Abstract
Objectives
To investigate the diagnostic performance of the coronavirus disease 2019 (COVID-19) Reporting and Data System (CO-RADS) for detecting COVID-19.
Methods
We searched PubMed, EMBASE, MEDLINE, Web of Science, Cochrane Library, and Scopus database until September 21, 2021. Statistical analysis included data pooling, forest plot construction, heterogeneity testing, meta-regression, and subgroup analyses.
Results
We included 24 studies with 8382 patients. The pooled sensitivity and specificity and the area under the curve (AUC) of CO-RADS ≥ 3 for detecting COVID-19 were 0.89 (95% confidence interval (CI) 0.85–0.93), 0.68 (95% CI 0.60–0.75), and 0.87 (95% CI 0.84–0.90), respectively. The pooled sensitivity and specificity and AUC of CO-RADS ≥ 4 were 0.83 (95% CI 0.79–0.87), 0.84 (95% CI 0.78–0.88), and 0.90 (95% CI 0.87–0.92), respectively. Cochran’s Q test (p < 0.01) and Higgins I2 heterogeneity index revealed considerable heterogeneity. Studies with both symptomatic and asymptomatic patients had higher specificity than those with only symptomatic patients using CO-RADS ≥ 3 and CO-RADS ≥ 4. Using CO-RADS ≥ 4, studies with participants aged < 60 years had higher sensitivity (0.88 vs. 0.80, p = 0.02) and lower specificity (0.77 vs. 0.87, p = 0.01) than studies with participants aged > 60 years.
Conclusions
CO-RADS has favorable performance in detecting COVID-19. CO-RADS ≥ 3/4 might be applied as cutoff values given their high sensitivity and specificity. However, there is a need for more well-designed studies on CO-RADS.
Key Points
• CO-RADS shows a favorable performance in detecting COVID-19.
• CO-RADS ≥ 3 had a high sensitivity 0.89 (95% CI 0.85–0.93), and it may prove advantageous in screening the potentially infected people to prevent the spread of COVID-19.
• CO-RADS ≥ 4 had high specificity 0.84 (95% CI 0.78–0.88) and may be more suitable for definite diagnosis of COVID-19.
Keywords: CO-RADS, COVID-19, Diagnosis, Sensitivity, Specificity
Introduction
The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a global threat [1–3]. Early and quick diagnosis of COVID-19 is crucial for isolating patients, preventing infection spread, and early patient intervention [4].
Currently, the reference standard for COVID-19 is the reverse transcription polymerase chain reaction (RT-PCR) test using respiratory tract specimens [5]. However, RT-PCR tests have limited sensitivity depending on symptom duration [6], viral load [7, 8], test sample types [9], delays in test access or processing times [10], and insufficient testing capacity [11].
Computed tomography (CT) is a widely available, time-saving, and non-invasive approach for detecting COVID-19 [12]. It could be an efficient tool for diagnosing COVID-19 with high sensitivity and monitoring disease course [10, 13–15]. Accordingly, it has been recommended for identifying patients with COVID-19 [16–18]. Nevertheless, the use of CT scanning for diagnosing COVID-19 given the lack of accurate, consistent evaluation across radiologists and medical centers is controversial [19]. There has been heterogeneity among previous studies on chest CT for detecting COVID-19, with sensitivity and specificity of 0.19 to 1.00 [20] and 0.25 to 0.70 [21], respectively. This was mainly attributed to the lack of a specific reporting system in various centers with different radiologists [20–23].
The COVID-19 Reporting and Data System (CO-RADS) was established by the Dutch Radiological Society to standardize the assessment scheme and simplify reporting using a 5-point scale of suspicion (from 1 [very low suspicion] to 5 [very high suspicion]) for COVID-19 pneumonia on chest CT images [24]. CO-RADS has excellent diagnostic performance for COVID-19 through interpretation of chest CT images, with moderate to substantial interobserver agreement [24–26]. A meta-analysis on the diagnostic performance of CO-RADS in detecting COVID-19 [27] only included six studies up to August 24, 2020, with a small sample size. This systematic review and meta-analysis aimed to provide an updated overview of the diagnostic performance of CO-RADS in COVID-19, with a focus on the possible practical application of CO-RADS.
Materials and methods
This meta-analysis was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analysis [28], with a standardized review and data extraction protocol. The primary study outcome was the diagnostic performance of CO-RADS for COVID-19. Additionally, we discussed the clinical application of CO-RADS.
Search strategy and selection criteria
A computerized search of PubMed, EMBASE, MEDLINE, Web of Science, Cochrane Library, and Scopus was performed up to September 21, 2021, without restrictions regarding regions or publication types. We manually reviewed the reference lists of all retrieved studies, review articles, and conference abstracts to identify additional potential studies. We used the following search terms: ([COVID-19 reporting-and-data system] or [COVID-RADS] or [CO-RADS]) AND ([COVID-19] or [SARS-CoV-2] or [coronavirus disease 2019] or [severe acute respiratory syndrome coronavirus 2]).
Inclusion and exclusion criteria
The inclusion criteria were (1) studies using CO-RADS in COVID-19 diagnosis, (2) studies using RT-PCR as the reference standard, (3) original scientific articles, and (4) sufficient data for reconstructing the 2 × 2 tables for diagnostic accuracy. The exclusion criteria were (1) duplication of publications; (2) non-original studies, including editorials, letters to the editor, review articles, and case reports; (3) non-English articles; and (4) studies with insufficient information for reconstructing a 2 × 2 table.
Data extraction
We extracted data from the included studies using a standardized form: (1) true positives, false positives, true negatives, false negatives; (2) study characteristics, including first author, publication year, country, study design, cutoff values, study duration, recruitment, number of centers, number of readers, setting, and sample size; and (3) patient characteristics including age, sex ratio, and symptoms. When different readers in a single study reported more than one result, we extracted the consensus result or the average result if there was no consensus result.
Quality assessment
The methodological quality of the studies was assessed using the Quality Assessment of Diagnostic Accuracy Studies-2 tool [29]. Each study was rated with respect to the following domains: patient selection, index test, reference standard, and flow and timing. Each domain was assessed based on the risk of bias and the first three domains, including applicability.
Statistical analysis
We created 2 × 2 tables to calculate the pooled sensitivity, specificity, and corresponding 95% confidence interval (CI) using a bivariate random-effects model; further, we calculated the diagnostic odds ratio (DOR). The results were graphically shown in the forest plots. We constructed hierarchical summary receiver operating characteristic (HSROC) models; further, we calculated the area under the curve (AUC). We performed Deeks’ funnel plot asymmetry test to evaluate the possible presence of publication bias, with p < 0.10 indicating a possibility of publication bias [30]. The heterogeneity of the included studies was evaluated using the inconsistency index (I2) and Q statistic of the chi-square value test [31].
Heterogeneity was further explored through meta-regression by adding the following covariates to bivariate model: (1) study design (prospective vs. retrospective), (2) study enrollment (consecutive vs. unavailable, NA), (3) number of centers (< 2 vs. ≥ 2), (4) number of readers (< 3 vs. ≥ 3), (5) study setting (emergency department, ED vs. NA), and (6) publication year (2020 vs. 2021). Finally, we performed subgroup analyses according to the age, sex, and symptoms.
Statistical analyses were performed using STATA 16.1 and RevMan 5.3. Statistical significance was set at p < 0.05.
Results
Selection and data extraction
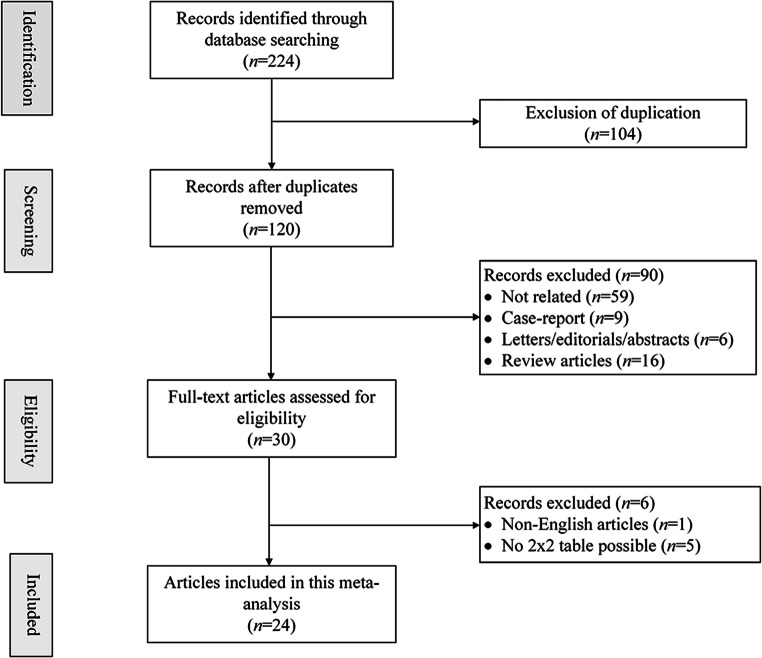
The systematic search initially yielded 224 results; among them, 104 were removed for duplication. After manually screening of titles and abstracts, 90 results were excluded, including articles not related (n = 59), case reports (n = 9), letters/editorials/abstracts (n = 6), and review articles (n = 16). After full-text reviews of the remaining 30 articles, we excluded one non-English article [32] and five articles without sufficient data for constructing 2 × 2 tables for lacking information on specificity of CO-RADS [33–37]. Finally, we included 24 studies with 8382 patients [12, 24, 25, 38–58] (Fig. 1).
Figure 1.
PRISMA flow chart of article selection process
Characteristics of included studies
The 24 included studies comprised 8382 patients. More than half of studies were published in 2021. The studies were conducted in various countries, including Italy, French, the Netherlands, and China. There were 3 and 17 prospective and retrospective studies, respectively, with the remaining 4 studies not providing information on study design. Fourteen studies consecutively enrolled participants, while the remaining 10 studies did not describe participant enrollment. Among the 24 studies, over half were single-center studies while six were multi-center studies and three studies did not provide relevant information. Regarding the number of readers, 12 and 4 studies included ≥ 3 and < 3 radiologists, respectively, while 8 studies lacked information. Ten studies were conducted in the ED. Five studies included both symptomatic and asymptomatic patients while 19 studies only included symptomatic patients. The sample size significantly varied from 40 to 1997, with all studies recruiting both male and female participants. Five studies did not report the age of all included patients; nonetheless, the age of patients ranged from 8 to 99 years. Moreover, all included studies used RT-PCR as the standard reference for COVID-19 (Table 1).
Table 1.
characteristics of 16 included studies
| First author | Year | Country | Study design | Cutoff value | Study duration | Patient recruitment | Number of centers | Number of readers | Setting | Symptoms | Sample size | Age* (year) |
Sex (M/F) |
Reference standard |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Turcato [38] | 2021 | Italy | R | 2, 3, 4, 5 | 2020/3/25–2020/4/25 | C | 1 | NA | ED | S | 120 | 68 (60–78) | 73/47 |
51 RT-PCR + 69 RT-PCR - |
| Sun [39] | 2021 | French | R | 2, 3, 4, 5 | 2020/3/18–2020/5/10 | NA | 1 | 1 | NA | B | 313 | 62 (50–70) | 156/157 |
22 RT-PCR + 291 RT-PCR - |
| Schalekamp [40] | 2021 | Netherlands | R | 2, 3, 4, 5 | 2020/3/20–2020/4/3 | C | 6 | NA | ED | S | 1070 | 66 (54–75) | 626/444 |
536 RT-PCR + 534 RT-PCR - |
| O' Neill [41] | 2020 | Canada, Iran, South Korea, China | R | 2, 3, 4, 5 | 2020/2/2–2020/5/16 | C | 4 | 3 | NA | S | 279 | 60.1 ± 17.3 | 143/136 |
194 RT-PCR + 85 RT-PCR - |
| Prokop [24] | 2020 | Netherlands | NA | 2, 3, 4, 5 | 2020/3/14–2020/3/25 | C | 7 | 8 | ED | S | 105 | 62 ± 16 | 61/44 |
53 RT-PCR + 52 RT-PCR - |
| Özel [42] | 2021 | Turkey | R | 2, 3, 4, 5 | 2020/3/20–2020/4/20 | NA | 1 | 2 | ED | S | 280 | 45.9 ± 15.9 | 150/130 |
111 RT-PCR + 169 RT-PCR - |
| Ossareh [43] | 2020 | Iran | NA | 2 | 2020/2/20–2020/4/20 | NA | 1 | NA | NA | B | 178 | 58.9 ± 16.5 (22–99) | 122/56 |
27 RT-PCR + 151 RT-PCR - |
| Liu [12] | 2021 | China | R | 4 | 2020/2/16–2020/3/20 | C | 2 | 2 | NA | S | 40 | NA | 23/17 |
25 RT-PCR + 25 RT-PCR - |
| Lieveld [44] | 2020 | Netherlands | P | 2, 3, 4, 5 | 2020/3/19–2020/5/28 | C | 2 | NA | ED | S | 741 | 62.1 ± 17.2 | 417/324 |
235 RT-PCR + 506 RT-PCR - |
| Lessmann [45] | 2021 | Netherlands | R | 2, 3, 4, 5 | 2020/3–2020/4 | C | 2 | 8 | ED | S | 262 | 64 ± 16 | 154/108 |
179 RT-PCR + 83 RT-PCR - |
| Hermans [46] | 2020 | Netherlands | P | 2, 3, 4, 5 | 2020/3/27–2020/4/20 | C | 1 | NA | ED | B | 319 | 59 (50–68) | 157/162 |
133 RT-PCR + 186 RT-PCR - |
| Gross [47] | 2021 | Germany | R | 2, 3, 4, 5 | 2020/3/22–2020/4/7 | C | NA | NA | ED | S | 96 | 64 | 50/46 |
20 RT-PCR + 76 RT-PCR - |
| Fujioka [48] | 2020 | Japan | R | 2, 3, 4, 5 | 2020/1–2020/6 | NA | 1 | 4 | NA | S | 154 | 61.3 ± 18.8 (21–93) | 101/53 |
76 RT-PCR + 78 RT-PCR - |
| De Smet [25] | 2021 | Belgium | P | 2, 3, 4, 5 | 202/3/19–2020/4/20 | C | 1 | 2 | NA | B | 1997 | 70 (52–81) | 1031/966 |
418 RT-PCR + 1579 RT-PCR- |
| Bellini [49] | 2020 | Italy | R | 2, 3, 4, 5 | 2020/3/9–2020/5/3 | C | 1 | 12 | NA | S | 572 | 63 ± 20 | 329/243 |
142 RT-PCR+ 430 RT-PCR- |
| Shohei [50] | 2020 | Japan | R | 2, 3, 4, 5 | 2020/1/30–2020/6/30 | NA | 1 | 8 | ED | S | 100 | NA | 68/32 |
50 RT-PCR+ 50 RT-PCR- |
| Wakfie [51] | 2021 | Spain | NA | 4, 5 | 2020/2/19–2020/5/29 | NA | NA | NA | NA | B | 41 | NA | 22/19 |
13 RT-PCR+ 28 RT-PCR- |
| Vicini [52] | 2021 | Italy | R | 2, 3, 4, 5 | 2020/3/18–2020/7/15 | C | 1 | 6 | NA | S | 714 | 64 ± 19 (8–96) | 419/295 |
263 RT-PCR+ 451 RT-PCR- |
| Van [53] | 2021 | Belgium | R | 2, 3, 4, 5 | 2020/3/7–2020/4/13 | NA | 1 | NA | NA | S | 200 | 67.30 ± 17.11 | 96/104 |
68 RT-PCR+ 132 RT-PCR- |
| Nair [54] | 2021 | Qatar | R | 2, 3, 4, 5 | 2020/4/1–2020/7/30 | C | 1 | 6 | NA | S | 164 | 49 ± 15 | 139/25 |
101 RT-PCR+ 63 RT-PCR- |
| Kurokawa [55] | 2021 | Japan | R | 2, 3, 4, 5 | 2020/4/1–2020/5/21 | NA | 1 | 6 | NA | S | 154 | NA | 108/46 |
26 RT-PCR+ 128 RT-PCR- |
| Tunç [56] | 2021 | Turkey | R | 2, 3, 4, 5 | 2020/3–2020/5 | NA | 1 | 4 | NA | S | 209 | NA | 115/94 |
149 RT-PCR+ 60 RT-PCR- |
| Dilek [57] | 2021 | Turkey | R | 4 | 2020/3/11–2020/5/11 | NA | 1 | 4 | ED | S | 178 | 47 ± 16 (14–86) | 102/76 |
143 RT-PCR+ 35 RT-PCR- |
| De Jaegere [56, 58] | 2020 | Netherlands | R | 2, 3, 4, 5 | 2020/3/12–2020/3/23 | C | 1 | 3 | NA | S | 96 | 70 (29–94) | 61/35 |
45 RT-PCR+ 51 RT-PCR- |
Abbreviations: R, retrospective; P, prospective; C, consecutive; ED, emergency department; S, symptomatic; B, both symptomatic and asymptomatic; NA, not available
*Age (year): median age (range) or average age
Quality assessment
Figure 2 shows the quality variables of 24 included studies. Ten studies showed an unclear risk of bias since there was no information suggestive of consecutive or random sampling. Regarding the index test domain, only one study had an unclear risk of bias since it did not provide information regarding blinding to the reference standard. In the reference standard domain, two studies showed a high risk for bias since the reference standard was not blinded to the index test results. Additionally, four studies had an unclear risk of bias since they did not mention whether the reference standard was performed blinded to the index test results. Regarding the flow and timing domain, four studies had a high risk of bias since some patients were not included in the analysis while seven studies had an unclear risk of bias since they did not provide clear information regarding the time interval between the reference standard test and index test. Due to concerns regarding applicability, unclear risk ratings were assigned in two, one, and two studies in the patient selection, index test, and reference standard domains, respectively.
Fig. 2.
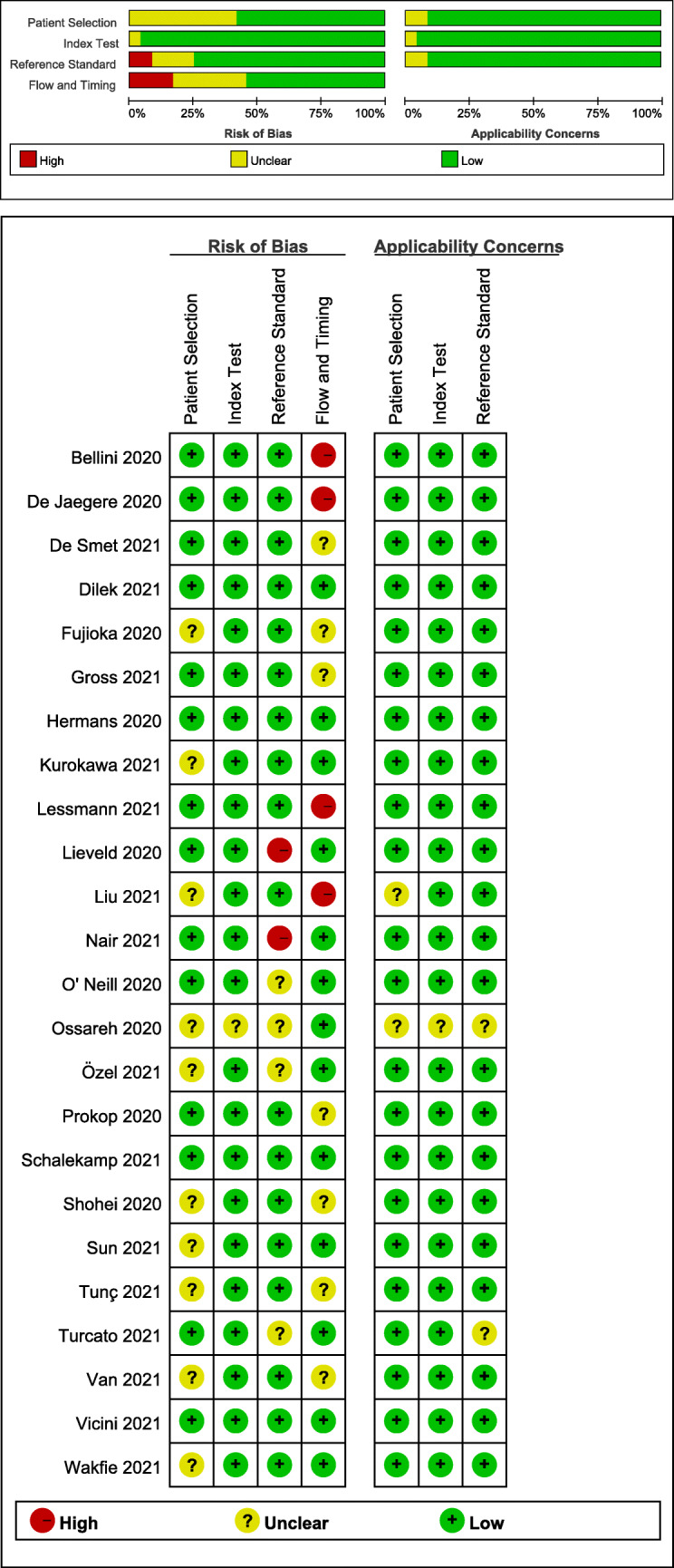
Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) criteria for the 24 included studies
Diagnostic accuracy
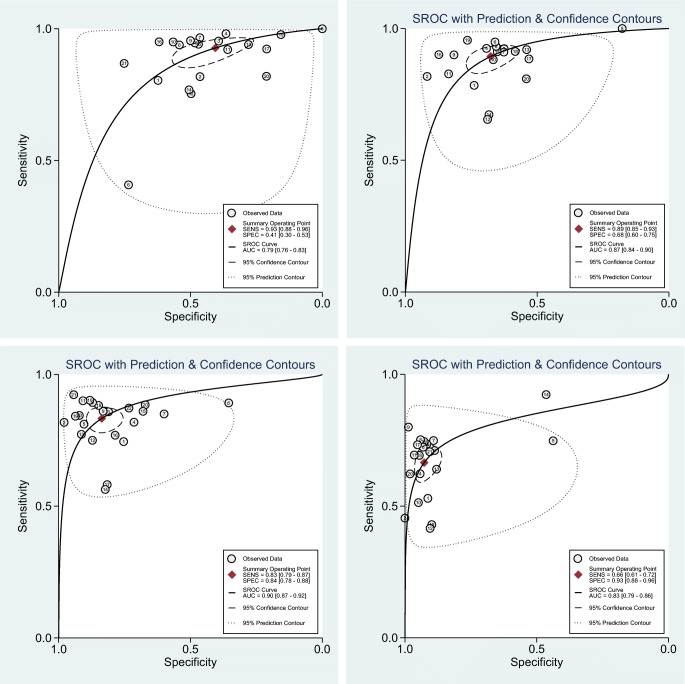
The pooled sensitivity and specificity for 21 studies using CO-RADS ≥ 2 were 0.93 (95% CI 0.88–0.96) and 0.41 (95% CI 0.30–0.53), respectively. In 20 studies using CO-RADS ≥ 3, the pooled sensitivity and specificity were 0.89 (95% CI 0.85–0.93) and 0.68 (95% CI 0.60–0.75), respectively. In 23 studies using CO-RADS ≥ 4, the pooled sensitivity and specificity were 0.83 (95% CI 0.79–0.87) and 0.84 (95% CI 0.78–0.88), respectively. In 21 studies using CO-RADS ≥ 5, the pooled sensitivity and specificity were 0.66 (95% CI 0.61–0.72) and 0.93 (95% CI 0.88–0.96), respectively (Fig. 3). Moreover, the DOR of CO-RADS using cutoff 2/3/4/5 was 9 (95% CI 5–14), 18 (95% CI 12–26), 26 (95% CI 16–40), and 25 (95% CI 15–42), respectively (Table 2).
Fig. 3.
Coupled forest plots of pooled sensitivity and specificity
Table 2.
Sensitivity, specificity, LR+, LR-, and DORs of CO-RADS for the detection of COVID-19
| Cutoff | 2 | 3 | 4 | 5 |
|---|---|---|---|---|
| Sensitivity (95% CI) | 0.93 (0.88–0.96) | 0.89 (0.85–0.93) | 0.83 (0.79–0.87) | 0.66 (0.61–0.72) |
| Specificity (95% CI) | 0.41 (0.30–0.53) | 0.68 (0.60–0.75) | 0.84 (0.78–0.88) | 0.93 (0.88–0.96) |
| LR+ (95% CI) | 1.6 (1.3–1.9) | 2.8 (2.2–3.4) | 5.1 (3.7–7.0) | 9.1 (5.7–14.7) |
| LR- (95% CI) | 0.18 (0.12–0.27) | 0.16 (0.11–0.22) | 0.20 (0.16–0.25) | 0.36 (0.31–0.42) |
| DOR (95% CI) | 9 (5–14) | 18 (12–26) | 26 (16–40) | 25 (15–42) |
Abbreviations: LR+, likelihood ratio positive; LR-, likelihood ratio negative; DOR, diagnostic odds ratio; CI, confidence interval
The AUCs of CO-RADS ≥ 2/3/4/5 were 0.79 (95% CI 0.76–0.83), 0.87 (95% CI 0.84–0.90), 0.90 (95% CI 0.87–0.92), and 0.83 (95% CI 0.79–0.86), respectively. The large difference between the 95% confidence and prediction regions presented in the HSROC curve indicated substantial among-study heterogeneity (Fig. 4).
Fig. 4.
Hierarchical summary receiver operating characteristic curve (HSROC) of the diagnostic performance of CO-RADS for detecting COVID-19
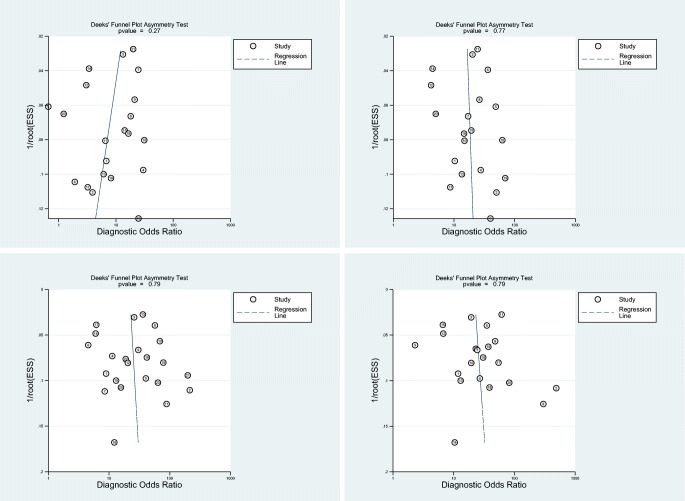
There was considerable among-study heterogeneity according to Cochran’s Q test (p < 0.01) and Higgins I2 heterogeneity index (Fig. 3). Deeks’ funnel plots (Fig. 5) revealed no major publication bias in CO-RADS ≥ 2/3/4/5 (p > 0.10).
Fig. 5.
Deeks’ funnel plot used to evaluate potential publication bias
Exploration of heterogeneity
Meta-regression
We performed meta-regression to explore the heterogeneity causes among 20 and 23 studies using CO-RADS ≥ 3 and CO-RADS ≥ 4, respectively (Table 3). Study heterogeneity was independently associated with publication year, study design, study enrollment, number of centers, number of readers, and setting.
Table 3.
Results of the meta-regression analysis of CO-RADS for the detection of COVID-19
| Cutoff | Covariates | Category | Studies (n) | Meta-analytic summary estimates | |||
|---|---|---|---|---|---|---|---|
| Sensitivity (95% CI) | p | Specificity (95% CI) | p | ||||
| 3 | Publication year | 2021 | 14 | 0.89 (0.84–0.93) | 0.00 | 0.69 (0.61–0.78) | 0.55 |
| 2020 | 6 | 0.91 (0.85–0.97) | 0.63 (0.49–0.77) | ||||
| Study design | R | 16 | 0.89 (0.84–0.93) | 0.03 | 0.66 (0.58–0.75) | 0.17 | |
| P | 3 | 0.92 (0.84–0.99) | 0.73 (0.56–0.90) | ||||
| Enrollment | C | 13 | 0.87 (0.82–0.92) | 0.00 | 0.71 (0.63–0.80) | 0.80 | |
| NA | 7 | 0.93 (0.88–0.97) | 0.60 (0.47–0.73) | ||||
| Centers (n) | < 2 | 14 | 0.89 (0.84–0.94) | 0.00 | 0.68 (0.59–0.77) | 0.56 | |
| ≥. | 5 | 0.91 (0.85–0.97) | 0.63 (0.47–0.79) | ||||
| Readers (n) | < 3 | 3 | 0.94 (0.86–1.00) | 0.88 | 0.70 (0.49–0.91) | 0.78 | |
| ≥ 3 | 11 | 0.87 (0.80–0.94) | 0.66 (0.54–0.78) | ||||
| Setting | ED | 9 | 0.93 (0.90–0.97) | 0.02 | 0.62 (0.50–0.73) | 0.02 | |
| NA | 11 | 0.85 (0.79–0.91) | 0.72 (0.63–0.81) | ||||
| 4 | Publication year | 2021 | 16 | 0.84 (0.80–0.88) | 0.00 | 0.84 (0.78–0.90) | 0.02 |
| 2020 | 7 | 0.82 (0.75–0.88) | 0.84 (0.74–0.93) | ||||
| Study design | R | 18 | 0.83 (0.79–0.87) | 0.00 | 0.82 (0.76–0.88) | 0.02 | |
| P | 3 | 0.86 (0.78–0.94) | 0.89 (0.80–0.99) | ||||
| Enrollment | C | 14 | 0.81 (0.77–0.86) | 0.00 | 0.85 (0.79–0.91) | 0.05 | |
| NA | 9 | 0.87 (0.82–0.92) | 0.81 (0.72–0.90) | ||||
| Centers (n) | < 2 | 15 | 0.82 (0.78–0.87) | 0.00 | 0.84 (0.78–0.91) | 0.13 | |
| ≥. | 6 | 0.85 (0.80–0.91) | 0.81 (0.69–0.92) | ||||
| Readers (n) | < 3 | 4 | 0.84 (0.75–0.93) | 0.03 | 0.82 (0.66–0.97) | 0.22 | |
| ≥ 3 | 12 | 0.81 (0.76–0.87) | 0.84 (0.76–0.92) | ||||
| Setting | ED | 10 | 0.87 (0.84–0.91) | 0.00 | 0.82 (0.74–0.90) | 0.00 | |
| NA | 13 | 0.79 (0.74–0.84) | 0.85 (0.78–0.91) | ||||
Abbreviations: ED, emergency department; NA, not available; CI, confidence interval
Among the 20 studies using CO-RADS ≥ 3, studies published in 2020 had a higher sensitivity than those published in 2021 (0.91, 95% CI 0.85–0.97 vs. 0.89, 95% CI 0.84–0.93; p < 0.01). Prospective studies had a higher sensitivity than retrospective studies (0.92, 95% CI 0.84–0.99 vs. 0.89, 95% CI 0.84–0.93; p = 0.03). Studies with unclear information regarding patient enrollment had a significantly higher sensitivity (0.93, 95% CI 0.88–0.97) than those with consecutive enrollment (0.87, 95% CI 0.82–0.92; p < 0.01). Multi-center studies had higher sensitivity than single-center studies (0.91, 95% CI 0.85–0.97 vs. 0.89, 95% CI 0.84-0.94; p < 0.01). Studies conducted in the ED had a higher sensitivity (0.93, 95% CI 0.90–0.97) and lower specificity (0.62, 95% CI 0.50–0.73) than those without setting information (0.85, 95% CI 0.79–0.91; p = 0.02 and 0.72, 95% CI 0.63–0.81; p = 0.02, respectively).
In 23 studies using CO-RADS ≥4, the sensitivity differed among the various potential covariates. Studies published in 2021 had a higher sensitivity than those published in 2020 (0.84, 95% CI 0.80-0.88 vs. 0.82, 95% CI 0.75–0.88; p < 0.01). Studies that consecutively enrolled participants had lower sensitivity than those without related information (0.81, 95% CI 0.77–0.86 vs. 0.87, 95% CI 0.82–0.92; p < 0.01). Prospective studies had a higher sensitivity than retrospective studies (0.86, 95% CI 0.78–0.94 vs. 0.83, 95% CI 0.79–0.87; p < 0.01). Multi-center studies had a higher sensitivity than single-center studies (0.85, 95% CI 0.80–0.91 vs. 0.82, 95% CI 0.78–0.87; p <0.01). Studies using < 3 readers had higher sensitivity than those using ≥ 3 readers (0.84, 95% CI 0.75–0.93 vs. 0.81, 95% CI 0.76–0.87; p = 0.03). Studies conducted in the ED had a higher sensitivity (0.87, 95% CI 0.84–0.91) than those without setting information (0.79, 95% CI 0.74–0.84; p < 0.01). Prospective studies had a higher sensitivity than retrospective studies (0.89, 95% CI 0.80–0.99 vs. 0.82, 95% CI 0.76–0.88; p = 0.02). Studies in the ED had lower specificity than those with unclear information on setting (0.82, 95% CI 0.74–0.90 vs. 0.85, 95% CI 0.78–0.91; p < 0.01).
Subgroup analysis
Table 4 summarizes the results of subgroup analyses of the age, sex, and symptoms.
Table 4.
Results of subgroup analyses of CO-RADS for the detection of COVID-19
| Cutoff | Covariates | Subgroup | Studies (n) | Meta-analysis summary estimates | |||
|---|---|---|---|---|---|---|---|
| Sensitivity (95% CI) | p | Specificity (95% CI) | p | ||||
| 3 | Age# | < 60 | 3 | 0.96 (0.92–1.00) | 0.81 | 0.57 (0.36–0.79) | 0.10 |
| ≥ .1 | 14 | 0.87 (0.81–0.92) | 0.72 (0.64–0.80) | ||||
| Sex* | < 50% | 3 | 0.90 (0.80–1.00) | 0.16 | 0.77 (0.63–0.92) | 0.87 | |
| ≥ 50% | 17 | 0.89 (0.85–0.94) | 0.66 (0.58–0.74) | ||||
| Symptoms | S | 17 | 0.90 (0.86–0.94) | 0.26 | 0.64 (0.56–0.72) | 0.00 | |
| B | 3 | 0.87 (0.75–0.99) | 0.83 (0.72–0.93) | ||||
| 4 | Age# | < 60 | 4 | 0.88 (0.82–0.94) | 0.02 | 0.77 (0.62–0.92) | 0.01 |
| ≥ .0 | 14 | 0.80 (0.75–0.85) | 0.87 (0.82–0.92) | ||||
| Sex* | < 50% | 3 | 0.88 (0.80–0.96) | 0.07 | 0.92 (0.86–0.99) | 0.70 | |
| ≥ .70 | 20 | 0.83 (0.79–0.86) | 0.82 (0.76–0.87) | ||||
| Symptoms | S | 19 | 0.83 (0.79–0.87) | 0.01 | 0.81 (0.75–0.87) | 0.00 | |
| B | 4 | 0.83 (0.74–0.92) | 0.92 (0.86–0.98) | ||||
Abbreviations: S, symptomatic; B, both symptomatic and asymptomatic; CI, confidence interval
#Age: median age or average age
*Sex: proportion of male subjects, including two groups: < 50% male subjects and ≥ 50% male subjects
In studies using CO-RADS ≥ 3, studies including both symptomatic and asymptomatic patients had a higher specificity than those only including symptomatic patients (0.83, 95% CI 0.72–0.93 vs. 0.64, 95% CI 0.56–0.72; p < 0.01). In studies using CO-RADS ≥ 4, studies with participants aged < 60 years had higher sensitivity (0.88, 95% CI 0.82–0.94 vs. 0.80, 95% CI 0.75–0.85; p=0.02) and lower specificity (0.77, 95% CI 0.62–0.92 vs. 0.87, 95% CI 0.82–0.92; p = 0.01) than those with participants aged > 60 years. Studies on both symptomatic and asymptomatic patients and those on only symptomatic patients had the same pooled sensitivity (0.83, 95% CI 0.74–0.92 vs. 0.83, 95% CI 0.79–0.87); however, the p value was < 0.01. Studies including both symptomatic and asymptomatic patients had higher specificity than those only including symptomatic patients (0.92, 95% CI 0.86–0.98 vs. 0.81, 95% CI 0.75–0.87; p < 0.01).
Discussion
Inconsistent reports regarding sensitivity and specificity impede our ability to determine the diagnostic accuracy of CO-RADS for detecting COVID-19. Few relevant systematic reviews or meta-analyses have assessed the diagnostic utility of CO-RADS for COVID-19 [27]. This is the first systematic review and meta-analysis to directly evaluate the accuracy of CO-RADS for COVID-19 and discuss the practical application of CO-RADS. Our findings demonstrated that CO-RADS has great utility in diagnosing COVID-19; moreover, CO-RADS ≥ 3/4 showed outstanding ability.
The AUC of CO-RADS revealed that CO-RADS ≥ 3/4 displayed favorable performance in detecting COVID-19. CO-RADS ≥ 3 might be preferred when priority is placed on the sensitivity of the examination. In case COVID-19 outbreaks re-occur in some countries, there is a need to rapidly screen potential infected patients to prevent disease spread. Contrastingly, CO-RADS ≥ 4 might be more suitable when it is more important to yield a definite diagnosis. In regions with low COVID-19 threat, there is a need to identify whether patients were infected with COVID-19 or other diseases to avoid unnecessary isolation and treatment.
Considerable among-study heterogeneity was observed. When the cutoff value was 3, there were significant differences in sensitivity according to publication year, study design, enrollment, number of centers, and setting; moreover, there were significant differences in specificity according to the setting. This suggests that the diagnostic performance of CO-RADS ≥ 3 may have been overestimated in studies with characteristics such as publication in 2020, failure to mention enrollment and setting, and multi-center settings, which yielded limitations in this meta-analysis. This suggests that CO-RADS ≥ 3 might facilitate quick detection of COVID-19 without geographical limitations in case of reoccurrence of COVID-19 outbreak. Additionally, the performance of CO-RADS ≥ 3 in readers has been relatively consistent which demonstrates the consistency and stability of CO-RADS ≥ 3. However, multi-center studies had significantly higher sensitivity than single-center studies (0.91 vs. 0.89, p < 0.01). Our findings provide additional evidence for exploring the consistency of CO-RADS ≥ 3 among different centers.
Meta-regression analysis of CO-RADS ≥ 4 revealed an association of study heterogeneity with publication year, study design, enrollment, number of centers, number of readers, and study settings, which indicated that the diagnostic performance of CO-RADS ≥ 4 may have been overestimated to some degree in studies published in 2021 without setting information, which led to several limitations in this study. Specifically, we focused on the consistency among various centers with different readers. Multi-center studies had significantly higher sensitivity than single-center studies (0.85 vs. 0.82, p < 0.01); moreover, studies with ≥ 3 readers showed significantly lower sensitivity than studies with < 3 readers (0.81 vs. 0.84; p = 0.03). Our findings provide additional evidence for exploring the consistency of CO-RADS ≥ 4 among different centers and whether the cutoff value of CO-RADS ≥ 4 should be recommended when there are < 3 readers. All the included studies recruited patients between January and June 2020, when there were rapid changes in the COVID-19 epidemic in different counties [59, 60], which may cause detection difficulties and finally lead to errors or heterogeneity. Moreover, not all studies were well-designed. Therefore, there is a need for better-designed studies on CO-RADS.
Subgroup analysis according to age revealed lower sensitivity in the older group with CO-RADS ≥ 4. Age affects the diagnostic performance for COVID-19 since older adults often have chronic lung diseases, including chronic obstructive pulmonary disease and interstitial pulmonary fibrosis [61]. Further, COVID-19 presentation in older patients is often atypical with less unspecific symptoms [62, 63], which may impede diagnosis through CO-RADS. Subgroup analysis according to sex revealed that most studies included more male participants, which is consistent with the sex-based susceptibility for COVID-19 infection [64–66]. Studies that included fewer male participants had higher sensitivity and specificity than those with more male participants, which may have translated to potential factor, including smoking, that affects CT scanning [66, 67], but there is a need for more well-designed studies to confirm the lack of statistical significance. Regarding the symptom subgroup, there was a significant difference in the sensitivity of CO-RADS ≥ 4; however, we could not determine the group with higher sensitivity. Further related studies are warranted. Moreover, studies enrolling both symptomatic and asymptomatic patients showed higher specificity for CO-RADS ≥ 3/4 than those only enrolling symptomatic patients, which is consistent with a previous report of higher specificity in asymptomatic patients (0.89) than in symptomatic patients (0.73) [25]. This could be attributed to patients with chronic lung diseases, who have respiratory symptoms similar to those of COVID-19, being included in the symptomatic group, which decreases the specificity [25, 68]. Moreover, this result could be attributed to subjective factors among observers and the local COVID-19 prevalence [24].
This study has several limitations. First, there was considerable heterogeneity among the included studies. Although we performed both meta-regression and subgroup analyses, there might have been insufficient exploration. Therefore, heterogeneity might be a concern. Second, some potential factors may remain unknown. Third, although we included 24 studies on CO-RADS, the available studies in each category were < 24 since some studies only provided data on one category. Finally, most of the included studies were retrospective, which led to a high risk of bias in the patient selection domain.
Our findings demonstrated that CO-RADS has favorable performance in detecting COVID-19. CO-RADS ≥ 3 and CO-RADS ≥ 4 might be suitable in different situations given their high sensitivity and specificity, respectively. However, future better-designed studies are warranted to further explore the consistency of CO-RADS among different centers and readers.
Abbreviations
- CO-RADS
COVID-19 Reporting and Data System
- COVID-19
Coronavirus disease 2019
- RT-PCR
Reverse transcription polymerase chain reaction
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
Funding
This study was supported by (1) 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZY2016104), and (2) Chinese National Natural Science Foundation (31801066).
Declarations
Guarantor
The scientific guarantor of this publication is Jiaming Liu.
Conflict of Interest
The authors declare no competing interests.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
None; this study is a systematic review and meta-anlysis.
Ethical approval
Institutional Review Board approval was not required because this studies was not related with human nor animal.
Methodology
systematic review
meta-analysis
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Guina Liu and Yuntian Chen contributed equally to this work
References
- 1.Wilder-Smith A, Chiew CJ, Lee VJ. Can we contain the COVID-19 outbreak with the same measures as for SARS? Lancet Infect Dis. 2020;20:e102–e107. doi: 10.1016/S1473-3099(20)30129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tahamtan A, Ardebili A. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization (2021) COVID-19 Clinical management: living guidance. In: World Health Organization. Available via https://www.who.int/publications/i/item/WHO-2019-nCoV-clinical-2021-1
- 6.Byrne AW, McEvoy D, Collins AB, et al. Inferred duration of infectious period of SARS-CoV-2: rapid scoping review and analysis of available evidence for asymptomatic and symptomatic COVID-19 cases. BMJ Open. 2020;10:e039856. doi: 10.1136/bmjopen-2020-039856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Porte L, Legarraga P, Vollrath V, et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int J Infect Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang W, Xu Y, Gao R et al (2020) Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 323:1843–1844 [DOI] [PMC free article] [PubMed]
- 10.Ai T, Yang Z, Hou H, et al. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology. 2020;296:E32–e40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Udugama B, Kadhiresan P, Kozlowski HN, et al. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 12.Liu H, Ren H, Wu Z, et al. CT radiomics facilitates more accurate diagnosis of COVID-19 pneumonia: compared with CO-RADS. J Transl Med. 2021;19:29. doi: 10.1186/s12967-020-02692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herpe G, Lederlin M, Naudin M, et al. Efficacy of Chest CT for COVID-19 Pneumonia Diagnosis in France. Radiology. 2021;298:E81–e87. doi: 10.1148/radiol.2020202568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20:425–434. doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan F, Ye T, Sun P, et al. Time Course of Lung Changes at Chest CT during Recovery from Coronavirus Disease 2019 (COVID-19) Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zu ZY, Jiang MD, Xu PP, et al. Coronavirus Disease 2019 (COVID-19): A Perspective from China. Radiology. 2020;296:E15–e25. doi: 10.1148/radiol.2020200490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang Z, Li X, Zhou S. Recommendation of low-dose CT in the detection and management of COVID-2019. Eur Radiol. 2020;30:4356–4357. doi: 10.1007/s00330-020-06809-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jalaber C, Revel MP, Chassagnon G, et al. Role of upfront CT pulmonary angiography at admission in COVID-19 patients. Thromb Res. 2020;196:138–140. doi: 10.1016/j.thromres.2020.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai HX, Hsieh B, Xiong Z, et al. Performance of Radiologists in Differentiating COVID-19 from Non-COVID-19 Viral Pneumonia at Chest CT. Radiology. 2020;296:E46–e54. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu B, Xing Y, Peng J, et al. Chest CT for detecting COVID-19: a systematic review and meta-analysis of diagnostic accuracy. Eur Radiol. 2020;30:5720–5727. doi: 10.1007/s00330-020-06934-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams HJA, Kwee TC, Yakar D, Hope MD, Kwee RM. Systematic Review and Meta-Analysis on the Value of Chest CT in the Diagnosis of Coronavirus Disease (COVID-19): Sol Scientiae, Illustra Nos. AJR Am J Roentgenol. 2020;215:1342–1350. doi: 10.2214/AJR.20.23391. [DOI] [PubMed] [Google Scholar]
- 22.Kim H, Hong H, Yoon SH. Diagnostic Performance of CT and Reverse Transcriptase Polymerase Chain Reaction for Coronavirus Disease 2019: A Meta-Analysis. Radiology. 2020;296:E145–e155. doi: 10.1148/radiol.2020201343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khatami F, Saatchi M, Zadeh SST, et al. A meta-analysis of accuracy and sensitivity of chest CT and RT-PCR in COVID-19 diagnosis. Sci Rep. 2020;10:22402. doi: 10.1038/s41598-020-80061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prokop M, van Everdingen W, van Rees VT, et al. CO-RADS: A Categorical CT Assessment Scheme for Patients Suspected of Having COVID-19-Definition and Evaluation. Radiology. 2020;296:E97–e104. doi: 10.1148/radiol.2020201473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Smet K, De Smet D, Ryckaert T, et al. Diagnostic Performance of Chest CT for SARS-CoV-2 Infection in Individuals with or without COVID-19 Symptoms. Radiology. 2021;298:E30–e37. doi: 10.1148/radiol.2020202708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bellini D, Panvini N, Rengo M, et al. Diagnostic accuracy and interobserver variability of CO-RADS in patients with suspected coronavirus disease-2019: a multireader validation study. Eur Radiol. 2021;31:1932–1940. doi: 10.1007/s00330-020-07273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwee RM, Adams HJA, Kwee TC. Diagnostic Performance of CO-RADS and the RSNA Classification System in Evaluating COVID-19 at Chest CT: A Meta-Analysis. Radiol Cardiothorac Imaging. 2021;3:e200510. doi: 10.1148/ryct.2021200510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberati A, Altman DG, Tetzlaff J et al (2009) The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 339:b2700 [DOI] [PMC free article] [PubMed]
- 29.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 30.Deeks JJ, Macaskill P, Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol. 2005;58:882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 31.Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327:557–560 [DOI] [PMC free article] [PubMed]
- 32.Dofferhoff ASM, Swinkels A, Sprong T et al (2020) [Diagnostic algorithm for COVID-19 at the ER]. Ned Tijdschr Geneeskd 164 [PubMed]
- 33.Abdel-Tawab M, Basha MAA, Mohamed IAI, et al. Comparison of the CO-RADS and the RSNA chest CT classification system concerning sensitivity and reliability for the diagnosis of COVID-19 pneumonia. Insights Imaging. 2021;12:55. doi: 10.1186/s13244-021-00998-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fonseca E, Loureiro BMC, Strabelli DG, et al. Evaluation of the RSNA and CORADS classifications for COVID-19 on chest computed tomography in the Brazilian population. Clinics (Sao Paulo) 2021;76:e2476. doi: 10.6061/clinics/2021/e2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palomar-Lever A, Barraza G, Galicia-Alba J, et al. Hepatic steatosis as an independent risk factor for severe disease in patients with COVID-19: A computed tomography study. JGH Open. 2020;4:1102–1107. doi: 10.1002/jgh3.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Çomoğlu Ş, Öztürk S, Topçu A et al (2021) The Role of CO-RADS Scoring System in the Diagnosis of COVID-19 Infection and its Correlation with Clinical Signs. Curr Med Imaging. 10.2174/1573405617666210827150937 [DOI] [PubMed]
- 37.Abdelsalam M, Althaqafi RMM, Assiri SA, et al. Clinical and Laboratory Findings of COVID-19 in High-Altitude Inhabitants of Saudi Arabia. Front Med (Lausanne) 2021;8:670195. doi: 10.3389/fmed.2021.670195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turcato G, Zaboli A, Panebianco L, et al. Clinical application of the COVID-19 Reporting and Data System (CO-RADS) in patients with suspected SARS-CoV-2 infection: observational study in an emergency department. Clin Radiol. 2021;76(74):e23–74.e29. doi: 10.1016/j.crad.2020.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun R, Achkar S, Ammari S et al (2021) Systematic screening of COVID-19 disease based on chest CT and RT-PCR for cancer patients undergoing radiotherapy in a coronavirus French hotspot. Int J Radiat Oncol Biol Phys. 10.1016/j.ijrobp.2021.02.022 [DOI] [PMC free article] [PubMed]
- 40.Schalekamp S, Bleeker-Rovers CP, Beenen LFM, et al. Chest CT in the Emergency Department for Diagnosis of COVID-19 Pneumonia: Dutch Experience. Radiology. 2021;298:E98–e106. doi: 10.1148/radiol.2020203465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ON SB, Byrne D, Müller NL et al (2020) Radiological Society of North America (RSNA) Expert Consensus Statement Related to Chest CT Findings in COVID-19 Versus CO-RADS: Comparison of Reporting System Performance Among Chest Radiologists and End-User Preference. Can Assoc Radiol J. 10.1177/0846537120968919:846537120968919 [DOI] [PubMed]
- 42.Özel M, Aslan A, Araç S (2021) Use of the COVID-19 Reporting and Data System (CO-RADS) classification and chest computed tomography involvement score (CT-IS) in COVID-19 pneumonia. Radiol Med. 10.1007/s11547-021-01335-x:1-9 [DOI] [PMC free article] [PubMed]
- 43.Ossareh S, Bagheri SM, Abbasi M, Abolfathi S, Bohlooli A (2020) Role of screening for COVID-19 in hemodialysis wards, results of a single center study. Iran J Kidney Dis 14:389–398 [PubMed]
- 44.Lieveld AWE, Azijli K, Teunissen BP et al (2020) Chest CT in COVID-19 at the ED: Validation of the COVID-19 Reporting and Data System (CO-RADS) and CT severity score. Chest. 10.1016/j.chest.2020.11.026 [DOI] [PMC free article] [PubMed]
- 45.Lessmann N, Sanchez CI, Beenen L, et al. Automated assessment of COVID-19 reporting and data system and chest CT severity scores in patients suspected of having COVID-19 using artificial intelligence. Radiology. 2021;298:E18–E28. doi: 10.1148/radiol.2020202439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hermans JJR, Groen J, Zwets E, et al. Chest CT for triage during COVID-19 on the emergency department: myth or truth? Emerg Radiol. 2020;27:641–651. doi: 10.1007/s10140-020-01821-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gross A, Heine G, Schwarz M, Thiemig D, Gläser S, Albrecht T. Structured reporting of chest CT provides high sensitivity and specificity for early diagnosis of COVID-19 in a clinical routine setting. Br J Radiol. 2021;94:20200574. doi: 10.1259/bjr.20200574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujioka T, Takahashi M, Mori M et al (2020) Evaluation of the Usefulness of CO-RADS for Chest CT in Patients Suspected of Having COVID-19. Diagnostics (Basel) 10 [DOI] [PMC free article] [PubMed]
- 49.Bellini D, Panvini N, Rengo M et al (2020) Diagnostic accuracy and interobserver variability of CO-RADS in patients with suspected coronavirus disease-2019: a multireader validation study. Eur Radiol. 10.1007/s00330-020-07273-y:1-9 [DOI] [PMC free article] [PubMed]
- 50.Inui S, Kurokawa R, Nakai Y, et al. Comparison of Chest CT Grading Systems in Coronavirus Disease 2019 (COVID-19) Pneumonia. Radiol Cardiothorac Imaging. 2020;2:e200492. doi: 10.1148/ryct.2020200492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wakfie-Corieh CG, Ferrando-Castagnetto F, Blanes Garcia AM et al (2021) Incidental findings suggestive of COVID-19 pneumonia in oncological patients undergoing 18F-FDG PET/CT studies: association between metabolic and structural lung changes. J Nucl Med. 10.2967/jnumed.121.261915 [DOI] [PMC free article] [PubMed]
- 52.Vicini S, Panvini N, Bellini D et al (2021) Radiographers and COVID-19 pneumonia: Diagnostic performance using CO-RADS. Radiography (Lond). 10.1016/j.radi.2021.04.010 [DOI] [PMC free article] [PubMed]
- 53.Van Berkel B, Vandevenne J, Coursier K, et al. Chest CT Diagnosis of COVID-19: Accuracy using CO-RADS and CT-Involvement Scoring. J Belg Soc Radiol. 2021;105:17. doi: 10.5334/jbsr.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nair AV, McInnes M, Jacob B et al (2021) Diagnostic accuracy and inter-observer agreement with the CO-RADS lexicon for CT chest reporting in COVID-19. Emerg Radiol. 10.1007/s10140-021-01967-6:1-10 [DOI] [PMC free article] [PubMed]
- 55.Kurokawa R, Inui S, Gonoi W, et al. Standardized reporting systems of chest computed tomography in a population with low coronavirus disease 2019 prevalence: A retrospective comparative study. Heliyon. 2021;7:e07743. doi: 10.1016/j.heliyon.2021.e07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koşar Tunç M, Kiş N, İnce O, Kurtul Yildiz H, Önder H (2021) Diagnostic Performance of COVID-19 Reporting and Data System Classification Across Residents and Radiologists: A Retrospective Study. J Comput Assist Tomogr. 10.1097/rct.0000000000001172 [DOI] [PubMed]
- 57.Dilek O, Kaya O, Akkaya H et al (2021) Diagnostic performance and interobserver agreement of CO-RADS: evaluation of classification in radiology practice. Diagn Interv Radiol. 10.5152/dir.2021.201032 [DOI] [PMC free article] [PubMed]
- 58.de Jaegere TMH, Krdzalic J, Fasen B, Kwee RM. Radiological Society of North America Chest CT Classification System for Reporting COVID-19 Pneumonia: Interobserver Variability and Correlation with Reverse-Transcription Polymerase Chain Reaction. Radiol Cardiothorac Imaging. 2020;2:e200213. doi: 10.1148/ryct.2020200213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Flora S, La Maestra S. Growth and decline of the COVID-19 epidemic wave in Italy from March to June 2020. J Med Virol. 2021;93:1613–1619. doi: 10.1002/jmv.26499. [DOI] [PubMed] [Google Scholar]
- 60.Tu H, Tu S, Gao S, Shao A, Sheng J. Current epidemiological and clinical features of COVID-19; a global perspective from China. J Infect. 2020;81:1–9. doi: 10.1016/j.jinf.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho SJ, Stout-Delgado HW. Aging and Lung Disease. Annu Rev Physiol. 2020;82:433–459. doi: 10.1146/annurev-physiol-021119-034610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lauretani F, Ravazzoni G, Roberti MF, et al. Assessment and treatment of older individuals with COVID 19 multi-system disease: Clinical and ethical implications. Acta Biomed. 2020;91:150–168. doi: 10.23750/abm.v91i2.9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dai WC, Zhang HW, Yu J, et al. CT Imaging and Differential Diagnosis of COVID-19. Can Assoc Radiol J. 2020;71:195–200. doi: 10.1177/0846537120913033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abbasi-Oshaghi E, Mirzaei F, Farahani F, Khodadadi I, Tayebinia H. Diagnosis and treatment of coronavirus disease 2019 (COVID-19): Laboratory, PCR, and chest CT imaging findings. Int J Surg. 2020;79:143–153. doi: 10.1016/j.ijsu.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moradi B, Ghanaati H, Kazemi MA, et al. Implications of Sex Difference in CT Scan Findings and Outcome of Patients with COVID-19 Pneumonia. Radiol Cardiothorac Imaging. 2020;2:e200248. doi: 10.1148/ryct.2020200248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abate BB, Kassie AM, Kassaw MW, Aragie TG, Masresha SA. Sex difference in coronavirus disease (COVID-19): a systematic review and meta-analysis. BMJ Open. 2020;10:e040129. doi: 10.1136/bmjopen-2020-040129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wenham C, Smith J, Morgan R. COVID-19: the gendered impacts of the outbreak. Lancet. 2020;395:846–848. doi: 10.1016/S0140-6736(20)30526-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Halpin DMG, Criner GJ, Papi A, et al. Global Initiative for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease. The 2020 GOLD Science Committee Report on COVID-19 and Chronic Obstructive Pulmonary Disease. Am J Respir Crit Care Med. 2021;203:24–36. doi: 10.1164/rccm.202009-3533SO. [DOI] [PMC free article] [PubMed] [Google Scholar]