Highlights
-
•
LncRNA GIHCG facilitated the proliferation and migration while suppressed apoptosis of RCC cells.
-
•
LncRNA GIHCG directly sponged miR-499a-5p and down-regulated its expression.
-
•
MiR-499a-5p negatively regulated XIAP expression.
-
•
LncRNA GIHCG knockdown inhibited the growth and metastasis of RCC in nude mice.
-
•
LncRNA GIHCG contributed to the development of RCC through miR-499a-5p/XIAP axis.
Keywords: Renal cell carcinoma, Proliferation, Migration, LncRNA GIHCG, miR-499a-5p, XIAP
Abstract
Background
Our previous study demonstrated that lncRNA GIHCG is upregulated in renal cell carcinoma (RCC) and that knockdown of lncRNA GIHCG suppresses the proliferation and migration of RCC cells. However, the mechanism of lncRNA GIHCG in RCC needs further exploration.
Methods
The proliferation, cell cycle, migration, and apoptosis of RCC cells were tested using CCK-8, flow cytometry, wound healing and Annexin-V/-FITC/PI flow cytometry assays, respectively. Dual-luciferase reporter and RNA pull-down or RNA immunoprecipitation assays (RIPs) were performed to analyze the interactions among lncRNA GIHCG, miR-499a-5p and XIAP. A tumour xenograft study was conducted to verify the function of lncRNA GIHCG in RCC development in vivo.
Results
Knockdown of lncRNA GIHCG inhibited cell proliferation and migration and induced G0/G1 arrest while promoting apoptosis. Overexpression of lncRNA GIHCG led to the opposite results. LncRNA GIHCG sponged miR-499a-5p and downregulated its expression in RCC cells. MiR-499a-5p overexpression suppressed RCC cell growth. MiR-499a-5p targeted XIAP and inhibited its expression. LncRNA GIHCG knockdown reduced the growth of tumour xenografts in vivo and the expression of XIAP while increasing miR-499a-5p levels.
Conclusion
LncRNA GIHCG accelerated the development of RCC by targeting miR-499a-5p and increasing XIAP levels.
Introduction
Renal cell carcinoma (RCC) is one of the most unpredictable tumours of the genitourinary system and causes incalculable losses to society [1]. Statistically, the global incidence of RCC was approximately 400,000 cases in 2019 [2]. RCC has become a severe public health issue that cannot be ignored, particularly in developed countries. The primary treatment of localized RCC is currently nephron-sparing approaches, while treatment of metastatic RCC remains challenging. Although cytoreductive nephrectomy combined with drug therapy has achieved some progress, patients with metastatic RCC have a poor prognosis [3]. Therefore, identifying novel and feasible biomarkers will be key to developing better therapies and improving survival in RCC patients [4]. It is therefore crucial to explore the pathogenic mechanism of RCC.
Long noncoding RNAs (lncRNAs) play crucial roles in regulating chromatin dynamics, gene expression and differentiation [5], and a number of publications in recent years have demonstrated that lncRNAs participate in the pathophysiological process of many different types of cancer [6], including RCC. LncRNA GIHCG, an oncogene, has attracted increasing attention in recent years. For example, one study showed that lncRNA GIHCG expression was increased in oesophageal cancer, and lncRNA GIHCG silencing impeded tumour growth [7]. Remarkably, our previous study demonstrated that lncRNA GIHCG was upregulated in RCC tissues and that increased expression of lncRNA GIHCG predicted a poor prognosis; more interestingly, lncRNA GIHCG knockdown obstructed RCC cell proliferation and migration [8]. However, the specific molecular mechanism of lncRNA GIHCG in RCC needs further investigation. The emergence of microRNAs (miRNAs) has been one of the defining developments in tumour biology in the past decade, providing a novel perspective for cancer treatment [9,10]. Currently, a valuable mechanism has been increasingly reported in which lncRNAs function as competing endogenous RNAs (ceRNAs) to sponge related miRNAs to promote downstream gene expression [11,12]. For example, miR-499a-5p exhibits decreased expression and anticancer effects in some tumours [13]. Zhao et al. demonstrated that miR-499a-5p was decreased in non-small cell lung cancer, and miR-499a-5p overexpression suppressed cell proliferation and arrested cells at the G0/G1 phase [14]. Furthermore, lncRNA SCAMP1 knockdown restrained glioma cell growth and metastasis by targeting miR-499a-5p[15]. Nevertheless, the function of miR-499a-5p in RCC remains to be investigated. Interestingly, bioinformatics analysis from StarBase indicated the presence of binding sites between lncRNA GIHCG and miR-499a-5p. Nevertheless, the interaction between and roles of lncRNA GIHCG and miR-499a-5p in RCC remain unclear.
X-linked inhibitor of apoptosis protein (XIAP) is a well-known cancer-promoting gene that represses levels of caspase-9 and the key proteases caspase-3 and caspase-7 as well as mitochondria-dependant apoptosis pathways, exerting an antiapoptotic role in tumours [16,17]. A previous study suggested that the XIAP levels were increased in RCC, especially in stage III/IV RCC, and was 2.5-fold higher than that in the normal kidney [18]. XIAP knockdown restored cellular sensitivity to apoptotic stimulation [19], further demonstrating that XIAP might promote the development of RCC. In addition, XIAP is regulated by combining with miR-212 and mediates the tumour-suppressive roles of miR-212 in RCC [20]. Interestingly, we identified binding sites between miR-499a-5p and XIAP, indicating a possible targeted relationship between miR-499a-5p and XIAP. However, this regulatory mechanism has not been reported at present, so further investigation is necessary.
In this study, we found that lncRNA GIHCG silencing suppresses RCC cell proliferation and migration while promoting apoptosis through the miR-499a-5p/XIAP axis, providing a feasible theoretical basis for the treatment of RCC.
Materials and methods
Cell collection and transfection
HK-2, Caki-1, 786-O, A498 and SN12C-PM6 cells obtained from the American Type Culture Collection (Manassas, VA, USA) were cultured in Dulbecco's modified Eagle medium (Thermo Fisher Scientific, Inc. Waltham, MA, USA) supplemented with 10% foetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and grown in a humidified atmosphere with 5% CO2 at 37 °C.
The short hairpin RNA (shRNA) was used to silence lncRNA GIHCG levels (sh-GIHCG in the Pglvu6/Puro vector). The full-length lncRNA GIHCG or XIAP coding sequence was amplified and cloned into a pcDNA3.1 (Invitrogen, Carlsbad, CA, USA) vector to achieve overexpression (oe-GIHCG, oe-XIAP). MiR-499a-5p mimics were purchased from GenePharma Company (Shanghai, China). The DNA fragment encoding the mutant lncRNA-GIHCG was synthesized by Sangon Biotech (Shanghai, China), inserted into the pcDNA3.1 vector and named oe-GIHCG-MUT. Cells were transfected with the above plasmids or their corresponding controls using Lipofectamine 3000 (Thermo). After 48 h, cells were harvested for subsequent experiments.
CCK-8 assay
Cells were plated into 96-well plates at 5 × 103 cells/well. After incubation for the indicated time, 10 µL CCK-8 solution was added to each well and incubated for 1 h. The absorbence of each well was subsequently measured at 450 nm using a Multiskan™ GO microplate spectrophotometer (Thermo, USA).
Cell cycle analysis
Cells were plated into 6-well plates and cultured for 24 h at 37 °C. Cells were centrifuged at 1000 g/min for 10 min after washing with PBS. Next, the cells were resuspended in PBS and fixed in 70% ethanol. Cells were washed with PBS again and subjected to PI staining for 30 min in the dark. Next, the percentage of cells in G1/G0, S and G2/M phases was determined using flow cytometry (BD Biosciences, BD).
Annexin-V/-FITC/PI flow cytometry
The apoptosis rate was evaluated using an Annexin V-FITC apoptosis kit (Sigma, USA). Cells were centrifuged at 500 g for 5 min after two washes in binding buffer. Next, the cells were incubated in the dark in 100 μL of 1x binding buffer supplemented with 5 μL of Annexin V-FITC and 10 μL of propidium iodide (PI). Apoptosis was assessed using a flow cytometer (BD). Early-stage (Annexin V+/PI−) and late-stage (Annexin V+/PI+) apoptosis rates were calculated.
Wound healing assay
A wound healing assay was performed to measure the cell migration capability. Briefly, the indicated cells were collected and incubated in serum-free medium overnight for starvation treatment. Then, cells plated in 6-well plates were cultured at 37 °C until reaching 100% confluence, followed by scratching a straight wound onto the surface of the cell monolayer using a sterile pipette. Next, debris on the cellular surface was removed by washing the cells twice with PBS and incubating them for 24 h. Images of migrating cells were acquired 0 and 24 h after the wound using a phase-contrast microscope.
Dual-luciferase reporter assay
StarBase 2.0 (http://starbase.sysu.edu.cn) was adopted to predict target genes of lncRNA GIHCG and miR-499a-5p. To construct lncRNA GIHCG reporter plasmids, a fragment of lncRNA GIHCG with 719 base pairs (NW_003315939) containing the upstream and downstream miR-499a-5p binding sites was amplified. The fragment was cloned into the pmirGLO vector (Promega, Madison, WI, USA) to establish the recombinant luciferase reporter plasmid GIHCG-WT. The same fragments with mutations at the miR-499a-5p binding sites were amplified and cloned into the pmirGLO vector and referred to as GIHCG-MUT. To construct the reporter plasmids for XIAP, the XIAP sequence (NM_001167, 8558 base pairs) containing 300 base pairs upstream and downstream of the 3′-UTR conserved binding site of miR-499a-5p was amplified and cloned into the pmirGLO vector (Promega, Madison, WI, USA), and the recombinant reporter plasmid was called XIAP-WT. The same fragments with mutations in the miR-499a-5p binding sites were amplified and cloned into the pmirGLO vector and called XIAP-MUT. Then, cells were cotransfected with the indicated recombinant luciferase plasmids and miR-499a-5p mimics or mimics NC. Forty-eight hours after cotransfection, luciferase intensity was measured using the Dual-Luciferase Reporter Assay System (Promega).
RNA pull-down assay
Biotin-labelled miR-499a-5p or antisense RNA was transcribed using Biotin RNA Labelling Mix and T7/SP6 RNA polymerase (Roche Diagnostics, USA) and then incubated with RNase-free DNase I (Roche). After purification using the RNeasy Mini Kit (Roche), biotin-labelled RNAs were mixed with extracted Caki-1 and 786-O cell nuclear proteins and subsequently subjected to incubation with streptavidin agarose beads. The enrichment of lncRNA GIHCG was analyzed using qRT–PCR.
RNA immunoprecipitation assay (RIP)
A Magna RIP™ Kit (Millipore, Bedford, MA, USA) was used in the RIP assay. Briefly, cells were lysed in RIP lysis buffer and then incubated overnight with anti-IgG and anti-Ago2 coated on magnetic beads. Then, the magnetic bead-bound complexes were immobilized using a magnet. The precipitated RNAs were quantified and purified and then analyzed using qRT–PCR.
Tumour xenograft and metastasis models
The 45 BALB/c-nude mice (female, 22–24 g, 6 weeks) used in this study were provided by the Animal Experiment Centre of the Chinese Academy of Sciences (Shanghai, China). Animals were randomly divided into three groups (n = 9): control, sh-NC and sh-GIHCG. In the sh-NC and sh-GIHCG groups, 786-O cells were transfected with sh-NC and sh-GIHCG, respectively. The above cell suspension (5 × 106 cells/tumour, 100 μL) was injected subcutaneously into the right flank of mice to perform tumorigenesis assays. In the control group, nude mice were injected with 786-O cells incubated with PBS. Tumour volume was measured every 7 days and calculated using the following formula: tumour volume (mm3) = 0.5 × (width)2 × (length). Mice were euthanized on day 28, and the tumours were excised, weighed and processed for subsequent experiments.
For the metastasis assay, animals were randomly divided into three groups (n = 6): control, sh-NC and sh-GIHCG. 786-O cells (5 × 107 cells/tumour, 200 μL) transfected with sh-GIHCG and sh-NC were injected into mice through the tail vein. In the control groups, nude mice were injected with 786-O cells incubated with PBS. After 4 weeks, the lungs of mice were harvested and fixed in 10% neutral-buffered formalin overnight followed by paraffin embedding. Then, sections were stained with haematoxylin and eosin (Vector Laboratories, Inc., Burlingame, CA, USA). Metastatic foci within mouse lungs were observed and quantified using a light microscope (Olympus Corporation, Tokyo, Japan). All animal experiments were approved by the Animal Ethics Committee of The Second Affiliated Hospital of Nanchang University (No., ncdxsydwfll-2020-26; Date, November15, 2020).
Quantitative real-time PCR (qRT–PCR) assay
Total RNA was extracted from tissues and cell lines using TRIzol reagent (Invitrogen), and RNA quality was assessed using a NanoDrop 2000c (Thermo). Next, the RNAs were converted into cDNAs using a RevertAid First Strand cDNA Synthesis kit (Thermo), and qRT–PCR was conducted on an ABI 7900 system using SYBR Green Real-Time PCR master mixes (Thermo) with the following reaction conditions: 95 °C for 3 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 75 °C for 30 s. GAPDH served as an internal control. Primers were synthesized by RiboBio (Guangzhou, China). The transcriptional levels of targeted genes were analyzed using the 2−ΔΔCt method. The sequences of all primers are as follows:
-
•
LncRNA GIHCG (F): 5′- GCTCCTCACTGTCGCTTCTC −3′.
-
•
LncRNA GIHCG (R): 5′- GCCTGTCTGCTCTGTGTCTG −3′.
-
•
miR-499a-5p (F): 5′- GAGTGTGGCGGGGAATCT −3′.
-
•
miR-499a-5p (R): 5′- GCGTGGTGCAGTCGTGTG −3′.
-
•
XIAP (F): 5′- CATCCATGGCAGATTATGAAGCA −3′.
-
•
XIAP (R): 5′- CTTCACTGGGCTTCCAATCAGTTAG −3′.
-
•
U6 (F): 5′- CTCGCTTCGGCAGCACA −3′.
-
•
U6 (R): 5′- AACGCTTCACGAATTTGCGT −3′.
-
•
GAPDH (F): 5′- CCAGGTGGTCTCCTCTGA −3′.
-
•
GAPDH (R): 5′- GCTGTAGCCAAATCGTTGT −3′.
Western blot
Protein extraction was performed using RIPA buffer (Beyotime). Protein samples were separated by 10% SDS–PAGE and then transferred to polyvinylidene difluoride membranes (Millipore, USA). Next, 5% skim milk (2 h) was used to block the nonspecific binding sites of the membranes. The membranes were subsequently incubated overnight with the indicated primary antibodies (Abcam, USA) against Ki67 (1:5000, ab92742), MMP-9 (1:1000, ab76003), cleaved caspase 9 (1:1000, ab2324), cleaved caspase 3 (1:500, ab2302), cleaved caspase 7 (1:1000, ab256469) and XIAP (1:1000, ab229050). The membranes were washed with PBS three times and then incubated with the corresponding secondary antibodies at 37 °C for 2 h. Signals were visualized using ECL reagent (Millipore), and protein expression was analyzed using ImageJ software 6.0 (National Institutes of Health, USA). β-actin (1:1000, ab8226) was used as an internal control.
Statistical analysis
All experimental data are expressed as the mean ± standard deviation (SD). Statistical analyzes were conducted using SPSS 20.0 (IBM, NY, USA), and all experiments were performed at least three times. Student's t-test was utilized to analyze differences between two groups, and one-way ANOVA with Tukey's post hoc test was performed to evaluate differences among 3 or more groups. A P value less than 0.05 was considered statistically significant.
Results
LncRNA GIHCG promoted the proliferation and migration of RCC cells
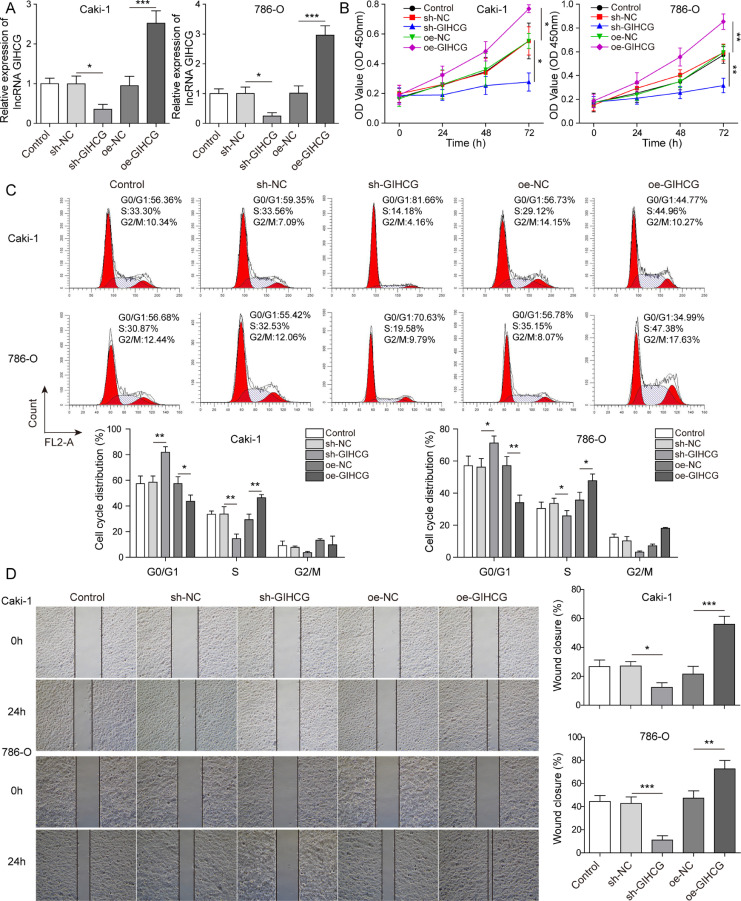
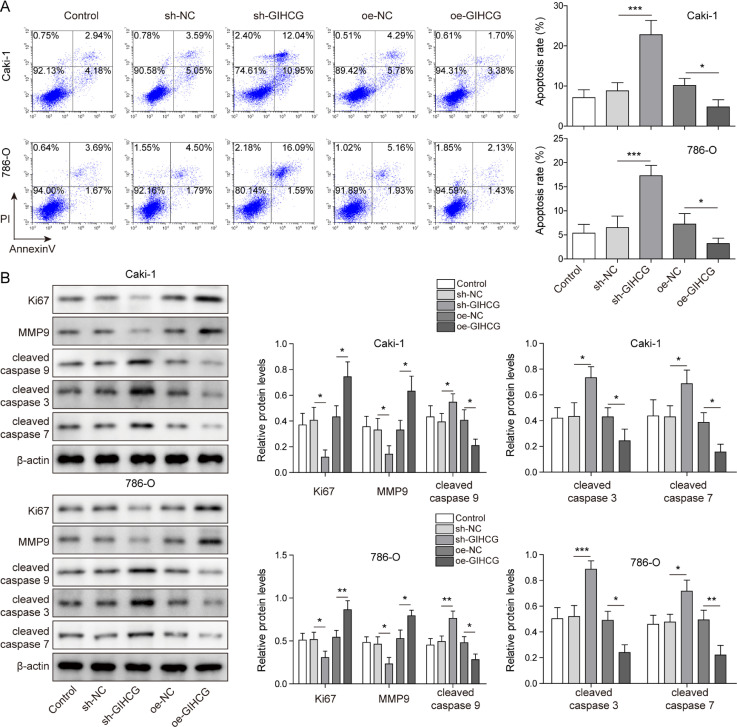
To explore the underlying mechanism of lncRNA GIHCG in RCC, we assessed the expression of lncRNA GIHCG in HK-2 cell and RCC cell lines (Caki-1, 786-O, A498 and SN12C-PM6). The results revealed that lncRNA GIHCG expression was significantly increased in Caki-1 and 786-O cells compared to HK-2 cells (Fig. S1A). Therefore, we silenced and overexpressed the lncRNA GIHCG in Caki-1 and 786-O cells, respectively (Fig. 1A). Knockdown of lncRNA GIHCG inhibited cell proliferation, while overexpression of lncRNA GIHCG enhanced proliferation (Fig. 1B). Meanwhile, silencing of lncRNA GIHCG clearly elevated the cell percentage in G0/G1 phase and reduced that in S and G2/M phase, while lncRNA GIHCG overexpression led to the opposite results, suggesting that lncRNA GIHCG accelerates the cell cycle in RCC cells (Fig. 1C). Additionally, the migration ability of RCC cells was repressed in the sh-GIHCG group, while overexpression of lncRNA GIHCG promoted the migration of RCC cells (Fig. 1D). As shown in Fig. 2A, lncRNA GIHCG silencing increased the apoptotic rate of RCC cells, while lncRNA GIHCG overexpression led to a reduction in apoptosis. After lncRNA GIHCG knockdown, the proliferation-associated protein Ki67 and migration-associated protein MMP-9 were downregulated, while the apoptotic proteins cleaved caspase 9, cleaved caspase 3 and cleaved caspase 7 were upregulated. The opposite results were observed in the oe-GIHCG groups (Fig. 2B). Moreover, as shown in Fig. S1A, lncRNA GIHCG level was upregulated in RCC cells (Caki-1, 786-O, SN12C-PM6 and A498), while we observed the least amount of upregulation in A498 cells. Therefore, we overexpressed GIHCG in A498 cells to further evaluate the impact of lncRNA GIHCG in RCC. The results indicated that overexpression of GIHCG promoted cell proliferation, reduced the percentage of cells in G0/G1 phase and facilitated cell migration (Fig. S1A–D). GIHCG overexpression suppressed apoptosis and upregulated the expression of Ki67 and MMP-9 (Fig. S2A,B). Moreover, there was no significant difference between the oe-GiHCG-MUT group and the oe-NC group (Figs. S1 and S2). These data verified the oncogenic effect of lncRNA GIHCG in RCC.
Fig. 1.
LncRNA GIHCG promoted RCC cell proliferation and migration. Caki-1 and 786-O cells were transfected with sh-NC, sh-GIHCH, oe-NC and oe-GIHCG. (A) LncRNA GIHCG expression was measured using qRT–PCR. (B–D) Cell proliferation, cycle and migration of treated cells were analyzed using CCK-8, flow cytometry and wound healing assays, respectively. The experimental data are shown as the mean ± SD. A–D used one-way ANOVA (Tukey's Post-Hoc Test). P < 0.05, ** P < 0.01, *** P < 0.001.
Fig. 2.
LncRNA GIHCG inhibited the apoptosis of RCC cells. (A) RCC cell apoptosis was analyzed using Annexin-V/-FITC/PI flow cytometry. (B) Proliferation-associated protein Ki67, migration-associated protein MMP-9 and apoptotic protein (cleaved caspase 9, cleaved caspase 3, cleaved caspase 7) levels were measured using western blot. Data are shown as the mean ± SD. A and B used one-way ANOVA (Tukey's Post-Hoc Test). *P < 0.05, ** P < 0.01, *** P < 0.001.
LncRNA GIHCG sponged miR-499a-5p and negatively regulated its expression in RCC cells
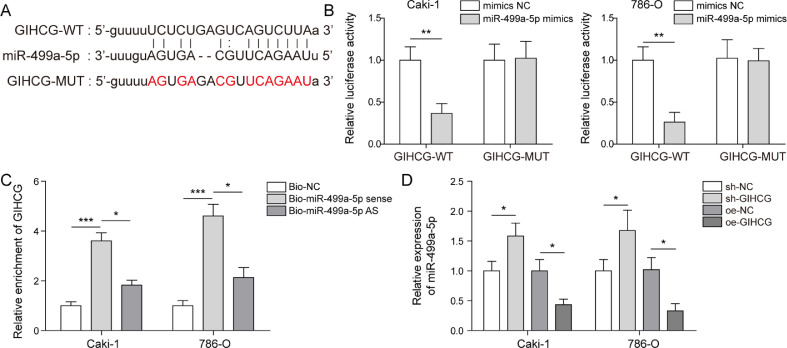
To further investigate the regulatory mechanism of lncRNA GIHCG in RCC cells, starBase was used to predict potential miRNAs that bind to lncRNA GIHCG. The following 5 potential miRNAs were predicted: miR-95-3p, miR-361-5p, miR-499a-5p, miR-345-5p and miR-3611 (Fig. 3A). Next, we assessed expression of these miRNAs in Caki-1 and 786-O cells after knocking down lncRNA GIHCG. The results indicated that expression of miR-499a-5p and miR-361-3p was decreased (Fig. S3). MiR-361–3p has been reported to be a target gene of lncRNA-ARAP1-AS1 in clear cell RCC [21]. Therefore, we chose miR-499a-5p as the research object. Subsequently, the results showed that the luciferase activities of GIHCG-WT were reduced by miR-499a-5p mimics but had no effect on the luciferase activities of GIHCG-MUT (Fig. 3B). LncRNA GIHCG was specifically enriched in miR-499a-5p probe detection compared to the NC and antisense probe groups (Fig. 3C), further confirming the targeted relationship between GIHCG and miR-499a-5p. Moreover, miR-499a-5p expression was increased in response to silencing lncRNA GIHCG, while it was decreased after lncRNA GIHCG overexpression (Fig. 3D). Taken together, our findings suggested that lncRNA GIHCG targeted miR-499a-5p and negatively regulated its expression.
Fig. 3.
LncRNA GIHCG sponges miR-499a-5p and negatively regulates its expression in RCC cells. (A) The binding sites between Five potential miRNAs (miR-95-3p, miR-361-5p, miR-499a-5p, miR-345-5p and miR-3611) and lncRNA GIHCG were predicted by starBase. (B,C) The targeted relationship was further confirmed using dual-luciferase assays and RNA pull-down. (D) Expression of miR-499a-5p was measured using qRT-PCR. Data are shown as the mean ± SD. B used Student's t-test; C and D used one-way ANOVA (Tukey's post hoc test). *P < 0.05, ** P < 0.01, *** P < 0.001.
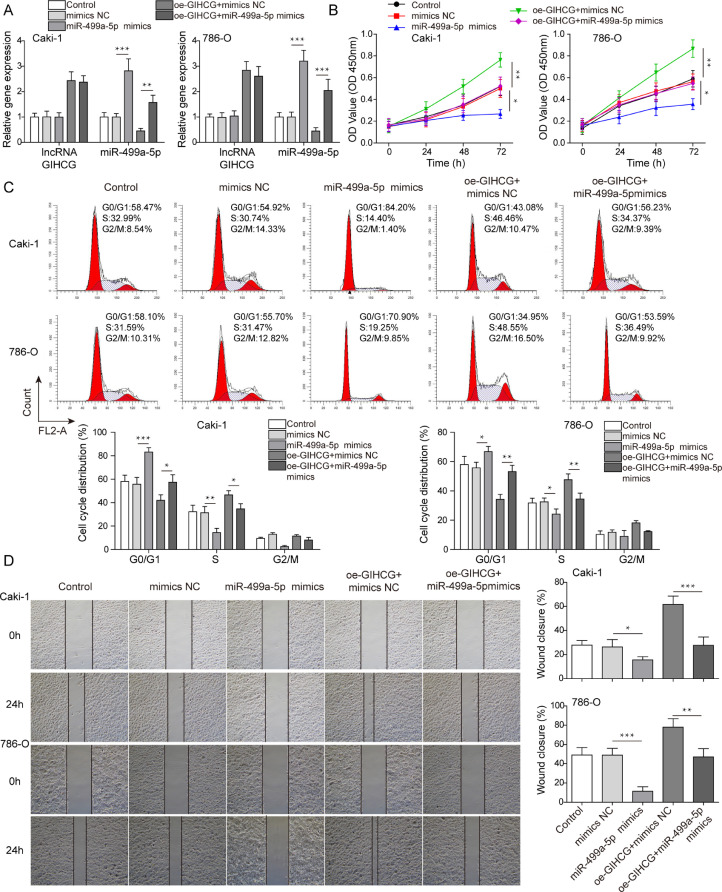
LncRNA GIHCG promoted RCC cell growth and migration by regulating miR-499a-5p
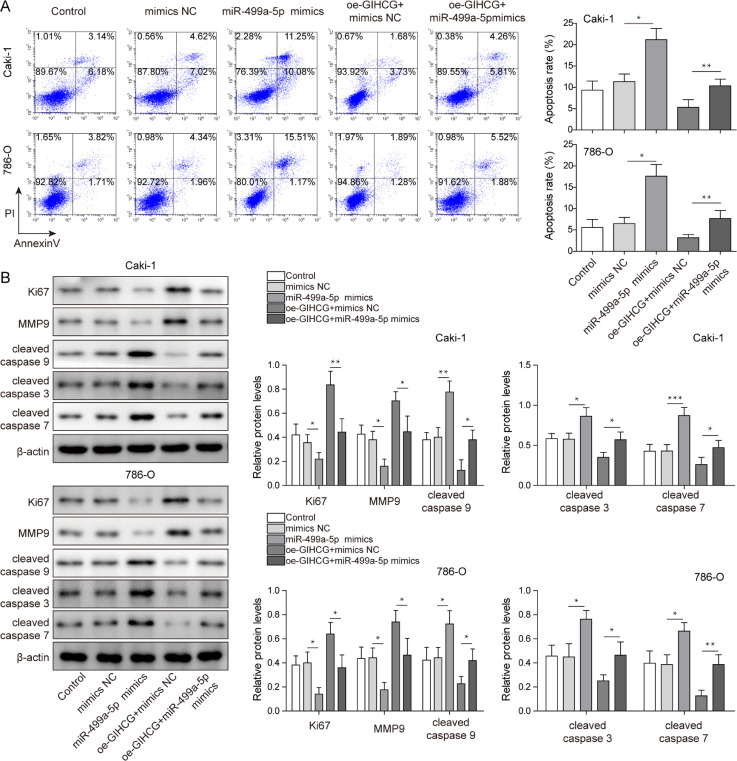
To explore whether lncRNA GIHCG plays a role in RCC by regulating miR-499a-5p expression, miR-499a-5p was successfully overexpressed in RCC cells, and rescue experiments were performed by transfection using the following plasmid combinations: oe-GIHCG + mimics NC and oe-GIHCG + miR-499a-5p mimics. We observed that lncRNA GIHCG expression exhibited no significant change after upregulating miR-499a-5p levels. Moreover, compared to overexpression of lncRNA GIHCG alone, no changes in lncRNA GIHCG expression and increased miR-499a-5p expression were observed after transfection with oe-GIHCG + miR-499a-5p mimics (Fig. 4A). As shown in Fig. 4B, overexpression of miR-499a-5p impeded RCC cell proliferation, and the promoting effect of lncRNA GIHCG overexpression on cell proliferation was blocked by miR-499a-5p overexpression. Cells were arrested in G0/G1 phase in the miR-499a-5p mimics group, and the decreased distribution of G0/G1 phase cells caused by oe-GIHCG was blocked by miR-499a-5p overexpression (Fig. 4C). Similarly, miR-499a-5p overexpression suppressed cell migration and blocked the promoting role of oe-GIHCG on cell migration (Fig. 4D). Furthermore, miR-499a-5p overexpression facilitated cell apoptosis, which weakened the inhibition effect of lncRNA GIHCG overexpression in cell apoptosis (Fig. 5A). Expression of Ki67 and MMP-9 was decreased, and cleaved caspase 9, cleaved caspase 3 and cleaved caspase 7 were increased in the miR-499a-5p mimics group, which reversed the effect of lncRNA GIHCG overexpression on the above proteins (Fig. 5B). Taken together, these findings indicated that lncRNA GIHCG contributed to RCC progression by regulating miR-499a-5p in vitro.
Fig. 4.
LncRNA GIHCG promoted RCC cell proliferation and migration by regulating miR-499a-5p. Caki-1 and 786-O cells were transfected with mimics NC, miR-499a-5p mimics, oe-GIHCG + mimics NC and oe-GIHCG + miR-499a-5p mimics. (A) LncRNA GIHCG and miR-499a-5p levels were analyzed by qRT–PCR. (B-D) The roles of miR-499a-5p enhancement on RCC cell proliferation, cell cycle progression and migration were analyzed using CCK-8, flow cytometry and wound healing assays, respectively. Data are shown as the mean ± SD. A–D used one-way ANOVA (Tukey's Post-Hoc Test). *P < 0.05, ** P < 0.01, *** P < 0.001.
Fig. 5.
LncRNA GIHCG inhibited RCC cell apoptosis by regulating miR-499a-5p. (A) Apoptosis was measured using Annexin-V/-FITC/PI flow cytometry. (B) Protein levels of Ki67, MMP-9, cleaved caspase 9, cleaved caspase 3 and cleaved caspase 7 in RCC cells were measured by western blot. Data are shown as the mean ± SD. A and B used one-way ANOVA (Tukey's Post-Hoc Test). *P < 0.05, ** P < 0.01, *** P < 0.001.
MiR-499a-5p negatively regulated XIAP expression in RCC cells
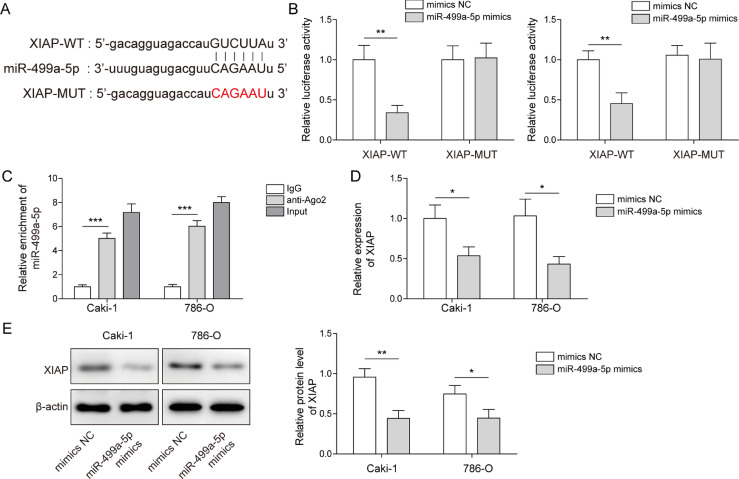
Next, downstream molecules of miR-499a-5p were explored. StarBase analysis indicated the existence of binding sites between miR-499a-5p and XIAP (Fig. 6A). Subsequently, the luciferase activities of XIAP-WT were decreased after miR-499a-5p mimics (Fig. 6B), and the results from RIP assay revealed that compared to IgG, miR-499a-5p was relatively enriched in Ago2 (Fig. 6C). These data further confirmed the interaction between miR-499a-5p and XIAP. In addition, we found that mRNA and protein levels of XIAP were decreased in response to miR-499a-5p overexpression (Fig. 6D,E). Therefore, we concluded that miR-499a-5p directly targeted XIAP and negatively regulated its expression.
Fig. 6.
MiR-499a-5p negatively regulated XIAP expression in RCC cells. (A–C) StarBase, dual-luciferase and RIP assays were used to predict and verify the targeted relationship between miR-499a-5p and XIAP. (D-E) XIAP expression was measured in RCC cells using qRT-PCR and western blot. Data are shown as the mean ± SD. B, D and E used Student's t-test; C used one-way ANOVA (Tukey's post hoc test). *P < 0.05, ** P < 0.01, *** P < 0.001.
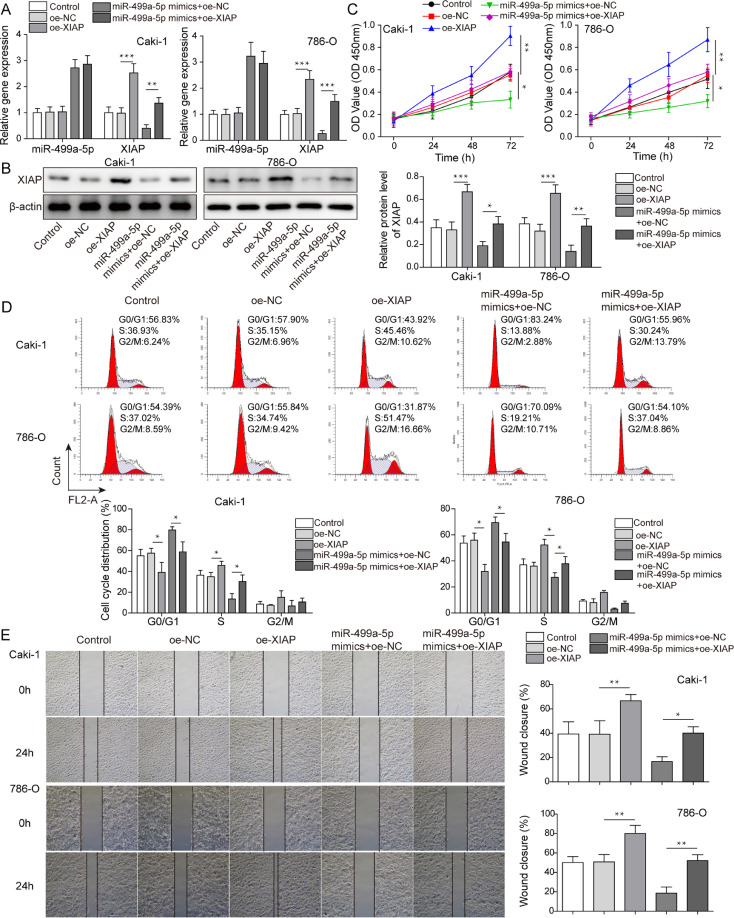
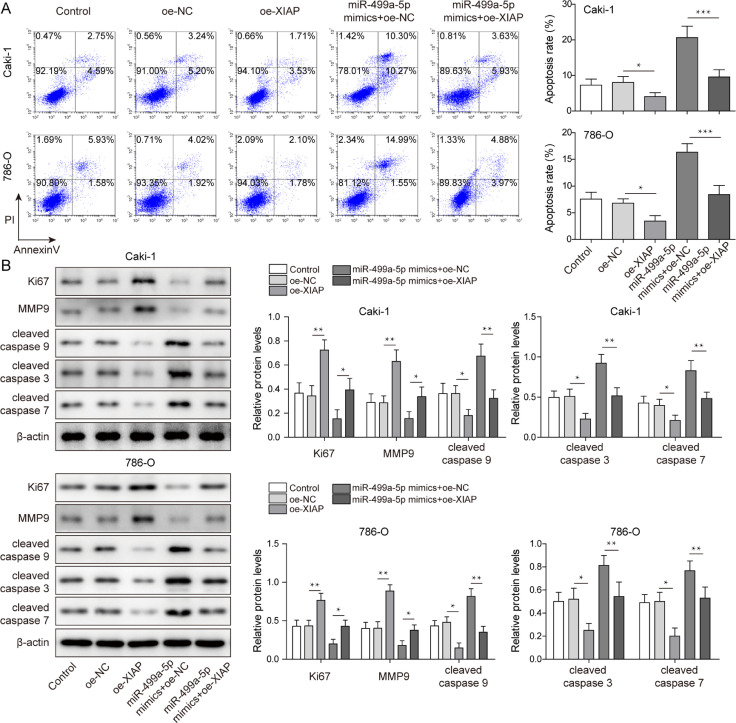
Overexpression of XIAP reversed the effect of miR-499a-5p overexpression on RCC cell growth and migration
To further explore whether RCC cell growth was regulated by the interaction between miR-499a-5p and XIAP, XIAP was successfully overexpressed in vitro to perform rescue assays, and we observed that XIAP overexpression did not change miR-499a-5p levels (Fig. 7A,B). XIAP overexpression enhanced cell proliferation and reversed the inhibition of proliferation caused by miR-499a-5p overexpression in vitro (Fig. 7C). Overexpression of XIAP decreased the proportion of G0/G1 phase cells while increasing the proportion of S phase cells, which blocked the inhibition of the cell cycle caused by miR-499a-5p mimics (Fig. 7D). The migrationcapability of RCC cells was strengthened by XIAP overexpression, which reversed the reduced migration capability caused by miR-499a-5p mimics (Fig. 7E). In contrast, apoptosis was suppressed in the oe-XIAP group, with XIAP overexpression blocking the proapoptotic effect of miR-499a-5p overexpression (Fig. 8A). As shown in Fig. 8B, Ki67 and MMP-9 were upregulated, while cleaved caspase 9, cleaved caspase 3 and cleaved caspase 7 were downregulated in the oe-XIAP group, which reversed the effect of miR-499a-5p overexpression on the above proteins. In conclusion, miR-499a-5p regulated RCC cell growth and migration by regulating XIAP.
Fig. 7.
Overexpression of XIAP reversed the effect of miR-499a-5p overexpression on the growth and migration of RCC cells. Caki-1 and 786-O cells were transfected with oe-NC, oe-XIAP, miR-499a-5p mimics + oe-NC and miR-499a-5p mimics + oe-XIAP. (A,B) MiR-499a-5p and XIAP levels were measured by qRT–PCR and western blot. (C–E) RCC cell proliferation, cell cycle progression and migration were measured after enhancing miR-499a-5p and XIAP expression by CCK-8, flow cytometry and wound healing assays, respectively. Data are shown as the mean ± SD. A-E used one-way ANOVA (Tukey's Post-Hoc Test). *P < 0.05, ** P < 0.01, *** P < 0.001.
Fig. 8.
Overexpression of XIAP reversed the effect of miR-499a-5p overexpression on the apoptosis of RCC cells. (A) Apoptosis was analyzed by Annexin-V/-FITC/PI flow cytometry. (B) Protein levels of Ki67, MMP-9, cleaved caspase 9, cleaved caspase 3 and cleaved caspase 7 in RCC cells were measured by western blot. Data are shown as the mean ± SD. A and B used one-way ANOVA (Tukey's Post-Hoc Test). *P < 0.05, ** P < 0.01, *** P < 0.001.
Knockdown of lncRNA GIHCG inhibited the development of RCC in nude mice
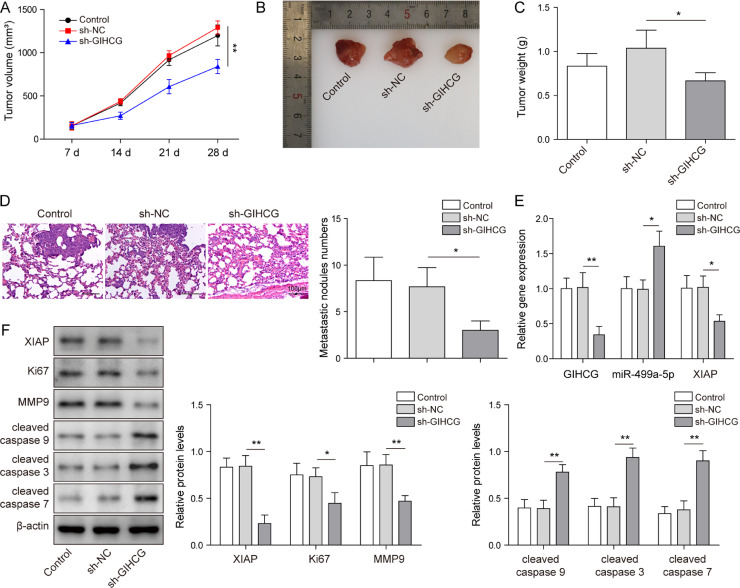
We next further examined whether the effects of lncRNA GIHCG are retained in vivo. According to previous studies, 786-O cells can be used for subcutaneous tumorigenesis in nude mice [22,23]. Briefly, 786-O cells stably expressing sh-NC or sh-GIHCG were injected into nude mice; the control group was comprised of nude mice injected with 786-O cells incubated in PBS. We observed that compared to the control and sh-NC groups, tumour volume and weight exhibited significant growth inhibition in response to silencing lncRNA GIHCG in vivo (Fig. 9A–C). Next, cells treated with control (incubated with PBS), sh-NC or sh-GIHCG were injected into nude mice through the tail vein to establish the lung metastasis model. Subsequently, HE staining results showed that the formation of metastasis was reduced after GIHCG knockdown (Fig. 9D). After lncRNA GIHCG knockdown, XIAP expression was decreased, while miR-499a-5p expression was increased in RCC tumour tissues (Fig. 9E). Furthermore, Ki67 and MMP-9 were downregulated, while cleaved caspase 9, cleaved caspase 3 and cleaved caspase 7 were upregulated in RCC tumour tissues after lncRNA GIHCG silencing (Fig. 9F). Taken together, these findings indicated that lncRNA GIHCG participated in RCC in vivo via the miR-499a-5p/XIAP axis.
Fig. 9.
Knockdown of lncRNA GIHCG inhibited the development of RCC in nude mice. 786-O cells stably expressing sh-GIHCG or sh-NC were injected into the right flank or tail vein of nude mice to establish mouse xenograft or tumour metastasis models. (A–C) The tumour volume, images of tumours and tumour weight were observed and analyzed, N = 9. (D) HE staining was used to evaluate the effect of lncRNA GIHCG knockdown on lung metastasis in nude mice, N = 6. (E) LncRNA GIHCG, miR-499a-5p and XIAP levels in RCC tumour tissues were detected by qRT–PCR, N = 3. (F) Protein levels of Ki67, MMP-9, cleaved caspase 9, cleaved caspase 3 and cleaved caspase 7 in RCC tumour tissues were measured using western blot, N = 3. Data are shown as the mean ± SD. A, C, D, E and F used one-way ANOVA (Tukey's post hoc test). *P < 0.05, ** P < 0.01, *** P < 0.001).
Discussion
The present study is the first direct exploration of the biological function of lncRNA GIHCG in RCC. We observed that lncRNA GIHCG facilitates cell proliferation and migration in vitro and in vivo while inhibiting apoptosis. Mechanistically, lncRNA GIHCG sponges miR-499a-5p and negatively regulates its expression in RCC cells. Moreover, miR-499a-5p targets XIAP and downregulates its levels. Based on these findings, we concluded that lncRNA GIHCG promotes RCC progression by upregulating XIAP through targeting miR-499a-5p.
Accumulating research suggests that lncRNAs play a vital role in RCC carcinogenesis [24], [25], [26]. LncRNA GIHCG has been reported to be overexpressed and to promote proliferation and migration in a variety of tumours, such as breast cancer [27], colorectal cancer [28] and gastric cancer [29]. Similarly, our previous study demonstrated that lncRNA GIHCG is upregulated in RCC, and knockdown of lncRNA GIHCG represses cell proliferation and migration [8]. Based on these findings, we further verified that lncRNA GIHCG silencing displays the same tumour-suppressive effects in vivo and that lncRNA GIHCG overexpression accelerates RCC cell proliferation and migration, demonstrating that lncRNA GIHCG functions as an oncogene in vitro and in vivo and participates in RCC progression. MiR-499a-5p plays an anticancer role in cervical cancer [30], osteosarcoma [31] and non-small cell lung cancer [14]. Nevertheless, the mechanism of miR-499a-5p in RCC remains unclear. Our data demonstrated for the first time that lncRNA GIHCG negatively regulates miR-499a-5p expression, and overexpression of miR-499a-5p suppresses RCC cell growth and migration, which reversed the promoting effect on RCC cells induced by lncRNA GIHCG overexpression. Therefore, we concluded that lncRNA GIHCG might promote the growth and migration of RCC by regulating miR-499a-5p.
Proteins from the IAP (inhibitors of apoptosis) family suppress caspases and promote cell survival, especially XIAP, which plays the most potent antiapoptotic role during apoptosis [32,33]. Yang et al. demonstrated that XIAP silencing enhances the sensitivity of RCC cells to apoptosis, indicating that XIAP is directly involved in the growth of RCC cells [34]. Furthermore, XIAP is involved in cancer progression as a downstream gene of miRNAs. For example, miR-142–5p has been reported to sensitize ovarian cancer cells to cisplatin-induced apoptosis by inhibiting the levels of XIAP [35]. We identified binding sites between miR-499a-5p and XIAP, and the targeted relationship between these factors was verified for the first time in this study. Functional experiments revealed that XIAP overexpression blocks the reduction in cell proliferation and migration and the enhancement of apoptosis induced by miR-499a-5p mimics, demonstrating that miR-499a-5p regulates RCC cell growth by targeting XIAP. Furthermore, we observed that lncRNA GIHCG silencing increases miR-499a-5p levels while decreasing XIAP levels in RCC tumour tissues, suggesting that the lncRNA CIHCG/miR-499a-5p/XIAP axis might also be involved in the progression of RCC in vivo.
Conclusion
In summary, our findings demonstrated that lncRNA GIHCG promotes the growth and migration of RCC by upregulating XIAP expression via miR-499a-5p. These data illustrate the therapeutic potential of lncRNA GIHCG in RCC treatment.
Fig. S1. GIHCG overexpression promoted the proliferation and migration of A498 cells (A) LncRNA GIHCG expression was measured by qRT–PCR in RCC cells (A498, Caki-1, 786-O and SN12C-PM6). A498 cells were treated with control, oe-NC, oe-GIHCG and oe-GIHCG-MUT. LncRNA GIHCG expression was measured by qRT–PCR. (B-D) Cell proliferation, cycle, and migration of cells were analyzed using CCK-8, flow cytometry and wound healing assays, respectively. Data are shown as the mean ± SD. A-D used one-way ANOVA (Tukey's post hoc test). *P < 0.05, ** P < 0.01, *** P < 0.001.
Fig. S2. GIHCG overexpression inhibited the apoptosis of RCC cells. RCC cell apoptosis was analyzed using Annexin-V/-FITC/PI flow cytometry. (B) Proliferation-associated protein Ki67, migration-associated protein MMP-9 and apoptotic protein (cleaved caspase 9, cleaved caspase 3, cleaved caspase 7) levels were measured using western blot. Data are shown as the mean ± SD. A and B used one-way ANOVA (Tukey's post hoc test). *P < 0.05, ** P < 0.01, *** P < 0.001.
Fig. S3. MiR-95–3p, miR-361–5p, miR-499a-5p, miR-345–5p and miR-3611 levels in Caki-1 and 786-O cells were measured by qRT–PCR after silencing lncRNA GIHCG. Data are shown as the mean ± SD. S3 used one-way ANOVA (Tukey's post hoc test). *P < 0.05, ** P < 0.01, *** P < 0.001.
CRediT authorship contribution statement
Shu-Ying Zhu: Conceptualization, Methodology, Formal analysis, Writing – original draft. Hong-Chang Zou: Project administration. Ming-Ming Gao: Data curation, Resources. Yan-Xia Chen: Supervision. Man Xu: Investigation, Software, Visualization. Xiao-Hua Qin: Funding acquisition, Validation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We would like to give our sincere gratitude to the reviewers for their constructive comments.
Funding
This work was supported by Natural Science Foundation of Jiangxi Province (20202BABL206030).
Ethical approval
All animal experiments were approved by the Animal Ethics Committee of The Second Affiliated Hospital of Nanchang University.
Consent for publication
Not Applicable. This article does not contain any studies with human participants performed by any of the authors.
Availability of data and material
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101356.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J. Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. 2016. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J. Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. 2019. [DOI] [PubMed] [Google Scholar]
- 3.Jonasch E., Gao J., Rathmell W.K. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deleuze A., et al. Immunotherapy in renal cell carcinoma: the future is now. Int. J. Mol. Sci. 2020;21(7) doi: 10.3390/ijms21072532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 6.Gutschner T., Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao W., et al. LncRNA GIHCG promotes the development of esophageal cancer by modulating miR-29b-3p/ANO1 axis. Onco Targets Ther. 2020;13:13387–13400. doi: 10.2147/OTT.S282348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He Z.H., et al. Long noncoding RNA GIHCG is a potential diagnostic and prognostic biomarker and therapeutic target for renal cell carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2018;22(1):46–54. doi: 10.26355/eurrev_201801_14099. [DOI] [PubMed] [Google Scholar]
- 9.Sassen S., Miska E.A., Caldas C. MicroRNA: implications for cancer. Virchows Arch. 2008;452(1):1–10. doi: 10.1007/s00428-007-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y.S., Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang L., et al. Long noncoding RNA (lncRNA)-mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int. J. Mol. Sci. 2019;20(22) doi: 10.3390/ijms20225758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu L., et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29(5):653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Jing L., et al. Exosomal miR-499a-5p inhibits endometrial cancer growth and metastasis via targeting VAV3. Cancer Manag. Res. 2020;12:13541–13552. doi: 10.2147/CMAR.S283747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao L., et al. Downregulation of miR-499a-5p predicts a poor prognosis of patients with non-small cell lung cancer and restrains the tumorigenesis by targeting fibroblast growth factor 9. Technol. Cancer Res. Treat. 2020;19 doi: 10.1177/1533033820957001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zong Z., et al. Knockdown of LncRNA SCAMP1 suppressed malignant biological behaviors of glioma cells via modulating miR-499a-5p/LMX1A/NLRC5 pathway. J. Cell. Mol. Med. 2019;23(8):5048–5062. doi: 10.1111/jcmm.14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Y., et al. XIAP overexpression promotes bladder cancer invasion in vitro and lung metastasis in vivo via enhancing nucleolin-mediated Rho-GDIbeta mRNA stability. Int. J. Cancer. 2018;142(10):2040–2055. doi: 10.1002/ijc.31223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X.B., et al. MiR-142 inhibits lung cancer cell proliferation and promotes apoptosis by targeting XIAP. Eur. Rev. Med. Pharmacol. Sci. 2019;23(17):7430–7437. doi: 10.26355/eurrev_201909_18852. [DOI] [PubMed] [Google Scholar]
- 18.Mizutani Y., et al. Overexpression of XIAP expression in renal cell carcinoma predicts a worse prognosis. Int. J. Oncol. 2007;30(4):919–925. [PubMed] [Google Scholar]
- 19.Chen C., et al. Potential biological process of X-linked inhibitor of apoptosis protein in renal cell carcinoma based upon differential protein expression analysis. Oncol. Lett. 2018;15(1):821–832. doi: 10.3892/ol.2017.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu C., et al. MicroRNA-212 inhibits the proliferation, migration and invasion of renal cell carcinoma by targeting X-linked inhibitor of apoptosis protein (XIAP) Oncotarget. 2017;8(54):92119–92133. doi: 10.18632/oncotarget.20786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong L., Zhong X. Long non-coding RNA ARAP1-AS1 contributes to cell proliferation and migration in clear cell renal cell carcinoma via the miR-361-3p/placental growth factor axis. Bioengineered. 2021;12(1):6629–6642. doi: 10.1080/21655979.2021.1975019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Z., et al. lncRNA MSC-AS1 activates Wnt/beta-catenin signaling pathway to modulate cell proliferation and migration in kidney renal clear cell carcinoma via miR-3924/WNT5A. J. Cell. Biochem. 2020;121(10):4085–4093. doi: 10.1002/jcb.29594. [DOI] [PubMed] [Google Scholar]
- 23.Zhai X., et al. Long noncoding RNA LINC01133 promotes the malignant behaviors of renal cell carcinoma by regulating the miR-30b-5p/Rab3D axis. Cell Transplant. 2020;29 doi: 10.1177/0963689720964413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton M.J., et al. HOTAIRM1 lncRNA is downregulated in clear cell renal cell carcinoma and inhibits the hypoxia pathway. Cancer Lett. 2020;472:50–58. doi: 10.1016/j.canlet.2019.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michal F., et al. Long non-coding RNAs and renal cell carcinoma. Klin. Onkol. 2020;33(5):340–349. doi: 10.14735/amko2020340. [DOI] [PubMed] [Google Scholar]
- 26.Liu S., et al. LncRNA RP11-567G11.1 accelerates the proliferation and invasion of renal cell carcinoma through activating the notch pathway. Eur. Rev. Med. Pharmacol. Sci. 2020;24(9):4738–4744. doi: 10.26355/eurrev_202005_21162. [DOI] [PubMed] [Google Scholar]
- 27.Fan L.Y., et al. LncRNA GIHCG regulates microRNA-1281 and promotes malignant progression of breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2019;23(24):10842–10850. doi: 10.26355/eurrev_201912_19788. [DOI] [PubMed] [Google Scholar]
- 28.Jiang X., et al. Long noncoding RNA GIHCG induces cancer progression and chemoresistance and indicates poor prognosis in colorectal cancer. Onco Targets Ther. 2019;12:1059–1070. doi: 10.2147/OTT.S192290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu G., et al. Lnc-GIHCG promotes cell proliferation and migration in gastric cancer through miR- 1281 adsorption. Mol. Genet. Genom. Med. 2019;7(6):e711. doi: 10.1002/mgg3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gu X., et al. MiR-499a-5p inhibits proliferation, invasion, migration, and epithelial-mesenchymal transition, and enhances radiosensitivity of cervical cancer cells via targeting eIF4E. Onco Targets Ther. 2020;13:2913–2924. doi: 10.2147/OTT.S241631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J., et al. MicroRNA-499a-5p inhibits osteosarcoma cell proliferation and differentiation by targeting protein phosphatase 1D through protein kinase B/glycogen synthase kinase 3beta signaling. Oncol. Lett. 2018;15(4):4113–4120. doi: 10.3892/ol.2018.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silke J., Meier P. Inhibitor of apoptosis (IAP) proteins-modulators of cell death and inflammation. Cold Spring Harb. Perspect. Biol. 2013;5(2) doi: 10.1101/cshperspect.a008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki Y., et al. X-linked inhibitor of apoptosis protein (XIAP) inhibits caspase-3 and -7 in distinct modes. J. Biol. Chem. 2001;276(29):27058–27063. doi: 10.1074/jbc.M102415200. [DOI] [PubMed] [Google Scholar]
- 34.Yang W.Z., Zhou H., Yan Y. XIAP underlies apoptosis resistance of renal cell carcinoma cells. Mol. Med. Rep. 2018;17(1):125–130. doi: 10.3892/mmr.2017.7925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X., et al. miR-142-5p enhances cisplatin-induced apoptosis in ovarian cancer cells by targeting multiple anti-apoptotic genes. Biochem. Pharmacol. 2019;161:98–112. doi: 10.1016/j.bcp.2019.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.