Abstract
The aadA genes, encoding resistance to streptomycin and spectinomycin, have been found as gene cassettes in different gram-negative and gram-positive bacterial species. The present study has revealed the sequence of a new gene, aadA5, integrated as a gene cassette together with the trimethoprim resistance gene dfr7 in a class 1 integron. The integron was located on a plasmid and was identified in a pathogenic porcine Escherichia coli isolate.
Hollingshead and Vapnek (12) were the first to describe aadA [also called ant(3")−1], which encodes an adenylylation enzyme that modifies streptomycin and spectinomycin. Sundström et al. (20) sequenced aadA1 as a gene cassette and the class 1 integron from Tn21 (GenBank accession no. 12870). They found that the cassette could be translated, and they recognized the target of a site-specific recombination site (59-bp element). The gene cassettes identified so far have been in the same orientation, and it is possible that the 59-bp element has a role in determination of the orientation when gene cassettes are integrated (19). The cassettes are expressed from the common promoter Pant, located on the 5′ conserved segment (CS) of the integron (7, 10), which also contains the gene for an integrase that catalyzes integration and excision of the cassettes (6). As part of the 3′ CS downstream from the gene cassettes, qacEΔ1 and sul1 are often found; they determine resistance to quaternary ammonium compounds and sulfonamides, respectively (16).
The aadA genes are the only characterized genes that encode both streptomycin and spectinomycin resistance, and many of these genes are found as gene cassettes (3, 5, 9, 17, 20) (GenBank accession no. AF047479 and Z50802.3). The present study characterizes the nucleotide sequence and expression of a novel streptomycin and spectinomycin resistance gene located as a gene cassette in an integron.
A gentamicin-resistant Escherichia coli 9516014-1 serotype 101 isolate was collected in a previous study from a case of diarrhea in a Danish pig herd. Salmonella enterica serotype Typhimurium DT104 no. 9720921 (18) was used as a positive control for integron PCRs. The recipient for conjugation was E. coli K-12 J62-2 (F− pro lac trp his), which is rifampin resistant (15). All strains were stored at −80°C.
A plasmid DNA preparation from 9516014-1 was made according to Kado and Liu (13), and conjugation of the plasmid that contains the class 1 integron was performed by overnight plate mating (1:10 ratio of donor to recipient) with E. coli K-12 J62-2 as the recipient. Luria-Bertani (LB) plates, to select for transconjugants, contained rifampin at 50 μg/ml and streptomycin at 30 μg/ml (Sigma Chemical Co., St. Louis, Mo.). Plasmid DNA was prepared from the transconjugants (13), and the size was determined. Susceptibility to antimicrobial agents was demonstrated as MICs by using the Sensititre system according to standard methods (Sensititre Ltd., West Sussex, United Kingdom). Antimicrobial agents tested were gentamicin (0.25 to 32 μg/ml), neomycin (0.5 to 64 μg/ml), apramycin (0.5 to 64 μg/ml), streptomycin (2 to 256 μg/ml), spectinomycin (1 to 128 μg/ml), nalidixic acid (1 to 128 μg/ml), enrofloxacin (0.06 to 8 μg/ml), trimethoprim (0.25 to 32 μg/ml), sulfamethoxazole (2 to 256 μg/ml), chloramphenicol (1 to 128 μg/ml), tetracycline (0.5 to 32 μg/ml), ampicillin (0.5 to 32 μg/ml), and colistin (1 to 64 μg/ml). Rifampin (0.5 to 64 μg/ml) (Sigma Chemical Co.) was tested under the same conditions but was not included in the Sensititre system.
PCR amplification was used to investigate the presence of integrons and gene cassettes in the strains; DNA preparation and PCR conditions were as described previously, and primers used are shown in Table 1 (18). Temperatures used in PCR amplification for denaturation, annealing, and elongation were 95°C for 45 s, 60°C for 45 s, and 72°C for 6 min, respectively, all of which was repeated for 31 cycles after an initial 3 min of denaturation at 94°C.
TABLE 1.
PCR primers for cloning and sequencing of a novel gene cassette isolated from a porcine E. coli strain
| Designationa | Sequence | Accession no.b | Position (bp) | Reference or source |
|---|---|---|---|---|
| Pant-F | GTG GAA ACG GAT TAA GGC ACG | L06418 | 1007–1027 | 10 |
| qacEΔ1-B | CAA GCT TTT GCC CAT GAA GC | X12869 | 788–769 | 20 |
| dfrA7-F | CTG TTC ACG TTG AGG TTG AAG | U31119 | 943–964 | 4 |
| aadA5-F | CGG AAC ATG GCG ACC GCT GG | AF137361 | 303–322 | This study |
F, forward sequence (5′ to 3′); B, backward sequence (3′ to 5′).
Accession numbers are from published sequences in the GenBank database.
The PCR amplification product from primers Pant-F and qacEΔ1-B was prepared for sequencing with the QIAquick purification kit (Qiagen, Hilden, Germany), and the nucleotide sequence was determined using a cycle sequencer as described previously (18). For comparison to known sequences, the FASTA search program and the advanced BLAST search program were used (1a, 14).
Cloning was performed with Pfu DNA polymerase (Stratagene Ltd., Cambridge, United Kingdom) and the Zero Blunt PCR cloning kit (Invitrogen Corporation, San Diego, Calif.) by using the pCR-Blunt plasmid as the vector and E. coli TOP10 (Invitrogen) as the recipient. The unknown sequence was amplified with the Pant-F and qacEΔ1-B primers; all other reagents were used according to the manufacturers’ instructions. To select for vectors with inserts in the appropriate orientation, trimethoprim at 200 μg/ml was incorporated into LB agar. Phenotypic susceptibilities to the antimicrobial agents listed above were determined with the Sensititre system to confirm expression of the cloned gene. The E. coli TOP10 recipient including the cloned vector was designated DS 160.
E. coli 9516014-1 was mated with E. coli K-12 J62-2 to determine whether the integron was transferable. The plasmid profile and resistance pattern of the transconjugant confirmed the presence of a single conjugative plasmid. The class 1 integron, including the drf7 (dhfrVII) and aadA5 cassettes, was located on this 22-kb transferable plasmid, which specified resistance to streptomycin, spectinomycin, trimethoprim, and sulfonamide only. The presence of the integron was confirmed by PCR amplification with transconjugant DNA as the template. Although the integron and gene cassettes were shown to be plasmid located, the possibility that a copy of the integron structure could also be located on the chromosome cannot be excluded.
Antimicrobial resistance was determined for the wild-type strain E. coli 9516014-1, the cloned DS160, the recipient J62-2, and the transconjugant DS175. The MICs of trimethoprim (>32 μg/ml), streptomycin (>256 μg/ml), and spectinomycin (>128 μg/ml) were the same for E. coli DS160 as for E. coli 9516014-1. This indicates successful cloning of the two gene cassettes and that the promoter expressed both cassettes. However, the sulfamethoxazole MIC for E. coli DS160 was 4.0 μg/ml; the strain did not contain the 3′ CS of the integron and was therefore not expected to contain the sul1 gene, which is a normal part of a class 1 integron. The MICs of the antimicrobial agents streptomycin, spectinomycin, trimethoprim, and sulfamethoxazole, >256 μg/ml, were the same for the transconjugant DS175 as for the wild-type strain.
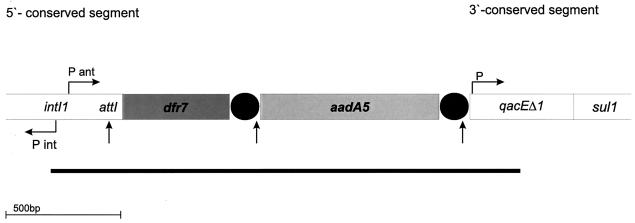
With the Pant-F and qacEΔ1-B primers from both conserved segments for amplification of the gene cassettes, a 2,100-bp amplicon was obtained. This product was sequenced (Fig. 1) in both directions and found to contain a known trimethoprim resistance gene, dfr7, and an additional open reading frame. The structure of the gene cassette and the integron boundaries are shown in Fig. 1.
FIG. 1.
Positions of gene cassettes and integron CSs of E. coli 9516014-1. Designations are as follows: intI1, integrase; attI, recombination site; dfr7, dihydrofolate reductase (trimethoprim resistance) gene; aadA5, streptomycin and spectinomycin resistance gene; qacEΔ1, quaternary ammonium compound resistance gene; sulI, sulfonamide resistance gene. Horizontal arrows indicate putative promoters (P) and direction of translation, and vertical arrows show recombination hot spots where the gene cassettes integrate. Black circles indicate the 59-bp elements, and the line beneath the integron structure indicates the sequenced area.
The sequences of the integrase (intI), the common promoter (Pant) (5), the attachment site (attI), and the trimethoprim resistance gene dfr7 (GenBank accession no. X58425) (4, 21) were located upstream from aadA5. In Danish veterinary practice, the combination of trimethoprim and sulfonamides is widely utilized to treat infectious diseases. That specific selection pressure would favor the acquisition or maintenance of a trimethoprim cassette by a class 1 integron containing sul1 in the 3′ region.
The initiation codon, ATG, of the novel streptomycin and spectinomycin resistance gene cassette is located at bp 64, and the gene continues for the next 789 bp, or 263 amino acids. The gene was designated aadA5 because it has the same features as the known and characterized aadA genes but a different sequence (Table 2) (3, 5, 8, 9, 12, 20). The nucleotide sequence contains a stop codon at position 850, which also can be found in an aadA3 gene sequenced by Adrian et al. (1) (GenBank accession no. Z50802.3). A comparison of the novel aadA5 resistance gene with previously sequenced aadA genes is shown in Table 2.
TABLE 2.
Comparison of the novel aadA5 gene cassette with previously published adenylylation enzyme genes encoding streptomycin and spectinomycin resistancea
| Gene | GenBank accession no.b | Reference or source | Origin | Position (length) (bp) | % Identity to aadA5 (no. identical/total)
|
|
|---|---|---|---|---|---|---|
| Nucleotide sequence (bp) | Amino acid sequence | |||||
| aadA1a | X12870 | 20 | Tn21 | 1299–2090 (792) | 60 (447/745) | 57 (149/261) |
| aadA1b | M95287 | 9 | R46 | 2324–3115 (792) | 59 (443/745) | 57 (149/261) |
| aadA2 | X68227 | 3 | pSA | 166–945 (780) | 59 (441/741) | 56 (141/250) |
| aadA3 | AF047479 | 8a | NR79 | 1296–2087 (792) | 62 (489/789) | 55 (141/254) |
| aadA3c | Z50802.3 | 1 | E. coli | 1306–2094 (789) | 95 (750/789) | 95 (249/262) |
| aadA5 | Af137361 | This study | E. coli | 64–852 (789) | 100 | 100 |
Comparisons between known aadA genes and aadA5 were done with the advanced BLAST search program (1a, 14).
GenBank accession numbers are examples of the particular genes inserted as gene cassettes.
The aadA3 gene submitted to GenBank by Adrian et al. in 1999 has the same designation as the gene submitted by Gravel et al. in 1998 despite different nucleotide sequences. The designation of GenBank accession no. Z50802.3 may be changed in the future.
The gene cassettes have two imperfect inverted recombination core sites flanking the central imperfect repeat within the 59-bp element. The imperfect inverted repeat core sites mark the point of insertion of the gene cassettes into the integron (6, 19). The main part of the aadA5 59-bp element is located at bp 898 to 949, and the 5′ end of the inserted gene cassette recombination core site for the site-specific insertion GTTRRRY was found at bp 54 to 60, which could be interpreted as the 5′-end cassette boundary. The recombination site is between the G and the first T, making the gene cassette begin at position 55 with the TTRRRY sequence derived from the 59-bp element (11). The gene cassette is bounded by an inverse core site (RYYYAAC) adhering to the 3′ end of the resistance gene, and this sequence is located at position 899 to 905. The 59-bp element ends at bp 949, where the next GTTRRRY inverted recombination core site begins, which is interpreted as the 3′ end of the gene cassette. A notable feature is the presence of an additional copy of the inverted repeat core site at bp 848 to 854.
Downstream from the 59-bp element at position 1057, the start codon and coding sequence of the qacEΔ1 gene of the 3′ CS were found. Both CSs showed 100% identity to a number of class 1 integrons previously sequenced (2, 11). The 3′ CS begins at bp 950 and continues downstream.
The present study has characterized a novel streptomycin and spectinomycin resistance gene cassette found in a class 1 integron. The distribution of this gene is still to be investigated. The novel gene cassette is present in a class 1 integron, which is again located on a transferable plasmid. These factors enhance the transfer possibilities for the aadA5 gene, and the conditions of transfer and the molecular epidemiology of gene cassettes will need further attention in the future.
Nucleotide sequence accession number.
The nucleotide sequence of the E. coli aadA5 gene has been deposited in the GenBank database under accession no. AF137360.
Acknowledgments
Thanks are due to Niels Einar Jensen, Danish Veterinary Laboratory, for supplying aminoglycoside-resistant E. coli. Many thanks are due to Joanna Zeitman Amenuvor, Danish Veterinary Laboratory, for inspiring discussion and technical assistance and to David Platt, Glasgow Royal Infirmary, for the cosmic onion and critical reading of the manuscript.
This study was supported by a grant from the Danish Agricultural and Veterinary Research Council (grant 9600012).
REFERENCES
- 1.Adrian, K. Unpublished data.
- 1a.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bissonnette L, Champetier S, Buisson J-P, Roy P H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol. 1991;173:4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bito A, Susani M. Revised analysis of aadA2 gene of plasmid pSa. Antimicrob Agents Chemother. 1994;38:1172–1175. doi: 10.1128/aac.38.5.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnside J M, Groot Obbink D J. Plasmid pDGO100 contains a second integron with the trimethoprim resistance gene dfrA7 as the inserted cassette. Plasmid. 1996;35:67–70. doi: 10.1006/plas.1996.0008. [DOI] [PubMed] [Google Scholar]
- 5.Clark N C, Olsvik O, Swenson J M, Spiegel C A, Tenover F C. Detection of a streptomycin/spectinomycin adenylyltransferase gene (aadA) in Enterococcus faecalis. Antimicrob Agents Chemother. 1999;43:157–160. doi: 10.1128/aac.43.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collis C M, Hall R M. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol Microbiol. 1992;6:2875–2885. doi: 10.1111/j.1365-2958.1992.tb01467.x. [DOI] [PubMed] [Google Scholar]
- 7.Collis C M, Hall R M. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fling M E, Kopf J, Richards C. Nucleotide sequence of the transposon Tn7 gene encoding an aminoglycoside-modifying enzyme, 3"(9)-O-nucleotidyltransferase. Nucleic Acids Res. 1985;13:7095–7105. doi: 10.1093/nar/13.19.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Gravel, V. Unpublished data.
- 9.Hall R M, Vockler C. The region of the IncN plasmid R46 coding for resistance to beta-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposon. Nucleic Acids Res. 1987;15:7491–7501. doi: 10.1093/nar/15.18.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall R M, Stokes H W. The structure of a partial duplication in the integron of plasmid pDGO100. Plasmid. 1990;23:76–79. doi: 10.1016/0147-619x(90)90047-g. [DOI] [PubMed] [Google Scholar]
- 11.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 12.Hollingshead S, Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenyltransferase. Plasmid. 1985;13:17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- 13.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Center for Biotechnology Information. 1999. Advanced BLAST. [Online.] National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md. http://www.ncbi.nlm.nih.gov/blast/blast.cgi. [4 June 1999, last date accessed.]
- 15.Obaseiki-Ebor E E. Rifampicin curing of plasmids in Escherichia coli K12-rifampicin resistant host. J Pharm Pharmacol. 1984;36:467–470. doi: 10.1111/j.2042-7158.1984.tb04428.x. [DOI] [PubMed] [Google Scholar]
- 16.Paulsen I T, Littlejohn T G, Rådström P, Sundström L, Sköld O, Swedberg G, Skurray R A. The 3′ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother. 1993;37:761–768. doi: 10.1128/aac.37.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 18.Sandvang D, Aarestrup F M, Jensen L B. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol Lett. 1997;157:177–181. doi: 10.1111/j.1574-6968.1997.tb12770.x. [DOI] [PubMed] [Google Scholar]
- 19.Stokes H W, O’Gorman D B, Recchia G D, Parsekhian M, Hall R M. Structure and function of the 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 20.Sundström L, Rådström P, Swedberg G, Sköld O. Site-specific recombination promotes linkage between trimethoprim and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 21.Sundström L, Swedberg G, Sköld O. Characterization of transposon Tn5086, carrying the site-specifically inserted gene dhfrVII mediating trimethoprim resistance. J Bacteriol. 1993;175:1796–1805. doi: 10.1128/jb.175.6.1796-1805.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]