.Zhernakova A, Kurilshikov A, Bonder MJ, et al. Population-Based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016;352:565–9. 10.1126/science.aad3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Deschasaux M, Bouter KE, Prodan A, et al. Depicting the composition of gut microbiota in a population with varied ethnic origins but shared geography. Nat Med 2018;24:1526–31. 10.1038/s41591-018-0160-1 [DOI] [PubMed] [Google Scholar]
- 63.He Z, Shao T, Li H, et al. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog 2016;8:64. 10.1186/s13099-016-0146-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goodrich JK, Davenport ER, Beaumont M, et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 2016;19:731–43. 10.1016/j.chom.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsilimigras MCB, Fodor AA. Compositional data analysis of the microbiome: fundamentals, tools, and challenges. Ann Epidemiol 2016;26:330–5. 10.1016/j.annepidem.2016.03.002 [DOI] [PubMed] [Google Scholar]
- 66.Luo XM, Edwards MR, Mu Q, et al. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl Environ Microbiol 2018;84:e02288–17. 10.1128/AEM.02288-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y, Wang H-F, Li X, et al. Disordered intestinal microbes are associated with the activity of systemic lupus erythematosus. Clin Sci 2019;133:821–38. 10.1042/CS20180841 [DOI] [PubMed] [Google Scholar]
- 68.Sun ZD, Wei F, H F X. Study on the changes of intestinal flora in patients with systemic lupus erythematosus. Int J Immunol 2019;42:579–82. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lupus-2021-000599supp001.pdf (79.6KB, pdf)
lupus-2021-000599supp002.pdf (631.7KB, pdf)
Data Availability Statement
Data are available upon reasonable request.